Abstract
Neutrophils kill bacteria generally through oxidative and nonoxidative mechanisms. Whereas much research has focused on the enzymes essential for neutrophil killing, little is known about the regulatory molecules responsible for such killing. In this study, we investigated the role of olfactomedin 4 (OLFM4), an olfactomedin-related glycoprotein, in neutrophil bactericidal capability and host innate immunity. Neutrophils from OLFM4−/− mice have increased intracellular killing of Staphylococcus aureus and Escherichia coli in vitro. The OLFM4−/− mice have enhanced in vivo bacterial clearance and are more resistant to sepsis when challenged with S. aureus or E. coli by i.p. injection. OLFM4 was found to interact with cathepsin C, a cysteine protease that plays an important role in bacterial killing and immune regulation. We demonstrated that OLFM4 inhibited cathepsin C activity in vitro and in vivo. The cathepsin C activity in neutrophils from OLFM4−/− mice was significantly higher than that in neutrophils from wild-type littermate mice. The activities of three serine proteases (neutrophil elastase, cathepsin G, and proteinase 3), which require cathepsin C activity for processing and maturity, were also significantly higher in OLFM4−/− neutrophils. The bacterial killing and clearance capabilities observed in OLFM4−/− mice that were enhanced relative to wild-type mice were significantly compromised by the additional loss of cathepsin C in mice with OLFM4 and cathepsin C double deficiency. These results indicate that OLFM4 is an important negative regulator of neutrophil bactericidal activity by restricting cathepsin C activity and its downstream granule-associated serine proteases.
Introduction
Neutrophils are a major component of innate immunity, providing a crucial first barrier against bacterial infection. It has been well documented that NADPH-mediated oxidative burst acts to kill certain pathogens (1). Other than reactive oxygen species, neutrophils also produce a multitude of antimicrobial molecules in their granules. The granules fuse with the phagolysosomes and contain defensins, bactericidal permeability-increasing protein, as well as serine proteases such as elastase, cathepsin G, and proteinase 3. The serine proteases generally degrade bacterial proteins and are major antimicrobial components. It has been shown that serine proteases, when activated, are primarily responsible for the destruction of bacteria (1). Recent studies suggest that bacteria are not killed by a common mechanism (2).
Cathepsin C (also called dipeptidyl peptidase I) is a lysosomal cysteine peptidase belonging to the papain family (3) and has numerous biological functions. In addition to a proposed degradative role as a lysosomal exopeptidase, cathepsin C is essential for activating granule-associated serine protease in neutrophils (elastase, cathepsin G, and proteinase 3) (4). These serine proteases require the activity cathepsin C to remove prodipeptide to become active. Serine proteases are mainly stored in azurophil granules in their active forms until they are released after neutrophil exposure to active stimuli and have broad biological effects including intracellular microbial killing and modulation of inflammatory-cell recruitment (4).
OLFM4 (also called hGC-1 and GW112) is a member of the olfactomedin family, which is characterized by the presence of an olfactomedin domain in the C terminus (5). Olfactomedin-related proteins have distinct tissue expression patterns and diverse cellular functions (5). OLFM4 was first cloned from G-CSF–stimulated human myeloid precursor cells (6). Although most of the other members are preferentially expressed in nervous tissues, OLFM4 is mainly expressed in hematopoietic myeloid cells and in the gastrointestinal tract (6). OLFM4 is involved in regulation of multiple cellular functions including apoptosis (7, 8), differentiation (8), and growth (9). OLFM4 expression is upregulated in some inflammatory diseases, such as chronic inflammatory bowel diseases (10), and in Helicobacter pylori-infected patients (11). It was recently reported that OLFM4 downregulates mouse host innate immunity to H. pylori infection (12). However, little is known about the physiological functions of phylogenetically conserved OLFM4 in host defense against a broader range of bacteria. Moreover, its expression, localization, and potential biological functions in mature myeloid cells (neutrophils) remain elusive. In this study, we investigated whether OLFM4 regulates host defense against a broader range of bacteria including Gram-positive Staphylococcus aureus and Gram-negative Escherichia coli using an OLFM4−/− mouse model.
Materials and Methods
Mice, cells, and bacteria
OLFM4−/− mice have been described previously (12) and backcrossed six generations into a C57BL/6 background. Cathepsin C target mutant mice (CatC−/−) in the C57BL/6 background were provided from Washington University School of Medicine (St. Louis, MO). Animals were kept in a specific pathogen-free facility at the National Institutes of Health (Bethesda, MD) and used according to protocols approved by the National Heart, Lung, and Blood Institute Animal Care and Use Committee. All experiments were conducted with animals 10–12 wk of age. HEK 293T cells were purchased from American Type Culture Collection (ATCC; Manassas, VA) and maintained as previously described (13). S. aureus ATCC 10390 and E. coli ATCC 10536 were purchased from ATCC. Salmonella enterica was obtained from Dr. William Coleman’s laboratory (National Institute of Diabetes and Digestive and Kidney Disease/National Institutes of Health, Bethesda, MD). All bacteria were grown in tryptic soy broth or on tryptic soy agar plates (Teknova, Hollister, CA).
Yeast two-hybrid library screening
Full-length human OLFM4 cDNA (bait) was cloned into pCWX200 and pLexA expression plasmid. This bait plasmid was transformed into yeast strain Y304 and screened using a TetR system against a HeLa cDNA library in pYESTrp2 containing 1 × 107 independent clones in strain EGY42. The positive colonies (those appearing blue) were retested for phenotypes. The plasmids from positive colonies were then isolated after transformation into E. coli, and the interactions were confirmed in yeast by mating. Plasmids from verified colonies were sequenced.
Isolation of neutrophils
Human peripheral blood neutrophils were isolated from healthy donors using HetaSep (Stem Cell Technologies, Vancouver, BC, Canada) sedimentation and Ficoll-Paque PLUS density gradient separation followed by hypotonic lysis of erythrocytes. Human fresh bone-marrow neutrophils were obtained from AllCells (Emeryville, CA). Mouse neutrophils were isolated from bone marrow. The femur was removed from mice, then flushed with PBS containing 0.5% BSA. The mouse bone marrow neutrophils were purified through a three-layer gradient of 78, 69, and 52% Percoll (Amersham Pharmacia Biotech, Uppsala, Sweden) as previously described (14). Mouse bone marrow-derived neutrophils were used in our study based on the finding that these neutrophils are in a resting state and are functionally competent (14).
Subcellular fractionation of neutrophil granules
Neutrophil granules were enriched by centrifugation on a three-layer Percoll density gradient as previously described (15).
Immunoprecipitation and Western blotting
Immunoprecipitation and Western blot analysis were performed as previously described (13). OLFM4 polyclonal Ab (16) and cathepsin C Ab (Santa Cruz Biotechnology, Santa Cruz, CA) were used as primary Abs (1:1000). HRP-conjugated donkey anti-rabbit or sheep anti-mouse Abs (1:4000; Amersham, Piscataway, NJ) were used as secondary Abs.
Quantitative reverse transcriptase PCR
Total RNA preparation and quantitative reverse transcriptase PCR was performed as previously described (8).
Bactericidal activity
Mouse neutrophils were isolated from the bone marrow. In a 24-well plate, neutrophils were resuspended in DMEM (106 cell/ml) and seeded in wells for 30 min. Nonadherent cells were removed by gentle rinsing. Bacteria were preopsonized with mouse serum for 30 min at 37°C. Bacteria (107) were added onto the monolayer of cells. After incubation at 37°C for 15 min, the media were gently removed, and neutrophils were lysed in PBS containing 0.1% Triton X-100 after incubation for 30 min in the presence or absence of gentamicin (100 μg/ml) or lysostaphin (50 μg/ml). The number of viable bacteria was determined using the standard plate method (17).
Peritoneum bacterial clearance and neutrophil influx
Mice were injected i.p. with a sublethal dose of E. coli (5 × 103) or S. aureus (5 × 104). Peritoneal cavities were lavaged with 5 ml PBS 2 h after inoculation. The number of viable bacteria was determined using the standard plate method. The percentages of blood granulocytes and mononuclear cells were determined by differential counting on cytospin preparations. The number of neutrophils was determined.
Survival and bacterial dissemination post-i.p. infection
E. coli and S. aureus were grown in tryptic soy broth to the exponential phase. Mice of each genotype between 10 and 12 wk of age were injected i.p. with varying amounts of bacteria (CFU). An optimal concentration to induce sepsis and mortality within 72 h was determined for E. coli (1.5 × 108 CFU) and S. aureus (3.5 × 108 CFU). In survival experiments using this concentration, survival was monitored every 6 h. To determine the bacterial dissemination to large organs during sepsis, liver and lung tissues were harvested 24 h after S. aureus or E. coli infection. The tissues were homogenized with PBS, and aliquots of serially diluted homogenate were plated on tryptic soy agar. The colonies were enumerated the following day.
OLFM4 protein purification
Human OLFM4 proteins were purified using the LEXSY system (Jena Bioscience, Jena, Germany) from the protozoan host Leishmania tarentolae. The OLFM4 ORF was PCR-amplified from pcDNA3.1/OLFM4-V5 vector (13) and inserted into the inducible LEXSY expression vector pLEXSY_I-neo2 in-frame to the LEXSY signal peptide and C-terminal HexaHis. The LEXSY host T7-TR was transfected with an expression plasmid for inducible secretory expression. Human OLFM4 proteins were purified by NiNTA affinity chromatography from LEXSY culture supernatants. Elutes were dialyzed into TBS (50 mM Tris pH 7.5, 150 mM NaCl).
Protease activity assays
Bone marrow-derived neutrophils were washed twice with PBS and lysed with cathepsin C lysis buffer (25 mM MES, pH 6.0, 50 mM NaCl, 5 mM DTT, 0.2% Nonidet P-40, for cathepsin C assays) or with serine protease lysis buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.2% Nonidet P-40, for serine protease assays). Debris was removed by centrifugation at 15,000 × g for 10 min, and the supernatants were retained. Cathepsin C activities were assayed in 25 mM MES, pH 6.0, 50 mM NaCl, 5 mM DTT, 0.1% PEG 3350 using Gly-Arg-AMC (Bachem, Torrance, CA) at 10 μM. Reaction progress was monitored continuously with product (AMC) on a FLUOstar Optima Fluorimeter (BMG, Gary, NC) with 380-nm excitation and 460-nm emission wavelength filters. Serine protease activities were assayed in 100 mM Tris-HCl, pH 7.5, and 50 mM NaCl supplemented with a peptide substrate specific for the serine protease assessed: for neutrophil elastase, 600 μM methoxysuccinyl-Ala-Ala-Pro-Val-p-nitroanilide (Elastin Products Company, Owensville, MO); for cathepsin G, 600 μM N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (Sigma, St. Louis, MO); and for proteinase 3, 100 μM Boc-Ala-Ala-Nva-SBzL (Elastin Products Company). Release of p-nitroanilide was measured by absorbance at 410 nm. Release of SBzL was measured by absorbance at 320 nm. Background was subtracted. For in vitro cathepsin C activity assays, recombinant human cathepsin C (R&D Systems, Minneapolis, MN) was activated by cathepsin L (R&D Systems) in the activation buffer (25 mM MES, 5 mM DTT, pH 6.0) at room temperature for 1 h. Various amounts of purified OLFM4 proteins were incubated with activated cathepsin C in the activation buffer (25 mM MES, 50 mM NaCl, 5 mM DTT, pH 6.0) for 30 min at room temperature. Cathepsin C activity was measured as described earlier.
Oxidative burst assay
Superoxide production was determined using the Diogenes Cellular Luminescence Enhancement System for Superoxide Detection according to the manufacturer’s instructions (National Diagnostics, Atlanta, GA).
Myeloperoxidase activity assay
Myeloperoxidase (MPO) activity was quantitatively measured by the method of Suzuki et al. (18) with some modifications. Neutrophils were adjusted to 5 × 106 cells/ml and incubated with N-formyl-Met-Leu-Phe (1 μM) and cytochalasin B (5 μg/ml) for 10 min at 37°C. Triton X-100 (final concentration, 0.1%) was added to the cell suspensions for total MPO release. Aliquots of the cell extracts were incubated with the TMB liquid substrate system, and the activity was detected spectrophotometrically at 370 nm.
Phagocytosis
Phagocytosis was assessed by flow cytometry as previously described (19). FITC-labeled S. aureus or E. coli particles (Molecular Probes, Eugene, OR) were reconstituted and opsonized with opsonizing reagent (Molecular Probes) according to the manufacturer’s instructions. Opsonized particles (107) were washed and incubated with neutrophils (106) (multiplicity of infection = 10). The threshold (forward-scattered light) was set to exclude particles not associated with neutrophils. After measuring the total number of neutrophils with bound/ingested particles, samples were quenched with 0.4% trypan blue to determine the percentage of neutrophils with ingested particles. The percentage phagocytosis was determined by the percentage of FITC+ neutrophils observed after quenching.
Statistical analysis
Significance of differences between experimental groups was determined by a two-way Student t test. Differences were considered significant when p < 0.05. Survival statistics were performed with the Kaplan–Meier log-rank test (GraphPad Prism version 4.0).
Results
OLFM4 is a neutrophil granule protein that responds to bacteria infection
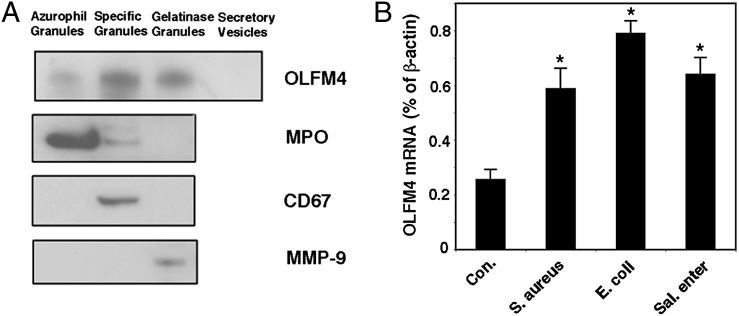
OLFM4 is a secreted glycoprotein (7) and is also retained inside neutrophils (8, 13). We first determined whether OLFM4 is a neutrophil granule protein. Neutrophil granule fractions were separated from human peripheral blood neutrophils using Percoll density gradient (15). OLFM4 protein expression was detected in all three neutrophil granule subsets, with high abundance in specific granules, modest levels in gelatinase and azurophil granules, and absence in secretory vesicles (Fig. 1A). Previously, it has been shown that OLFM4 is upregulated under inflammatory (10) and H. pylori infection (11) conditions. In this article, we demonstrated that OLFM4 mRNA expression in human neutrophils was upregulated in response to a broad range of bacterial infections, including Gram-positive S. aureus and Gram-negative E. coli and S. enterica infections (Fig. 1B). These results indicate that OLFM4 is a novel neutrophil granule protein that exhibits enhanced expression to a broad range of bacterial infections.
FIGURE 1.
OLFM4 is a neutrophil granule protein that responds to bacterial infection. (A) Western blotting was performed to detect OLFM4, MPO, CD67, and MMP-9 in human peripheral blood neutrophil granule subsets. (B) Quantitative reverse transcriptase PCR was performed to detect OLFM4 mRNA expression in human peripheral blood neutrophils incubated in vitro in the presence or absence of S. aureus, E. coli, or S. enterica (Sal. enter) infection for 6 h (neutrophil/bacteria = 1:10). Data represent expression relative to β-actin expression and are expressed as mean ± SD (n = 3). *p < 0.05 when compared with control (Con.).
OLFM4 deficiency in neutrophils leads to an increased bactericidal capacity, enhanced in vivo bacterial clearance, and resistance to sepsis
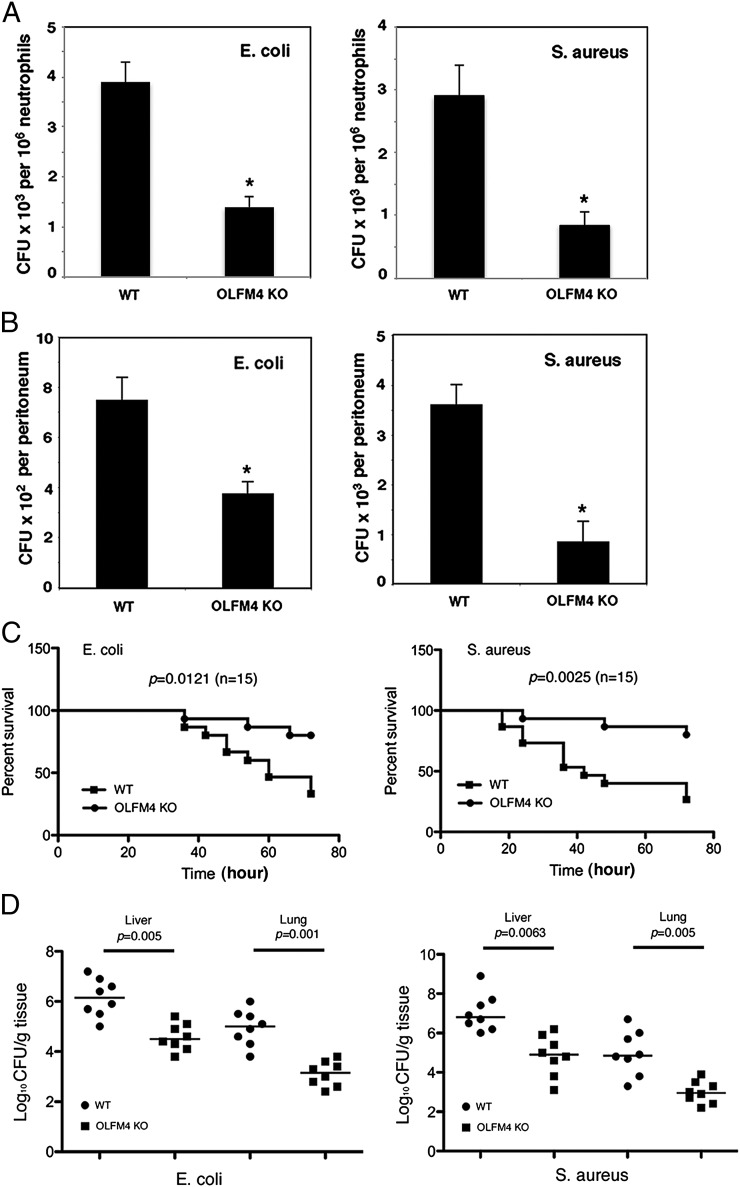
To determine the mode of action of OLFM4 in combating bacterial infections, we compared the ability of neutrophils from OLFM4−/− mice and their wild-type (WT) littermates to recognize and kill bacteria in vitro. We exposed monolayers of neutrophils to S. aureus or E. coli. After 15 min, unbound bacteria were gently removed, and the number of viable bacteria associated with the neutrophil monolayer was assessed. The number of viable bacteria for OLFM4−/− and WT neutrophil groups at 15 min was comparable (Supplemental Fig. 1A). This observation suggests that OLFM4−/− and WT neutrophils were able to recognize and bind bacteria equally well. After incubation for an additional 30 min in the presence of gentamicin or lysostaphin that eliminates extracellular E. coli or S. aureus, respectively, the intracellular bactericidal activity was assessed. Under these conditions, OLFM4−/− neutrophils exhibited more bactericidal activity than WT neutrophils (Fig. 2A).
FIGURE 2.
OLFM4−/− mice display enhanced intracellular bacterial killing, increased peritoneal bacterial clearance, and resistance to sepsis. (A) Neutrophils (106) derived from the bone marrow of WT and OLFM4−/− (OLFM4 KO) mice were incubated with preopsonized E. coli (107) or S. aureus (107) in a 24-well plate. After 15 min, the media were gently removed. The cells were treated with gentamicin (100 μg/ml) for E. coli or lysostaphin (50 μg/ml) for S. aureus for additional 30 min. The viable bacteria were counted on tryptic soy agar plates. Data are expressed as mean ± SD (n = 4). *p < 0.05. (B) Mice were challenged i.p. with E. coli (5 × 103) or S. aureus (5 × 104). After 2 h, the peritoneal cavity was lavaged and the number of bacterial colonies was determined on plates. Data are expressed as mean ± SD (n = 4). *p < 0.05. (C) Mice (10–12 wk old, n = 15) were infected i.p. with E. coli (1.5 × 108 CFU) or S. aureus (3.5 × 108 CFU), and survival was monitored every 6 h over 72 h. (D) Mice (n = 8) were treated as in (C). Mice were euthanized 24 h after bacterial infection, and liver and lung tissues were collected. Homogenized tissues were serially diluted and plated, and colonies were enumerated the following day.
We next determined the capability of OLFM4−/− and WT mice to clear bacteria from the site of infection in vivo. Mice were challenged i.p. with S. aureus or E. coli. After 2 h, the number of viable bacteria in the peritoneal cavities of OLFM4−/− mice was significantly less than that in WT mice (Fig. 2B). Neutrophil recruitment into the peritoneal cavity was comparable for OLFM4−/− and WT mice as determined by neutrophil counts and MPO activity (Supplemental Fig. 1B, 1C).
Furthermore, we compared the susceptibility of OLFM4−/− and WT mice to sepsis induced by i.p. challenge with S. aureus or E. coli. We first determined an optimal bacterial concentration to induce lethal sepsis over 72 h in WT mice, then used this concentration to examine survival. OLFM4−/− mice were found to be much more resistant to sepsis than WT mice (Fig. 2C). Consistent with the survival assay, the number of bacteria disseminated to the liver and lung in OLFM4−/− mice was significantly lower than that in WT mice postinfection for 24 h (Fig. 2D). These data suggested that endogenous OLFM4 is a critical negative regulator of intracellular bactericidal activity in neutrophils and thus dampens mice host immune response to bacterial infection.
The NADPH oxidative burst, MPO activity, and phagocytosis are not affected in OLFM4−/− neutrophils
The NADPH oxidase-mediated superoxide (O2−) burst (Supplemental Fig. 2), MPO activity (Supplemental Fig. 3), and phagocytosis of S. aureus and E. coli (Supplemental Fig. 4) were found to be comparable in OLFM4−/− and WT neutrophils. We also found that purified recombinant human OLFM4 protein had no direct effect on the growth of E. coli and S. aureus in vitro (data not shown). These data suggest that OLFM4 regulation of bactericidal activity is not mediated directly through the oxidative pathway, and OLFM4 protein does not have a direct effect on bacteria growth.
OLFM4 binds cathepsin C and inhibits cathepsin C activity in vitro and in vivo
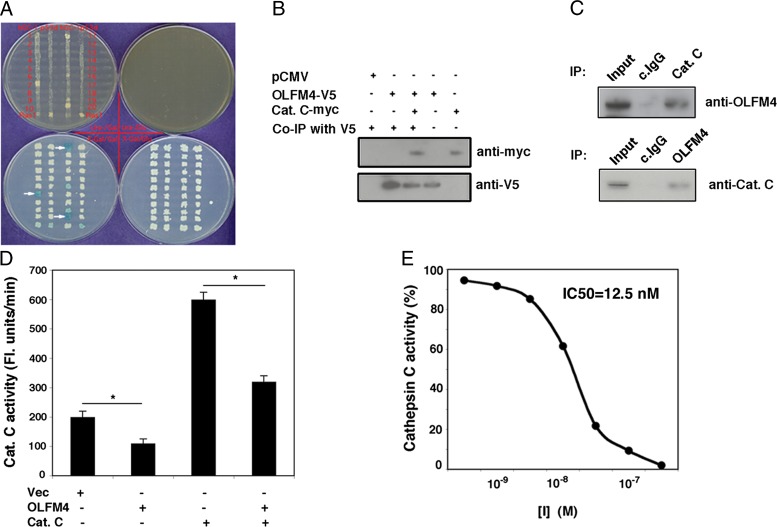
To further investigate the molecular mechanism of OLFM4 in regulating neutrophil killing of bacteria, we performed a yeast two-hybrid assay to identify the potential binding partner of human OLFM4. Cathepsin C, a cysteine protease, was shown to interact with OLFM4 (Fig. 3A). A coimmunoprecipitation assay confirmed that OLFM4 binds to cathepsin C in HEK 293T cells that overexpress full-length OLFM4 and cathepsin C (Fig. 3B). A reciprocal coimmunoprecipitation assay demonstrated endogenous association of OLFM4 with cathepsin C in human bone marrow neutrophils (Fig. 3C). The physical association between OLFM4 and cathepsin C raised the possibility that OLFM4 plays a role in modulating cathepsin C activity. Expression of OLFM4 in HEK 293T cells suppressed the activity of endogenous cathepsin C and of overexpressed cathepsin C activity (Fig. 3D). Purified OLFM4 protein effectively inhibited recombinant human cathepsin C activity in vitro (Fig. 3E), with an IC50 of 12.5 nM. These results indicate that OLFM4 is an effective inhibitor of cathepsin C in vitro and in vivo.
FIGURE 3.
OLFM4 binds cathepsin C and is an inhibitor of cathepsin C in vitro and in vivo. (A) A yeast two-hybrid assay was performed to identify potential binding partners of OLFM4. The blue colonies (indicated by white arrows) were isolated and confirmed to be cathepsin C by sequencing. (B) Expression plasmids of OLFM4-V5 and cathepsin C-myc were transfected individually or cotransfected into HEK 293T cells. After 24 h, the total cell lysates were immunoprecipitated with V5 Ab and probed with myc or V5 Ab by Western blotting. (C) Total cell lysates from normal human bone marrow neutrophils were immunoprecipitated with cathepsin C Ab (Cat. C) or control IgG Ab (c.IgG) and probed with OLFM4 Ab by Western blotting (top panel). Total cell lysates from normal human bone marrow neutrophils were immunoprecipitated with OLFM4 Ab or c.IgG and probed with Cat. C Ab by Western blotting (bottom panel). Lysate before immunoprecipitation was loaded as a control (Input). (D) HEK 293T cells were transfected with OLFM4-V5, cathepsin C-myc, or control vector (Vec) expression plasmid, or cotransfected with OLFM4-V5 and cathepsin C-myc expression plasmids. Cathepsin C activity was analyzed by hydrolyzing Gly-Arg-AMC. Results are expressed as mean ± SD (n = 4) fluorescence units per minute. *p < 0.05. (E) Cathepsin C (250 ng/ml) was incubated with purified OLFM4 protein at various concentrations as indicated at room temperature for 30 min, and the cathepsin C activity was analyzed. The percentage of cathepsin C activity was demonstrated.
Cathepsin C and serine protease activities are enhanced in neutrophils of OLFM4−/− mice
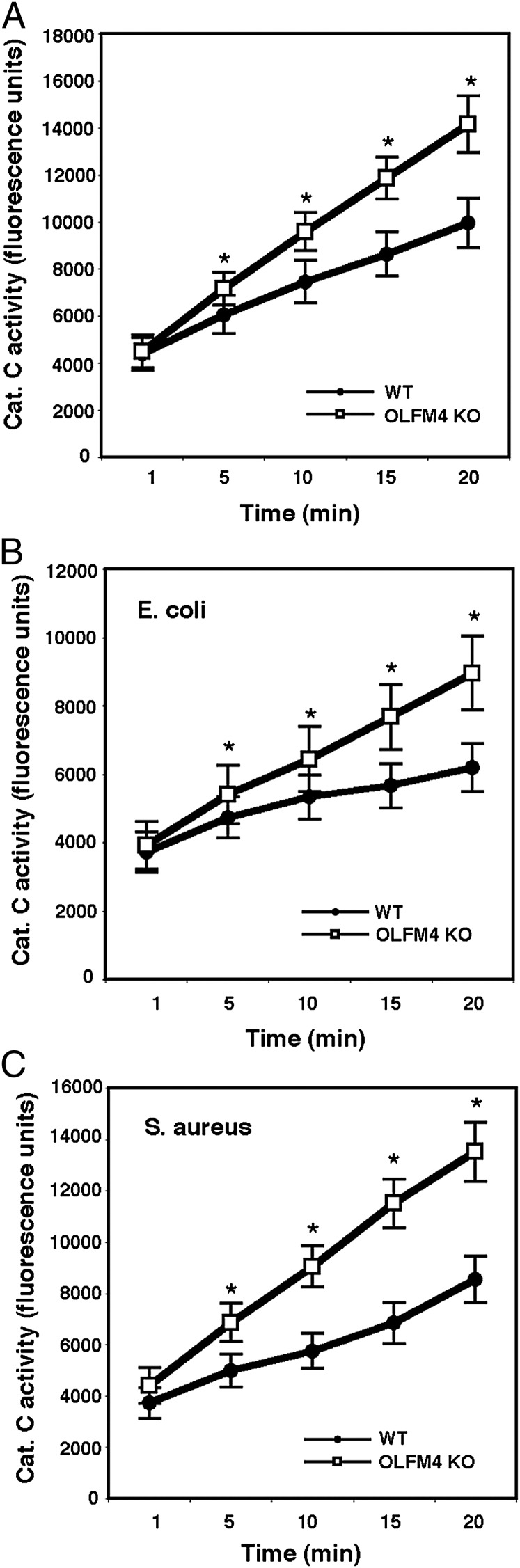
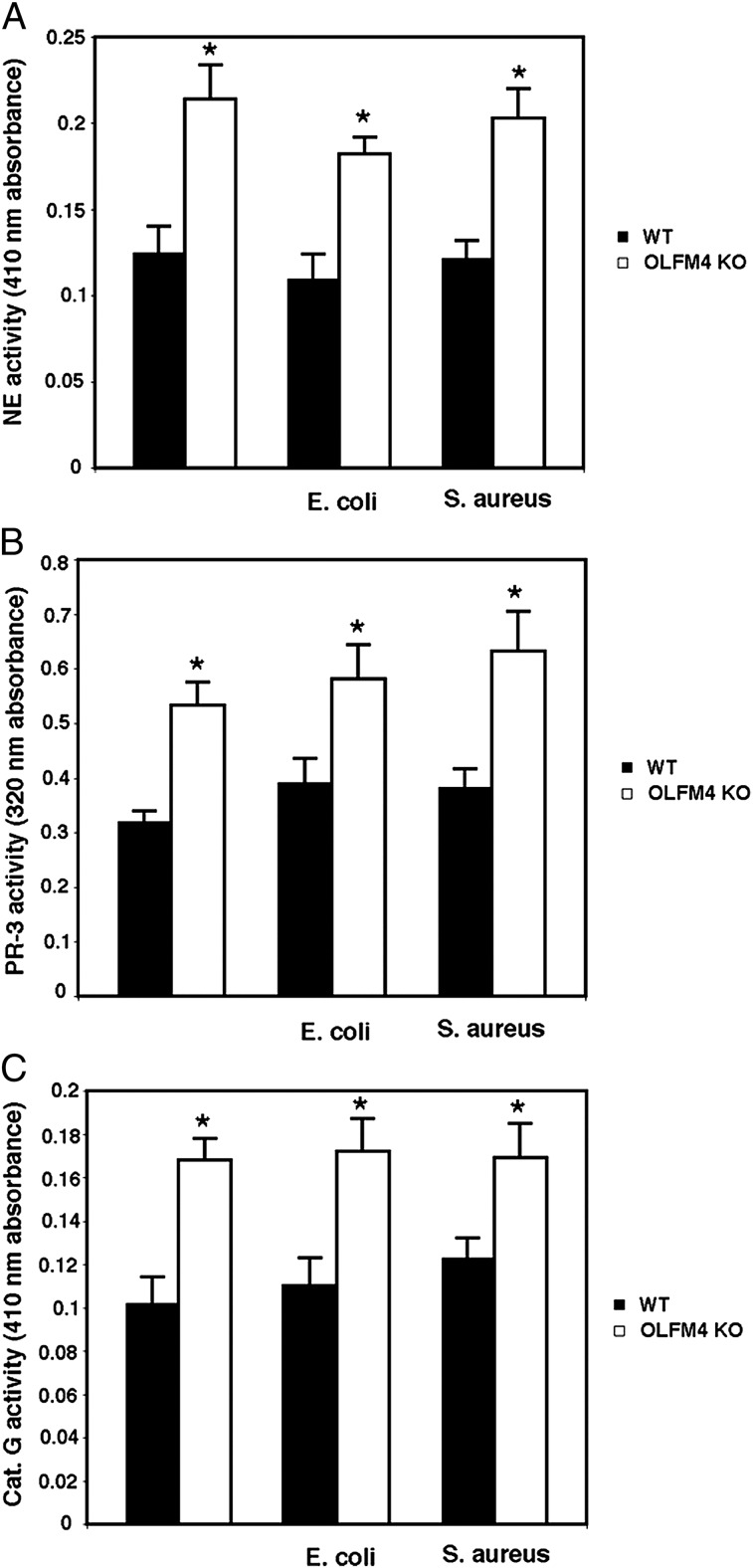
It has been recently reported that neutrophil serine proteases play much more important roles in killing bacteria than previously thought (1). The processing and activation of serine proteases require cathepsin C activity (4). Having demonstrated that OLFM4 is associated with cathepsin C in neutrophils and suppresses its activity, we investigated whether the enhanced neutrophil bacteria-killing capability in OLFM4−/− mice was accompanied by enhanced cathepsin C activity. The cathepsin C activity in neutrophils from OLFM4−/− mice was significantly increased relative to that of neutrophils from WT mice under both noninfection and infection conditions (Fig. 4). The activities of the serine proteases neutrophil elastase (Fig. 5A), proteinase 3 (Fig. 5B), and cathepsin G (Fig. 5C) were also significantly higher in neutrophils from OLFM4−/− mice compared with those isolated from WT mice. These results suggest that loss of OLFM4 in neutrophils leads to enhanced cathepsin C and serine protease activities, thereby increasing bacterial killing.
FIGURE 4.
Neutrophils from OLFM4−/− mice display enhanced activities of cathepsin C. Neutrophils (5 × 106) derived from bone marrow of WT and OLFM4−/− (OLFM4 KO) mice were incubated in vitro without (A) or with E. coli (5 × 107) (B) or S. aureus (5 × 107) (C) for 1 h. Cells were lysed in cathepsin C lysis buffer, and equal amounts of lysates were used for cathepsin C activity assays. The cathepsin C activities in neutrophils from OLFM4 KO mice were significantly higher than those from WT mice (*p < 0.01) in the absence or presence of bacterial challenge. Data are expressed as mean ± SD (n = 3).
FIGURE 5.
Neutrophils from OLFM4−/− mice display enhanced activities of serine proteases. (A) Cells were lysed in serine protease lysis buffer, and equal amounts of lysates were used for neutrophil elastase (NE) activity assay using methoxysuccinyl-Ala-Ala-Pro-Val-p-nitroanilide as a substrate at room temperature for 30 min. The absorbance at 410 nm was measured. *p < 0.05 when compared with WT. (B) Cells were lysed in serine protease lysis buffer, and equal amounts of lysates were used for proteinase-3 (PR-3) activity assay using Boc-Ala-Ala-Nva-SBzL as a substrate at room temperature for 30 min. The absorbance at 320 nm was measured. *p < 0.05 when compared with WT. (C) Cells were lysed in serine protease lysis buffer, and equal amounts of lysates were used for cathepsin G (Cat. G) activity assay using N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide as a substrate at room temperature for 30 min. The absorbance at 410 nm was measured. *p < 0.05 when compared with WT. All data are expressed as mean ± SD (n = 3).
Cathepsin C contributes to enhanced bacterial killing in OLFM4−/− mice
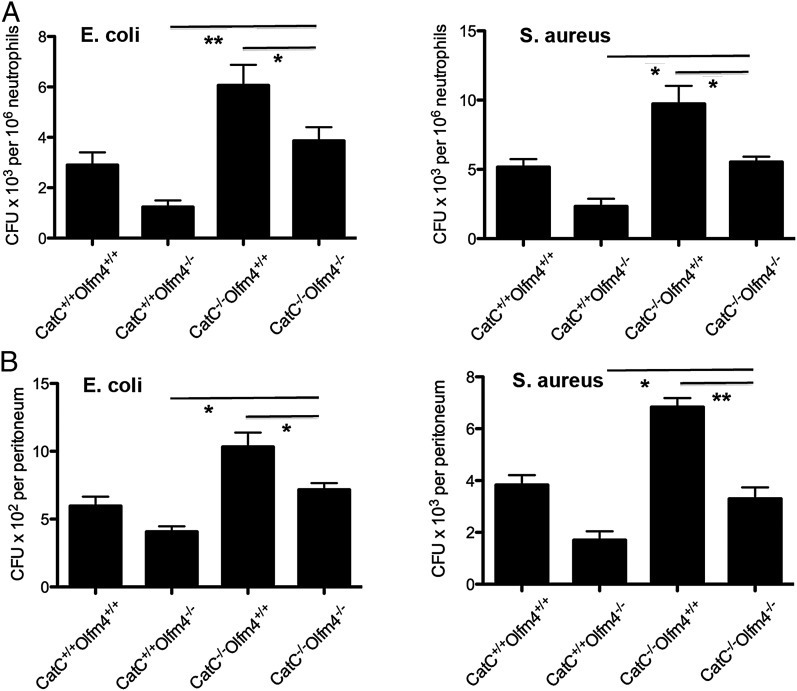
To confirm that cathepsin C plays an essential role in OLFM4’s downregulation of neutrophil bacterial killing, we created OLFM4 and cathepsin C double-deficient mice (OLFM4−/−CatC−/−). The intracellular bacterial killing activity observed in OLFM4−/− mice that was enhanced relative to WT mice was significantly compromised by the additional loss of cathepsin C in mice with OLFM4 and cathepsin C double deficiency (OLFM4−/−CatC−/−; Fig. 6A). Similarly, double deficiency of OLFM4 and cathepsin C also significantly reduced the in vivo bacterial clearance observed in OLFM4−/− mice that was enhanced relative to WT mice (Fig. 6B). In agreement with a previous report that cathepsin C-deficient mice have impaired bacterial clearance (20), the intracellular bacteria killing and clearance of S. aureus and E. coli were found to be significantly impaired (Fig. 6A, 6B). That the additional loss of cathepsin C in OLFM4−/− mice (OLFM4−/−CatC−/−) led to a compromised bacterial clearance, but not to the extent of defect in CatC−/− mice (Fig. 6A, 6B), suggested that OLFM4 modulates bacterial clearance through both a cathepsin C-dependent and -independent mechanism. These data provide direct evidence that there is a functional interplay between OLFM4 and cathepsin C gene, and OLFM4 restriction of neutrophil bacterial killing is mediated, at least partially, through interaction with cathepsin C.
FIGURE 6.
Enhanced in vitro intracellular bacterial killing and in vivo clearance observed in OLFM4−/− mice are compromised in cathepsin C and OLFM4 double-deficient mice. (A) Bone-marrow neutrophils (106) from different genotype mice as indicated were incubated with preopsonized E. coli (107) or S. aureus (107) in a 24-well plate. After 15 min, the media were gently removed. The cells were treated with gentamicin (100 μg/ml) for E. coli or lysostaphin (50 μg/ml) for S. aureus for an additional 30 min. The viable bacteria were counted on tryptic soy agar plates. Data are expressed as mean ± SD (n = 3). *p < 0.05, **p < 0.01. (B) Mice were challenged i.p. with E. coli (5 × 103) or S. aureus (5 × 104). After 2 h, the peritoneal cavity was lavaged and the number of viable bacteria was determined using the standard plate method. Data are expressed as mean ± SD (n = 3). *p < 0.05, **p < 0.01.
Discussion
S. aureus and E. coli infections are still major threats to human health, especially because of emerging antimicrobial resistance (21). Neutrophils play essential roles in killing invading bacteria. Therefore, it is important to understand the molecular mechanisms underlying neutrophil activity to better combat bacteria infection. In this study, we demonstrate that OLFM4 is a critical regulator of neutrophil’s ability to kill S. aureus and E. coli. This function is associated with restriction of cathepsin C and downstream serine proteases, but not with NADPH-mediated oxidative burst.
We demonstrate that OLFM4 physically associates with cathepsin C in neutrophils. We show that OLFM4 is localized in multiple granule compartments with most abundance in specific granules, but also in azurophilic granules where cathepsin C and serine proteases are mostly localized (15). As previously described, there is a functional interplay between different granule subsets (22). For example, neutrophil gelatinase-associated lipocalin that is normally found in the specific granules is able to complex with gelatinase (23). Proteases from azurophil granules may activate the cathelicidin present in specific granules (22). Therefore, OLFM4 abundantly present in specific granules may also interact with cathepsin C in azurophil granules, especially when azurophil granules and specific granules are fused with phagosomes to form phagolysosomes after bacteria phagocytosis by neutrophils.
Recent studies showed that cathepsin C is involved in multiple pathological processes, particularly in neutrophils where it plays an important role in bacterial clearance (20) and the inflammatory response (24). Thus, cathepsin C represents a potential therapeutic target for the treatment of inflammatory disease. Many small peptides have been selected to be potent cathepsin C inhibitors, but they have poor stability and/or pharmacokinetics (25). Cathepsin C is also inhibited by protein inhibitors, such as rat stefin A and chicken cystatin F, two important inhibitors of cysteine peptidases from the cystatin superfamily (26, 27). Cystatin F is expressed primarily in CD8+ T cells and in CD56+ NK cells, and has been shown to complex with cathepsin C in human monocytic and NK cells (27). Our study showed that OLFM4, a granule protein, was associated with cathepsin C in neutrophils, and strongly and specifically inhibited cathepsin C both in vitro and in vivo. Moreover, the data from the OLFM4−/− mouse model demonstrated that OLFM4 also affects multiple serine protease activities indirectly. Thus, OLFM4 represents a novel protein inhibitor of cathepsin C, specifically in neutrophils.
Our results show that enhanced bacteria killing activity in OLFM4-deficient neutrophils is associated with increased cathepsin C and downstream serine protease activities. It has been reported that bacterial clearance is impaired in cathepsin C−/− mice using a cecal ligation and puncture model of septic peritonitis (20), demonstrating that cathepsin C is an important regulator for bacteria control and clearance. Although a regulatory role of cathepsin C on specific bacteria such as S. aureus and E. coli used in this study has not been reported, mice clearance of E. coli and S. aureus are impaired in absence of active neutrophil elastase and cathepsin G, which requires activation from cathepsin C (1). The mode of action of OLFM4 through cathepsin C is further confirmed by experiments in OLFM4 and cathepsin C double-deficient mice. The enhanced bactericidal abilities in the neutrophils of OLFM4−/− mice were compromised, but not eliminated, by further deletion of cathepsin C gene, confirming the involvement of cathepsin C in the OLFM4-mediated effect. This suggests that the increased bactericidal activity in OLFM4−/− neutrophils is, at least in part, due to the enhanced cathepsin C activity. However, these data do not rule out possible involvement of other mechanisms in the observed phenotypes. For example, it was previously reported that OLFM4 negatively regulates the nucleotide-binding oligomerization domain (Nod) 1 and Nod2 (12). Nod1 has a broad influence on neutrophil function, activating both oxidative and nonoxidative mechanisms of killing (28). Therefore, the increased bactericidal activity in OLFM4-deficient neutrophils may also be partially caused by subsequent enhanced Nod1 activity in neutrophils.
OLFM4 and other olfactomedin-related proteins have a high m.w. multimer structure with intramolecular and intermolecular disulfide bonds (13). It has been previously reported that OLFM4 complexes with cadherin (13) and Nods (12) proteins. These observations together with this study suggest a possibility that OLFM4 may be a component of multiple protein complexes as an adaptor or scaffold protein. The notion that olfactomedin class of proteins may be scaffolds for proteases and their substrates was also recently suggested by Inomata and coworkers (29) based on their finding that a Xenopus secreted olfactomedin 1 recruits the Tolloid proteases to their substrate chordin and is required for normal chordin degradation.
In summary, we demonstrate that OLFM4 is a neutrophil granule protein, which proved to be an important negative regulator of bacteria killing as evidenced by that neutrophils from OLFM4−/− mice have increased capability to kill S. aureus and E. coli, and OLFM4−/− mice are more resistant to systemic sepsis. Taken together with the previous study (12), which demonstrated that OLFM4 downregulates host innate immunity against H. pylori, OLFM4 appears to be an important regulator of host innate immunity against a broad array of bacterial infections. OLFM4 may prove to be a potential target for therapeutic augmentation of host innate immunity in genetic immune-deficient patients such as those with chronic granulomatous disease.
Supplementary Material
Acknowledgments
We thank Dr. Christine T.N. Pham, Washington University School of Medicine, for providing the CatC−/− mice. We thank Drs. John I. Gallin and Kol A. Zarember, Clinical Center, National Institutes of Health, for helpful discussions and careful review of the manuscript. We also thank Dr. Daniel Wright, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, for critical review of the manuscript.
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Disease and Inter-Agency Agreement Y3-DK-3521-07 with the National Institute on Minority Health and Health Disparities, National Institutes of Health.
The online version of this article contains supplemental material.
- ATCC
- American Type Culture Collection
- MPO
- myeloperoxidase
- Nod
- nucleotide-binding oligomerization domain
- OLFM4
- olfactomedin 4
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Segal A. W. 2005. How neutrophils kill microbes. Annu. Rev. Immunol. 23: 197–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Standish A. J., Weiser J. N. 2009. Human neutrophils kill Streptococcus pneumoniae via serine proteases. J. Immunol. 183: 2602–2609 [DOI] [PubMed] [Google Scholar]
- 3.Dolenc I., Turk B., Pungercic G., Ritonja A., Turk V. 1995. Oligomeric structure and substrate induced inhibition of human cathepsin C. J. Biol. Chem. 270: 21626–21631 [DOI] [PubMed] [Google Scholar]
- 4.Pham C. T. 2006. Neutrophil serine proteases: specific regulators of inflammation. Nat. Rev. Immunol. 6: 541–550 [DOI] [PubMed] [Google Scholar]
- 5.Tomarev S. I., Nakaya N. 2009. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol. Neurobiol. 40: 122–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J., Liu W. L., Tang D. C., Chen L., Wang M., Pack S. D., Zhuang Z., Rodgers G. P. 2002. Identification and characterization of a novel member of olfactomedin-related protein family, hGC-1, expressed during myeloid lineage development. Gene 283: 83–93 [DOI] [PubMed] [Google Scholar]
- 7.Zhang X., Huang Q., Yang Z., Li Y., Li C. Y. 2004. GW112, a novel antiapoptotic protein that promotes tumor growth. Cancer Res. 64: 2474–2481 [DOI] [PubMed] [Google Scholar]
- 8.Liu W., Lee H. W., Liu Y., Wang R., Rodgers G. P. 2010. Olfactomedin 4 is a novel target gene of retinoic acids and 5-aza-2′-deoxycytidine involved in human myeloid leukemia cell growth, differentiation, and apoptosis. Blood 116: 4938–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L., Li H., Liu W., Zhu J., Zhao X., Wright E., Cao L., Ding I., Rodgers G. P. 2011. Olfactomedin 4 suppresses prostate cancer cell growth and metastasis via negative interaction with cathepsin D and SDF-1. Carcinogenesis 32: 986–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinozaki S., Nakamura T., Iimura M., Kato Y., Iizuka B., Kobayashi M., Hayashi N. 2001. Upregulation of Reg 1alpha and GW112 in the epithelium of inflamed colonic mucosa. Gut 48: 623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mannick E. E., Schurr J. R., Zapata A., Lentz J. J., Gastanaduy M., Cote R. L., Delgado A., Correa P., Correa H. 2004. Gene expression in gastric biopsies from patients infected with Helicobacter pylori. Scand. J. Gastroenterol. 39: 1192–1200 [DOI] [PubMed] [Google Scholar]
- 12.Liu W., Yan M., Liu Y., Wang R., Li C., Deng C., Singh A., Coleman W. G., Jr., Rodgers G. P. 2010. Olfactomedin 4 down-regulates innate immunity against Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA 107: 11056–11061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W., Chen L., Zhu J., Rodgers G. P. 2006. The glycoprotein hGC-1 binds to cadherin and lectins. Exp. Cell Res. 312: 1785–1797 [DOI] [PubMed] [Google Scholar]
- 14.Boxio R., Bossenmeyer-Pourié C., Steinckwich N., Dournon C., Nüsse O. 2004. Mouse bone marrow contains large numbers of functionally competent neutrophils. J. Leukoc. Biol. 75: 604–611 [DOI] [PubMed] [Google Scholar]
- 15.Lominadze G., Powell D. W., Luerman G. C., Link A. J., Ward R. A., McLeish K. R. 2005. Proteomic analysis of human neutrophil granules. Mol. Cell. Proteomics 4: 1503–1521 [DOI] [PubMed] [Google Scholar]
- 16.Liu W., Zhu J., Cao L., Rodgers G. P. 2007. Expression of hGC-1 is correlated with differentiation of gastric carcinoma. Histopathology 51: 157–165 [DOI] [PubMed] [Google Scholar]
- 17.Malaviya R., Ross E. A., MacGregor J. I., Ikeda T., Little J. R., Jakschik B. A., Abraham S. N. 1994. Mast cell phagocytosis of FimH-expressing enterobacteria. J. Immunol. 152: 1907–1914 [PubMed] [Google Scholar]
- 18.Suzuki K., Ota H., Sasagawa S., Sakatani T., Fujikura T. 1983. Assay method for myeloperoxidase in human polymorphonuclear leukocytes. Anal. Biochem. 132: 345–352 [DOI] [PubMed] [Google Scholar]
- 19.Bjerknes R., Bassøe C. F. 1984. Phagocyte C3-mediated attachment and internalization: flow cytometric studies using a fluorescence quenching technique. Blut 49: 315–323 [DOI] [PubMed] [Google Scholar]
- 20.Mallen-St Clair J., Pham C. T., Villalta S. A., Caughey G. H., Wolters P. J. 2004. Mast cell dipeptidyl peptidase I mediates survival from sepsis. J. Clin. Invest. 113: 628–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Duijn P. J., Dautzenberg M. J., Oostdijk E. A. 2011. Recent trends in antibiotic resistance in European ICUs. Curr. Opin. Crit. Care 17: 658–665 [DOI] [PubMed] [Google Scholar]
- 22.Borregaard N., Cowland J. B. 1997. Granules of the human neutrophilic polymorphonuclear leukocyte. Blood 89: 3503–3521 [PubMed] [Google Scholar]
- 23.Kjeldsen L., Johnsen A. H., Sengeløv H., Borregaard N. 1993. Isolation and primary structure of NGAL, a novel protein associated with human neutrophil gelatinase. J. Biol. Chem. 268: 10425–10432 [PubMed] [Google Scholar]
- 24.Pham C. T. 2008. Neutrophil serine proteases fine-tune the inflammatory response. Int. J. Biochem. Cell Biol. 40: 1317–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guay D., Beaulieu C., Percival M. D. 2010. Therapeutic utility and medicinal chemistry of cathepsin C inhibitors. Curr. Top. Med. Chem. 10: 708–716 [DOI] [PubMed] [Google Scholar]
- 26.Dahl S. W., Halkier T., Lauritzen C., Dolenc I., Pedersen J., Turk V., Turk B. 2001. Human recombinant pro-dipeptidyl peptidase I (cathepsin C) can be activated by cathepsins L and S but not by autocatalytic processing. Biochemistry 40: 1671–1678 [DOI] [PubMed] [Google Scholar]
- 27.Hamilton G., Colbert J. D., Schuettelkopf A. W., Watts C. 2008. Cystatin F is a cathepsin C-directed protease inhibitor regulated by proteolysis. EMBO J. 27: 499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke T. B., Davis K. M., Lysenko E. S., Zhou A. Y., Yu Y., Weiser J. N. 2010. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16: 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inomata H., Haraguchi T., Sasai Y. 2008. Robust stability of the embryonic axial pattern requires a secreted scaffold for chordin degradation. Cell 134: 854–865 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.