Abstract
Genome-wide disease association studies contrast genetic variation between disease cohorts and healthy populations to discover single nucleotide polymorphisms (SNPs) and other genetic markers revealing underlying genetic architectures of human diseases. Despite scores of efforts over the past decade, many reproducible genetic variants that explain substantial proportions of the heritable risk of common human diseases remain undiscovered. We have conducted a multispecies genomic analysis of 5,831 putative human risk variants for more than 230 disease phenotypes reported in 2,021 studies. We find that the current approaches show a propensity for discovering disease-associated SNPs (dSNPs) at conserved genomic positions because the effect size (odds ratio) and allelic P value of genetic association of an SNP relates strongly to the evolutionary conservation of their genomic position. We propose a new measure for ranking SNPs that integrates evolutionary conservation scores and the P value (E-rank). Using published data from a large case-control study, we demonstrate that E-rank method prioritizes SNPs with a greater likelihood of bona fide and reproducible genetic disease associations, many of which may explain greater proportions of genetic variance. Therefore, long-term evolutionary histories of genomic positions offer key practical utility in reassessing data from existing disease association studies, and in the design and analysis of future studies aimed at revealing the genetic basis of common human diseases.
Keywords: phylomedicine, GWAS, heritability
In genetic disease association studies, up to millions of genomic loci are genotyped across large population samples of disease (case) and healthy (control) individuals to elucidate genetic basis of diseases. Genetic associations are determined by estimating the significance (P value) and effect size (odds ratio) of the statistical relationship between alleles at genetic loci and a disease trait (Feero et al. 2010). To date, thousands of putative disease-associated genetic variants (disease-associated single nucleotide polymorphisms [dSNPs]) underlying complex disease phenotypes have been identified (Hindorff et al. 2011). However, discovered dSNPs vary among studies and explain relatively small fractions of the total heritability of the respective disease trait (Manolio et al. 2009). Nonadditive effects of epistatic interactions, effects of structural variants, synthetic associations with rare alleles, epigenetics, and gene–environment interactions are among many hypotheses put forward to explain these phenomena (Dickson et al. 2010; Eichler et al. 2010; McClellan and King 2010; Patel et al. 2010).
Instead, we take a phylogenetic approach to investigating and solving the problem of reproducibility and discovery of dSNPs. A long-term evolutionary history (phylogenetic) approach has not been explored likely due to the widespread realization that genetic variants underlying complex diseases will not impact fecundity because they occur relatively later in life (Thomas 2004; Blekhman et al. 2008; Cai et al. 2009). Still, molecular evolutionary patterns inform functional importance of genomic positions, as functionally important positions are likely to be more conserved and will directly impact the frequency of segregating alleles within populations under the neutral theory of molecular evolution (Kimura 1983; Barreiro et al. 2008; Kumar et al. 2009). However, the common practice of direct comparison of association statistics (e.g., P values and odds ratios) across genomic positions in individual studies does not explicitly account for these evolutionary differences among genomic positions when identifying variants with the most significant disease associations.
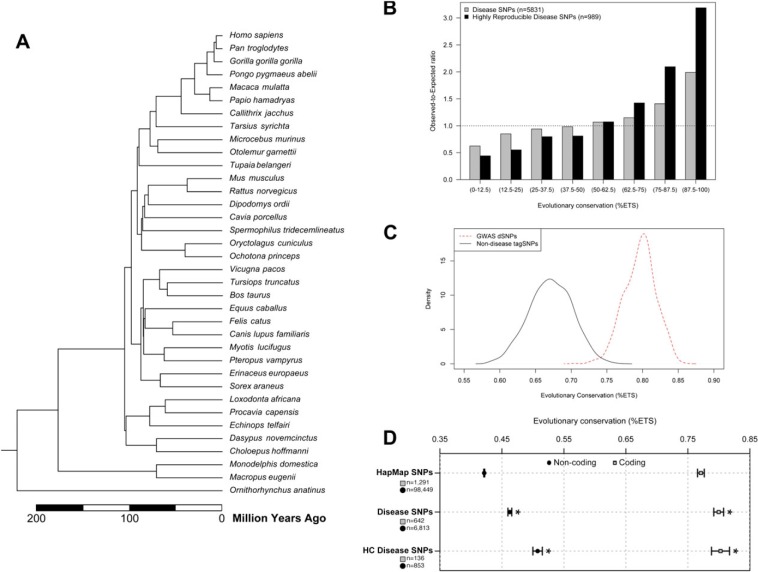
Therefore, we systematically investigated the relationship between the evolutionary anatomies of positions harboring disease-associated variants for 5,831 SNPs (dSNPs) reported to be associated with more than 230 disease phenotypes (2,021 published studies) representing a broad range of complex disease categories (Chen et al. 2010) (Table 1). We tested the null hypothesis that the discovery of dSNPs is not biased by the long-term evolutionary properties of genomic locations harboring dSNPs, which are inferred from multispecies alignments from diverse mammals (fig. 1A; Materials and Methods). Figure 1B shows the distribution of evolutionary conservation, estimated here as the percent evolutionary time span (ETS; see Materials and Methods) over which the position is maintained in the mammalian genomes, for positions harboring statistically significant risk variants. The preponderance of dSNPs discovered in positions with higher ETS is greater than that expected based on the distribution of HapMap3 (International HapMap Consortium et al. 2010) population SNPs (dotted line). This result is more pronounced for dSNPs replicated in three or more studies in three or more distinct populations (highly replicated [HR]-dSNPs, black bars) than those reported in at least one study (gray bars). A similar pattern is observed in an analysis of only those dSNPs that have reported significant (P < 10−7) associations in the National Human Genome Research Institute (NHGRI) genome-wide association studies (GWASs) catalog (http://www.genome.gov/gwastudies/) (fig. 1C). This pattern is also observed in separate analysis of coding dSNPs and noncoding dSNPs (fig. 1D). Therefore, dSNPs have been discovered disproportionately at positions that have been highly conserved over evolutionary time.
Table 1.
Summary of Major Disease Categories Represented by Variants Used in This Study.
| Disease Category | Number of Studies | Number of SNPs | Distinct Diseases |
|---|---|---|---|
| Neoplasms | 132 | 514 | 29 |
| Cardiovascular diseases | 86 | 370 | 24 |
| Nervous system diseases | 89 | 748 | 20 |
| Digestive system diseases | 84 | 376 | 19 |
| Eye diseases | 61 | 102 | 13 |
| Musculoskeletal diseases | 70 | 749 | 13 |
| Mental disorders | 73 | 1311 | 11 |
| Nutritional and metabolic diseases | 120 | 414 | 9 |
| Female urogenital diseases and pregnancy complications | 12 | 29 | 8 |
| Skin and connective tissue diseases | 58 | 795 | 6 |
| Respiratory tract diseases | 20 | 127 | 5 |
| Hemic and lymphatic diseases | 9 | 23 | 5 |
| Otorhinolaryngologic diseases | 3 | 21 | 3 |
| Stomatognathic diseases | 5 | 12 | 3 |
| Bacterial infections and mycoses | 3 | 18 | 3 |
| Endocrine system diseases | 2 | 15 | 2 |
| Virus diseases | 3 | 51 | 2 |
NOTE.—dSNPs used in this study were organized into high-level disease categories using the Medical Subject Headings (MeSH) annotating their associated disease phenotype in the VARIMED database. dSNPs were placed into top-level MeSH categories using the MeSH hierarchy. In cases where the dSNPs mapped to more than one top-level disease category, the dSNP was counted once in each category. If the dSNP phenotype did not have an associated MeSH annotation (37 dSNPs in this study), it was not represented in this table.
FIG. 1.
Patterns of evolutionary conservation of positions harboring disease-associated SNPs. (A) A time tree of 36 mammalian species used for deriving evolutionary information for each SNP. Species divergence times were obtained from www.timetree.org (Hedges et al. 2006). (B) Relationship of the observed-to-expected numbers of disease-associated SNPs, dSNPs, at human genomic positions preserved with different degrees over time (high-to-low is given left-to-right). Results from all dSNPs (gray bars) and high-confidence (HC) dSNPs (black bars) are shown. Expected numbers were estimated using HapMap3 SNPs. The right axis indicates the fraction of total SNPs in each dSNP category that fall into the conservation bins defined on the bottom axis. (C) The distribution of evolutionary conservation for GWAS dSNPs associated at a stringent significance threshold of P < 5 × 10−7 or lower in two or more studies (red, dashed) is compared with the distribution of evolutionary conservation of 100,000 randomly selected tagSNPs chosen from two of the most popular GWAS genotyping platforms (black). The mean evolutionary conservation for GWAS dSNPs is significantly higher than that of tagSNPs (t-test P < 5 × 10−20). (D) Comparison of average conservation of coding (gray squares) and noncoding (circles) dSNPs and HC dSNPs to HapMap3 SNPs. The HapMap3 distributions are estimated from a representative random sample of 100,000 HapMap3 SNP loci. As expected, coding dSNPs occur at more highly conserved positions than noncoding dSNPs, but the trend toward more conserved positions at disease-associated loci is observed in both cases. SNPs in noncoding regions are overall found at much less conserved positions, however, the mean conservation for noncoding is significantly more conserved than noncoding HapMap SNPs (error bars: standard error of mean) (* indicates t-test P value < 10−8 compared with HapMap3).
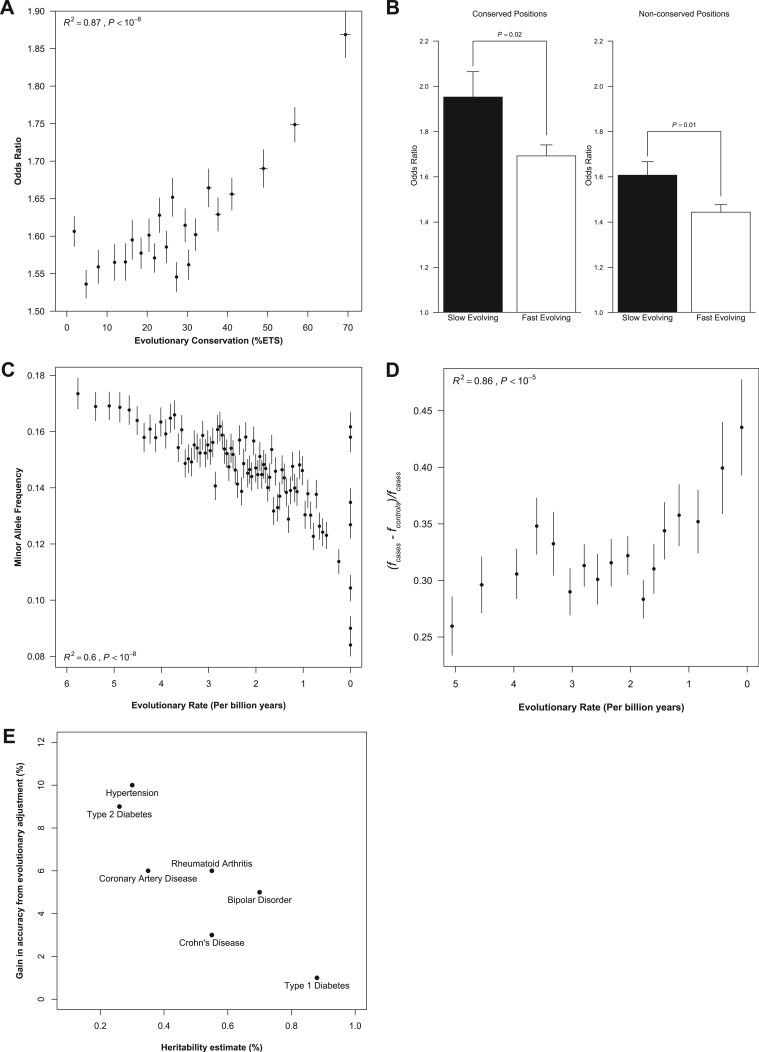
To assess the impact of this evolutionary trend in explaining the genetic variance of a disease trait, we investigated the relationship between dSNP association odds ratio and evolutionary conservation, because the proportion of the genetic variance of a disease trait explained by a variant relates to the effect size (odds ratio) of association (Park et al. 2010). We find that the reported effect size of disease-associated variants is strongly related to the evolutionary conservation of its genomic position (fig. 2A, R2 = 0.87, P < 10−8). The rate of long-term evolutionary substitution of nucleotides also differentiates the odds ratio distributions for dSNPs found at positions with high (top 25% ETS) as well as low (bottom 25% ETS) degrees of positional conservation among species (fig. 2B). The quartile of the slowest evolving positions harbor dSNPs with higher odd ratios in association studies as compared with the quartile of the fastest evolving positions (P < 0.05). These observations provides one possible fundamental explanation for the preferential discovery of lower-frequency variants with high odds ratios, as slower evolving positions are expected to have lower minor allele frequencies (MAFs) due to stronger purifying selection (e.g., Kumar et al. 2009) (fig. 2C). This is confirmed in an analysis of 3,372 dSNPs reported in 515 independent case-control GWAS, where we observe a strong negative relationship between the evolutionary rate and the normalized difference in the dSNP risk allele frequencies between case and control populations in individual GWAS studies (fig. 2D; R2 = 0.86, P < 10−5).
FIG. 2.
The effect size of disease variants relates to the evolutionary anatomy of genomic positions. (A) Relationship between the odds ratio reported for disease-associated variants and the evolutionary conservation of the genomic position harboring the variant. All reported odds ratios were normalized toward disease risk estimation by taking the exponent of the absolute loge of odds ratios < 1. Each point represents the mean of nonoverlapping bins of n = 200 associated loci ordered by increasing %ETS. The trend was best described by a second order polynomial (R2 = 0.87, P < 10−8; Pearson r = 0.86, P < 10−4) (error bars: standard error of mean [SEM]). (B) Slowly evolving sites (bottom 25% of evolutionary rates) at highly conserved positions (top 25% of evolutionary conservation) exhibit higher average odds ratios than faster-evolving sites (top 25% of evolutionary rates) at both conserved (top 25% ETS) and nonconserved (bottom 25% ETS) positions (error bars: SEM). (C) Relationship of the multispecies evolutionary rate with the MAFs in human populations. Each point is estimated as the average of evolutionary rate and MAFs for 100,000 SNPs randomly sampled from HapMap 3 CEU population data (second order polynomial R2 = 0.6, P < 10−8; Pearson r = −0.76; P < 10−4) (error bars: SEM). (D) The influence of evolutionary rate on the risk allele frequency disparities between cases and controls. Δf is the difference in risk allele frequency between cases (fcases) and controls (fcontrols) divided by fcontrols to control for the MAF of the risk allele in healthy populations; Δf = (fcases − fcontrols)/ fcontrols. Each point represents the mean of nonoverlapping bins of n = 200 associated loci ordered by increasing evolutionary rate (third order polynomial R2 = 0.86, P < 10−5; Pearson r = 0.74, P < 10−3) (error bars: SEM). (E) For each of the diseases measured in the WTCCC, the gain in predictive accuracy (i.e., difference between the evolutionary adjusted P value (E-rank) AUC and the “raw” P value (P-rank) AUC) is plotted against the total heritability estimate for the disease (Sofaer 1993; Poulsen et al. 1999; Katzmarzyk et al. 2000; Smoller and Finn 2003; Agarwal et al. 2005; Harney et al. 2008; Ounissi-Benkalha and Polychronakos 2008).
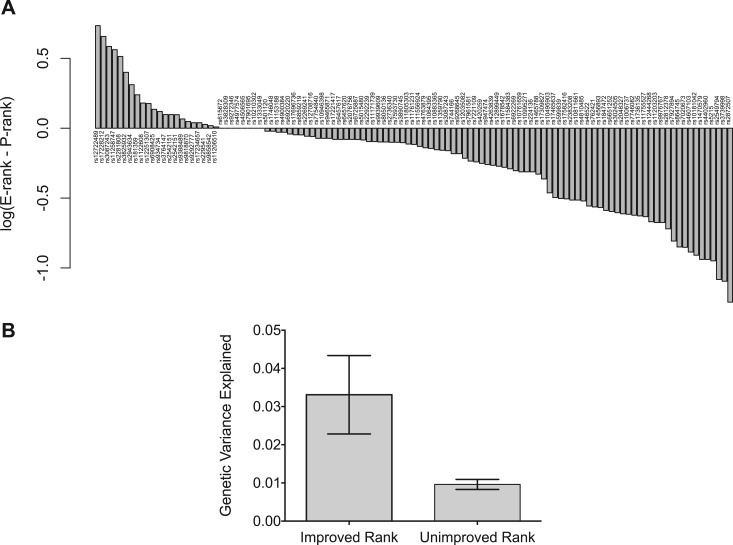
Our results indicate the need to use evolutionary conservation scores as priors in evaluating relative importance of SNPs in disease association studies. Therefore, we integrate evolutionary conservation score along with the allelic P value of association for each SNP in GWAS to generate an evolutionary-adapted ranking (“E-rank”; see Materials and Methods). As designed, the E-rank ameliorates the effect of evolutionary bias in disease association discovery by prioritizing putative dSNPs that have reached relatively high population frequencies at positions with high ETS. To demonstrate and assess the utility of this approach in prioritizing dSNPs in individual GWAS studies, we applied the E-rank method to the original association data for 500,000 loci profiled across seven common diseases by Wellcome Trust Case Control Consortium (WTCCC)(2007). Figure 3A shows that a majority of significant dSNPs (NHGRI GWAS catalog P < 10−7) represented in the WTCCC study have improved E-ranks relative to their classical P value based rank (“P-rank”). Overall, the dSNPs whose ranks are improved by E-rank explain significantly more of the genetic variance of the disease trait in the measured population relative to those dSNPs with unimproved ranks (fig. 3B).
FIG. 3.
Characteristics of evolutionary ranking (E-rank) of disease-associated variants in WTCCC. (A) The difference in the original P value rank (P-rank) versus the evolutionary adjusted rank (E-rank) is shown for a large set of established disease-associated variants that were measured in the WTCCC study. E-rank generally improves the rank of bona fide disease associations in the WTCCC data. (B) Although the E-rank method does not improve the ranks of all established disease-associated variants in the WTCCC data, it tends to improve the ranks of SNP loci that explain significantly more of the genetic variance of the disease trait compared with the SNP loci that are not improved by E-rank (t-test P < 1 × 10−5; error bars = standard error of mean).
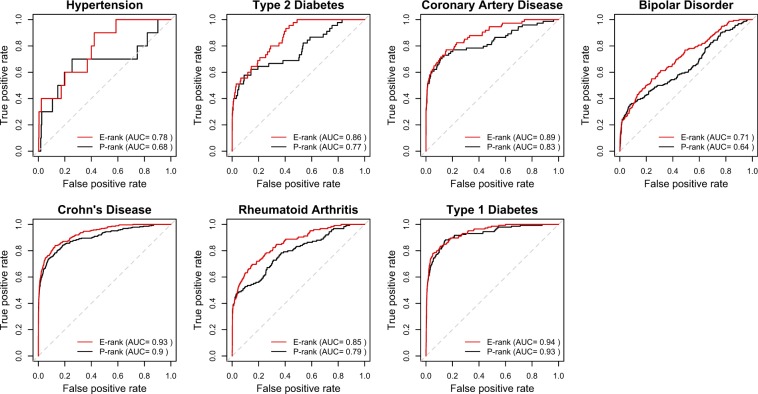
Similarly, we found the E-rank approach to perform better in discriminating dSNPs that have been replicated in three or more independent studies (HR-dSNPs) or by large-scale meta-analysis, which is used in this study as an indicator of likely true positive association (NCI-NHGRI Working Group on Replication in Association Studies et al. 2007; Wei et al. 2009). For this analysis, we identified 859 HR-dSNPs in our data set of 8,963 and compared the performance of E-ranks versus P-ranks in discriminating reproducible disease associations from the original WTCCC data. Figure 4 shows the resulting Receiver Operating Characteristic (ROC) curves, where the area under the curve (AUC) represents the accuracy of P-rank (black line) and E-rank (red line) to predict replicated associations for the seven diseases in WTCCC data. In every case, E-rank performs better than P-rank, with the greatest improvements found for diseases that have been previously estimated to have relatively low degrees of heritability. For example, both hypertension and Type 2 diabetes gain 10% and 9% accuracy, respectively, using the E-rank approach, yet both traits are individually estimated to have total heritability below 30% (Poulsen et al. 1999; Agarwal et al. 2005).
FIG. 4.
Evolutionary adjustment improves discriminatory power to identify reproducible associations. ROC curves representing the accuracy, represented by the AUC, to predict associations subsequently replicated in three or more independent association studies are shown for each of the seven disease association studies represented in the Wellcome Trust Case Control Consortium (WTCCC) data. The black line indicates the predictive accuracy of the original allelic association P values estimated by comparison of cases versus controls in the WTCCC study (P-rank). The red line indicates the predictive accuracy after prioritizing SNP loci using the E-rank approach. The gray diagonal line represents random performance (AUC = 0.5).
On the other end of the spectrum, the heritability of Type 1 diabetes is estimated to be near 90%, and evolutionary priors do not provide significant additional gains above the high predictive accuracy (AUC = 0.94) from P-ranks (Wei et al. 2009). Across all seven diseases, we observe that E-rank improvements track closely with the contemporary knowledge of the heritability of the diseases (fig. 2E), which suggests that evolutionary anatomies of disease-associated risk variants could inform on the nature and complexity of the allelic architecture underlying common diseases. Future efforts to develop more sophisticated evolutionary methods for disease association analysis may realize even greater gains in both predictive power and our understanding of the genetic architectures of diseases.
In summary, our results demonstrate the utility and potential clinical relevance of evolutionary properties derived from cross-species genome analysis, highlighting the need and importance of sequencing the genomes of species both closely and distantly related to humans in evolutionary time, to enable and improve the fidelity of evolutionary inferences bearing on human health and disease (Kumar et al. 2011). Our approach is dependent on the availability of comprehensive genomic sequence data for extant mammalian genomes, which dictates the accuracy of estimating ETS and evolutionary rate. Therefore, future efforts to increase the availability of high-quality genome assemblies for nonhuman mammals will improve methods for evolutionary assessment of human disease–associated variation. Furthermore, the results presented here have implications for evaluating rare versus common-variant hypotheses concerning genetic susceptibility to common diseases (Bodmer and Bonilla 2008; Goldstein 2009), which is of great practical significance in clinical genomics. We have also shown that position-specific evolutionary information can enhance discovery in individual association studies. Therefore, evolutionarily informed analyses of existing and future association data would likely enhance discovery of genetic disease susceptibility variants, and offer further crucial insights into the genetic basis of human diseases.
Materials and Methods
SNP Data Sets
We used data from the VARIMED database of quantitative human disease–SNP associations curated from the full text and supplementary info of 3,333 published human genetics papers recording more than 100 features per SNP association, including the disease name, specific phenotype, study population, case and control population information, genotyping technology, major/minor/risk alleles, odds ratio, 95% confidence interval of the odds ratio, published P value, and genetic model (further details in references Ashley et al. 2010 and Chen et al. 2010). For this study, we only considered single locus associations and excluded variants for which information on the effect size (odds ratio) was not available in the published results. We selected disease associations with reported odds ratio values and association P value <5 × 10−3. These criteria yielded a set of 5,831 variants associated with 230 disease phenotypes obtained from 2,021 published studies.
We also obtained data from a public catalog of disease associations curated from published GWAS, which is provided in downloadable format from http://www.genome.gov/gwastudies/ (accessed 3 May 2011). Although the VARIMED database is more comprehensive in its annotation and representation of published disease-associated variants, data from the NHGRI GWAS catalog was included in the analysis because it is a widely used and an accepted resource for disease association data. All available data from HapMap3 was retrieved from the HapMap Project FTP server (ftp://ftp.ncbi.nlm.nih.gov/hapmap/). All SNPs were mapped to their genomic locations on hg19 release using NCBI dbSNP (hs130) identifiers.
Evolutionary Anatomies of SNP Loci
For each SNP, we estimated evolutionary conservation score and the rate of substitution using mammalian nucleotide sequence alignments obtained from the University of California at Santa Cruz Genome Browser resource (Kent et al. 2002). “Evolutionary conservation” score quantifies the fraction of evolutionary time among species for which the given human position has existed in the evolutionary history of the mammalian lineage (%ETS; fig. 1). In this way, the evolutionary conservation relates to the retention of the genomic position or positional conservation (Kumar et al. 2009). In addition, we estimated the “evolutionary rates” of nucleotide change at each site by dividing the total number of substitution in the mammalian phylogeny by the total time elapsed on the tree (substitutions per site per billion years) (Kumar et al. 2009). For each position, species containing alignment gaps or missing data were pruned from the tree before calculating substitution rates.
Definition of the E-Rank Method to Prioritize Disease-Association Statistics
Because evolutionary information can be estimated from multispecies alignments for each position in the human genome independent of the population data a priori, its use is particularly attractive to prioritize loci with segregating alleles in a disease association study to help identify genuine dSNPs. Based on the empirical observations concerning the evolutionary properties of dSNPs revealed by this study, we developed an evolutionary ranking method (E-rank) in which the allelic P value of association (P) for an SNP was modified using the evolutionary conservation of the position harboring the allele and the MAF of the tagSNP: E-rank = (P/MAF) × (1/[Kr·Kt]), where Kr is the rank of the evolutionary rate of the position and Kt is the rank of the evolutionary time span of the position. The rationale for this approach is that segregating dSNPs found at positionally conserved loci, quantified by Kr, should be given a better rank (higher ordinal rank) than segregating dSNPs at less retained positions. Furthermore, dSNPs at faster-evolving positions, quantified by Kr, should be given a better rank (higher ordinal rank) because these positions are more likely to harbor high-frequency dSNPs explaining greater proportions of the genetic variance of a trait. At positions with high ranks of Kr and Kt, the MAF adjusted P value of association (P/MAF) will become smaller by multiplication with the inverse product of Kr and Kt, which will improve its overall priority position in the ranking of association results.
The P is divided by MAF because P is a function of both the MAF and the effect size (i.e., odds ratio) of the associated allele. For example, if we analyze the Wellcome Trust Case Control Consortium (2007) results for Crohn's disease, across 500,000 loci, the Pearson correlation between the log(P value) and the log(odds ratio) is rather weak in effect (R = 0.22, P < 10−15). However, the correlation between log(P value)/MAF and the log(odds ratio) is much stronger (R = 0.73, P < 10−15) with the binomial variance from the allele frequency accounted for. Therefore, we divide by MAF to normalize the effect of the control allele frequency on the probability of rejecting the null hypothesis given the same sample size, and then perform evolutionary adjustment on the remaining component. We applied E-rank to association data for 500,000 loci profiled across seven common diseases by the WTCCC.
Estimation of Genetic Variance Explained by a Disease-Associated SNP
Using the same approach taken by Park et al. (2010), we estimated the percent genetic variance (GV) explained by a disease-associated SNP, i, as GVi = 2 × log(ORi)2 × MAFi(1 − MAFi), where ORi is the odds ratio of the association between SNP i and its respective disease trait, and MAFi is the MAF of SNP i in the measured population. This method provides an estimate of GVi under the assumption of an additive polygenic model.
Determination of a “Gold Standard” of Reproducible Disease Associations
To determine one set of reproducible SNPs, we queried the VARIMED disease SNP database to identify SNP loci that were reported to be significantly associated (P < 0.05) with the relevant disease phenotype in three or more independent studies measured from Caucasian populations, which was the primary ethnicity measured by the WTCCC.
Definition of the SNP Classification Problem
SNPs measured in the WTCCC study that mapped to a “gold standard” replicated SNP by dbSNP identifier were assigned to the “Replicated” class and all others were assigned to the “Unreplicated” class. Note that this includes the directly measured SNP and any other SNP that could serve as a proxy SNP determined by the LD threshold (r2 > 0.8). Accuracy in distinguishing Replicated SNPs from Unreplicated was determined for each disease using the original allelic association P value and the E-rank by estimating the area under the ROC curve (AUC) using the ROCR package (Sing et al. 2005). In this way, the AUC relates to the likelihood that the study will rank a randomly selected locus with reproducible association (Replicated class) higher than a randomly selected locus with nonreproducible association (Unreplicated class).
Acknowledgments
J.T.D. was supported by the National Library of Medicine (NLM) Biomedical Informatics Training Grant to Stanford University (T15 LM007033). S.K. is supported by funding from NLM (R01 LM010834) and the National Human Genome Research Institute (R01 HG002096). A.J.B. is supported by funding from the Lucile Packard Foundation for Children's Health, National Library of Medicine (K22 LM008261), NLM (R01 LM009719), Howard Hughes Medical Institute, and the Pharmaceutical Research and Manufacturers of America Foundation. The authors thank Philip Hedrick, Yuseob Kim, Li Liu, Jay Taylor, Marina Sirota, and Dmitri Petrov for useful comments during early stages of the study.
References
- Agarwal A, Williams GH, Fisher ND. Genetics of human hypertension. Trends Endocrinol Metab. 2005;16:127–133. doi: 10.1016/j.tem.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Ashley EA, Butte AJ, Wheeler MT, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- Blekhman R, Man O, Herrmann L, Boyko AR, Indap A, Kosiol C, Bustamante CD, Teshima KM, Przeworski M. Natural selection on genes that underlie human disease susceptibility. Curr Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai JJ, Borenstein E, Chen R, Petrov DA. Similarly strong purifying selection acts on human disease genes of all evolutionary ages. Genome Biol Evol. 2009;1:131–144. doi: 10.1093/gbe/evp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Davydov EV, Sirota M, Butte AJ. Non-synonymous and synonymous coding SNPs show similar likelihood and effect size of human disease association. PLoS One. 2010;5:e13574. doi: 10.1371/journal.pone.0013574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB, Hastie N. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feero WG, Guttmacher AE, Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- Harney SM, Vilarino-Guell C, Adamopoulos IE, et al. Fine mapping of the MHC Class III region demonstrates association of AIF1 and rheumatoid arthritis. Rheumatology (Oxford) 2008;47:1761–1767. doi: 10.1093/rheumatology/ken376. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Hindorff L, Junkins H, Hall P, Mehta J, Manolio T. A catalog of published genome-wide association studies. [cited 2011 Dec 1] 2011. Available from: www.genome.gov. [Google Scholar]
- International HapMap Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmarzyk PT, Perusse L, Rice T, Gagnon J, Skinner JS, Wilmore JH, Leon AS, Rao DC, Bouchard C. Familial resemblance for coronary heart disease risk: the HERITAGE Family Study. Ethn Dis. 2000;10:138–147. [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. New York: Cambridge University Press; 1983. [Google Scholar]
- Kumar S, Dudley JT, Filipski A, Liu L. Phylomedicine: an evolutionary telescope to explore and diagnose the universe of disease mutations. Trends Genet. 2011;27:377–386. doi: 10.1016/j.tig.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Suleski MP, Markov GJ, Lawrence S, Marco A, Filipski AJ. Positional conservation and amino acids shape the correct diagnosis and population frequencies of benign and damaging personal amino acid mutations. Genome Res. 2009;19:1562–1569. doi: 10.1101/gr.091991.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J, King M-C. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- NCI-NHGRI Working Group on Replication in Association Studies, Chanock SJ, Manolio T, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Ounissi-Benkalha H, Polychronakos C. The molecular genetics of type 1 diabetes: new genes and emerging mechanisms. Trends Mol Med. 2008;14:268–275. doi: 10.1016/j.molmed.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Park J-H, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CJ, Bhattacharya J, Butte AJ. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PLoS One. 2010;5:e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance—a population-based twin study. Diabetologia. 1999;42:139–145. doi: 10.1007/s001250051131. [DOI] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123C:48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- Sofaer J. Crohn's disease: the genetic contribution. Gut. 1993;34:869–871. doi: 10.1136/gut.34.7.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD. Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: evolutionary evidence for differences in molecular effects. Proc Natl Acad Sci U S A. 2004;101:15398–15403. doi: 10.1073/pnas.0404380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Wang K, Qu HQ, et al. (15 co-authors) From disease association to risk assessment: an optimistic view from genome-wide association studies on type 1 diabetes. PLoS Genet. 2009;5:e1000678. doi: 10.1371/journal.pgen.1000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]