Abstract
Activation-induced cytidine deaminase (AID) initiates DNA double strand breaks (DSBs) in the immunoglobulin heavy chain gene (Igh) to stimulate isotype class switch recombination (CSR), and widespread breaks in non-Igh (off-target) loci throughout the genome. Because the DSBs that initiate class switching occur during the G1 phase of the cell cycle, and are repaired via end joining, CSR is considered a predominantly G1 reaction. By contrast, AID-induced non-Igh DSBs are repaired by homologous recombination. Although little is known about the connection between the cell cycle and either induction or resolution of AID-mediated non-Igh DSBs, their repair by homologous recombination implicates post-G1 phases. Coordination of DNA breakage and repair during the cell cycle is critical to promote normal class switching and prevent genomic instability. To understand how AID-mediated events are regulated through the cell cycle, we have investigated G1-to-S control in AID-dependent genome-wide DSBs. We find that AID-mediated off-target DSBs, like those induced in the Igh locus, are generated during G1. These data suggest that AID-mediated DSBs can evade G1/S checkpoint activation and persist beyond G1, becoming resolved during S-phase. Interestingly, DSB resolution during S-phase can promote not only non-Igh break repair, but also immunoglobulin CSR. Our results reveal novel cell cycle dynamics in response to AID-initiated DSBs, and suggest that the regulation of the repair of these DSBs through the cell cycle may ensure proper class switching while preventing AID-induced genomic instability.
Introduction
B-cell development and maturation requires multiple rounds of programmed, tightly regulated genomic DNA rearrangements that generate antibody diversity, modify affinity for antigen, and alter immunoglobulin effector function (1-4). In mature B-cells, immunoglobulin class switch recombination (CSR) and somatic hypermutation (SH) are both initiated by Activation Induced Cytidine Deaminase (AID, encoded by Aid) (5-11). The programmed, AID-dependent DNA double strand breaks (DSBs) that drive immunoglobulin CSR, are targeted to actively transcribed, repetitive sequences within Igh, termed switch (S) regions (1, 12). Class-switch initiating DSBs are detectable by LM-PCR preferentially during G1, and are resolved by end-joining mechanisms that are functional during G1 (13-19). These data demonstrate CSR to be a G1 process, and it has been proposed that restriction of CSR to G1 minimizes risk of AID-induced genomic instability. However, CSR is also known to be division-linked, with increasing class switching presumably due to increasing AID levels (20, 21).
Unlike site-specific endonucleases such as the RAG1/2 complex, AID lacks known target site specificity (12). In this context, several recent studies have shown that AID can generate point mutations at multiple non-immunoglobulin genes (22, 23). Moreover, it has been demonstrated that AID generates widespread DSBs at non-Igh locations (24-26). Whereas the mechanisms that target AID to Igh are at least partly understood, little is known about the mechanisms or the cell-cycle dynamics governing AID non-Igh (so-called “off-target”) DNA damage and repair. One model suggests that non-Igh DSBs occur at sites of DNA replication, either by passage of a fork over an AID-initiated single strand break (SSB), or by direct attack on the fork by AID itself (22, 27). In either case, off-target DSBs would mostly arise at DNA replication sites during S-phase. Consistent with this model, we found recently that homologous recombination (HR) is crucial for the repair of AID off-target DSBs. Activated B-cells deficient in XRCC2, a member of the RAD51 family of HR factors, are acutely sensitive to AID-activity, accumulate numerous off-target DSBs per cell, and can incur massive chromosomal instability, and ultimately die (25).
HR is a high-fidelity post-G1 DSB repair pathway with roles in general genome stability maintenance in a range of organisms and cell types (28, 29). Our previous findings were the first to highlight HR as an important genome-protective pathway in B-cells, required specifically to survive the promiscuous genome damaging activity of AID. There is growing evidence that AID-mediated genomic DSBs, if not rapidly and correctly resolved, are substrates for potentially oncogenic chromosomal translocations in stimulated B-cells (13, 14, 17, 26, 30-32). Thus, the mechanisms that regulate AID activity at Igh versus off-target locations, as well as the processes that govern DNA repair at on- versus off-target locations, are of considerable importance to understanding normal immune development and the etiology of B-cell malignancies.
Here we investigate the relationship between AID-mediated Igh (on-target) and non-Igh (off-target) DSBs, in the context of the cell cycle. We find that off-target AID-mediated DSBs arise during G1, prior to the onset of S-phase, similar to on-target CSR initiating DSBs in the Igh locus. Importantly, we demonstrate that AID-mediated off-target DSBs can arise independently of DNA replication. We also provide evidence that repair of these off-target DSBs, which arise during G1, is largely delayed until S-phase. Interestingly, we find that CSR itself can also take place during S-phase. Taken together, we propose that AID generates DSBs broadly throughout the genome during G1 and that, when this occurs, delayed repair provides an opportunity to repair off-target DSBs by homologous recombination, while concomitantly ensuring CSR by end-joining. We suggest that this phenomenon represents a unique cellular mechanism that allows B-cells to generate isotype diversity, while simultaneously protecting the genome from chromosomal instability.
Materials and Methods
Mice
C57BL/6J (stock # 000664, The Jackson Laboratory), Atm-/-, Trp53-/- (33) and Aid-/- (11) mouse colonies were maintained in pressurized, individually ventilated caging and fed standard lab diet. All mouse work was carried out according to IACUC approved protocols. B-cells were isolated from spleens of 4 to 8-week old mice following euthanasia.
B-cell isolation and culture
Splenic B-cells were isolated by magnetic bead-based cell sorting using anti-CD43 (Ly-48) microbeads (Miltenyi, Cat # 130-049-801) according to the manufacturer's protocol. Isolated cells were tested for purity by flow cytometry analysis, staining for B220, CD43, and IgM (see flow cytometry method). Purified B-cells were cultured with RPMI 1640 with L-Glutamine (Gibco, Cat # SH30034.01) supplemented with 10% (vol/vol) heat inactivated FBS (Atlanta Biologicals, Cat. # S11150). Purified B-cells at a concentration of 1×106 cells per mL were stimulated with 1μg/mL anti-CD40 antibody (BD Pharmingen, clone HM40, Cat. # 3553721) and 25ng/mL IL-4 (Peprotech, Cat # 214-14) for 2 days, then readjusted to 1×106 cells/mL and restimulated with an additional 1 μg/mL anti-CD40 and 25ng/mL of IL-4. In some assays, the DNA polymerase inhibitor aphidicolin (APH, Sigma, Cat. # A0781), dissolved in DMSO and stored as 10mM aliquots, was used. Aphidicolin was added to a final concentration of 0.4μM or 1.2μM at the same time anti-CD40 and IL-4 were administered. For cell proliferation assays, B-cells were incubated for 10 minutes with 5μM Carboxyfluorescein succinimidyl ester (CFSE), then transferred to fresh media and activated with or without aphidicolin (as described). Lentiviral transduction, of total splenocytes or purified B-cells, with Xrcc2 shRNA or control constructs was performed as previously described (25).
EdU incorporation assays
To determine the extent of colocalization of EdU and γ-H2AX foci, activated (22-hours) Aid+/+ and Aid-/- B-cells were pulse labeled with 10μM EdU (5-ethynyl-2’-deoxyuridine) for 15 minutes, incubated for 1 hour on coverslips, fixed and stained for γ-H2AX as described below, for 1 hour. The cells were stained for EdU according to manufacturer's protocol (Life technologies, Click-iT EdU® Alexa Fluor 488® Imaging Kit, Cat. # C-10337,). Samples were imaged using a Leica SP-2 spectral confocal microscope with a 63X 1.4 NA oil objective. Images were manually segmented using Imaris software (Imaris 7.2.0, Bitplane) and were scored for co-association of γ-H2AX foci and EdU labeled chromatin. To determine the extent EdU is incorporated in activated Aid+/+ and Aid-/- B-cells, the cells were incubated with 10μM EdU for 2 days and stained for EdU according to manufacturer's protocol (Life technologies, Click-iT® Alexa Fluor 488® Flow Cytometry Assay Kit, Cat # C-10425). The extent of EdU incorporation was measured by flow cytometry (below).
Immunofluorescence
Cells were adhered to poly-L-lysine (Sigma, Cat. #P8920-100ML) coverslips, fixed with 3% Neutral Buffered Formalin (NBF) with 2% sucrose in PBS for 10 minutes, permeabilized with 0.1%Triton-X100 in PBS for 10 minutes, and blocked with 3% FBS for 1 hour. Fixed cells were incubated with primary antibody (anti-phosphorylated γ-H2AX, 1:400 dilution, Bethyl Cat. #A300-081A; anti-phosphorylated S1981 ATM, 1:500 dilution, Rockland Cat. # 600-401-398) overnight at 4°C, followed by secondary antibodies conjugated with either Alexa 488 secondary (1:1000 dilution, Invitrogen Cat. #A11-008) or TRITC secondary (1:1000 dilution, Jackson ImmunoResearch Cat. # 111-26-003) antibodies for 30 minutes at room temperature. Samples were mounted with Vectashield containing DAPI counterstain (Vector Laboratories, Inc, Cat. #H-1200), imaged by epifluorescence wide-field microscopy using a Nikon 90i upright microscope and analyzed with NIS Elements software.
Flow cytometry
For flow cytometry, 1×105 to 1×106 cells were stained for 1 hour at 4°C with an antibody-fluorophore cocktail containing phycoethrythrin-indotricarbocyanine-anti-B220 (1:100 dilution; ebioscience Cat. # 15-0452-82), phycoerythrin-anti IgG1 (1:100 dilution; Southern Biotech Cat. #1070-09), and fluorescein isothiocyanate-IgM (1:100 dilution, Pharmingen Cat. # 553408) for analysis of class switch recombination. To measure for B-cell purity after magnetic bead isolation, cells were stained with phycoethrythrin-anti CD43 (1:400 dilution, BD Biosciences Cat.# 553271), fluorescein isothiocyanate-anti-CD19 (1:800 dilution, BD Biosciences, Cat. # 559785) phycoethrythrin-indodicarbocyanine-anti-B220 (1:100 dilution, BD Biosciences, Cat. # 561879), and Allophycocyanin-anti-IgM (1:240 dilution, eBioscience, Cat. # 17-5790-82). The cells were washed with 2% FBS (in PBS) before analysis. Cell-cycle analysis was performed as previously described, after staining genomic DNA with 20μg/mL propidium iodide (Sigma, Cat. #P4170) (34). For flow cytometric analysis of intracellular ATM phospho-serine 1981, cells were fixed, permeabilized, and blocked, in suspension, as described for immunofluorescence above. The cells were then incubated with anti-phosphorylated S1981 ATM (1:500 dilution, Rockland Cat. # 600-401-398) overnight at 4°C, followed by secondary antibodies conjugated with either Alexa 488 secondary (1:1000 dilution, Invitrogen Cat. #A11-008) for 1 hour at room temperature. The cells were washed with 1% FBS in PBS and analyzed. All flow cytometry analysis was performed with FACSCaliber using CellQuest Pro acquisition software (BD Bioscience) and was analyzed with Flowjo software (V.8.4.6; Treestar Inc.)
Neutral Comet Assay
B-cells were activated with αCD40 plus IL-4 and treated with aphidicolin for 2 days, or were exposed to either 0 or 5 Gy irradiation (controls). Cell cultures were then washed with PBS, mixed with 1.5 mL 1% low melting point agarose (40°C) and spread on frosted microscope slides (Fisher, Cat. # 12-544-5CY). After agarose polymerization, slides were immersed in 0.5% SDS with 30mM EDTA (pH 8) for 4 hours at 50°C, stored in 1 × TBE (90mM Tris, 90mM Boric Acid, 2mM EDTA, pH 8,5) overnight, then electrophoresed for 25 minutes in TBE at 7 mA. Slides stained with 2.5 μg/mL propidium iodide for 20 minutes, and imaged by epifluorescence microscopy as described above. Cells exhibiting comet tails were quantified as a fraction of the total cells imaged.
Immunoblot Analysis
Whole cell extracts were prepared from total splenocytes or purified splenic B-cells as described (25). 20 μg of whole cell extracts, quantified by Bio-Rad DC (Bio-Rad, Cat. # 500-0114), were electrophoresed in 10% SDS polyacrylamide Tris-Glycine gels (SDS-PAGE, Lonza, Cat. # 58502), and the proteins were blotted onto a PVDF membrane (Millipore Immobilon-P, Cat. # IPVH00010) for Western analysis according to ECL Plus Western Blotting Detection protocol (GE Healthcare, Cat # RPN2132). The following primary antibodies were used: anti-p53, R-19 p53 (1:200 dilution, Santa Cruz, Cat. # sc1313,); anti-phospho-p53, Ser15 (Cat. #9284, 1:1000, Cell Signaling); anti-ATM, D2E2 (dilution 1:1000, Cell Signaling, Cat. # 2873); anti-phospho-ATM, Ser1981 clone 10H11.E12 (1:2000 dilution, Millipore, Cat. # 05-740); anti-HSP90 (1:1000 dilution, Enzo Life Sciences, Cat. # ADI-SPA-835); and anti Actin (1:5000 dilution, Abcam, Cat. Ab8227-50). Horseradish peroxidase-labeled secondary antibody was used to detect the primary antibodies (anti-rabbit, 1:10000 dilution, GE Healthcare, Cat. # NA934V; anti-rat, dilution 1:5000, Jackson Immuno Research Cat. #112-035-003; and anti-mouse, dilution 1:15000, Thermo Scientific, Cat. # 32230). Detection of the immunoblots was performed using ECL Plus system (GE Healthcare, Cat # RPN2132). The blot was exposed to X-OMAT Blue XB film (Kodak, Cat. # NEF596) to detect the protein bands.
FISH
Fluorescence in situ hybridization (FISH) was carried as previously described (25, 34). After fixation and permeabilization, samples were alkali denatured, and stored in 70% ethanol. Samples were blocked with salmon sperm DNA-tRNA-formamide, and then hybridized with indocarbocyanine-labelled bacterial artificial chromosome (BAC) probes. Coverslips were washed with 50% (wt/vol) formamide in 2X SSC and were then stained for γ–H2AX, and imaged as described above.
PCR Analysis of CSR Products
Genomic DNA was prepared by standard protocols. 200-500 ng of input DNA was used per reaction. PCR was performed using GoTaqGreen (Promega M172A), with PCR parameters: 94°C2 minutes followed by 35 cycles of 94°C for 15 seconds, 64°C for 30 seconds, 72°C for 2 minutes, and a final incubation at 72°C for 10 min. The primers used were: SμF primer (primer 1): CTGGACTCAACTGGGCTGGCTGATGGGATC, SμR primer (primer 2): ATGGTTGAATGGCCTGCTGCTGGGCTGG(35), Sγ1 primer (primer 3): GTATAAGGTACCAGGCTGAGCAG. Xrcc2 PCR was done as previously described (36).
Statistics
Unless otherwise indicated, quantitative data are represented as means derived from at least three independent biological samples. Confidence intervals (95% CI) are indicated as error bars, and p-values were calculated by the Student t-test.
Results
HR repairs recurrent, AID-initiated DSBs at both Igh and non-Igh target sites
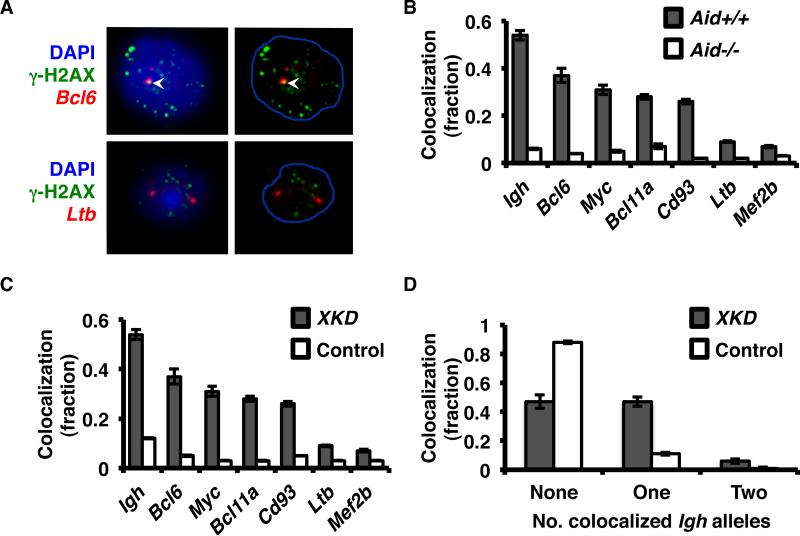
Several recent studies have identified a number of non-immunoglobulin (non-Igh) genes that are recurrently mutated by AID in activated B-cells (5, 6, 22-24). While we demonstrated that AID generates multiple, non-Igh (off-target) DSBs, it was not initially known whether off-target DSBs arise at specific, recurrent locations (25). To answer these questions, we identified Bcl6, Myc, Bcl11a, and CD93 as a set of candidates genes, previously published as frequent targets for AID-dependent hypermutation, to test for recurrent AID -dependent DSBs (23, 37). Gene-specific FISH plus immunofluorescence for phospho (γ)-H2AX foci was used to measure the fraction of AID-initiated DSBs at each of these loci in Aid+/+ (wild-type) versus Aid-/- activated B-cells (Fig. 1A-D). These assays were performed either in B-cells with or without an XRCC2-specific shRNA knockdown construct (XKD) that we have previously validated for inhibition of XRCC2-mediated HR in primary B-cells (25). Igh, as the major known physiological target for AID, was used as a positive control; and Ltb and Mef2b, which are rarely targeted for point mutations, were used as negative controls. Cells were evaluated for co-localization of γ-H2AX and FISH signals, and the percentage of cells showing at least one co-localization event was determined for each gene (Fig. 1B-D). We observed the highest rate of AID-dependent DSBs at the Igh locus as expected, and recurrent DSBs, at varying frequencies, at Bcl6, Myc, Bcl11a, and CD93 (Fig. 1B-C). The frequency of γ-H2AX foci at all locations was at background level in Aid-/- B-cells, confirming AID dependency for both on- and off-target breaks. Interestingly, the frequency of γ-H2AX foci localizing to the Igh locus was significantly elevated in cells with Xrcc2 knockdown, suggesting a role for HR in the repair of Igh, as well as non-Igh, DSBs. Because HR is dispensable for immunoglobulin class switch recombination, this result suggests that HR can catalyze error-free repair of DSB within Igh, as it does for DSBs at other genomic locations. These data, together with our previously published results, clearly establish that AID-dependent supernumerary γ–H2AX foci (those numbering more than 2 per cell) included recurrent, non-Igh DSBs at multiple specific locations. This corroborates our earlier findings of a key role for HR in the repair of off-target DSBs, and now suggests that HR is able to access DSBs in the Igh locus as well (25).
Figure 1.
Homologous recombination repairs AID-mediated DSBs at Igh and non-Igh sites. A, Representative widefield images showing fluorescence in situ hybridization (FISH) of BAC probes to either Bcl6 or Ltb (red) plus γ-H2AX foci (green) in activated XKD B-cells. DNA was stained with DAPI (left panels, blue) and used to segment the nuclear periphery (right panels). Arrowheads represent co-localization of FISH signal and γ-H2AX foci. B, Bar chart showing the fraction of total Aid+/+ XKD (gray bars) versus Aid-/- XKD (white bars) activated B-cells exhibiting co-localization of at least one gene specific BAC signal with γ-H2AX foci as observed in (A). Additional BAC probes (and corresponding genes analyzed) included RP23-109B20 (Igh), RP23-127N11 (Bcl6), RP23-307D14 (Myc), RP23-150J18 (Bcl11a), RP23-459O5 (Cd93), RP23-110M12 (Ltb), and RP23-320H10 (Mef2b). (n > 100 cells per gene per genotype per experiment). The data shown is an average of 3 experiments. Error bars indicate 95% CI). C, Bar chart showing the fraction of total XKD (gray bars) (from B) and versus Control B-cells (all Aid+/+) with co-localization of at least one gene specific BAC signal and γ-H2AX foci. D, Bar chart showing the fraction of cells with γ-H2AX foci colocalized with none, one or two Igh alleles in XKD and Control cells.
AID-dependent genomic breaks occur, but are not resolved, before the G1/S transition
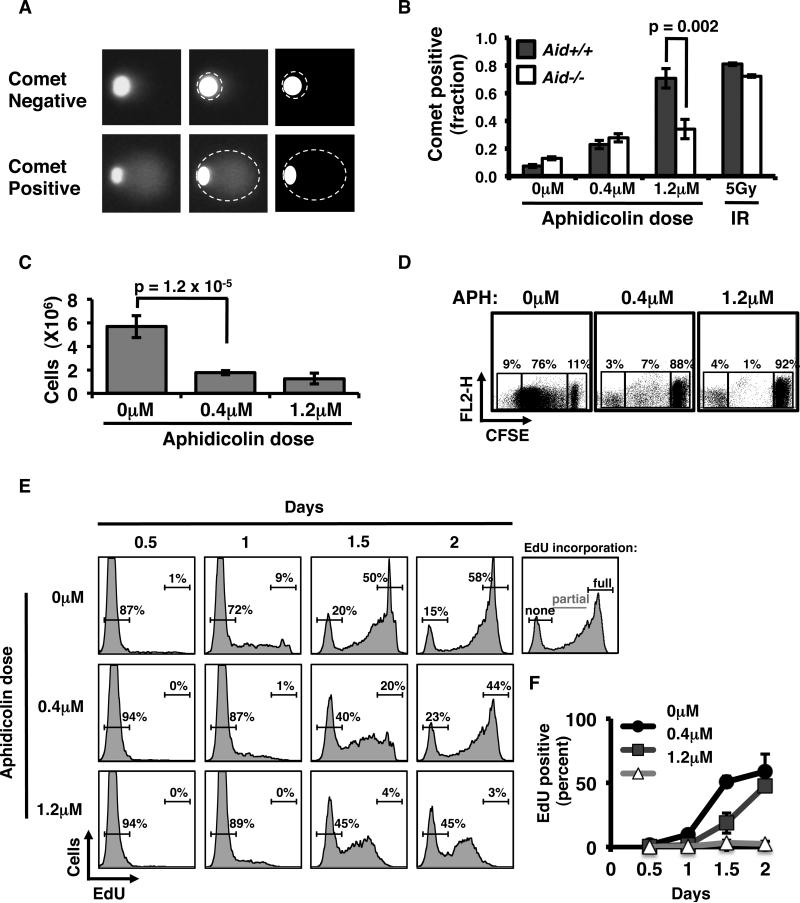
It has been definitively shown that AID-mediated Igh DSBs are generated during the G1 phase of the cell cycle (15, 38). While there has been no previous analysis of cell cycle dependency for repair of off-target DSBs, the requirement for HR suggests that off-target repair occurs, at least in part, in the S- or G2-phase. To determine the cell cycle dependency of AID-induced DSBs, we developed an assay system to distinguish G1 from S-phase chromosomal damage in mouse B-cells (Fig. 2A-B). Either cell cycle arrest at the G1/S boundary, or S-phase enrichment without complete arrest, was experimentally induced by exposure to different doses of the DNA polymerase inhibitor aphidicolin (APH). Exposure to1.2μM APH induced abrupt arrest at the G1/S transition point and led to accumulation of cells with a G1 DNA content, whereas, 0.4μM APH induced an initial accumulation of cells in G1-phase, and slowed but did not halt S-phase progression (Fig. 2C-F; Supplementary Fig. 1A-B). Exposure to either 0.4μM or 1.2μM APH inhibited overall B-cell expansion (Fig. 2C). However, both CFSE dilution and EdU incorporation analyses confirmed that 0.4μM-treated B-cells continued to cycle but to slowly transit S-phase, eventually completing replication; whereas 1.2μM–treated B-cells arrested with minimal DNA replication or cellular expansion (Fig. 2D-F). B-cells arrested at the G1/S boundary (1.2μM) showed a 10-fold increase in comet tails relative to untreated controls, reaching levels comparable to 5Gy irradiation (Fig. 2A-B). This effect was not due to general inhibition of DSB repair by APH, as exposure to neither 0.4μM nor 1.2μM APH impaired overall cell viability, or altered DSB repair kinetics in irradiated B-cells (Supplementary Fig. 1B-F). Importantly, after treatment with 1.2μM APH, Aid+/+ B-cells showed roughly a 2-fold higher frequency of comet tails than Aid-/- B-cells (0.71 versus 0.34, respectively). This demonstrates AID-dependency for the genomic damage, and confirms that AID induced DSBs arise, but do not resolve, prior to the onset of S-phase. In this context, cells treated with 0.4μM APH, which do not arrest at the G1/S boundary but transit S-phase slowly, exhibit a markedly lower frequency of AID-dependent comet tails. (Fig. 2B). Taken together, these data show that AID induces significant genomic damage prior to the G1/S boundary, and suggests that this damage may be repaired as cells transit S-phase.
Figure 2.
AID initiated DSBs occur in G1-phase of the cell cycle. A, Representative wide-field images from neutral comet assays showing comet negative (top panels), or comet positive (bottom panels) cells. Comet tails are segmented with dashed lines. B, Bar chart showing quantification of comet positive Aid+/+ versus Aid-/- purified B-cells, 2 days after activation. As a control, purified Aid+/+ or Aid-/- B-cells were exposed to 5Gy ionizing irradiation, and subjected to comet analysis. C, Average number of cells in culture after 4 days of activation; data represent 5 independent experiments, with 3 - 5 mice per experiment. D, Activated, splenic B-were cultured with 0μM, 0.4μM, or 1.2μM aphidicolin and loaded with 5μM Carboxyfluorescein succinimidyl ester (CFSE). Dilution of CFSE after 5 days was measured by flow cytometry. . Boxes show, from right to left, undivided, CFSE-diluted, and unlabeled cells, respectively. The percent of the population within each gate is shown. Data shown represent B-cells pooled from 3 independent mice. E, Purified B-cells were incubated with 10μM EdU and assayed for incorporation via flow cytometry every 0.5 days. The cells were analyzed for no, partial or full EdU incorporation. Representative histograms showing percent of EdU incorporation from each condition at 0.5, 1, 1.5 and 2 days. F, Line graph showing the average percent with full EdU incorporation (average from 3 independent tests). Error bars represent 95% CI; p-values were calculated using t-tests.
Unrepaired off-target DSBs are detectable during S-phase but are independent of DNA replication sites
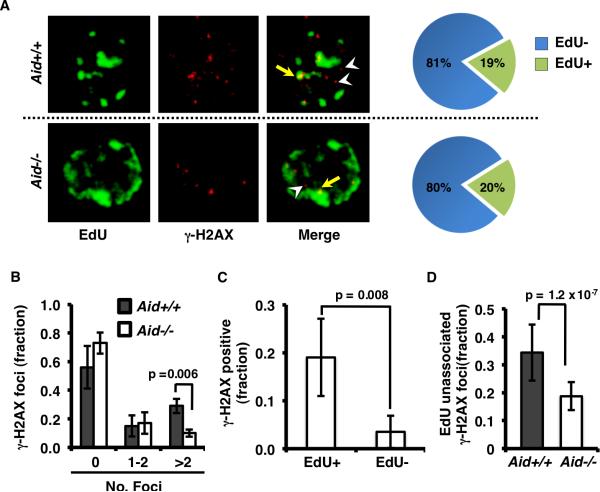
One model for the generation of off-target DSBs by AID suggests that they are DNA replication dependent, arising either by passage of a replication fork over an AID-generated single strand break (SSB) or by AID attack of the replication fork itself (23, 27). Either mechanism predicts that AID-dependent supernumerary (>2) DSBs should occur largely during S-phase, and coincide with sites of active DNA replication. To directly test these predictions, we evaluated co-localization of AID-mediated DSB foci with regions of nucleotide incorporation (replicating DNA). XKD B-cells from either Aid+/+ or Aid-/- mice were activated, pulse-labeled with EdU, and co-stained for γ-H2AX foci and EdU incorporation (Fig. 3A) (39). Consistent with our published results, XRCC2 deficient Aid+/+ B-cells showed a significant increase in supernumerary (>2) γ–H2AX foci relative to Aid-/- B-cells. (Fig. 3B) (25). This is consistent with our previously published results, showing that AID induces widespread off-target genomic DSBs. Co-stained cells were imaged by 3D confocal microscopy to quantify co-association of γ-H2AX foci with EdU-labeled DNA (Fig. 3C). We observed a higher fraction of AID-mediated DSBs in EdU-positive (replicating) cells than in EdU-negative cells, indicating that supernumerary DSBs accumulate in the S- or G2-phase (Fig. 3C). Surprisingly however, a significant fraction of the AID-initiated γ-H2AX foci present in the EdU+ cells, were not co-localized with the EdU labeling (Fig. 3C-D). This indicates that, although off-target DSBs accumulate during the S-phase of the cell cycle, they are frequently unassociated with sites of active nucleotide incorporation. While we do not rule out that some AID-dependent DSBs may coincide with DNA replication sites, these results demonstrate that AID-induced genomic breaks often occur independent of DNA replication forks, and thus form γ-H2AX foci that are not coincident with replication sites. This, together with prior reports that AID is physiologically active during G1, supports a model for off-target DSBs in which AID initiates bona fide DSBs at multiple non-Igh loci during the G1-phase of the cell cycle, analogous to CSR-initiating DSBs (15, 24, 38).
Figure 3.
AID-dependent DSB foci are present in S-phase, but are independent of replication forks. A, Aid+/+ or Aid-/- activated XKD B-cells were cultured for 1 day and pulsed labeled with EdU prior to fixation. EdU was detected following conjugation to AlexaFluor-488 (green) and γ-H2AX was detected with TRITC (red) conjugated antibody, and imaged by confocal immunofluorescence microscopy. Three dimensional (3D) image reconstruction was used to analyze cells for co-localization of EdU with γ-H2AX foci. Examples of γ-H2AX foci co-localized (arrows) and not co-localized (arrowheads) with EdU are highlighted in single optical sections. Pie charts indicate the percent of cells with EdU-positive (green) verses EdU-negative cells. B, Aid+/+ and Aid-/- XKD B-cells were scored for the number of γ-H2AX foci and grouped into bins of no foci (none), 1-2 foci, or >2 foci (supernumerary). Bar chart shows the fraction of cells within each bin for Aid+/+ (grey bars) verses Aid-/- (white bars) B-cells. C, Bar chart showing the fraction of activated Aid+/+ B-cells that were either EdU+ or EdU-. D, Bar chart showing the fraction of γ-H2AX foci per Aid+/+ or Aid-/- cell that were unassociated with EdU-labeled DNA. For each condition 20 individual cells were imaged (22-51 optical sections per cell); error bars represent 95% CI of the mean and the p-values were calculated using t-tests
AID-induced DSBs do not strongly activate the G1/S checkpoint
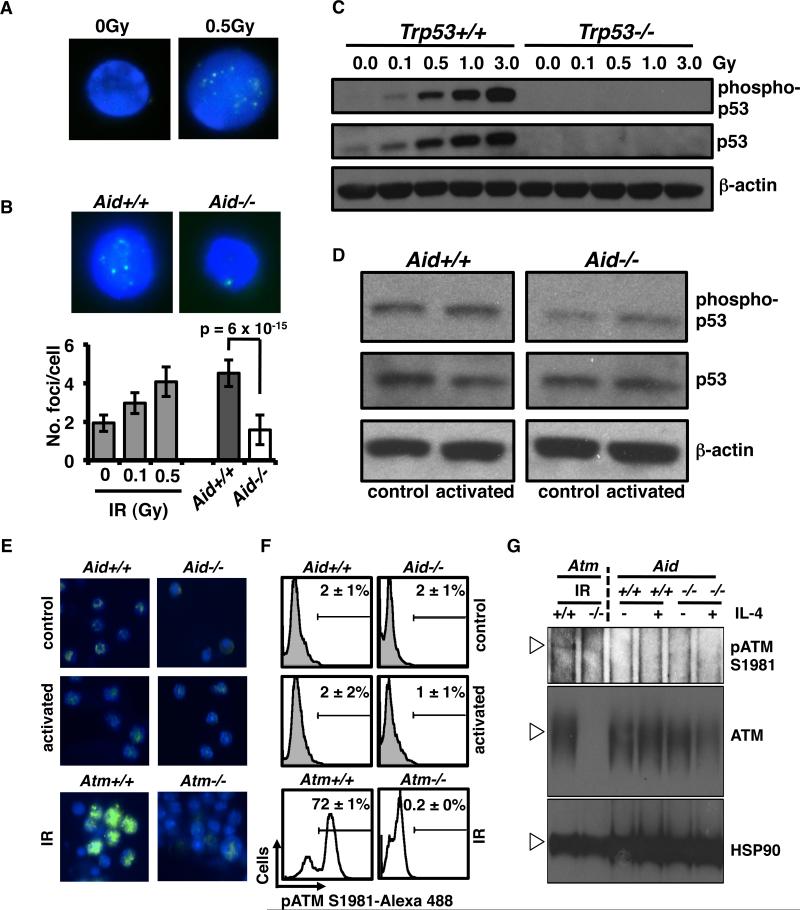
Our data suggest that AID-initiated DSBs, which arise in G1, can persist until S-phase. This implies that these DSBs do not activate conventional G1/S DNA damage checkpoints, and thus avoid repair until traversal into S-phase. To directly test this, we measured activation of p53 and ATM, both key components of the G1/S DNA damage checkpoint, in activated B-cells (Figure 4). As a positive control, splenic B-cells were exposed to ionizing radiation (IR) at doses ranging from 0.1 to 3.0Gy, confirming that even at the lowest tested doses, non-specific DNA double strand breaks could induce checkpoint activation (Fig. 4A-B). Western blot analysis of either total or phosphorylated p53 confirmed both an increase in total protein levels and in the activated phospho-p53 even at the lowest IR level tested (0.1Gy), with stronger activation at higher IR doses (Figure 4B). These data show that primary splenic B-cells are sensitive to exogenous DSBs and are able to activate p53 in response to total levels of DNA breakage. By contrast, activation of AID in wild-type splenic B-cells by αCD40 plus IL-4, which induced γ-H2AX foci roughly equivalent to 0.5Gy IR, did not strongly induce p53 or phospho-ATM (Fig. 4C-D). While we did observe slightly higher levels of phospho-p53 in Aid+/+ than Aid-/- B-cells, this occurred in both αCD40 and αCD40+IL-4 treated cells, suggesting that it was not directly related to CSR or AID (Fig. 4D). We did not observe differences in phospho-ATM expression between Aid+/+ and Aid-/- B-cells by immunofluorescence, intracellular flow cytometry, or Western blot assays, irrespective of activation (Figure 4E-G, Supplementary Fig. 2). This suggests that the observed p53 activation was non-specific, and likely associated with general proliferation induced by αCD40 rather than AID-induced DSBs. These results, together with recent studies that have investigated ATM and p53 in germinal center B-cells, imply that AID-initiated off-target DSBs, induced after αCD40 exposure, do not strongly activate the G1/S checkpoint, at least immediately following their induction (40-44).
Figure 4.
AID induced DSBs do not strongly activate G1/S checkpoint factors. A, Representative images of Aid+/+ or Aid-/- B-cell nuclei stained for γ-H2AX (green) after 0Gy or 5Gy ionizing irradiation. DNA was DAPI counterstained (blue). B, Representative images of Aid+/+ or Aid-/- B-cell nuclei stained for γ-H2AX (green) after activation. DNA was DAPI counterstained (blue). Bar chart (below) shows average number of γ-H2AX foci per cell after 0Gy, 0.1Gy, or 0.5Gy ionizing irradiation. Also shown are the average numbers of μ-H2AX foci per cell in activated XKD Aid+/+ versus Aid-/- B-cells. Error bars represent 95%CI the p-value was calculated by t-tests. For each assay 20 individual cells were imaged. C, Western blot analysis of total p53 (encoded by Trp53) and p53 phosphorylated at serine 15 (phospho-p53) in extracts from Trp53+/+ and Trp53-/- B-cells after ionizing irradiation at doses from 0 to 3.0 Gy. Top panel shows phospho-p53, middle panel shows total p53, and lower panel shows β-actin (loading control). D, Western blot analysis of phospho-p53 and total p53 from Aid+/+ and Aid-/-B-cells cultured with αCD40 plus IL-4 (activated) or αCD40 alone (control). β-actin, loading control. E, Representative images from untreated, or activated Aid+/+ and Aid-/- B-cells stained for ATM phosphorylated at serine 1981(green). Atm+/+ and Atm-/- B-cells treated with 0 or 3 Gy of ionizing irradiation were used as controls. DNA was counterstained with DAPI (blue).F, Histograms of untreated, or activated Aid+/+ and Aid-/- B-cells intracellularly stained for phospho-ATM serine 1981 and detected by Alexa 488 conjugated secondary antibody. Average perctent phosphor-ATM positive cells, determined from 3 independent experiments is indicated. Atm+/+ and Atm-/- B-cells treated with 0 or 3 Gy of ionizing irradiation were used as controls. G, Western blot analysis of total ATM and phospho-ATM (pATM S1981) from Aid+/+ and Aid-/- B-cells cultured with αCD40 plus IL-4 (activated) or αCD40 alone (control). HSP90 was used as loading control.
G1/S transition and class switch recombination
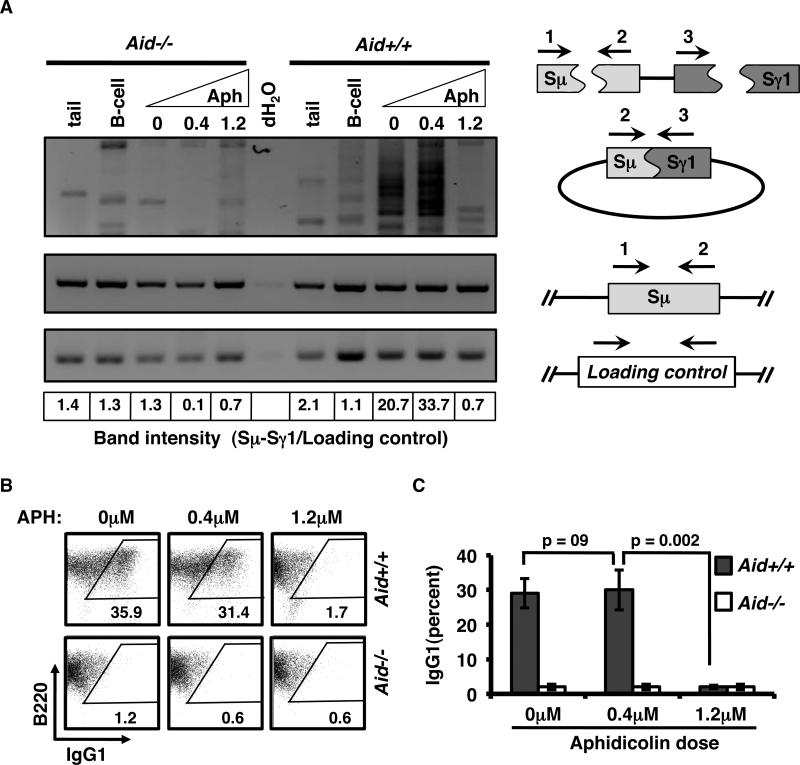
Previously published data have shown that CSR-initiating DSBs within Igh are generated during G1 and rapidly dissipate as cells traverse into and through S-phase (15, 24, 38). Because G1/S arrest appears to forestall resolution of AID-dependent off-target DSBs, the effect of G1/S arrest on resolution of AID-dependent CSR-initiating DSBs was also evaluated (Fig. 5; Supplementary Fig. 3). Activated B-cells were treated with 0μM, 0.4μM, or 1.2μM APH, and class switch recombination was quantified by a PCR assay to directly measure recombination products between the Sμ and Sγ1 elements of the Igh locus (Fig. 5A). As negative controls for Sμ-Sγ1 recombination, PCR analysis was also carried out for either unstimulated B-cells or for non-lymphoid DNA isolated from mouse tail. As a CSR specificity control, all assays were carried out in parallel in Aid+/+ and Aid-/- samples. Recombination products were readily detectable in both untreated and 0.4μM APH treated B-cells isolated from Aid+/+ mice (Fig. 5A). By contrast, recombination efficiency in 1.2μM APH treated B-cells was reduced approximately 30-fold relative to untreated controls, showing background levels similar to Aid-/- B-cells (Fig. 5A; Supplementary Fig. 3). Consistent with this finding, we found that exposure to 1.2μM APH completely abrogated both immunoglobulin class switching (Fig. 5B-C). This effect was not due to impaired proliferation, as the B-cells treated with 0.4μM APH showed dramatically reduced cellular proliferation, but underwent CSR as efficiently as untreated samples (Fig. 5B). Furthermore, the absence of recombination products or switching in 1.2μM APH treated B-cells was not due to AID inhibition (Supplementary Fig. 4) These results suggest that, at least under some conditions, CSR-initiating DSBs, though clearly generated by AID during G1, can be recombined in cells that traverse the G1/S boundary and enter into S-phase (3, 5, 15, 24, 38).
Figure 5.
CSR can occur during S-phase. A, The presence of Sμ to Sγ recombination products, was detected by PCR from genomic DNA isolated from activated Aid+/+ or Aid-/- B-cells treated with 0 μM, 0.4μM or 1.2μM APH. Internal Sμ and Xrcc2 (loading control) products were amplified as controls. Negative controls included B-cells treated with αCD40 alone, and genomic DNA prepared from mouse tails. Band intensity of the recombination products was determined by line scans (Supplementary Fig. 3) and the sum of intensities for the recombination products were quantified relative to loading control. Ratios are shown below the gel images. B, Representative FACS plots showing CD45R/B220 and IgG1 expression of activated Aid+/+ and Aid-/- B-cells, treated with 0μM, 0.4μM or 1.2μM APH after 4 days in culture. C, Bar chart showing the percent IgG1+ B-cells from activated Aid+/+ (grey bars) and Aid-/- (white bars) cultures treated with 0μM, 0.4μM or 1.2μM APH after 4 days in culture. The error bars are the 95% CI from 5 independent experiments (3 - 5 mice per experiment); p-values determined by t-test.
Discussion
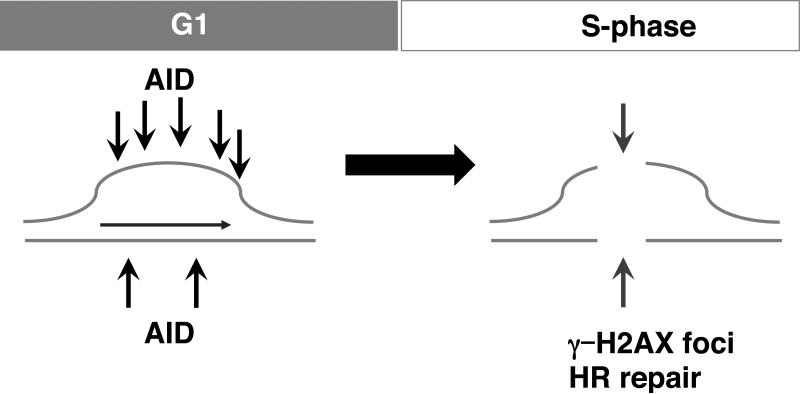
Here we have shown that AID initiates non-Igh (off-target) DSBs during the G1 phase of the cell cycle and these breaks can persist until S-phase for repair. We further demonstrate that AID initiated DSBs arise independently of DNA replication. We provide evidence that AID-mediated DNA breaks are generated during G1, but initially avoid activating the G1/S checkpoint, and then become sensed and repaired during S-phase. Interestingly, we find that CSR-initiating DSBs within Igh may, at least under some circumstances, undergo bona fide class switch recombination during S-phase. We propose that these cell cycle dynamics allow for coordination of repair at both Igh and non-Igh locations in cells harboring widespread damage from AID (Fig. 6). This would promote immunoglobulin class switching while simultaneously reducing risk of deleterious chromosomal rearrangements.
Figure 6.
Model for cell cycle modulation and coordinated repair of AID initiated DSBs. AID initiates DSBs in G1 phase of the cell cycle. DSBs that fail to resolve can bypass the G1/S checkpoint, and proceed into S-phase for resolution.
There is a growing body of evidence that shows AID induces both point mutations and DSBs at numerous non-Igh (off-target) locations throughout the genome (23-25). While the Igh locus is recognized as the preferred, physiological target for AID, recent studies have suggested that other, non-Igh genes may also be targeted by AID for point mutations, albeit at significantly lower frequencies (23, 26, 37, 45). AID has also been implicated in DSBs at non-Igh genes, although the precise mechanisms, frequency, and relationship to off-target point mutations has been unresolved (24, 25). We now show that, in addition to the Igh locus, AID initiates recurrent DSBs at a multiple other sites throughout the genome. We further demonstrate that homologous recombination defective cells accumulate unrepaired DNA breaks in Igh and in various non-Igh sites, demonstrating a role for HR in the resolution of both on- and off-target damage. Because productive class switch recombination is known to require DNA end-joining, we suggest that HR mediates high fidelity repair of AID-induced Igh DSBs leading to restoration of the original template without class switching. Although XRCC2 mediated HR is known to promote avian immunoglobulin pseudogene conversions, this is the first study to document a role for HR within the Igh locus in mammalian cells (46).
Two possible models for AID-initiated off-target DSBs can be envisaged. First, AID may generate base-pair mismatches that are directly converted into DSBs via processing of the mutated bases. Excision of two closely apposed bases, leading to juxtaposed single strand nicks, would generate a DSB. Alternatively, induction of single strand nicks could produce fragile sites that are converted to frank DSBs by mechanical stress. In either case, AID-mediated DSBs could arise independent of cell cycle stage. The second model, by contrast, suggests that off-target DSBs are replication dependent, arising either by passage of a replication fork over an AID-initiated single strand break (SSB), or by direct attack on the fork itself by AID (22, 27). By this model, AID-initiated off-target DSBs should largely coincide with sites of active nucleotide incorporation during S-phase. We now demonstrate that AID-dependent off-target DSBs, like those occurring in Igh, are generated during the G1 phase of the cell cycle and occur largely independently of DNA replication sites. Our findings indicate that off-target DSBs arise by mechanisms analogous to those that induce CSR within the Igh locus. In this context, non-Igh DSBs could be considered the products of an ectopic CSR-like process.
This poses an interesting dichotomy. CSR-initiating DSBs within Igh resolve predominantly in G1 via classical non-homologous end joining (NHEJ) or micro-homology mediated end joining (MMEJ). By contrast, off-target DSBs are preferentially repaired by HR, which is repressed during G1 but active in post-G1 cells (25). We now find that both off-target DSBs and Igh breaks are enhanced by HR deficiency (Figure 1B-C), suggesting a multi-phasic response to AID-initiated DSBs. We speculate that, under normal circumstances, the majority of Igh S regions DSBs are resolved via end-joining during G1 (15). However, we show that S region DSBs can be resolved during S-phase. Together, these findings suggest a cellular fail-safe mechanism — S region DSBs that fail to recombine in G1 can then be recombined later in S-phase. Moreover, we now demonstrate that HR can function in the Igh locus. We propose that S region DSBs that fail to undergo a switch recombination (in either G1 or S), can be repaired by high-fidelity HR, ensuring cell survival and providing another chance to switch in the next cell cycle.
How some AID-mediated DSBs that arise during G1 evade canonical checkpoint activation and persist until S-phase for repair is not understood. One possibility is that there are too few AID-dependent DSBs to reach a threshold level for checkpoint activation. While we cannot definitively rule out this possibility, previous reports have documented p53 activation in B-cells stimulated by LPS plus anti-δ-dextran, demonstrating checkpoint competency in mature splenic B-cells (40). Consistent with these, we show that splenic B-cells are acutely sensitive to checkpoint activation by IR-induced DSBs (Figure 4). Thus, another possibility is that checkpoint initiation by AID-mediated DSBs is context dependent, such that some stimuli (e.g. TLR signaling after LPS exposure) initiate checkpoints, while others (e.g. CD40 signaling) do not. This may be physiologically important, as CD40 signaling is key in germinal center formation, while TLR signaling is not.
Given that we find neither ATM nor p53 to be initially activated by AID-induced damage, Atm or Trp53 mutant B-cells might be expected to class switch at least as efficiently as wild-type. This seems to be partly true in the case of p53, although the effect of p53 on class switching appears to be mediated by its antioxidant role, rather than its checkpoint function (3). However, defects in Atm generally impinge upon CSR, suggesting that, although ATM initially escapes activation by AID-mediated DSBs activation, there are subsequently direct roles for ATM in promoting class switching. The partial inhibition of CSR seen in AT patients further suggests a direct role for ATM in promoting class switching (47). Taken together, these diverse observations support the conclusion that class switching B-cells mount a multifactorial response to AID-induced DNA damage, which includes checkpoint modulation and dynamic cell cycle regulation.
Why, and under what circumstances, should activated B-cells delay the repair of AID-initiated DSBs until S-phase? One possibility is that, in cells incurring numerous AID-initiated breaks, it is advantageous to delay repair until S-phase to ensure that both homologous and non-homologous modes of repair are available to properly repair appropriate targets. By this model, delayed repair would prevent resolution ofzs off-target, non-Igh DSBs by end-joining mechanisms, which could increase the likelihood of deleterious translocations in cells harboring numerous AID-induced DSBs (Figure 6). Delaying repair until S-phase would allow homology-mediated repair of off-target DSBs, but preserve the opportunity for CSR by end-joining processes. Another intriguing possibility is that some CSR events may involve S-phase, even in the absence of supernumerary DSBs at non-Igh locations. Previous studies have shown that physiological class switching is cell-division linked, where increased efficiency of class switching correlates with increased number of cellular divisions. Our data now show that proliferation per se is not strictly necessary for B-cells to class switch. Because cells treated with 0.4μM aphidicolin, and therefore enriched in S-phase and prevented from undergoing multiple rounds of cell division, are able to switch as efficiently as untreated wild-type B-cells, we suggest that S-phase transit is sufficient to promote class switch recombination, but that proliferation enhances CSR and amplifies switched subclones within a B-cell population. In this context, both S-phase progression and overall proliferation combine to magnify physiological class switching.
Mechanistically, S-phase may promote long-range recombination, which occurs at an unexpectedly high rate in class switching B-cells (35, 48). Finally, it is clear that both end-joining and homologous recombination modes of DSB repair are used for different purposes in activated B-cells. A major question is how repair pathway choice is regulated and how specific modes of repair might be spatially directed to particular target sites. While this question is not yet resolved, it is likely that S-phase progression influences the balance between end-joining and homology mediated modes of repair. Repair choice may be particularly relevant in B-cells undergoing CSR. In support of this notion, 53BP1, a key regulator of DNA end resection, modulates both the type of end-joining repair within switch regions, and the likelihood of ATM activation by persistent switch region breaks (47, 49-51).
In summary, we have shown here that AID initiates recurrent DNA double strand breaks at numerous locations throughout the genome, that these DSBs can arise during G1 independently of DNA replication, and that HR preferentially influences off-target DSB repair. This highlights a dichotomous response to Igh versus non-Igh DSBs in activated B-cells. Overall, our findings indicate a tightly regulated and highly orchestrated response to AID-initiated genomic damage that involves both cell cycle regulation and DNA repair pathway choice. Elucidating the detailed mechanisms that link AID, cell cycle dynamics, and DSB repair pathways provides insight into normal adaptive immune development and the etiology of B-cell neoplasms associated with genomic instability.
Supplementary Material
Acknowledgements
The authors thank Kristin Lamont, Caroline McPhee, and Rick Maser for discussion technical assistance, and critical reading of the manuscript; Ted Duffy and Will Schott for flow cytometry support; and Rafael Casellas (NIH/NIAMS) and Eric Brown (University of Pennsylvania) for advice and discussion and sharing of unpublished data.
This work was supported by National Institutes of Health Grants R01 CA138646 (K.D.M), P20RR018789 (D. Wojchowski, PI; K.D.M Project Leader), and P30 CA034196 (NCI/CCSG).
References
- 1.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, Manis J, Alt FW. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 3.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Ann. Rev. Immunol. 2008;26:261–92. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin. Immunol. 2004;16:257–275. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Stavnezer J. Complex regulation and function of activation-induced cytidine deaminase. Trends Immunol. 2011;32:194–201. doi: 10.1016/j.it.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pavri R, Nussenzweig MC. AID targeting in antibody diversity. Adv. Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- 7.Nussenzweig MC, Alt FW. Antibody diversity: one enzyme to rule them all. Nat Med. 2004;10:1304–1305. doi: 10.1038/nm1204-1304. [DOI] [PubMed] [Google Scholar]
- 8.Ramiro AR, Nussenzweig MC. Immunology: aid for AID. Nature. 2004;430:980–991. doi: 10.1038/430980b. [DOI] [PubMed] [Google Scholar]
- 9.Chua KF, Alt FW, Manis JP. The function of AID in somatic mutation and class switch recombination: upstream or downstream of DNA breaks. J. Exp. Med. 2002;195:F37–41. doi: 10.1084/jem.20020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storb U, Stavnezer J. Immunoglobulin genes: generating diversity with AID and UNG. Curr Biol. 2002;12:R725–727. doi: 10.1016/s0960-9822(02)01247-2. [DOI] [PubMed] [Google Scholar]
- 11.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription- targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 13.Boboila C, Yan C, Wesemann DR, Jankovic M, Wang JH, Manis JP, Alt FW. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J Exp Med. 2010;207:417–427. doi: 10.1084/jem.20092449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, Alt FW. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci U S A. 2010;107:3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J. Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 16.Franco S, Murphy MM, Li G, Borjeson T, Boboila C, Alt FW. DNA-PKcs and Artemis function in the end-joining phase of immunoglobulin heavy chain class switch recombination. J. Exp. Med. 2008;205:557–564. doi: 10.1084/jem.20080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 18.Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 19.O'Driscoll M, Cerosaletti KM, Girard PM, Dai Y, Stumm M, Kysela B, Jeggo P. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 20.Rush JS, Liu M, Odegard VH, Unniraman S, Schatz DG. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proc. Natl. Acad. Sci. U S A. 2005;102:13242–13247. doi: 10.1073/pnas.0502779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodgkin PD, Lee JH, Lyons AB. B cell differentiation and isotype switching is related to division cycle number. J. Exp. Med. 1996;184:277–281. doi: 10.1084/jem.184.1.277. 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 2009;30:173–181. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 24.Staszewski O, Baker RE, Ucher AJ, Martier R, Stavnezer J, Guikema JE. Activation-induced cytidine deaminase induces reproducible DNA breaks at many non-Ig Loci in activated B cells. Mol Cell. 2011;41:232–242. doi: 10.1016/j.molcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasham MG, Donghia NM, Coffey E, Maynard J, Snow KJ, Ames J, Wilpan R, He Y, King BL, Mills KD. Widespread genomic breaks generated by activation- induced cytidine deaminase are prevented by homologous recombination. Nat Immunol. 2010;11:820–826. doi: 10.1038/ni.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Nussenzweig MC. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–38. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen HM. Activation-induced cytidine deaminase acts on double-strand breaks in vitro. Mol. Immunol. 2007;44:974–983. doi: 10.1016/j.molimm.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 28.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartlerode AJ, Scully R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J. 2009;423:157–168. doi: 10.1042/BJ20090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2011;29:319–350. doi: 10.1146/annurev-immunol-031210-101329. [DOI] [PubMed] [Google Scholar]
- 31.Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 32.Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Nussenzweig MC. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr., Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 34.Caddle LB, Hasham MG, Schott WH, Shirley BJ, Mills KD. Homologous recombination is necessary for normal lymphocyte development. Mol. Cell. Biol. 2008;28:2295–303. doi: 10.1128/MCB.02139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dudley DD, Manis JP, Zarrin AA, Kaylor L, Tian M, Alt FW. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc. Natl. Acad. Sci. U S A. 2002;99:9984–9989. doi: 10.1073/pnas.152333499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc. Natl. Acad. Sci. U S A. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio M, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, Redon C, Reid T, Bonner WM, Honjo T, Nussenzweig MC, Nussenzweig A. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 40.Guikema JE, Schrader CE, Brodsky MH, Linehan EK, Richards A, El Falaky N, Stavnezer J. p53 represses class switch recombination to IgG2a through its antioxidant function. J. Immunol. 2010;184:6177–6187. doi: 10.4049/jimmunol.0904085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–9. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 42.Shansab M, Selsing E. p21 is dispensable for AID-mediated class switch recombination and mutagenesis of immunoglobulin genes during somatic hypermutation. Mol. Immunol. 2011;48:973–978. doi: 10.1016/j.molimm.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherman MH, Kuraishy AI, Deshpande C, Hong JS, Cacalano NA, Gatti RA, Manis JP, Damore MA, Pellegrini M, Teitell MA. AID-induced genotoxic stress promotes B cell differentiation in the germinal center via ATM and LKB1 signaling. Mol. Cell. 2010;39:873–885. doi: 10.1016/j.molcel.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Starczynski J, Simmons W, Flavell JR, Byrd PJ, Stewart GS, Kullar HS, Groom A, Crocker J, Moss P, Reynolds G, Glavina-Durdov M, Malcolm A, Taylor R, Fegan C, Stankovic T, Murray PG. Variations in ATM protein expression during normal lymphoid differentiation and among B-cell-derived neoplasias. Am. J. Pathol. 2003;163:423–432. doi: 10.1016/S0002-9440(10)63672-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride K, Klein I, Stone G, Eisenreich TR, Reid T, Nussenzweig A, Nussenzweig MC. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol. Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sale JE, Calandrini DM, Takata M, Takeda S, Neuberger MS. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- 47.Pan Q, Petit-Frere C, Lahdesmaki A, Gregorek H, Chrzanowska KH, Hammarstrom L. Alternative end joining during switch recombination in patients with ataxia-telangiectasia. Eur. J. Immunol. 2002;32:1300–1308. doi: 10.1002/1521-4141(200205)32:5<1300::AID-IMMU1300>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 48.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv. Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 49.Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J. Exp. Med. 2010;207:855–65. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1-/- B cells. Eur. J. Immunol. 2007;37:235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- 51.Lumsden JM, McCarty T, Petiniot LK, Shen R, Barlow C, Wynn TA, Morse HC, III, Gearhart PJ, Wynshaw-Boris A, Max EE, Hodes RJ. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J Exp Med. 2004;200:1111–1121. doi: 10.1084/jem.20041074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.