Abstract
Members of the BET (bromodomain and extra terminal motif) family of proteins have been shown to be chromatin-interacting regulators of transcription. We previously generated a mutation in the testis-specific mammalian BET gene Brdt (bromodomain, testis-specific) that yields protein lacking the first bromodomain (BRDTΔBD1) and observed disrupted spermiogenesis and male sterility. To determine whether BRDTΔBD1 protein results in altered transcription, we analyzed the transcriptomes of control versus BrdtΔBD1/ΔBD1 round spermatids. Over 400 genes showed statistically significant differential expression, and among the up-regulated genes, there was an enrichment of RNA splicing genes. Over 60% of these splicing genes had transcripts that lacked truncation of their 3′-untranslated region (UTR) typical of round spermatids. We selected four of these genes to characterize: Srsf2, Ddx5, Hnrnpk and Tardbp. The 3′-UTRs of Srsf2, Ddx5 and Hnrnpk mRNAs were longer in mutant round spermatids and resulted in reduced protein levels. Tardbp was transcriptionally up-regulated and a splicing shift toward the longer variant was observed. All four splicing proteins were found to complex with BRDT in control and mutant testes. We thus suggest that, along with modulating transcription, BRDT modulates gene expression as part of the splicing machinery. These modulations alter 3′-UTR processing in round spermatids; importantly, the BD1 is essential for these functions.
INTRODUCTION
The BET (bromodomain and extra terminal motif) family of proteins, so-called because they contain two N-terminal bromodomains and a C-terminal extra terminal motif, has been widely implicated in transcriptional regulation (1). Although these proteins do not bind DNA directly, they can regulate transcription through modulating chromatin by binding to specific acetylated lysine residues on histone tails (2–5). The mammalian BET proteins BRD2, BRD3 and BRD4 are ubiquitously expressed (6) and each has been implicated in modulation of transcription but through distinct mechanisms. BRD2 has been shown to recruit the TATA box binding protein (TBP) into the E2F transcriptional complex and initiate transcription of cell cycle genes (7). BRD3 has been shown to bind to the acetylated form of the transcription factor GATA1 and target it to chromatin (8,9). Both BRD2 and BRD3 have been shown to bind to acetylated chromatin in the coding regions of genes and thereby aid in transcriptional elongation (10). BRD4 has been broadly implicated in transcriptional regulation by recruiting the positive transcription elongation factor b (P-TEFb) to genes poised to be transcribed (11–14). BRD4 also has a P-TEFb-independent role in transcriptional regulation that involves recruiting the histone–lysine N-methyltransferase NSD3 to promoter regions (15).
The highly regulated program of spermatogenesis involves a correspondingly highly regulated transcriptional program (16), with many layers of differentiation stage-specific transcriptional controls (17–20). Many of the transcription factors responsible for this regulation are expressed in somatic tissues as well, but in the testis, they produce unique transcripts and splice variants (21–25). In fact, the use of tissue-specific promoters and alternative splicing is prevalent in the testis (25–30), although the mechanisms regulating these processes are poorly understood. Modification of the 3′-untranslated region (UTR) of many transcripts, specifically selecting a truncated version in favor of the previous longer transcript, is a novel form of transcriptional control that is seen frequently in spermatogenic cells (17,25,31–34). This modification is particularly prevalent in round spermatids (31) and has also been observed in some cancers (35). Although it is apparent that 3′-UTR truncation occurs due to selection of a more proximal polyadenylation site; the molecular mechanisms underlying this process are not known (31,34,36).
Brdt is a testis specific member of the BET family (6,37). It is the only BET protein highly expressed throughout the transcriptionally active pachytene stage of meiotic prophase (38) and is still expressed in haploid round spermatids along with BRD2 and BRD3 (38,39). Among the mammalian BETs, BRDT is most similar to BRD4 in that both have a long C-terminal region which contains the protein-interacting C-terminal domain (CTD) (1,14). It is through this domain that BRD4 binds P-TEFb, and in cell lines the CTD of BRDT has been shown to also be capable of this interaction (40). However, to date, no BRDT-dependent transcriptional modulation has been reported, although we previously observed that BRDT binds to a repressive element in the promoter of the gene histone cluster 1, H1t (Hist1h1t) (39). Truncated BRDT protein lacking the first bromodomain (BRDTΔBD1) did not bind the promoter and homozygous BrdtΔBD1/ΔBD1 spermatocytes and spermatids exhibited increased HIST1H1T expression.
In the present study, we have investigated the role of BRDT in multiple aspects of transcriptional regulation during spermatogenesis. We found that BRDT complexes not only regulate the transcription of a large number of genes in a BD1-dependent manner, but that BRDT is also a component of the splicing machinery in pachytene spermatocytes and round spermatids. This is of particular interest since the yeast BET gene, BDF1, has been implicated in regulating the transcription of small nuclear RNAs that are important components of splicing pathways (41,42). We also uncovered a function for BRDT in the novel 3′-UTR truncation of specific mRNAs that characterizes the post-meiotic spermatid transcriptome.
MATERIALS AND METHODS
Germ cell separation
Preparation of enriched populations of pachytene spermatocytes and round spermatids was carried out according to our laboratory’s established protocol (43) (see ‘Supplementary Materials and Methods’ section). The purity of the cellular populations was assessed by flow cytometric analysis (44–46). Every third fraction from the gradient was stained with propidium iodide (Sigma cat#P4170) and examined on a Becton Dickinson FACScan Flow Cytometer. Red fluorescence emitted after excitation with a 488-nm argon laser was recorded from approximately 5000 cells per sample. Results were analyzed using CellQuest Pro software.
RNA purification and microarray analysis
Total RNA was extracted, purified and concentrated from eight independent pools of round spermatid populations (four control, four mutant) using Trizol reagent (Invitrogen) and the RNeasy MinElute Cleanup kit (Qiagen) according to the manufacturer’s protocol. Gene expression analysis was carried out using Affymetrix GeneChip Mouse Genome 430 2.0 microarrays according to the manufacturer’s protocols (see ‘Supplemental Materials and Methods’ section).
Signal intensities were ascertained and statistical analysis was performed within the R\Bioconductor statistical framework (47,48). We used the limma package (49) to pre-process the raw data and perform the quality controls. Expression intensities were background corrected, normalized and summarized using the Gene Chip Robust Multiarray Algorithm (GC-RMA) (50,51). The raw and processed data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE33132. See ‘Supplementary Materials and Methods’ section for microarray quality controls and differential expression quantification (Supplementary Figure S1).
Quantitative real-time PCR
To perform quantitative real-time PCR, new samples of purified control and mutant round spermatids were prepared, and RNA was extracted with Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Real-time PCR was carried out according to our laboratory’s standard protocol (52). The values were normalized to the expression of β-actin (Actb) as an internal control. The primers were designed using the Primer3 program (http://frodo.wi.mit.edu/primer3) and synthesized by Integrated DNA Technologies (see ‘Supplementary Materials and Methods’ section).
Northern blot hybridization analysis
RNA was prepared for northern blotting from purified control and mutant pachytene spermatocyte populations and a third set of control and mutant purified round spermatid populations using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Northern blot hybridization analysis was performed according to a modified version of our laboratory’s standard protocol (38) (see ‘Supplementary Materials and Methods’ section). Probes were constructed by ligating PCR products of 400–600 bp of the coding region of each gene into the pGEM®-T Easy vector (Promega) and after excision, 32P-labeled RNA probes were transcribed (see ‘Supplementary Materials and Methods’ section). The blot was visualized using a GE Healthcare Typhoon Trio phosphorimager.
In situ hybridization
Adult control and BrdtΔBD1/ΔBD1 mutant testes were fixed in 4% paraformaldehyde (PFA) and paraffin imbedded. Histological sections were prepared according to our laboratory’s standard protocol (38,53). In situ hybridization was performed according to a of our laboratory’s standard protocol (38) (see ‘Supplementary Materials and Methods’ section). Sense and anti-sense Srsf2 digoxigenin (Dig) labeled probes were transcribed from the plasmids described in the northern blot hybridization analysis.
Immunoblot analysis
For immunoblot analysis, lysates of whole testes from control and mutant animals were used and our previously published procedures were followed (52) (see ‘Supplementary Materials and Methods’ section).
Immunofluorescence
Immunofluorescent staining was carried out according to a modified version of our laboratory’s standard protocols (39,52,54) (see ‘Supplementary Materials and Methods’ section).
Flag-BRDT pull down
Full-length Brdt sequence with a fused C-terminal Flag-tag was inserted into the pBabe vector which was then transfected into 293T cells using Lipofectamine reagent (Invitrogen, Inc.). After 2 days puromycin was added and drug-resistant Flag-BRDT colonies were selected. Of the puromycin-resistant colonies, two were identified by immunoblotting with Flag antibody (Sigma cat# F7425) and BRDT antibody as stable Flag-BRDT expressing cell lines. Cells grown from one colony were lysed in homogenization buffer [10 mM Tris (pH 8.0), 10 mM KCl, 2 mM MgCl2, 50 mM β-glycerol phosphate, 0.1% NP40 and a protease inhibitor cocktail (Roche)] and then centrifuged for 5 min at 2500g at 4°C. Pelleted nuclei were resuspended and allowed to lyse in nuclei buffer [20 mM Tris (pH 8.0), 420 mM NaCl, 1 mM EDTA, 50 mM β-glycerol phosphate, 25% glycerol and a protease inhibitor cocktail (Roche)] on ice for 30 min. Nuclear extracts were cleared by centrifugation at 18 000g for 10 min at 4°C. Flag-tagged BRDT-containing complexes were purified by incubating nuclear extracts with EZview red anti-FLAG M2 affinity gel (Sigma cat#F-2426) overnight at 4°C. The beads were loaded onto a column and washed extensively with buffer BC-0 [20 mM HEPES (pH 7.9), 20% glycerol, 2 mM EDTA, 1 mM DTT and 0.5 mM PMSF] supplemented with 0.15 M KCl, followed by a wash with buffer BC-0.3 (BC-0 containing 300 mM KCl). Bound proteins were eluted in BC-0.1 with Flag peptide (Sigma cat#F-3290). Protein microsequencing by mass spectrometry was performed by Columbia University’s Protein Core facility.
Co-immunoprecipitation
Nuclear extracts from adult control, heterozygous and mutant animals were prepared as outlined above, and pre-cleared with Protein A agarose beads (Roche, cat#11134515001) at 4°C for 1 h. Pre-cleared lysates were then incubated with primary antibody or IgG control for 4 h with gentle agitation at 4°C. Protein A agarose beads were added and incubation continued overnight. The beads and immunoprecipitated complexes were pelleted by a 10 s centrifugation at 500g, and then washed in wash buffer [20 mM Tris (pH 8.0), 150 mM NaCl, 0.1% NP40] four times at 4°C. Final pellets were resuspended in 1× SDS loading buffer and boiled for 5 min. The supernatant was run on an 8 or 10% SDS–PAGE gel and immunoblotting was performed as described above. In addition to the antibodies listed above, we also used a C-terminal anti-BRDT antibody made and characterized by our laboratory (39) at 1:3000. For all rabbit antibodies, Clean-Blot® IP detection reagent HRP (Thermo Scientific, cat# 21230) was used as the secondary antibody at 1:500.
RESULTS
Altered gene expression in mutant round spermatids
In our initial study of BrdtΔBD1/ΔBD1 mutant mice, a panel of 20 candidate genes expressed during spermatogenesis was examined for altered transcription (39). Only the gene Hist1h1t showed a significant change in expression levels: it was up-regulated in the mutant testis. To obtain a more complete picture of the role of BRDT in transcriptional regulation during spermatogenesis, we carried out microarray analysis on enriched populations of control and BrdtΔBD1/ΔBD1 mutant round spermatids using Affymetrix GeneChip® Mouse Genome 430 2.0 microarrays. Each array used RNA from round spermatids (≥92% purity) pooled from seven adult animals. Four independent control and four independent mutant arrays were carried out and the results were analyzed using R\Bioconductor. Raw and normalized gene expression levels were deposited in the GEO under accession number GSE33132.
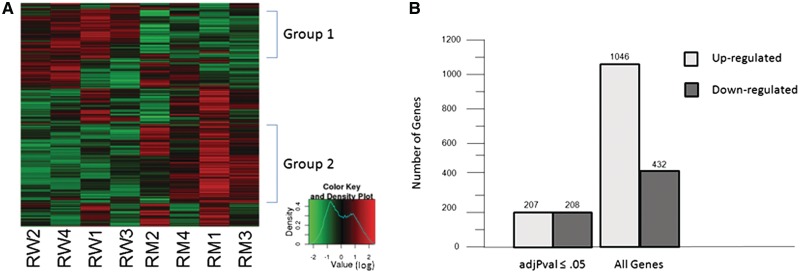
Unsupervised hierarchical clustering analysis, displayed as a heat map of all eight arrays, showed that there is a cluster of genes whose expression is down-regulated (Figure 1A, group 1) and a cluster of genes that are up-regulated (Figure 1A, group 2) in mutant round spermatids. In order to examine changes in transcription, the expression values for each probe set from the four chips of the same genotype were averaged and the log base 2-fold change (log FC) between mutant and control was calculated. A positive log FC corresponds to elevated transcription in the mutant. For this analysis, genes with a log FC ≥1 or ≤ −1 were considered to have altered expression. Due to the large numbers of probes involved, an adjusted P-value (adjPVal) was calculated for each probe set and used in place of P-value to assess the significance of these changes. At an adjPVal of ≤0.05, 456 probe sets showed a statistically significant change in expression. Of those, 234 probe sets corresponded to 207 unique genes that were up-regulated and 222 probe sets corresponded to 208 unique genes that were down-regulated (Figure 1B, Supplementary Data and Supplementary Table S1).
Figure 1.
Microarray analysis of RNA from purified control and BrdtΔBD1/ΔBD1 mutant round spermatids reveals genes that are both up-regulated and down-regulated in the mutant cells. (A) Heat map of the eight microarrays showing two groups of genes, one up-regulated and one down-regulated in mutant round spermatids. (B) A total of 415 genes exhibited statistically significant (adjPval ≤0.05) altered expression (logFC ≥1 or ≤−1) in mutant round spermatids. When the criteria were relaxed to include all differential expression, 1478 genes with altered expression were observed.
Transcription of splicing genes is up-regulated in mutant round spermatids
The functional annotation tool of the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 Software (55,56) was used to look for enrichment of genes in a specific pathway or a specific process. Of the 208 up-regulated genes, 9 were components of the spliceosome and 12 genes, 5 of which were not in the spliceosome list (14 genes in total), have been shown to have a role in mRNA splicing (Table 1, upper section). These enrichments were statistically significant (≤0.05), with FDR-corrected P-values of 0.017 and 0.0074, respectively (57) (Supplementary Data and Supplementary Table S2). The down-regulated genes did not show a statistically significant enrichment for any particular pathway or process (Supplementary Data and Supplementary Table S3). Although using adjPVal is helpful in removing false positive expression changes, it can also remove genuinely differentially expressed genes. Thus, we performed the same analysis, but used any probe sets that had a log FC ≥ 1 or less than or equal to −1 regardless of the adjPval. This resulted in 1281 probes sets corresponding to 1046 genes that were up-regulated and 492 probe sets corresponding to 432 genes that were down-regulated (Figure 1B, Supplementary Data and Supplementary Table S1). Again, the down-regulated genes did not show any statistically significant enrichment (Supplementary Data and Supplementary Table S5), but 23 of the up-regulated genes were components of the spliceosome and 36 genes, 18 of which were not in the spliceosome list (41 total genes), have been shown to have roles in mRNA splicing (FDR corrected P-values of 1.7 × 10−4 and 7.7 × 10−7, respectively) (Table 1, Supplementary Data and Supplementary Figure S4).
Table 1.
Transcription and processing of genes encoding components of the spliceosome and functioning in RNA splicing are altered in mutant round spermatids
| Pathway/gene ontology | Symbol | Gene name | adjPval ≤ 0.05 | Specificty of up-regulation | Interacts with Brdt |
|---|---|---|---|---|---|
| Spliceosome | Ddx5 | DEAD (Asp–Glu–Ala–Asp) box polypeptide 5 | Yes | Distal UTR | Yes |
| Spliceosome | Hnrnpk | Heterogeneous nuclear ribonucleoprotein K | Yes | Distal UTR | Yes |
| Spliceosome | Hnrnpm | Heterogeneous nuclear ribonucleoprotein M | Yes | Unknown | No |
| Spliceosome | Hspa8 | Heat shock protein 8 | Yes | Coding | Yes |
| Spliceosome | Srsf1 | splicing factor, Arginine/serine-rich 1 (ASF/SF2) | Yes | Coding | No |
| Spliceosome | Srsf2 | splicing factor, Arginine/serine-rich 2 (SC-35) | Yes | Distal UTR | Yes |
| Spliceosome | Srsf6 | splicing factor, Arginine/serine-rich 6 | Yes | Distal UTR | No |
| Spliceosome | Srsf9 | splicing factor, Arginine/serine rich 9 | Yes | Unknown | No |
| Spliceosome | U2af1 | U2 small nuclear ribonucleoprotein Auxiliary factor (U2AF) 1 | Yes | Coding | No |
| RNA Splicing | Celf1 | CUG triplet repeat, RNA binding protein 1 | Yes | Distal UTR | No |
| RNA Splicing | Hnrnpf | Heterogeneous nuclear ribonucleoprotein F | Yes | Distal UTR | No |
| RNA Splicing | Ppp1r8 | Protein phosphatase 1, regulatory (inhibitor) subunit 8 | Yes | Distal UTR | No |
| RNA Splicing | Rbm39 | RNA-binding motif protein 39 | Yes | Distal UTR | No |
| RNA Splicing | Sfpq | Splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated); similar to PTB-associated splicing factor | Yes | Distal UTR | Yes |
| Spliceosome | Hnrnpa3 | Heterogeneous nuclear ribonucleoprotein A3 | No | Distal UTR | No |
| Spliceosome | Lsm3 | LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) | No | Unknown | No |
| Spliceosome | Lsm5 | LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) | No | Unknown | No |
| Spliceosome | Lsm6 | LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) | No | Coding | No |
| Spliceosome | Lsm8 | LSM8 homolog, U6 small nuclear RNA associated (S. cerevisiae) | No | Unknown | No |
| Spliceosome | Pcbp1 | Poly(rC) binding protein 1 | No | Unknown | Yes |
| Spliceosome | Rbm25 | RNA binding motif protein 25 | No | Distal UTR | No |
| Spliceosome | Sf3b14 | Splicing factor 3B, 14 kDa subunit | No | Coding | No |
| Spliceosome | Snrnp40 | Small nuclear ribonucleoprotein 40 (U5) | No | Distal UTR | No |
| Spliceosome | Snrpb | Small nuclear ribonucleoprotein B | No | Coding | No |
| Spliceosome | Srsf3 | Splicing factor, arginine/serine-rich 3 (SRp20) | No | Distal UTR | No |
| Spliceosome | Srsf7 | Splicing factor, arginine/serine-rich 7 | No | Distal UTR | No |
| Spliceosome | Srsf10 | FUS interacting protein (serine–arginine rich) 1 | No | Distal UTR | No |
| Spliceosome | Thoc4 | THO complex 4 | No | Coding | No |
| RNA Splicing | Celf2 | CUG triplet repeat, RNA binding protein 2 | No | Distal UTR | No |
| RNA Splicing | Tardbp | TAR DNA binding protein | No | Coding/Variant | Yes |
| RNA Splicing | Wbp4 | WW domain binding protein 4 | No | Distal UTR | No |
| RNA Splicing | Wtap | Wilms’ tumor 1-associating protein | No | Distal UTR | No |
The expression of 41 splicing genes is altered at the level of RNA in mutant round spermatids. Eight genes have elevated levels of transcription, 1 gene has a shift as to which variant is transcribed, 1 gene has both elevated transcription and a shift as to which variant is transcribed, 24 genes have a loss of truncated 3′-UTRs, and for 7 genes the nature of the change in RNA expression could not be determined. Ten of these genes encode proteins that were also shown to interact with Flag-tagged BRDT in 293T cells.
Changes in transcription reflect a loss of mRNAs with a short 3′-UTR
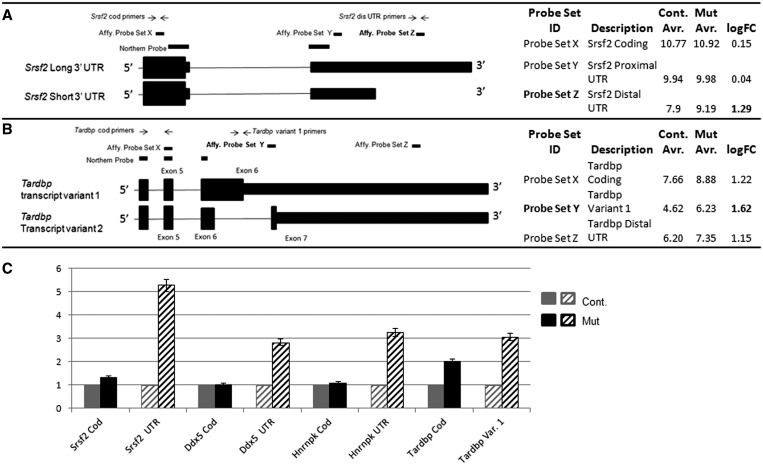
In the course of our analysis, we made an interesting observation regarding differential processing of the 3′-UTR of specific transcripts. This emerged by examining in detail changes in relative levels of different transcripts from the same gene. That is, closer examination of the microarray results from the 14 statistically significantly up-regulated splicing genes revealed that 11 of the genes had multiple probe sets. Genes that have multiple probe sets can be investigated for changes in mRNA processing as well as changes in total transcription (35,58,59). Of those 11 genes, 9 (Ddx5, Hnrnpk, Srsf2, Srsf6, Celf1, Hnrnpf, Ppp1r8, Rbm39 and Sfpq) were detected by probes that did not indicate an overall increase in transcription in the mutant, but rather an increase in transcription of the most distal part of the 3′-UTR (Table 1, upper section). Probes in the coding region and proximal 3′-UTR showed high expression but no change between the control and mutant, and only probes in the distal 3′-UTR were detected as up-regulated in the mutant. This is illustrated in cartoon form for the gene Srsf2 in Figure 2A. Inefficient 3′- to 5′-reverse transcription, required for making the cRNA used for hybridization to the array, can lead to distal 5′ probes appearing to be absent or expressed at very low levels while the 3′ probe shows high levels of transcription. In the case of these nine genes, however, probes closer to the 5′-end actually showed higher expression than the distal 3′ probes (Figure 2A), suggesting that poor 3′- to 5′-reverse transcription is not an issue.
Figure 2.
Detailed examination of Affymetrix probe sets yields unexpected information about mRNA processing as well as expected changes in total transcription. (A and B) Cartoon schematics of two genes, Srsf2 and Tardbp, showing the location of the Affymetrix probes, designed qPCR primers and northern blot probes. The qPCR primer location mirrors the positions of the Affymetrix probes, and northern blot probes are chosen to hybridize to all transcripts. (A) Three Affymetrix probes to the Srsf2 gene are depicted: one in the coding region, one in the proximal 3′-UTR and one in the distal 3′-UTR. The probe in the distal 3′-UTR would uniquely hybridize to the long transcript of Srsf2. In control spermatids, hybridization of the Srsf2 distal UTR probe is low compared to the coding and proximal UTR probes demonstrating lack of the Srsf2 long transcript. The long transcript is present in mutant spermatids. (B) Three Affymetrix probes to the Tardbp gene are depicted: one in the coding region, one unique to transcript variant 1 and one in the distal 3′-UTR. Tardbp transcription is up-regulated in mutant spermatids, but the possibility exists that there may be a specific up-regulation of transcript variant 1. (C) Q-PCR of four selected genes. Overall levels of transcription of Srsf2, Ddx5 and Hnrnpk are not altered in mutant round spermatids. However, the presence of normally truncated 3′-UTR sequence is increased. In contrast, overall levels of Tardbp transcription are increased in mutant round spermatids, in particular, for transcript variant 1.
Wide-spread loss of 3′-UTR-truncated short transcripts occurs in mutant round spermatids
The occurrence of transcripts in which the length of the 3′-UTR is truncated relative to the full-length transcripts is prevalent in the testis, particularly in round spermatids (25,31). In control round spermatids, the coding-region or proximal 3′-UTR probes of the nine above mentioned genes showed 2- to 5-fold higher levels of expression than the probes in the distal 3′-UTR, suggesting that these mRNAs normally have a short 3′-UTR that does not include the region containing the distal probe (Figure 2A). Since only the distal 3′-UTR probes showed up-regulation in mutant round spermatids, it is likely that these genes are not transcriptionally up-regulated but rather that the normally truncated 3′-UTR transcripts are now full-length.
Of the 14 statistically significantly up-regulated genes that were involved in splicing, 9 showed this retention of the longer 3′-UTR (64%) and three showed transcriptional up-regulation in the coding region of the gene (21%). The two other genes had only a single probe set which spanned the 3′-UTR and the coding region and thus no information about possible UTR truncation could be gleaned (14%) (Table 1, upper section). Of the 41 total splicing genes (regardless of adjPVal), 24 showed loss of 3′-UTR truncation (59%), 9 showed up-regulation in the coding region (22%) and 7 had a single uninformative probe set (17%). Two genes, one of which also showed up-regulation in the coding region, had a specific increase in one transcriptional variant, but not another (Table 1). In most cases of alternative splicing, expression microarrays are not informative (59), but for both of these genes, the probe sets nearly completely uniquely overlapped with a specific variant (example in Figure 2B). When all 1046 up-regulated genes were considered, the percentages shifted to 25% with loss of 3′-UTR truncation, 17% with up-regulation in the coding region and 58% with a single uninformative probe set. Whether considering only the statistically significant subset or the full database of up-regulated genes, our observations suggest that loss of the BD1 of BRDT has a general effect on 3′-UTR processing in spermatids.
Validation of a loss of 3′-UTR truncation and indication of a shift in alternative splicing
To validate the results of the microarray experiment, four genes whose role in splicing is well characterized, and which were identified in a pull-down screen for BRDT interacting proteins (see results below), were selected for further analysis by quantitative real-time PCR and northern blot hybridization: Srsf2, Ddx5, Hnrnpk and Tardbp. From the microarray data, in mutant round spermatids Srsf2, Ddx5 and Hnrnpk would be predicted to express higher levels of their longer transcripts, i.e. exhibit a loss of truncation of the 3′-UTR of some transcripts. As Tardbp showed up-regulation in the coding region and UTR, it would be predicted that there would be increased overall expression and specifically, more of the longest transcripts (Figure 2B). For Srsf2 and Ddx5, primers were designed for both the coding region and distal 3′-UTR (Figure 2A). Mouse Hnrnpk has seven variant transcripts that would have been detected by the microarray probes. Three of the variants have a short 3′-UTR, and four have a full-length UTR. The variants also differ in the transcriptional start site and the inclusion of an extra exon; however, the microarray probes did not differentiate among these various isoforms. As such, we made probes to detect only the coding region and distal 3′-UTR of Hnrnpk. There are five mouse protein-coding alternative splice forms of Tardbp transcripts in the EST database (60). Although they are mostly overlapping, the longest isoform, transcript variant 1, has only six exons instead of seven as what was once intron 6 is now part of the spliced mRNA (see cartoon in Figure 2B). A probe set in our microarray (Probe Set Y in Figure 2B) corresponded to the 3′-end of intron 6 and the 5′-end of exon 7 and thus partially corresponds to the unique region of Tardbp transcript variant 1. This probe set showed a slightly greater up-regulation than did the probe set in the coding region (Figure 2B).
To investigate whether this difference was real and in fact due to a specific increase in the long isoform, we designed primers for the coding region and the region unique to transcript variant 1 (Figure 2B). Real-time PCR revealed that in the mutant spermatid RNA, there was no change in expression of the coding regions of Srsf2, Ddx5 or Hnrnpk but all three genes yielded increased levels of transcripts containing the distal 3′-UTR (Figure 2C). For Tardbp, there was an overall increased expression and indeed the long isoform was increased at a level even greater than the total transcriptional up-regulation (Figure 2C).
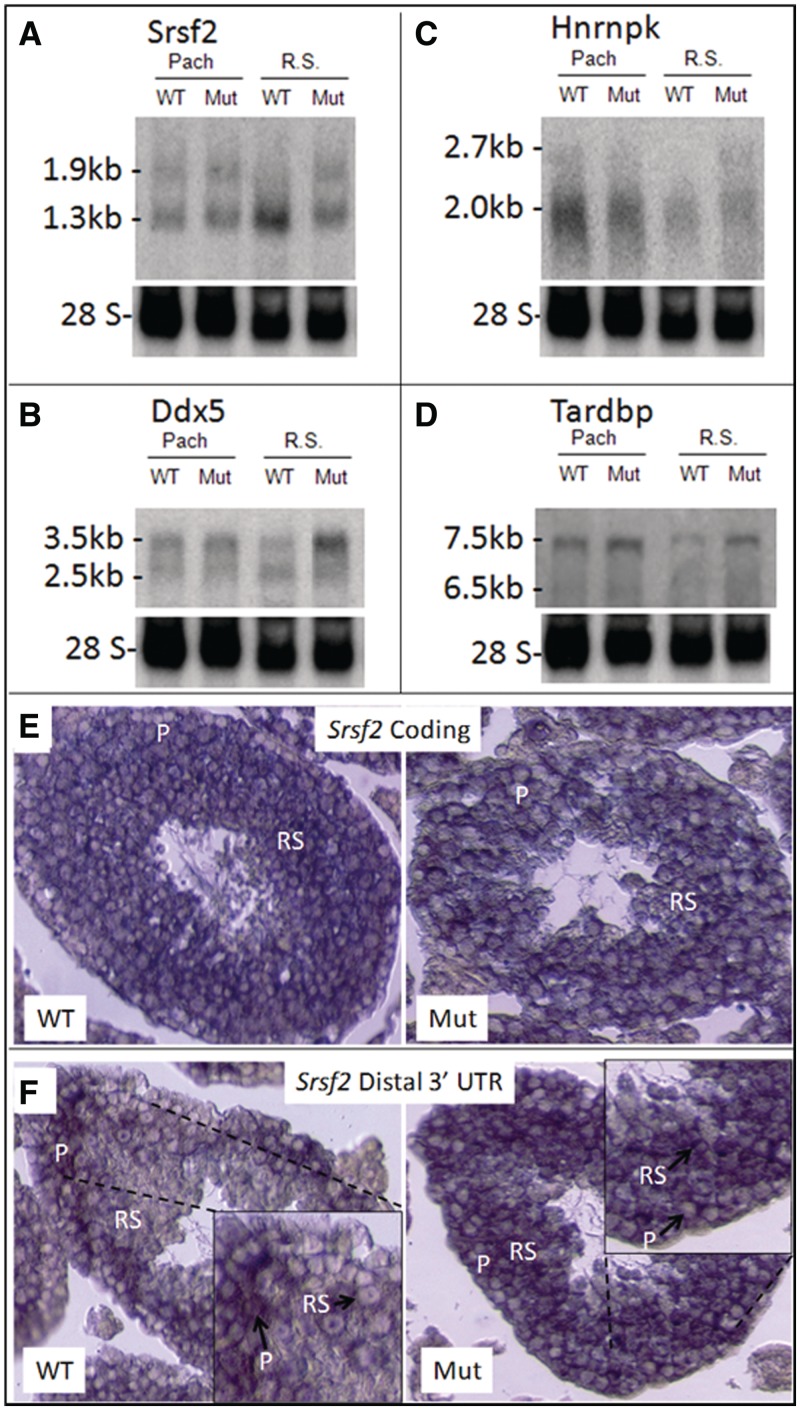
Since BRDT is expressed in pachytene spermatocytes as well as round spermatids (38), RNA from a cellular suspension of enriched pachytene spermatocytes was also examined for alterations in mRNA processing. Northern blot hybridization probes were designed that would recognize all known variant transcripts of the four genes (see ‘Supplemental Materials and Methods’ section) and were used to analyze RNAs from spermatocytes and spermatids from mutant and control testes. Srsf2 produces transcripts of 1.9 and 1.3 kb in length which are detected in both control and mutant pachytene spermatocytes and in mutant round spermatids. However, in control round spermatids, we cannot detect any of the longer mRNA (Figure 3A). The sizes of these transcripts correlate with full-length Srsf2 transcripts and transcripts that are polyadenylated at a more proximal site in the 3′-UTR (61). To visualize this change in prevalence of transcript variants at the cellular level, we carried out in situ hybridization using probes that would detect either the coding region or only the distal 3′-UTR region of Srsf2 transcripts. The coding region probe localized to spermatocytes and round spermatids in both the mutant and control (Figure 3E). In contrast, while the distal 3′-UTR probe hybridized in control and mutant spermatocytes and mutant round spermatids, there was no signal in control round spermatids (Figure 3F). Sense probes for both the Srsf2 coding region and the Srsf2 distal 3′-UTR failed to hybridize (Supplementary Data and Figure S2).
Figure 3.
Loss of truncation of the 3′-UTR in mRNAs from BrdtΔBD1/ΔBD1 round spermatids. (A–D) Northern blot hybridization analysis of total RNA isolated from control and mutant pachytene spermatocytes and round spermatids. (A) Srsf2 long and short mRNA transcripts are present in control and mutant pachytene spermatocytes. mRNA expression is comparable in these two populations. Only short transcripts are present in control round spermatids, indicating that all Srsf2 transcripts have a truncated 3′-UTR in this cell type. Mutant spermatids express long and short Srsf2 transcripts and the total mRNA expression is comparable to that of the spermatocyte populations, indicating that there is a loss of truncation of some mRNAs. (B) Ddx5 long and short mRNA transcripts are present in control and mutant pachytene spermatocytes and control round spermatids. mRNA expression is similar in these three populations, but short transcripts make up a greater percentage of all Ddx5 transcripts in control spermatids. Mutant spermatids also express long and short transcripts, but the long transcript is much more abundant relative to the short as compared to control spermatids. (C) Hnrnpk is mostly expressed as short transcripts in control and mutant pachytene spermatocytes although there is some expression of long transcripts. There is a slight reduction in short transcripts in mutant spermatocytes. Control round spermatids only express short Hnrnpk transcripts, but mutant spermatids still express the long Hnrnpk transcripts. (D) The long transcript variant 1 of Tardbp is highly expressed in control pachytene spermatocytes along with lower expression of the shorter variants. In mutant pachytene spermatocytes, there is a general increase in expression of Tardbp mRNA, in particular, for the long variant 1 transcript. The same patterns of Tardbp expression are seen in control and mutant round spermatids. (E and F) In situ hybridization of adult control and mutant testicular sections with Srsf2 probes. (E) A probe to the coding region of Srsf2 hybridized in control and mutant pachytene spermatocytes (P) and control and mutant round spermatids (RSs). (F) A probe to the distal end of the Srsf2 3′-UTR hybridized in control and mutant pachytene spermatocytes, but only in mutant round spermatids. Control round spermatids do not expressed Srsf2 mRNA with a long 3′-UTR.
There is no record of a truncated mouse Ddx5 mRNA, in addition to the 3.5 kb full-length transcript, in the EST database. However, the rat Ddx5 gene is 94% identical to mouse Ddx5 (and 89% identical in the 3′-UTR), and the EST database includes a rat transcript polyadenylated at a more proximal site in the 3′-UTR. Therefore, mouse Ddx5 likely also has a proximal polyadenylation site. Our northern blot hybridization analysis supports this notion, as indeed we detected two transcripts, a full-length 3.5 kb transcript and a 2.5 kb transcript which is the approximate length of the rat transcript. Both transcripts are present in control and mutant spermatocytes and round spermatids. In spermatocytes, the ratio of long to short transcript is the same in the control and the mutant. However, in mutant round spermatids, as compared to control, there is a shift to greater abundance of the longer transcript (Figure 3B).
Three of the seven known transcriptional variants of Hnrnpk are between 1.9 and 2.0 kb in length and have short 3′-UTRs; the other four range in length from 2.6 to 2.9 kb and have longer 3′-UTRs. Our northern blot hybridization analysis was not able to distinguish transcripts that differ in 300 nt or less; however, we were still able to visualize the shorter and longer groups of transcripts. Although clearly Hnrnpk transcripts were present predominantly as the shorter isoforms, some longer transcripts were detected in control and mutant spermatocytes and also in mutant round spermatids. However, control round spermatids only expressed the shorter forms of Hnrnpk (Figure 3C).
Of the alternative splice variants of Tardbp, only the long 7.5 kb transcript variant 1 can be uniquely distinguished on our northern blots. The other four transcripts range from 6.4 to 6.5 kb in length and cannot be uniquely resolved. Both the longer and shorter isoforms are expressed in control and mutant spermatocytes and round spermatids. Mutant pachytene spermatocytes appear to have increased transcription of Tardbp and specifically an increase of the longer mRNA. These changes are also apparent in mutant round spermatids when compared to control spermatids (Figure 3D).
Loss of 3′-UTR truncation results in lowered protein expression in mutant round spermatids
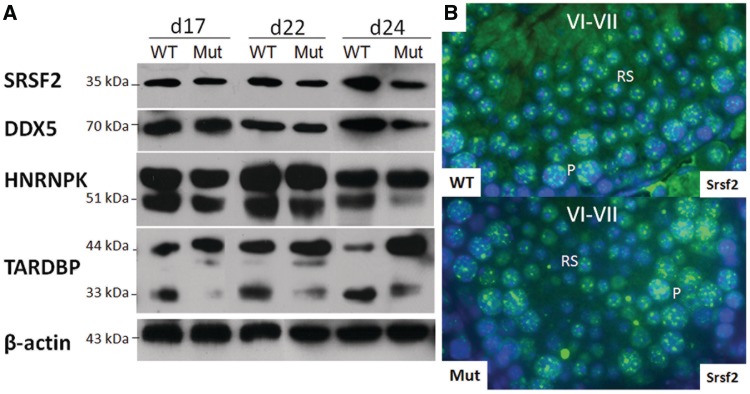
The significance of 3′-UTR-truncated transcript variants of specific genes in the testis has yet to be elucidated, but the 3′-UTR is a known target for RNA binding proteins (62,63) and microRNAs (64), which can modulate translation (65,66). We therefore wished to test the hypothesis that a failure to produce these truncated transcripts in the BrdtΔBD1/ΔBD1 mutant might affect the levels of protein made. We therefore examined protein extracts from control and mutant testis of 17-, 22- and 24-day-old mice for the expression of SRSF2, DDX5 and HNRNPK proteins using immunoblot analysis. Day 17 testes do not yet have round spermatids present, and from the northern blot analysis, we would expect that there would be no change in SRSF2, DDX5 or HNRNPK protein expression. At Day 22, round spermatids are now present in the testis and by Day 24, they make up an even greater proportion of the cells. If loss of shorter UTR-containing mRNA transcripts affects translation, the results of the northern blot hybridization analysis would predict that we would observe altered protein levels.
SRSF2 protein was comparably expressed at Day 17 in control and mutant testis, but at Days 22 and 24, there was noticeably less SRSF2 expression in the mutant (Figure 4A). To be certain that the changes at Days 22 and 24 are indeed due to loss of protein expression specifically in mutant round spermatids, histological sections from adult control and mutant testes were immunostained for SRSF2. Expression of SRSF2 in pachytene spermatocytes was comparable in the control and mutant, but its expression in mutant round spermatids was indeed reduced as compared to control cells (Figure 4B). DDX5 also showed a similar pattern of expression, with comparable levels at Day 17 and less protein present in the Days 22 and 24 mutants (Figure 4A). Immunostaining with this antibody yielded very high background in the cytoplasm and was thus uninformative.
Figure 4.
Changes in protein expression correlate with alterations in 3′-UTR processing and alternative splicing. (A) Immunoblot analysis of protein extracted from whole control and BrdtΔBD1/ΔBD1 mutant testes of mice at post natal age 17, 22 and 24 days. At Day 17, no round spermatids are present in the testis. At Day 22 and even more so at Day 24, round spermatids compose a large proportion of the cells of the testis. SRSF2, DDX5 and HNRNPK protein expression is unchanged in Day 17 mutants. In Day 22 mutants, there is a slightly lower expression of these proteins, and this reduction in expression is even more pronounced at Day 24. The higher molecular weight band above HNRNPK is mouse IgG. Total TARDPB expression is increased relative to control expression at all time points. The ratio of longest isoform to the shortest isoforms is also increased in all mutant samples. An unknown ∼40 kDa isoform of TARDBP is weakly expressed in Day 22 control testis. Expression of this isoform is increased in the Day 22 mutant testis and its expression newly appears in the Day 17 mutant testis. (B) Immunostaining of SRSF2 in adult control and mutant testicular sections. SRSF2 protein is highly expressed in control pachytene spermatocytes and round spermatids and at a comparably high level in mutant spermatocytes. SRSF2 expression in mutant spermatids is noticeably reduced.
As previously mentioned, Hnrnpk has seven variant transcripts: four encode a protein of ∼51 kDa, two encode a protein of ∼48 kDa and one encodes a protein of ∼42 kDa protein. Immunoblot analysis detected two broad bands migrating at ∼54 and 51 kDa. The higher migrating band appears to be denatured mouse IgG, as use of Clean-Blot® IP detection reagent HRP in place of secondary goat-anti-mouse IgG-HRP resulted in no detection of this band. However, as the Clean-Blot reagent is not ideally meant to detect even natural mouse IgG, the HNRNPK band at ∼51 kDa, although present, was weak (data not shown). Although it is certain that there is no protein of ∼42 kDa (data not shown), it may be that the broad band observed at ∼51 kDa is actually a doublet of the 51 and 48 kDa bands. At Day 17, the HNRNPK band of protein(s) is comparable in the mutant testis but is highly reduced by Day 24 (Figure 4A).
Changes in alternative splicing lead to altered ratios of protein isoforms
The longest transcript of Tardbp encodes a ∼44 kDa protein while the other four known shorter transcripts encode proteins of ∼34 and 33 kDa. We observed a readily detected band at 44 kDa and what appears to be a protein doublet at ∼33–34 kDa in all analyzed samples. A previously undescribed putative TARDBP isoform of ∼40 kDa was also observed, but only in 17 and 22 day testes and at higher levels in the mutant. It is possible that this band is a degradation product of the 44 kDa isoform. There appears to be an overall increase in total TARDBP protein expression in the mutant starting at Day 17. Additionally, the amounts of the larger proteins are increased and the shorter isoforms are decreased as compared to control (Figure 4A). Thus, it appears that the changes in transcription and alternative splicing yield corresponding changes at the protein level.
Flag-tagged BRDT co-purifies with splicing proteins in cell lines
As bromodomains, the ET domain and the CTD are all protein interaction motifs, concomitant studies in our lab involve the identification of possible BRDT-interacting proteins using a BRDT pull-down approach in cultured 293T cells (L. Wang and D.J. Wolgemuth, manuscript in preparation). In this study, we focused on identifying interacting proteins that might explain mechanistically how BRDT is involved in mRNA processing. The proteins identified in our pull-down experiments were analyzed using the functional annotation tool of the DAVID v6.7 Software, and 10 were characterized as encoding components of the spliceosome, which represented a statistically significant enrichment (FDR corrected P ≤ 3.4 × 10−5) and 19 genes, 12 of which were not in the spliceosome list, were characterized as having a role in RNA splicing (FDR corrected P ≤ 3.5 × 10−10). This gave a total of 22 proteins that were involved in splicing events and might interact with BRDT (Table 2). Of those 22, 8 also showed a change in expression in the microarray data (Table 1) suggesting that perhaps this BRDT-containing spliceosome complex autoregulates some of its components on the transcriptional level.
Table 2.
Splicing proteins that co-precipitate with BRDT in 293T cell lines
| Pathway/gene ontology | Gene symbol | Gene name |
|---|---|---|
| Spliceosome | Ddx5 | DEAD (Asp–Glu–Ala–Asp) box polypeptide 5 |
| Spliceosome | Ddx39b | DEAD (Asp–Glu–Ala–Asp) box polypeptide 39b |
| Spliceosome | Hnrnpa1 | Heterogeneous nuclear ribonucleoprotein A1 |
| Spliceosome | Hnrnpk | Heterogeneous nuclear ribonucleoprotein K |
| Spliceosome | Hspa1a | Heat shock 70 kDa protein 1A; heat shock 70 kDa protein 1B |
| Spliceosome | Hspa1l | Heat shock 70 kDa protein 1 like |
| Spliceosome | Hspa8 | Heat shock 70 kDa protein 8 |
| Spliceosome | Pcbp1 | Poly(rC) binding protein 1 |
| Spliceosome | Srsf2 | Splicing factor, arginine/serine-rich 2 |
| Spliceosome | Snrpd3 | Small nuclear ribonucleoprotein D3 polypeptide 18 kDa |
| RNA splicing | Clns1a | Chloride channel, nucleotide sensitive, 1A |
| RNA splicing | Hnrnpa2b1 | Heterogeneous nuclear ribonucleoprotein A2/B1 |
| RNA splicing | Hnrnph1 | Heterogeneous nuclear ribonucleoprotein H1 (H) |
| RNA splicing | Hnrnpl | Heterogeneous nuclear ribonucleoprotein L |
| RNA splicing | Khsrp | KH-type splicing regulatory protein |
| RNA splicing | Nono | Non-POU domain containing, octamer-binding |
| RNA Splicing | Pcbp2 | Poly(rC) binding protein 2 |
| RNA Splicing | Ppp2r1a | Protein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform |
| RNA splicing | Prmt5 | Protein arginine methyltransferase 5 |
| RNA splicing | Sfpq | Splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) |
| RNA splicing | Tardbp | TAR DNA-binding protein |
| RNA splicing | Wdr77 | WD repeat domain 77 |
Full-length and truncated BRDT can complex with SRSF2, DDX5, HNRNPK and TARDBP in the testis
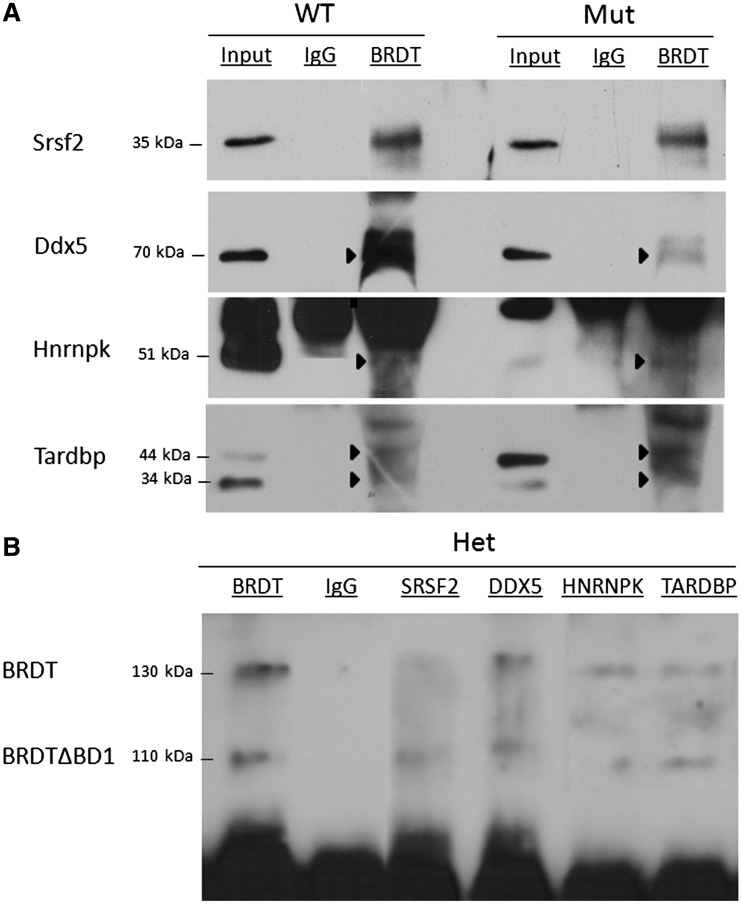
Each of the four genes whose expression changes, we investigated as described above are on the list of splicing proteins pulled down with BRDT (Table 2). We therefore confirmed that they indeed interact with BRDT by co-immunoprecipitation (Co-IP) using BRDT antibody and lysates from both control and mutant testes, followed by immunoblotting. All four proteins co-immunoprecipitated with both control and mutant BRDT but not with IgG (Figure 5A). The reciprocal Co-IP was performed using SRSF2, DDX5, HNRNPK and TARDBP antibodies and nuclear lysates from heterozygous BrdtΔBD1/+ testes, as this would allow simultaneous confirmation of interaction with both the full-length and truncated BRDT proteins. In each case, both full-length and mutant BRDT protein was co-immunoprecipitated (Figure 5B). Immunoprecipitation with BRDT antibody and IgG were used as positive and negative controls.
Figure 5.
Full-length and truncated BRDT protein can interact with SRSF2, DDX5, HNRNPK and TARDBP. (A) BRDT-containing complexes were precipitated from control and BrdtΔBD1/ΔBD1 mutant whole testis lysates using an anti-BRDT antibody or control rabbit IgG. Immunoblotting of the co-precipitated proteins was carried out with SRSF2, DDX5, HNRNPK and TARDBP antibodies. These four proteins all co-precipitate with both full-length and truncated BRDT protein, but not with IgG. IgG can be seen in all lanes at 55 kDa. (B) Co-immunoprecipitation using anti-SRSF2, anti-DDX5, anti-HNRNPK and anti-TARDBP antibodies and control anti-BRDT antibody and IgG with whole heterozygous BrdtΔBD1/+ testis lysates. Both full-length and truncated BRDT protein co-precipitates with all four proteins, but not IgG.
DISCUSSION
BET family genes have been widely implicated in transcriptional regulation (1). BRD4, in particular, has been shown to interact with the general transcription machinery via its CTD (11,40). As BRD4 is not expressed in the testis after the spermatogonial stages, BRDT is the only CTD-containing BET protein expressed in both meiotic and postmeiotic stages (14,38). We have previously suggested that BRDT may act as a transcriptional repressor of Hist1h1t expression and that this repression is achieved through BRDT binding to chromatin in the promoter of the gene via its BD1 (39), but there are no other reports to date about the possible role of BRDT in modulating gene expression during spermatogenesis.
To obtain a broader understanding of the role of BRDT and its BD1, in particular, in regulation of transcription, we performed gene expression microarray analysis with a focus on purified populations of control and BrdtΔBD1/ΔBD1 mutant round spermatids for several reasons. First, these cells are transcriptionally active and directly precede the step IX spermatids in which striking abnormalities in chromatin architecture and head morphology are observed in BrdtΔBD1/ΔBD1 mutant mice (39). Second, purified populations of cells were used instead of whole testes to enhance the sensitivity of detection of changes in expression in a specific population of cells, without the background of the rest of the testicular cell types. That is, the use of purified populations obviates detecting changes in gene expression that would merely reflect the changes in cellularity of the mutant and wild-type testis. Although BRDT could also regulate transcription in pachytene spermatocytes, this population of cells was not used for microarray analysis in this study because of technical difficulties in isolating adequate numbers of sufficiently pure populations of these cells.
It should be noted that the BrdtΔBD1 mutant allele is most probably hypomorphic, as the second bromodomain, ET domain and CTD are all intact and the truncated protein is as abundant as the full length protein (39). It is thus possible that there exists an entire set of genes that is only misregulated when cells lack the full-length BRDT protein. Nevertheless, our microarray analysis found that transcription of a large number of genes was both up-regulated and repressed in the absence of BD1 alone.
Interestingly, we observed that the genes that were normally repressed by BRDT, i.e. appearing to be up-regulated in the microarray of mutant Brdt spermatids, were enriched with genes involved in mRNA splicing. For some of these genes, the apparent up-regulation was genuinely at the level of transcription. For example, the transcriptional repressor (67) and splice regulator (68) gene Tardbp was up-regulated in mutant pachytene spermatocytes and round spermatids. However, for the majority of the splicing genes (63%), overall transcription did not change significantly, but rather the 3′-UTR processing in these genes was altered. That is, the enhanced signal detected in the mutant population reflected enhanced detection by probes covering regions in the 3′-UTR that are normally missing in wild type spermatids (31). The choice of polyadenylation site is regulated by a multitude of molecular factors and determines the length of the 3′-UTR (69). Many of these factors are shared by the spliceosome pathway (70). It is thus possible that the spliceosome uses 3′-UTR truncation as a mechanism for auto-regulation in haploid spermatids.
The physiological consequences of 3′-UTR truncation of specific mRNAs are not fully understood. The 3′-UTR is a known regulatory region as it can be bound by RNA-binding proteins (62,63) and microRNAs (64–66) which can modulate translation. Further, sequences in the 3′-UTR have been implicated in modulating translatability of stored meiotic and post meiotic transcripts (71,72). UTR truncation may be one mechanism by which transcripts are translationally regulated. Indeed, for the three genes we tested, the presence of a full-length 3′-UTR resulted in a drop in the corresponding protein levels in mutant round spermatids.
Although expression microarrays are normally not very informative about alterations in mRNA splicing, some probes will uniquely hybridize to alternative regions of transcripts (59). Tardbp was an example of this phenomenon, as there was a probe on the array that hybridized uniquely to intron 6, which when retained yields a longer transcriptional variant. In the mutant, splicing of Tardbp transcripts shifted such that there was a noticeable increase in the long variant and a decrease in the shorter ones. This shift was reflected on the protein level as there was an increase in the larger protein isoform and a decrease in the smaller ones.
Interestingly, we have demonstrated that not only can BRDT form a complex with the splicing proteins SRSF2 (Sc-35), DDX5, HNRNPK and TARDBP, but that it may also be involved in the regulation of expression of a number of other splicing factors that may regulate levels of alternative transcripts. The interaction with SRSF2 is of particular significance as it is one of the major splicing factors required for general mRNA splicing (73–75) but has heretofore not been found in complex with bromodomain-containing proteins. The fact that BRDT is part of the spliceosome complex may thus explain, at least in part, the changes in alternative splicing observed in the mutant testis.
It should be noted that, although not the focus of the present studies, our analysis also revealed differences in transcription levels, splicing, and 3′-UTR processing in BrdtΔBD1/ΔBD1 mutant pachytene spermatocytes. For example, there was increased transcription and altered splicing of Tardbp in mutant spermatocytes. These data, coupled with our previous observations on Hist1h1t (39), suggest that BRDT plays a role in transcriptional regulation and splicing from its first appearance in pachytene spermatocytes through the round spermatid stage. 3′-UTR truncation occurs in pachytene spermatocytes, but generally to a lesser extent than in round spermatids (31). However, for the genes we examined, this process was unchanged in mutant spermatocytes. In the BrdtΔBD1/ΔBD1 mutant testis, the loss of truncation was only detected in round spermatids; this yielded mutant spermatids with an overall mRNA composition that looked nearly identical to spermatocytes. Collectively, these observations suggest that the unique processes that result in increased 3′-UTR-truncated mRNAs in round spermatids are tightly regulated by BRDT and BD1-dependent, as they are clearly dis-regulated in the BrdtΔBD1/ΔBD1 mutant.
It is also possible that BRDT has a direct role in polyadenylation site choice and 3′-UTR processing itself, as several spliceosome components have been implicated in this process (70). However, this seems unlikely to be a critical function of BRDT, as the polyadenylation machinery is present in spermatocytes but no defects in 3′-UTR formation were detected in these BrdtΔBD1/ΔBD1 mutant cells. We suggest that, more likely, BRDT regulates the transcription and/or splicing of genes that are specifically involved in the formation of the shorter 3′-UTRs that are prevalent in round spermatids.
Finally, it is important to note that the BD1 of BRDT may be dispensable for BRDT–spliceosome complex formation since, at least as assessed by pull-down assays, the truncated protein can interact with components of the spliceosome complex. Rather, the BD1 of BRDT could be involved in targeting the spliceosome to specific chromatin locations. Interestingly, the spliceosome component SRSF2 has been implicated along with P-TEFb in transcriptional elongation (76) and reciprocally, P-TEFb has been implicated in regulating splicing (77). As mentioned above, BRD4 is known to bind to P-TEFb: we now speculate that BET genes which contain a CTD (BRD4, BRDT), may act as a scaffold to bring the spliceosome and P-TEFb together and target both of them to the promoters of genes which are to be transcribed. Although the CTD of BRDT was shown to bind P-TEFb in cell lines (40), it is not known if P-TEFb has a role during spermatogenesis. It will be interesting to determine whether BRDT targets P-TEFb to genes that are regulated during meiosis or spermiogenesis in vivo. Additionally, whether BRD4 shares BRDT’s role in the spliceosome will be of equal if not greater interest.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5, Supplementary Figures 1 and 2, Supplementary Materials and Methods and Supplementary References [78,79].
FUNDING
Funding for open access charge: National Institutes of Health [GM081767 to D.J.W.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Claire Egan, Justin DeGrazio and Dr Marcia Manterola for their help in maintaining the BrdtΔBD1 mouse colony. We also are grateful to Xiangyuan Wang for preparing histological sections. Additionally, we wish to thank Dr Sanny Chung, Dr Ana Vasileva and Dr Sunil Panigrahi for technical advice and Dr Manterola for critical comments and suggestions on the manuscript.
REFERENCES
- 1.Florence B, Faller DV. You bet-cha: a novel family of transcriptional regulators. Front. Biosci. 2001;6:D1008–D1018. doi: 10.2741/florence. [DOI] [PubMed] [Google Scholar]
- 2.Chiang CM. Brd4 engagement from chromatin targeting to transcriptional regulation: selective contact with acetylated histone H3 and H4. F1000 Biol. Rep. 2009;1:98. doi: 10.3410/B1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- 4.Huang H, Zhang J, Shen W, Wang X, Wu J, Wu J, Shi Y. Solution structure of the second bromodomain of Brd2 and its specific interaction with acetylated histone tails. BMC Struct. Biol. 2007;7:57. doi: 10.1186/1472-6807-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 6.Taniguchi Y, Suzuki H, Ohtsuka M, Kikuchi N, Kimura M, Inoko H. Isolation and characterization of three genes paralogous to mouse Ring3. Nucleic Acids Res. Suppl. 2001;2001:247–248. doi: 10.1093/nass/1.1.247. [DOI] [PubMed] [Google Scholar]
- 7.Peng J, Dong W, Chen L, Zou T, Qi Y, Liu Y. Brd2 is a TBP-associated protein and recruits TBP into E2F-1 transcriptional complex in response to serum stimulation. Mol. Cell Biochem. 2007;294:45–54. doi: 10.1007/s11010-006-9223-6. [DOI] [PubMed] [Google Scholar]
- 8.Lamonica JM, Deng W, Kadauke S, Campbell AE, Gamsjaeger R, Wang H, Cheng Y, Billin AN, Hardison RC, Mackay JP, et al. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc. Natl Acad. Sci. USA. 2011;108:E159–E168. doi: 10.1073/pnas.1102140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamsjaeger R, Webb SR, Lamonica JM, Billin A, Blobel GA, Mackay JP. Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3. Mol. Cell Biol. 2011;31:2632–2640. doi: 10.1128/MCB.05413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol. Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol. Cell. Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 15.Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bettegowda A, Wilkinson MF. Transcription and post-transcriptional regulation of spermatogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:1637–1651. doi: 10.1098/rstb.2009.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lui,W.C., CY. (2008). Transcription regulation in spermatogenesis. In: Cheng CY (ed). Molecular Mechanisms in Spermatogenesis. Springer, New York, Vol. 636, pp. 115–132. [DOI] [PubMed]
- 18.Hogeveen KN, Sassone-Corsi P. Regulation of gene expression in post-meiotic male germ cells: CREM-signalling pathways and male fertility. Hum. Fertil. 2006;9:73–79. doi: 10.1080/14647270500463400. [DOI] [PubMed] [Google Scholar]
- 19.Wolgemuth DJ, Chung SS. Retinoid signaling during spermatogenesis as revealed by genetic and metabolic manipulations of retinoic acid receptor alpha. Soc. Reprod. Fertil. Suppl. 2007;63:11–23. [PMC free article] [PubMed] [Google Scholar]
- 20.Chalmel F, Rolland AD, Niederhauser-Wiederkehr C, Chung SS, Demougin P, Gattiker A, Moore J, Patard JJ, Wolgemuth DJ, Jegou B, et al. The conserved transcriptome in human and rodent male gametogenesis. Proc. Natl Acad. Sci. USA. 2007;104:8346–8351. doi: 10.1073/pnas.0701883104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaspers S, Gellersen B, Kempf R, Samalecos A, Bergmann M, Steger K. Functional characterization of male germ cell-specific CREM isoforms. J. Androl. 2007;28:59–66. doi: 10.2164/jandrol.106.000976. [DOI] [PubMed] [Google Scholar]
- 22.Don J, Stelzer G. The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis. Mol. Cell Endocrinol. 2002;187:115–124. doi: 10.1016/s0303-7207(01)00696-7. [DOI] [PubMed] [Google Scholar]
- 23.Freiman RN. Specific variants of general transcription factors regulate germ cell development in diverse organisms. Biochim. Biophys. Acta. 2009;1789:161–166. doi: 10.1016/j.bbagrm.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gan Q, Chepelev I, Wei G, Tarayrah L, Cui K, Zhao K, Chen X. Dynamic regulation of alternative splicing and chromatin structure in Drosophila gonads revealed by RNA-seq. Cell Res. 2010;20:763–783. doi: 10.1038/cr.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eddy EM. Male germ cell gene expression. Recent Prog. Horm. Res. 2002;57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- 26.Elliott DJ, Grellscheid SN. Alternative RNA splicing regulation in the testis. Reproduction. 2006;132:811–819. doi: 10.1530/REP-06-0147. [DOI] [PubMed] [Google Scholar]
- 27.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venables JP. Alternative splicing in the testes. Curr. Opin. Genet. Dev. 2002;12:615–619. doi: 10.1016/s0959-437x(02)00347-7. [DOI] [PubMed] [Google Scholar]
- 29.Hecht N. Gene expression during male germ cell development. In: Ewing L, Desjardins C, editors. Cell and Molecular Biology of the Testis. Oxford: Oxford University Press; 1993. pp. 464–503. [Google Scholar]
- 30.Eddy EM, O'Brien DA. Gene expression during mammalian meiosis. Curr. Top. Dev. Biol. 1998;37:141–200. [PubMed] [Google Scholar]
- 31.Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007;35:234–246. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Sartini BL, Millette CF, Kilpatrick DL. A developmental switch in transcription factor isoforms during spermatogenesis controlled by alternative messenger RNA 3′-end formation. Biol. Reprod. 2006;75:318–323. doi: 10.1095/biolreprod.106.052209. [DOI] [PubMed] [Google Scholar]
- 33.O'Brien DA, Welch JE, Fulcher KD, Eddy EM. Expression of mannose 6-phosphate receptor messenger ribonucleic acids in mouse spermatogenic and Sertoli cells. Biol. Reprod. 1994;50:429–435. doi: 10.1095/biolreprod50.2.429. [DOI] [PubMed] [Google Scholar]
- 34.McMahon KW, Hirsch BA, MacDonald CC. Differences in polyadenylation site choice between somatic and male germ cells. BMC Mol. Biol. 2006;7:35. doi: 10.1186/1471-2199-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res. 2009;69:9422–9430. doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauschendorf MA, Zimmer J, Hanstein R, Dickemann C, Vogt PH. Complex transcriptional control of the AZFa gene DDX3Y in human testis. Int. J. Androl. 2011;34:84–96. doi: 10.1111/j.1365-2605.2010.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones MH, Numata M, Shimane M. Identification and characterization of BRDT: a testis-specific gene related to the bromodomain genes RING3 and Drosophila fsh. Genomics. 1997;45:529–534. doi: 10.1006/geno.1997.5000. [DOI] [PubMed] [Google Scholar]
- 38.Shang E, Salazar G, Crowley T, Wang X, Lopez RA, Wang X, Wolgemuth DJ. Identification of unique, differentiation stage-specific patterns of expression of the bromodomain-containing genes Brd2, Brd3, Brd4, and Brdt in the mouse testis. Gene Expr. Patterns. 2004;4:513–519. doi: 10.1016/j.modgep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Shang E, Nickerson HD, Wen D, Wang X, Wolgemuth DJ. The first bromodomain of Brdt, a testis-specific member of the BET sub-family of double-bromodomain-containing proteins, is essential for male germ cell differentiation. Development. 2007;134:3507–3515. doi: 10.1242/dev.004481. [DOI] [PubMed] [Google Scholar]
- 40.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl Acad. Sci. USA. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lygerou Z, Conesa C, Lesage P, Swanson RN, Ruet A, Carlson M, Sentenac A, Seraphin B. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res. 1994;22:5332–5340. doi: 10.1093/nar/22.24.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattaj IW, Tollervey D, Seraphin B. Small nuclear RNAs in messenger RNA and ribosomal RNA processing. FASEB J. 1993;7:47–53. doi: 10.1096/fasebj.7.1.8422974. [DOI] [PubMed] [Google Scholar]
- 43.Wolgemuth DJ, Gizang-Ginsberg E, Engelmyer E, Gavin BJ, Ponzetto C. Separation of mouse testis cells on a Celsep (TM) apparatus and their usefulness as a source of high molecular weight DNA or RNA. Gamete Res. 1985;12:1–10. doi: 10.1002/mrd.1120120102. [DOI] [PubMed] [Google Scholar]
- 44.Lotan G, Golan R, Efrati Y, Vigodner M, Lewin LM, Shochat L, Klin B. An experimental study of the effect of two distinct surgical techniques of orchiopexy on spermatogenesis and testicular damage in cryptorchid testes. Fertil. Steril. 2005;84:749–755. doi: 10.1016/j.fertnstert.2005.02.039. [DOI] [PubMed] [Google Scholar]
- 45.Golan R, Lewin LM, Soffer Y, Lotan G, Shochat L, Vigodner M. Evaluation of spermatogenesis using flow cytometry and confocal microscopy. Isr. Med. Assoc. J. 2003;5:536. [PubMed] [Google Scholar]
- 46.Janca FC, Jost LK, Evenson DP. Mouse testicular and sperm cell development characterized from birth to adulthood by dual parameter flow cytometry. Biol. Reprod. 1986;34:613–623. doi: 10.1095/biolreprod34.4.613. [DOI] [PubMed] [Google Scholar]
- 47.Team RDC. R: a language and environment for statistical computing. Vienna Austria R Foundation for Statistical Computing. 2008;1:ISBN 3–900051-900007-900050. [Google Scholar]
- 48.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 50.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 51.Binder H, Preibisch S, Berger H. Calibration of microarray gene-expression data. Methods Mol. Biol. 2010;576:375–407. doi: 10.1007/978-1-59745-545-9_20. [DOI] [PubMed] [Google Scholar]
- 52.Berkovits BD, Wolgemuth DJ. The first bromodomain of the testis-specific double bromodomain protein Brdt is required for chromocenter organization that is modulated by genetic background. Dev. Biol. 2011;360:358–368. doi: 10.1016/j.ydbio.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chapman DL, Wolgemuth DJ. Identification of a mouse B-type cyclin which exhibits developmentally regulated expression in the germ line. Mol. Reprod. Dev. 1992;33:259–269. doi: 10.1002/mrd.1080330305. [DOI] [PubMed] [Google Scholar]
- 54.Chung SS, Wolgemuth DJ. Role of retinoid signaling in the regulation of spermatogenesis. Cytogenet. Genome Res. 2004;105:189–202. doi: 10.1159/000078189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 56.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 58.D'Mello V, Lee JY, MacDonald CC, Tian B. Alternative mRNA polyadenylation can potentially affect detection of gene expression by affymetrix genechip arrays. Appl. Bioinformatics. 2006;5:249–253. doi: 10.2165/00822942-200605040-00007. [DOI] [PubMed] [Google Scholar]
- 59.Cui X, Loraine AE. Consistency analysis of redundant probe sets on affymetrix three-prime expression arrays and applications to differential mRNA processing. PLoS One. 2009;4:e4229. doi: 10.1371/journal.pone.0004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang HY, Wang IF, Bose J, Shen CK. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–139. doi: 10.1016/s0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- 61.Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 63.Idler RK, Yan W. Control of messenger RNA fate by RNA binding proteins: an emphasis on mammalian spermatogenesis. J. Androl. 2011 doi: 10.2164/jandrol.111.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 65.Yu Z, Raabe T, Hecht NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol. Reprod. 2005;73:427–433. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]
- 66.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 67.Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bose JK, Wang IF, Hung L, Tarn WY, Shen CK. TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J. Biol. Chem. 2008;283:28852–28859. doi: 10.1074/jbc.M805376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 70.Martinson HG. An active role for splicing in 3′-end formation. Wiley Interdiscip. Rev. RNA. 2011;2:459–470. doi: 10.1002/wrna.68. [DOI] [PubMed] [Google Scholar]
- 71.Kleene KC. A possible meiotic function of the peculiar patterns of gene expression in mammalian spermatogenic cells. Mech. Dev. 2001;106:3–23. doi: 10.1016/s0925-4773(01)00413-0. [DOI] [PubMed] [Google Scholar]
- 72.Steger K. Haploid spermatids exhibit translationally repressed mRNAs. Anat. Embryol. 2001;203:323–334. doi: 10.1007/s004290100176. [DOI] [PubMed] [Google Scholar]
- 73.Moen PT, Jr, Smith KP, Lawrence JB. Compartmentalization of specific pre-mRNA metabolism: an emerging view. Hum. Mol. Genet. 1995;4 doi: 10.1093/hmg/4.suppl_1.1779. (Spec No), 1779–1789. [DOI] [PubMed] [Google Scholar]
- 74.Bourgeois CF, Lejeune F, Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:37–88. doi: 10.1016/S0079-6603(04)78002-2. [DOI] [PubMed] [Google Scholar]
- 75.Blencowe BJ, Bowman JA, McCracken S, Rosonina E. SR-related proteins and the processing of messenger RNA precursors. Biochem. Cell Biol. 1999;77:277–291. [PubMed] [Google Scholar]
- 76.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lenasi T, Barboric M. P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms. RNA Biol. 2010;7:145–150. doi: 10.4161/rna.7.2.11057. [DOI] [PubMed] [Google Scholar]
- 78.Brettschneider J, Collin F, Bolstad BM, Speed TP. Quality assessment for short oligonucleotide microarray data. Technometrics. 2008;50:241–264. [Google Scholar]
- 79.Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics: a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.