Abstract
DNA cytosine methylation is a widespread epigenetic mark. Biological effects of DNA methylation are mediated by the proteins that preferentially bind to 5-methylcytosine (5mC) in different sequence contexts. Until now two different structural mechanisms have been established for 5mC recognition in eukaryotes; however, it is still unknown how discrimination of the 5mC modification is achieved in prokaryotes. Here we report the crystal structure of the N-terminal DNA-binding domain (McrB-N) of the methyl-specific endonuclease McrBC from Escherichia coli. The McrB-N protein shows a novel DNA-binding fold adapted for 5mC-recognition. In the McrB-N structure in complex with methylated DNA, the 5mC base is flipped out from the DNA duplex and positioned within a binding pocket. Base flipping elegantly explains why McrBC system restricts only T4-even phages impaired in glycosylation [Luria, S.E. and Human, M.L. (1952) A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol., 64, 557–569]: flipped out 5-hydroxymethylcytosine is accommodated in the binding pocket but there is no room for the glycosylated base. The mechanism for 5mC recognition employed by McrB-N is highly reminiscent of that for eukaryotic SRA domains, despite the differences in their protein folds.
INTRODUCTION
Modification of genomic DNA by methylation is an important epigenetic signal. In mammals and other vertebrate, DNA methylation occurs at the C5 position of cytosine, resulting in 5-methylcytosine (5mC), mostly within CpG dinucleotides (1,2). In plants DNA methylation is found in the symmetric CG and CHG contexts (where H = A, T or C) and the asymmetric CHH context (3). Since 5mC is related to repressed chromatin state and inhibition of transcription (1,2), an establishment and maintenance of the methylation pattern is of key importance for the cell functions. To interpret DNA methylation status proteins must faithfully discriminate between the non-modified C and the 5mC bases that differ only by a single methyl group. Structural studies revealed that eukaryotic methyl-DNA-binding proteins employ two different strategies for the 5mC recognition. The methyl-CpG-binding domain from the transcriptional repressor MeCP2 recognizes the specific hydration pattern of methylated DNA rather than cytosine methylation by itself (4). The SRA domain from the mammalian UHRF1 employs a radically different strategy: it flips out the 5mC of the DNA duplex and positions the extruded base within the protein pocket for discrimination (5–7). The SRA domain from the Arabidopsis SUVH5 follows a dual flip-out mechanism and extrudes both the methylated base and neighbouring base in the complementary DNA strand (8).
The evidence from comparative genomics suggests that mechanisms of 5mC discrimination may have emerged in the bacterial world as a result of on-going arms race between viruses and bacteria (9). In bacteria, methylated DNA bases are employed by restriction–modification systems to discriminate between self and foreign epigenetic modification patterns (10,11). The lack of the specific methyl-tag in the foreign DNA which enters the cell serves as a signal that triggers the restriction endonuclease cleavage. To protect their DNA from cleavage bacteriophages acquired an ability to incorporate modified bases such as 5mC and 5′-hydroxymethylcytosine (5hmC) in their genomes during DNA replication (12). It is thought that bacteria responded by developing methylation-dependent restriction endonucleases Mcr (for Modified cytosine restriction) to cut the modified foreign DNA [see (10) for a review]. To obtain resistance against the Mcr cleavage, T4 even phages extended C base modification through the glycosylation of the 5hmC residues (13–15). Actually, the first phage restriction phenomenon observed in the studies of T-even phage mutants impaired in glycosylation (16) was due to the Mcr cleavage of 5mC/5hmC-containing viral DNA. In Escherichia coli K12 strain there are at least two chromosomally encoded restriction endonucleases that act on the 5mC/5hmC containing DNA: McrA and McrBC (17,18). In contrast to the eukaryotic methyl-DNA-binding proteins, structural mechanisms leading to the 5mC/5hmC recognition by Mcr systems and other bacterial endonucleases acting on the methylated DNA (19) are as yet unknown.
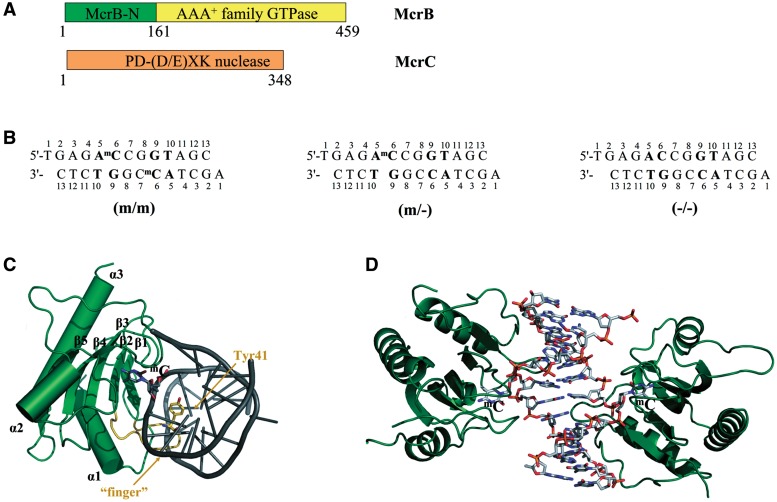
To understand the structural mechanisms of the 5mC/5hmC recognition in prokaryotes, we focused on the E. coli McrBC (18,20). The McrBC restriction system consists of two subunits: McrB (53 kDa) and McrC (40.5 kDa) (21) (Figure 1A). The nuclease active site resides in McrC (22), while McrB is responsible for DNA binding and GTP hydrolysis (20,23). The C-terminal part of McrB harbours the signature motifs of AAA+ (ATPases associated with various cellular activities) protein family (24,25). As most of AAA+ family proteins which usually function as ring-shaped oligomers (26), McrB was shown to assemble into heptameric ring structures and tetradecamers (27). DNA cleavage by McrBC requires GTP and occurs between two well-separated (30- to 3000-bp apart) recognition sites, 5′-RmC (where R = A or G) triggered by encounter of two DNA-translocating McrBC complexes (18,28,29). While two recognition elements at two well-separated locations are required for cleavage, McrB can bind substrates with recognition elements at a single location (30,31). The domain responsible for recognition of methylated DNA was shown to reside in the N-terminal part (residues 1–161) of the McrB subunit (Figure 1A) (30,32).
Figure 1.
(A) Schematic representation of the McrBC restriction system. McrBC is composed of two subunits: McrB harbours an N-terminal DNA-binding domain (McrB-N) and GTP-ase motifs, while McrC contains a nuclease active site (20,22,23). (B) The oligoduplexes used for crystallization. The 5′-AmC (5′-AC) sequences are shown in bold. (C) Closer view of the McrB-N monomer bound to DNA. The ‘finger’ loop penetrating into the minor groove is highlighted in orange. (D) Overall structure of the two McrB-N monomers bound to the di-methylated (m/m) oligoduplex. The DNA is bent by ∼29° and both the 5-methylcytosines are flipped out.
In this report we present the crystal structure of the McrB N-terminal domain (McrB-N) in complex with di-methylated, hemi-methylated and non-methylated DNA. To our knowledge it represents the first structure of a prokaryotic 5mC-binding domain in the DNA-bound form.
MATERIALS AND METHODS
Protein expression and purification
The region coding for the 1–161 residues of McrB was PCR-amplified from the pBBI/McrB template, introducing a hexahistidine tag at the C-terminus, and inserted into the pBAD24 vector. The McrB-N protein was expressed in E. coli ER2267 cells grown in the LB medium. The expression was induced by cultivation for 3–4 h at 37°C in presence of 0.2% arabinose. The cells were re-suspended in a buffer A [20 mM sodium phosphate (pH 7.4), 0.5 M NaCl] and sonicated. The soluble fraction was loaded on a HiTrap chelating column (GE Healthcare) and eluted by an imidazole gradient. The target protein was dialysed against a buffer B [10 mM potassium phosphate (pH 7.0), 0.05 M KCl, 1 mM EDTA, 7 mM 2-mercaptoethanol, 10% (v/v) glycerol] and loaded on a heparin–sepharose (GE Healthcare) column. The final preparation was stored at −20°C in a storage buffer [10 mM Tris–HCl (pH 7.5 at 25°C), 0.2 M KCl, 0.1 mM EDTA, 1 mM DTT, 50% (v/v) glycerol].
Crystallization and data collection
McrB-N was concentrated in a buffer C [10 mM Tris–HCl (pH 7.5 at 25°C), 0.2 M KCl, 0.1 mM EDTA, 1 mM DTT, 0.02% NaN3] and mixed with the oligoduplex (Figure 1B). The final concentrations of protein and DNA were 375 and 206 µM, respectively. Crystals were grown at 19°C by the sitting drop vapour diffusion method. The crystallization conditions for each complex are presented in Supplementary Table S1. For data collection, the crystals were transferred into a reservoir solution supplemented with 25% PEG400 for 30 min prior to flash cryo-cooling at 100 K. A Pt-derivative was obtained by soaking in 10 mM of K2PtCl4 for 1 h. The diffraction data (Supplementary Table S1) were collected at the EMBL beamlines at the DORIS storage ring, DESY, Hamburg or on a rotating anode source. The MOSFLM (33), SCALA (34) and TRUNCATE (35) programs were used to process the data.
Structure determination
The structure was solved by combining MAD phases (Pt-peak and Pt-infl datasets, Supplementary Table S1) with the SIRAS phases (Pt-peak and dataset-III, Supplementary Table S1) by SIGMAA (36). FOM for the combined phases before density modification was 0.62. The MAD phases were calculated using the SHELXD and SHELXE programs (37), the SIRAS phasing was carried out via Auto-Rickshaw pipeline (38). The experimental map at 2.7 Å showed clear protein-solvent boundaries and several secondary structure elements. The phases were improved by NCS-averaging using the DM program (36).
The manually built partial model of the Pt-derivative was used as a search model for molecular replacement to solve the native structure (dataset-I, Supplementary Table S1), as the datasets were non-isomorphous. After molecular replacement followed by NCS-averaging, a high quality map at 2.1 Å was obtained. This allowed automatic rebuilding of most of the protein residues by ARP/wARP (39). The DNA was built manually. The structures of complexes containing hemi- or non-methylated DNA (dataset-II and dataset-IV, Supplementary Table S1) were solved by molecular replacement using the protein model as a search target. Molecular replacement was carried out with MOLREP (40); manual building was carried out with Coot (41); CNS (42) and REFMAC (43) were used for refinement. The quality of the structures was analysed with the MOLPROBITY (44) program. The figures were prepared using PyMol (The PyMOL Molecular Graphics System, Schrödinger, LLC) and NUCPLOT (45).
The refinement statistics for the final models are presented in the Table 1. The N-terminal methionine and several C-terminal residues including the hexahistidine tag are not visible in the structures. The electron densities for the DNA bases outside the central palindromic 5′-ACCGGT sequence were very poor and did not allow determine the preferred orientation of the asymmetric oligoduplex. Thus, two alternative conformations were modelled.
Table 1.
Refinement statistics
| Structure | McrB-N/ (m/m) | McrB-N/ (m/−) | McrB-N/ (−/−) |
|---|---|---|---|
| Resolution range (Å) | 35–2.1 | 19–2.2 | 39–2.7 |
| Number of reflections | 18102 | 16474 | 9412 |
| R-factor | 20.4 | 21.1 | 21.6 |
| Free R-factor (10% set) | 26.0 | 26.3 | 29.2 |
| Number of atoms | 3755 | 3597 | 3477 |
| Average B-factors (Å2) | 25.4 | 39.2 | 22.9 |
| R.m.s. deviations from ideal values: | |||
| Bonds (Å) | 0.008 | 0.008 | 0.005 |
| Angles (°) | 1.1 | 1.1 | 0.9 |
| Ramachandran analysis: | |||
| Favoured (%) | 96.0 | 96.7 | 98.0 |
| Allowed (%) | 100 | 100 | 100 |
Gel mobility shift assay for DNA binding
The oligoduplexes (Figure 1B) were radio-labelled using [γ-33P]ATP and T4 polynucleotide kinase (Fermentas). The DNA was incubated with McrB-N in a binding buffer [30 mM MES (pH 6.5), 30 mM histidine, 10% (v/v) glycerol, 0.2 mg/ml of bovine serum albumin] for 10 min at room temperature. Binding mixes contained 2 nM of the 33P-labelled oligoduplex, 100 nM of the unlabelled oligoduplex and 20–20 000 nM of the protein monomer. The samples were electrophoresed through 8% polyacrylamide gels in a running buffer [30 mM MES (pH 6.5), 30 mM histidine] for 2 h at 6 V/cm. The radio-labelled DNA was detected using a Cyclone Storage Phosphor System with the OptiQuant program.
Pyrrolo-dC fluorescence measurements
Steady-state fluorescence measurements were acquired in photon counting mode on a Fluoromax-3 (Jobin Yvon) spectrofluorometer equipped with Xe lamp. Sample temperatures were maintained at 25°C. Oligoduplexes p/p or p/m with pyrrolo-dC (PC) substitutions (Table 2) were purchased from IBA. Emission spectra (400–600 nm) were recorded at an excitation wavelength of λex = 350 nm with excitation and emission bandwidths of 5 nm. At least four scans were averaged for each spectrum. The samples contained 5 µM of McrB-N and 1 µM of DNA in 30 mM MES (pH 6.5), 30 mM histidine. A 5-fold excess of the protein was used to ensure the complete binding of the DNA. Control spectra used for the background subtraction corrections were collected under identical conditions except that duplex m/m-30 was used instead of the fluorescent DNA. The fluorescence emission value of the corrected spectrum was determined at the emission maximum for each sample.
Table 2.
Oligoduplexes used for fluorescence measurements
| Oligoduplex | Sequence |
|---|---|
| p/p | 5′-ATCTCTCTATTCApCCG GTATTCTCTCTTTC-3′ |
| 3′-TAGAGAGATAAGT GGCpCATAAGAGAGAAAG-5′ | |
| p/m | 5′-ATCTCTCTATTCApCCG GTATTCTCTCTTTC-3′ |
| 3′-TAGAGAGATAAGT GGCmCATAAGAGAGAAAG-5′ | |
| m/m-30 | 5′-ATCTCTCTATTCAmCCG GTATTCTCTCTTTC-3′ |
| 3′-TAGAGAGATAAGT GGCmCATAAGAGAGAAAG-5′ |
pC: pyrrolocytosine (pyrrolo-dC), mC: 5-methylcytosine.
RESULTS
To understand the molecular mechanism for the 5mC recognition by the McrB N-terminal domain we have solved three crystal structures of McrB-N in complex with di-methylated, hemi-methylated and non-methylated DNA, respectively. Since 5′-AmCCGGT sequence is bound by McrB-N more tightly than other 5′-RmC containing sequences (30), three different oligoduplexes containing the methylated (m/m and m/−) and non-methylated (−/−) 5′-ACCGGT sequence were used for crystallization experiments (Figure 1B). (i) In the di-methylated oligoduplex (m/m) the first C within the 5′-ACCGGT sequence is methylated in both strands to yield two 5′-AmC sites separated by 2 bp; (ii) in the hemi-methylated oligoduplex (m/−) the first C residue within the 5′-ACCGGT sequence is methylated only in one DNA strand to generate a single 5′-AmC site; (iii) in the oligoduplex (−/−) there are two non-methylated 5′-AC sites on the opposite DNA strands. McrB-N gave crystals belonging to the same P212121 group in presence of any of these DNA fragments. The structure of a platinum derivative of the McrB-N/hemi-methylated DNA complex was solved using both the anomalous and isomorphous signal (see ‘Experimental procedures’ section for more details). The structures of McrB-N bound to di-methylated, hemi-methylated and non-methylated DNA were solved by molecular replacement to the resolutions of 2.1, 2.2 and 2.7 Å, respectively. In all the structures there are two McrB-N molecules and one oligoduplex in the asymmetric unit. The protein components of the three independent structures are highly similar (r.m.s.d. of <1 Å when comparing protein monomers). Here we will describe the structure of McrB-N bound to the di-methylated DNA and will discuss the differences between the three structures.
Overall structure of protein–DNA complex
The N-terminal recognition domain of McrB folds into three α-helices and five β-strands with a topological order: α1(4–18), β1(33–39), β2(49–54), β3(63–71), β4(75–83), α2(102–111), β5(121–128) and α3(134–153) (Figure 1C). The five β-strands are arranged into a single anti-parallel β-sheet. The N-terminal α1 and the C-terminal α3 helices pack on the convex side of the β-sheet, while the concave side is oriented towards the DNA. Three loops that emanate from the β-sheet grip the DNA from the minor groove side. In the asymmetric unit there are two McrB-N monomers bound to 5′-AmC sites on different strands within di-methylated DNA (m/m) (Figure 1D). In crystal, these two McrB-N monomers do not contact each other. The DNA is heavily distorted, with both 5mC residues flipped out of the DNA duplex and positioned in the binding pockets within the protein. According to the analysis by the CURVES program (46), the minor groove is significantly wider in comparison to the canonical B-DNA, and the oligoduplex is bent by an overall angle of 29° towards the major groove. The loop β1β2(40–48), which we named ‘finger’, protrudes into the widened minor groove and the side chain of Tyr41 intercalates into the DNA base stack replacing the flipped out 5′-methylcytosine (Figure 1C).
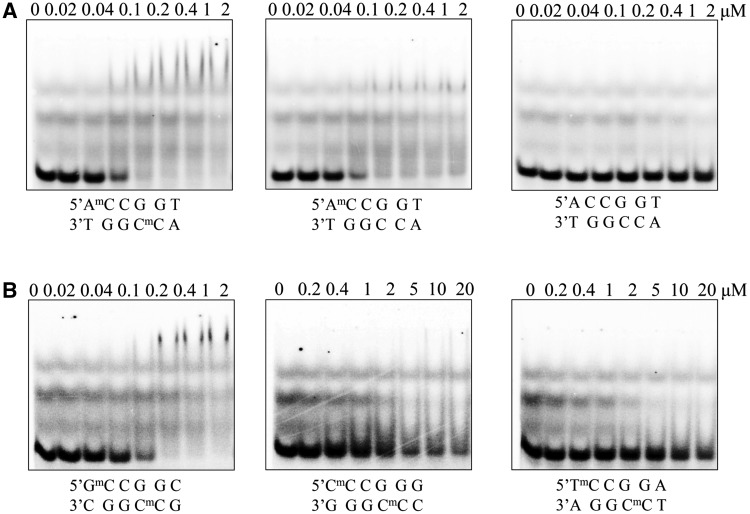
Both the structure of the protein and the conformation of the DNA are nearly identical in the complexes of McrB-N with hemi-methylated (m/−) and di-methylated (m/m) oligoduplexes (Supplementary Figure S1). In the case of the hemi-methylated oligoduplex (m/−) there are two McrB-N monomers bound to separate 5′-RmC/5′-RC sites, the DNA is bent, and both the methylated and non-methylated C bases are flipped out (we could not determine the preferred orientation of the oligoduplex (m/−) in the McrB-N–DNA complex, thus two alternative DNA conformations with the methyl-group positioned on different DNA strands were modelled). According to the gel-mobility shift assay, McrB-N binds the hemi-methylated (m/−) and di-methylated (m/m) oligoduplexes with the same affinity (Figure 2A). These results are in good agreement with the biochemical data that DNA methylated in one strand can be efficiently recognized by the McrB protein (18,20). The stoichiometry of the McrB-N–DNA complexes could not be determined in the gel-shift assay since the complexes were purely resolved.
Figure 2.
Gel mobility shift analysis for McrB-N binding to DNA. (A) McrB-N binding to the oligoduplexes used for crystallization. McrB-N at concentrations indicated above the relevant lanes was mixed with the di-methylated (m/m), hemi-methylated (m/−) or non-methylated (−/−) DNA (Figure 1B) at the final concentration of 100 nM. The samples were electrophoresed through 8% PAA gels under native conditions. (B) Dependence of the McrB-N binding on the sequence context 5′-upstream of 5mC McrB-N at concentrations indicated above the relevant lanes was mixed with the di-methylated (m/m) DNA containing either G, C or T nucleotide upstream of the 5mC. The samples were analysed as described in (A).
Surprisingly, in the structure of the McrB-N bound to the non-methylated oligoduplex (−/−) two cytosine residues adjacent to the A base are also flipped out. The overlay of the McrB-N structures bound to the di-methylated (m/m) and non-methylated (−/−) oligoduplexes does not show significant differences neither in protein nor in DNA conformation (Supplementary Figure S2). However, according to the gel mobility shift assay, the lack of the methyl group on the cytosine decreases the stability of the protein–DNA complex, since the McrB-N binding to the oligoduplex (−/−) is undetectable in the gel (Figure 2A). One cannot exclude that the complex between McrB-N and oligoduplex (−/−) is stabilized by crystal packing forces.
Protein–DNA interactions
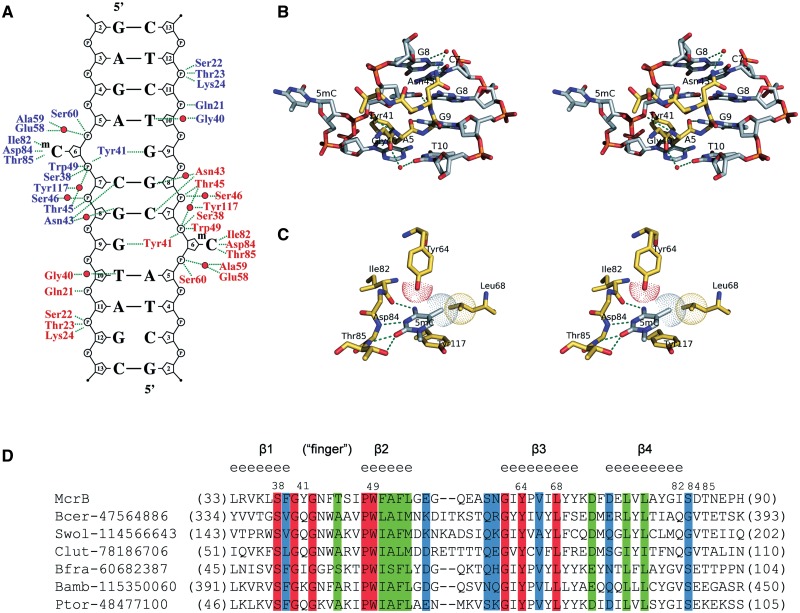
The contacts made by the two McrB-N monomers bound to the di-methylated oligoduplex (m/m) are identical (Figure 3A) therefore we will further discuss protein–DNA interactions made by one monomer. The McrB-N approaches DNA from the minor groove side (Figure 1D) and three neighbouring loops α1β1(19–32), β1β2(40–48) (‘finger’) and β2β3(55–62) emanating from the β-sheet contribute amino acid residues for protein–DNA interactions. Ser38, Thr45, Ser46, Trp49, Glu58, Ala59 and Ser60 make direct and water-mediated interactions with 3′- and 5′-phosphates of the extrahelical 5mC base, while the residues on the α1β1(19–32) loop (Gln21, Ser22, Thr23 and Lys24) contact the 3′- and 5′-phosphates of C11 on the opposite strand (Figure 3A). The only direct contacts to the bases in the DNA duplex are made by the Tyr41 and Asn43 residues. Asn43 is involved in the hydrogen bond interactions with the two C:G base-pairs interspaced between the two 5′-RmC sequences. The backbone N atom of Asn43 donates a hydrogen bond to the O2 atom of C7 while its side-chain interacts via a water-molecule with G8 (Figure 3B). Tyr41 occupies the space left by the flipped out 5mC, and its backbone O atom accepts a hydrogen bond from the amino group of the orphaned guanine G9 (Figure 3B).
Figure 3.
Detailed protein–DNA interactions in the McrB-N complex with di-methylated DNA. (A) Schematic representation of hydrogen bond interactions. Residues from different McrB-N monomers are shown in blue and red, water molecules are shown as red spheres. In the crystal there are two ways for the DNA duplex binding to McrB-N. Crystallographic quality parameters indicate that both binding modes are possible, and therefore both the DNA duplexes were modelled. Only one of two possible DNA orientations is shown in the picture. The protein–DNA interactions are almost identical in the McrB-N complexes with hemi-methylated and non-methylated DNA (Supplementary Figure S3). (B) Close-up stereo-view of the ‘finger’ loop penetrating into the minor DNA groove. (C) Close-up stereo-view of the McrB-N interactions with the flipped out 5mC. (D) Multiple sequence alignment of the McrB-N family proteins. NPS@ server (48) was used for analysis. Secondary structure for McrB-N is displayed above the sequence. Identical residues are indicated by red shading. Sequence names are composed of organism abbreviation and the GI number.
McrBC cuts DNA between two 5′-RmC sites separated by ∼30–2000 bp but it does not cleave at 5′-YmC sites (18,28). Thus, McrBC is able to discriminate purines versus pyrimidines 5′-upstream of the 5mC. Indeed, our data demonstrate that McrB-N affinity to the oligoduplexes containing 5′-AmC or 5′-GmC sites is ∼100 times higher in comparison to the oligoduplexes containing 5′-CmC or 5′-TmC sites (Figure 2B). Surprisingly, in the crystal structure there are no direct contacts to the A5 residue 5′-upstream of the 5mC residue. The Gly40 residue preceding the intercalating Tyr41 interacts via a water molecule with the A5 partner thymine T10 base (Figure 3B), but this contact does not explain the specificity for a purine residue in the vicinity of the 5mC. To test whether stacking interactions between the 5′-purine and aromatic ring of Tyr41 intercalating into DNA contribute to the target sequence discrimination by McrB-N, Tyr41 was replaced by site-directed mutagenesis by Ala and Gln which is present in McrB homologues (Figure 3D). The Tyr41Ala and Tyr41Gln mutants’ ability to bind methylated DNA was compromised (Supplementary Figure S4) indicating the importance of Tyr41 in McrB-N–DNA interactions. On the other hand, an indirect readout due to DNA susceptibility for deformation may also contribute to the Pu versus Py discrimination similarly to the EcoRV restriction enzyme (47).
Recognition of 5mC in the binding pocket
The extrahelical 5mC is positioned in the protein-binding pocket and adopts an anti-conformation. The walls of the binding pocket for 5mC are made by the side chains of Trp49, Leu68 and Tyr117 on one side and the side chains of Tyr64, Ala59, Ser60 and backbone atoms of 82–85 residues on the opposite side. The compromised DNA-binding activity of the Trp49Cys mutant (29) confirms the importance of the binding pocket residues in the McrB-N function. Tyr117 stacks with the flipped out base, while the residues Ile82, Asp84 and Thr85 are involved in hydrogen bonds interactions with the donor and acceptor atoms on the Watson–Crick edge of the extrahelical base (Figure 3C). More specifically, the O atom of Ile82 accepts a hydrogen bond from the exocyclic 4-amino-group; the backbone N of Asp84 donates a hydrogen bond to the N3 atom, and the backbone N and the side-chain OG atom of Thr85 are engaged in the hydrogen bond interactions with the O2 atom of 5mC. Since most of the hydrogen bond interactions are made by the backbone atoms of β4α2(83–102) loop, it is not surprising that Ile82, Asp84 and Thr85 residues are not conserved in the McrB-N homologues (Figure 3D). The direct hydrogen bond interactions in the binding pocket of McrB-N are compatible with C or m5C residues but exclude T, A and G bases. Discrimination between the extrahelical 5mC and C bases in the binding pocket is achieved by van der Waals interactions between the 5-methyl group and the side-chains of the Tyr64 and Leu68 residues (Figure 3C). The importance of these interactions is supported by the conservation of Tyr64 and Leu68 residues in protein sequences of putative McrB-N homologues (Figure 3D).
In the crystal structure of McrB-N bound to the non-methylated DNA, the cytosine is also flipped out of the base stack and accommodated in the binding pocket. However, due to the lack of the methyl group the extrahelical cytosine makes fewer contacts with protein atoms in comparison to 5mC. Van der Waals’ interactions with the methyl group of 5mC presumably make an important contribution to the McrB-N–DNA complex stability, since McrB-N complex with the non-methylated oligoduplex (−/−) is much less stable than complexes with the di- and hemi-methylated DNA.
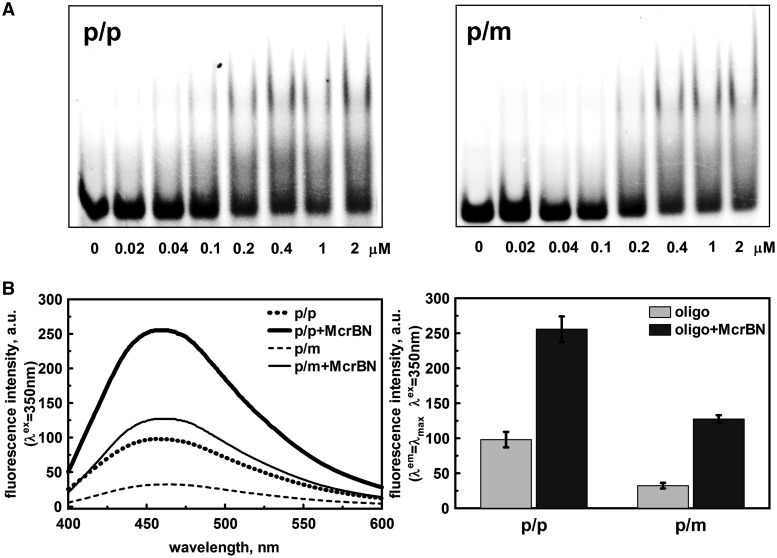
Base flipping in solution
The crystal structures revealed that two McrB-N monomers bind to opposite strands of the 5′-ACCGGT sequence and flip out methylated, hemi-methylated or non-methylated C bases in the vicinity of 5′-A. To probe whether base flipping by McrB-N occurs in solution, we used a fluorescent cytidine analogue, pyrrolo-dC (PC). The quantum yield of PC fluorescence is sensitive to base unstacking (49), therefore PC is used to probe DNA structural changes (50–52) similarly to 2-aminopurine (53,54). Incorporation of the PC instead of the first C base in the 5′-ACCGGT sequence in one or both DNA strands did not significantly disturb McrB-N binding (Figure 4A). Mixing of McrB-N with the p/p and p/m oligoduplexes, resulted in the 2.6- and ∼4-fold increase of the PC fluorescence at ∼460 nm, presumably due to the base unstacking occuring in solution upon McrB binding to PC containing DNA (Figure 4B). This finding is consistent with the nucleotide flipping observed in the crystal structure.
Figure 4.
McrB-N interaction with PC DNA. (A) McrB-N binding to the oligoduplexes containing one (p/m) or two PC (p/p) bases (Table 2). The binding reactions contained 33P-labelled 30-bp oligoduplex (100 nM) and the protein at concentrations as indicated by each lane. The samples were electrophoresed through 8% PAA gels under native conditions. (B) PC steady state fluorescence measurements in solution. Left panel: corrected PC emission spectra of McrB-N–DNA complexes. Reactions contained 5 μM protein and 1 μM oligoduplex p/p or p/m (see ‘Materials and Methods’ section for the details). Right panel: diagrams illustrate the maximum fluorescence intensity (456 nm for p/p, 459 nm for McrB-N–p/p, 466 nm for p/m and 463 nm for McrB-N–p/m) values of the corrected fluorescence emission spectra presented in left panel.
Structural similarity to other proteins
According to the DALI server search (55), McrB-N is most similar to 2a8e (z = 6.2), 2ffg (z = 6.0) and 3mgz (z = 5.5) structures, which are not yet functionally characterized. Therefore, it remains to be determined whether these proteins possess DNA-binding activity similar to McrB-N. The DALI search also identified another related structure: the E. coli protein yjbR (2fki, z = 4.9), which reportedly contains a ‘double wing’ DNA-binding motif. It is proposed that yjbR protein differently from the McrB-N interacts predominantly with the DNA major groove (56,57); however, the crystal structure of yjbR in the DNA bound form is required to support this structural model.
The recognition domain of the McrBC restriction endonuclease shows no structural similarity neither to the methyl-CpG-binding domain of MeCP2 (4) nor to the SRA domains from the mammalian UHRF1 and the Arabidopsis SUVH5 (5–8). Therefore, to our knowledge, McrB-N represents a third distinct fold that was adapted for the 5mC recognition.
DISCUSSION
Distinct fold but similar mechanism
Within eukaryotes there are three currently known distinct families of proteins that bind methylated DNA: the methyl-binding domain (MBD) family, Kaiso and Kaiso-like proteins, and SRA domain proteins (58). MBD family and SRA domain proteins have unrelated folds and mechanisms for 5mC recognition, while the structural characterization of Kaiso has not been yet reported. The structure of the McrB-N in complex with methylated DNA provides the first structural glimpse of a prokaryotic methylated DNA-binding domain in DNA-bound form. The key feature of the mechanism of 5mC recognition by the N-terminal domain of McrB is the base flipping.
The structure analysis shows that three different steps ensure a unique discrimination of the 5mC against other bases by McrB-N. First, the methylated cytosine is flipped out of the double helix and positioned in the well defined binding pocket within the protein. The size of the pocket pre-selects pyrimidines against purines as a purine base would not fit in. Second, direct readout of the donor and acceptor atoms on the Watson–Crick edge of the flipped out base discriminates cytosine against thymine. The pattern of the hydrogen bond donors and acceptors provided by the three amino acid residues within the binding pocket of McrB-N is compatible to that of C or 5mC but not of other heterocyclic bases in their typical conformation. Finally, van der Waals interactions of the conserved binding pocket residues Tyr64 and Leu68 residues with the methyl group discriminates 5mC against cytosine.
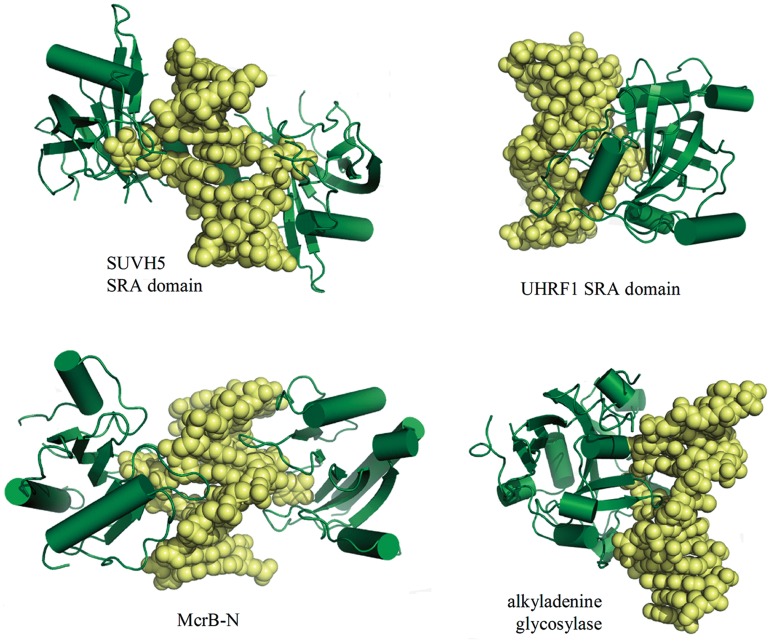
The protein fold of the McrB-N is distinct from those of eukaryotic methyl-CpG-binding domains of UHRF1 and SUVH5 (5–8) that flip out the methylated base. Surprisingly, despite of the differences in the protein fold and DNA sequence context, the mechanism of the 5mC discrimination by McrB-N show similarities to that of the SRA domains of mammalian UHRF1 (5–8) and Arabidopsis SUVH5 (Figure 5). Indeed, all three proteins flip out 5mC from DNA helix to achieve discrimination of 5mC against C. Furthermore, in all the cases the extrahelical base is accommodated in the protein pocket, which provides an interface for the direct readout of the donor and acceptor atoms on the Watson–Crick edge of the extrahelical C although different sets of amino acid residues are used for hydrogen bonding interactions. Finally, van der Waals interactions with the methyl group in the binding pocket play a key role in discriminating of the 5mC against C. Thus, despite of the distinct folds and different sequence context (AmC for McrB-N, mCG for mammalian UHRF1 and mCG/mCHG/mCHH for Arabidopsis SUVH5, respectively) both the prokaryotic McrB-N and eukaryotic methyl-binding SRA domains use base flipping mechanism for 5mC discrimination in the binding pocket although details still vary among different proteins. The SRA domain from mammalian UHRF1 binds to a hemimethylated CpG dinucleotide as a monomer (Figure 5) and flips the methylated cytosine into a binding pocket (5–7). The SRA domain from Arabidopsis SUVH5 binds to fully or hemimethylated CG, fully methylated CHG and methylated CHH, and hemimethylated CHG (8). In contrast to mammalian UHRF1, the plant SUVH5 domain follows a double-flip mechanism (8) extruding simultaneously two bases in the complementary DNA strands and positioning both extrahelical bases within binding pockets of individual protein domains. In the McrB-N complexes with di-methylated, hemi-methylated and non-methylated oligoduplexes, two bases from opposite strands (5mC/5mC, 5mC/C and C/C) also occupy extrahelical positions. However, there is no biochemical evidence that such ‘double flip’ mechanism would be important for McrBC function. Most likely, ‘double flip’ results from independent interactions between two McrB-N monomers and two 5′-AC targets in the palindromic 5′-ACCGGT sequence. Furthermore, differently from the SUVH5 and UHRF1 SRA domains that do not disturb B-DNA conformation except of base flipping (5–8), McrB-N bends DNA nearly by 30°. In this respect McrB-N mechanistically resembles the human alkyladenine glycosylase that approaches DNA form the minor groove side, flips out the damaged base and severely distorts DNA conformation (59) (Figure 5). Intriguingly, ROS1 DNA glycosylase which catalyses active DNA demethylation also uses base flipping to discriminate 5mC (60).
Figure 5.
McrB-N in comparison to other base-flipping proteins. Two monomers of SUVH5 SRA domain (3Q0C) or McrB-N are bound to the same DNA duplex, while in the case of the UHRF SRA domain (2ZKD) a single monomer is bound to the DNA. The DNA retains almost canonical B-DNA structure, except for the flipped out nucleotide, in the SRA domain complexes, while McrB-N bends DNA similarly to the human alkyladenine glycosylase (1BNK).
Implications for McrBC function
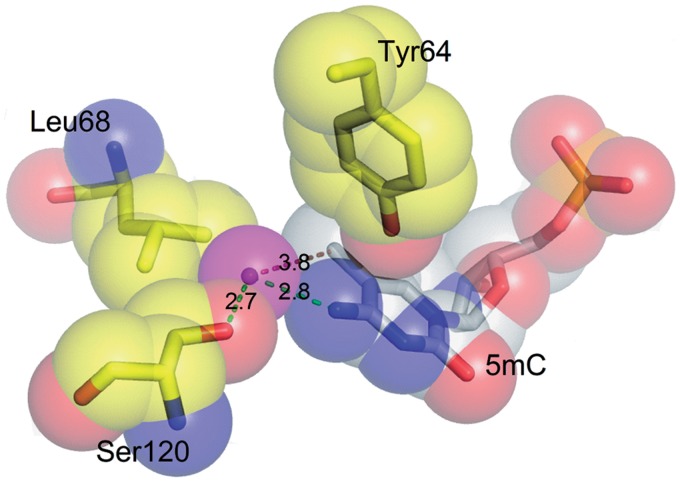
Genetic experiments indicate that McrBC restriction endonuclease alongside with the 5mC-modified DNA also restricts DNA containing N4-methylcytosine or 5hmC but does not cleave glycosylated DNA of T-even phages (13–16,61). The McrB-N structures in complex with di-methylated and hemi-methylated DNA show that N4-methylcytosine or 5hmC could fit into the binding pocket. Indeed, the free space in the pocket filled with 5mC is large enough to accommodate the hydroxyl group bound to the C5 atom in the case of 5hmC or the methyl group of N4-methylcytosine. Moreover, in one of the protomers a water molecule fills in this space. The water molecule is sandwiched between the N4 of 5mC and OG atoms of the Ser120 residue (Figure 6) and could be replaced by a hydroxyl group of 5hmC or by the methyl group of N4-methylcytosine (but not by both). On the other hand, glycosylation of the 5hmC by T-even phages should create a steric obstacle for the accommodation of the extrahelical base in the binding pocket and interfere with the McrBC cleavage.
Figure 6.
Close-up view of the water molecule in the vicinity of the methyl group in the McrB-N complex with di-methylated DNA. The hydroxyl group of 5hmC or methyl group of the N4-methylcytosine may occupy the space filled by the water molecule (shown in magenta).
The crystal structures of McrB-N in complex with the dimethylated and non-methylated DNA are nearly indistinguishable but the cytosine methylation is absolutely required for the McrBC cleavage (20,31). Van der Waals interactions are critical for the methyl group discrimination in the 5mC and presumably make an important contribution to the McrB-N–DNA complex stability. Flipping of the cytosine which makes fewer contacts with protein atoms in the binding pocket may result in a more dynamic McrB-N–DNA complex. It is likely that McrB interrogating for 5mC flips out bases in the 5′-RC context; however, cleavage occurs only when methylated C is bound in the pocket.
The McrB protein assembles into heptameric ring structures and tetradecamers in presence of GTP (27). The methylated DNA-binding domains McrB-N most likely are located on the surface of the heptameric rings (27). The crystal structure shows that McrB-N binds to 5-AmC site approaching DNA from the minor groove. In principle, a heptameric McrB assembly could bind seven 5-RmC sites independently. It would be interesting to see whether binding of methylated DNA results in DNA wrapping around the McrB heptamer and packaging into the nucleosome-like particles as it was earlier suggested (31).
McrBC systems are widespread among prokaryotes. In silico analysis suggests that McrB-N homologues may recognize 5mC in different sequence context (11). Interestingly, McrB-N, which is specific for the 5′-RmC sequence (20,31), does not form direct hydrogen bonds neither to A base nor to the partner T base 5′-upstream of the 5mC. The single water-mediated hydrogen bond to the partner T base cannot explain McrB specificity for purine residues preceding 5mC. The preference for a purine residue may be determined by an indirect readout mechanism, e.g. susceptibility to DNA deformation and/or stacking interactions with the aromatic ring of the intercalating Tyr41. However, in contrast to phosphate-clamping ‘finger’ loop residues Ser38 and Trp49, the DNA intercalating Tyr41 residue is not conserved (Figure 3D). It is tempting to speculate that the amino acid residues replacing intercalating Tyr41 in the McrB-N homologues may contribute to the differences in sequence specificity.
ACCESSION NUMBERS
The coordinates and structure factors are deposited in the Protein Data Bank under the accession codes: 3SSC (McrB-N bound to di-methylated DNA), 3SSD (McrB-N bound to hemi-methylated DNA) and 3SSE (McrB-N bound to non-methylated DNA).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
Funding for open access charge: European Social Fund under the Global Grant measure [project R100].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Prof. Alfred Pingoud (Justus-Liebig-Universität, Giessen) for a generous gift of the pBBI/McrB plasmid. The authors are grateful to Andrea Schmidt, Manfred Weiss, Santosh Panjikar and Paul Tucker (EMBL Hamburg) for their expert assistance at the beamlines. The authors thank Giedre Tamulaitiene, Lena Manakova, Dmitrij Golovenko and Giedrius Sasnauskas for valuable discussions.
REFERENCES
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Mol. Biotechnol. 2010;44:71–81. doi: 10.1007/s12033-009-9216-2. [DOI] [PubMed] [Google Scholar]
- 3.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 4.Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol. Cell. 2008;29:525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 5.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 6.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajakumara E, Law JA, Simanshu DK, Voigt P, Johnson LM, Reinberg D, Patel DJ, Jacobsen SE. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 2011;25:137–152. doi: 10.1101/gad.1980311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- 10.Bickle TA, Kruger DH. Biology of DNA restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa K, Fukuda E, Kobayashi I. Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems. DNA Res. 2010;17:325–342. doi: 10.1093/dnares/dsq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warren RA. Modified bases in bacteriophage DNAs. Annu. Rev. Microbiol. 1980;34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- 13.Hattman S, Fukasawa T. Host-induced modification of T-even phages due to defective glucosylation of their DNA. Proc. Natl Acad. Sci. USA. 1963;50:297–300. doi: 10.1073/pnas.50.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shedlovsky A, Brenner S. A chemical basis for the host-induced modification of T-even bacteriophages. Proc. Natl Acad. Sci. USA. 1963;50:300–305. doi: 10.1073/pnas.50.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raleigh EA, Trimarchi R, Revel H. Genetic and physical mapping of the mcrA (rglA) and mcrB (rglB) loci of Escherichia coli K-12. Genetics. 1989;122:279–296. doi: 10.1093/genetics/122.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luria SE, Human ML. A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol. 1952;64:557–569. doi: 10.1128/jb.64.4.557-569.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiom K, Sedgwick SG. Cloning and structural characterization of the mcrA locus of Escherichia coli. J. Bacteriol. 1991;173:7368–7373. doi: 10.1128/jb.173.22.7368-7373.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutherland E, Coe L, Raleigh EA. McrBC: a multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 1992;225:327–348. doi: 10.1016/0022-2836(92)90925-a. [DOI] [PubMed] [Google Scholar]
- 19.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA Restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruger T, Wild C, Noyer-Weidner M. McrB: a prokaryotic protein specifically recognizing DNA containing modified cytosine residues. EMBO J. 1995;14:2661–2669. doi: 10.1002/j.1460-2075.1995.tb07264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dila D, Sutherland E, Moran L, Slatko B, Raleigh EA. Genetic and sequence organization of the mcrBC locus of Escherichia coli K-12. J. Bacteriol. 1990;172:4888–4900. doi: 10.1128/jb.172.9.4888-4900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pieper U, Pingoud A. A mutational analysis of the PD…D/EXK motif suggests that McrC harbors the catalytic center for DNA cleavage by the GTP-dependent restriction enzyme McrBC from Escherichia coli. Biochemistry. 2002;41:5236–5244. doi: 10.1021/bi0156862. [DOI] [PubMed] [Google Scholar]
- 23.Pieper U, Brinkmann T, Kruger T, Noyer-Weidner M, Pingoud A. Characterization of the interaction between the restriction endonuclease McrBC from E. coli and its cofactor GTP. J. Mol. Biol. 1997;272:190–199. doi: 10.1006/jmbi.1997.1228. [DOI] [PubMed] [Google Scholar]
- 24.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:2743. [PubMed] [Google Scholar]
- 25.Pieper U, Schweitzer T, Groll DH, Gast FU, Pingoud A. The GTP-binding domain of McrB: more than just a variation on a common theme? J. Mol. Biol. 1999;292:547–556. doi: 10.1006/jmbi.1999.3103. [DOI] [PubMed] [Google Scholar]
- 26.White SR, Lauring B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8:1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- 27.Panne D, Muller SA, Wirtz S, Engel A, Bickle TA. The McrBC restriction endonuclease assembles into a ring structure in the presence of G nucleotides. EMBO J. 2001;20:3210–3217. doi: 10.1093/emboj/20.12.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart FJ, Raleigh EA. Dependence of McrBC cleavage on distance between recognition elements. Biol. Chem. 1998;379:611–616. [PubMed] [Google Scholar]
- 29.Panne D, Raleigh EA, Bickle TA. The McrBC endonuclease translocates DNA in a reaction dependent on GTP hydrolysis. J. Mol. Biol. 1999;290:49–60. doi: 10.1006/jmbi.1999.2894. [DOI] [PubMed] [Google Scholar]
- 30.Gast FU, Brinkmann T, Pieper U, Kruger T, Noyer-Weidner M, Pingoud A. The recognition of methylated DNA by the GTP-dependent restriction endonuclease McrBC resides in the N-terminal domain of McrB. Biol. Chem. 1997;378:975–982. doi: 10.1515/bchm.1997.378.9.975. [DOI] [PubMed] [Google Scholar]
- 31.Stewart FJ, Panne D, Bickle TA, Raleigh EA. Methyl-specific DNA binding by McrBC, a modification-dependent restriction enzyme. J. Mol. Biol. 2000;298:611–622. doi: 10.1006/jmbi.2000.3697. [DOI] [PubMed] [Google Scholar]
- 32.Pieper U, Schweitzer T, Groll DH, Pingoud A. Defining the location and function of domains of McrB by deletion mutagenesis. Biol. Chem. 1999;380:1225–1230. doi: 10.1515/BC.1999.155. [DOI] [PubMed] [Google Scholar]
- 33.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 34.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 35.French GS, Wilson KS. On the treatment of negative intensity observations. Acta Crystallogr. 1978;A34:517–525. [Google Scholar]
- 36.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 37.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 38.Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. Auto-Rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. D Biol. Crystallogr. 2005;61:449–457. doi: 10.1107/S0907444905001307. [DOI] [PubMed] [Google Scholar]
- 39.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP's model-building algorithms. I. The main chain. Acta Crystallogr. D Biol. Crystallogr. 2002;58:968–975. doi: 10.1107/s0907444902005462. [DOI] [PubMed] [Google Scholar]
- 40.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luscombe NM, Laskowski RA, Thornton JM. NUCPLOT: a program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res. 1997;25:4940–4945. doi: 10.1093/nar/25.24.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 47.Martin AM, Sam MD, Reich NO, Perona JJ. Structural and energetic origins of indirect readout in site-specific DNA cleavage by a restriction endonuclease. Nat. Struct. Biol. 1999;6:269–277. doi: 10.1038/6707. [DOI] [PubMed] [Google Scholar]
- 48.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 49.Berry DA, Jung K, Wise DS, Sercel DA, Pearson WH, Mackie H, Randolph JB, Somers RL. Pyrrolo-dC and pyrrolo-C: fluorescent analogs of cytidine and 2′-deoxycytidine for the study of oligonucleotides. Tetrahedron Lett. 2004;45:2457–2461. [Google Scholar]
- 50.Liu C, Martin CT. Fluorescence characterization of the transcription bubble in elongation complexes of T7 RNA polymerase. J. Mol. Biol. 2001;308:465–475. doi: 10.1006/jmbi.2001.4601. [DOI] [PubMed] [Google Scholar]
- 51.Dash C, Rausch JW, Le Grice SF. Using pyrrolo-deoxycytosine to probe RNA/DNA hybrids containing the human immunodeficiency virus type-1 3′ polypurine tract. Nucleic Acids Res. 2004;32:1539–1547. doi: 10.1093/nar/gkh307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tinsley RA, Walter NG. Pyrrolo-C as a fluorescent probe for monitoring RNA secondary structure formation. RNA. 2006;12:522–529. doi: 10.1261/rna.2165806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holz B, Klimasauskas S, Serva S, Weinhold E. 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 1998;26:1076–1083. doi: 10.1093/nar/26.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamulaitis G, Zaremba M, Szczepanowski RH, Bochtler M, Siksnys V. Nucleotide flipping by restriction enzymes analyzed by 2-aminopurine steady-state fluorescence. Nucleic Acids Res. 2007;35:4792–4799. doi: 10.1093/nar/gkm513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li N, Sickmier EA, Zhang R, Joachimiak A, White SW. The MotA transcription factor from bacteriophage T4 contains a novel DNA-binding domain: the ‘double wing’ motif. Mol. Microbiol. 2002;43:1079–1088. doi: 10.1046/j.1365-2958.2002.02809.x. [DOI] [PubMed] [Google Scholar]
- 57.Singarapu KK, Liu G, Xiao R, Bertonati C, Honig B, Montelione GT, Szyperski T. NMR structure of protein yjbR from Escherichia coli reveals ‘double-wing’ DNA binding motif. Proteins. 2007;67:501–504. doi: 10.1002/prot.21297. [DOI] [PubMed] [Google Scholar]
- 58.Defossez PA, Stancheva I. Biological functions of methyl-CpG-binding proteins. Prog. Mol. Biol. Transl. Sci. 2010;101:377–398. doi: 10.1016/B978-0-12-387685-0.00012-3. [DOI] [PubMed] [Google Scholar]
- 59.Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 60.Ponferrada-Marin MI, Parrilla-Doblas JT, Roldan-Arjona T, Ariza RR. A discontinuous DNA glycosylase domain in a family of enzymes that excise 5-methylcytosine. Nucleic Acids Res. 2011;39:1473–1484. doi: 10.1093/nar/gkq982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raleigh EA, Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc. Natl Acad. Sci. USA. 1986;83:9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.