Abstract
Heterogeneity in the content and function of subcellular organelles on the intercellular and intracellular level plays an important role in determining cell fate. These variations extend to normal- and disease-state cellular functions and responses to environmental stimuli, such as oxidative stress and therapeutic drugs. Analytical tools to characterize variation in all types of organelles are essential to provide insights that can lead to advances in medicine, such as therapies targeted to specific subcellular regions. In this review, we discuss analytical techniques for interrogating individual intact organelles (e.g. mitochondria and synaptic vesicles) and lysates in a high-throughput manner, including a recently developed nanoscale fluorescence-activated subcellular sorter and techniques based on capillary electrophoresis with laser-induced fluorescence detection. We then highlight the advantages that droplet microfluidics offers for probing subcellular heterogeneity.
Introduction
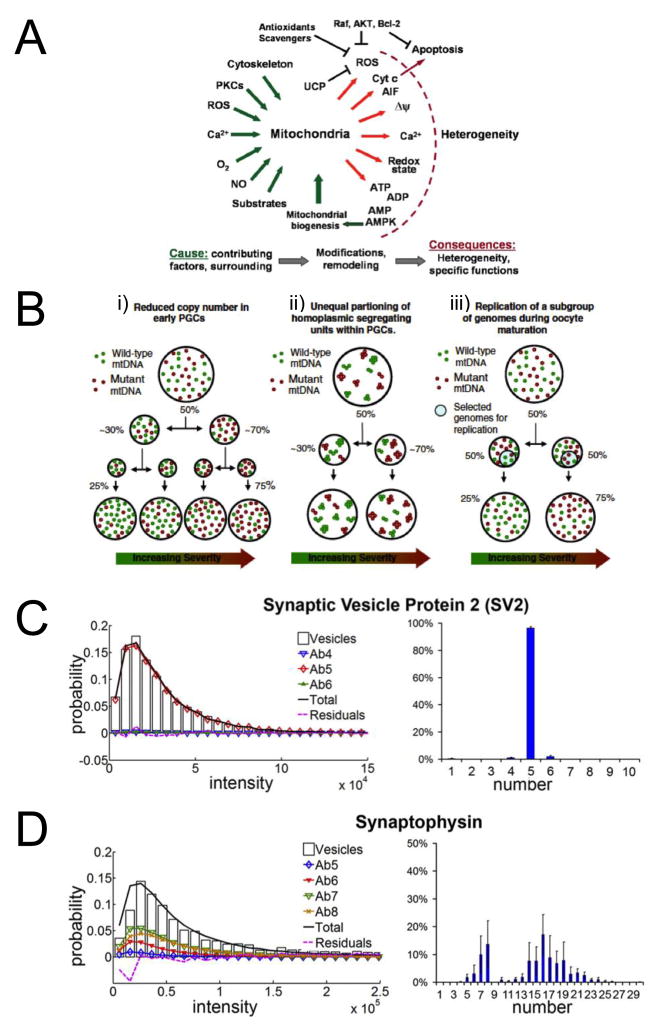
The existence of heterogeneity among individual cells of a given tissue type or even within a clonal cell line is well established; such variations are thought to be biologically meaningful, affecting outcomes at the organism level [1–5]. Similarly, because intercellular and intracellular variation exists in the composition and function of each type of organelle, ensemble analytical techniques do not adequately capture the details. Organelles, like the cells in which they reside, are versatile in function, adaptive to internal and external stressors, and subject to dysfunction. Within organelle populations, therefore, considerable heterogeneity exists as a result of factors such as tissue type, pathology, and location within the cell. Superimposed on these patterns are time-dependent changes related to aging (e.g. formation of giant mitochondria [6]) as well as stochastic variation in all cellular processes over time [1,2,7]. Figure 1 highlights heterogeneity in mitochondria and synaptic vesicles, two of the organelles examined by techniques reviewed here.
Figure 1.
A–D) Schemes and plots relating some of the current research interests in organelle heterogeneity. A) Scheme summarizing the specific questions and hypotheses regarding origin and possible mechanisms contributing to the heterogeneity of mitochondria and mitochondrial function. Reproduced with permission from Ref. [27]. B) Proposed mechanisms for the mtDNA genetic bottleneck. i. Variation in heteroplasmy levels are generated through the unequal partitioning (segregation) of mutant and wild-type genotypes during cell division. This leads to accelerated drift in heteroplasmy levels which occurs principally as a consequence of the dramatic reduction in mtDNA copy number immediately prior to primordial germ cell (PGC) population expansion. ii. Variation in heteroplasmy generated through the unequal segregation of homoplasmic mucleoids in PGCs. Each nucleoid contains multiple copies of mtDNA which are identical. iii. Variation in heteroplasmy generated through the replication of a subpopulation of mitochondrial genomes during oocyte maturation in post-natal life. Increasing severity refers to the clinical consequences of a higher percentage level of mutant mtDNA. Reproduced with permission from Ref. [28]. Quantification of C) monodispersed and D) polydispersed synaptic vesicle proteins. Plots of representative fits for each protein is shown on the left, and histograms showing the percentage of vesicles containing n number of the indicated vesicle protein averaged across ~10,000–20,000 vesicles is shown on the right. Error bars, SE between different datasets. Reproduced with permission from Ref. [18].
Techniques capable of high-throughput, high-sensitivity analysis of organelles are essential for gaining a deeper understanding of cellular function and can lead to advancements in medicine. For example, the elucidation of the normal- and disease-state function of organelles may aid in development of targeted therapies. There is a broad range of analytical targets in subcellular particles. All organelles contain proteins; most contain or generate small molecules; and some, like mitochondria, carry their own genetic material.
In this brief review, we will focus on a few examples of analyses of organelle heterogeneity and use these examples to address current issues in the field. We will not cover single-cell analytical tools in great detail although many are capable of determining some aspects of intracellular variation in organelles and are necessary to examine organelle function in its native environment. For example, Zhang et. al developed an array of carbon microfiber electrodes for monitoring exocytosis from multiple sites on a single cell simultaneously [8]. We acknowledge the relevant and meritorious work of many researchers that is not described here. The examples covered in the current review include separation of subpopulations of synaptic vesicles by fluorescence-activated sorting (P Schiro et al., unpublished) and separation of mitochondria by capillary electrophoresis (CE) [••9]. We will then briefly describe current challenges in subcellular analysis and the potential of droplet microfluidics to address some of these challenges.
Single-cell analysis: the same toolbox, a different level of challenge
High-throughput analyses of individual organelles would be premature without first dealing with single-cell heterogeneity. There is now an abundance of single-cell analytical techniques that probe living cells and cell lysates, some with a relatively high throughput [3]. These techniques have been extensively reviewed elsewhere [1–5]. For example, recent advances have enabled sampling of selected cells for single-cell CE [10], mass spectrometry imaging (MSI) with submicron resolution [••11], multi-component and long-term studies [3,5], cytotoxicity screening [12], and gene expression analysis [13,14] as well as bioinformatics approaches to handling the large amount of data generated in these studies [1]. Many of these single-cell techniques rely on microfluidic platforms [3,15].
The toolbox available for single-cell sampling, sorting, manipulation and interrogation can be adapted for characterization of individual organelles; but the challenges in certain aspects, such as sampling, throughput, sensitivity, and possible contamination, are amplified. For example, mass spectrometry (MS) is sufficiently sensitive to probe the metabolome of a single cell (low femtomoles of metabolites per few pLs of cell volume) and many protein targets; but MS cannot be used for direct detection of some low-abundance proteins, which are present at <1000 copies per cell [3,5]. In contrast to cells, individual organelle volumes are atto- to femto- liters, and contain attomolar or fewer quantities of metabolites and proteins, even as low as single copy numbers. However, advancements in MS sampling and amplification schemes have enabled more in-depth profiling of single cells and organelles. For example, a strategy for quantifying low-copy-number proteins is mass cytometry (CYTOF), which is a recently developed single-cell analysis technique [••16]. In CYTOF, single cells are stained with tens of antibodies conjugated to distinct metal isotope-containing polymers prior to time-of-flight MS detection. Using this signal amplification technique, proteins from single cells were detected with sensitivity approaching that of fluorescence techniques [••16]. In a separate experiment, Sweedler and coworkers reported matrix-assisted laser desorption/ionization (MALDI) MS detection of peptides from single secretory vesicles sampled from Aplysia californica [17].
Several effective strategies exist to probe low abundance targets, including amplification schemes and detection methods with single-molecule sensitivity such as certain fluorescence and electrochemistry techniques. For example, our group developed a total-internal-reflectance-fluorescence (TIRF) microscopy technique to count low-copy-number membrane proteins in synaptic vesicles [•18,19], which release neurotransmitters to propagate signals between neurons. We employed single-molecule intensity distributions and statistical analysis methods to account for fluorescence variability and to accurately describe population distributions [•18,19]. We found some vesicle proteins to be highly monodisperse in copy number (e.g. SV2) and exhibit little variation between vesicles; in contrast, other vesicle proteins are highly polydisperse in copy number (e.g. synaptophysin). This polydispersity may represent variability between classes of synaptic vesicles, influencing the probability that neurotransmitters will be released from these vesicles into the synaptic cleft [•18]. Figures 1c and 1d show examples of each a monodispersed and polydispersed protein, respectively.
Sample preparation by fractionation of subcellular organelles
The current approach in proteomics is to examine organelle fractions (rather than cell homogenates) by MS, thereby breaking the complex human proteome into simpler components and providing information about protein subcellular localization, which is key to understanding functionality [20,21]. From these studies, we now know that some proteins, for instance, are located in multiple types of organelles and are transported between different areas of the cell [20,21].
The quality of the data depends mainly on the purity of the organelle fraction. Classical methods of fractionation include, most prominently, differential centrifugation and density gradient centrifugation [20,21]. A number of more recently developed methods generate fractions of greater purity, especially for organelles that normally sediment at the same rate as other organelles or contaminants and are therefore harder to purify [20,21]. These methods are listed in Table 1 along with a short description of each. These fractionation strategies can be used to purify and enrich a single type of organelle before interrogating them individually.
Table 1.
| Method | Description | Strengths and weaknesses |
|---|---|---|
| Free-flow electrophoresis | Subcellular components (often pre-fractionated by centrifugation) are flowed through a perpendicular electric field, which separates components into different flow laminae based on their individual electrophoretic mobilities. | High selectivity for many organelle types Appropriate for preparative scale separation Viability of organelles maintained |
| Alteration in buoyant density | Plasma membranes are treated with dilute detergent (lysis detergent such as Digitonin), thus decreasing their density, prior to density gradient centrifugation. Phagocytosis (ingestion and encapsulation into membrane-bound structures called phagosomes) of latex beads causes phagosomes to float in a sucrose gradient. |
Simple and selective yet limited to a few specific organelles |
| Fluorescence-activated organelle sorting (FAOS) | Derived from fluorescence-activated cell sorting (FACS), subcellular components are labeled with organelle-specific fluorescent antibodies and flowed passed a detector. Fluorescent events trigger sorting of labeled organelles into a collection stream. The collection stream may be integrated to instrumentation or a microwell plate for downstream analysis. | Highly selective and rapid (kHz) Provides the means to measure several characteristics of each subcellular particle Maintains viability although some organelles may be damaged by shear stress Requires special instrumentation |
| Immunoaffinity purification | A solid support or beads* are modified with an antibody specific to an antigen at the surface of the organelle of interest allowing its extraction from cell homogenate or pre-fractionated organelles. The captured organelle fraction is washed to remove non-specifically adsorbed components, and then the antibody-antigen interaction is interrupted to recover the purified fraction. *In the case of magnetic beads, the beads are exposed to cell homogenate and then extracted magnetically onto a solid support. |
Highly selective Gentle, maintaining viability Low capacity for retained material Relatively expensive due to reagent costs |
Separating subpopulations based on fluorescence-activated sorting: Nanoscale fluorescence-activated subcellular sorter (nFASS)
Of the fractionation strategies listed in Table 1, several can be adapted to probe organelle heterogeneity. For example, fluorescence-activated organelle sorting (FAOS), much like its better-known cell-sorting counterpart (FACS), is a flow sorter triggered by fluorescent events. Organelles are labeled with fluorescently tagged antibodies that interact with a highly expressed surface protein specific to the organelle of interest. Because of this label-based sorting, FAOS can generate highly pure fractions, especially at slower sorting rates. FAOS has been adapted for protein [20] as well as mtDNA [22] quantification. However, current FAOS sorting rates are too low for conveniently processing the large number of organelles required for population statistics, and the sensitivity of current FAOS systems is too low for detection of a single dye molecule.
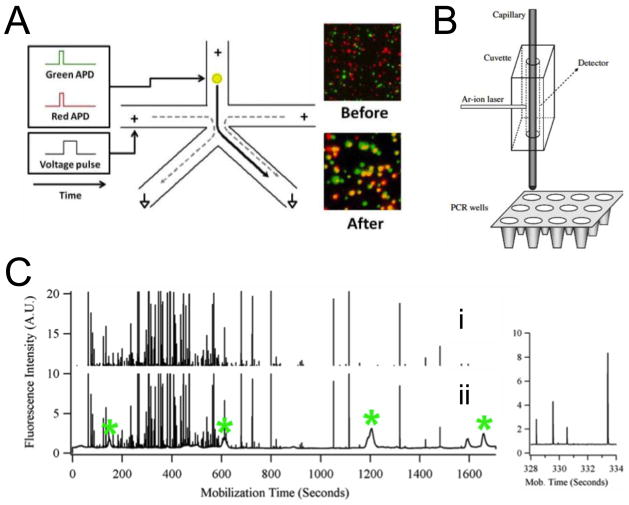
Our group recently developed nFASS, which is capable of sorting subcellular organelles at rates of ~10–20 kHz (91% recovery, 99.7% purity) and with single molecule sensitivity (Figure 2a; P Schiro et al., unpublished). nFASS uses nanoscale channel dimensions and electroosmotic-flow switching to dramatically increase sorting speed. Laser-induced fluorescence (LIF) with beam shaping decreases probe volume to improve sensitivity by decreasing background fluorescence. The advantages of the technique were demonstrated by sorting SV2- and VAMP-labeled synaptic vesicles. Multi-color sorting enables selection or rejection based on multiple protein targets. This technique is a powerful tool for enriching organelle subpopulations and can be adapted for many types of organelles. Most importantly, its rapidity allows collection of sufficient data for statistical description of population variance (P Schiro et al., unpublished). However, in its current design, nFASS can sort organelles into only two subpopulations at a time. This limitation may be overcome by simply running sequential experiments or nFASS devices in series. Furthermore, with its continuous-flow design, certain organelle analyses that involve reactions that generate freely diffusing fluorescent species (e.g. enzyme activity assays or RT-PCR) cannot be accomplished. This latter problem may be addressed by droplet encapsulation [23].
Figure 2.
A) Schematic representation of the nanoscale fluorescence-activated sorter (nFASS) and TIRF microscopy images showing synaptic vesicles labeled with SV2 (red) and VAMP (green). A fluorescent event in the top (inlet) channel for a synaptic vesicle labeled with both SV2 and VAMP is detected at both red and green avalanche photodiodes (APDs). The detection event triggers a voltage pulse (left side-channel), which causes electroosmotic flow sorting of the detected vesicle to the collection channel (bottom right). ‘Before’ image demonstrates a low rate of co-localization of SV2 and VAMP before sorting. ‘After’ image demonstrates co-localization (shown in yellow) after only those vesicles bound with both antibodies triggered a sorting voltage pulse and were collected (P. Schiro et al., unpublished). B) On-column detection and collection of mitochondria. Mitochondria are detected individually by on-column LIF. A section of a PCR plate is placed on a collection tray that allows positioning of the capillary inside PCR wells to collect mitochondria. Mitochondrial particles observed in ~10 s time windows were collected in the same vial as they reached the end of the capillary. Reproduced with permission from Ref. [24]. C) Signal processing to select mitochondrial events. i. Mitochondrial events selected after application of median filter and clipping of the lower signal. There are 360 mitochondrial events detected. The inset at the right is an expanded view of a crowded section of the electropherograms showing the resolution between individual spikes. ii. Signal during mobilization step of focused mitochondria and pI marker internal standards. Narrow spikes represent individual mitochondrial events; broad peaks represent pI markers or fluorescent species from the carrier ampholytes. The pI markers are indicated with an asterisk (*). Reproduced with permission from Ref. [9].
Capillary electrophoresis with laser-induced fluorescence as a tool for segregating and analyzing unknown subpopulations
Arriaga and coworkers have developed techniques based on capillary electrophoresis with laser-induced fluorescence (CE-LIF) that enrich subpopulations of mitochondria [••9,•24,25]. Heterogeneity of mitochondria is the focus of many research areas related to disease and aging [26–28]. Figure 1a shows several causes and the many resulting manifestations of mitochondrial heterogeneity [27]. Besides supplying energy to the cell via oxidative phosphorylation, mitochondria are involved in cell-signaling pathways. There can be thousands of mitochondria in a eukaryotic cell and the number is roughly proportional to the cell’s energy demands [26,27]. Mitochondria are organized in networks within the cell. There is evidence that mitochondria communicate within these networks and are organized into clusters with specialized functionality depending on their location within the cell with respect to other organelles, such as the nucleus [27].
Given their wide-range of functionality and complex organization, mitochondrial populations can be divided into many subpopulations. Electrophoretic techniques offer the advantage of separating organelles with high resolution based on physical differences linked to composition and function. For example, Arriaga and coworkers recently developed a capillary isoelectric focusing (cIEF) technique [••9], which separated mitochondria labeled with 10-N-acridine orange (NAO) by isoelectric point. Isoelectric point is dependent on membrane composition of lipids and proteins. In experiments by Arriaga and coworkers, each cIEF run generated 400–500 mitochondrial events with high resolving power (ΔpI = 0.01) (Figure 2c). In a different study by this group, the CE-LIF setup was integrated with a microwell plate for real-time quantitative polymerase chain reaction (qRT-PCR) of mitochondrial DNA (mtDNA) [•24] (Figure 2b). However, Arriaga and coworkers reported that microwells contained a range of numbers of mitochondria (0–64) and that false positive results in empty wells indicated contamination [•24].
The advantages of CE-LIF techniques are that they provide high-resolution separation of a large and unknown number of mitochondrial subpopulations based on relevant characteristics such as membrane composition and organelle size; offer high-sensitivity detection of fluorescent labels; and are easily integrated with the segregation of isolated organelles into microwell plates for downstream analysis. The current shortcomings of these CE-LIF techniques are that the separation criteria rely on physical parameters that are not unique to a given organelle size and membrane composition and do not reveal which specific proteins or lipids comprise the membrane and in what proportions. However, as downstream analyses are performed, a clearer correlation between these physical parameters and composition, and ultimately organelle function, may develop.
We have demonstrated high-throughput CE-LIF analysis of the contents of individual mitochondria in a microfluidic device [29]. In this study, we employed a membrane-permeable amine-reactive dye, Oregon Green diacetate succinimidyl ester, to derivatize the amine contents of the mitochondria prior to rapid laser photolysis and CE-LIF separation and detection. Diffusional broadening of the mitochondrial contents was countered by rapid CE separation (a few milliseconds). The resulting electropherograms indicated a multi-modal distribution with three distinct subpopulations [29]. The advantage of this approach is that the data are detailed, which allows multiple components to be measured and correlated.
Droplets as a tool for high-throughput/high-sensitivity analysis of organelle contents
The primary motivation for droplets in organellar analysis is compartmentalization and confinement. A droplet can contain lysis products during examination of intraorganellar species; provide a reaction vessel for assays (e.g. ELISA or PCR); act as storage containers; and prevent cross-contamination prior to analysis [30,31]. Microfluidic devices are capable of rapidly generating droplets [31], and provide a platform for the large number of experiments that are required for measuring stochastic variations in organelles and for collecting relevant statistical data. The droplets can be made highly monodisperse (<1% variation in diameter [31]), and the femtoliter to picoliter size range is appropriate for the encapsulation of individual organelles (for example, mitochondria have diameters ranging from 0.5 μm to 1.0 μm, and the droplets can range from tens of μm to a few μm in size).
Microfluidic devices excel at handling droplets [30,31] and perform a variety of functions, which include generating droplets with a single particle per droplet, sorting (fluorescence-activated droplet sorting [23]), merging, splitting, storage, and integrating with instrumentation (e.g. MS or CE) via merging droplets with a continuous-flow aqueous phase or direct injection [30,31,32].
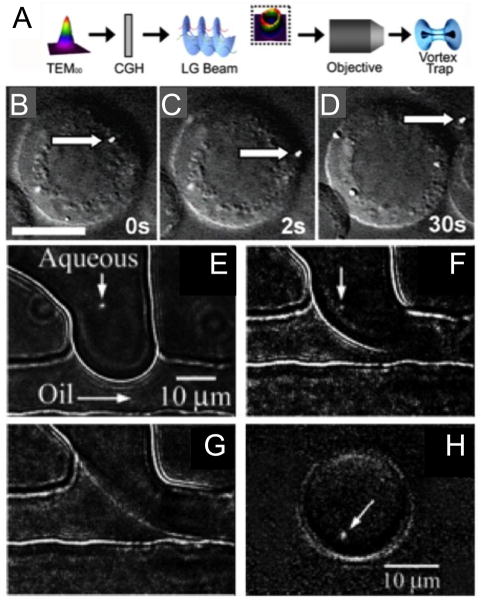
Our group has developed many unique droplet-handling strategies that can be used for subcellular analysis [30] including in-flow droplet freezing, droplet size and concentration manipulation, and particle sizing in droplets. The sizing of subcellular organelles in droplets is useful for probing heterogeneity in organelle morphology and the concentration of analytes [33]. We have also demonstrated several organelle-handling strategies, including single-cell nanosurgery (Figure 3a–d) [34] and droplet encapsulation of individual mitochondria (Figure 3e–3h) [35]. While these handling techniques are not currently high-throughput, they offer exquisite control and precision, and further automation and parallelization would make them powerful tools for examining organelle heterogeneity.
Figure 3.
A) Schematic showing the conversion of a Gaussian beam (TEM00) into that of an optical vortex beam (Laguerre-Gaussian (LG) beam) by using a computer generated hologram (CGH) and B–D) a sequence of images showing the removal of a fluorescent lysosome (stained with Lysotracker Green dye) from a B-lymphocyte. Scale bar = 10 μm in panel B. Reproduced with permission from Ref. [34]. E–H) Sequence of images showing the droplet encapsulation of a single mitochondrion. We visualized under fluorescence and optically manipulated a single mitochondrion stained with Mitotracker Green FM at the interface of the two fluids (E, F). Upon application of a pressure pulse to the microchannels (F, G), the mitochondrion was carried away by the flow as the droplet was sheared off (H). Reproduced with permission from Ref. [35].
To illustrate the advantages of droplet microfluidics in organelle analysis, consider the mitochondrial heteroplasmy problem, which is the coexistence of mutant and wild-type mtDNA in an organism and the cause of a number of intractable mitochondrial diseases.
Approximately 5–10 double stranded circular mtDNA molecules are contained in each of several DNA-protein complexes called nucleoids in the lumen of mitochondria [•24,36]. The mtDNA codes for unique mitochondrial proteins essential to cellular respiration as well as tRNAs and rRNAs necessary for its own transcription [28]. The mtDNA is not protected by compartmentalization and tight winding around histones, as is nuclear DNA. Its close proximity to the apparatus for oxidative phosphorylation (ATP generation) subjects mtDNA to constant mutagenic exposure to reactive oxygen species [28]. Therefore, mutations in mtDNA are common. The existence and severity of mitochondrial disease traits are dependent on the ratio of mutant to wild-type mtDNA in the cell (referred to as the degree of heteroplasmy) [28].
Distribution of heteroplasmy at the subcellular level is also relevant to questions of mtDNA inheritance [28]. Mitochondrial diseases are heritable but do not follow the same pattern of inheritance, as does genomic DNA. The mtDNA in offspring is solely maternal in origin. However, the level of heteroplasmy in the mother often does not match that of her offspring, leading researchers to search for a bottleneck at which some mtDNA is lost during embryogenesis. The location of this genetic bottleneck is debated, and certain studies even suggest that the loss process is selective, creating polarization of the offspring into high and low disease-load groups. Figure 1b shows three proposed mechanisms causing the bottleneck during the proliferation of primordial germ cells (PGCs, the precursors to oocytes) in a developing embryo. All three mechanisms assume the basic units of inheritance are either single mtDNA molecules or homoplasmic nucleoids [28]. Recent studies suggest that nucleoids are probably homoplasmic [•24,37]. Importantly, knowing the level at which heteroplasmy exists helps to narrow the possible mechanisms for inheritance patterns.
Using the tools available for droplet microfluidics, we can envision a process flow to address questions of heteroplasmy. An integrated microfluidic device could be used to analyze a mitochondrial fraction as follows. First, individual mitochondria are encapsulated in droplets using on-demand droplet generation triggered by the presence of each organelle. Second, the droplets are merged with a stream of droplets containing reagents for qRT-PCR to analyze the wild-type and mutant mtDNA strands. Third, the pairs of droplets are fused (either passively or by electrocoalescence). Fourth, laser photolysis bursts open the mitochondria. Then, the droplets are transported through heated and cooled regions of the chip to facilitate several rounds of the PCR. Finally, the droplets undergo LIF detection of the RT-PCR products revealing the degree of heteroplasmy in each organelle.
However, it is important to note that droplet microfluidics has not yet gained widespread acceptance for characterizing organelle heterogeneity because several advancements are yet to be made. In particular, droplets are a challenge for device integration to common instrumentation, such as MS, that are needed in downstream analysis of droplet contents. Furthermore, such instrumentation is often not sufficiently sensitive for measurements at the single organellar level. Most significantly, the many droplet-handling techniques that have emerged over the last decade have yet to be integrated seamlessly with one another [31], owing to cross-talk between modules and increased complexity.
Future outlook and conclusions
The most pressing challenge in high-throughput analyses of organelles is the need for strategies to segregate organelles into subpopulations based on information rich and relevant characteristics and to achieve a level of discrimination that accurately represents the level of heterogeneity among organelles. Fluorescent labeling provides high-specificity, but comparisons based on biological activity (e.g. from enzymatic assays) may yield more relevant information. Droplet microfluidics promises to achieve these goals by providing reaction vessels for probing organelle activity and intraorganellar contents. However, integration of droplet microfluidic techniques with one another and with external instrumentation has yet to be fully realized.
Highlights.
Characterization of heterogeneity and variation in subcellular organelles is key to understanding their normal-and disease-state functions and responses to external stimuli (e.g. oxidative stress or therapeutic agents).
New analytical tools are required to segregate organelles into subpopulations and to perform high-throughput, high-sensitivity, and information-dense measurements of their contents and processes.
Nanoscale fluorescence-activated subcellular sorting (nFASS) and capillary electrophoresis with laser-induced fluorescence (CE-LIF) separation and detection of organelles are two recently developed and promising techniques for characterizing organelle heterogeneity.
Droplet microfluidics has the ability to overcome many of the challenges in subcellular analysis but requires advancements in downstream analytical techniques to make full use of its strengths.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Roach KL, King KR, Uygun BE, Kohane IS, Yarmush ML, Toner M. High throughput single cell bioinformatics. Biotechnol Prog. 2009;25:1772–1779. doi: 10.1002/btpr.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kovarik ML, Allbritton NL. Measuring enzyme activity in single cells. Trends Biotechnol. 2011;29:222–230. doi: 10.1016/j.tibtech.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin H, Marshall D. Microfluidics for single cell analysis. Curr Opin Biotech. 2012;23:110–119. doi: 10.1016/j.copbio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Lindström S, Andersson-Svahn H. Overview of single-cell analyses: microdevices and applications. Lab Chip. 2010;10:3363–3372. doi: 10.1039/c0lc00150c. [DOI] [PubMed] [Google Scholar]
- 5.Wu M, Singh AK. Single-cell protein analysis. Curr Opin Biotech. 2012;23:83–88. doi: 10.1016/j.copbio.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navratil M, Terman A, Arriaga EA. Giant mitochondria do not fuse and exchange their contents with normal mitochondria. Exp Cell Res. 2008;314:164–172. doi: 10.1016/j.yexcr.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 7.Kalisky T, Blainey P, Quake SR. Genomic analysis at the single-cell level. Annu Rev Genet. 2011;45:431–445. doi: 10.1146/annurev-genet-102209-163607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, Adams KL, Luber SJ, Eves DJ, Heien ML, Ewing AG. Spatially and temporally resolved single-cell exocytosis utilizing individually addressable carbon microelectrode arrays. Anal Chem. 2008;80:1394–1400. doi: 10.1021/ac702409s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Wolken GG, Kostal V, Arriaga EA. Capillary isoelectric focusing of individual mitochondria. Anal Chem. 2011;83:612–618. doi: 10.1021/ac102712r. Intact mitochondria are separated via capillary isoelectric focusing with laser-induced fluorescence detection demonstrating the high degree of heterogeneity in outer membrane composition of the hundreds of mitochondria analyzed per run. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cecala C, Sweedler JV. Sampling techniques for single-cell electrophoresis. Analyst. 2012 doi: 10.1039/c2an16211c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Lanni EJ, Rubakhin SS, Sweedler JV. Mass spectrometry imaging and profiling of single cells. J Proteomics. 2012 doi: 10.1016/j.jprot.2012.03.017. An excellent review of mass spectrometric imaging and profiling techniques for single cells with subcellular resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rotheberg JM, Link DR, Perrimon N, Samuels ML. Droplet microfluidic technology for single-cell high-throughput screening. PNAS. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White AK, VanInsberghe M, Petriv OI, Hamidi M, Sikorski D, Marra MA, Piret J, Aparicio S, Hansen CL. High-throughput microfluidic single-cell RT-qPCR. PNAS. 2011;108:13999–14004. doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong JF, Chen Y, Marcus JS, Scherer A, Quake SR, Taylor CR, Weiner LP. A microfluidic processor for gene expression profiling of single human embryonic stem cells. Lab Chip. 2008;8:68–74. doi: 10.1039/b712116d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zare RN, Kim S. Microfluidic Platforms for Single-Cell Analysis. Annu Rev Biomed Eng. 2010;12:187–201. doi: 10.1146/annurev-bioeng-070909-105238. [DOI] [PubMed] [Google Scholar]
- 16••.Bendall SC, Simonds EF, Qiu P, Amir E-aD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. The authors report a single-cell analysis technique, in which proteins are labeled with metal-isotope conjugated antibodies for ultra-sensitive high-throughput screening. This technique holds promise for analysis of individual organelles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubakhin SS, Garden RW, Fuller RR, Sweedler JV. Measuring the peptides in individual organelles with mass spectrometry. Nat Biotech. 2000;18:172–175. doi: 10.1038/72622. [DOI] [PubMed] [Google Scholar]
- 18•.Mutch SA, Kensel-Hammes P, Gadd JC, Fujimoto BS, Allen RW, Schiro PG, Lorenz RM, Kuyper CL, Kuo JS, Bajjalieh SM, et al. A quantitative single vesicle imaging technique reveals a subset of synaptic vesicle membrane proteins are trafficked with high precision. J Neurosci. 2010;31:1461–1470. doi: 10.1523/JNEUROSCI.3805-10.2011. The authors use TIRF microscopy to quantify the average and variance in protein copy number for seven target proteins in synaptic vesicles. Importantly, this method is able to quantifying few to tens of proteins in subcellular particles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mutch SA, Gadd JC, Fujimoto BS, Kensel-Hammes P, Schiro PG, Bajjalieh SM, Chiu DT. Determining the number of specific proteins in cellular compartments by quantitative microscopy. Nature Protocol. 2011;6:1953–1968. doi: 10.1038/nprot.2011.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauthier DJ, Lazure C. Complementary methods to assist subcellular fractionation in organellar proteomics. Expert Rev Proteomics. 2008;5:603–617. doi: 10.1586/14789450.5.4.603. [DOI] [PubMed] [Google Scholar]
- 21.Lee YH, Tan HT, Chung MCM. Subcellular fractionation methods and strategies for proteomics. Proteomics. 2010;10:3935–3956. doi: 10.1002/pmic.201000289. [DOI] [PubMed] [Google Scholar]
- 22.Cavelier L, Johannisson A, Gyllensten U. Analysis of mtDNA copy number and composition of single mitochondrial particles using flow cytometry and PCR. Exp Cell Res. 2000;259:79–85. doi: 10.1006/excr.2000.4949. [DOI] [PubMed] [Google Scholar]
- 23.Baret J-C, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, Rick C, Samuels ML, Hutchison JB, Agresti JJ, et al. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab Chip. 2009;9:1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- 24•.Poe BG, 3rd, Duffy CF, Greminger MA, Nelson BJ, Arriaga EA. Detection of heteroplasmy in individual mitochondrial particles. Anal Bioanal Chem. 2010;397:3397–3407. doi: 10.1007/s00216-010-3751-3. Capillary electrophoresis with laser-induced fluorescence is interfaced with a microwell plate for PCR of mitochondrial DNA and detection of heteroplasmy in single to few mitochondria per well. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Taylor TH, Arriaga EA. Analysis of the bioactivity of magnetically immunoisolated peroxisomes. Anal Bioanal Chem. 2012;402:41–49. doi: 10.1007/s00216-011-5476-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuznetsov AV, Hermann M, Saks V, Hengster P, Margreiter R. The cell-type specificity of mitochondrial dynamics. Int J Biochem Cell Biol. 2009;41:1928–1939. doi: 10.1016/j.biocel.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Kuznetsov AV, Margreiter R. Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. Int J Mol Sci. 2009;10:1911–1929. doi: 10.3390/ijms10041911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carling PJ, Cree LM, Chinnery PF. The implications of mitochondrial DNA copy number regulation during embryogenesis. Mitochondrion. 2011;11:686–692. doi: 10.1016/j.mito.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Allen PB, Doepker BR, Chiu DT. High-throughput capillary-electrophoresis analysis of the contents of a single mitochondria. Anal Chem. 2009;81:3784–3791. doi: 10.1021/ac900099y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorenz RM, Chiu DT. Chemistry and biology in femtoliter and picoliter volume droplets. Acc Chem Res. 2008;42:649–658. doi: 10.1021/ar8002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theberge AB, Courtois F, Schaerli Y, Fischlechner M, Abell C, Hollfelder F, Huck WTS. Microdroplets in microfluidics: an evolving platform for discoveries in chemistry and biology. Angew Chem Int Ed. 2010;49:5846–5868. doi: 10.1002/anie.200906653. [DOI] [PubMed] [Google Scholar]
- 32.Pei J, Li Q, Kennedy RT. Rapid and label-free screening of enzyme inhibitors using segmented flow electrospray ionization mass spectrometry. J Am Soc Mass Spectrom. 2010;21:1107–1113. doi: 10.1016/j.jasms.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Gadd JC, Kuyper CL, Fujimoto BS, Allen RW, Chiu DT. Sizing subcellular organelles and nanoparticles confined within aqueous droplets. Anal Chem. 2008;80:3450–3457. doi: 10.1021/ac8000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeffries GDM, Edgar JS, Zhao YQ, Shelby JP, Fong C, Chiu DT. Using polarization-shaped optical vortex traps for single-cell nanosurgery. Nano Lett. 2007;7:415–420. doi: 10.1021/nl0626784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He MY, Edgar JS, Jeffries GDM, Lorenz RM, Shelby JP, Chiu DT. Selective encapsulation of single cells and subcellular organelles into picoliter- and femtoliter-volume droplets. Anal Chem. 2005;77:1539–1544. doi: 10.1021/ac0480850. [DOI] [PubMed] [Google Scholar]
- 36.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilkerson RW, Schon EA, Hernandez E, Davidson MM. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol. 2008;181:1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]