Abstract
In this work, we describe the results of a comprehensive structural bioinformatics analysis of the spliceosomal proteome. We used fold recognition analysis to complement prior data on the ordered domains of 252 human splicing proteins. Examples of newly identified domains include a PWI domain in the U5 snRNP protein 200K (hBrr2, residues 258–338), while examples of previously known domains with a newly determined fold include the DUF1115 domain of the U4/U6 di-snRNP protein 90K (hPrp3, residues 540–683). We also established a non-redundant set of experimental models of spliceosomal proteins, as well as constructed in silico models for regions without an experimental structure. The combined set of structural models is available for download. Altogether, over 90% of the ordered regions of the spliceosomal proteome can be represented structurally with a high degree of confidence. We analyzed the reduced spliceosomal proteome of the intron-poor organism Giardia lamblia, and as a result, we proposed a candidate set of ordered structural regions necessary for a functional spliceosome. The results of this work will aid experimental and structural analyses of the spliceosomal proteins and complexes, and can serve as a starting point for multiscale modeling of the structure of the entire spliceosome.
INTRODUCTION
The spliceosome is a eukaryotic macromolecular ribonucleoprotein (RNP) complex that performs the excision of introns (non-coding sequences) from pre-mRNAs following transcription. In humans, two forms of the spliceosome exist. The major spliceosome, which excises >99% of human introns, is composed primarily out of four stable small nuclear ribonucleoprotein (snRNP) particles (subunits), named after their small nuclear RNA (snRNA) components: U1, U2, U4/U6 and U5. The minor spliceosome, which is absent in many species and which in human excises the remaining <1% introns, contains a U5 snRNP identical to the one from the major spliceosome, as well as two other snRNPs: U11/U12, and U4atac/U6atac. The U11/U12, and U4atac/U6atac di-snRNPs are distinct from, but structurally and functionally analogous to, the U1 and U2, and U4/U6 di-snRNP, respectively (1). The major human spliceosome contains 45 distinct proteins in its snRNP subunits in addition to around 80 abundant non-snRNP proteins (2). These proteins, together with the snRNAs, may be considered to be an experimental approximation of the ‘core’ of the spliceosome, that is the set of structural elements necessary for the procession of the splicing reaction. Proteomics analyses of spliceosomal proteomes from various species yield also up to over 100 non-abundant splicing proteins (2–8), which may be active e.g. in certain instances of splicing. Out of the 45 distinct snRNP proteins, only seven, the so-called Sm proteins, are present in more than one copy. The Sm proteins form heteroheptamers with a toric shape, one per each of the U1, U2, U4 and U5 snRNPs. In each snRNP, the Sm heteroheptamer forms a platform that supports the respective snRNA. A similar platform associated with the U6 snRNA is composed of a set of seven related ‘like-Sm’ proteins (9).
Splicing-related proteins may also participate in other cellular events, including mRNA transcription (10,11), 5′ capping, 3′ cleavage and polyadenylation, as well as mRNA export, localization and decay (12,13) and box C/D snoRNP formation (14). While the majority of non-snRNP proteins are independent factors, some associate into non-snRNP protein complexes, which include the hPrp19/CDC5L (NTC) complex (15), the exon-junction complex (EJC) (16), the cap-binding complex (CBP) (17), the retention-and-splicing complex (RES) (18), and the transport-and-exchange complex (TREX) (19). These complexes may also have non-splicing functions (16,20).
A characteristic feature of the spliceosome is its extraordinary dynamism, as the snRNP composition of a spliceosome entity bound to the substrate pre-mRNA changes depending on the stage of the splicing reaction. For the major spliceosome, an E (entry) complex spliceosome contains U1 snRNP, an A complex contains U1 and U2 snRNP, a B complex contains U1 and U2 snRNP in addition to a tri-snRNP entity composed of the U4/U6 and U5 snRNPs, called U4/U6.U5, while the activated B (B-act) and catalytic (C) complexes contain U2, U5 and U6 snRNPs. After the splicing catalysis occurs and the mRNA is released, the initial configuration of the snRNPs (U1, U2 and U4/U6 and U5 separately) is recycled (21). Each stage-specific configuration of the snRNP subunits is also associated with a different non-snRNP protein complement. As a result, just like the snRNP composition, the non-snRNP composition of a given instance of the spliceosome also varies (2). In recent years, evidence has surfaced that ubiquitin-based (22–24) and intrinsic disorder-based (25) systems may contribute to the regulation of splicing assembly and dynamics.
To further the studies of the spliceosome and the association between splicing and other cellular processes, it is useful to determine the domain architecture and the three-dimensional structures of spliceosomal proteins. Detailed knowledge of protein structure can help determine how molecules perform their biological functions. Structure can also aid in understanding the effects of variations, resulting, e.g. from SNPs or from alternative splicing, which may have implications for disease. Besides, identification of structural similarities can reveal distant evolutionary relationships between proteins that cannot be detected from a comparison of their sequences alone (26). Of particular importance is the structural analysis of components of larger systems and complexes that have eluded high-resolution structural characterization. For instance, it has been suggested that high-resolution models of individual snRNP components may be fit into molecular envelopes created by low-resolution cryo-electron microscopy (cryo-EM) maps (27) to construct structures of the spliceosome at different stages of its action (28). Thereby, structural characterization of individual components of the spliceosome can bring us closer to modeling the structure and function of the entire system.
There are two main potential gaps in our understanding of the structure of the protein components of the spliceosome. The first one lies in recognizing the protein architecture at the primary level, e.g. the detection of conserved/structured domains and disordered regions. Most structural domains of splicing proteins are annotated by automated inferences in protein sequence databases such as UniProt (29). Many domains, especially those of the ‘core’ splicing proteins, have also been characterized in literature. However, automated annotations are limited in that they can only either spread information that is already available in the system (such as through homology inferences) or information that conforms to tight preset standards (such as in the detection of domains that conform to PFAM domain profiles) (30). Hence, at times, elements of protein architecture remain undetected throughout automated annotation, and can only be determined through additional analyses and human interpretation of other data.
The second gap lies in the lack of structural representation. Partial or complete structures have been determined for many splicing-related proteins and their complexes. These include a nearly complete U1 snRNP (31), U4 snRNP core with the Sm ring (32), several complexes associated with the spliceosome such as the human EJC (33) or the human CBP (34) and various protein–protein and protein–RNA complexes, such as the human U2 snRNP protein p14 (SF3b14a) bound to a region of SF3b155 (35). In total, as of December 2011, data from the Protein Data Bank (PDB) (36) show that at least 340 structures have been determined by X-ray crystallography and NMR for human spliceosomal proteins or their domains, either alone or in various complexes. Many of these structural models are redundant because they represent the same regions of the same proteins. However, for many regions, no three-dimensional models are available.
As an essential step towards enhancing our current understanding of the spliceosome, we have carried out a systematic structural bioinformatics analysis of the proteins of the human spliceosomal proteome, with a dual focus on characterizing their ordered parts and modeling their structures. In an effort to help set the priorities for future modeling of the entire spliceosome, we also compared the human spliceosomal proteome with the proteome of the parasitic diplomonad Giardia lamblia, known for its genomic minimalism. We put forward the set of structural regions common for human and G. lamblia as an attractive target for future studies. This analysis complements a parallel study of the unstructured part of the proteins of the spliceosome (I.K. and J.M.B., submitted for publication), and runs alongside efforts of many research groups to characterize the structure of spliceosomal RNAs and map out the interactions between the spliceosomal components.
MATERIALS AND METHODS
Collection and classification of spliceosome proteins
A total of 244 proteins found in the proteomics analyses of the major human spliceosome [sourced from one or more of the following references (2,4,8,37–41)], and 8 proteins specific to the U11/U12 di-snRNP subunit of the minor spliceosome (Supplementary Table S1) (42), were downloaded from the NCBI Protein (nr) database. Proteins were classified as ‘abundant’ and ‘non-abundant’ according to (2), and they were assigned into groups based mainly on (2), followed by references (4,38–40). Proteins classified here as ‘miscellaneous’ were classified in primary sources, variably, as ‘miscellaneous proteins’, ‘miscellaneous splicing factors’, ‘additional proteins’, ‘proteins not reproducibly detected’ and ‘proteins not previously detected’. We disclaim any responsibility for the factual accuracy of the association of proteins with the relevant groups beyond the point of following the primary sources.
Sequence searches, alignments and clustering
Searches of protein homologs in the NCBI Protein (nr) database were carried out at the NCBI using BLASTP/PSI-BLAST (43) with default parameter settings. Putative homology was validated by reciprocal BLASTP searches against the Protein database with ‘human’ (NCBI taxon id: 9606) as a taxon search delimiter. Sequence alignments were calculated using the MAFFT server using the Auto strategy (http://mafft.cbrc.jp/alignment/server/) (44). Clustering analysis of helicase sequences was performed with CLANS (45).
Identification and description of structural regions of proteins
Identification of intrinsically ordered and disordered regions of proteins, prediction of protein secondary structure and domain boundaries, as well as fold-recognition (FR) analyses, were carried out via the GeneSilico MetaServer gateway (for references to the original methods, see https://genesilico.pl/meta2) (46). In non-trivial cases (usually when putative modeling templates returned by FR scored low and/or various methods disagreed on the best template), FR alignments to the top-scoring templates from the PDB were compared, evaluated and ranked by the PCONS server (47), and the PCONS result was used to identify region boundaries. Additional searches were performed on the HHPRED server (48).
SCOP database (49) IDs used for the purposed of structural domain identification were either extracted from the Protein Data Bank or from the SCOP parseable files on the SCOP website (http://scop.mrc-lmb.cam.ac.uk/scop/parse/index.html) or assigned using the fastSCOP server (http://fastscop.life.nctu.edu.tw/) (50). PFAM domain names were assigned on the PFAM website (http://pfam.sanger.ac.uk/). SCOP v. 1.75 and PFAM v. 25.0 were used. Structural similarity was compared using the DALI server (51).
Assignment of models to structural regions of proteins
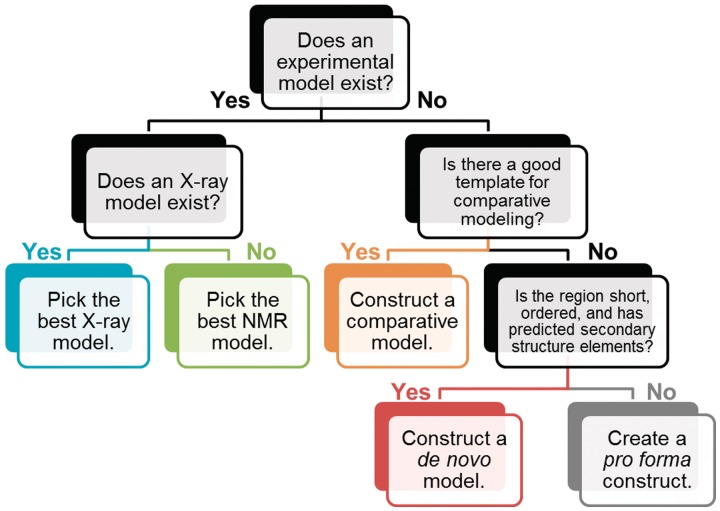
In assigning structural models to regions, we followed a four-step procedure (Figure 1). Whenever a high-resolution experimental structural model (either X-ray or NMR structure) was available, we assigned it to the corresponding sequence region. If a structural similarity to a protein of known structure was predicted for a given region by fold-recognition algorithms (see below for details), we constructed a model for this region by a comparative (template-based) modeling technique, using the detected experimental structures as templates. In the absence of confidently predicted templates, we used de novo folding methods for relatively small fragments likely to form globular domains. For the remaining regions (those without experimentally solved structures and for which the current modeling methodology cannot provide confident predictions of the 3D structure), we generated pro forma models, in which only the primary and (predicted) secondary structure was represented explicitly, while the tertiary arrangement was arbitrary. Pro forma models are not supposed to be reliable at the tertiary level and were constructed for the sake of further analyses (e.g. to initialize protein folding analyses that require some kind of a structural representation as an input).
Figure 1.
Rules for selecting and producing structural representations of protein regions. From left to right, structural representations decrease in the average confidence.
For regions with multiple solved structures in the Protein Data Bank, the following criteria of preference were used: (i) structures of the region in complex with other proteins and/or nucleic acids (i.e. in a potentially ‘active’ or ‘functionally relevant’ state) were given priority over structures of the region in isolation, (ii) crystallographic structures were given priority over NMR structures, (iii) higher-resolution crystallographic structures were given priority over lower-resolution structures and (iv) more complete structures were given priority over less complete structures. The following experimental artifacts were removed from experimental structure files or corrected by standard modeling procedures: non-native sequences added to aid in the protein expression and structure determination process (e.g. affinity tags), non-standard amino acids (e.g. selenomethionine was replaced by methionine), and gaps in sequences (e.g. short disordered loop fragments were added). Single chains only were retained if the original PDB file contained multiple chains of the same protein.
Comparative models were constructed by default with MODELLER (52) based on templates identified in the fold-recognition process. Selected challenging models were constructed using the I-TASSER server (53). Selected models were also adjusted with ROSETTA 3.0/3.1 using the loop modeling mode (54). De novo models were produced with the ROSETTA 3.0/3.1 AbInitioRelax application and clustered with the Rosetta 3.0/3.1 Cluster Application, following the protocols set out in the ROSETTA User Guide for version 3.1. (http://www.rosettacommons.org/manual_guide) (54). De novo folding was attempted if the following conditions were fulfilled: the region was ≤125 residues in length, predicted to be completely ordered and predicted to contain secondary structure elements. These conditions correspond to the current practical limit of utility of this type of methods (55). Artificial pro forma spatial representations of protein chains of unknown/uncertain structure or predicted to lack a stable structure were built with UCSF Chimera (v.1.4/1.5) using the Tools>Structure Editing>Build Structure command (56). Pro forma constructs reflect only the known primary and predicted secondary structure of the corresponding regions, while their tertiary structure should be regarded as unassigned (and remains to be modeled in the future). Miscellaneous manipulations of structures and models of molecules during this stage were performed in UCSF Chimera (56) and Swiss-PdbViewer v. 4.0.1 (57).
Protein model quality assessment
Assessment of model quality was performed with MetaMQAPII [https://genesilico.pl/toolkit/unimod?method=MetaMQAPII, an updated version of a method described in (58)] and QMEAN [http://swissmodel.expasy.org/qmean/ (59)].
MetaMQAP predicts the deviation of the query model from the (unknown) native structure and expresses it as the predicted global root mean square deviation (RMSD) and the predicted global distance test total score (GDT_TS) (60). The lower the predicted RMSD and the higher the predicted GDT_TS score, the better the model.
QMEAN first calculates an internal score, and then the QMEAN Z-score indicates by how many standard deviations the QMEAN score of the model differs from expected values for experimental structures that have a similar length to the model. High quality models are expected to have positive QMEAN Z-scores, and good models are expected to have a QMEAN Z-score above −2.0. Indicators of accuracy of individual residues were generated by MetaMQAPII and are supplied as B-factor values inside the model files available from the SpliProt3D database website (see below). They can be visualized with the UCSF Chimera command Render By Attribute > (attributes of residues: average B-factor) or with equivalent commands in other molecular visualization programs. Mean values and standard deviations of the QMEAN Z-scores for the six QMEAN contributing factors are provided with this publication (Supplementary Table S4) and the values for all models are provided with the model files. Models of low quality are expected to have a strongly negative QMEAN Z-score, but also strongly negative Z-scores for most of the contributing terms.
As MetaMQAPII is not capable of evaluating multimeric models, for models of protein complexes (11 X-ray models and 2 NMR models) only the quality of the longest chain was evaluated by MetaMQAPII.
Website/database of models
Models and additional data, including alignments of representative sequences annotated with predictions of order/disorder, secondary structure, binding disorder, solvent accessibility and coiled coils, as well as and annotations of sites of post-translational modification from UniProt (29), are available via the SpliProt3D web server at http://iimcb.genesilico.pl/spliprot3D. The entire archive of files available for download has approximately 250 MB.
Visualization of sequence alignments and molecular structures
Sequence alignments were visualized with Jalview v. 2.6.1 (61), while molecular structure graphics were produced with UCSF Chimera (56).
RESULTS AND DISCUSSION
Identification of structural domains of splicing proteins
Our main priorities in identifying structural domains of splicing proteins were to check and correct previously reported domain boundaries and to identify and characterize domains that were not available in UniProt and other databases. We focused on 252 proteins of the human spliceosome, including 244 proteins found in the results of proteomics analyses of the major human spliceosome and 8 proteins specific to the U11/U12 subunits of the minor spliceosome (see ‘Materials and Methods’ section for references to protein sources and Supplementary Table S1 for protein GIs). We did not find any references to U4atac/U6atac-specific proteins either in literature or in the Gene Ontology (GO) database [http://geneontology.org (62)]. A total of 118 proteins were classified as ‘abundant’ as in (2); other proteins were classified as ‘non-abundant’. ‘Abundant’ proteins are suggested to be the most important for the correct action of the spliceosome (2).
Using a combination of protein fold-recognition and sequence conservation-based domain identification methods, we identified 465 ordered structural domains in the 252 proteins, including 80 domains in the snRNP proteins of the major human spliceosome (Table 1 and Supplementary Table S2). Ordered structural domains cover >80% of the ordered regions of the proteins, and ∼50% of all residues in the splicing proteins. Correspondingly, close to a half of the human spliceosomal proteome is predicted to be intrinsically disordered. The analysis of various structural and functional types of intrinsic disorder in the spliceosome brought about a quantity of data whose presentation is beyond the scope of this article and that has been consequently made the subject of an independent article (I.K. and J.M.B., submitted for publication).
Table 1.
Statistics of structural domains detected in the human spliceosomal proteome
| Feature | Major spliceosome snRNP | All proteins |
|---|---|---|
| Number of proteins | 45 | 252 |
| Number of residues | 20 390 | 133 040 |
| Number of ordered residues | 13 427 | 63 242 |
| Number of ordered structural domains | 80 | 465 |
| Number of suspected ordered structural domains | 7 | 25 |
| Number of domains predicted to be disordered, but found to be ordered in experimentally determined structures | 3 | 9 |
| Fraction of ordered residues covered by ordered structural domains (%) | 89.6 | 90.3 |
| Fraction of total number of residues covered by ordered and disordered structural domains (%) | 61.0 | 43.4 |
Based on the predicted order/disorder boundaries and the presence/absence of predicted secondary structure elements, we also detected 25 regions that we termed ‘suspected domains’. This category included two groups of regions. The first group were domain-length (>40 residues) regions without a recognized fold that were the only ordered regions of otherwise highly intrinsically disordered proteins (≥70% residues predicted to be disordered). The second group were present in proteins with low-to-middle intrinsic disorder content (<70% residues predicted to be disordered) that contained other ordered structural domains. The ‘suspected domains’ in these proteins were ordered regions that had clear order/disorder boundaries and contained predicted secondary structure elements, but lacked a PFAM domain assignment (30) and showed no clear relationship to any known folds according to protein fold-recognition analyses.
Ordered domains of splicing proteins classified in the SCOP (49) catalogue belong to classes a–e and g, with an over-representation of class d, which contains superfamily d.58.7 (RNA-binding domain, RRM (RBD), which usually corresponds to PFAM domain PF00076, RRM_1; Table 2). RRM is present in the 252 proteins in as many as 117 copies. This means that roughly each fourth to fifth domain in the spliceosomal proteome is an RRM. As RRM is a small domain that usually binds single-stranded RNA (63,64), this reflects the key character of protein–RNA interactions in the splicing process.
Table 2.
Statistics of ordered structural domains of the human spliceosome according to the SCOP classification
| SCOP ID | Description | Number of domains |
|---|---|---|
| a | All α | 79 |
| b | All β | 83 |
| c | α and β (a/b) | 53 |
| d | α and β (a + b) | 159 |
| e | Multi-domain (α and β) | 1 |
| g | Small | 49 |
Other common types of ordered protein regions found in the human spliceosomal proteome include other small RNA-binding domains, large α- and β-repeat-based protein-binding domains, small protein disorder-binding domains, ubiquitin-related domains and stable multidomain RNA helicase architectures (Table 3). Repeat-based domains are often found as building blocks of protein complexes, while some of the ubiquitin-related domains have been shown to be part of a putative ubiquitin-based system of controlling spliceosome assembly and dynamics (22,65).
Table 3.
Common types of ordered structural domains in the human spliceosomal proteome
| Domain type | Example PFAM domains | Number of copies | Examples of proteins |
|---|---|---|---|
| Small RNA-binding domains | RRM_1a, PWI, KH_1, S1, KOW, dsrm, G-patch, Surpb, SAP, zf-CCCH, zf-U1c, zf-metc, zf-C2H2_jazc, zf-U11-48K, zf-CCHC, FYVE | ≥201 | U1-A, U1-70K, U1-C |
| Small protein disorder-binding domains | WW, FHA, FF, GYF, SMN, SH3_1 | ≥24 | FBP11, U5-52K (CD2BP2) |
| Repeat-based protein-binding domains | Arm, TPR/HAT, HEAT, LRR_4, WD40 repeats | ≥28 | U4/U6-60K (hPrp4), U5-102K (hPrp6), SF3b155, U2-A' |
| Ubiquitin-related domains | Ubiquitin, U-box, zf-UBP, UCH, Rtf2, zf-C3HC4, ZZ, DWNN, RWD, JAB + PROCT | ≥19 | SF3a120, U4/U6.U5-65K, RNF113A |
| Heat shock-related | DnaJ, HSP70, HSP20, CS | ≥6 | CCAP1 |
| Proline isomerase | Pro_isomerase | 8 | U4/U6-20K (PPIH) |
| Stable helicase architectures | DEAD + Helicase_C, DEAD + Helicase_C + HA2 + OB_NTP_bind, (DEAD + Helicase_C + Sec63) × 2, Upf1p-like | ≥19 | hPrp43 (DHX15), U5-200K (hBrr2), KIAA0560 (AQR) |
| Small domains that act as ligands | U1snRNP70_N, SF3b1, PRP4, SF3a60_bindingd | ≥6 | SF3b155, U4/U6-60K (hPrp4) |
| Sm/Lsm domains | LSM | 14 | Sm, Lsm proteins |
aSome RRM domains bind peptide ligands (66).
bThe Surp domain is predicted to bind RNA. However, in the only single structure of a Surp domain in complex (PDB ID: 2DT7), the Surp domain binds a peptide ligand.
cSome zf-C2H2 domains mediate protein binding.
In addition to ordered domains, we found nine regions with an expected independent function that were predicted to be disordered, but that were either found in experimental structures or could be confidently modeled due to strong sequence matches to known domains. We considered these nine regions to be putative disordered domains that undergo a transition to order upon entering a complex. We discuss the features of these domains in an independent article that focuses specifically on intrinsic disorder in the spliceosomal proteome (I.K. and J.M.B., submitted for publication). Here, we will only note that, in general, the identification of disordered structural domains is currently a non-trivial task in comparison with the identification of ordered structural domains, as fewer experimentally validated examples of disorder exist in databases and the properties of disorder make automated identification and propagation more difficult.
Non-redundant set of experimental and theoretical structural models
Following the identification of domains, we constructed a non-redundant set of experimental and theoretical structural models of regions in splicing proteins. As the utility and credibility of models, both experimental and theoretical, depends on their accuracy, we set some simple heuristic rules of preference to increase the chance that we chose the models with the best quality. We preferred experimental models over theoretical models, X-ray experimental models over NMR experimental models and comparative theoretical models over de novo theoretical models (Figure 1). The lowest tier in the hierarchy was pro forma constructs, in which only the primary and secondary structure were represented explicitly, while the tertiary arrangement was arbitrary. As a result, we mapped 104 non-redundant experimental models to the sequences of the spliceosomal proteins, and created 255 comparative and 43 de novo models (Table 4 and Supplementary Table S3), as well as over 500 constructs. The 104 non-redundant experimental models include 23 models of (nucleo)protein complexes, of which 13 complexes have residues from more than one spliceosome-associated protein. While models of complexes tend to have lower accuracy than models of isolated chains, we considered them to be more informative about the protein functional than models of isolated chains. This was the only instance where we favored the availability of additional information over plain accuracy of the structure.
Table 4.
Structural representations of regions of proteins of the human spliceosomal proteome
| Feature | Major spliceosome snRNP | All proteins |
|---|---|---|
| Number of proteins | 45 | 252 |
| Number of residues | 20 390 | 133 040 |
| Number of ordered residues | 13 427 | 63 242 |
| Number of non-redundant experimental models | 20 | 104 |
| Number of non-redundant X-ray models | 11 | 43 |
| Mean resolution of X-ray models (Å) | 2.20 | 2.08 |
| Number of non-redundant NMR models | 9 | 61 |
| Number of non-redundant theoretical models | 49 | 297 |
| Number of non-redundant comparative models | 37 | 255 |
| Number of non-redundant de novo models | 13 | 43 |
| Total number of non-redundant representations | 139 | 803 |
| Number of experimental models containing residues of more than one splicing protein (X-ray/NMR) | 9 (8/1) | 13 (11/2) |
| Total fraction of structural order covered (%) | 91.2 | 92.7 |
| Total fraction of combined protein sequence covered (%) | 64.3 | 48.7 |
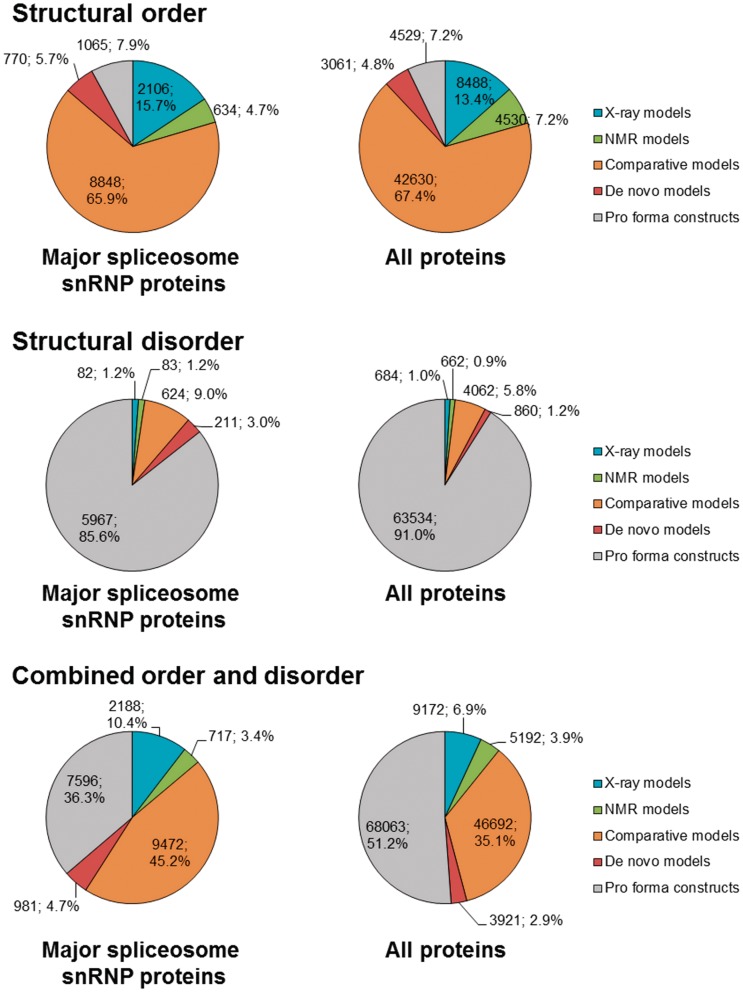
Over 90% of ordered regions of splicing proteins can be associated with experimental structural information or with comparative and de novo models (Figure 2). This value is similar for the proteins of the snRNP subunits of the major spliceosome and other proteins associated with the human spliceosome. Between different types of structural representations, experimentally determined structural models cover 20.6% of all ordered residues, the comparative models we generated cover 67.4% of all ordered residues, and the de novo models cover 4.8% of all ordered residues. Hence, our theoretical models cover three times the length of ordered protein sequence covered by experimental models.
Figure 2.
Coverage of structural order and disorder with different types of structural models. The values displayed on the graph are the number of residues covered by a given type of structural model, followed by percentage value.
X-ray crystallography is useful for the structure determination of large proteins (>30 kDa) and protein complexes, while NMR is well-suited for the structure determination of relatively small proteins. Not surprisingly, the ratio of the number of ordered residues in proteins from snRNP subunit structures solved by X-ray crystallography versus NMR is ∼3:1 (15.7%:4.7%), while this ratio for all splicing proteins is ∼1.77:1 (13.4%:7.2%). The main reason for this is that small domains are statistically more populous in the general set of splicing proteins compared to the snRNP subunits. Contrariwise, most structures of protein–protein complexes available for splicing proteins include regions from snRNP proteins. Since the resolution (and hence accuracy) of experimentally determined structures is typically inversely correlated with the molecule or complex size, X-ray models of snRNP proteins have on average a slightly worse resolution (mean 2.20 Å) than X-ray models of all spliceosomal proteins (mean 2.08 Å).
For predicted disordered regions, confident structural coverage is very low in comparison to ordered regions. Less than 2% of residues predicted to be disordered are covered by experimental models, and even together with our theoretical models, we could only cover 8.9% of all disordered residues. Moreover, most of the residues covered belong to linkers between ordered structural domains or short regions in protein termini. This low coverage of intrinsically disordered regions by structural models may be in the future a considerable challenge in producing a comprehensive structural model of the spliceosome.
Assessment of model quality
For all models except pro forma constructs, we also independently evaluated their accuracy to determine how credible they were. To do this, we used two methods: MetaMQAPII (58) and QMEAN (59). Both of them provide a global score for the entire model (predicted RMSD for MetaMQAPII, QMEAN Z-score for QMEAN) as well as a local score for individual residues (in this analysis, only the MetaMQAPII score was used). Functionally relevant and evolutionarily conserved regions (e.g. binding interfaces) are typically predicted with a higher than average accuracy, in particular when comparative modeling is used. Consequently, even a model with a poor global score can be useful for functional considerations, if its functionally important parts are scored well and are likely to be accurate. Some readers may also be interested in scores that describe only the model’s quality with respect to a particular feature (e.g. secondary structure). To help describe different features of models, we recorded the mean values and standard deviations of QMEAN Z-scores for six QMEAN contributing factors. These values for all models are provided with the manuscript (Supplementary Table S4).
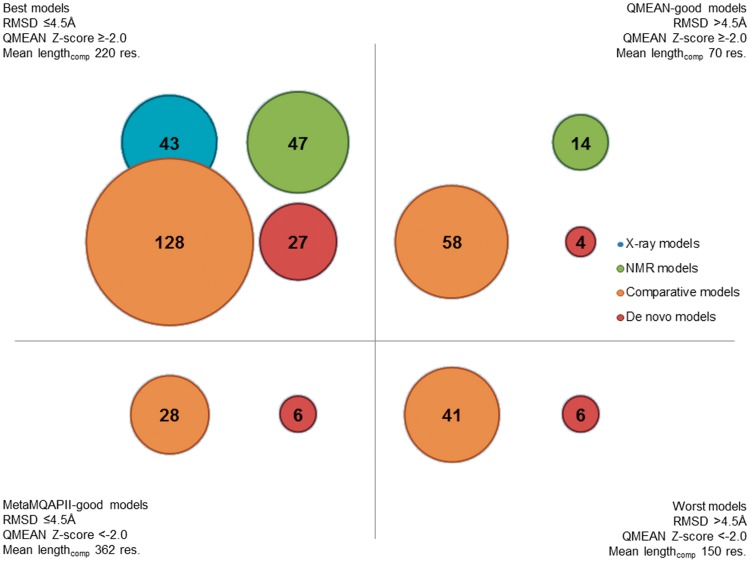
For comparison with theoretical models, we ‘predicted’ the global quality of experimentally determined structures (Supplementary Figure S1). Expectedly, both X-ray and NMR models we selected for our data set are highly scored by both MetaMQAPII and QMEAN, which is an indicator of the high accuracy of these structures (Table 5; for RMSD, the lower the score, the better the model; for the QMEAN Z-score good models are scored higher). Mean QMEAN Z-scores for models of both types (0.42 for X-ray and 0.08 for NMR) compare favorably to mean QMEAN Z-scores of models across the entire PDB (−0.58 and −1.19, respectively) (67). As X-ray models in our database were scored slightly better than NMR models, we used scores for X-ray models as a benchmark with which to classify theoretical models into those ‘likely to be globally accurate’ or ‘unlikely to be globally accurate’. The worst-scored X-ray models in our data set have a predicted RMSD of 4.5 Å (PDB ID 2ok3, resolution 2.0 Å) and a QMEAN Z-score of −1.99 (PDB ID 2qfj, resolution 2.10 Å). Consequently, we divided all non-X-ray models into four classes depending on passing one or both thresholds: predicted RMSD ≤4.5 Å and QMEAN Z-score ≥−2.0 (Figure 3).
Table 5.
Predicted quality of models of regions of human spliceosomal proteins
| Feature | X-ray | NMR | Comparative | De novo |
|---|---|---|---|---|
| Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | |
| Number of models | 43 | 61 | 255 | 43 |
| Predicted RMSD (MetaMQAPII) | 1.90 (0.84) | 3.85 (1.82) | 4.53 (1.96) | 4.02 (1.50) |
| Predicted GDT_TS (MetaMQAPII) | 78.56 (12.78) | 55.94 (19.45) | 47.28 (21.35) | 45.59 (15.85) |
| QMEAN total score | 0.805 (0.087) | 0.744 (0.110) | 0.585 (0.164) | 0.562 (0.132) |
| QMEAN Z-score | 0.42 (0.87) | 0.08 (0.86) | −1.30 (1.43) | −1.42 (1.33) |
Figure 3.
Models of regions of human splicing proteins divided by quality. This bubble graph displays the numbers of models of different types that belong to different classes of quality. Mean lengthcomp is the mean length of a comparative model of a given quality class.
The majority of both NMR and theoretical models belong to the most reliable class (i.e. ‘scored not worse than the worst crystal structures in the data set’). These models are expected to be generally correct, although their local accuracy may vary. Models scored well only by one method should be treated with more caution than models scored well by both methods. However, poor scoring by one method may also be due to the model being either very short or very long. Models that are scored poorly by MetaMQAPII, but are scored well according to the QMEAN Z-score are usually short, while models that are scored high by MetaMQAPII and low by QMEAN are usually long. The mean length of a model scored well by both methods is 220 residues, but the mean length of a model scored well only by QMEAN is 70 residues and the mean length of a model scored well only by MetaMQAPII is 362 residues. Therefore, we urge the reader to consider the length of the model before while using models scored poorly by only one method.
Over 40 models are scored poorly by both MetaMQAPII and QMEAN. These models may have been built on remotely related templates or did not fold well when modeled de novo, and are to be expected to have various errors. Based on our previous experience, we believe that some of these cases may represent new protein folds or interesting variations of known folds that present considerable challenge for protein modeling methods. Hence, while we regard these models as unreliable, we propose the corresponding proteins or domains as attractive targets both for experimental protein structure determination, and for protein modeling with other advanced techniques.
Database
The entire non-redundant set of representations (including selected representative models determined by experimental methods, and all theoretical models built with computational methods) is available as an online database SpliProt3D at http://iimcb.genesilico.pl/SpliProt3D. The web server allows for browsing, selecting and downloading the models. Proteins are also associated with sequence alignments annotated with predictions of intrinsic order versus disorder, predictions of secondary structure, protein-binding disorder, solvent accessibility and coiled-coils, as well as the positions of post-translational modifications. The database will be curated and new entries will be added and obsolete ones archived following the progress in structure determination of new spliceosomal proteins and/or publication of new theoretical models with better predicted accuracy. We would like to encourage structural biologists working on structure determination or prediction for spliceosomal proteins to contact us to have their models included and referenced in our database.
Comparison of predictions with the experimentally determined SF3A structure
After submission of this article for review, a crystal structure of the yeast U2 snRNP SF3A sub-complex was published (68), giving us an opportunity to compare some of our predictions with the independently determined experimental structure.
The structure of the yeast SF3A complex includes, in addition to several regions composed of individual secondary structure elements, three ordered domains for which an experimental structure had not been published before. One domain in the yeast protein Prp9 is >200 residues long (its counterpart in the human protein SF3a60 is situated roughly between residues 1–77, 129–244 and 310–372); it features a novel helical architecture. Originally, we made no tertiary structural predictions for this domain (i.e. our database contained only constructs), and it is highly unlikely that the structure of this domain could have been predicted accurately by a standard bioinformatics approach. Another domain in the yeast Prp9 is a zf-C2H2 zinc finger inserted into the long helical domain, whose counterpart in the human protein SF3a60 lacks the Zn-binding residues and is closely neighbored by another insertion, of a SAP domain. Despite these differences, in our original model of this domain (with a predicted RMSD of 8.8 Å and QMEAN Z-score of −1.93), we correctly predicted the fold and the position of nearly all residues in this zinc finger. We also correctly predicted the boundaries and the fold of an all-β domain in the human protein SF3a66, a counterpart of the yeast protein Prp11. The original comparative model of this domain had a predicted RMSD of 4.7 Å and a QMEAN Z-score of −0.92, with a medium reliability of the fold prediction. In practice, upon comparison, this translated to predicting the position of approximately a half of the residues in the domain correctly. This analysis demonstrates the utility of the predictions, and that even models with a predicted relatively low accuracy can, in fact, exhibit correct folds, spatial shapes and locations of some of the functionally important residues.
Given the availability of the new template, we generated new models for the human counterparts of the SF3A crystal structure, using the comparative approach. We also generated a new comparative model for a domain in the C-complex-related protein cactin (NY-REN-24/C19orf29, gi: 126723149) as this protein is predicted to have a domain with the same all-β fold as the SF3a66 domain. The new models have been deposited in the database, while the old models have been moved to the archive of the ‘obsolete’ entries and are still available for analysis.
Ubiquitin-related domains are most common in the proteins of the late stages of splicing
Given the known role of ubiquitin in controlling spliceosome assembly and dynamics (21,22), and the fact that ubiquitin-related domains are one of the largest groups of domains in splicing proteins, we were interested in learning how these domains were distributed across the different groups of splicing proteins. We found 19 potential or known ubiquitin-related domains in 15 splicing-related proteins, including 12 abundant proteins of the major spliceosome and one protein of the U11/U12 di-snRNP subunit of the minor spliceosome (Table 6 and Figure 4). These domains cover most of the main classes of ubiquitin-related domains, including ubiquitin fold domains, RING zinc finger/U-box domains that may act as ubiquitin ligases, a ubiquitin conjugating enzyme-like domain, a ubiquitin carboxyl-terminal hydrolase domain and the JAB1/MPN domain of protein U5-220K (hPrp8) described in (23). In several cases, such as that of the abundant C-complex-specific protein FLJ35382 (C1orf55) and the TREX complex protein THOC5, only similarity of a protein region to a known ubiquitin-related fold could be detected.
Table 6.
Ubiquitin-related regions in the spliceosomal proteome
| Type of domain | SCOP ID | PFAM ID | Protein | Protein region | Protein group |
|---|---|---|---|---|---|
| Ubiquitin | d.15.1 | Ubiquitin | SF3a120a | 689,785 | U2 snRNP |
| d.15.1 | Ubiquitin | U11/U12-25K (C16orf33) | 41,132 | U11/U12 di-snRNP | |
| d.15.1 | SAP18 | SAP18a | 18,140 | EJC | |
| d.15.1 | ubiquitin | UBL5 | 1,73 | B complex | |
| d.15.1 | FLJ35382 (C1orf55)a | 7,74 | C complex | ||
| d.15.1 | XAP5 | XAP-5 (FAM50A)a | 197,283 | C complex | |
| DWNN | d.15.2 | DWNN | RBQ-1 | 3,77 | Miscellaneous |
| RING zinc finger/U-box | g.44.1 | zf-UBP | U4/U6.U5-65K (USP39)a | 97,200 | U4/U6.U5 trisnRNP |
| g.44.1 | U-box | hPRP19a | 1,60 | hPrp19 / CDC5L | |
| g.44.1 | Rtf2 | Cyp-60a | 36,94 | B-act complex | |
| g.44.1 | Rtf2 | Cyp-60a | 101,161 | B-act complex | |
| g.44.1 | zf-C3HC4 | RNF113Aa | 256,319 | B-act complex | |
| g.44.1 | Rtf2 | NOSIPa | 33,79 | C complex | |
| g.44.1 | Rtf2 | NOSIPa | 217,286 | C complex | |
| g.44.1 | DUF572 (ZZ) | CCDC130 | 43,117 | C complex | |
| g.44.1 | U-box | RBQ-1 | 258,312 | Miscellaneous | |
| UCH | d.3.1 | UCH | U4/U6.U5-65K (USP39)a | 220,556 | U4/U6.U5 trisnRNP |
| UBC-like (RWD) | d.20.1 | THOC5 | 468,640 | TREX | |
| JAB1/MPN | c.97.3 | JAB+PROCT | U5-220K (Prp8)a | 2064,2335 | U5 snRNP |
aAbundant protein.
Figure 4.
Ubiquitin-related structural regions of human splicing proteins. (A) Ubiquitin-fold region of protein FLJ35382 (C1orf55; residues 1–80). Predicted RMSD 3.5 Å, QMEAN Z-score −1.33. (B) RWD-like region of protein THOC5 (residues 458–641). Predicted RMSD 3.9 Å, QMEAN Z-score −1.85.
Ubiquitin-related domains are more abundant in proteins active in the late stages of splicing (B, B-act and C complexes). The ubiquitin-fold domain of protein SF3a120 is the only ubiquitin-related domain found in the U2 snRNP (its counterpart is found in the U11/U12 di-snRNP). On the other hand, as many as three proteins of the B/B-act complex (UBL5, Cyp-60 and RNF113A) and four proteins of the C complex (FLJ35382/C1orf55, XAP-5/FAM50A, NOSIP and CCDC130) contain ubiquitin-related domains, in addition to a domain in the U5 snRNP (the JAB1/MPN of U5-220K) and a protein in the U4/U6.U5 tri-snRNP (U4/U6.U5-65K). In summary, this distribution suggests that the late stages of splicing are probably under a stricter ubiquitin-based control than the early stages. This may be due to the fact that the earlier stages of splicing, such as intron/exon definition, are more dependent on weak, disorder-based interactions, while the later catalytic stages require precise subunit rearrangements.
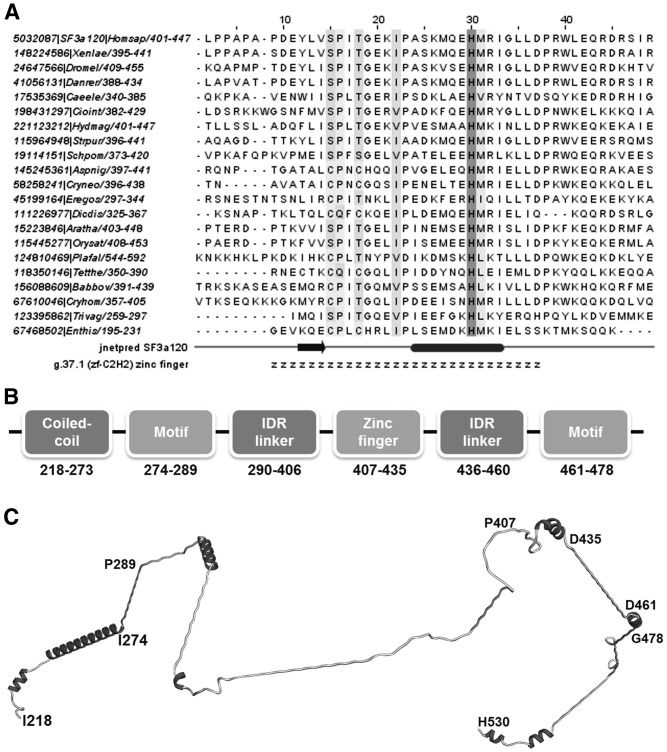
Zinc finger-like domains flanked by conserved intrinsically disordered regions in U2 snRNP SF3a120 and other splicing proteins
Our FR analysis detected that the human SF3A sub-complex contains, in addition to the zinc finger in protein SF3a60, another degenerate C2H2 (g.37.1)-type zinc finger in the middle conserved region of protein SF3a120 (conserved region: residues 217–530, PFAM domain PRP21_like_P; zinc finger: residues 407–435). In Saccharomyces cerevisiae, this zinc finger is absent entirely. However, in the majority of non-animal species, especially other fungi, amoeba and Apicomplexa, this zinc finger retains some of the cysteine and histidine zinc-binding residues (Figure 5A). The zinc finger remnant is surrounded on both sides by intrinsically unstructured regions that are in part predicted to form helical (potentially coiled-coil) structures. The short motifs lying on the distal ends of the disordered linkers are conserved. An additional coiled-coil region connects the N-terminal conserved motif with the previously described (69) second Surp module of SF3a120. Thus, the PRP21_like_P module consists of three motifs, the second of which is a zinc-finger remnant, connected by flexible linkers, with an N-terminal coiled coil that connects the N-terminal motif to the Surp region (Figure 5B). Structural modules of this type usually serve to simultaneously contact a binding partner of the protein in several locations. In the particular case of SF3a120, it has been suggested that both the U2 snRNA and a so far, unidentified splicing protein are potential partners (69).
Figure 5.
Architecture of the conserved middle region of protein SF3a120 (residues 217–530). (A) Alignment of the residues of a zinc-finger domain in the middle part of SF3a120 (residues 407–435). The ‘g.37.1’ annotation row displays residues predicted to form a part of a g.37.1 (zf-C2H2) zinc finger. The ‘jnetpred SF3a120’ annotation row displays predicted secondary structure elements of the human of the human SF3a120 (ovals represent α-helices, while arrows represent β-strands). (B) Architecture of the middle region of SF3a120; disordered linkers denoted as ‘IDR linker’ (intrinsically disordered region-linker). (C) Model of the middle region.
Through a systematic search, we found several other examples of zinc finger and zinc finger-like domains embedded in conserved disordered regions in the spliceosomal proteome (Table 7). Alternatively, tandem zinc fingers can be separated, e.g. by predicted coiled-coil regions. The new zinc-finger domains we found belong usually to the zf-C2H2 (g.37.1)-type, which can bind RNA and/or mediate protein–protein interactions. The pre-mRNA/mRNA-binding protein ARS2 contains a ZZ RING zinc finger, while the C complex protein NOSIP contains two RING zinc finger/U-box-like regions.
Table 7.
Zinc-finger domains flanked by or embedded in predicted disordered regions
| PFAM domain | Protein | Protein group | Region | SCOP superfamily ID | PFAM domain of template | SCOP description | Confidence | Region- superfamily similarity |
|---|---|---|---|---|---|---|---|---|
| PRP21_like_P | SF3a120a | U2 snRNP SF3A | 406,435 | g.37.1 | zf-U11-48K | β–β–α zinc fingers | High | High |
| LUC7 | LUC7B1 | A complex | 30,74 | g.66.1 | zf-CCCH | CCCH zinc finger | High | High |
| LUC7 | LUC7B1 | A complex | 186,232 | g.37.1 | zf-C2H2_jaz | β–β–α zinc fingers | High | High |
| DUF572 | CCDC130 | C complex | 43,117 | g.44.1 | ZZ | RING/U-box | High | High |
| Rtf2 | NOSIPa | C complex | 33,79 | g.44.1 | RING | RING/U-box | High | High |
| Rtf2 | NOSIPa | C complex | 217,286 | g.44.1 | zf-C3HC4 | RING/U-box | High | High |
| Fra10Ac1 | Fra10Ac1 | C complex | 166,220 | d.325.1 | Ribosomal_L28 | L28p-like | Lowb | Low |
| ARS2 | ASR2Ba | pre-mRNA/mRNA-binding | 714,738 | g.37.1 | zf-C2H2 | β–β–α zinc fingers | High | High |
aAbundant protein.
bAlternative templates: FYVE, fn1.
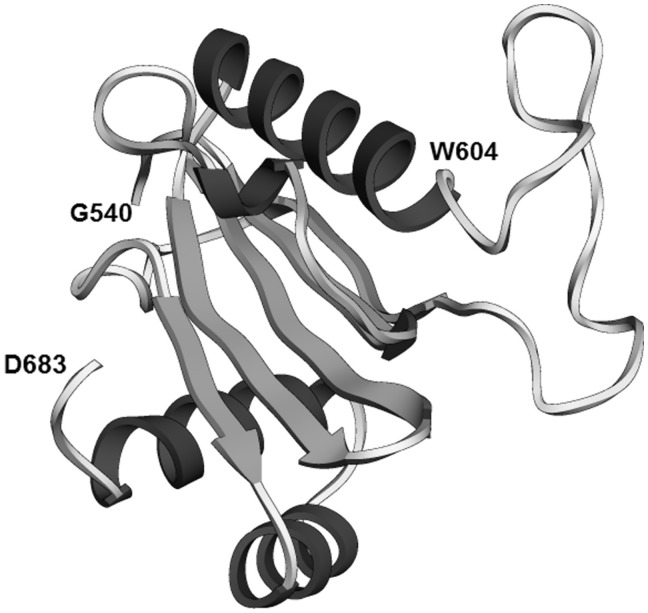
BLUF-like domain (DUF1115) of the U4/U6 di-snRNP protein 90K (hPrp3)
The C-terminal ordered domain of protein U4/U6-90K (hPrp3), which corresponds to PFAM domain DUF1115 (PFAM ID: PF06544; residues 540–683), was predicted in our analysis to have a ferredoxin-like fold. It is predicted to be related to the acylphosphatase/BLUF domain-like superfamily (SCOP ID: d.58.10). BLUF family domains have two additional helices in the C-terminus compared to acylphosphatase family domains. These helices are present in the DUF1115 domain, and so this domain is predicted to be a BLUF-like domain (Figure 6). This is an unusual assignment, because the BLUF domain is a FAD/FMN-binding blue light photoreceptor domain found primarily in bacteria. In Eukaryota, it is found almost exclusively in euglenids and Heterolobosea. On the other hand, DUF1115 is found exclusively in eukaryotes. However, very high scores of BLUF domain templates yielded by FR methods for the hPrp3 DUF1115 sequence suggest that this protein is definitely homologous to the BLUF family.
Figure 6.
BLUF-like region of protein U4/U6-90K (hPrp3) (domain DUF1115, residues 540–683). The position of the conserved residue W604 is displayed. Predicted RMSD 3.7 Å, QMEAN Z-score −3.06.
Nevertheless, DUF1115 differs from BLUF domains in some key features. The conserved FAD/FMN-binding residues are not conserved in DUF1115, and nor is a tryptophan residue whose position is altered depending on the excitement state of the photoreceptor (70) (Supplementary Figure S2). On the other hand, DUF1115 contains a disordered loop between the second α-helix and the fifth β-strand. The presence of this loop, though not its length, is conserved in DUF1115 domains. Moreover, a conserved tryptophan residue, W604 in hPrp3, is located next to the disordered loop.
Based on biochemical data, the DUF1115 domain may be a region of interaction of hPrp3 with the U5 snRNP protein hPrp6 and/or the U4/U6.U5 tri-snRNP protein U4/U6.U5-110K (SART-1) (71). However, it is also possible that this interaction proceeds through the disordered PRP3 domain of this protein (71). A possible alternative role for DUF1115 is suggested by the fact that, apart from proteins from the hPrp3 family, it is found only in a family of proteins containing the RWD domain. The RWD domain belongs to the ubiquitin conjugating enzyme superfamily (72). Hence, the hPrp3 DUF1115 may be a part of the spliceosomal ubiquitin-based system.
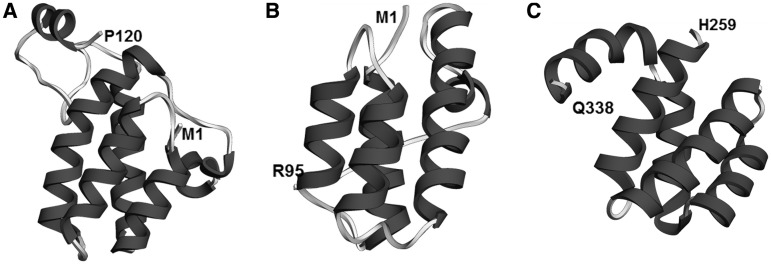
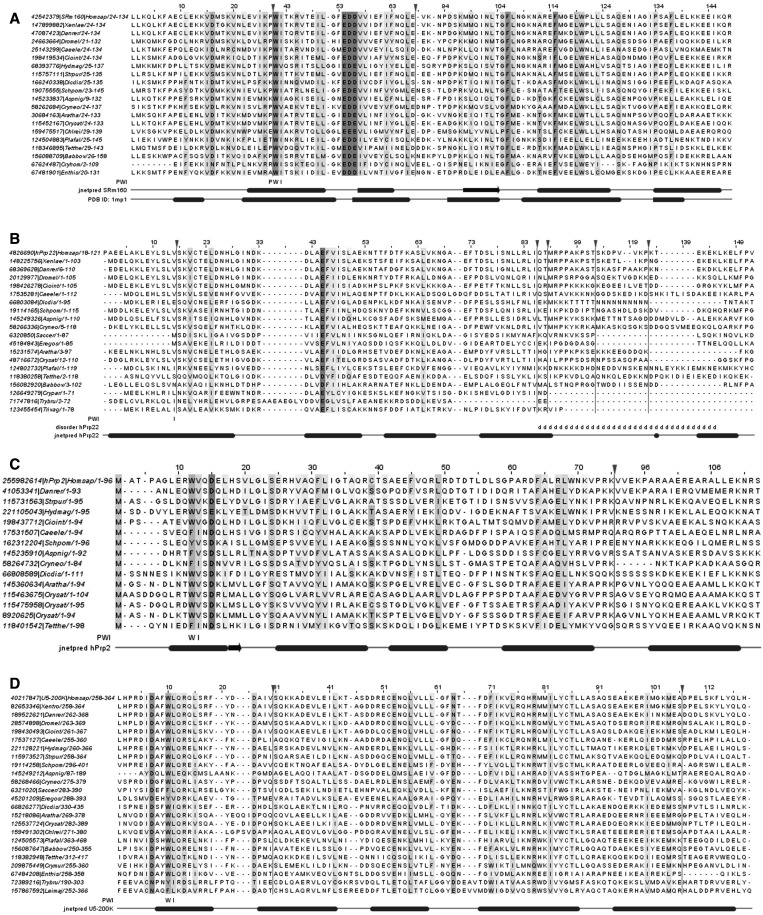
N-terminal PWI-like domains of the helicases hPrp22 (DHX8), hPrp2 (DHX16) and hBrr2 (U5-200K)
hPrp22 (DHX8) and hPrp2 (DHX16) are RNA helicases that function in the remodeling of the spliceosome (6). According to our predictions, these two helicases contain N-terminal ordered helical bundles with a PWI superfamily fold (SCOP superfamily a.188.1) and similarity to the PFAM PWI domain (Figures 7 and 8). PWI is a nucleic acid-binding domain first described in the splicing protein SRm160 (73,74). PWI is also found in the animal protein U4/U6-90K (hPrp3). The hPrp22 and hPrp2 PWI-like bundles (hPrp22: residues 1–92 or 1–120; hPrp2: 1–95) are not found in a search with the profile of the PFAM PWI domain, possibly because their eponymous PWI tripeptide motifs are degenerated. In hPrp22 and its homologs, only the third position of this motif is conserved: [x][x][IV], while in hPrp2 and its homologs, the second and third positions are usually conserved: [x][WFY][IV]. However, PFAM displays several putative hPrp2/hPrp22 homologs when queried for proteins that contain PWI domains. Furthermore, stable binding to nucleic acids by PWI requires an adjacent basic-rich region (74). We found potential candidates for such ancillary regions both in hPrp22 and in hPrp2 (hPrp22: residues: 93–116; hPrp2: residues 120–132).
Figure 7.
PWI-like regions of splicing helicases. (A) hPrp22 (DHX8; residues 1–120 shown, but domain may end at residue 92). Predicted RMSD 2.4 Å, QMEAN Z-score −2.76. (B) hPrp2 (DHX16; residues 1–95). Predicted RMSD 5.8 Å, QMEAN Z-score −2.19. (C) U5-200K (hBrr2; residues 259–338). Predicted RMSD 3.8 Å, QMEAN Z-score −0.79.
Figure 8.
The PWI domain and PWI-like regions in splicing helicases. In all alignments, the ‘PWI’ annotation row displays the residues of the PWI motif conserved in a given protein. The ‘jnetpred (…)’ annotation row displays secondary structure elements predicted in the relevant human proteins (ovals represent α-helices, while arrows represent β-strands). Vertical lines indicate hidden columns (inserted residues present in only one or two sequences in the alignment). (A) Alignment of a ‘canonical’ PWI domain from protein SRm160. The ‘PDB ID: 1mp1’ annotation row displays the actual secondary structure elements found in the structure of the PWI domain of the human protein SRm160. (B) PWI-like region from protein hPrp22 (DHX8). The ‘disorder’ annotation row displays the position of a disordered region in the hPrp22 protein. (C) PWI-like region from protein hPrp2 (DHX16). (D) PWI-like region from protein U5-200K (hBrr2).
We also found a PWI-like helical bundle in the N-terminus of the human protein U5-200K (hBrr2; residues 258–338; Figure 7). This helical bundle is conserved across the majority of eukaryotes, and is found, for instance, in the S. cerevisiae Brr2. The PWI-like domain of U5-200K retains a relatively well conserved second and third position of the tripeptide PWI motif: [x][WFY][ILV]. Notably, if correct, this prediction represents the first case when a PWI-like domain is located in the middle of a protein. Usually, as is the case of SRm160, hPrp3, hPrp22 and hPrp2, a PWI domain is located either in the immediate N-terminus or in the immediate C-terminus of a protein. There are at least three candidate basic-rich regions in the vicinity of the U5-200K PWI-like domain (residues 254–259; 343–349; 373–386).
Sequences of proteins from the hPrp22 (DHX8) and hPrp2 (DHX16) families are very similar, to the effect that we could not easily separate them in a clustering analysis (Supplementary Figure S3). The most important discriminant between the two families appears to be the presence of an S1 RNA-binding domain (PDB ID: 2eqs; DOI:10.2210/pdb2eqs/pdb, manuscript to be published) between the N-terminal PWI-like bundle and the C-terminal helicase domains. This domain is present in hPrp22 and its homologs, but not in hPrp2 and its homologs. This led us to the hypothesis that Prp2, with the PWI-like domain, was the ancestral protein, which then underwent the insertion of the S1 domain. Nevertheless, the PWI-like domains of hPrp22 and hPrp2 differ in several aspects.
The first difference lies in the above-mentioned degree of degeneration of the tripeptide PWI motif, which is larger in hPrp22 and its homologs than in hPrp2 and its homologs. In an extreme case, the N-terminus of the Prp22 protein of S. cerevisiae and the related organism Eremothecium (Ashbya) gossypii is located inside the motif, which is therefore incomplete. The degeneration of the PWI motif may be offset by the heavy conservation of a [DE][FY] motif in the second helix of the bundle. The main reason for the conservation of the PWI motif in canonical PWI domains is that it stabilizes the structure of the PWI domain (74). It is possible that the conservation of the [DE][FY] motif is sufficient to guarantee the stabilization of the bundle in conjunction with the conservation of the third position of the PWI motif.
Second, there is also a possible difference in either the number or the arrangement of helices comprising the PWI domain. SCOP describes superfamily a.188.1 as a ‘four-helix bundle’. However, in the structure of the PWI domain from protein SRm160, the bundle is followed by an additional short α-helix orthogonal to the bundle (PDB ID: 1mp1) (74). The presence of this α-helix is also predicted for the hPrp3 PWI domain, although it is missing from the available experimental structure (PDB ID: 1x4q; DOI:10.2210/pdb1x4q/pdb, manuscript to be published). Similarly, secondary structure predictions for hPrp2 also indicated that this protein is likely to contain an additional α-helix. However, for hPrp22, predictions of domain boundaries are less decisive. The hPrp22 PWI-like domain is either predicted to be a four-helix bundle (in which case it is confined to residues 1–92), or to contain an additional α-helix, but separated from the bundle by an intrinsically disordered region (in which case the domain spans residues 1–120). In either case, the helix arrangement is predicted to be different than in hPrp2. To note, the U5-200K PWI-like domain is predicted to be a five-helix domain.
Third, the pattern of evolutionary conservation of the PWI-like domains is different in hPrp22 and hPrp2. Fewer putative and confirmed hPrp2 homologs from different species have the PWI-like domain than do hPrp22 homologs. For instance, the functional analog of hPrp2 in S. cerevisiae, Prp2, is considered to be its homolog, but lacks the PWI-like domain. The Prp22 combination of PWI + S1 appears to be retained, while the Prp2 PWI is missing, also in putative homologs in organisms, such as kinetoplastids (Trypanosoma brucei, Leishmania major), some Apicomplexa (Plasmodium falciparum, Babesia bovis, but not Tetrahymena thermophila, which has both), Trichomonas vaginalis and Entamoeba histolytica.
Altogether, the PWI-like domain of hPrp22 is more diverged from the canon, but more often retained, while the PWI-like domain of hPrp2 is less diverged from canon, but more often completely lost. This result does not contradict the hypothesis that the Prp22 protein was formed in the insertion of the S1 domain into the ancestral Prp2. It rather suggests the possibility that some property of the ‘degenerated’ PWI-like domain ensured its retention in evolution. An in-depth structural study of this region may elucidate the reason why.
As hinted above, the U5-200K PWI-like domain is in many respects a ‘canonical’ PWI-like domain similar to that of hPrp2,it retains two out of three of the positions of the tripeptide PWI motif, and is predicted to be a five-helix domain. However, U5-200K is in general highly conserved, and unlike in hPrp2, this conservation also applies to its PWI-like domain.
The N-termini of S. cerevisiae Prp2 and Prp22 are dispensable for splicing (75,76), while the N-terminus of S. cerevisiae Brr2 was shown not to contact any of the proteins of the U4/U6.U5 tri-snRNP (71). Hence, the N-terminal PWI-like domains of hPrp2, hPrp22 and U5-200K are likely to have only a supporting role in splicing, one that is not revealed in the activity of the yeast proteins. We suggest that they may help in the correct positioning of the C-terminal helicase domains on the relevant snRNAs. Nevertheless, we could not find any data on the activity of the N-termini of hPrp2, hPrp22 and U5-200K. Furthermore, no experimental model of a PWI domain bound to RNA exists, to which we could compare the mode of binding of the hPrp2, hPrp22 and U5-200K PWI-like domains. Hence, as far as this publication is concerned, the question of what is bound to the PWI-like domains of the splicing helicases remains open.
An N-terminal domain of the hPrp8 protein (U5-220K)
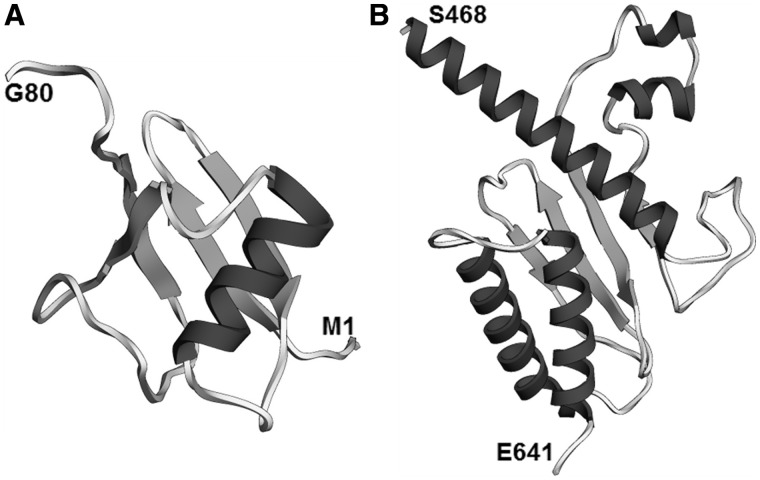
We could not confirm a published prediction of a bromo-domain encompassing hPrp8 residues 127–242 (a part of the N-terminal PFAM domain PRO8NT), originally made for yeast Prp8 residues 200–315 (77). In our view, the bromo-domain assignment does not command a consistent evolutionary conservation pattern. It encompasses 20 residues universally conserved in Prp8 homologs from all known species and nearly 100 residues conserved only in some eukaryotic Prp8 homologs. On the other hand, we were able to construct a de novo model for the most conserved part (residues 86–150) of the PRO8NT domain (Supplementary Figure S4). Quality evaluation indicates that the model of the putative Prp8 bromo-domain described in (77) has low predicted accuracy (predicted RMSD 8.7 Å, QMEAN Z-score −4.25) compared to our de novo model of residues 86–150 (predicted RMSD of 2.4 Å, QMEAN Z-score −1.93). Altogether, although we cannot exclude the possibility that PRO8NT encases a bromo-domain, we suggest that further studies (ideally: experimental structure determination) will be required to provide a confident structural model of this region.
Other previously uncharacterized structural regions of abundant splicing proteins
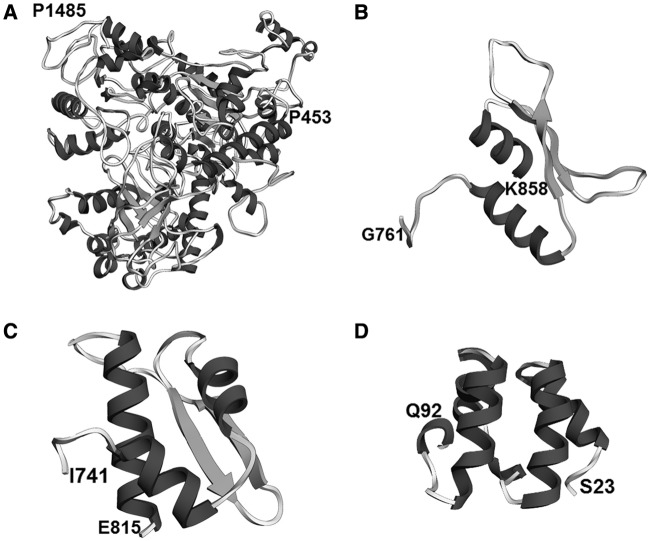
We found several other new types of structured regions in abundant splicing proteins that we were able to assign to known folds and/or are similar to existing structures, with varying degree of confidence (Table 7). For instance, a region in the C-terminus of the hPrp19/CDC5L-related protein KIAA0560 (IBP160/Aquarius homolog; residues 453–1485) has a helicase architecture similar to the nonsense-mediated decay protein Upf1p (Figure 9). KIAA0560 is a 1485-residue-long protein, whose binding to pre-mRNA introns is necessary for the successful deposition of the exon junction complex on the pre-mRNA (78) and for successful release of box C/D snoRNAs (small nucleolar RNAs) from introns (14). Upf1p contains two RNA helicase domains (c.37.1), the first of which is interrupted twice by two insertions: an all-β and an all-α domain insertion (79). In KIAA0560, this first c.37.1 domain is interrupted three times: both of the original insertions are kept, but a third insertion, largely disordered, has appeared between them.
Figure 9.
Other previously uncharacterized structural regions of the spliceosomal proteome. (A) The C-terminus of protein KIAA0560 (AQR), structurally similar to protein Upf1p (residues 453–1485). RMSD 3.3 Å, QMEAN Z-score −4.97. (B) Dsrm-like region of protein TFIP11 (residues 701–838). Predicted RMSD 4.5 Å, QMEAN Z-score −2.28. (C) The G-patch domain of LUCA15 (residues 741–815). Predicted RMSD 3.0 Å, QMEAN Z-score −1.22. (D) HTH-like region of protein hnRNP R (residues 23–92). Predicted RMSD 1.3 Å, QMEAN Z-score 0.12.
Another previously not described region lies in the C-terminus of the B complex protein TFIP11 (homolog of the yeast protein Spp382). The results of our FR analysis suggest that region is a potential double-stranded RNA binding domain (dsRBD) (Figure 9). In other splicing proteins, such as the non-abundant A complex protein DHX9, dsRBD domains often occur in tandem, but the TFIP11 region does not have a partner. However, TFIP11 contains also another previously structurally uncharacterized region with a putative RNA-binding function, a G-patch domain. While the G-patch domain does not show sequence similarity to any other known domains, a highly scoring de novo model of this domain shows structural similarity to a dsRBD domain (Figure 9). In fact, in the non-abundant splicing-related protein SON, the G-patch domain occurs in tandem with a dsRBD domain partner. If the G-patch domain has a dsRBD-like fold, the TFIP11 G-patch domain could provide the functionality of a second tandem dsRBD-like domain for the not described suspected domain of TFIP11.
We were also able to construct highly scored de novo models with a clear structural similarity to known folds for ordered helical regions located on the N-termini of proteins hnRNP R and Q. No known structural domain is assigned to these regions, but our de novo models of these regions exhibit fairly high scores (predicted RMSD 1.3 Å, QMEAN Z-score 0.12) for the region in protein hnRNP R. Based on structural similarity scores yielded by the DALI server (51), these may be helix-turn-helix domains (Figure 9).
Other new putative structural domains are described in Table 8.
Table 8.
New types of predicted structural regions in the human spliceosomal proteome that can be classified into known superfamilies
| PFAM domain | Protein | Protein group | Region | SCOP superfamily ID | PFAM domain of template | SCOP description | Confidence | Region- superfamily similarity |
|---|---|---|---|---|---|---|---|---|
| KIAA0560 (A) | hPrp19/ CDC5L-related | 1,452 | a.118.1 | Arm repeats | ARM repeat | Medium | Medium | |
| KIAA0560 (A) | hPrp19/ CDC5L-related | 453,1348 | Upf1pa | High | High | |||
| TFIP11 | B-complex | 771,837 | d.50.1 | dsrm | dsRNA-binding domain-like | Mediumb | High | |
| G-patch | LUCA15 (A) | A-complex | 741,815 | d.50.1 | dsrm | dsRNA-binding domain-like | Mediumc | High |
| hnRNP R | hnRNP | 28,92 | a.4.14 | KorB (clan HTH) | KorB DNA-binding domain-like | Mediumd | High | |
| DUF2414 | ELG | pre-mRNA/ mRNA-binding | 124,182 | d.58.7 | RNA_bind | RNA-binding domain, RBD | High | High |
| DUF1604 | Q9BRR8 | C-complex | 28,53 | b.34.2 | SH3_1 | SH3-domain | High | High |
| CTK3 | SR140 | U2 snRNP-related | 534,680 | a.118.9 | DUF618 | ENTH/VHS domain | High | High |
| Slu7 | hSlu7 (A) | step 2 factors | 424,457 | BTK motif | Lowe | High | ||
| PRP38 | hPrp38 (A) | B-complex | 26,206 | a.96.1 | HhH-GPD | DNA-glycosylase | Lowf | Medium |
| TRAP150 (A) | A-complex | 861,934 | Btz | Highg | High | |||
| BCLAF1 | pre-mRNA/ mRNA-binding | 827,899 | Btz | Highg | High | |||
| DZF | NFAR | A-complex | 82,177 | d.218.1 | NTP_transf_2 | Nucleotidyl transferase | Highh | High |
| DZF | NFAR | A-complex | 194,325 | a.160.1 | OAS1_C | PAP/OAS1 substrate- binding domain | Highh | High |
aProtein.
bHighly scored alternative template TcpQ (bacterial).
cDe novo model, highly scored, structural similarity only (1DI2_B).
dDe novo model, highly scored, structural similarity only (1R71_A).
eShort; BTK motif always found C-terminal to PH domains, which is not found in Slu7.
fAlternative templates: HtH motifs.
gPredicted disordered region.
hDZF is a member of clan NTP_transf.
Comparison of the human and Giardia lamblia spliceosomal proteome: setting priorities for spliceosome structure modeling
The human spliceosome, with its 119 abundant proteins, represents a fairly challenging target for both experimental and theoretical structural analyses. To round-off our analysis, we wanted to put forth a candidate minimum set of structural regions in a functional spliceosome that, in our opinion, should be prioritized during the modeling of the structure of the complex.
In general, eukaryotic species with fewer introns have fewer splicing proteins. The yeast Saccharomyces cerevisiae has homologs of only 61 of the human abundant splicing-related proteins (2). On the other hand, S. cerevisiae has also some Saccharomycetes-specific splicing proteins, such as Prp24 (41), which do not appear in other fungi. In the search of a ‘minimum’ set of regions to include in the model of a functional spliceosome, we turned to the extremely intron-scarce (80,81) parasitic organism G. lamblia, which is also known for its genome minimalism (82). This organism apparently underwent a reversed process with respect to the diversified and specialized human spliceosomal proteome, namely the loss of many genes encoding spliceosomal proteins.
The genome of G. lamblia ATCC50803 encodes homologs of only 30 human abundant splicing proteins (Table 9). Two more proteins can be found in G. lamblia P15. However, not all of these homologs may be involved in splicing. For instance, G. lamblia ATCC50803 possesses orthologs of U4/U6-15.5K and EIF4A3. In humans, U4/U6-15.5K is a component of the U4/U6 di-snRNP, where it binds to U4/U6-61K (hPrp31) (83), while EIF4A3 is a protein of the EJC (33). U4/U6-61K and all EJC proteins save EIF4A3 are missing in G. lamblia. However, the human U4/U6-15.5K protein also participates in box C/D snoRNP formation (83), where it binds a different protein, which does have a G. lamblia homolog, and the human EIF4A3 is an isoform of the eukaryotic translation initiation factor 4A. It is therefore possible that their orthologs in G. lamblia perform only these splicing-unrelated functions.
Table 9.
Human spliceosomal proteins with potential G. lamblia homologs, and these potential homologs
| Protein group | Human protein | GI of G. lamblia homolog | Human protein architecture | Giardia lamblia protein architecture |
|---|---|---|---|---|
| Sm | Sm-B/B′ | 159117899 | LSM + G-rich disorder + poly-P disorder | LSM |
| Sm | Sm-D1 | 159116502 | LSM + G-rich disorder | LSM |
| Sm | Sm-D2 | 159111944 | LSM | LSM |
| Sm | Sm-D3 | 159107430 | LSM + G-rich disorder | LSM |
| Sm | Sm-E | 159110758 | LSM | LSM |
| Sm | Sm-F | 159114826 | LSM | LSM |
| Lsm | Lsm2 | 159109501 | LSM | LSM |
| Lsm | Lsm3 | 159118879 | LSM | LSM |
| Lsm | Lsm4 | 159110729 | LSM + G-rich disorder | LSM |
| U1 snRNP/U2 snRNP | U1-A/U2-B″ | 253745584 | (RRM_1) × 2 | RRM_1 |
| U1 snRNP | U1-C | 308158556 | zf-U1 + poly-P disorder | zf-U1a |
| U2 snRNP | U2-A′ | 159115402 | (LRR_4) × 2 | (LRR_4) × 2 |
| U2 snRNP | SF3a66 | 159112716 | PRP4 + zf-met + b.15.1 + poly-P disorder | zf-met + b.15.1 |
| U2 snRNP | SF3a60 | 159115731 | SF3a60_bindingd + SAP + g.37.1 + g.37.1 | zf-met (g.37.1) + g.37.1b |
| U2 snRNP | SF3b155 | 253747536 | ULM + SF3b1 + a.118.1 (HEAT) repeats | a.118.1 repeatsc |
| U2 snRNP | SF3b145 | 159118535 | SAP + poly-P disorder + RS-like disorder + DUF382 + PSP | DUF382 + PSP |
| U2 snRNP | SF3b130 | 308162520 | WD40 repeats + CPSF_A | CPSF_Ad |
| U2 snRNP | SF3b49 | 159117358 | (RRM_1) × 2 + poly-P disorder | (RRM_1) × 2 |
| U2 snRNP | PHF5A | 159114698 | PHF5 | PHF5 |
| U2 snRNP-related | U2AF35 | 159112951 | zf-CCCH + RRM_1 + zf-CCCH + G-rich disorder | zf-CCCH + RRM_1 + zf-CCCH |
| U4/U6 di-snRNP | NHP2L1 | 159112698 | Ribosomal_L7Ae | Ribosomal_L7Aee |
| U4/U6 di-snRNP | NHP2L1 | 159111753 | Ribosomal_L7Ae | Ribosomal_L7Aee |
| U5 snRNP | U5-15K | 159116909 | DIM1 | DIM1 |
| U5 snRNP | U5-200K | 159109491 | a.188.1 + (DEAD + Helicase_C + Sec63) × 2 | DEAD + Helicase_C + Sec63 |
| U5 snRNP | U5-220K | 159109144 | PRO8NT + PROCN + RRM_4 + U5_2-snRNA_bdg + U6-snRNA_bdg + PRP8_domainIV+c.97.3 (JAB + PROCT) | PRO8NT + PROCN + RRM_4 + U5_2-snRNA_bdg + U6-snRNA_bdg + PRP8_domainIV + d.15.3f |
| U2 snRNP-related | hPrp43 (DHX15) | RS-like disorder + DEAD + Helicase_C + HA2 + OB_NTP_bind | g | |
| B-act complex | hPrp2 (DHX16) | a.188.1 + DEAD + Helicase_C + HA2 + OB_NTP_bind | g | |
| step 2 factors | hPrp22 (DHX8) | a.188.1 + RS-like disorder + S1 + DEAD + Helicase_C + HA2 + OB_NTP_bind | g | |
| step 2 factors | hPrp16 (DHX38) | RS-like disorder + DEAD + Helicase_C + HA2 + OB_NTP_bind | g | |
| 159108899 | ATP11 + DEAD + Helicase_C + HA2g,h | |||
| 159113861 | DEAD + Helicase_C + HA2 + OB_NTP_bindg | |||
| 159117264 | DEAD + Helicase_C + HA2g,h | |||
| B complex | hPrp38A | 159116389 | PRP38 + RS-like disorder | PRP38 |
| B-act complex | RNF113A | 159114937 | zf-CCCH + zf-C3HC4 | zf-CCCH |
| hPrp19/CDC5L | CCAP2 | 159115167 | Cwf_Cwc_15 | |
| EJC | EIF4A3 | 159117719 | DEAD + Helicase_C | DEAD + Helicase_Ci |
Only abundant human splicing proteins with homologs in G. lamblia are shown. Predicted disordered regions with an independent function are included in italics. Ordered structural regions are usually described with their PFAM domains; SCOP IDs are used if the structural region does not correspond to a PFAM domain.
aOnly in G. lamblia P15.
bSAP domain insertion is limited to animals and plants.
cSimilarity to human SF3b155 only in C-terminal region (human SF3b155: 998–1304).
dOnly in G. lamblia P15; WD40 repeat-like domain may be found via FR.
eMay not participate in splicing (other possible human homologs: ribosomal protein L7, 15.5K).
fUbiquitin-like fold (d.15) found in protein instead of c.97.3 domain.
gThe human splicing helicases hPrp43, hPrp2, hPrp22 and hPrp16 and potential G. lamblia homologs cannot be unequivocally assigned to one another.
hOB_NTP_bind found via FR.
iMay not participate in splicing (other possible human homolog: initiation factor EIF4A).
There is a pattern to the presence and absence of abundant splicing-related proteins and/or their domains and disordered regions in the G. lamblia proteome. Almost all the proteins of the U2 snRNPs are present in G. lamblia, as well as a homolog of U2AF35K, but only some core proteins of the U5 snRNP, such as Prp8 and Brr2. Snu114, which, according to the current understanding, is in other organisms the third part of the troika of U5 proteins essential to splicing (21), is an important absentee. Many proteins of the U1 snRNP and U4/U6 di-snRNP proteome are missing, as well as are all proteins specific to the human U4/U6.U5 tri-snRNP. The set of Step 2 factors is reduced to three RNA helicases, and these helicases are reduced to C-terminal regions of their human counterparts, with a common architecture. The G. lamblia helicases are also impossible to assign unambiguously to their human or yeast counterparts. Clustering analysis of helicase sequences from different organisms places the G. lamblia helicases away from any major cluster (Supplementary Figure S3). Finally, G. lamblia has very few homologs of human proteins of the auxiliary complexes, and only two non-snRNP stage-specific proteins (PRP38 and RNF113A) are present in this organism.
The snRNP protein homologs present in the G. lamblia proteome are shorter than their human counterparts. Three main types of structural features that are common for human spliceosomal proteins are largely absent from the G. lamblia spliceosomal proteome:
intrinsically disordered proteins or disordered regions with possibly autonomous function (long protein disorder that does not form inter-domain linkers, including compositionally biased disorder and some regions of disorder with preformed structural elements); consequently, highly disordered proteins, such as the U4/U6.U5-specific proteins U4/U6.U5-110K and U4/U6.U5-27K;
short peptide regions that act as ligand partners for other splicing proteins (PRP4, SF3a60_bindingd, SF3b1 and the ULM-containing region of protein SF3b155); and their partners (PRP4 partner: U4/U6-20K; SF3a60_bindingd partner: second Surp domain of protein SF3a120. This protein is missing entirely (see below); SF3b1 partner: p14; SF3b155 ULM partner: U2AF65K);
ubiquitin-related domains. This includes: the entire protein SF3a120 (which contains an ubiquitin domain in addition to the Surp domains); the U4/U6.U5-specific protein U4/U6.U5-65K, which contains the ubiquitin hydrolase domains zf-UBP and UCH; the zf-C3HC4 RING zinc finger of protein RNF113A. In contrast, the zf-CCCH zinc finger of RNF113A, which is a putative RNA-binding domain, is present.
In our analysis of intrinsic disorder in the human spliceosomal proteome (I.K and J.M.B., submitted for publication), we discuss how disordered regions of splicing proteins are tied to functions of dynamics, assembly and regulation of the spliceosome. This is also the function of known ubiquitin-related regions. Hence, it appears that G. lamblia is missing most proteins and/or protein regions primarily responsible for splicing regulation and dynamics. On the other hand, G. lamblia retained pre-mRNA and snRNA-binding proteins and/or regions, as well as proteins that directly assist in splicing, such as the catalytic factor helicases. It also appears that this parasitic organism’s ubiquitin-based system of splicing control is reduced, rather than entirely missing. The C-terminal Mov34/MPN/JAB1 domain present in Prp8 from human or yeast (SCOP superfamily c.97.3), which may be implicated in an ubiquitin-based system (65), is absent from the G. lamblia Prp8 (84), but the corresponding region in the latter protein is predicted by FR analysis to be a domain with a ubiquitin-like fold (SCOP superfamily d.15.1).
It is possible, that, like yeast, G. lamblia evolved its own specialized splicing proteins, which would not be detected in sequence similarity searches done with proteins from other organisms. Since G. lamblia is a parasite, it is also possible that it supplements some of its missing proteins (such as Snu114) from the host. Finally, it is also possible that some information was missed by our bioinformatics analysis but may be uncovered by an in-depth experimental analysis. With the caveat of the possibility of gaps in data (such as, possibly, Snu114), these are not single proteins that are missing, reduced or degenerated, but entire systems. The cropped set of proteins remaining in our G. lamblia spliceosomal proteome data set, corresponds to a system much less dynamical than the human spliceosome, less precisely regulated and less able to adapt to variable conditions. However, such a spliceosome may still be functional. Hence, we propose that from a practical standpoint, the set of structural regions with homologs in G. lamblia is a good starting point for the higher order structural modeling of the spliceosome, as well as constitutes an attractive list of targets for experimental structural determination.
CONCLUSIONS AND FUTURE PROSPECTS
This work has been intended to review the existing structural information about human spliceosomal proteins and to fill in gaps, providing a framework of reference for future structural analyses of the spliceosome. We used protein structure prediction methods to identify ordered spliceosomal protein structural elements either not characterized at all on the structural level or characterized insufficiently, and thus underreported in databases and literature. Examples of such un-/under-characterized elements include the zinc-finger domain in protein SF3a120 of the U2 snRNP, PWI-like domains in the essential splicing helicases hPrp22 (DHX8), hPrp2 (DHX16) and the U5 snRNP protein hBrr2 (U5-200K), and several ubiquitin-related regions in abundant splicing proteins. In the latter case, by combining database data with our results, we determined that ubiquitin processing-related domains are common especially in non-snRNP splicing factors active in the later stages of the splicing reaction. Having completed the characterization of ordered domains of splicing proteins, we constructed a minimum non-redundant set of experimental structural representations of the proteins of the human spliceosome and modeled most of the (potentially) ordered structural elements without experimental structural models. Confident high-resolution structural models can be assigned to over 90% of structural order in the spliceosome proteins, which corresponds to about 50% of all amino acid residues.
We analyzed the spliceosomal proteome of the intron-poor organism G. lamblia to determine a candidate minimum set of structural elements present in a functional spliceosome. We found that the G. lamblia spliceosome does not contain the majority of disordered regions found in the human splicing proteome, and has retained only a vestigial ubiquitin-based system of control. Overall, the G. lamblia spliceosome appears to be much simpler than the human or the yeast one, in accordance with this organism’s overall genomic minimalism and its genome’s intron-poorness.
The results of our analysis of the structural domains in proteins of the human spliceosome may be used to guide experimental characterization of these regions. The characterization of the reduced G. lamblia spliceosome may help set priorities in selecting the structural regions for experimental structural determination, and those to be included in a first draft of a model of a functional spliceosome. We suggest that in the event of modeling the structure of a functional spliceosome, the ordered protein regions found in G. lamblia proteins should take priority. Finally, as long as the corresponding structural information is absent, the models we constructed may be used in further structural studies, for instance in modeling the structure of the entire spliceosome. Models of non-‘core’ proteins can be used to broaden our understanding of alternative splicing. Our models, domain characterizations and suggested priorities thus form a framework of reference for future structural studies of the spliceosome, and in particular, for the modeling of the structure of the functional spliceosome.
Following the (near) completion of the parts list of the spliceosome, we are also advancing our understanding of the structure of these parts. This work provides working structural models for a majority of the parts that appear to be ordered regardless of their functional state. While experimental determination of high-resolution structures for all of these elements would be desirable, theoretical models can be used to design experiments or perform calculations/simulations that require protein structure as a basis. The next step in the structural analysis the spliceosome would be to use integrative modeling techniques to generate three-dimensional pictures of the splicing machinery, in analogy to the previous work on the nuclear pore complex (85,86). The even greater challenge ahead will be to model the dynamics of the splicing cycle, for which even greater union of experimental and theoretical techniques will be required.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1–4.
FUNDING
EU 6th Framework Programme Network of Excellence EURASNET [EU FP6 contract no LSHG-CT-2005-518238]. J.M.B. has been additionally supported by the 7th Framework Programme of the European Commission [EC FP7, grant HEALTHPROT, contract number 229676], by the European Research Council [ERC, StG grant RNA + P = 123D] and by the ‘Ideas for Poland’ fellowship from the Foundation for Polish Science. Computing power has been provided in part by the Interdisciplinary Centre for Mathematical and Computational Modeling of the University of Warsaw [grant number G27-4]. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the article. Funding for open access charge: EC FP7 contract number 229676 (HEALTHPROT) and by ERC (RNA + P = 123D).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Łukasz Kozłowski, Albert Bogdanowicz, Marcin Pawłowski, Geoff Barton, Jim Procter and Pascal Benkert for help with their software. We also thank Reinhard Lührmann, El ż bieta Purta, Łukasz Kozłowski, Joanna Kasprzak, and Anna Czerwoniec for critical reading of the article, useful comments and suggestions.
REFERENCES
- 1.Tarn WY, Steitz JA. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 2.Agafonov DE, Deckert J, Wolf E, Odenwalder P, Bessonov S, Will CL, Urlaub H, Luhrmann R. Semi-quantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol. Cell Biol. 2011;31:2667–2682. doi: 10.1128/MCB.05266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 4.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 5.Luz Ambrosio D, Lee JH, Panigrahi AK, Nguyen TN, Cicarelli RM, Gunzl A. Spliceosomal proteomics in Trypanosoma brucei reveal new RNA splicing factors. Eukaryot. Cell. 2009;8:990–1000. doi: 10.1128/EC.00075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valadkhan S, Jaladat Y. The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine. Proteomics. 2010;10:4128–4141. doi: 10.1002/pmic.201000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren L, McLean JR, Hazbun TR, Fields S, Vander Kooi C, Ohi MD, Gould KL. Systematic two-hybrid and comparative proteomic analyses reveal novel yeast pre-mRNA splicing factors connected to Prp19. PLoS One. 2011;6:e16719. doi: 10.1371/journal.pone.0016719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessonov S, Anokhina M, Krasauskas A, Golas MM, Sander B, Will CL, Urlaub H, Stark H, Luhrmann R. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. Rna. 2010;16:2384–2403. doi: 10.1261/rna.2456210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veretnik S, Wills C, Youkharibache P, Valas RE, Bourne PE. Sm/Lsm genes provide a glimpse into the early evolution of the spliceosome. PLoS Comput. Biol. 2009;5:e1000315. doi: 10.1371/journal.pcbi.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. Rna. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander R, Beggs JD. Cross-talk in transcription, splicing and chromatin: who makes the first call? Biochem. Soc. Trans. 2010;38:1251–1256. doi: 10.1042/BST0381251. [DOI] [PubMed] [Google Scholar]
- 12.Hsu SN, Hertel KJ. Spliceosomes walk the line: splicing errors and their impact on cellular function. RNA Biol. 2009;6:526–530. doi: 10.4161/rna.6.5.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 14.Hirose T, Ideue T, Nagai M, Hagiwara M, Shu MD, Steitz JA. A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Mol. Cell. 2006;23:673–684. doi: 10.1016/j.molcel.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Hogg R, McGrail JC, O'Keefe RT. The function of the NineTeen Complex (NTC) in regulating spliceosome conformations and fidelity during pre-mRNA splicing. Biochem. Soc. Trans. 2010;38:1110–1115. doi: 10.1042/BST0381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Lewis JD, Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- 18.Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 2004;23:4847–4856. doi: 10.1038/sj.emboj.7600482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katahira J. Regulation of nuclear export and cytoplasmic localization of mRNAs by NXF family proteins. Tanpakushitsu Kakusan Koso. 2009;54:2109–2113. [PubMed] [Google Scholar]
- 20.Zhang N, Kaur R, Lu X, Shen X, Li L, Legerski RJ. The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J. Biol. Chem. 2005;280:40559–40567. doi: 10.1074/jbc.M508453200. [DOI] [PubMed] [Google Scholar]
- 21.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Bellare P, Small EC, Huang X, Wohlschlegel JA, Staley JP, Sontheimer EJ. A role for ubiquitin in the spliceosome assembly pathway. Nat. Struct. Mol. Biol. 2008;15:444–451. doi: 10.1038/nsmb.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pena V, Liu S, Bujnicki JM, Luhrmann R, Wahl MC. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol. Cell. 2007;25:615–624. doi: 10.1016/j.molcel.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 24.Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW, Rape M. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 2010;24:1434–1447. doi: 10.1101/gad.1925010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathew R, Hartmuth K, Mohlmann S, Urlaub H, Ficner R, Luhrmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat. Struct. Mol. Biol. 2008;15:435–443. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski RA, Thornton JM. Understanding the molecular machinery of genetics through 3D structures. Nat. Rev. Genet. 2008;9:141–151. doi: 10.1038/nrg2273. [DOI] [PubMed] [Google Scholar]
- 27.Stark H, Luhrmann R. Cryo-electron microscopy of spliceosomal components. Annu. Rev. Biophys. Biomol. Struct. 2006;35:435–457. doi: 10.1146/annurev.biophys.35.040405.101953. [DOI] [PubMed] [Google Scholar]
- 28.Jurica MS. Detailed close-ups and the big picture of spliceosomes. Curr. Opin. Struct. Biol. 2008;18:315–320. doi: 10.1016/j.sbi.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database. 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung AK, Nagai K, Li J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature. 2011;473:536–539. doi: 10.1038/nature09956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Luhrmann R, Glover JN, MacMillan AM. Crystal structure of a core spliceosomal protein interface. Proc. Natl Acad. Sci. USA. 2006;103:1266–1271. doi: 10.1073/pnas.0508048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Luhrmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 38.Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 41.Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Luhrmann R. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 42.Will CL, Schneider C, Hossbach M, Urlaub H, Rauhut R, Elbashir S, Tuschl T, Luhrmann R. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA. 2004;10:929–941. doi: 10.1261/rna.7320604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 46.Kurowski MA, Bujnicki JM. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lundstrom J, Rychlewski L, Bujnicki J, Elofsson A. Pcons: a neural-network-based consensus predictor that improves fold recognition. Protein Sci. 2001;10:2354–2362. doi: 10.1110/ps.08501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 49.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 50.Tung CH, Yang JM. fastSCOP: a fast web server for recognizing protein structural domains and SCOP superfamilies. Nucleic Acids Res. 2007;35:W438–W443. doi: 10.1093/nar/gkm288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 53.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das R, Baker D. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 55.Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, Meiler J. Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry. 2010;49:2987–2998. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 58.Pawlowski M, Gajda MJ, Matlak R, Bujnicki JM. MetaMQAP: a meta-server for the quality assessment of protein models. BMC Bioinformatics. 2008;9:403. doi: 10.1186/1471-2105-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benkert P, Kunzli M, Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37:W510–W514. doi: 10.1093/nar/gkp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zemla A, Venclovas, Moult J, Fidelis K. Processing and evaluation of predictions in CASP4. Proteins. 2001;(Suppl 5):13–21. doi: 10.1002/prot.10052. [DOI] [PubMed] [Google Scholar]
- 61.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 64.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 65.Bellare P, Kutach AK, Rines AK, Guthrie C, Sontheimer EJ. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA. 2006;12:292–302. doi: 10.1261/rna.2152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kielkopf CL, Lucke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin PC, Xu RM. Structure and assembly of the SF3a splicing factor complex of U2 snRNP. EMBO J. 2012;31:1579–1590. doi: 10.1038/emboj.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kramer A, Ferfoglia F, Huang CJ, Mulhaupt F, Nesic D, Tanackovic G. Structure-function analysis of the U2 snRNP-associated splicing factor SF3a. Biochem. Soc. Trans. 2005;33:439–442. doi: 10.1042/BST0330439. [DOI] [PubMed] [Google Scholar]
- 70.Yuan H, Anderson S, Masuda S, Dragnea V, Moffat K, Bauer C. Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling. Biochemistry. 2006;45:12687–12694. doi: 10.1021/bi061435n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu S, Rauhut R, Vornlocher HP, Luhrmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA. 2006;12:1418–1430. doi: 10.1261/rna.55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andersen KM, Hofmann K, Hartmann-Petersen R. Ubiquitin-binding proteins: similar, but different. Essays Biochem. 2005;41:49–67. doi: 10.1042/EB0410049. [DOI] [PubMed] [Google Scholar]
- 73.Blencowe BJ, Ouzounis CA. The PWI motif: a new protein domain in splicing factors. Trends Biochem. Sci. 1999;24:179–180. doi: 10.1016/s0968-0004(99)01387-0. [DOI] [PubMed] [Google Scholar]
- 74.Szymczyna BR, Bowman J, McCracken S, Pineda-Lucena A, Lu Y, Cox B, Lambermon M, Graveley BR, Arrowsmith CH, Blencowe BJ. Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing. Genes Dev. 2003;17:461–475. doi: 10.1101/gad.1060403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edwalds-Gilbert G, Kim DH, Silverman E, Lin RJ. Definition of a spliceosome interaction domain in yeast Prp2 ATPase. RNA. 2004;10:210–220. doi: 10.1261/rna.5151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schneider S, Schwer B. Functional domains of the yeast splicing factor Prp22p. J. Biol. Chem. 2001;276:21184–21191. doi: 10.1074/jbc.M101964200. [DOI] [PubMed] [Google Scholar]
- 77.Dlakic M, Mushegian A. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA. 2011;17:799–808. doi: 10.1261/rna.2396011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ideue T, Sasaki YT, Hagiwara M, Hirose T. Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes Dev. 2007;21:1993–1998. doi: 10.1101/gad.1557907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 80.Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 81.Nixon JE, Wang A, Morrison HG, McArthur AG, Sogin ML, Loftus BJ, Samuelson J. A spliceosomal intron in Giardia lamblia. Proc. Natl Acad. Sci. USA. 2002;99:3701–3705. doi: 10.1073/pnas.042700299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 83.Liu S, Li P, Dybkov O, Nottrott S, Hartmuth K, Luhrmann R, Carlomagno T, Wahl MC. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 2007;316:115–120. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 84.Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 86.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.