Abstract
Removal of substrate (+)-camphor from the active site of cytochrome P450cam (CYP101A1) results in nuclear magnetic resonance-detected perturbations in multiple regions of the enzyme. The 1H,15N correlation map of substrate-free diamagnetic Fe(II) CO-bound CYP101A permits these perturbations to be mapped onto the solution structure of the enzyme. Residual dipolar couplings (RDCs) were measured for 15N-1H amide pairs in two independent alignment media for the substrate-free enzyme and used as restraints in solvated molecular dynamics (MD) simulations to generate an ensemble of best-fit structures of the substrate-free enzyme in solution. NMR-detected chemical shift perturbations reflect changes in the electronic environment of the NH pairs, such as hydrogen bonding and ring current shifts, and are observed for residues in the active site as well as in hinge regions between secondary structural features. RDCs provide information regarding relative orientations of secondary structures, and RDC-restrained MD simulations indicate that portions of a β-rich region adjacent to the active site shift so as to partially occupy the vacancy left by removal of substrate. The accessible volume of the active site is reduced in the substrate-free enzyme relative to the substrate-bound structure calculated using the same methods. Both symmetric and asymmetric broadening of multiple resonances observed upon substrate removal as well as localized increased errors in RDC fits suggest that an ensemble of enzyme conformations are present in the substrate-free form.
Over the years, the concept of molecular recognition between enzyme and substrate has evolved significantly. The early idea of lock-and-key recognition (1), in which rigid body shape complementarity and specific interactions were assumed to dominate the process, later gave way to the concept of induced fit, in which the free energy change associated with binding drives conformational changes favoring transition state stabilization (2). In recent years, improved simulation methods (3), variable-temperature X-ray crystallography (4, 5) and nuclear magnetic resonance (NMR) (6, 7) have demonstrated that proteins exist as ensembles of conformers that interconvert on a range of timescales. This has lead to the concept of pre-existing enzyme conformational ensembles, in which substrate binds to the appropriate enzyme conformers present in solution, thereby shifting the equilibrium of the remaining substrate-free conformations towards those that most effectively stabilize bound substrate (8). The distinction between the induced-fit and pre-existing equilibrium models is subtle, but important. Induced-fit implies that the free energy change upon substrate binding is sufficiently large to drive changes in protein conformation, while the equilibrium ensemble model assumes only that the productive binding conformer must be stabilized upon substrate binding relative to other members of the ensemble. Implicit in this argument is that in order to destabilize the ground state of bound substrate relative to the transition state of the reaction to be catalyzed, further conformational changes are required in order to reach the catalytically competent enzyme conformation. These further changes could be the result of low-frequency (high-barrier) motions intrinsic to the bound form, or might be driven by the binding of a second species (effector) to the enzyme-substrate complex.
We have used NMR to detect such a high-barrier conformational change in substrate-bound cytochrome P450cam (CYP101A1) that is driven by the binding of an effector protein putidaredoxin (Pdx) (9). Pdx is a required component of the reconstituted CYP101A1 enzyme system: Even if all other components are present, no turnover is observed in the absence of Pdx (10). Other groups have also reported the formation of a transient species upon addition of Pdx to substrate-bound CYP101A1 under turnover conditions (11). We have provided experimental evidence that Pdx binding converts a substrate-bound but catalytically inactive form of the enzyme to the catalytically competent form via the isomerization of a single Ile-Pro peptide bond from trans or distorted trans to cis (12, 13). Recently, we described the use of one-bond 1DNH residual dipolar couplings (RDCs) as restraints in solvated molecular dynamics simulations of reduced substrate- and carbonmonoxy-bound CYP101A1 to characterize the accessible substrate-bound conformations of the pre-catalytic form of CYP101A1 in solution (14).
In the current work, we turn our attention to the ensemble of conformations accessible to this enzyme in the absence of substrate. We used chemical shift perturbations observed in TROSY-15N, 1H HSQC spectra of substrate-depleted samples of reduced and CO-bound CYP101A1 to identify regions of the enzyme structure that are sensitive to substrate binding. We then measured 1DNH RDCs for these samples in two independent aligned media (liquid crystal and phage). Using these RDCs as restraints in fully solvated molecular dynamics simulations, we identified ensembles of equilibrium conformations for the substrate-free enzyme with reasonable fits to the experimental restraints. A subset of RDCs, often assigned to NH pairs near the ends of helices or in regions of poorly defined secondary structure, were outliers from the best-fit alignment tensors, but could not themselves be reliably fit to alternative tensors, suggesting that some secondary structures are subject to fraying in the substrate-free enzyme.
Experimental
Expression and purification of CYP101A1
The C334A CYP101A1 mutant plasmid was transformed in E. coli NCM533 competent cells by electroporation. The C334A mutant is spectroscopically and enzymatically identical to the wild type but does not form dimers in solution, and so is more suitable for NMR characterization than the wild type enzyme (15). Uniformly 15N-labeled CYP101A1 was expressed in M9 minimal media containing 15NH4Cl as the sole nitrogen source and trace metals at 37 °C. Kanamycin (50 μg/mL) and chloroamphenicol (25 μg/mL) were used for strain selection and plasmid maintenance, respectively. The heme precursor δ-aminolevulinic acid (70 mg/L) and camphor (3 mM) were added to the medium 10 min prior to induction of CYP101A1 expression by addition of solid IPTG (isopropyl β-D-1-thiogalactopyranoside) to a final concentration of 1 mM. The cells were incubated for an additional 12 h after induction at 37 °C. The collected cells were suspended in buffer (50 mM Tris•HCl (pH 7.4), 100 mM KCl, and 1 mM (1R)-(+)-camphor) and lysed via sonication. The protein purification that followed included a protamine sulfate cut to precipitate nucleic acids and a 70% ammonium sulfate cut to precipitate CYP101A1. The resulting precipitate was dissolved in fresh buffer and desalted by dialysis against buffer containing 50 mM Tris•HCl (pH 7.4), 100 mM KCl, and 1 mM (1R)-(+)-camphor at 4 °C for 18 hours total (1 liter buffer for 6 hours followed by transfer to another liter of fresh buffer for an additional 12 hours). The protein was then passed through an anion-exchange column (DEAE sepharose fast flow resin) and eluted with a salt gradient (50 mM to 300 mM KCl). Fractions with A391/A280 ≥ 0.8 were collected and concentrated before passing the protein through a P-100 size exclusion column at 4°C equilibrated with a buffer containing 50 mM Tris•HCl (pH 7.4), 100 mM KCl, and 1 mM (1R)-(+)-camphor. Further details concerning protein expression and purification details have been published previously (16). Protein purity was assessed spectroscopically with a A391/A280 ratio of at least 1.3.
Removal of substrate and sample preparation for NMR experiments
Samples of camphor-free CYP101A1 were prepared as follows. 400 μL of CYP101A1 (0.5 mM) in standard camphor-containing buffer was diluted to 15 mL in camphor-free buffer (50 mM Tris•HCl (pH 7.4), 50 mM KCl) and then re-concentrated to a final volume of ≈400 μL. This process of dilution and concentration into a camphor-free buffer is performed a total of 5 times. The removal of camphor from the enzyme is facilitated by reducing KCl concentration. In the final dilution step, NMR buffer was used for dilution (50 mM Tris•HCl (pH 7.4) and 50 mM KCl in 90:10 H2O:D2O). The protein was then transferred to a septum-sealed reaction vial and flushed with CO for 10 minutes. The protein was then reduced with a freshly prepared solution of 250 mM Na2S2O4 in 1M Tris•HCl buffer (pH 8.0) that was added in 1 μL aliquots. Typically a total of 8-9 μL of the 250 mM Na2S2O4 solution would be added to completely reduce ~350 μL of 0.45 mM protein. The protein has a cherry-red color once it is CO-bound and reduced. The reduced protein was anaerobically transferred to a susceptibility-matched NMR tube (Shigemi, Inc., Allison Park, PA).
Alignment of CYP101A1 in C12E5/hexanol for RDC measurements
A 10% (w/w) solution of pentaethylene glycol monododecyl ether (C12E5, 98+%, Sigma-Aldrich) in degassed NMR buffer (50 mM Tris•HCl (pH 7.4) and 50 mM KCl in 90:10 H2O:D2O) was prepared and vortexed to insure homogeneity. From this stock solution 180 μL was transferred into a septum-sealed reaction vial and the aliquot was flushed with carbon monoxide for 10 minutes. Normal hexanol was added (anhydrous, 99+%, Sigma-Aldrich) in small aliquots (as small as 0.5 μL when nearing the nematic phase) and the solution vortexed after each addition. Over the course of hexanol addition the solution turned from clear to biphasic, then to clear again, and finally the solution becomes opalescent when the proper molar ratio of n-hexanol to C12E5 is reached. The final addition of hexanol, and resulting transition to opalescence, is dependent on temperature. As such, it is important to keep the temperature of the solution at 298 K (the temperature used for the NMR experiments) during the period of addition and after reaching the proper C12E5/hexanol molar ratio. The final C12E5/hexanol molar ratio was determined to be 0.85, consistent with literature (17). Reduced and CO bound WT CYP101A1 (0.4 mM, 180 μL), in NMR buffer, was added to the C12E5/hexanol mixture (1:1, v/v) under nitrogen in a glove bag. (Note: at this point it was sometimes necessary to flush the vial containing the mixture with CO and to add an additional 2-3 μL of 250 mM Na2S2O4 in 1M Tris•HCl buffer (pH 8.0) to the sample to ensure that the protein remains reduced and CO bound.) After mixing, the solution was transferred into a susceptibility-matched NMR tube and the sample was placed into a 298 K constant temperature bath for 15 minutes to ensure proper equilibration. All steps involving the C12E5/hexanol mixture were performed at room temperature (typically 20-25 °C). The sample was then placed in the NMR magnet, and alignment was monitored using the residual quadrupolar splitting of the buffer 2H signal (28.8 Hz after ~1/2 hour), and data acquisition begun after no further changes occurred in observed splitting.
Alignment of CYP101A1 in Pf1 filamentous phage for RDC measurement
The Pf1 phage/protein solution was prepared by first flushing a septum-sealed reaction vial containing 100 μL of a 50 (± 4) mg/mL solution of Pf1 bacteriophage (Asla Biotech Ltd., Latvia) with CO for 10 minutes. Then, in an anaerobic chamber, reduced and CO-bound WT CYP101A1 (0.4 mM, 410 μL), in NMR buffer (50 mM Tris•HCl (pH 7.4) and 50 mM KCl in 90:10 H2O:D2O), was added to the phage solution. The mixture was again flushed with CO and an additional 3 μL of 250 mM Na2S2O4 in 1M Tris•HCl buffer (pH 8.0) was added to ensure that the protein remained both reduced and CO-bound. Finally, the phage/protein sample was gently mixed by rotation before being transferred anaerobically to a susceptibility-matched NMR tube. The final phage concentration was 9.8 mg/mL. Alignment was monitored using the residual quadrupolar splitting of the buffer 2H signal (8.0 Hz after ~1/2 hour), and data acquisition begun after no further changes occurred in observed splitting.
NMR experiments and data analysis
All NMR data was acquired on a Bruker Avance spectrometer operating at 800.13 MHz (1H) and 81.08 MHz (15N) equipped with a TXI cryoprobe and pulsed field gradients. NMR data acquisition and analysis were performed as described previously (14). RDCs were measured in both alignment media using the combined results of TROSY and semi-TROSY experiments as adapted from Weigelt et al. (18), in which peaks are offset from each other by 1JNH in the 1H dimension of two-dimensional 1H,15N correlation experiments. A data set acquired in the absence of alignment media was used to obtain the reference coupling for a given correlation. Previously published assignments for CYP101A1 were used to identify peaks in each spectrum (13). Datasets were processed identically in TopSpin (Bruker Biospin), and imported and analyzed in SPARKY(19).
Molecular dynamics simulations
In our previous work (14) we applied RDCs measured in two independent media as restraints in fully solvated molecular dynamics (MD) simulations to calculate solution structural ensembles for reduced substrate- and carbonmonoxy-bound cytochrome CYP101A1. The conformation with the lowest total violation energy was identified and the coordinates were deposited as RCSB Protein Data Bank entry 2L8M. That structure, with substrate camphor removed, was used as the starting point for the current work. The substrate-free 2L8M structure was solvated with 22170 TIP3P water molecules, and sufficient K+ and Cl- ions to maintain the same ionic strength used in the NMR experiments. A 30 Å distance was maintained between the enzyme and the solvent box edge. Simulations were performed with the Amber 10 package (20), using the ff03 force field as detailed previously (14). A preliminary refinement employed 552 total RDC N-H restraints, 277 measured in Pf1 medium and 275 in C12E5/hexanol, representing approximately 70% of the polypeptide backbone. During the refinement a +/− 3Hz uncertainty was imposed by penalizing calculated couplings outside that range, with a penalty function proportional to (Dcalc - Dobs)2. Alignment tensors were calculated by minimizing the initial structure, while keeping the atoms with restraints frozen. After minimization, the temperature was increased in three cycles, from 0K to 50K, 50K to 100K and 100K to 300K, with 5 kcal/mol/Å2 harmonic constant. The density was equilibrated through a fixed pressure run with a relaxation time of 0.2 ps. Once volume and pressure were equilibrated, a 100 ps constant volume molecular dynamics was performed. Then a 2 ns molecular dynamics simulation at 300K was performed. The largest deviations from experimental RDC values in the resulting ensemble were identified and cross-checked with experimental datasets, in order to rule out assignment errors or incorrect RDC measurements. This process was repeated iteratively until violations plateaued with a final set of 478 RDC restraints (250 corresponding to Pf1 medium and 228 to C12E5/hexanol medium).
The final refinement was performed as described above, starting from the results of equilibration runs performed using the final RDC restraint set (14). A 2 ns production molecular dynamics simulation at 300K was performed. During this simulation, trajectories were saved every ps, yielding an ensemble of 2000 conformers. RDCs values were calculated every 200 ps, these values were averaged and the averages compared with the set of experimental values.
Results
Chemical shift perturbations upon removal of substrate from CYP101A1
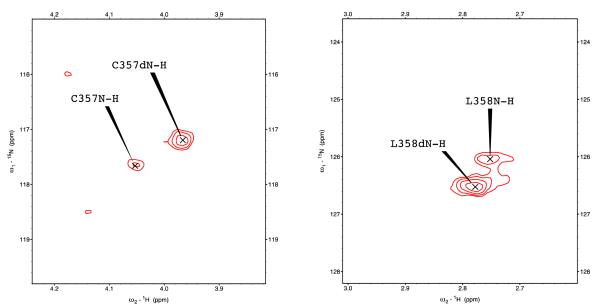
Two-dimensional 1H, 15N TROSY-HSQC correlation experiments were performed with both oxidized and reduced samples of CYP101A1 from which substrate was removed in order to identify those regions of the enzyme that are most affected by substrate. The results of experiments with oxidized substrate-free enzyme will be reported elsewhere, but Figure 1 provides a summary of the observed perturbations as a function of sequence for the reduced CO-bound enzyme, while Figure 2 shows the largest perturbations superimposed on the structure. Figure 3 shows examples of the observed shifts for two residues, the axial heme iron ligand Cys 357 and the adjacent Leu 358. Both of these correlations are shifted well upfield of typical amide 1H shifts by ring current shielding from the adjacent heme porphyrin macrocycle. These residues are among those that exhibit significant shifts upon substrate removal (Fig. 1). Where distinct correlations can be assigned for a particular residue to the substrate-free and bound forms, there does not appear to be a lower chemical shift limit beyond natural line width and digital resolution allowing distinguishability, meaning that chemical exchange of substrate between free and bound forms is slow on the 1H chemical shift time scale (< 30 s-1). Where comparison is possible, the half-height 1H line width of the substrate-free correlation typically exceeds that of the substrate-bound form by ~10 Hz. Furthermore, the broadening of substrate-free correlations is often asymmetric, suggesting that multiple conformers with different populations at slow exchange on the chemical shift time scale are contributing to the observed correlation in the substrate-free form. These observations will be addressed more completely in a report on local dynamics as measured by H/D exchange and 15N relaxation in oxidized and reduced substrate-free CYP101A1 (Young et al., in preparation).
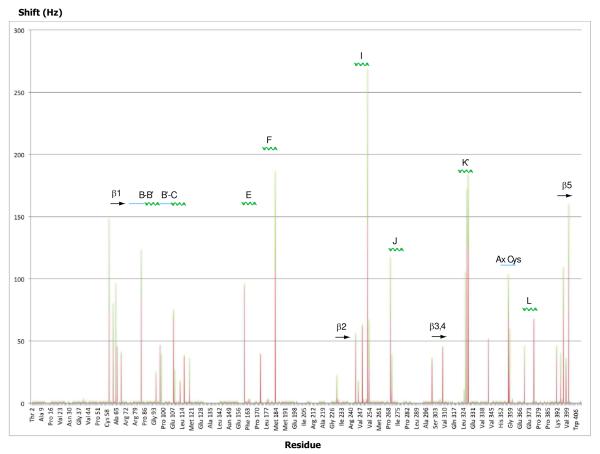
Figure 1.
Chemical shift perturbations observed in 1H,15N TROSY-HSQC spectra upon removal of substrate from reduced and CO-bound CYP101A1 displayed as a function of sequence and secondary structure. Both 1H (orange bars) and 15N (green bars) shifts are shown, so the y-axis value is the sum of perturbations in sec-1 (Hz) observed in both dimensions for a given correlation. Secondary structural features with which perturbed residues are associated are indicated above, with solid arrow representing β structure, zig-zag green lines for helix, and straight blue lines for loop or irregular structure. Secondary structural features are named using the conventions of Poulos et al. (21). The Ax Cys label refers to Cys 357 and Leu 358.
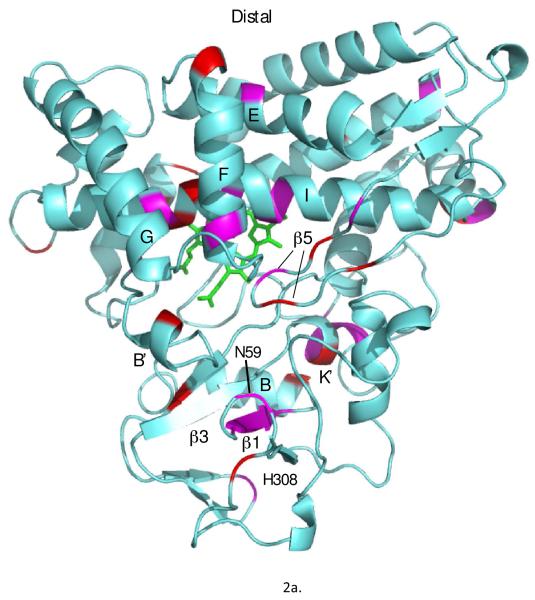
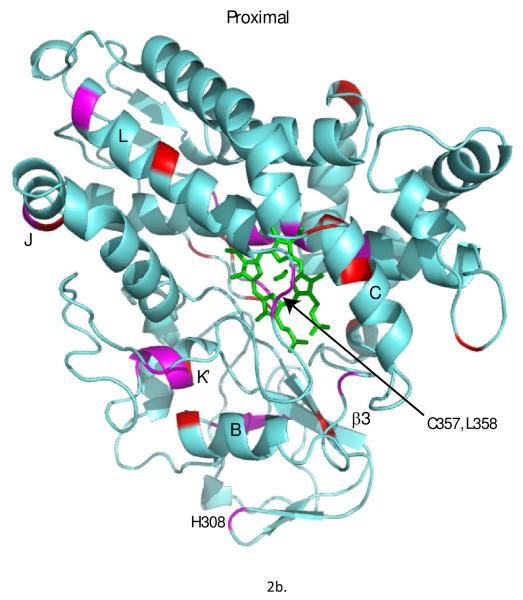
Figure 2.
Chemical shift perturbations observed in 1H,15N TROSY-HSQC spectra upon removal of substrate from reduced and CO-bound CYP101A1 superimposed on the solution structure 2L8M (ref. (14)). Only those residues for which N-H correlations distinct from those of the substrate-bound form are observed are highlighted. Residues highlighted in magenta exhibit shifts > 40 Hz in either the 1H or 15N dimensions, while smaller shifts are represented in red. Top: distal view (active site cavity towards the reader) with perturbed secondary structural features or residue IDs labeled using the conventions of Poulos et al. (21). Bottom: Proximal view, rotated slightly for clarity.
Figure 3.
Chemical shift perturbations observed in 1H,15N TROSY-HSQC spectra in camphor-depleted samples of reduced CO-bound CYP101A1 for heme axial cysteine ligand C357 (left) and adjacent L358 (right). Correlations marked C357N-H and L358N-H are at the positions observed for camphor-bound CYP101A1 and are assigned to residual camphor-bound enzyme. Those peaks marked as C357dN-H and L358dN-H are shifted relative to their positions in camphor-bound enzyme and are assigned to the camphor-free form.
The structural distribution of amide NH correlations that are affected by the absence of substrate is intriguing (Fig. 3). As expected, many of the affected residues are in or adjacent to the active site, with the largest identifiable perturbations to residues in the I helix that form the I helix “kink”, into which the axial CO (or O2) fits in the closed structure (21). Perturbations in the I helix are quite localized, with Gly 243, Leu 244, Gly 248 and Thr 252, all of which lie on the active site edge of the I helix, being strongly affected, while Leu 245, Val 247, Gly 249 and Asp 251 show smaller perturbations. Residues at the C-terminal end of the B’ helix and B’-C loop are also affected, although to a lesser extent. Within the B’ helix, only Ala 95 shows clear evidence of doubling, while Phe 98 and Ile 99, which form part of the B’-C turn-loop, are more strongly perturbed. However, Asp 97, the C-terminal residue of the B’ helix, is severely broadened in the absence of substrate, suggesting that this end of the B’ helix is sampling multiple conformations. This is in agreement with crystallographic studies showing fraying and/or loss of the B’ helix in substrate-free structures (22).
The secondary structure forming part of the active site of CYP101A1 that exhibits the most widespread chemical shift perturbations is the β5 sheet (Fig. 2a), which was found to be more disordered in solution than in crystallographic structures even in the presence of substrate (23). Substrate-free CYP101A1 crystallographic structures also show a less ordered β5 region than is observed in substrate-bound forms (21,22). Although only the side chain of Ile 395, which lies on the turn between the two β5 strands, directly contacts substrate in crystallographic structures, residues on both strands, including His 391, Gly 394, Val 396, Gly 398 and Gln 400, are perturbed in the absence of substrate.
External to the active site, sizable (i.e., > 40 Hz in either dimension) chemical shift perturbations are most often found in hinge regions and points of contact between secondary structural elements that are themselves affected by the absence of substrate as determined by RDC-restrained MD simulations (vide infra). Such “hinge” residues include Ala 65 (β1-B helix) Ser 83 (B-B’ loop), Ile 160, which sits on the E helix at the juncture between the F and I helices, Glu 172 and Gln 183 (beginning of the F helix and F helix-FG loop junction, respectively), His 270 (between the I and J helices) and Thr 376 (near the end of the L helix). However, not all observed perturbations fall into this category. The largest perturbations to residues not directly adjacent to the active site are observed in the K’ helix, including Leu 324, Ser 325, Gly 326 and Leu 327. The K’ helix acts as a flexible link between the β3 sheet bordering the active site and the proximal β-meander, and was also found to be the most responsive to the binding of modified substrate outside of the active site (24).
Besides the K’ helix, the other secondary structure external to the active site with resonances most uniformly perturbed by substrate removal is in β1 sheet. Asn 59, which forms part of the turn between two strands of the β1 sheet, shows the greatest sensitivity to absence of substrate, and forms part of the proposed entrance to the active site (12, 25, 26). Asn 59 is spatially adjacent to Pro 89 at the base of the B’ helix and to Tyr 29, which stabilizes the otherwise un-solvated cis conformation of the Ile 88-Pro 89 bond by a hydrogen bond between the Tyr 29 side chain phenolic OH and the carbonyl oxygen of Ile 88 (21). We have proposed that the same Tyr 29 phenolic OH stabilizes the transition state for effector-induced isomerization of the Ile 88-Pro 89 bond, modulating access to the active site (12, 13). Also perturbed in the β1 sheet are His 61, Ile 64 and Ala 65. The strand from His 61-Ala 65 is adjacent to the β3 sheet, and chemical shift perturbations in this region indicate changes in hydrogen bonding within the β1 sheet or with the adjacent β3 strand. As will be seen (vide infra) the β3 sheet is among the most structurally displaced secondary structural features in CYP101A1.
RDC-restrained molecular dynamics simulations
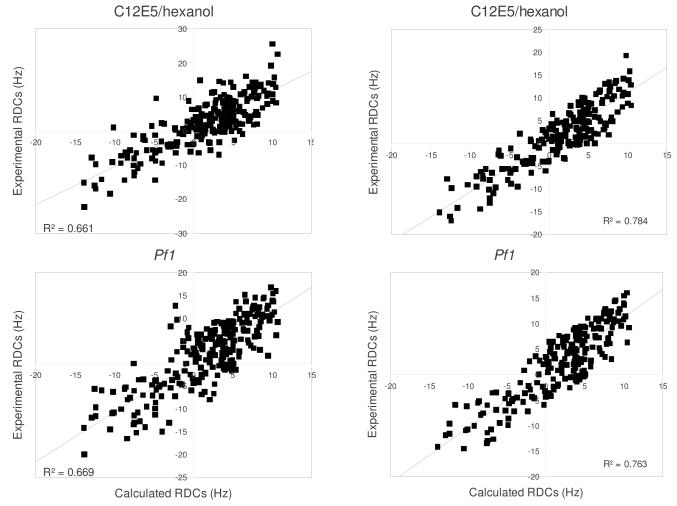
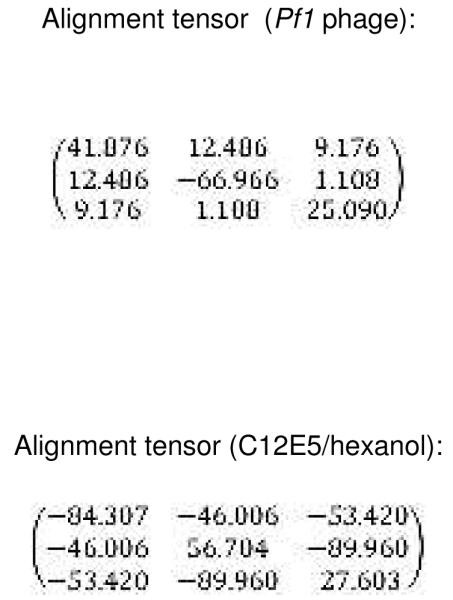
A total of 250 RDCs determined from alignment in Pf1 and 228 RDCs from C12E5/hexanol were used as restraints in fully solvated molecular dynamics simulation. During the 2 ns production MD simulation, RDC values were calculated from saved trajectories every 200 ps, the values were averaged at the end of the run and the averages compared with experimental RDCs. Figure 4 shows the fit between experimental and values calculated using independent alignment tensors for each medium (Table 1). In both media the fit is fairly good, with a linear R2 correlation of 0.66 in both media when all RDC restraints are considered. Removal of selected restraints with the largest restraint violations (> 6 Hz) from the calculation (31 from Pf1 and 37 from C12E5/hexanol) yields considerably better fits for both data sets (R ~ 0.77) (See Fig. 4). Careful re-examination of experimental data confirms that this group of outlying RDC values were correctly assigned and measured, and those with the largest violations appear to be local perturbations, with many of them located at the beginning or end of helices. These include Ala 95 at the C-terminus of the B’ helix, Glu 107 initiating helix C, Thr 192 initiating the G helix and Ala 284, which forms part of a bend near the N-terminus of the K helix. Other NH correlations showing significant deviations from predicted 1DNH values include Glu 262 in the I helix at a pivot point involving the β5 sheet, Val 295 in the β3 sheet, Arg 342 in the β-meander and Leu 356 immediately preceding the axial heme ligand Cys 357. A simulation restrained only by the subset of large-violation RDCs performed (i.e., with the main body of RDC restraints turned off) did not result in a reasonable fit of those RDCs to a single set of alignment tensors, so it is unlikely that they represent a single alternative conformational ensemble. Instead, the positions of many residues showing larger violations near the beginning or end of helices or at pivot points between secondary structures suggests that fraying or movement may be occurring on the time scale of the RDC measurement. Only five RDCs assigned to a given residue from both data sets were excluded in both improved fits (residues 34 and 36 prior to the A helix, residue 59 in the β1 loop, residue 70 near the N-terminus of the B helix and residue 230 in the H-I loop). However, the arbitrary cutoff used for determining which RDCs would not be included in the improved fit shown in Fig. 4 may disguise correlations between outliers in the two data sets. The absence of significant chemical shift perturbations associated with the presence of the aligning medium would seem to rule out an alternative explanation of the anomalous RDC values, that is, direct interaction between the aligning medium and the protein at these sites.
Figure 4.
Fitting of experimental RDCs versus RDC values calculated from MD simulations. Top left, fitting of all 228 RDCs measured in C12E5/hexanol medium used in MD simulations. Top right, fitting after removal of outliers with deviations > 6 Hz from calculated values (see text). All RDC values are in sec-1 (Hz). Bottom left, fitting of all 228 RDCs measured in Pf1 phage medium used in MD simulations. Bottom right, fitting after removal of outliers with deviations > 6 Hz from calculated values (see text).
Table 1.
Alignment tensors calculated for structure REP1 and used for fitting of RDCs as shown in Figure 4. Cartesian tensor elements are scaled by 10^5 according to AMBER convention. For a complete description of the fitting procedure and generation of alignment tensors, see Amber 10 manual (Ref. 20).
|
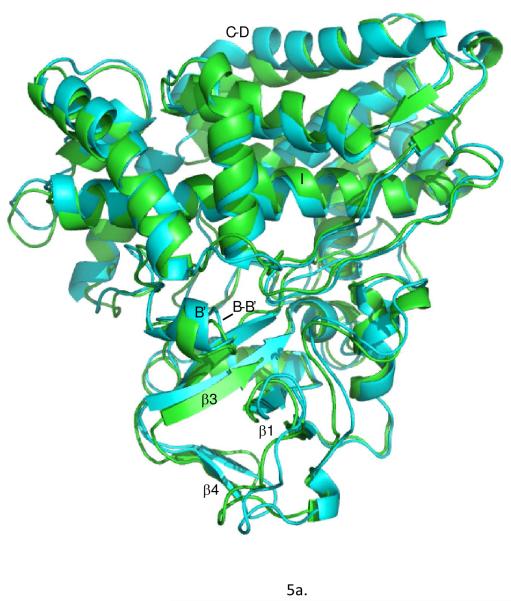
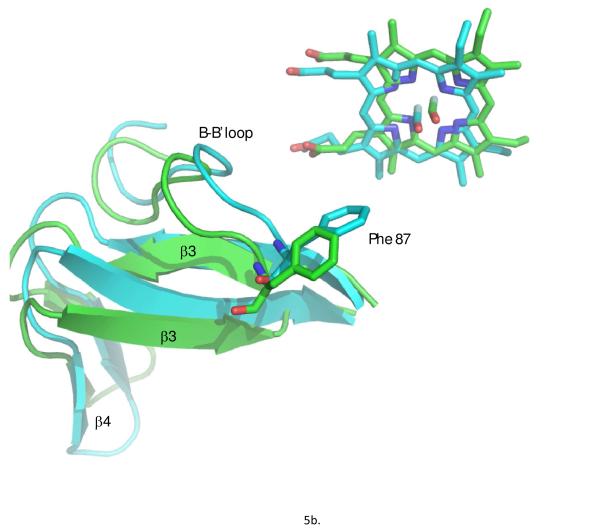
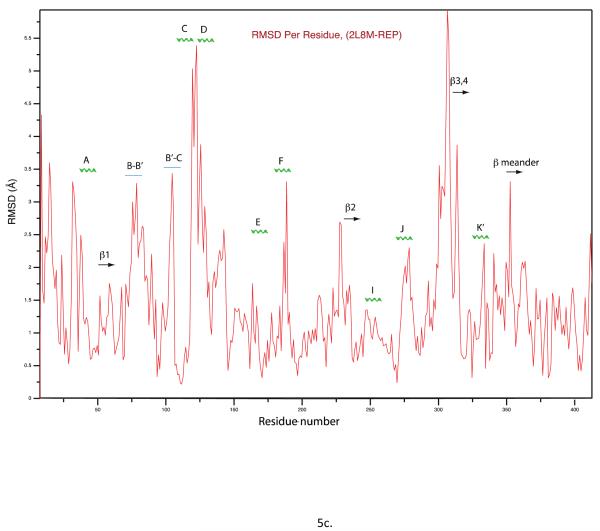
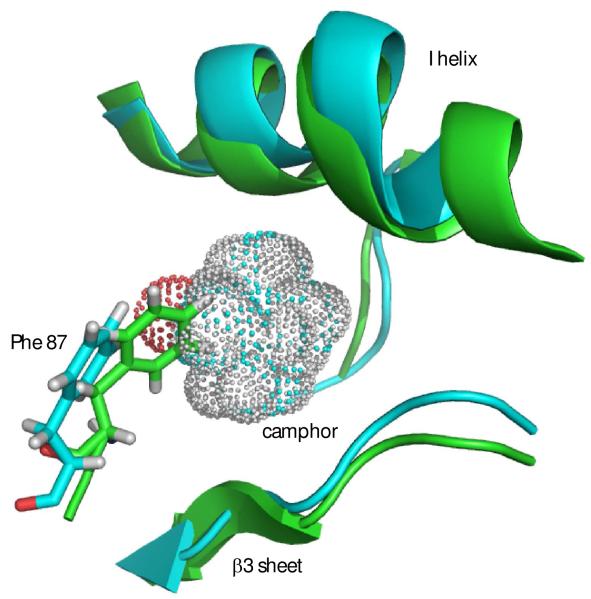
From the ensemble, the conformer with lowest restraint violation energy was identified (REP1). The rmsd of REP1 with the other members of the ensemble is 0.9 Å, while the rmsd with respect to the average structure is 0.6 Å, so one can safely consider REP1 as representative of the ensemble. In Figure 5a, REP1 is superimposed on substrate-bound structure 2L8M. Residues in REP1 showing significant structural deviations from 2L8M include Ser 32-Leu 34 and Gln 39 in turn region preceding helix A and Pro 105-Glu 107 in the B’-C loop. At the N-terminus of the D helix, residues Met 121,Val 123 and Val 124 show significant displacements upon removal of camphor, and these displacements are transmitted to a lesser extent along the D helix and the C-terminus of the C helix. However, the largest overall displacement of secondary structure (Fig. 5c) occurs for residues Tyr 78 through Phe 87 in in the B-B’ loop in tandem with the β3-β4 sheet complex, residues 299-315. These structures are displaced as a unit, moving the side chain of Phe 87 into the active site (Fig. 5b). In the camphor-bound 2L8M (and other camphor-bound structures), the side chain of Phe 87 is a primary substrate contact. In the substrate-free form, the side chain of Phe 87 occupies some of the volume normally occupied by substrate (see Figure 6). To a lesser extent, the side chains of I helix residues Leu 244 and Val 247, B’-C loop residue Phe 98 and F helix Met 184 also shift to occupy some of the volume occupied by camphor in 2L8M. The net result of these displacements is that the active site is considerably smaller in REP1 than in 2L8M, and much of the volume occupied by camphor in 2L8M is occupied in REP1 by hydrophobic groups that line the active site cavity. Estimation of the active site volume in REP1 based on inter-atomic distances of residues lining the active site yields an active site volume 30% smaller than that of 2L8M.
Figure 5.
a) Substrate-bound 2L8M structure (green) aligned by Cα carbon on REP1 (cyan), identified as the substrate-free structure with lowest RDC restraint violation energy. b) Close-up view of the same superposition highlighting the displacements of the heme, the B-B’ loop including Phe 87 and the β3-β4 domain. RMSD values between the two structures per residue are shown in the Figure 5c.
Figure 6.
Contraction of active site in REP1 (green) from best-fit backbone alignment to 2L8M (cyan). Camphor from 2L8M is shown as a van der Waals dot surface. Camphor-bound 2L8M structure is shown in cyan. Note that the side chain of Phe 87 partially occupies volume vacated by camphor. The side chains of Leu 244 and Val 247 also partially occlude the space occupied by camphor (see text). Unless otherwise noted, structural figures are generated using PyMOL (35).
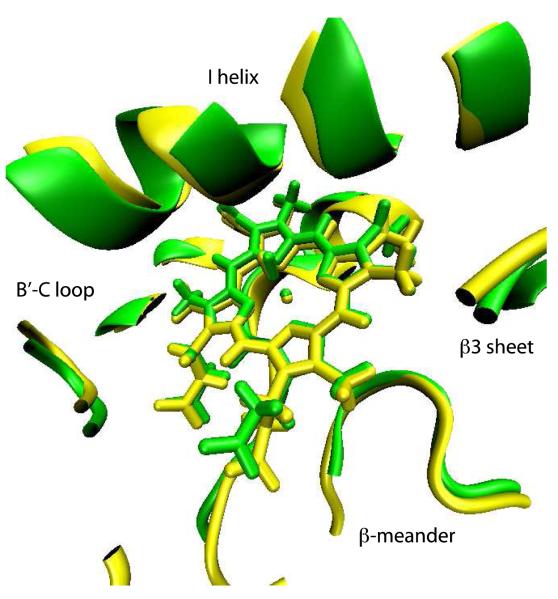
The guanidinium group of β3 residue Arg 299, which forms a salt bridge with the heme propionate in all published structures of CYP101A1, maintains that interaction in REP1. However, the inward displacement of the B-B’ loop and the β3-β4 sheet complex results in an apparent ~10° clockwise rotation of the heme around the heme normal as viewed from the distal (active site) face of the enzyme (Fig. 7). This rotation is either followed or enforced by the side chain of Arg 299, thereby maintaining the salt bridge with the heme propionate. Given that we have no direct experimental restraints for the heme, we cannot say with certainty that the observed heme rotation is not an artifact of the simulations. However, the heme rotation is consistent with the inward movement of the B-B’ loop and the β3-β4 sheet complex and slight lateral displacement of the B’ helix, as well as chemical shift perturbations to the NH correlations of heme-facing residues of the I helix as well as Cys 357 and Leu 358. It is expected that heme rotation would give rise to changes in ring-current shift patterns sufficient to explain these observations, as well as the high degree of localization of the chemical shift changes noted above. Furthermore, the chemical shift sensitivity of residues in the β1 strand (noted above) adjacent to the β3 sheet suggests that this portion of the β1 sheet acts as a hinge for the observed displacement of the B-B’ loop and the β3-β4 sheet complex.
Figure 7.
2L8M (green) and REP1 (yellow) structures aligned using the heme macrocycle as the reference. Residues at distances less than 6 Å from the heme are shown using a cartoon representation.
Discussion
The complex catalytic cycle of CYP101A1 requires multiple changes in spin, oxidation and ligation states of the heme prosthetic group to achieve turnover (27). Our research has been directed towards understanding the role of protein structure and dynamics in directing these changes as a function of oxidation state and effector binding, with a focus upon the role of the effector Pdx in driving the formation of the closed, catalytically competent form of the substrate-bound enzyme. However, recent publications dealing with the structural consequences of substrate removal on CYP101A1 demonstrate that substrate binding also plays an important role in conformational selection (22, 28). In the current work, we have measured NH chemical shift perturbations in the substrate-free enzyme in order to determine which regions of the CYP101A1 structure are affected by the absence of substrate. We have also used RDCs measured in two independent alignment media to restrain molecular dynamics simulations of the substrate-free enzyme, using methodology described previously (14). The results of these simulations are an ensemble of best-fit structures that provide insight into the conformations accessible to the substrate-free enzyme in solution on the timescale of the simulation (2 ns). We observe no major deviations from the ensemble average after ~1 ns, suggesting that the structure has equilibrated sufficiently to provide a realistic representation of the ensemble giving rise to the observed RDCs.
We have chosen the reduced diamagnetic CO-bound form of the substrate-free enzyme for structural characterization because we have the most complete set of resonance assignments for this form: Paramagnetic broadening in the oxidized form results in a considerable loss of information in and around the active site (29). It could be argued that the biologically relevant form for substrate-free enzyme is in fact the oxidized form, as the physiological redox partner of CYP101A1, Pdx, does not reduce the enzyme unless substrate is bound (27). However, the most significant differences that we have identified between oxidized and reduced CYP101A1 with substrate bound are dynamic, with the oxidized enzyme exhibiting higher frequency and larger amplitude motions than the reduced form (29). Therefore, it is likely that the structural ensemble reported here is a subset of those occupied by the substrate-free oxidized enzyme in solution. A report comparing the dynamics of oxidized substrate-free and substrate-bound CYP101A1 is in preparation.
Comparison of RDC-derived substrate-free and substrate-bound structures
We observe generally good agreement between secondary structural features found to be perturbed by chemical shift mapping and those that are structurally displaced in RDC-restrained simulations of the camphor-free and bound forms. However, we note that residues undergoing the largest physical displacements within those structural features are typically not those that show the greatest chemical shift perturbations. There is a good reason for this: Chemical shift perturbations are indicative of a changed electronic environment (hydrogen bonds changing length, change in ring current shift patterns, etc.), whereas secondary structural features can be displaced with little or no change in hydrogen bonding patterns except at junctions with other features (which is where the largest chemical shift displacements outside the active site are often seen).
Active site of CYP101A1 in the absence of substrate in solution and comparison with crystallographic results
The displacement of the side chain phenyl group Phe 87 on the B-B’ loop into the active site cavity described above is also seen, to a lesser extent, in the first crystallographic structure of substrate-free CYP101A1 (PDB entry 1PHC) (30). Similarly, the side chain of Tyr 96, which projects into the active site from the B’ helix, is only marginally displaced in REP1 compared to the substrate-bound 2L8M, an observation also made in reference to 1PHC. However, the fairly major movements of the B-B’ loop and the β3-β4 sheet complex that accompany the Phe 87 displacement are not observed in any crystallographic structure of substrate-free CYP101A1 and deserves some comment. The first reported structure of substrate-free CYP101A1 was obtained using crystals which were grown in the presence of dithiothreitol bound in place of camphor in the active site, which was subsequently removed by soaking in DTT-free buffer (30). This structure showed minimal displacement of backbone atom positions relative to substrate-bound forms. Recently, Goodin and co-workers described a crystallographic structure of an open substrate-free form of CYP101A1 determined using crystals grown in the absence of camphor and potassium ion in which the active site is connected to the solvent by a fairly wide “throat” containing 12 ordered water molecules (22). The opening in that structure (PDB entry 3L62) is generated by displacement of the F-G loop and disordering of the B’ helix while maintaining the cis conformation of the Ile 88-Pro 89 amide bond. The absence of the B’ helix in this case is likely due in part at least to the absence of K+ ion, which stabilizes the helix in other crystallographic structures (31). The presence of an ordered water molecule hydrogen-bonded to the amide of Ile 88 in 3L62 indicates that this region is more solvent-accessible in 3L62 than in other CYP101A1 structures. We note that recently the crystal structure of an open form of the related CYP101C1 has been determined in which the X-Pro bond preceding the intact B’ helix is found to be in the trans conformation (32). Comparison of 3L62 with camphor-bound structure 3L63 also shows quite large rmsd (> 8 Å) displacements, although instead of the B-B’ loop and the β3-β4 sheet being the primary site of displacement, as we observe, the displacements in the crystallographic structures are localized in the F and G helices, the F-G loop and the “sprung” B’ helix (22). In the current work, enzyme conformations are not constrained by crystal packing, and it appears the preferred conformers select against a large void volume in the active site, which would require ordered water within the site to maintain. The crystals used for determining the 3L62 structures were of oxidized protein grown without a pre-determined iron axial ligand present, so it is possible that the oxidized form, with more available positive charge density, is better able to stabilize ordered water within the active site.
I helix “kink”
The I helix “kink” is one of the most recognizable and conserved features of cytochrome P450 structure. In CYP101A1, the kink is formed by the interruption of regular α-helix i → i+4 hydrogen bonding between the carbonyl of Gly 248 and amide NH of Thr 252. The resulting gap in the I helix forms the niche in which O2 sits when ligated to the heme iron (33). The I helix kink was reported by Poulos and co-workers in the first CYP101A1 structures (21), and is found in some fashion in almost every P450 structure of any type published to date (23). While the kink is maintained in the current structure, it is somewhat narrower in the absence of substrate, as evidenced by the shorter distance between the carbonyl oxygen of Gly 248 and the Thr 252 NH in REP1 than in the substrate-bound 2L8M (Table 1). Furthermore, the two i → i+5 hydrogen bonds that stabilize the kink in 2L8M (Val 247 C=O ---- HN of Thr 252 and Gly 248 C=O ---- HN of Val 253) are both somewhat elongated in REP1 (see Table 2).
Table 2.
Comparison of hydrogen bond distances between 2L8M and REP1 in the I helix and the β3-K’ helix junction regions. Differences (Δ) are reported as 2L8M - REP1, so a negative value for Δ indicates closer approach in REP1 (substrate-free form).
| 2L8M (distance in Å) | REP1 (distance in Å) | Δ | |
|---|---|---|---|
| I helix | |||
| Leu 244 C=O → Gly 248 NH | 1.85 | 1.97 | +0.12 |
| Leu 245 C=O → Gly 249 NH | 1.80 | 2.10 | +0.30 |
| Leu 246 C=O → Leu 250 NH | 2.49 | 1.81 | −0.68 |
| Val 247 C=O → Asp 251 NH | 2.14 | 2.12 | −0.02 |
| Val 247 C=O → Thr 252 NH | 2.21 | 2.46 | +0.25 |
| Gly 248 C=O → Thr 252 NH | 3.16 | 2.68 | −0.48 |
| Gly 248 C=O → Val 253 NH | 2.25 | 2.49 | +0.24 |
| β3-K’ helix | |||
| Pro 321 C=O → Leu 324 NH | 2.09 | 2.14 | +0.05 |
| Pro 321 C=O → Ser 325 NH | 2.93 | 2.22 | −0.71 |
| Gln 322 C=O → Ser 325 NH | 2.03 | 2.29 | +0.26 |
| Gln 322 C=O → Ser 325 OγH | 1.93 | 5.01 | +3.08 |
| Ser 325 OyH → Thr 348 Oγ | 2.59 | 1.83 | −0.76 |
| Ser 325 Oy → Thr 348 NH | 1.85 | 3.68 | +1.83 |
| Gln 322 C=O → Gly 326 NH | 1.92 | 3.60 | +1.68 |
| Met 323 C=O → Leu 327 NH | 1.97 | 3.30 | +1.33 |
| Leu 324 C=O → Asp 328 NH | 4.02 | 2.43 | −1.59 |
| Ser 325 C=O → Asp 328 NH | 2.24 | 2.93 | +0.69 |
K’ helix and K’-β3 linkage
Another region that shows extensive chemical shift perturbation upon removal of substrate is the K’ helix (Ser 325-Gly 326-Leu 327-Asp 328). As can be seen from Table 1, a number of hydrogen bonds that are present in 2L8M (substrate-bound) are missing in REP1. In particular, a hydrogen bond between the carbonyl oxygen of Gln 322 in the β3 sheet and the Ser 325 OγH in helix K’ (observed in 2L8M) is absent in REP1, and has been replaced by a new hydrogen bond between the Ser 325 OγH and the Oγ of Thr 348 in the turn region preceding the L helix. Conversely, the hydrogen bond between the amide NH of Thr 348 and the Oγ of Ser 325 found in 2L8M is missing in REP1. Hydrogen bonds forming the primary link between the β3 sheet and the K’ helix in 2L8M (Gln 322 C=O → Gly 326 NH and Met 323 C=O → Leu 327 NH) are absent in REP1. These structural differences between the RDC-derived structures are well supported by the observed chemical shift perturbations in this region (vide supra). We have previously found that NH correlations in the K’ helix are remarkably sensitive to the identity of the substrate bound in the active site, despite the fact that the amide nitrogen of Ser 325 is 14 Å from the nearest carbon of (+)-camphor bound in the crystallographic structure 2CPP (21). As a result of these observations, we proposed that the K’ helix acts as a molecular “spring”, changing in length by switching between a 3,10 helix (i →i+3) and α-helical (i → i+4) hydrogen bonding pattern (24). This proposed motion would move one end of the β3 sheet either further in or out of the active site, effectively adjusting the position of the β3 residue Val 295, a primary substrate contact, in response to substrate size. In the present case, changes within the K’ helix upon substrate removal appear to be coupled to the displacements of the β3-β4 complex, and the modified hydrogen bonding arrangements observed in REP1 relative to 2L8M (Table 1) for the K’ helix indicate that in the absence of substrate, the interactions between the β3 sheet and the K’ helix are attenuated.
Conclusions
The current results are consistent with the active site of CYP101A1 being more mobile than has been previously suspected. While active site plasticity has been observed in other P450 enzymes (23), the lack of large-scale backbone displacements in most CYP101A1 structures described to date, regardless of the presence or type of substrate, hinted at a preformed active site, ideally suited to the binding of camphor and more exclusive of other potential substrates or inhibitors. However, our results, along with those of Goodin et al. (22), show that portions of the protein structure that contribute to the shape and size of the CYP101A1 active site are involved in large-scale movements, and different populations can be stabilized depending upon environmental factors. That the crystallographic and solution structures differ significantly in the location of the largest displacements suggests that the enzyme samples a range of conformations in the absence of substrate, consistent with a pre-existing equilibrium ensemble of conformers. Which conformations are favored under particular conditions is determined by environmental factors (e.g., crystal packing, oxidation state and axial ligation of the heme). This suggests that the barrier between such conformations are relatively low, and relatively small perturbations might be expected to shift equilibrium: A small population of solution conformers with an open, water-filled active site may pack more efficiently into a crystal lattice, while the compact water-free active site that we observe may be favored entropically due to fewer restrictions on water molecules in bulk solvent (34). We are currently analyzing dynamic data that will provide insight into local dynamics in oxidized and reduced CYP101A1 as a function of substrate binding.
Supplementary Material
Footnotes
This work was supported in part by a grant from the U.S. National Institutes of Health (R01-GM44191, TCP).
The REP1 structure described here has been deposited with the RCSB Protein Data Base as entry 2LQD. RDC restraints and chemical shifts are deposited with the BMRB database as accession number 17415.
Supporting information available. PDB format structure file for REP1, and RDC restraints in Amber format are available free of charge via the Internet at http://pubs.acs.org.
References cited
- 1.Fischer E. Einfluss der Configuration auf die Wirkung der Enzyme. Ber. Deutsche Chem. Gessel. 1894;27:2985–2993. [Google Scholar]
- 2.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niesen MJM, Bhattacharya S, Vaidehi N. The role of conformational ensembles in ligand recognition in G-protein coupled receptors. J. Am. Chem. Soc. 2011;133:13197–13204. doi: 10.1021/ja205313h. [DOI] [PubMed] [Google Scholar]
- 4.Fraser JS, van den Bedem H, Samelson AJ, Lang PT, Holton JM, Echols N, Alber T. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc. Natl. Acad. Sci. USA. 2011;108:16247–16252. doi: 10.1073/pnas.1111325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frauenfelder H, Petsko GA, Tsernoglou D. Temperature-dependent X-ray diffraction as a probe of protein structural dynamics. Nature. 1979;280:558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- 6.Pochapsky TC, Kostic M, Jain N, Pejchal R. Redox-dependent conformational selection in a Cys(4)Fe(2)S(2) ferredoxin. Biochemistry. 2001;40:5602–5614. doi: 10.1021/bi0028845. [DOI] [PubMed] [Google Scholar]
- 7.Labeikovsky W, Eisenmesser EZ, Bosco DA, Kern D. Structure and dynamics of Pin1 during catalysis by NMR. J. Mol. Biol. 2007;367:1370–1381. doi: 10.1016/j.jmb.2007.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fetler L, Kantrowitz ER, Vachette P. Direct observation in solution of a preexisting structural equilibrium for a mutant of the allosteric aspartate transcarbamoylase. Proc. Natl. Acad. Sci. USA. 2007;104:495–500. doi: 10.1073/pnas.0607641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei JY, Pochapsky TC, Pochapsky SS. Detection of a high-barrier conformational change in the active site of cytochrome P450cam upon binding of putidaredoxin. J. Am. Chem. Soc. 2005;127:6974–6976. doi: 10.1021/ja051195j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipscomb JD, Sligar SG, Namtvedt MJ, Gunsalus IC. Autooxidation and hydroxylation reactions of oxygenated cytochrome P450cam. J. Biol. Chem. 1976;251:1116–1124. [PubMed] [Google Scholar]
- 11.Glascock MC, Ballou DP, Dawson JH. Direct observation of a novel perturbed oxyferrous catalytic intermediate during reduced putidaredoxin-initiated turnover of cytochrome P450cam - Probing the effector role of putidaredoxin in catalysis. J. Biol. Chem. 2005;280:42134–42141. doi: 10.1074/jbc.M505426200. [DOI] [PubMed] [Google Scholar]
- 12.OuYang B, Pochapsky SS, Dang M, Pochapsky TC. A functional proline switch in cytochrome P450cam. Structure. 2008;16:916–923. doi: 10.1016/j.str.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asciutto EK, Madura JD, Pochapsky SS, OuYang B, Pochapsky TC. Structural and dynamic implications of an effector-induced backbone amide cis-trans isomerization in cytochrome P450cam. J. Mol. Biol. 2009;388:801–814. doi: 10.1016/j.jmb.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asciutto EK, Dang M, Pochapsky SS, Madura JD, Pochapsky TC. Experimentally restrained molecular dynamics simulations for characterizing the open states of cytochrome P450cam. Biochemistry. 2011;50:1664–1671. doi: 10.1021/bi101820d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nickerson DP, Wong LL. The dimerization of Pseudomonas putida cytochrome P450cam: Practical consequences and engineering of a monomeric enzyme. Protein Eng. 1997;10:1357–1361. doi: 10.1093/protein/10.12.1357. [DOI] [PubMed] [Google Scholar]
- 16.Rui LY, Pochapsky SS, Pochapsky TC. Comparison of the complexes formed by cytochrome P450cam with cytochrome b5 and putidaredoxin, two effectors of camphor hydroxylase activity. Biochemistry. 2006;45:3887–3897. doi: 10.1021/bi052318f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruckert M, Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 2000;122:7793–7797. [Google Scholar]
- 18.Weigelt J. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J. Am. Chem. Soc. 1998;120:12706–12706. [Google Scholar]
- 19.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: 2008. [Google Scholar]
- 20.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani FV, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. AMBER 10. Univ. of California; San Francisco: 2008. [Google Scholar]
- 21.Poulos TL, Finzel BC, Howard AJ. High resolution crystal structure of cytochrome P450cam. J. Mol. Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 22.Lee YT, Wilson RF, Rupniewski I, Goodin DB. P450cam visits an open conformation in the absence of substrate. Biochemistry. 2010;49:3412–3419. doi: 10.1021/bi100183g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pochapsky TC, Kazanis S, Dang M. Conformational plasticity and structure/function relationships in cytochromes P450. Antioxidants & Redox Signaling. 2010;13:1273–1296. doi: 10.1089/ars.2010.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang M, Pochapsky SS, Pochapsky TC. Spring-loading the active site of cytochrome P450cam. Metallomics. 2011;3:339–343. doi: 10.1039/c0mt00065e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raag R, Poulos TL. The structural basis for substrate-induced changes in redox potential and spin equilibrium in cytochrome P450cam. Biochemistry. 1989;28:917–922. doi: 10.1021/bi00428a077. [DOI] [PubMed] [Google Scholar]
- 26.Ludemann SK, Lounnas V, Wade RC. How do substrates enter and products exit the buried active site of cytochrome P450cam? 1. Random expulsion molecular dynamics investigation of ligand access channels and mechanisms. J. Mol. Biol. 2000;303:797–811. doi: 10.1006/jmbi.2000.4154. [DOI] [PubMed] [Google Scholar]
- 27.Sligar SG. Coupling of spin, substrate and redox equilibria in cytochrome P450. Biochemistry. 1976;15:5399–5406. doi: 10.1021/bi00669a029. [DOI] [PubMed] [Google Scholar]
- 28.Markwick PRL, Pierce LCT, Goodin DB, McCammon JA. Adaptive accelerated molecular dynamics (Ad-AMD) revealing the molecular plasticity of P450cam. J. Phys. Chem. Lett. 2011;2:158–164. doi: 10.1021/jz101462n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pochapsky SS, Dang M, OuYang B, Simorellis AK, Pochapsky TC. Redox- dependent dynamics in cytochrome P450cam. Biochemistry. 2009;48:4254–4261. doi: 10.1021/bi900002k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poulos TL, Finzel BC, Howard AJ. Crystal structure of substrate-free Pseudomonas putida cytochrome P450. Biochemistry. 1986;25:5314–5322. doi: 10.1021/bi00366a049. [DOI] [PubMed] [Google Scholar]
- 31.OuYang B, Pochapsky SS, Pagani GM, Pochapsky TC. Specific effects of potassium ion binding on wild-type and L358P cytochrome P450(cam) Biochemistry. 2006;45:14379–14388. doi: 10.1021/bi0617355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma M, Bell SG, Yang W, Hao YM, Rees NH, Bartlam M, Zhou WH, Wong LL, Rao ZH. Structural analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444. Chembiochem. 2011;12:88–99. doi: 10.1002/cbic.201000537. [DOI] [PubMed] [Google Scholar]
- 33.Poulos TL. Structural biology of P450-oxy complexes. Drug Metab. Rev. 2007;39:557–566. doi: 10.1080/03602530701498240. [DOI] [PubMed] [Google Scholar]
- 34.Miao YL, Baudry J. Active site hydration and water diffusion in cytochrome P450cam: A highly dynamic process. Biophys. J. 2011;101:1493–1503. doi: 10.1016/j.bpj.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLano WL. The PyMOL Molecular Graphics System. 2008 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.