Abstract
Epstein-Barr Virus (EBV), which is associated with multiple human tumors, persists as a minichromosome in the nucleus of B-lymphocytes and induces malignancies through incompletely understood mechanisms. Here, we present a large-scale functional genomic analysis of EBV. Our experimentally generated nucleosome positioning maps and viral protein binding data were integrated with over 700 publicly available high-throughput sequencing data sets for human lymphoblastoid cell lines mapped to the EBV genome. We found that viral lytic genes are coexpressed with cellular cancer-associated pathways, suggesting that the lytic cycle may play an unexpected role in virus-mediated oncogenesis. Host regulators of viral oncogene expression and chromosome structure were identified and validated, revealing a role for the B-cell-specific protein Pax5 in viral gene regulation and the cohesin complex in regulating higher order chromatin structure. Our findings provide a deeper understanding of latent viral persistence in oncogenesis and establish a valuable viral genomics resource for future exploration.
Introduction
Viruses co-evolve with their hosts to establish stable and co-regulated genomes and gene expression programs (Iyer et al., 2006). DNA tumor viruses are distinguished by their ability to provide a selective growth advantage to host cells and typically establish long-term persistent intracellular infections (Moore and Chang, 2010). Among the most extensively characterized human tumor viruses is Epstein-Barr Virus (EBV), which has been implicated as a causative agent in multiple B-cell lymphomas, gastric carcinomas, and nasopharyngeal carcinomas (Rickinson and Kieff, 2007; Young and Rickinson, 2004). EBV is estimated to be responsible for ~1% of all human cancers and may also contribute to other disorders, including multiple sclerosis (Ascherio and Munger, 2010; Parkin, 2006). While EBV oncogenes and regulatory pathways have been characterized individually, there have been few studies that examine the network of virus-host interactions at a genomic scale.
Methods emerging from systems biology and functional genomics provide powerful approaches for elucidating these viral-host interaction networks (Aderem et al., 2011). Chronic infection by DNA tumor viruses, such as EBV, is particularly well suited for systems-level interrogation. EBV genomes persist as multicopy DNA episomes in the nucleus of human B-lymphocytes (Lieberman, 2006; Lindner and Sugden, 2007), which can be assayed by modern high-throughput sequencing methods. EBV infection of primary human B-lymphocytes leads to the efficient establishment of continuously proliferating genetically stable human lymphoblastoid cell lines (LCLs). Additionally, the EBV expression program in LCLs allows for the assay of the full repertoire of viral latency genes, which are able to drive cellular proliferation and survival in vitro and in vivo in immunocompromised hosts (Thorley-Lawson and Gross, 2004). LCLs have also been extensively characterized by human genetic studies, including large-scale consortium projects such as ENCODE and HapMap(ENCODE Consortium, 2007; International HapMap Consortium, 2010), which have created vast repositories of genotype, gene expression, and cellular phenotype data that can be leveraged to better understand host and viral regulatory networks. The contribution of EBV to LCL genome biology and the comprehensive mapping of functional elements of the EBV genome have not been evaluated in these prior studies.
Previous genomics studies of human viruses have been limited in scope, exploring novel virus discovery (Feng et al., 2008), environmental niche characterization (Breitbart et al., 2003; Reyes et al., 2010; Tadmor et al., 2011), in vitro protein interactions (Calderwood et al., 2007; Dyer et al., 2008; Pinney et al., 2009), and small-scale functional genomics. Many of these studies have been done in herpesviruses, such as a RNA-seq analysis of a Burkitt lymphoma cell line (Lin et al., 2010; Xu et al., 2010), a low density qPCR primer array for chromatin immunoprecipitation (ChIP) on a limited set of host factors (e.g. CTCF and a small set of histone modifications) (Tempera et al., 2010), and ChIP of the viral factors EBNA1(Dresang et al., 2009; Lu et al., 2010) and BZLF1(Bergbauer et al., 2010). There has also been recent exploration of the KSHV epigenome (Günther and Grundhoff, 2010; Stedman et al., 2008; Toth et al., 2010), though it is unclear if these epigenetic controls are conserved between gammaherpesviruses.
Here, we present a large-scale functional genomic analysis of EBV, which provides insights into viral pathogenesis and B-cell biology. We integrated our own experimentally generated nucleosome positioning maps and viral protein binding studies with over 700 publicly available high-throughput sequencing data sets for human LCLs that we have mapped to the EBV genome. Many of the cell lines we examined were created by the HapMap Project, and one of these was extensively assayed by the ENCODE Consortium (ENCODE Consortium, 2007; International HapMap Consortium, 2010). Using this massive repository of combined host and virus data, we create a comprehensive atlas of interactions between host factors and the EBV genome. We characterized genome-wide binding profiles of over 60 human transcription factors, which cluster into specific regulatory regions, suggesting combinatorial control of viral gene expression. We discovered host genes that are coexpressed with viral genes and cluster into B-cell proliferation pathways. We demonstrated that the host B-cell specificity factor Pax5 plays an unexpected role in regulating viral gene expression and chromatin organization. Finally, we characterized the Cohesin-mediated spatial conformation of the viral episome and demonstrate its relevance in gene regulation through knockdown of its structural components. Our study represents the exploration of functional elements and regulators of Epstein-Barr virus on a large scale. All raw and processed data are publicly available at http://ebv.wistar.upenn.edu.
RESULTS
Generating an atlas of functional elements in the EBV genome
To generate a comprehensive atlas of functional elements for the EBV genome, we analyzed many existing large-scale functional genomics data sets for EBV positive LCLs (Table S1). We examined 319 RNA-seq experiments from five separate studies (Cheung et al., 2010; ENCODE Consortium, 2007; Kasowski et al., 2010; Montgomery et al., 2010; Pickrell et al., 2010), covering LCLs from 143 donors, multiple RNA size ranges, cellular compartments and both poly(A)+ and poly(A)− transcripts. We explored DNA-binding proteins and histone modifications and variants by incorporating more than 300 ChIP-seq experiments from the ENCODE project (ENCODE Consortium, 2007). We also integrated cytokine peptide levels (Choy et al., 2008) and related independent functional genomic studies (Lee et al., 2011; Lu et al., 2010; Ramagopalan et al., 2010).
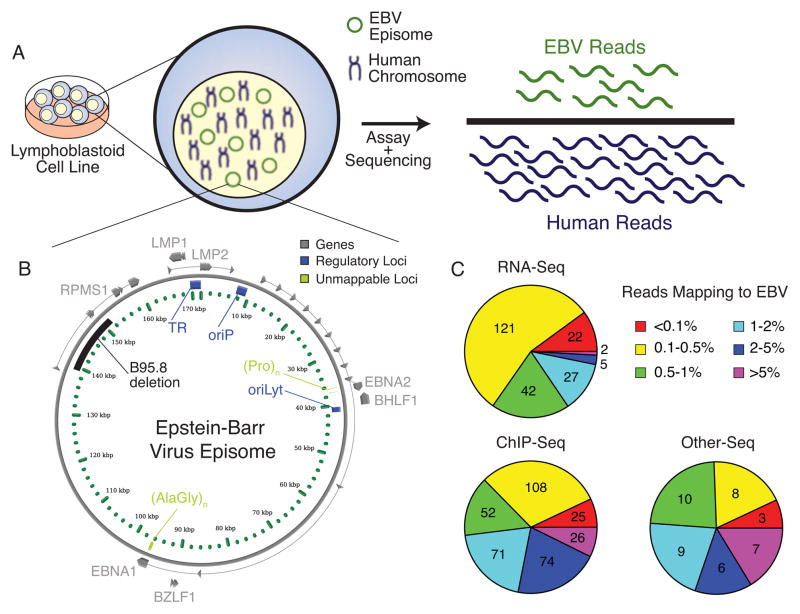
To ensure that we characterized reads of viral origin and excluded reads from homologous human regions, we subtracted reads mapping to the human genome prior to alignment against the EBV genome (Figure 1). This subtraction did not affect mappability of the EBV genome since only a small number of highly repetitive EBV elements are unmappable with reads longer than 24 nucleotides (Figures 1B and S1A–D). Alignments to EBV accounted for a significant number of reads in nearly all assays (Fig. 1C, and Table S2). In total, we aligned over 166 million reads to the EBV genome, which accounted for 1.2% of all mappable reads. As a negative control, we aligned reads from experiments performed in uninfected cell lines and primary tissue and found negligible alignment to EBV (Figure S1E and F).
Figure 1. Genomic profiling of Epstein-Barr virus in lymphoblastoid cell lines.
(A) EBV infected cell lines were subjected to functional genomics assays coupled with high throughput sequencing, giving rise to reads that map to the human host and viral genomes. (B) Known functional elements of the 172kbp viral episome examined in this study. While we provide all coordinates with respect to the 172kbp reference, most LCLs were generated with the B95.8 strain, which contains a 12kbp deletion (139–151kbp). Unmappable loci have highly similar regions in the human genome, corresponding to reads that are of ambiguous origin. (C) Short reads from hundreds of ChIP, RNA, and other (DNase, FAIRE, MNase, HiC, etc) sequencing experiments were mapped to the viral and host genomes. For each experiment, percentage of aligned viral reads was determined as a fraction of aligned human reads. An average of more than 1% of reads map to the viral genome, though the percentage of viral reads can vary across 4 orders of magnitude. Acronyms: EBNA: EBV nuclear antigen; LMP: Latent membrane protein; oriP: Latent origin of replication; oriLyt: Lytic origin of replication; TR: Terminal repeats; BHLF1: Viral lytic non-coding RNA, BamHI H leftward fragment 1; BHRF1: Viral BCL2 homologue, BamHI H rightward fragment 1; BZLF1: Viral lytic activating gene, BamHI Z leftward fragment 1; RPMS1: Viral gene and promoter for BART miRNAs; RPM: Reads per million. See also Figure S1 and Table S1.
Identification of regulatory and transcribed elements of Epstein-Barr virus
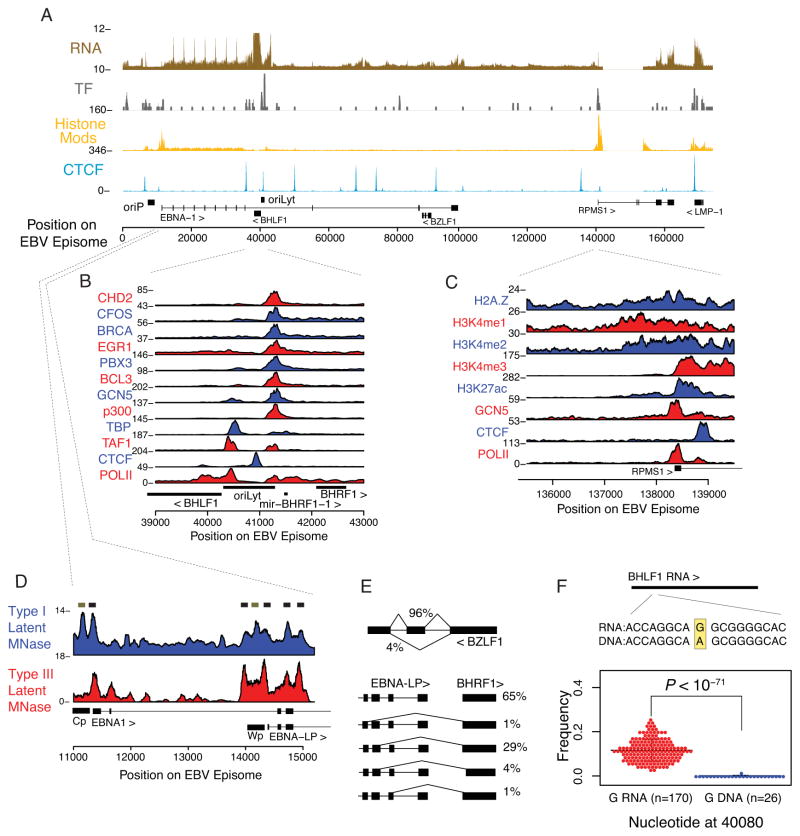
To explore the overall organization of the functional EBV genome, we summarized viral gene expression, transcription factor binding, histone modifications, and binding sites of the chromatin insulator CTCF (Figure 2A). An initial overview of the data provided several striking observations.
Figure 2. Transcribed and regulatory elements of the EBV genome.
(A) The average RNA expression profile (transformed by square root) shows unexpectedly high RNA reads of the lytic cycle associated transcript BHLF1, in addition to the canonical genes associated with type III latency. Also shown are the number of transcription factor binding sites and average signal of activating histone modifications at known and candidate regulatory regions of the episome. Furthermore, CTCF segments the genome into many more domains than previously recognized. (B) The lytic origin of replication is highly enriched in transcription factor occupancy. The locus acts as a divergent promoter for the lytic-associated BHLF1 and BHRF1 transcripts separated by a central CTCF binding site. (C) Histone modifications at the highly transcribed and multiply processed RPMS1 promoter. A peak for H3K4me3 was more proximal, while H3K4me1 was distal to the transcription start site. The histone variant H2A.Z can be seen at the −1 and +1 nucleosomes, and downstream histones are acetylated up to the CTCF site. (D) Nucleosome occupancy correlates with promoter silencing in different latency types. Nucleosomes have higher occupancy at Cp and Wp in type I latent cells (MutuI), where these promoters are transcriptionally silenced, relative to type III latent cells (LCLs). (E) RNA-seq reveals transcript isoforms of BZLF1 (top) and EBNA-LP joined to BHRF1 (bottom). The BZLF1 transcript can be alternatively spliced to exclude the coding region for the DNA binding domain, and multiple exons of EBNA-LP can act as donor splice sites to multiple acceptor splice sites in BHLF1. Percentages are given as total of all BZLF1 transcripts and EBNA-LP transcripts, as estimated from reads that cross alternative and canonical splice junctions. BHRF1 is also transcribed canonically from OriLyt. (F) The BHLF1 RNA transcript differs from the DNA template, suggesting RNA-editing by ADAR-mediated deamination. Position 40080 is templated to be an A but RNA-seq reveals a significant portion of reads across 170 distinct RNA-seq experiments contain a G (p < 1e-71, t-test; error bars show standard deviation). See also Figure S2.
Viral regulatory elements
We identified many candidate regulatory domains of the viral genome that were bound by transcriptional regulators and enriched in histone modifications. We analyzed binding profiles for over 60 human transcription factors, including factors involved in B cell development and viral response, such as Pax5, Irf3, Ebf1, and NFKB. We found that at least 26 transcription factors (TFs) cumulatively have at least 109 reproducible significant binding sites across the viral genome (Figure 2A, S2A, and Table S3). Furthermore, transcription factor binding sites cluster into regulatory loci that are reminiscent of “hotspot” regions identified in other model organisms(Gerstein et al., 2010; Moorman et al., 2006; Roy et al., 2010). Example clusters occurred at promoters for actively transcribed latent transcripts (Cp, RPMS1), as well as at the left lytic origin of replication (OriLytL). The divergent promoters at OriLytL are partitioned into two major clusters of binding factors (Figure 2B). The leftward transcript for BHLF1 has the highest RNA polymerase II peak in the viral genome over its initiation site, which also overlaps with known polymerase cofactors TBP and TAF1. The rightward promoter for BHRF1 (and associated viral miRNAs) contains a peak enriched with several factors, including p300, Gcn5, Bcl3, Pbx3, Egr1, Brca1, cFos and Chd2. We ensured that this locus did not have enrichment for IgG or input DNA sequencing controls and that the overlap was significant (Figures S2B and S2C). The prominent binding of the insulator protein CTCF separates these independent binding sites and their respective divergent promoters, consistent with prior findings (Tempera et al., 2010) (Figure S2D–F).
Histone modifications and chromatin boundary factors, like CTCF, are known to contribute to EBV latency regulation but have never been assayed at high resolution(Tempera et al., 2010). The highest measured histone modifications aggregated to a peak at the RPMS1 promoter region, which is the transcription initiation site for a cluster of EBV miRNAs and non-coding RNAs (Figure 2C). CTCF binds downstream of this promoter and appears to serve as a boundary for high-level promoter proximal histone modifications H3K27ac and H3K4me3 (Figure 2C). We also mapped nucleosome positions in two cell lines with different viral latency programs. Nucleosomes strongly occupied the transcriptionally silent Cp promoter in type I Burkitt lymphoma cell lines (which fail to express EBNA2 family genes), while nucleosomes did not occupy the transcriptionally active Cp in type III LCLs (which do express EBNA2 family genes) (Figure 2D). These findings are consistent with reports that active genes have nucleosome free promoter regions (Zhou et al., 2005).
Viral transcriptome
The EBV genome is a complex patchwork of densely packed, overlapping, and extensively alternatively spliced genes. As expected in LCLs, the coding and transcribed non-coding domains of the EBV genome were strongly enriched for RNA-seq reads mapping to the latency transcripts for the EBNA genes, latent membrane proteins, RPMS1, as well as the non-coding RNAs, and miRNAs (Table S4). Less expectedly, we detected very high read counts covering the BHLF1 transcript, which is typically associated with early stages of lytic cycle gene activation and is transcribed from the lytic origin of replication (OriLyt). We also detected other immediate early transcripts such as BZLF1 and BRLF1, though these were expressed at much lower levels. We were also able to verify a recently reported viral-encoded snoRNA (Hutzinger et al., 2009).
We characterized all RNA splice junctions by analysis of paired end sequencing assays (Table S5). We found many known isoforms and confirmed several recently reported isoforms (Austin et al., 1988; Kelly et al., 2009; Lin et al., 2010). Using deep sequencing from hundreds experiments across multiple labs, we were able to detect dozens of isoforms, including alternative splicing in BZLF1, the BART locus, and multiple 5′ sites that spliced to acceptors in BHRF1 and BHLF1 (Figure 2E). The extensive usage of alternative donor and acceptor splice sites is remarkable and may be relevant for virus regulation and function.
RNA editing occurs in numerous host transcripts and was recently shown to also occur in viral RNAs (Iizasa et al., 2010; Li et al., 2011). Analysis of the many RNA-seq experiments revealed that the BHLF1 RNA transcript contains a guanine nucleotide that differs from the expected adenine residue encoded in the template DNA. This is consistent with classic ADAR-mediated deamination editing (Iizasa et al., 2010). Specifically, ~10% of RNA-seq reads (as averaged across 170 experiments with >10× coverage) mapping to this locus contain the alternative base whereas only 1 read in 1 DNA sequencing experiment (out of n=26 with >5× coverage) contained this alteration (Figures 2F and S2G–H). BHLF1 RNA has been implicated in the initiation of viral DNA replication, and RNA-editing may contribute to this process (Rennekamp and Lieberman, 2011).
Heterogeneity of viral gene expression and lytic reactivation in LCLs
In proliferating LCLs, the EBV genome persists predominantly as a type III latent infection, where a limited set of viral genes and miRNAs are expressed and the genome is replicated exclusively by host-cell replication machinery. However, all LCLs contain subpopulations of cells undergoing various degrees of lytic cycle gene expression and replication, the extent of which varies among cell populations and culture conditions (Davies et al., 2010; Glaser et al., 1989).
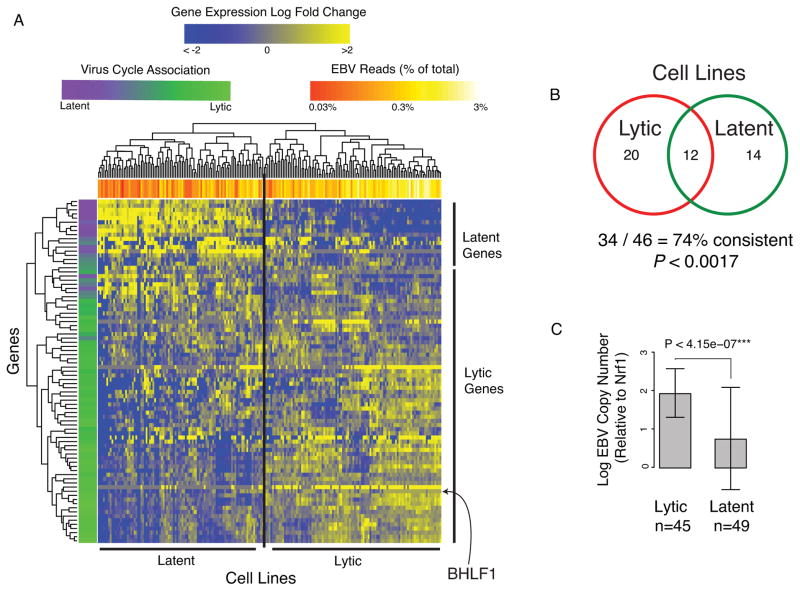
We analyzed the EBV gene expression profiles of over 300 LCLs and, after quality filtering, we clustered 201 viral expression profiles (Table S6 and Supplemental Material). The two main clusters correlated with canonical type III latent and lytic cycle patterns (Figures 3A, and S3). This clustering pattern was stable and consistent across multiple independent labs using independent LCL subclones (Figure 3B and S4). The gene expression clusters correlated with the average EBV load, as measured by the percentage of RNA-seq reads mapping to EBV (Figure 3A, top bar and Figure S3) and EBV episome copy number as assayed by an independent lab in independent subclones (Figure 3C)(Choy et al., 2008). Since lytic reactivation results in productive replication of the EBV episome, this correlation is an independent validation of lytic activity. However, we also observed disproportionately lower expression of EBV structural and packaging genes associated with infectious virion production (Table S4). This suggests that incomplete or abortive lytic gene reactivation occurs frequently in LCLs.
Figure 3. Lymphoblastoid cell lines cluster by viral reactivation propensity.
(A) We analyzed gene expression from 201 independent profiles of across 143 lymphoblastoid cell lines and found clustering into type III latency (upper left) or lytic (lower right) gene expression programs. The lytic cluster has a higher percentage of reads mapping to EBV than the latent cluster (bar along top of heatmap). The clustering of genes reveals association with latent and lytic cycle, which is quantified by the first principal component (bar to the left of heatmap; Supplemental Material). While many of the early lytic genes are actively expressed, very few of the late lytic genes required for virion formation are present, suggesting that most reactivation may be abortive. (B) Propensity for lytic reactivation persists in subclones. Forty-six cell line subclones were assayed in independent labs and have significantly similar latent and lytic viral expression profiles (P < 0.0017, binomial test; Supplemental Material). (C) EBV copy number, as assayed by qPCR in a separate lab from the RNA-seq experiments, correlates with lytic reactivation clustering and confirms the heritability of expression program. See also Figure S3.
B-cell proliferation pathways are coexpressed with viral lytic gene expression
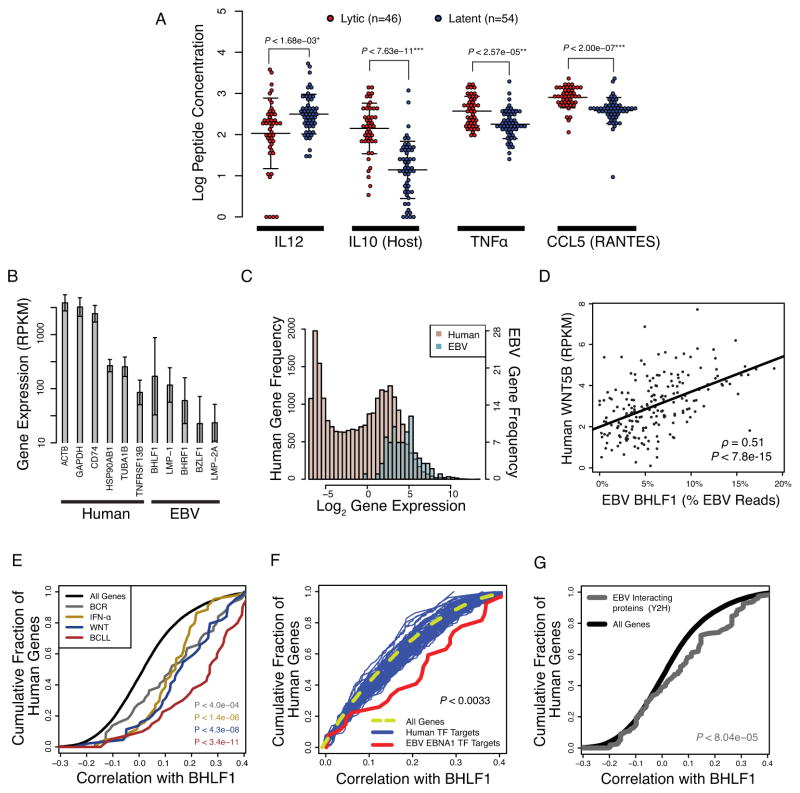
To better understand the dynamic intracellular environment during viral reactivation, we examined host gene expression and protein levels that correlate with lytic cycle. We correlated known cellular proteins, metabolic molecules, and surface markers with latent/lytic state. The cells lines with increased viral lytic cycle tended to have significantly higher ATP, more TNFα, more CCL5, less IL12, and more than double the levels of IL10 (Figures 4A and S4)(Choy et al., 2008). While these molecules are known B-cell regulatory factors, their role in lytic reactivation has been less well characterized.
Figure 4. Identification of host factors and genes that correlate with spontaneous lytic reactivation.
(A) Intercellular host cytokine concentrations, as measured by ELISA, are correlated with viral reactivation propensity (Holm corrected p-values, see Supplementary Material; error bars show standard deviation). (B) Specific EBV lytic and latent genes are expressed at similar levels relative to highly expressed human genes (error bars show standard deviation across the 201 samples). (C) The EBV transcriptome is expressed at similar levels to the human transcriptome. (D) The human WNT5B gene expression is highly correlated with the EBV lytic reactivation marker BHLF1. (E) Human genes that correlate with BHLF1 fall into distinct signaling pathways, including B cell receptor (BCR), interferon alpha (IFN-α), Wnt, and B cell chronic lymphocytic leukemia (BCLL) signaling pathways. These pathways represent an amalgamation of EBV-affected alterations and cellular anti-viral response mechanisms to increased viral load. (F) The viral transcription factor EBNA1 (EBV-encoded nuclear antigen 1) binds to genes that are activated during lytic reactivation. Correlation between BHLF1 and ChIP-seq derived high confidence targets of transcription factors is shown. (G) Host proteins that interact with EBV proteins have their RNA transcripts upregulated during lytic reactivation, as shown by increased correlation with BHLF1. See also Figure S4.
We next used RNA-seq data to simultaneously assay host and viral gene expression and identify cellular gene transcripts that correlate with viral lytic phenotypes (Tables S7, S8, and Figures S3). LCL RNA-seq was quantified using reads per kilobase of exon per million mapped sequence reads (RPKM) to compute a correlation between EBV lytic gene BHLF1 and human host genes. Many viral genes were expressed at surprisingly high levels relative to human transcripts (Figures 4B, C).
We found that EBV lytic gene expression correlated with many human transcripts, for example, the WNT5B transcript (Figure 4D and S4). Searching pathway databases (Subramanian et al., 2005), we found that several families of human genes were significantly correlated with EBV lytic gene expression, including pathways involved in B cell chronic lymphocytic leukemia, interferon-alpha (INFα), WNT, and B cell receptor signaling (Figure 4E; Supplemental Methods; Table S9). An aggregate analysis of all cellular genes bound by the viral transcription factor EBNA1, which is upregulated during lytic reactivation (Figure S4D), revealed a statistically significant increase in virally targeted genes during lytic viral expression, suggesting direct viral regulation of host genes (Figure 4F). We also found that cellular genes identified in two-hybrid protein interaction assays with EBV proteins as bait (Calderwood et al., 2007) were correspondingly upregulated with increased lytic gene expression (Figure 4G). Furthermore, we found a small subset of host genes that may be coregulated by multiple means. For instance, the WNT5B gene is bound by EBNA1, is a member of the WNT pathway, and is commonly dysregulated in human cancers, including chronic lymphocytic leukemia (Lu et al., 2004; Lu et al., 2010).
The host B-cell lineage regulator Pax5 represses viral oncogenes
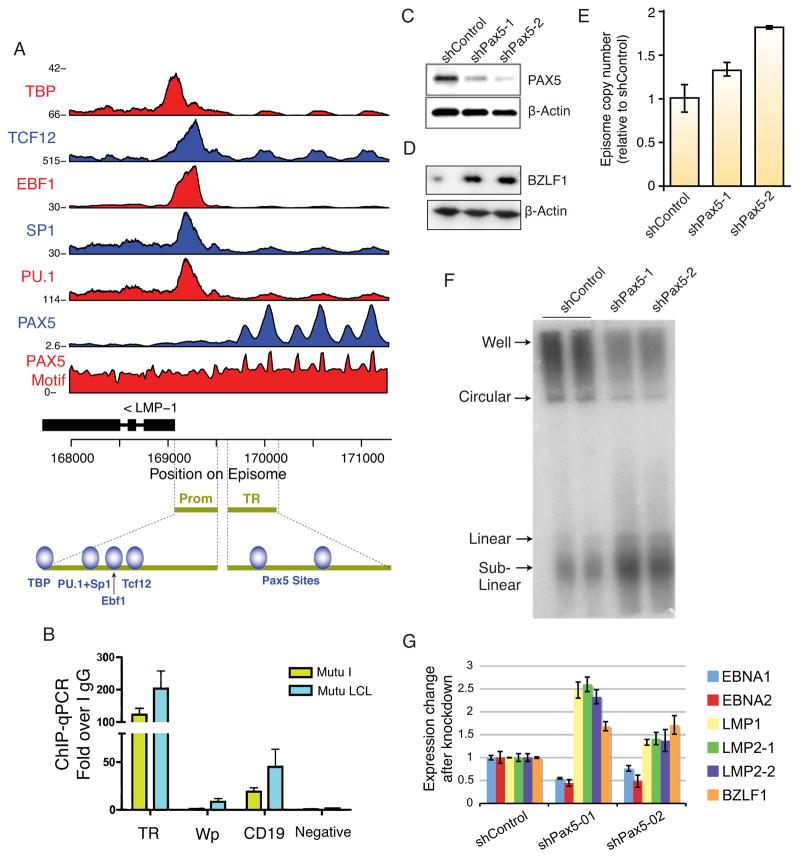
We found that Pax5 binds with high occupancy to the viral terminal repeats (Figure 5), which are known to regulate LMP1 through an unknown mechanism (Repic et al., 2010). Pax5 is a B-cell specific factor that plays an essential role in B-cell development and contributes to B-cell tropism of EBV latency through initial activation of the latency transcription program at Wp (Tierney et al., 2000; Tierney et al., 2007). ChIP-seq analysis reveals two major peaks of Pax5 in at least one, though possibly all, of the viral terminal repeats (Figure 5A). Pax5 binding was validated by real-time PCR with primers specific for terminal repeat (TR) DNA, and a Pax5 consensus motif was identified within the centers of the ChIP-seq peaks (Figures 5A, B, S5A). We also confirmed that Pax5 binds the TR in an alternative EBV strain (Mutu) and in both type I (MutuI) and type III (Mutu-LCL) latently infected cells (Figure 5B). Surprisingly, Pax5 binding at the TR did not overlap with RNA polymerase II initiation factors, suggesting that its function at the TR is more complex than proximal promoter transcription repression. The Pax5 site in the TR is distinct from the known binding sites for Pu.1 and Sp1 in the LMP1 promoter (Sjöblom et al., 1995), which are identified by ChIP-seq along with binding sites for Ebf1 and Tcf12, and are the known sites of EBNA2 transactivation.
Figure 5. Pax5 binds the viral terminal repeats and regulates viral oncogene expression.
(A) Pax5 and other transcriptional regulators bind the LMP control region. Pax5 binds the terminal repeats (TR) directly at two sites, as supported by ChIP-seq and sequence motif. In the promoter, EBF1, PU.1 and SP1 were previously documented to co-bind a GC-box element and regulate LMP1, while TCF12 has not been reported previously. (B) Pax5 binds with high occupancy at the TR locus compared with two known binding sites (EBV Wp promoter and human CD19 promoter) as determined by ChIP-qPCR. We also confirm that Pax5 binds the TR in multiple latency types and EBV strains. MUTU presents type I latency and does not express LMP1, whereas MUTU-LCL presents type III latency and expresses LMP1. (C) Pax5 protein expression is depleted by two independent lentiviral shRNA constructs. Western blot of LCL infected with shPax5-1 or shPax5-2 lentivirus and assayed 5 days post-infection followed by puromycin selection. (D) Western blot of BZLF1 and actin in shPax5-1 or shPax5-2 infected LCLs. (E) Real-time qPCR analysis of EBV genome copy number relative to cellular actin. Values are normalized to shControl infected LCLs. Error bars are standard deviation from the mean (n=3). (F) PFGE analysis of Raji cells infected with shControl, shPax5-1, or shPax5-2 for 5 days post-infection and then analyzed by Southern blot analysis. Circular (C), linear (L), and sub-linear (SL) forms of the viral genome are indicated. (G) Knockdown of Pax5 induces LMP1 and LMP2, while decreasing expression of EBNA1 and EBNA2. Pax5 depletion also leads to an increase in the BZLF1 lytic activator. All transcripts are quantified by qRT-PCR and normalized to cellular actin. Error bars are standard deviation from the mean (n=3). See also Figure S5.
To explore the potential function of Pax5 in regulation of EBV gene expression, we depleted Pax5 protein from LCLs using lentivirus-delivered shRNA. We identified two different shRNA targeting sequences that effectively deplete Pax5 protein at five days post lentivirus infection and selection (Figure 5C). We first assayed the effect of Pax5 on EBV lytic activation by monitoring BZLF1 protein expression by Western blot (Fig. 5D) and EBV genome copy number by qPCR (Fig. 5E). We found that Pax5 depletion modestly (<2 fold) increased BZLF1 protein expression (Fig. 5D) and viral genome copy number (Fig. 5E). To assess the effect of Pax5 depletion on EBV genome configuration, we assayed both LCL and Raji cells for circularized and linear viral episomes by pulse-field gel electrophoresis (PFGE) (Fig. 5F). PFGE analysis indicated that Pax5 depletion had only marginal effects on EBV genome copy number in LCL (Fig. S5B). In contrast, a percentage of viral episomes were converted to linear and sublinear genomes in Raji cells (Fig. 5F). Since Raji are defective for viral lytic replication, these findings suggest that Pax5 may be important for maintaining the circular episomal form of the genome during latency.
To assess the effect of Pax5 depletion on viral transcription, we measured EBV gene expression for LMP1, LMP2 isoform 1 (LMP2-1), LMP2 isoform 2 (LMP2-2), EBNA1, EBNA2, as well as the lytic immediate early gene BZLF1. We found that Pax5 depletion led to the activation of LMP1 and both isoforms of LMP2 while decreasing EBNA1 and EBNA2 transcript accumulation (Figure 5G). Pax5 depletion also increased BZLF1 transcription, consistent with Western blotting results (Fig. 5D). These findings indicate that Pax5 contributes to the regulation of EBV transcription during latent infection.
Cohesin mediates DNA looping of the viral episome
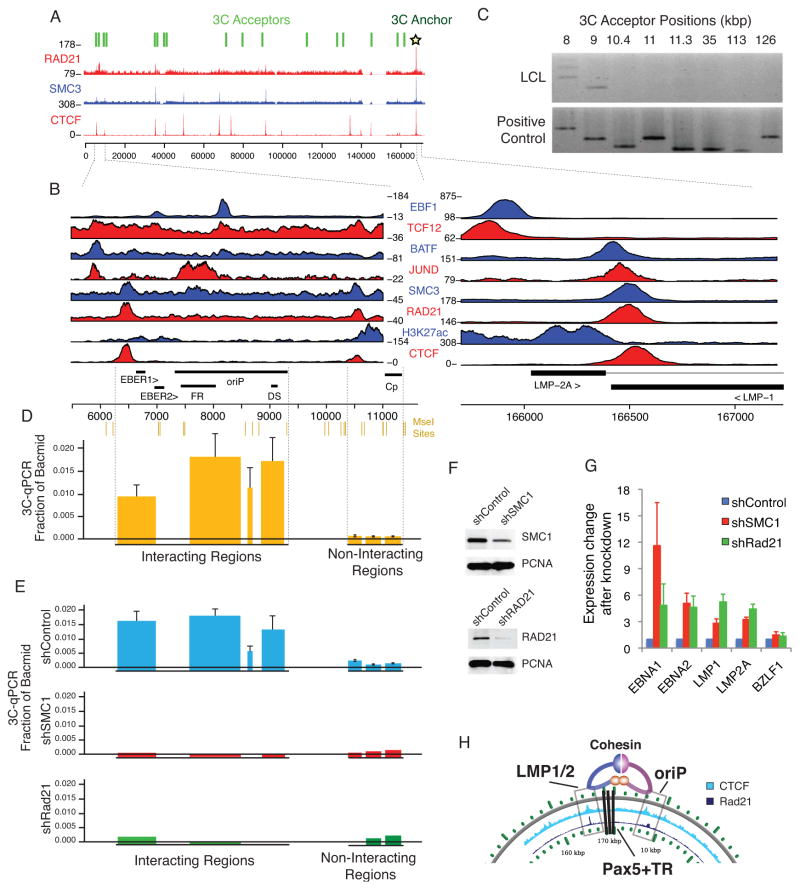
To further explore viral chromosome architecture, we analyzed ChIP-seq binding profiles for the chromatin structural factors CTCF, SMC3, and Rad21 and observed a pattern of colocalization (Figure 6A and S6). Most striking was the strong ChIP signal at the convergence of the LMP1 termination site and LMP2 first intron (Figure 6B). This positioning was highly reminiscent of the CTCF-cohesin site identified within the latency transcript of the KSHV genome(Stedman et al., 2008). Interestingly, Rad21 and SMC3 also colocalized at the origin of latent replication (OriP), which is known to be a central regulatory region for episome maintenance and has been shown to regulate LMP1 through a long-distance enhancer mechanism(Gahn and Sugden, 1995) (Figure 6B).
Figure 6. Cohesin regulates EBV chromosome conformation and latent cycle gene expression.
(A) ChIP-seq profiles for chromatin structure protein CTCF and Cohesin components (Smc3 and Rad21) demonstrate co-binding at several loci. Chromosome conformation capture (3C) anchor and acceptor primers (shown above) were used to determine DNA loop structures of the viral episome. (B) Co-occupancy of CTCF, Smc3, and Rad21 at LMP1 and OriP is accompanied by additional transcription factors JUND, BATF, TCF12, EBF1 and histone modifications. (C) Identification of a DNA loop between CTCF-cohesin at LMP1 (166.5kbp) and OriP (~6–9.5kbp). LCLs (top) subjected to 3C using MseI restriction digestion show specific interaction between oriP and 166.5kbp (anchor). Positive control (bottom) was performed on bacmid ligation products, where all permutations of acceptor sites and anchor are created. The assay is qualitatively specific to under 1kbp, as shown by the multiple negative regions proximal to OriP. (D) The long-range interaction is highly robust and specific. 3C-qPCR shows a highly specific quantitative interaction with the region surrounding OriP. Height of bars represents qPCR signal relative to positive control bacmid ligation products. Width of bars demonstrates the size of the MseI restriction enzyme fragment, which is also given at the top of the plot. Error bars show standard deviation from the mean (n=3). (E) Depletion of cohesin components through lentiviral shRNA results in loss of long-range interaction. 3C-qPCR as described in panel D, except LCLs were transduced with shControl (top panel), shSMC1 (middle panel), or shRad21 (lower panel). (F) Lentiviral shRNA constructs deplete protein expression for Smc1 and Rad21. (G) Depletion of cohesin components activates transcription of latency genes. Expression of EBNA1, EBNA2, LMP1, LMP2A, and BZLF1 mRNA was assayed by qRT-PCR in LCLs infected with shSmc1, shRad21, or shControl. mRNA expression is normalized to shControl. Error bars are standard deviation from the mean (n=3). (H) Model depicting a DNA-loop between the OriP and LMP loci, which is mediated by CTCF-cohesin binding sites and contains the Pax5 binding sites in the terminal repeats. Acronyms: OriP: latent origin of replication; FR: family repeats; DS: dyad symmetry repeats; EBER: EBV encoded RNA; Cp: BamHI C promoter. See also Figure S6.
As CTCF and cohesin have been implicated in long-distance DNA looping interactions, we used the chromatin conformation capture (3C) method to test whether the CTCF-cohesin sites physically linked OriP with the LMP1/LMP2 locus (Supplemental Methods; Table S10). Using anchor primers at the LMP1 locus and MseI digestion of the EBV genome, we found a strong 3C linkage formed between the anchor and positions centered around OriP (episome coordinates 6–9 kb) but not at several proximal control regions or other regions across the EBV genome (Figures 6C and 6D). These findings were corroborated by DNA sequencing of the PCR amplified junctions and by using different primer sets for real time qPCR.
To test whether cohesin subunits have any effect on EBV gene expression, we depleted SMC1 and Rad21 proteins using shRNA. LCLs were infected with lentivirus expressing Rad21 or SMC1 targeting shRNA and assayed by Western blot for knockdown efficiency (Figure 6F). Cohesin depletion led to a complete loss of the 3C loop formed between the OriP and LMP1/LMP2 control regions (Fig. 6E). qPCR analysis of EBV gene expression revealed that Rad21 or SMC1 depletion resulted in a general increase of latency transcripts, including LMP1 and LMP2 (Figure 6G). Cohesin depletion also led to a modest increase in BZLF1 expression. These results support the model that EBV chromatin forms higher order structures that include loop formation between OriP and the LMP1/LMP2 locus and help maintain viral latent cycle gene expression (Figure 6H).
Discussion
Our work integrates large genomic data sets into a comprehensive atlas of functional elements of a human pathogen. We used a strategy of large-scale computational analysis, hypothesis generation, and functional validation to dissect the complex factors implicated in the regulation of both virus and host genes. Our meta-analysis approach also reveals how large-scale projects such as ENCODE and HapMap can have far-reaching scientific impact in a field outside their original scope. We discovered examples of coordinated mechanisms of viral gene control through the combined analysis of nucleosome occupancy, histone modification patterning, combinatorial transcription factor binding, and long range DNA looping. Several of these mechanisms are reminiscent of those in the human genome and illustrate the functional similarity of a virus and its host.
We provide insights into host-virus interactions through combined genome-wide expression analyses. We aggregated hundreds of RNA-seq experiments and observed many cell lines displaying surprisingly high levels of early lytic cycle genes relative to late lytic genes. The correlation of this gene expression program with EBV copy number suggests that abortive lytic DNA replication may be an important mechanism for viral genome maintenance and host cell population fitness. Furthermore, analyses of virus and host transcriptomes revealed unexpected coordination between viral lytic reactivation and cellular pathways involved in B-cell expansion and tumor promotion. These findings are consistent with studies showing that viral lytic cycle gene expression correlates with EBV carcinogenesis in humans (Dardari et al., 2000; Hanto et al., 1983) and contributes to lymphomagenesis in a humanized mouse model (Ma et al., 2011).
We also examine host-virus interactions at individual regulatory loci and provide insights into the relationship between chromatin organization, gene regulation, and how developmental regulatory factors link these processes. The B-cell identity factor Pax5 was found to bind within the viral terminal repeats and modulate transcription of viral oncogenes from within the DNA loop connecting CTCF-cohesin peaks at LMP1 and OriP loci. These binding sites are reminiscent of a similar coordination of Pax5 and CTCF sites identified within the IgG locus, which function to direct class switching during B-cell maturation (Ebert et al., 2011). Pax5 mutations have been implicated in B-cell lymphomagenesis, which may result in dysregulation of LMP expression in certain EBV malignancies, including Hodgkins’s disease (McCune et al., 2006; Pasqualucci et al., 2001). Additionally, down-regulation of Pax5 during typical germinal center reaction may derepress LMP1 and LMP2, providing a mechanism for virus-mediated rescue from apoptosis of somatically hypermutated B cells that harbor tumorigenic translocations so commonly seen in EBV-associated malignancies(Bechtel et al., 2005; Klein and Dalla-Favera, 2008; Mancao et al., 2005; Roughan and Thorley-Lawson, 2009). These observations suggest that breakdown of host regulatory pathways for controlling EBV gene expression may contribute to tumorigenesis through dysregulation of viral oncogenes.
EBV was discovered as the first human tumor virus nearly fifty years ago and was among the first genomes sequenced. It is thus fitting that it serves as the prototype for functional genomic and epigenomic elements in a human tumor virus, providing a foundation for future work in viral genomics and the emerging field of systems virology (Peng et al., 2009). We anticipate that system-level and data-driven approaches will ultimately lead to more comprehensive models of viral persistence and its role in human cellular development and oncogenesis.
Experimental Procedures
Aligning reads to the viral and host genomes
Data was aggregated from a variety of sources (Table S1) and aligned to the EBV genome (Genbank accession NC_007605, March 2010) using the bowtie program allowing for one mismatch. We ignore all reads that aligned to the human genome, which ensured that all EBV reads are uniquely EBV and not actually from similar host sequence. To subtract human reads, we mapped all reads to hg19 (including all “Un” and “random” chromosomes), using the same parameters as when aligning to the viral genome. The subtraction of human-aligning reads resulted in small regions of the EBV-genome becoming unmappable (Figure S1).
Analysis of transcription factor binding sites
Transcription factor ChIP-seq peaks with significantly more reads than IgG and input control sequencing experiments were identified. We modeled reads in the control experiments by a Poisson distribution and a p-value of 1.72e-7 was used as a cutoff for peak calling, which represents a genome-wide familywise error rate of 0.01. Validation of a subset of ChIP-seq peaks was performed as previously described (Tempera et al., 2010). Briefly, cells were fixed in 1% formaldehyde for 30 minutes and DNA was sonicated to between 200 and 350 basepairs. ChIP DNA was amplified on a ABI Prism 7000 using SYBR green chemistry.
Estimation of viral and human gene expression
TopHat and Cufflinks were used to align reads and quantify known transcripts (Trapnell et. al., 2010). Transcript abundances were normalized by Reads Per Kilobase per Million (RPKM). Human transcripts were taken from RefSeq annotations for hg19. Multimapping reads were allowed to map to 30 regions in the transcriptome before being ignored. RPKM values were normalized to account for multi-mapping reads. Gene abundance was estimated as the sum of all transcript isoforms. The per-million normalization was done relative only to reads mapping to EBV or human, since this ensured independence of transcript quantification in EBV and Human. Use of absolute transcript quantities (normalized to total alignable reads in both EBV and human) is used only when noted. RNA-seq experiments with low read-count (less than 1 million alignable human reads) samples were removed and technical replicates were averaged. From the original 294 RNA-seq assays, we obtained 201 reliable and independent samples.
Chromatin Conformation Capture
Chromatin Conformation Capture (3C) was performed as previously described (Hagège et al. 2007 and Tempera et. al. 2011) with minor modifications. Briefly, cells were put through a 70μm filter to obtain single cells. 10M cells were fixed in 1% formaldehyde for 30 minutes and the reaction was quenched using 0.125M glycine. Cells were centrifuged, resuspended in lysis buffer, and lysed for 10 min. Nuclei were collected and digested with 500 U of MseI restriction enzyme overnight at 37C. The digestion was halted by incubation at 65C in 1.6% SDS on a 1200RPM shaker. The sample was diluted 10-fold, followed by ligation reaction containing 100 U T4 DNA ligase for 4 hours at 16C and 45 min at room temperature. The sample was digested by 300 μgr of proteinase K at 65C overnight, followed by RNase treatment for 1hr at 37C. DNA was phenol-chloroform extracted and ethanol precipitated and analyzed using PCR, qPCR, and sequencing. As a control, the EBV bacmid was MseI digested and ligated, thus creating all possible ligation products at background concentrations.
Micrococcal Nuclease (MNase) Digestion
Nucleis were isolated from 5 × 107 Mutu and Mutu-LCL cells using a Dounce homogenizer in 4 ml Lysis Buffer (0.3 M sucrose, 2mM magnesium acetate, 3 mM CaCl2, 1% Triton X-100, and 10 mM HEPES (pH7.9)). The lysate was centrifuged through a glycerol cushion (25% glycerol, 5 mM magnesium acetate, 0.1 mM EDTA, and 10 mM HEPES (pH 7.4)) at 1000g for 15 min at 4°C. The nuclei were incubated with micrococal nuclease I (500 U/ml) at 37 °C for 20 min in 200 μl digestion buffer (25 mM KCl, 4 mM MgCl2, 1 mM CaCl2, 50 mM Tris (pH 7.4) and 12.5% glycerol). The reaction was stopped by equal volume of Stop Buffer (2% SDS, 0.2 M NaCl, 10 mM EDTA, 10 mM EGTA, 50 mM Tris (pH 8.0), and proteinase K (100ug/ml)) for 2 hours at 50°C. MNase I resistant DNA was extracted by phenolchloroform and ethanol precipitation. The ~150 bp mononucleosomal DNA was isolated from 1.5% agarose gel and purified by QIAquick Gel extraction kit (QIAGEN) following the manufacturer’s protocol. Purified DNA was then subject to Solexa sequencing using manufacturers recommendations (Illumina, Inc.).
Lentiviral delivery of shRNA
Lentiviral shRNAs were obtained from the Sigma-Aldrich MISSION shRNA library. 4 million Mutu-LCL cells were passed through a 40μm filter, spundown, and resuspended in 2ml lentivirus suspension. Cells were spun at 500g for 90 minutes at 25C. Cell were cultured at ~500K cells/ml for 5 days with media changes in 1μg/ml of puromycin. Knockdown was confirmed by Western blot and qRT-PCR.
Pulse field gel electrophoresis
To resolve large viral genomic DNA-fragments, pulse field gel electrophoresis was performed as described previously (Dheekollu and Lieberman, 2011). Briefly, DNA migrated for a duration of 23 hours at 14C with the pulse ramping linearly every 60–120s through 120C using a Bio-Rad CHEF Mapper.
Supplementary Material
HIGHLIGHTS.
The EBV transcriptome and epigenome were analyzed by systems approaches
Viral lytic genes are coexpressed with cellular cancer-associated pathways
B-cell-specific Pax5 protein regulates viral oncoprotein expression
CTCF-cohesin mediate long-distance DNA interactions important for latency regulation
Acknowledgments
This work was supported by funds from NIH awards CA085678, CA093606, and DE017336 to PML, NIH award HG006798 to CL, and a K99AI099153 award from the National Institute Of Allergy And Infectious Diseases to IT. We thank Dr. Louise Showe and Priyankara Wikramasinghe from the Wistar Institute Genomics and Bioinformatics facilities and acknowledge the support of the Wistar Institute Cancer Center Core grant P30 CA10815. We also thank Chris Wawak and Joanne Edington for technical support.
Footnotes
Supplemental Materials: This text is accompanied by supplemental tables, figures, and text. All experiment data and results are available for viewing and downloading at http://ebv.wistar.upenn.edu.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aderem A, Adkins J, Ansong C, Galagan J, Kaiser S, Korth M, Law L, McDermott J, Proll S, Rosenberger C, et al. A systems biology approach to infectious disease research: innovating the pathogen-host research paradigm. mBio. 2011;2:e00325–00310. doi: 10.1128/mBio.00325-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascherio A, Munger K. Epstein-barr virus infection and multiple sclerosis: a review. Journal of Neuroimmune Pharmacology. 2010;5:271–277. doi: 10.1007/s11481-010-9201-3. [DOI] [PubMed] [Google Scholar]
- Austin PJ, Flemington E, Yandava CN, Strominger JL, Speck SH. Complex transcription of the Epstein-Barr virus BamHI fragment H rightward open reading frame 1 (BHRF1) in latently and lytically infected B lymphocytes. PNAS. 1988;85:3678–3682. doi: 10.1073/pnas.85.11.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtel D, Kurth J, Unkel C, Küppers R. Transformation of BCR-deficient germinal-center B cells by EBV supports a major role of the virus in the pathogenesis of Hodgkin and posttransplantation lymphomas. Blood. 2005;106:4345–4350. doi: 10.1182/blood-2005-06-2342. [DOI] [PubMed] [Google Scholar]
- Bergbauer M, Kalla M, Schmeinck A, Göbel C, Rothbauer U, Eck S, Benet-Pagès A, Strom T, Hammerschmidt W. CpG-methylation regulates a class of Epstein-Barr virus promoters. PLoS Pathogens. 2010;6:e1001114. doi: 10.1371/journal.ppat.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Hewson I, Felts B, Mahaffy J, Nulton J, Salamon P, Rohwer F. Metagenomic analyses of an uncultured viral community from human feces. Journal of Bacteriology. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood M, Venkatesan K, Xing L, Chase M, Vazquez A, Holthaus A, Ewence A, Li N, Hirozane-Kishikawa T, Hill D, et al. Epstein-Barr virus and virus human protein interaction maps. PNAS. 2007;104:7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V, Nayak R, Wang I, Elwyn S, Cousins S, Morley M, Spielman R. Polymorphic cis- and trans-regulation of human gene expression. PLoS Biology. 2010;8:e1000480. doi: 10.1371/journal.pbio.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E, Yelensky R, Bonakdar S, Plenge R, Saxena R, De Jager P, Shaw S, Wolfish C, Slavik J, Cotsapas C, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genetics. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardari R, Khyatti M, Benider A, Jouhadi H, Kahlain A, Cochet C, Mansouri A, El Gueddari B, Benslimane A, Joab I. Antibodies to the Epstein-Barr virus transactivator protein (ZEBRA) as a valuable biomarker in young patients with nasopharyngeal carcinoma. International Journal of Cancer. 2000;86:71–75. doi: 10.1002/(sici)1097-0215(20000401)86:1<71::aid-ijc11>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Davies M, Xu S, Lyons-Weiler J, Rosendorff A, Webber S, Wasil L, Metes D, Rowe D. Cellular factors associated with latency and spontaneous Epstein-Barr virus reactivation in B-lymphoblastoid cell lines. Virology. 2010;400:53–67. doi: 10.1016/j.virol.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Dheekollu J, Lieberman P. The Replisome Pausing Factor Timeless Is Required for Episomal Maintenance of Latent Epstein-Barr Virus. Journal of Virology. 2011;85:5853–5863. doi: 10.1128/JVI.02425-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresang L, Vereide D, Sugden B. Identifying sites bound by Epstein-Barr virus Nuclear Antigen 1 (EBNA1) in the human genome: Defining a position-weighted matrix to predict sites bound by EBNA1 in viral genomes. Journal of Virology. 2009;83:2930–2940. doi: 10.1128/JVI.01974-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MD, Murali TM, Sobral BW. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathogens. 2008;4:e32. doi: 10.1371/journal.ppat.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. The distal VH gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- ENCODE Consortium T. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore P. Clonal Integration of a Polyomavirus in Human Merkel Cell Carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahn TA, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. Journal of Virology. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser R, Tarr K, Dangel A. The transforming prototype of epstein-barr virus (B95-8) is also a lytic virus. International Journal of Cancer. 1989;44:95–100. doi: 10.1002/ijc.2910440118. [DOI] [PubMed] [Google Scholar]
- Günther T, Grundhoff A. The epigenetic landscape of latent Kaposi Sarcoma-associated herpesvirus genomes. PLoS Pathogens. 2010;6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nature Protocols. 2007 Jul;2(7):1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- Hanto DW, Gajl-Peczalska KJ, Frizzera G, Arthur DC, Balfour HH, McClain K, Simmons RL, Najarian JS. Epstein-Barr virus (EBV) induced polyclonal and monoclonal B-cell lymphoproliferative diseases occurring after renal transplantation. Clinical, pathologic, and virologic findings and implications for therapy. Annals of Surgery. 1983;198:356–369. doi: 10.1097/00000658-198309000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutzinger R, Feederle R, Mrazek J, Schiefermeier N, Balwierz P, Zavolan M, Polacek N, Delecluse HJ, Hüttenhofer A. Expression and Processing of a Small Nucleolar RNA from the Epstein-Barr Virus Genome. PLoS Pathogens. 2009;5:e1000547. doi: 10.1371/journal.ppat.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizasa H, Wulff BE, Alla N, Maragkakis M, Megraw M, Hatzigeorgiou A, Iwakiri D, Takada K, Wiedmer A, Showe L, et al. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. The Journal of Biological Chemistry. 2010;285:33358–33370. doi: 10.1074/jbc.M110.138362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium T. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Research. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak S, Habegger L, Rozowsky J, Shi M, Urban A, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly G, Long H, Stylianou J, Thomas W, Leese A, Bell A, Bornkamm G, Mautner J, Rickinson A, Rowe M. An Epstein-Barr virus anti-apoptotic protein constitutively expressed in transformed cells and implicated in Burkitt lymphomagenesis: The Wp/BHRF1 link. PLoS Pathogens. 2009;5:e1000341. doi: 10.1371/journal.ppat.1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nature Reviews Immunology. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Lee BK, Bhinge A, Iyer V. Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucleic Acids Research. 2011;39:3558–3573. doi: 10.1093/nar/gkq1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang I, Li Y, Bruzel A, Richards A, Toung J, Cheung V. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman P. Chromatin regulation of virus infection. Trends in Microbiology. 2006;14:132–140. doi: 10.1016/j.tim.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Lin Z, Xu G, Deng N, Taylor C, Zhu D, Flemington E. Quantitative and qualitative RNA-seq-based evaluation of Epstein-Barr virus transcription in type I latency Burkitt’s lymphoma cells. Journal of Virology. 2010;84:13053–13058. doi: 10.1128/JVI.01521-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, Sugden B. The plasmid replicon of Epstein-Barr virus: mechanistic insights into efficient, licensed, extrachromosomal replication in human cells. Plasmid. 2007;58:1–12. doi: 10.1016/j.plasmid.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Zhao Y, Tawatao R, Cottam H, Sen M, Leoni L, Kipps T, Corr M, Carson D. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. PNAS. 2004;101:3118–3123. doi: 10.1073/pnas.0308648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Wikramasinghe P, Norseen J, Tsai K, Wang P, Showe L, Davuluri R, Lieberman P. Genome-wide analysis of host-chromosome binding sites for Epstein-Barr virus Nuclear Antigen 1 (EBNA1) Virology Journal. 2010;7:262. doi: 10.1186/1743-422X-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SD, Hegde S, Young K, Sullivan R, Rajesh D, Zhou Y, Jankowska-Gan E, Burlingham W, Sun X, Gulley M, et al. A new model of Epstein-Barr virus infection reveals an important role for early lytic viral protein expression in the development of lymphomas. Journal of Virology. 2011;85:165–177. doi: 10.1128/JVI.01512-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancao C, Altmann M, Jungnickel B, Hammerschmidt W. Rescue of “crippled” germinal center B cells from apoptosis by Epstein-Barr virus. Blood. 2005;106:4339–4344. doi: 10.1182/blood-2005-06-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune RC, Syrbu SI, Vasef MA. Expression profiling of transcription factors Pax-5, Oct-1, Oct-2, BOB.1, and PU.1 in Hodgkin’s and non-Hodgkin’s lymphomas: a comparative study using high throughput tissue microarrays. Mod Pathol. 2006;19:1010–1018. doi: 10.1038/modpathol.3800622. [DOI] [PubMed] [Google Scholar]
- Montgomery S, Sammeth M, Gutierrez-Arcelus M, Lach R, Ingle C, Nisbett J, Guigo R, Dermitzakis E. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nature Reviews Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman C, Sun LV, Wang J, de Wit E, Talhout W, Ward LD, Greil F, Lu XJ, White KP, Bussemaker HJ, et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. PNAS. 2006;103:12027–12032. doi: 10.1073/pnas.0605003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin D. The global health burden of infection-associated cancers in the year 2002. International Journal of Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Küppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- Peng X, Chan E, Li Y, Diamond D, Korth M, Katze M. Virus-host interactions: from systems biology to translational research. Current Opinion in Microbiology. 2009;12:432–438. doi: 10.1016/j.mib.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J, Marioni J, Pai A, Degner J, Engelhardt B, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard J. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinney JW, Dickerson JE, Fu W, Sanders-Beer BE, Ptak RG, Robertson DL. HIV-host interactions: a map of viral perturbation of the host system. AIDS. 2009;23:549–554. doi: 10.1097/QAD.0b013e328325a495. [DOI] [PubMed] [Google Scholar]
- Ramagopalan S, Heger A, Berlanga A, Maugeri N, Lincoln M, Burrell A, Handunnetthi L, Handel A, Disanto G, Orton SM, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: Associations with disease and evolution. Genome Research. 2010;20:1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennekamp AJ, Lieberman PM. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. Journal of Virology. 2011;85:2837–2850. doi: 10.1128/JVI.02175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repic A, Shi M, Scott R, Sixbey J. Augmented latent membrane protein 1 expression from Epstein-Barr virus episomes with minimal terminal repeats. Journal of Virology. 2010;84:2236–2244. doi: 10.1128/JVI.01972-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly F, Heath A, Rohwer F, Gordon J. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickinson AB, Kieff E. Fields Virology. Lippincott Williams & Wilkins; 2007. Epstein-Barr virus; pp. 2655–2700. [Google Scholar]
- Roughan JE, Thorley-Lawson DA. The intersection of Epstein-Barr virus with the germinal center. Journal of Virology. 2009;83:3968–3976. doi: 10.1128/JVI.02609-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom A, Jansson A, Yang W, Laín S, Nilsson T, Rymo L. PU box-binding transcription factors and a POU domain protein cooperate in the Epstein-Barr virus (EBV) nuclear antigen 2-induced transactivation of the EBV latent membrane protein 1 promoter. J Gen Virol. 1995;76(Pt 11):2679–2692. doi: 10.1099/0022-1317-76-11-2679. [DOI] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha V, Mukherjee S, Ebert B, Gillette M, Paulovich A, Pomeroy S, Golub T, Lander E, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadmor AD, Ottesen EA, Leadbetter JR, Phillips R. Probing individual environmental bacteria for viruses by using microfluidic digital PCR. Science. 2011;333:58–62. doi: 10.1126/science.1200758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempera I, Wiedmer A, Dheekollu J, Lieberman P. CTCF prevents the epigenetic drift of EBV latency promoter Qp. PLoS Pathogens. 2010;6:e1001048. doi: 10.1371/journal.ppat.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley-Lawson DA, Gross A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N Engl J Med. 2004;350:1328–1337. doi: 10.1056/NEJMra032015. [DOI] [PubMed] [Google Scholar]
- Tierney R, Kirby H, Nagra J, Rickinson A, Bell A. The Epstein-Barr Virus Promoter Initiating B-Cell Transformation Is Activated by RFX Proteins and the B-Cell-Specific Activator Protein BSAP/Pax5. Journal of Virology. 2000;74:10458–10467. doi: 10.1128/jvi.74.22.10458-10467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney R, Nagra J, Hutchings I, Shannon-Lowe C, Altmann M, Hammerschmidt W, Rickinson A, Bell A. Epstein-Barr Virus Exploits BSAP/Pax5 To Achieve the B-Cell Specificity of Its Growth-Transforming Program. J Virol. 2007;81:10092–10100. doi: 10.1128/JVI.00358-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, Maglinte D, Lee S, Lee HR, Wong LY, Brulois K, Lee S, Buckley J, Laird P, Marquez V, et al. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathogens. 2010;6:e1001013. doi: 10.1371/journal.ppat.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams B, Pertea G, Mortazavi A, Kwan G, van Baren M, Salzberg S, Wold B, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Fewell C, Taylor C, Deng N, Hedges D, Wang X, Zhang K, Lacey M, Zhang H, Yin Q, et al. Transcriptome and targetome analysis in MIR155 expressing cells using RNA-seq. RNA. 2010;16:1610–1622. doi: 10.1261/rna.2194910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nature Reviews Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- Zhou J, Chau C, Deng Z, Shiekhattar R, Spindler MP, Schepers A, Lieberman P. Cell cycle regulation of chromatin at an origin of DNA replication. The EMBO Journal. 2005;24:1406–1417. doi: 10.1038/sj.emboj.7600609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.