Abstract
Duchenne muscular dystrophy (DMD) cardiomyopathy patients currently have no therapeutic options. We evaluated catheter-based transendocardial delivery of a recombinant adeno-associated virus (rAAV) expressing a small nuclear U7 RNA (U7smOPT) complementary to specific cis-acting splicing signals. Eliminating specific exons restores the open-reading frame resulting in translation of truncated dystrophin protein. To test this approach in a clinically relevant DMD model, golden retriever muscular dystrophy (GRMD) dogs received serotype 6 rAAV-U7smOPT via the intracoronary or transendocardial route. Transendocardial injections were performed with an injection-tipped catheter and fluoroscopic guidance using X-ray fused with MRI (XFM) roadmaps. Three months after treatment, tissues were analyzed for DNA, RNA, dystrophin protein, and histology. Whereas intracoronary delivery did not result in effective transduction, transendocardial injections, XFM guidance, enabled 30±10 non-overlapping injections per animal. Vector DNA was detectable in all samples tested and ranged from <1 to >3000 vector genome copies per cell. RNA analysis, western blot analysis, and immunohistology demonstrated extensive expression of skipped RNA and dystrophin protein in the treated myocardium. Left ventricular function remained unchanged over a three-month follow-up. These results demonstrated that effective transendocardial delivery of rAAV-U7smOPT was achieved using XFM. This approach restores an open reading frame for dystrophin in affected dogs and has potential clinical utility.
Keywords: Duchenne muscular dystrophy, rAAV, exon-skipping, cardiomyopathy
INTRODUCTION
Duchenne muscular dystrophy (DMD) is both the most common and one of the most severe inherited myopathies affecting up to 1 in 3500 live male births1,2. The genetically recessive myopathy results from mutations in the dystrophin gene, cytogenetically located on the X-chromosome at p21.13. Dystrophin is the largest known vertebrate gene in which an estimated one-third of DMD-causing mutations arise spontaneously. Mutations in the dystrophin gene, mostly due to out-of-frame deletions, lead to loss of functional dystrophin, which is a critical myocyte structural protein 4.
DMD has extensive systemic manifestations and substantial morbidity5. Cardiomyopathy develops in nearly all patients by adulthood, at which point it confers a grave prognosis with a 5-year survival of 50%6,7. Medical therapy may delay the progression of the cardiomyopathy but has not been shown to affect mortality8,9.
One novel therapeutic approach aims to convert the out-of-frame mutation responsible for the disease into an in-frame mutation, thereby restoring the open reading frame and translation of a truncated, and presumably, quasi-functional protein. Binding anantisense oligonucleotide, to specific cis-acting splicing signals in the primary transcript target sequence, interferes with recognition of that splicing substrate resulting in deletion, or skipping, of one or more exons. These specific deletions, by design, restore the translational reading frame. The anti-sense sequence can be delivered by a recombinant adeno-associated virus (rAAV) that contains an expression cassette producing a small nuclear (sn) U7 RNA modified with a short anti-sense sequence complementary to the cis-acting splicing signals of the targeted exon(s) (U7smOPT). This approach was proven effective in tissue culture10, and in dystrophin null (mdx strain) mice11,12. Translating this proof of concept to the clinic requires successful demonstration of this approach in a clinically relevant, large animal DMD model. The golden retriever muscular dystrophy dog is the best-established large animal DMD model13–15. The large mass of the GRMD dogs poses significant challenge in terms of viral quantities and delivery approaches needed in order to achieve effective gene therapy.
We have developed a large-scale rAAV production process yielding a vector with dual canine U7 smOPT exon skipping expression cassettes (Figure 1)16, and assessed the efficacy of various delivery methods into the myocardium of healthy and sick animals.
Figure 1. Diagram of the rAAVU7 smOPT E6E8 vector genome.
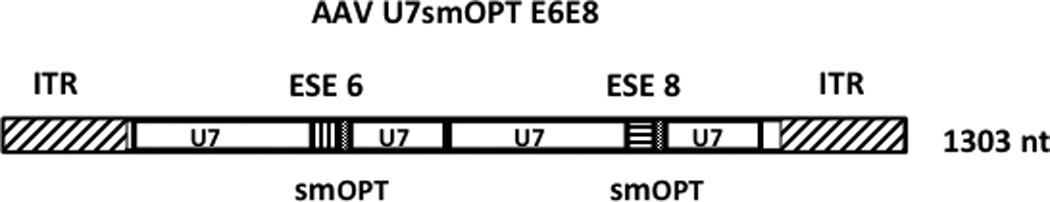
The 1303 nt single-stranded rAAVU7smOPT E6E8 vector genome is represented schematically. The AAV type 2-derived ITRs are represented by rectangles with diagonal slashes. The two U7smOPT expression cassettes are identical except for the short anti-sense sequences complementary to the exon 6 and exon 8 exon splice enhancers (ESE) of the canine dystrophin gene (vertical and horizontal slashes, respectively). Sequences homologous to the non-coding murine U7 snRNA are represented by rectangles labeled “U7” and the positions of the smOPT modification are indicated.
RESULTS
The rAAV6 was delivered to the left ventricular myocardium of the treated dogs either by intracoronary infusion (n=3) or by transendocardial injections (n=5). One homozygous female GRMD dog served as negative control and received sham injections. Both delivery methods were well tolerated without any hemodynamic changes or significant arrhythmias. Initial experience with intracoronary delivery of rAAV6 of either GFP (n=2) or U7 snRNA (n=1) showed minimal transduction efficiency as evidenced by low rAAV DNA in the myocardium. Therefore, subsequent experiments were performed using transendocardial injections.
X-ray fused with MRI (magnetic resonance imaging) (XFM) successfully guided the transendocardial injections by overlaying the myocardial contours and structures over the fluoroscopy images viewed by the operator (Figure 2) thus enabling 30±10 target injections per animal. By updating injection locations in real-time, XFM provided a 3D map for distributing the injections as uniformly as possible. Previously injected areas were avoided and untreated regions were identified and targeted enabling additional transendocardial injections at those locations (Figure 2). The extensive distribution of injections was demonstrated also by T2*-weighted MRI shortly after the injections based on the presence of the SPIO particles in the injected myocardium (Figure 3). Healthy animals (n=2) were survived for 31±3 days, and GRMD animals (n=3) were survived for 84±24 days after injections. The average number of injections to the basal areas was lower (1.2±1.1) than the mid and apical areas (2.1±1) due to technical difficulty in reaching the basal ventricle of small animals using an injection catheter with curves designed for larger human ventricles (Figure 4).
Figure 2. Transendocardial injections locations on X-ray fused with MRI (XFM) images.
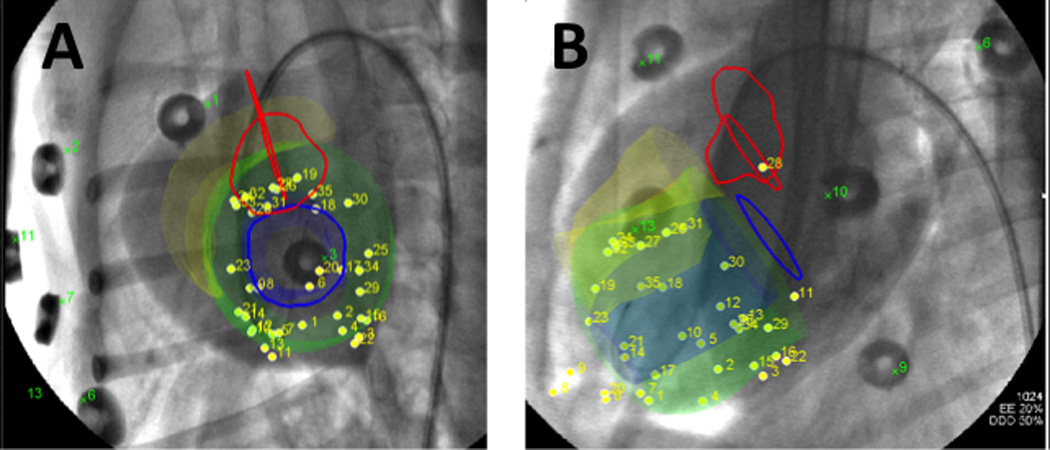
X-ray fused with MRI (XFM) overlays heart contours and structures onto X-ray fluoroscopy. In multiple projections this provides 3-dimensional orientation and guidance for transendocardial injections. In (A) a short axis and long axis (B) views of the heart are demonstrated with the XFM contours overlaid on the fluoroscopy. The green contour represents the left ventricular endocardium, orange represents the right ventricular endocardium, blue lines indicate the mitral valve, and red lines indicate the location of the aortic valve.
Figure 3. Locations of the rAAV-U7smOPT and the SPIO particles in the left ventricular myocardium.
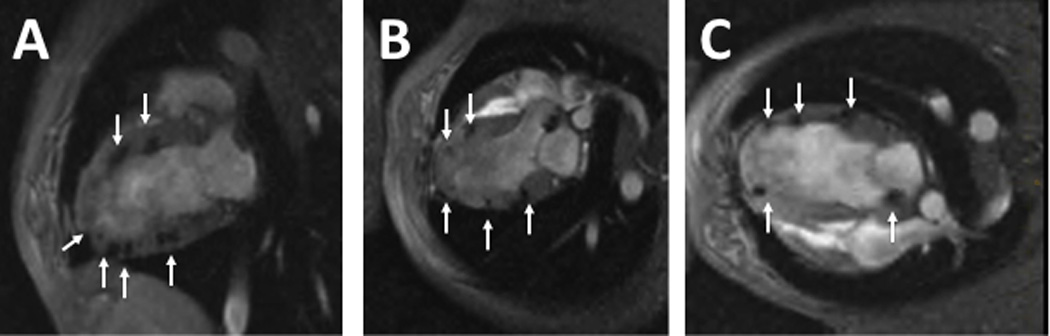
Representative images of 2-chamber (A), 3-chamber (B) and 4-chamber (C) T2*-weighted MRI immediately following the injection demonstrating wide distribution of the injections throughout the left ventricular wall. Arrows point to MRI signal voids, which reflect SPIO particles included in the virus injection mixture.
Figure 4.
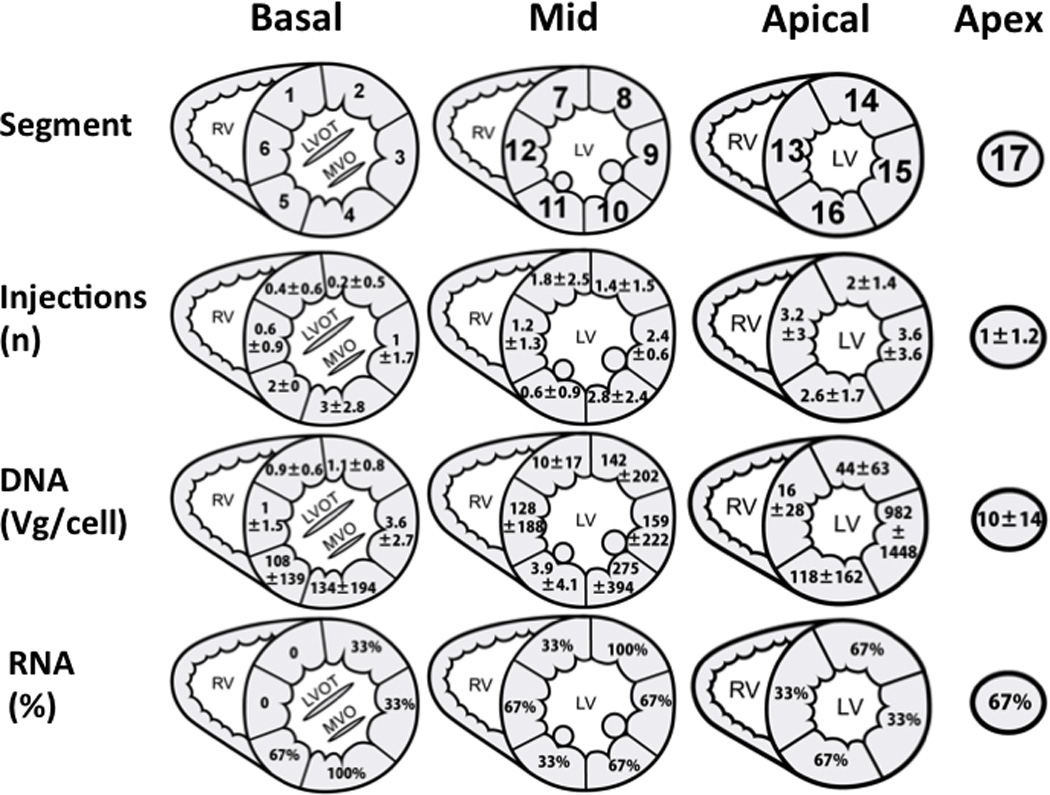
Distribution of injections, viral DNA and exon-skipped RNA following transendocardial delivery of rAAV6-U7smOPT. Panel A maps the myocardial segments numbering; The basal slice is represented by segments 1–6, the mid-ventricular slice is represented by segments 7–12, the apical slice is represented by segments 13–16 and the apical cap is represented by segment number 17. Panel B: Average ± standard deviation of injections per segment. Panel C: Amounts of viral DNA (Vg) per cell in myocardial segments 1–17. Panel D: Samples from all segments were analyzed for presence of alternative splicing. In this panel, the percentage of positive samples for alternative splicing is reported for each segment.
Vector DNA detection
In order to assess the efficacy of rAAV delivery we analyzed tissue samples for viral DNA. Vector DNA was detectable in all samples tested using nested PCR and averaged 126±346 vg copies per cell (Figure 5). Similar to the injection distribution which was limited in the basal areas, larger amounts of viral DNA were found in the mid or apical segments (172±282 vg/cell) compared to the basal segments (41±62) (Figure 4). There was correlation between the number of injections in a specific segment and the numbers of vector DNA identified (Figure 6, r=0.789).
Figure 5. Presence of viral DNA in the various myocardial segments.
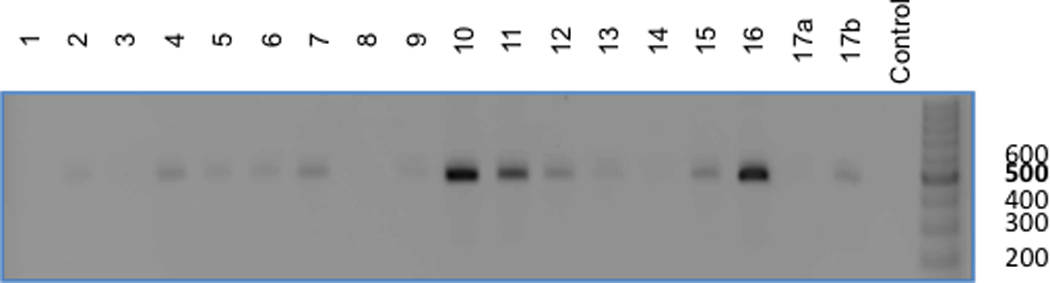
Demonstrated is a representative agarose gel analysis of a sample from each of the 17 myocardial segments (including 2 samples from segment 17, a+b) and a negative control from an untreated GRMD dog. This panel demonstrates a 500 base-pair PCR product on an ethidium bromide gel in the majority (13/17) of the sampled myocardial segments.
Figure 6. Correlation between the numbers of injections per myocardial segment and the amount of viral DNA.
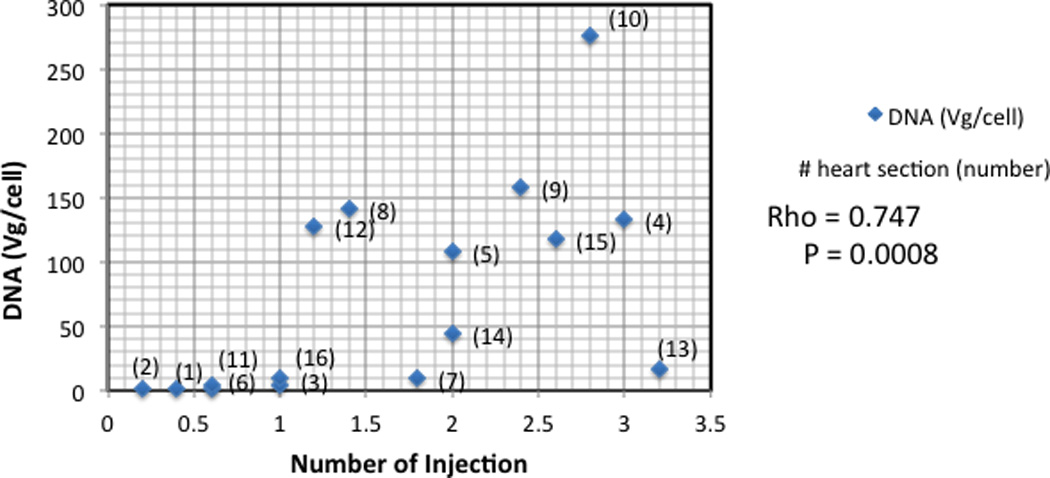
As demonstrated, a higher number of injections per myocardial segment correlated with larger amounts of viral DNA (vg/cell) (r=0.789).
Dystrophin RNA detection
The RT-PCR amplified region of the native RNA transcript of dystrophin in healthy canine has a predicted size of 911 bp whereas in an untreated GRMD dog, the same rtPCR amplified region generates a product of 792 bp due to exon 7 deletion (Figure 7). Induced exon skipping resulting from effective rAAV transduction of GMRD created an alternative splicing pattern with two unique RT-PCR products appearing as 478 bp and 305 bp bands. In samples with effective exon-skipping, the alternative splicing resulted also in diminished band intensity at the 792 nt un-skipped transcript location (Figure 7). The prevalence of exon skipping was associated with higher number of injections and rAAV DNA (Figure 4).
Figure 7. Sizes of dystrophin gene RNA transcript in the various myocardial segments.
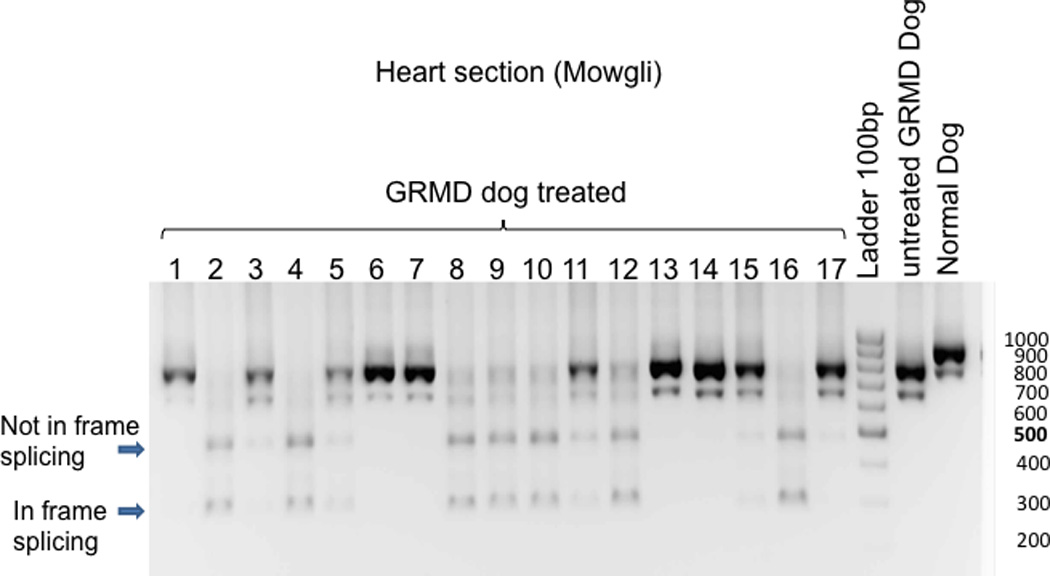
Sample from “Normal dog” demonstrate the predicted RNA transcript size band at 911 base-pair (bp) whereas an “Untreated GRMD dog” (natural occurrence of exon 7 skipping) produce a typical band at 792bp. The predicted size of bands for the in-frame skipping of treated dog with U7-ESE6/8 construct is 437bp for skipping of exons 6 to 8 and 305bp for skipping of exons 6 to 9. In the presented representative gel, two dominant new bands appear in samples 2, 4, 5, 8–12 and 16 of the treated dog. The lower band (300bp) corresponds to an in-frame skipping of exons 6 to 9, whereas the upper band (500pb) represents a non in-frame skipping.
The identities of the two prominent new RT-PCR products were determined by DNA sequencing (Figure 8A and B). Based on the sequence analysis, the 478 bp product resulted from dystrophin transcripts with deletions of exons 7, 8, and 9, whereas the 305 bp product resulted from dystrophin transcripts that have deletions of exons 6 – 9. Although the additional deletion of exon 9 does not alter the dystrophin open reading frame, transcripts retaining exon 6, i.e. transcripts with deletions of exons 7, 8, 9, results in a reading frame-shift that terminates translation at a nonsense codon in exon 10. However, in each sample in which the Δ7, 8, 9 RT-PCR product was produced the Δ6, 7, 8, 9 (reading-frame restored) product was also detected. Whereas, no RT-PCR products were generated corresponding to transcripts with Δ6, 7, 9 suggesting that the sequential order of splicing remains intact.
Figure 8. Purification and sequencing of nested RT PCR product.
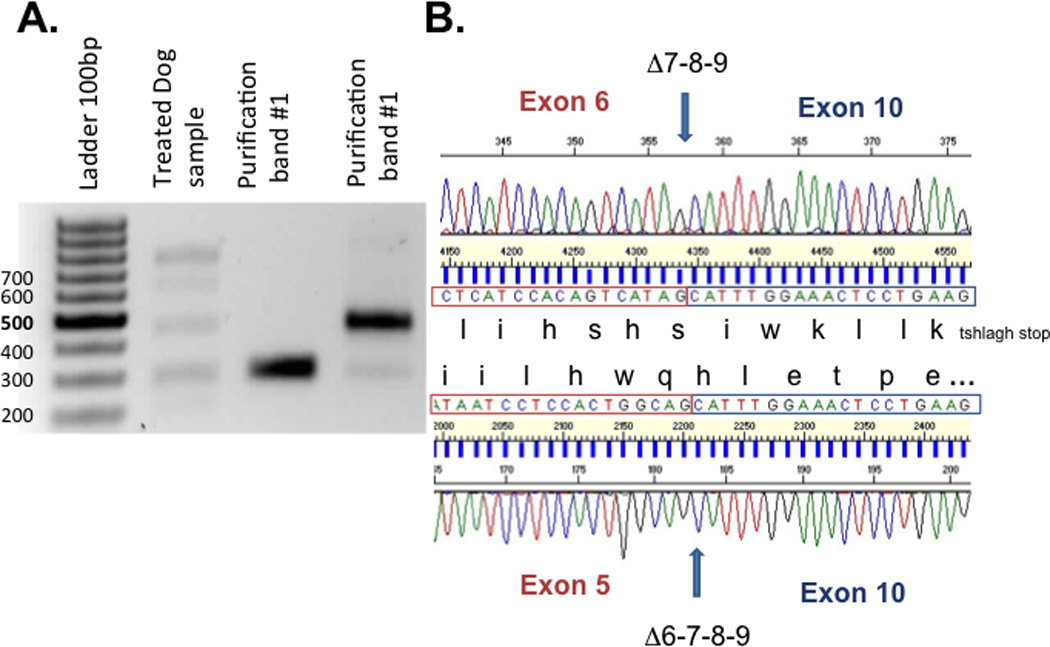
(A) The in-frame and the non in-frame skipping band have been extracted from the gel and were sequenced using the inner primer exon 3 and the inner primer exon 10 (purification band #1). (B) Sequences analysis demonstrate that the lower band (305bp) corresponds to skipping of exons 6 to 9 with restoration of open reading frame whereas the upper band (478pb) corresponds to skipping of exons 7 to 9 which did alter the open reading frame.
Dystrophin Western Blot Detection
Dystrophin protein was detected in protein extracted from treated heart samples by western blot analysis (Figure 9). The amount of dystrophin expressed in the treated heart samples corresponds to a relatively low level compared to normal heart samples. Every normal cardiomyocyte expresses dystrophin, however, only transduced myocytes are capable of dystrophin expression. The amount of dystrophin protein represents an average value of both transduced and untransduced cells that were present in each randomly selected sample.
Figure 9. Western blot analysis of heart protein.
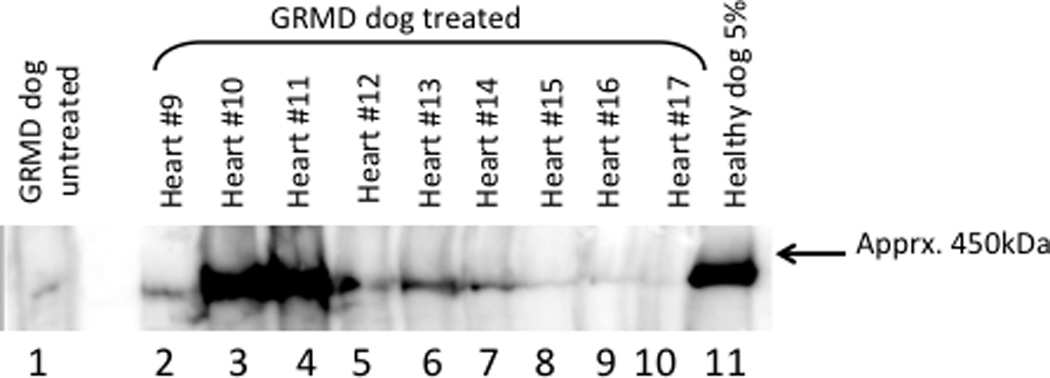
Soluble protein extracts prepared from randomly selected 20µm thick frozen sections were fractionated by SDS-polyacrylamide gel electrophoresis and transferred to PVDF membrane. The heart segments are indicated and the positive control consists of 5% normal dog heart protein extract in untreated GRMD heart protein extract. The negative control (lane 1) is untreated GRMD heart protein. The low level dystrophin protein signal was observed reproducibly and is likely the result of the so-called “revertant” myocytes in the GRMD dogs. Presumably, aberrant splicing restores the dystrophin open reading frame in a relatively small percentage of cells resulting in a natural form of exon skipping.
Histology and Immunohistochemistry
Immunohistochemistry (IHC) using anti-dystrophin immune serum demonstrated extensive expression of dystrophin protein throughout the treated heart of GRMD dogs (Figure 10). Control GRMD dogs had no detectable dystrophin staining in myocardial samples. The highest levels of IHC fluorescence corresponded to the myocardial segments that had the highest amounts of vector DNA and were also strongly positive for exon-skipped dystrophin rtPCR products. T-cell infiltration into the heart parenchyma is characterized using hemotoxylin and eosin (H&E) staining. H&E stained sections from normal heart, untreated GRMD heart, and treated GRMD heart, were examined by light microscopy. Treated GRMD heart sections tended to have more hematoxylin stained nuclei than untreated GRMD sections (Supplemental Figure 1). Because the GRMD pathology involves inflammation and fibrosis, and due to the variability between individual animal pathologies, it is difficult to conclude that the vector per se caused or worsened the inflammation. However, we have designed future study with an immunosuppressive regimen to simplify the data analysis.
Figure 10. Immunohistochemical staining to dystrophin proteins in GRMD canines treated with rAAV-U7smOPT.
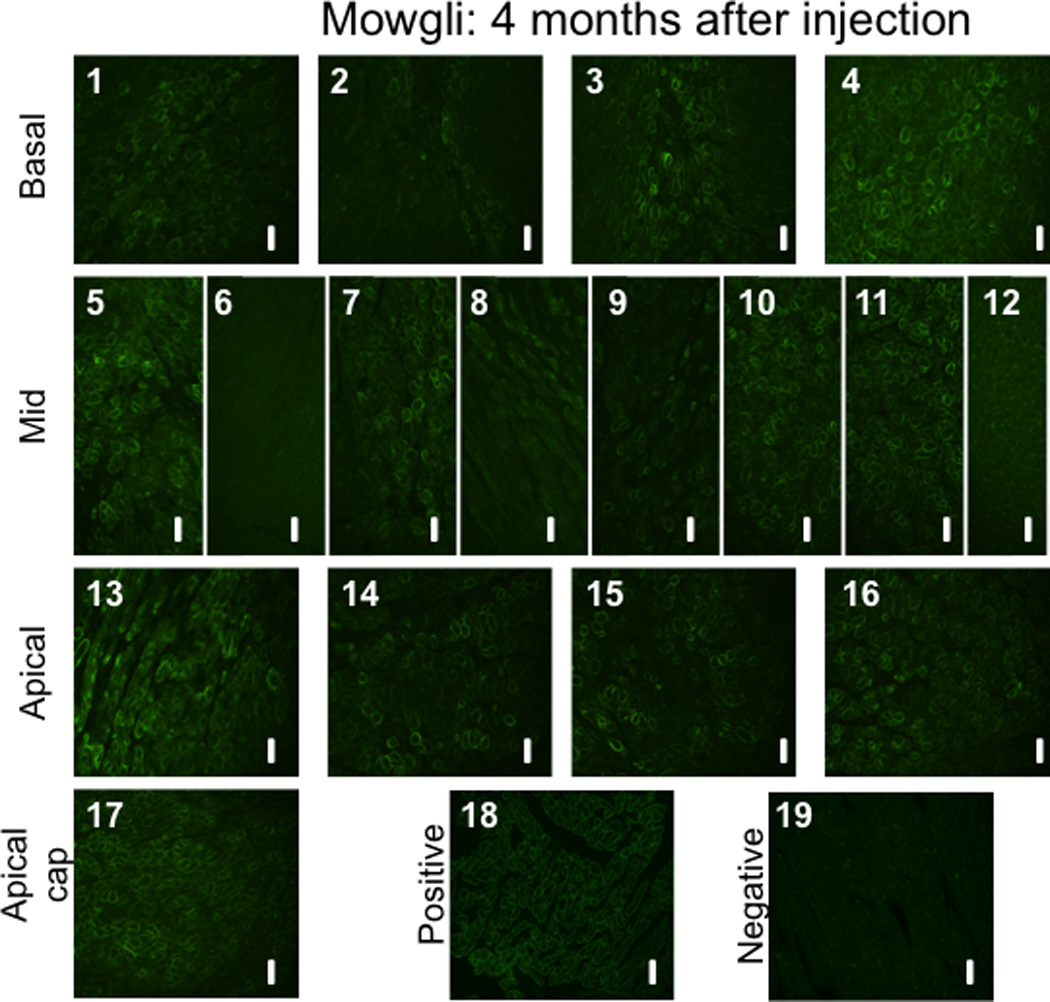
(1–17) Slides 1 to 17 (excluding slide 6) demonstrate various levels of positive staining for dystrophin in samples collected randomly from each of the 17 pre-defined left ventricular myocardial segments. The positive staining is typically located at the cell membrane, corresponding to the location of the functional dystrophin protein. As controls, tissues from (18) normal dog (positive control) and (19) untreated GRMD dog (negative control) were used. All images were acquired at a 20X magnification. The scale bars represents 50 µm.
Left ventricular function
GRMD animals used in the present study had advanced disease with clinical cardiomyopathy at the time of enrollment into the study, as indicated by the impaired LV function at baseline (LVEF 30.4±3.6) (Figure 11). However, throughout the follow up period, there was no additional deterioration of the LV function (−0.4±0.8) (Figure 11). In accordance to the weight gain of the animals throughout the follow up period, the LVEDV, LVESV and SV increased (Figure 11).
Figure 11. Left ventricular size and function at baseline and 3 months follow-up.
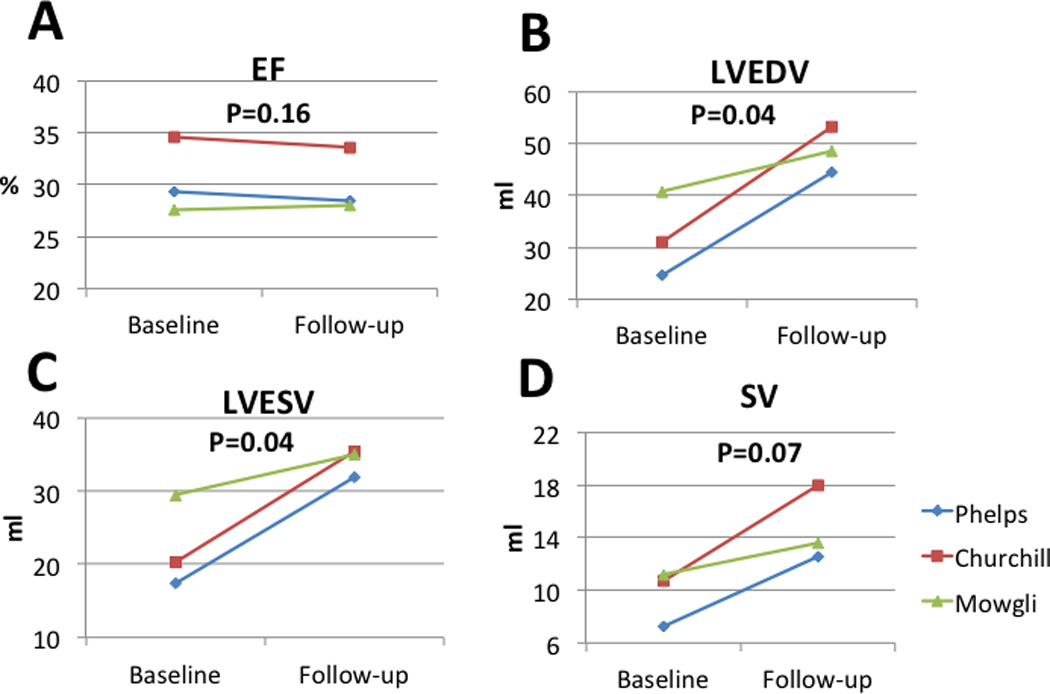
Left ventricular size and function were evaluated by cardiac MRI at baseline and at 3 months follow-up. As demonstrated, left ventricular ejection fraction remained unchanged during the study period. During the study period, left ventricular volumes and stroke volumes increased as the juvenile animals grew in size.
EF – Ejection fraction, LVEDV – Left ventricular end diastolic volume, LVESV – Left ventricular end systolic volume, SV – Stroke volume
DISCUSSION
Patients who develop Duchenne cardiomyopathy currently have a grave prognosis with no therapeutic options. We therefore evaluated a novel exon-skipping gene therapy approach for the treatment of Duchenne cardiomyopathy. This pre-clinical study was performed in GRMD canines which are the closest animal model prior to translation into clinical research. An injection catheter was introduced into the left ventricular cavity by means of percutaneous arterial access. This catheter was used retrograde across the aortic valve to deliver rAAV6-U7smOPT into the myocardium using MRI roadmaps co-registered with live X-ray fluoroscopy. This approach allowed wide transendocardial distribution of the injections, induced extensive exon skipping and rescued dystrophin expression in the diseased cardiomyocytes.
As a monogenic recessive disease, DMD is an attractive target for gene-based therapy. However, several aspects of DMD in particular, create challenges in developing effective gene therapy approaches. The large size of the dystrophin gene (approximately 2.5 Mbp) and the resulting mRNA of the full-length protein (approximately 12 knt) 4 preclude standard “gene” replacement approaches. An AAV-based approach to deliver mini- or micro- dystrophin cDNAs which can produce a truncated, but functional dystrphin protein, face difficulties because the size of the cDNAs exceed the length of wild-type AAV genomes resulting in less efficient vector genome encapsidation and poor vector yields. Exon-skipping provides an alternative approach for restoring the dystrophin open reading frame. This process blocks transcript and exon specific cis-acting recognition signals consisting of intronic elements (pyrimidine tract, branch point, etc.), or exonic splice enhancer element, and canonical splice donors and acceptors 17–21. Thus, by targeting a specific splicing signal with the complementary element, one or more exons may be deleted, or skipped, thereby restoring the translational reading frame. Initially developed using synthetic antisense oligonucleotides (AON), exon skipping has been adapted as a component of small nuclear RNAs (snRNA), either the spliceosomal U1 snRNA or a modified U7 snRNA that interacts with the spliceosome, U7 smOPT22. By introducing a short, complementary sequence into the 3’-end of the snDNA, the transcript can target a specific splicing signal in the dystrophin pre-mRNA. This approach is currently undergoing clinical evaluation 23–25. Although the cellular pathways for salvaging or degrading polynucleotides do not metabolize synthetic oligonucleotides, these oligonucleotides do not persist indefinitely either intracellularly or systemically. Thus, this approach requires repeated, systemic administrations. To overcome this limitation an alternative approach has been developed in which the snRNA expression cassette has been engineered into recombinant adeno-associated virus vector genome (rAAV). This approach was proven effective in skipping a single exon and restoring dystrophin expression in the mdx mouse, which due to a transition in exon 23, a stop codon prematurely terminates translation11,12.
While myocardial gene therapy demonstrated promising results in small animal models 26, translating these methods into large animal models and humans has been non-trivial. Several approaches to achieve effective transduction of the myocardium have been tested and reported with varying degrees of success in terms of histologic or phenotypic effects. Intracoronary infusion of the desired vector seems to be the most promising approach but has yielded varying results 27–29. To improve efficiency rates, several improvements have been proposed including retroinfusion of the vector through the coronary veins 27 or cardiac recirculation of the delivered vector 30. In the present study we failed to achieve detectable cardiomyocyte transduction following intracoronary infusion of the vector with proximal flow occlusion. Based on initial experience of transendocardial delivery of viral vectors 31–33 we have pursued a related approach. However, as the left ventricular cavity is a complex three dimensional structure an appropriate approach to efficiently target all myocardial locations would require guiding by an imaging modality that would be able to depict three-dimensional structures. Bish et al. 31,34 performed rAAV transendocardial injections with fluoroscopic guidance, this allowed 2-dimenstional guidance without the ability to identify any ventricular structures such as the papillary muscles or valves. While demonstrating efficient gene transfer in two areas (septum and left ventricular free wall), the robustness of the distribution with such approach is unknown. To address this limitation we used an advanced multimodality imaging methodology that combines MRI-derived contours of region of interest and overlaid these contours on top of the standard fluoroscopic views, these merged images accurately represented the relevant cardiac structures. This approach allowed accurate guidance and real-time assessment of injection locations. Indeed, in the present study, basal areas were less effectively targeted; however this limitation is attributed to sub-optimal catheter shapes which are intended for use in humans.
In the present study, three months post-treatment, the effects of vector-mediated exon-skipping were evident. The concentration of vector DNA in randomly selected samples ranged from 981 to less than 1 vg/cell and dystrophin expression was restored efficiently. As anticipated, there was good correlation between the amount of injections and the viral DNA in the tissue, however, because the DNA processed for PCR and RNA processed for rtPCR were extracted from different specimens, the exon-skipped RNA and vector genome copy numbers results did not correlate.
Using large-scale rAAV production, we were able to evaluate two routes of administration for treating dystrophic dogs. This capability is essential for translating treatments from small animal proof-of-concept studies to demonstrating feasibility in a clinically relevant, large animal model of DMD.
The phenotypic effect of this exon skipping gene therapy was modest, demonstrating stable left ventricular function without any significant improvement in three months follow up. The treated GRMDs were relatively old (7–11 months old), had advanced systemic disease, demonstrating dramatic muscle wasting, weakness and some required percutaneous gastrostomy feeding due to impaired swallowing mechanism and recurrent pulmonary aspiration of gastric contents. Accordingly, all had advanced cardiomyopathy with impaired LV function at the time of treatment. It is conceivable that even the most effective treatment would not be able to reverse LV dysfunction in this set of animals. Thus, the present results which demonstrate prevention of further deterioration in LV function might be viewed as encouraging results.
The present study has several limitations. Our attempts to deliver the rAAV via the coronary approach failed. It is possible that supplementing our techniques with the previously reported methods 27,30 might have increased the efficiency of gene transfer via the intracoronary approach. The rAAV6-U7smOPT vector restored dystrophin expression in most samples tested. Distributing the injections uniformly throughout the LV myocardium and prolonging the dwell time for each injection is difficult without inducing cardioplegia. Thus, it is conceivable that subsequent to withdrawing the needle from the injection site, the vector refluxes along the needle track and does not diffuse interstitially35. Despite the high doses of concentrated rAAV6-U7smOPT, it is likely that the heart contractions caused leakage of the vector back into the ventricular cavity, thereby limiting the amount of vector remaining in situ. Finally, due to the difficulty in breeding and availability of GRMD canines, the number of GRMD canines that were used in this study was relatively small. It is possible that lager number of animals and longer follow-up would have yielded more meaningful phenotypic data. No control dogs were used for comparison in the functional studies.
METHODS
Animals and study design
Animal procedures were approved by the institutional Animal Care and Use Committee and performed according to contemporary National Institutes of Health guidelines. A total of nine dogs were used, including four normal male beagles (11.9±1.1 kg) and five affected GRMD dogs (four male and one homozygous female) (12.1±3.1 kg). Of these, three dogs (two normal dogs and one GRMD dog) received intracoronary rAAV infusion, five (two normal dogs (beagles) and three GRMD dogs, same sire different dames) received transendocardial rAAV delivery and one homozygous female GRMD dog served as control (alittermate an experimental GRMD affected male dog). Animals were pretreated by oral amiodarone, atenolol and prophylactic cefazolin. Anesthesia was induced by midazolam, propofol and butorphanol and was maintained by sevoflurane inhalation and fentanyl infusion.
Vascular femoral access was achieved with a 4-Fr micropuncture kit followed by insertion of 6- or 8-Fr and 5-Fr sheaths in the right femoral artery and vein respectively. Vascular hemostasis was achieved by manual pressure and application of D-stat Dry (Vascular Solutions, Minneapolis, MN).
Bivalirudin (Angiomax, The Medicines Co., Parsippany, NJ) was used as a procedural anticoagulant due to the affinity of AAV6 capsids with heparin that effectively neutralizes the biological activity of rAAV vectors36,37. An initial bolus of 0.75 mg/kg was followed by continuous infusion of 1.75 mg/kg/hr adjusted to maintain an activated clotting time (Hemotec) > 250 seconds.
Treated animals were survived up to three months after the procedure. Immediately following euthanasia, hearts were explanted and were sliced systematically in a predefined mode through the short axis of the left ventricle into 17 segments at the basal (Segments 1–6), mid-cavitary (Segments 7–12), apical (Segments 13–16) and apical cap (Segment 17), according to standardized segmentation of the left ventricle 38. Random samples were taken from each segment and fresh frozen (−80°C) in cryopreservation medium (Tissue-Tek O.C.T. compound, Sakura, The Netherlands).
Adeno-associated virus vector
Recombinant adeno-associated virus serotype 6 (rAAV6) was produced in Spodoptera frugiperda (Sf9) insect cells using recombinant baculovirus expression vectors (BEV) as previously described 39. Briefly, cells were cultivated in a 250 liter single-use bioreactor (200 liter working volume) in serum-free insect cell medium (SFX Insect Cell Medium, HyClone, Logan, UT) at 28° C with 30% dissolved O2 (dO) (Integrity PadReactor, ATMI Lifesciences, Bloomington, MN). Temperature and dO set points were maintained during cell growth and vector production with control loops responding to outputs from the appropriate probes (TruViu and TruLogic Control System, Finesse Solutions, San Jose, CA). Late log-stage Sf9 cells from shake-flasks were diluted into the bioreactor’s minimum working volume (40 liters) and expanded to the target volume by serial dilution to the final working volume (200 liters). Two BEVS were required for rAAV production: one BEV for AAV-2 rep proteins and AAV6 cap proteins and the second BEV provided the ITR-bearing vector genome. Rather than using free, extracellular BEVS in solution for rAAV production, a pre-determined amount of concentrated, cryopreserved baculovirus infected insect cells (BIIC) were added to the 200 liter culture (BIIC : producer cell = 1:1000 ). The producer cells continued to divide until baculovirus cell cycle arrest occurred at the target cell density of ≈5×106 cells/ml. The downstream process for rAAV purification involved the following series of steps as previously described 39; Homogenization, nuclease digestion, clarification and microfiltration, immunoaffinity column chromatography, tangential flow filtration for concentration and diafiltration and final filtration and sterile dispensing into single-use vials. Depending on the homogeneity of the vector, gel exclusion chromatography step was performed (Superdex 200, GE LifeSciences).
The vector particles, consisting of AAV6 capsid proteins and a linear single-stranded DNA genome were characterized for composition and quantity. Following immunoaffinity chromatography, the vector genome concentration was determined by two methods: Polymerase chain reaction (PCR) using vector specific oligonucleotide primers, and by SYBR gold fluorescent dye binding to the vector-extracted DNA. Tangential flow filtration was used to achieve the final vector concentration and buffer exchange into PBS (Pellicon 2, Millipore Corp., Billerica, MA). The protein composition of the AAV6 capsid: VP1, VP2, and VP3, was analyzed by SDS - polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining (Pierce Thermo Fisher Scientific, Rockford, IL).
The vector genome has two cassettes, each consisting of a U7snRNA promoter, a modified U7snRNA (U7smOPT), and short sequence complementary to the exonic enhancement element (ESE) of exons 6 and 8 10. The terminal palindromes were derived from AAV2. (Supplemental Table 1, Figure 1).
Intra-coronary delivery
Coronary injections were performed in anticoagulant-treated animals during balloon occlusion to delay washout. Engaging coronary arteries in these small animals required 5Fr short pediatric transfemoral diagnostic catheters (JL1.5-2.0, Merrit Medical, South Jordan, UT). The left anterior descending and circumflex coronary arteries were injected separately using over-the-wire coronary angioplasty balloons (2.0–3.0mm diameter × 8.0–12.0mm as needed) and 0.014” coronary guidewires positioned in the mid left anterior descending or proximal circumflex segments. Preconditioning of the left ventricular myocardium (to help the animal tolerate prolonged ischemia) was accomplished by two consecutive cycles of two minute ischemia by coronary balloon inflation followed by five minute reperfusion (balloon deflation). Next, the solution containing the AAV vector (2ml × 3.5 × 1013 copies / ml) was delivered through the coronary balloon catheter lumen while inflating the balloon to occlude distal coronary artery flow (5 minutes). The same protocol was followed for the second vessel.
Transendocardial delivery
Transendocardial injections were performed under fluoroscopic guidance using X-ray fused with MRI (XFM). Injections were performed using a 7-Fr endomyocardial 27G injection needle catheter (Stiletto, Boston Scientific, Natik MA) 40. The Stiletto catheter was steered retrograde across the aortic valve inside the left ventricular cavity by means of two coaxial guiding catheters (8-Fr and/or 6-Fr IMA and JR guiding catheters) (Vista Brite-Tip, Cordis Corp., Bridgewater, NJ). The endocardium was injected repeatedly (0.2 ml per site × 20–40 sites for each procedure) with the intention to distribute the vector homogenously throughout the left ventricular myocardial mass. No attempts were made to inject into the right ventricle free wall.
The injection cocktail included virus and both MRI and X-ray contrast agents. Each 200µl injection was comprised of ≈1.5 ×1013 vg rAAV stock (1014 vg/ml), 50 µg of supraparamagnetic iron oxide particles (SPIO) (50mg/ml stock, BioMag 547, Bangs Laboratories Inc., Fishers IN), and 50µl iopamidol, a fluoroscopy contrast agent (Isovue 300, Bracco Therapeutics, Princeton, NJ). The control dog received identical solution; however the viral volume was replaced by saline. The delivery of this solution enabled correlation between real-time localization of the injection sites under XFM (iopamidol) with 3-dimensional, anatomic, early post-procedural analysis of the injections according to the SPIO particles void signal in T2* weighted MR images.
We were careful to identify, record and verify each injection site in order to distribute the virus widely. Because the injectate contained iopamidol, the 3D position of each injection was recorded under XFM to avoid overlap. The injectate also included SPIO MRI contrast, a MRI scan immediately after injections showed a complementary image of injections throughout the heart. Finally, these images were compared with histopathology.
X-ray fused with magnetic resonance imaging
XFM combines pre-acquired 3-dimensional MR image roadmaps of anatomic structures (the left ventricular myocardium in this experiment) and standard X-ray fluoroscopy images that provide high resolution images of the injection catheter and radiocontrast. This co-registration, or fusion enables 3-dimensional targeting of injections into soft tissue that is otherwise not visible using X-ray alone.
For XFM we combined MRI volumes of the heart and X-ray fluoroscopy as previously described 41. Briefly, a set of external fiducial multimodality markers (Beekley, Bristol, CT) were placed on the animal’s chest and back. T1-weighted gradient echo MR images and a set of X-ray fluoroscopy projections were used to register the two modalities. Relevant cardiac structures (e.g. left and right ventricles, aorta) were segmented from MR images and projected onto live X-ray fluoroscopy for real-time XFM guidance. Fusion images were displayed alongside live X-ray images on an external monitor. During the procedure injection sites were marked on two different X-ray projections and displayed on the fusion images as well as a 3D model of the heart.
Magnetic resonance imaging
Scans were performed in a 1.5T MRI scanner (Espree, Siemens Medical Solutions, Erlangen, Germany) using surface phased array receiver coils. Geometric and functional measurements of the LV used ECG-gated, segmented, breath-held, balanced steady state free precession (bSSFP) sequences incorporating parallel imaging with an acceleration factor of two, repetition time (TR)/echo time (TE) 3.2/1.4 ms; flip angle 80°; field of view (FOV) 300 × 300 mm; matrix 192 × 192 pixels; slice thickness 6 mm; bandwidth 930 Hz/pixel. Argus Function software (Siemens Medical Solutions) was used to calculate LV ejection fraction (I.M.B.). T2*-weighted, FLASH sequence with TR/TE 6.6/3.5 ms; flip angle 15°; FOV 250 × 160 mm; matrix 192×125 pixels; slice thickness 6mm; bandwidth 260 Hz/pixel was used to identify injection sites by means of the SPIO iron-induced signal voids. Images were analyzed using a commercial cardiac MRI workstation (Argus, Siemens).
DNA analysis
Total DNA from a random sample from each of the 17 pre-defined myocardium segments was isolated and purified using Gentra Puregene kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. PCR reactions were performed as described previously 42. Products were analyzed on ethidium bromide stained 2% agarose gels and sequenced. Six pg per diploid genome was used to calculate the vector copy number per cell using 2.5 × 109 bp × 2 as the approximate size of the canine diploid genome.
RNA analysis
Total RNA from each of the 17 pre-defined myocardium segments was extracted from randomly sampled specimens using a commercially available kit (RNeasy, Qiagen, Valencia, CA). The RNA was reverse transcribed to cDNA using Superscript I or II (RT) system (Invitrogen, Carlsbad, CA). Canine dystrophin specific exon 3 and exon 10 primers were for the reaction at 55°C for 30 minutes. The RT reaction was used for nested PCR reactions: First using a pair of external primers, and subsequently, with a pair of internal primers. The forward primers (F) for both external and internal primers were in exon 3 and the reverse primers (R) were in exon 10. External primers 42: F 5’GGAAGCAGCACATAGAGA; R 5’ TCACTTCTTCGACATCATTAG. Internal primers: F 5’ GAGACGCCTCCTAGACCT; R 5’ TCACTTCTTCGACATCATTAG. The PCR reactions were analyzed using ethidium bromide stained agarose gel electrophoresis. The open reading frames of the the isolated PCR products were determined by DNA sequence analysis
To increase the sensitivity of detecting the dystrophin mRNA, nested PCR using “outer” and “inner” set of primer pairs were used following reverse transcription of extracted cardiomyocyte RNA. Both sets of primers, specific to dystrophin exons 3 and 10, amplified the region targeted by the exon-skipping vector. The predicted sizes of the wild-type and GRMD (Δ7) rtPCR products are 911 bp and 792 bp, respectively (Figure 3, “Normal Dog” and “GRMD dog untreated” lanes). The rAAV-U7smOPT vector was designed to induce skipping of exons 6 and 8 generating a Δ6, 7, 8 transcript with a predicted size of 437 bp. However, exon 9 deletion occurs cryptically producing a Δ6, 7, 8, 9 dystrophin rtPCR product that is 305 nt.
Dystrophin western blot analysis
Western blot analyses were performed on total protein extracted from previously frozen O.C.T. embedded heart segment sections (10 µm thickness) mounted on uncoated glass slides. Protein was extracted using a modified SDS-protein gel loading buffer as previously described consisting of 4% SDS, 125 mM Tris-HCl (pH 8.8), 40% glycerol, phenylmethylsulfonyl fluoride (0.5mM), and 100 mM dithiothreitol 43. Equivalent volumes of the protein extracts were fractionated using pre-cast polyacrylamide gradient gel electrophoresis (NuPAGE Tris-Acetate SDS polyacrylamide gels, Invitrogen, Inc.) and electrophoretically transferred to polyvinylidene fluoride (PVDF) membranes in transfer buffer (NuPAGE Transfer Buffer) containing 20% methanol (constant 30V, overnight at 4°C). Protein loads were adjusted by digital analysis of common protein bands in Coomassie blue stained gels. Standard western blot conditions were used for membrane blocking, washing, staining, and chemiluminescence. Blocking buffers (BB): PBST - phosphate buffered saline (PBS = 50 mM Tris-HCl (pH 7.6) and 150 mM NaCl), 0.1% Tween-20, and 5% non-fat dry milk (w:v) (BB1) and BB2, PBST with 5% bovine serum albumin (BSA)(w:v). Washing buffer: phosphate buffered saline and 0.1% Tween-20. Chemiluminesence reagent: SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). After blocking the membranes (30 min at ambient temperature), the primary antibodies were diluted and added to the BB solutions. The anti-dystrophin-peptide antibody (rabbit polyclonal) (ab15277, Abcam Inc., Cambridge, MA USA) was diluted 1:200 in BB1, and incubated overnight at 4° on a rocking platform platform. Following incubation, the membranes were washed in the BB1, 3 × 10 min at room temperature, and the diluted secondary antibody, 1:5,000 goat anti-rabbit IgG – horse-radish peroxidase (HRP) (A5278, Sigma-Aldrich), was added to the BB1 for 1hr at room temperature with agitation. The solution with the secondary antibody was removed and the membrane was washed in PBST (3 × 10min). After the final wash, the luminol substrate was added (SuperSignal West Dura Extended Duration Substrate, Thermo Scientific) and the chemiluminescence signal was detected using a charged-coupled device (CCD) camera and digital imaging processor (G:BOX Chemi, Syngene, Frederick, MD USA). The data were analyzed using software (Gene Tools) provided with the imaging system.
Immunohistochemistry and histology
Tissue specimens were sectioned using a cryostatic microtome into 5 to 10µm thick slices and mounted onto silane–treated glass slides and fixed with 10% formalin for 10 minutes at room temperature then washed (3x) in phosphate buffered saline (PBS). For IHC, the formalin-fixed tissue sections were incubated in blocking buffer with either 5% goat serum or 5% bovine serum diluted in PBS, depending on the primary antiserum source, for 30 minutes at ambient temperature. The blocking buffer was removed and primary antiserum was applied. For dystrophin detection, a rabbit polyclonal antiserum (ab15277, Abcam) was diluted (1:1000) into fresh 2% goat serum in PBS and applied to the section and incubated overnight at 4°C in a humidified chamber. The antibody solutions were removed and the samples were washed with PBS (3x) at room temperature. Fluorescently labeled secondary antibodies were diluted in bovine serum - PBS and applied to the samples and incubated at room temperature for one hour in a dark plastic chamber to exclude light. The antibody solutions were then removed and washed with PBS. Coverslip mounting medium was applied (Vectashield Mounting Medium with DAPI) (Vector Laboratories Inc.) and a coverslip was placed over the tissue sample.
Data analysis
Statistical analysis used InStat (GraphPad Software, Inc., La Jolla, CA). Continuous parameters are reported as mean ± standard deviation, tested using two-tailed paired t-test. A p value less than 0.05 was considered significant. Correlation between number of injection and presence of viral DNA was assessed by Spearman’s rank correlation test.
Supplementary Material
Hematoxylin and Eosin stained heart tissue samples. Following euthanasia, the explanted hearts were cut sliced perpendicular to the long axis and divided into segments (Figure 4). Samples from each segment were processed for H&E staining. Representative stained samples are presented in the figure. The normal control dog heart sample (Top panel “Normal dog”) shows no extracellular nuclei. Stained sections from a treated GRMD dog heart (“Mowgli”), segments 14 and 16, show infiltrating T-cells and fibrotic lesions. Other sections from the same segments appear to have less inflammation. Overall, there is less pathology and inflammation observed in the untreated GRMD dog specimens, but the presence of infiltrating T-cells is observed. The frozen O.C.T. embedded specimens were cut into 6 to 10 µm thick sections and placed onto glass slides. The frozen sections were air-dried (5 min) then fixed in formalin (5min). Following the classic Harris hematoxylin and eosin staining protocol, the samples were dehydrated by immersion into a gradient of ethanol solutions (75% – 100%) then final xylene immersions (2x). The staining process was performed using a LEICA Auto Stainer XL. The samples were coverslips were applied and examined using a Leica CV 5030 microscope.
Acknowledgements
The authors are grateful to the NHLBI Laboratory of Animal Medicine and Surgery technologists for assistance with animal experiments, and the veterinarians and technicians of the Division of Veterinary Research, and Victor Wright for assistance with MRI scans. We appreciate the professional skills and advice of the Dr. Christian Combs, NHLBI Light Microscopy Core and Dr. Xu-Zi Yu, NHLBI Pathology Core. Dr. Victoria Joan Hoffman of Diagnostic & Research Services Branch, Division of Veterinary Resources, NIH, provide essential support and advice. Dr. H. Lee Sweeney provided helpful comments and discussion. We thank Boston Scientific for providing the injection catheters. Funding was provided by the National Heart, Lung, and Blood Institute, Division of Intramural Research, and the International Collaborative Effort (ICE) for Duchenne Muscular Dystrophy. The ICE consists of the Duchenne Parent Project France and the Association Monagasque Contre les Myopathies.
Footnotes
Disclosures
The authors have no financial conflicts of interest to disclose.
Supplementary information is available at Gene Therapy's website
References
- 1.Drousiotou A, et al. Neonatal screening for Duchenne muscular dystrophy: a novel semiquantitative application of the bioluminescence test for creatine kinase in a pilot national program in Cyprus. Genetic testing. 1998;2:55–60. doi: 10.1089/gte.1998.2.55. [DOI] [PubMed] [Google Scholar]
- 2.Emery AE. Population frequencies of inherited neuromuscular diseases--a world survey. Neuromuscular disorders : NMD. 1991;1:19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- 3.Koenig M, et al. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 5.Bushby K, et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurology. 2010;9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- 6.Connuck DM, et al. Characteristics and outcomes of cardiomyopathy in children with Duchenne or Becker muscular dystrophy: a comparative study from the Pediatric Cardiomyopathy Registry. Am Heart J. 2008;155:998–1005. doi: 10.1016/j.ahj.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nigro G, Comi LI, Politano L, Bain RJ. The incidence and evolution of cardiomyopathy in Duchenne muscular dystrophy. Int J Cardiol. 1990;26:271–277. doi: 10.1016/0167-5273(90)90082-g. [DOI] [PubMed] [Google Scholar]
- 8.Cohn RD, et al. Angiotensin II type 1 receptor blockade attenuates TGF-beta-induced failure of muscle regeneration in multiple myopathic states. Nature medicine. 2007;13:204–210. doi: 10.1038/nm1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duboc D, et al. Effect of perindopril on the onset and progression of left ventricular dysfunction in Duchenne muscular dystrophy. J Am Coll Cardiol. 2005;45:855–857. doi: 10.1016/j.jacc.2004.09.078. [DOI] [PubMed] [Google Scholar]
- 10.De Angelis FG, et al. Chimeric snRNA molecules carrying antisense sequences against the splice junctions of exon 51 of the dystrophin pre-mRNA induce exon skipping and restoration of a dystrophin synthesis in Delta 48–50 DMD cells. Proc Natl Acad Sci U S A. 2002;99:9456–9461. doi: 10.1073/pnas.142302299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denti MA, et al. Chimeric adeno-associated virus/antisense U1 small nuclear RNA effectively rescues dystrophin synthesis and muscle function by local treatment of mdx mice. Hum Gene Ther. 2006;17:565–574. doi: 10.1089/hum.2006.17.565. [DOI] [PubMed] [Google Scholar]
- 12.Goyenvalle A, et al. Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306:1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- 13.Cooper BJ, et al. The homologue of the Duchenne locus is defective in X-linked muscular dystrophy of dogs. Nature. 1988;334:154–156. doi: 10.1038/334154a0. [DOI] [PubMed] [Google Scholar]
- 14.Kornegay JN, Tuler SM, Miller DM, Levesque DC. Muscular dystrophy in a litter of golden retriever dogs. Muscle & nerve. 1988;11:1056–1064. doi: 10.1002/mus.880111008. [DOI] [PubMed] [Google Scholar]
- 15.Sharp NJ, et al. An error in dystrophin mRNA processing in golden retriever muscular dystrophy, an animal homologue of Duchenne muscular dystrophy. Genomics. 1992;13:115–121. doi: 10.1016/0888-7543(92)90210-j. [DOI] [PubMed] [Google Scholar]
- 16.Suter D, et al. Double-target antisense U7 snRNAs promote efficient skipping of an aberrant exon in three human beta-thalassemic mutations. Hum Mol Genet. 1999;8:2415–2423. doi: 10.1093/hmg/8.13.2415. [DOI] [PubMed] [Google Scholar]
- 17.Aartsma-Rus A, et al. Exploring the frontiers of therapeutic exon skipping for Duchenne muscular dystrophy by double targeting within one or multiple exons. Mol Ther. 2006;14:401–407. doi: 10.1016/j.ymthe.2006.02.022. [DOI] [PubMed] [Google Scholar]
- 18.Dunckley MG, Manoharan M, Villiet P, Eperon IC, Dickson G. Modification of splicing in the dystrophin gene in cultured Mdx muscle cells by antisense oligoribonucleotides. Hum Mol Genet. 1998;7:1083–1090. doi: 10.1093/hmg/7.7.1083. [DOI] [PubMed] [Google Scholar]
- 19.Fletcher S, Ly T, Duff RM, Mc CHJ, Wilton SD. Cryptic splicing involving the splice site mutation in the canine model of Duchenne muscular dystrophy. Neuromuscul Disord. 2001;11:239–243. doi: 10.1016/s0960-8966(00)00187-5. [DOI] [PubMed] [Google Scholar]
- 20.Gorman L, Suter D, Emerick V, Schumperli D, Kole R. Stable alteration of pre- mRNA splicing patterns by modified U7 small nuclear RNAs. Proc Natl Acad Sci U S A. 1998;95:4929–4934. doi: 10.1073/pnas.95.9.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilton SD, Fletcher S. Modification of pre-mRNA processing: application to dystrophin expression. Curr Opin Mol Ther. 2006;8:130–135. [PubMed] [Google Scholar]
- 22.Popplewell LJ, et al. Comparative analysis of antisense oligonucleotide sequences targeting exon 53 of the human DMD gene: Implications for future clinical trials. Neuromuscular disorders : NMD. 2010;20:102–110. doi: 10.1016/j.nmd.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 23.Cirak S, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goemans NM, et al. Systemic administration of PRO051 in Duchenne's muscular dystrophy. The New England journal of medicine. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 25.van Deutekom JC, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 26.Barbash IM, et al. Interventional magnetic resonance imaging for guiding gene and cell transfer in the heart. Heart. 2004;90:87–91. doi: 10.1136/heart.90.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boekstegers P, et al. Myocardial gene transfer by selective pressure-regulated retroinfusion of coronary veins. Gene Therapy. 2000;7:232–240. doi: 10.1038/sj.gt.3301079. [DOI] [PubMed] [Google Scholar]
- 28.Hayase M, et al. Catheter-based antegrade intracoronary viral gene delivery with coronary venous blockade. Am J Physiol Heart Circ Physiol. 2005;288:H2995–H3000. doi: 10.1152/ajpheart.00703.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright MJ, Wightman LM, Latchman DS, Marber MS. In vivo myocardial gene transfer: optimization and evaluation of intracoronary gene delivery in vivo. Gene Therapy. 2001;8:1833–1839. doi: 10.1038/sj.gt.3301614. [DOI] [PubMed] [Google Scholar]
- 30.Byrne MJ, et al. Recirculating cardiac delivery of AAV2/1SERCA2a improves myocardial function in an experimental model of heart failure in large animals. Gene Therapy. 2008;15:1550–1557. doi: 10.1038/gt.2008.120. [DOI] [PubMed] [Google Scholar]
- 31.Bish LT, et al. Percutaneous transendocardial delivery of self-complementary adeno-associated virus 6 achieves global cardiac gene transfer in canines. Molecular therapy : the journal of the American Society of Gene Therapy. 2008;16:1953–1959. doi: 10.1038/mt.2008.202. [DOI] [PubMed] [Google Scholar]
- 32.Sanborn TA, et al. Percutaneous endocardial transfer and expression of genes to the myocardium utilizing fluoroscopic guidance. Catheterization and cardiovascular interventions : official journal of the Society for Cardiac Angiography & Interventions. 2001;52:260–266. doi: 10.1002/1522-726x(200102)52:2<260::aid-ccd1061>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 33.Sylven C, et al. Catheter-based transendocardial myocardial gene transfer. Journal of interventional cardiology. 2002;15:7–13. doi: 10.1111/j.1540-8183.2002.tb01026.x. [DOI] [PubMed] [Google Scholar]
- 34.Bish LT, et al. Long-term Restoration of Cardiac Dystrophin Expression in Golden Retriever Muscular Dystrophy Following rAAV6-mediated Exon Skipping. Molecular therapy : the journal of the American Society of Gene Therapy. 2012;20:580–589. doi: 10.1038/mt.2011.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grossman PM, Han Z, Palasis M, Barry JJ, Lederman RJ. Incomplete retention after direct myocardial injection. Catheterization and cardiovascular interventions : official journal of the Society for Cardiac Angiography & Interventions. 2002;55:392–397. doi: 10.1002/ccd.10136. [DOI] [PubMed] [Google Scholar]
- 36.Levy HC, et al. Heparin binding induces conformational changes in Adeno-associated virus serotype 2. Journal of structural biology. 2009;165:146–156. doi: 10.1016/j.jsb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen WY, et al. Practical considerations of recombinant adeno-associated virus-mediated gene transfer for treatment of retinal degenerations. The journal of gene medicine. 2003;5:576–587. doi: 10.1002/jgm.375. [DOI] [PubMed] [Google Scholar]
- 38.Cerqueira MD, et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart: a statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation. 2002;105:539–542. doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
- 39.Cecchini S, Virag T, Kotin RM. Reproducible high yields of recombinant adeno-associated virus produced using invertebrate cells in 0.02- to 200-liter cultures. Human gene therapy. 2011;22:1021–1030. doi: 10.1089/hum.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naimark WA, et al. Adenovirus-catheter compatibility increases gene expression after delivery to porcine myocardium. Human gene therapy. 2003;14:161–166. doi: 10.1089/104303403321070856. [DOI] [PubMed] [Google Scholar]
- 41.Gutierrez LF, et al. Technology preview: X-ray fused with magnetic resonance during invasive cardiovascular procedures. Catheterization and cardiovascular interventions : official journal of the Society for Cardiac Angiography & Interventions. 2007;70:773–782. doi: 10.1002/ccd.21352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aartsma-Rus A, et al. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- 43.Cooper ST, Lo HP, North KN. Single section Western blot: improving the molecular diagnosis of the muscular dystrophies. Neurology. 2003;61:93–97. doi: 10.1212/01.wnl.0000069460.53438.38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hematoxylin and Eosin stained heart tissue samples. Following euthanasia, the explanted hearts were cut sliced perpendicular to the long axis and divided into segments (Figure 4). Samples from each segment were processed for H&E staining. Representative stained samples are presented in the figure. The normal control dog heart sample (Top panel “Normal dog”) shows no extracellular nuclei. Stained sections from a treated GRMD dog heart (“Mowgli”), segments 14 and 16, show infiltrating T-cells and fibrotic lesions. Other sections from the same segments appear to have less inflammation. Overall, there is less pathology and inflammation observed in the untreated GRMD dog specimens, but the presence of infiltrating T-cells is observed. The frozen O.C.T. embedded specimens were cut into 6 to 10 µm thick sections and placed onto glass slides. The frozen sections were air-dried (5 min) then fixed in formalin (5min). Following the classic Harris hematoxylin and eosin staining protocol, the samples were dehydrated by immersion into a gradient of ethanol solutions (75% – 100%) then final xylene immersions (2x). The staining process was performed using a LEICA Auto Stainer XL. The samples were coverslips were applied and examined using a Leica CV 5030 microscope.


