Abstract
Homologous recombination serves multiple roles in DNA repair that are essential for maintaining genomic stability. We here describe RI-1, a small molecule that inhibits the central recombination protein RAD51. RI-1 specifically reduces gene conversion in human cells while stimulating single strand annealing. RI-1 binds covalently to the surface of RAD51 protein at cysteine 319 that likely destabilizes an interface used by RAD51 monomers to oligomerize into filaments on DNA. Correspondingly, the molecule inhibits the formation of subnuclear RAD51 foci in cells following DNA damage, while leaving replication protein A focus formation unaffected. Finally, it potentiates the lethal effects of a DNA cross-linking drug in human cells. Given that this inhibitory activity is seen in multiple human tumor cell lines, RI-1 holds promise as an oncologic drug. Furthermore, RI-1 represents a unique tool to dissect the network of reaction pathways that contribute to DNA repair in cells.
INTRODUCTION
Homologous recombination (HR) is an evolutionarily conserved pathway that serves multiple roles in DNA repair including the repair of replication forks and DNA double strand breaks (DSBs). HR repairs damaged DNA by identifying a stretch of homologous sequence on an undamaged sister chromatid and using that chromatid as a template to guide the repair process in an error-free manner. This distinguishes HR from non-homologous end joining (NHEJ), which is an error-prone pathway of DSB repair. HR also facilitates cellular recovery from replication-blocking lesions or collapsed replication forks, such that cells with impaired HR exhibit profound sensitivities to several classes of chemotherapeutics including inter-strand DNA cross-linkers (1–3).
The early steps of HR involve 5′ to 3′ nuclease activity that generates a 3′ single-stranded DNA (ssDNA) tail at the site of damaged DNA. This tail becomes coated with replication protein A (RPA), which is subsequently replaced by a helical filament of RAD51 protein. This displacement of RPA by RAD51 appears to be facilitated by several mediator proteins, which include BRCA2, RAD52, RAD51 paralog complexes and other proteins (reviewed in Ref. 4). HR efficiency is reduced in cells harboring defects in mediator proteins, and overexpression of RAD51 protein can partially bypass these deficient mediator functions (3,5). Even in some situations where the mediator activity is thought to be intact, a growing body of evidence suggests that RAD51 overexpression may up-regulate HR function and promote cellular resistance to DNA-damaging agents (6–9).
RAD51 protein is highly expressed in many human cancers including breast, bladder, prostate, pancreas, soft tissue sarcoma, upper aerodigestive and lung (reviewed in Ref. 10). This high expression is largely driven at the transcriptional level, given that the RAD51 promoter is activated, an average of 840-fold in cancer cells relative to normal cells (11). Furthermore, emerging clinical studies have observed higher than expected levels of aggressive pathologic features (12,13) and unfavorable outcomes (14–16) in patients whose tumors strongly express RAD51. These observations suggest that human tumors may develop ‘addictions’ to abnormally high RAD51 levels and that RAD51 represents a potential therapeutic target in oncology drug development. This potential strategy is particularly appealing in light of reports showing that HR inhibition may promote preferential sensitization of tumor cells relative to normal cells (17,18).
We performed a high-throughput screen of a naïve library of 10 000 small molecules in search of compounds that modify the binding of RAD51 protein to ssDNA (19). We now present a RAD51-inhibitory compound called RI-1 that inactivates RAD51 by directly binding to a protein surface that serves as an interface between protein subunits in RAD51 filaments. Cell-based experiments demonstrate that RI-1 specifically inhibits HR efficiency and sensitizes human cancer cells to mitomycin C (MMC).
MATERIALS AND METHODS
Plasmids, proteins and cells
HsRAD51 and ScRAD51 were prepared as previously described (19,20). Escherichia coli RecA was purchased from New England Biolabs. Prior to use in experiments, the ScRAD51 and EcRecA proteins were precipitated in 2.3 M ammonium sulfate and resuspended in a DTT-free storage buffer. The SH2038 (+/− RAD51C) cells were provided by Helmut Hanenberg, who previously described the construction of these cells in detail (21). U2OS cells stably transfected with the DR-GFP reporter construct were provided by Maria Jasin, as were the I-SceI expressing pCβASce plasmid and empty vector control pCAG (22). All other cell lines were obtained from ATCC. Standard tissue culture media were supplemented with 0.2–0.5% dimethyl sulfoxide (DMSO) during treatment with RI-1 to minimize any potential concerns about compound solubility.
High-throughput screen and chemicals
Pharmacy-grade MMC was used (Ben Venue Laboratories). Methods and details of the chemical library and primary screen were previously described (19). For the initial screening experiments, RI-1 was purchased from ChemBridge Corporation (San Diego, CA). For all subsequent experiments, RI-1 was synthesized in our laboratories, based on methods detailed in the Supplementary Methods. RI-1 was dissolved in 100% DMSO immediately before use, and the concentration was confirmed by UV-VIS spectrometry using a molar extinction coefficient of 5128 cm−1 at 400 nm.
Quantification of HR and SSA efficiencies in cells
U2OS cells stably transfected with the DR-GFP reporter construct were previously described (22). The HEK293 cells stably transfected with SA-GFP reporter were also previously described (23). In total, 0.5–1.0 × 107 cells grown to 80% confluence were electroporated with 30–60 µg pCβASce (or pCAG) in 4-mm cuvettes, using the following settings: 240–350 V, 975 µF. Cells were transferred into the appropriate complete growth medium and allowed to grow for 24 h in the presence or absence of a candidate compound. Compounds were removed and cells were incubated for an additional 24 h in normal media, following which they were analyzed with a Becton–Dickinson FACScan. Live cells were collected based on size/complexity and 7-aminoactinomycyin D (7-AAD) exclusion. The fraction of cells exhibiting GFP positivity is displayed, and error bars denote standard error.
DNA binding assays
All reactions were performed in black non-binding polystyrene 384-well plates with reaction volumes of 30–100 μl. Purified DNA strand exchange proteins and chemical compounds were pre-incubated at room temperature for 5 min; they were then further incubated at 37°C for 30 min with 100 nM of ssDNA substrate, consisting of a 45-mer poly-dT tagged with Alexa 488 at the 5′ terminus (synthesized and purified by Integrated DNA Technologies). Reactions were performed in 20 mM HEPES pH 7.5, 10 mM MgCl2, 0.25 μM BSA, 2% glycerol, 30 mM NaCl, 4% DMSO and 2 mM ATP. Some conditions included DTT or TCEP (tris(2-carboxyethyl)phosphine) as indicated. DNA binding was measured as a function of fluorescence polarization (FP) with a Safire2 plate reader (Tecan), using the following settings: excitation 470 ± 5 nm, emission 530 ± 5 nm, 10 reads/well, Z height and G factor auto-calibrated from control wells. Displayed error bars represent standard deviation. For experiments involving a titration of protein concentrations, data are fit to an equation that accounts for the cooperative nature by which recombinase proteins bind DNA (24). For experiments involving a titration of RI-1, protein concentrations were selected to give an ∼80% saturation of the FP signal in the absence of RI-1.
D-loop assays
Strand assimilation methods were as previously described (19) with some modifications. Briefly, 10 μl reaction volumes that included 0.4 μM HsRAD51 were pre-incubated for 5 min at 37°C with 3.6 μM (nucleotide concentration) 32P-labeled oligonucleotide 306.90 in a reaction buffer containing 25 mM Tris–HCl (pH 7.5), 1 mM TCEP, 3 mM ATP, 5 mM MgCl2 and various concentrations of RI-1. Following this initial binding reaction, 1 μl of 220 μM (base pair concentration) supercoiled homolog-containing target plasmid DNA (pRS306) was added. DNA substrates were allowed form a homology-dependent association (or ‘D-loop’), and reactions were stopped with SDS and proteinase K. The resulting DNA was separated on 0.9% agarose gels, transferred onto positively charged nylon membranes and analyzed by phosphorimaging.
Mass spectrometry methods
Biotinylated HsRAD51 protein was expressed in E. coli as previously described (19). The resulting protein contains an N-terminal AviTagTM, which is linked to HsRAD51 by a peptide sequence recognized by the Tobacco Etch Virus (TEV) protease. The protein was purified from bacterial lysate by incubating with streptavidin-conjugated polyacrylamide beads (Ultralink, Pierce). The protein-bound beads were washed, incubated with RI-1 for 30 min at 37°C, and washed again. Protein was then eluted by incubating with TEV protease (gift from Phoebe Rice) for 2 h at 15°C. Resulting proteins were separated on a non-reducing SDS–PAGE, and cut from the coomassie-stained gel. Mass spectrometry was performed at the W.M. Keck Biotechnology Resource Laboratory (Yale University) using a Thermo Scientific LTQ-Orbitrap XL mass spectrometer and at the University of Illinois at Chicago using a Thermo Scientific Orbitrap Velos unit. Proteomics data was analyzed using Scaffold 3.1.4.1 available from Proteome Software (Portland, OR).
RAD51 focus cytology methods
In some experiments, the cells were treated with 5-ethynyl-2′-deoxyuridine (EdU) during the last 3 h prior to harvest, in order to mark S-phase cells. Cells were harvested and spotted onto poly-L-lysine coated slides. Cells were subsequently fixed with 3% paraformaldahyde/3.4% sucrose, and permeabilized with a standard buffer (20 mM HEPES pH 7.4, 0.5% Triton X-100, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose). Slides containing EdU-treated cells were processed with Click-iT® EdU Alexa Fluor® 647 Imaging Kit (Invitrogen) as per the manufacture’s protocol. Slides were subsequently stained with a rabbit polyclonal HsRAD51 antibody (1:1000 dilution) and/or a mouse monoclonal RPA antibody (1:1000 dilution), followed by Alexa 488-conjugated goat anti-rabbit and Alexa 594-conjugated goat anti-mouse secondary antibodies (Invitrogen, both 1:1000 dilution). Slides were viewed using a Zeiss Axio Imager.M1 microscope that allows high-resolution detection of foci throughout the entire nuclear volume. Images were recorded as a Z-axis series of 15 focal planes (over a 10–12 µm thickness) using a CCD camera. The 15 images were subsequently combined to form a single image using AxioVision V4.8 and NIH Image software packages. For each experimental condition, 50 randomly selected nuclei were quantified using NIH Image software.
Cell survival assays
Cytotoxicity was determined by loss of colony-forming ability. Experiments were performed in triplicate. Crystal violet stained colonies were imaged with a CCD camera and counted using NIH Image software. Error bars denote standard error.
Western blotting
Whole cell protein extracts were separated via SDS–PAGE and subjected to western blotting. Primary antibodies included protein A purified rabbit anti HsRAD51 (1:1000 dilution, gift of Akira Shinohara) and mouse anti α tubulin (1:5000 dilution, Ab-2 from Fitzgerald). Secondary antibodies consisted of HRP-conjugated anti-rabbit IgG (1:1000 dilution, GE healthcare) and HRP-conjugated anti-mouse IgG (1:2000 dilution, GE healthcare).
RESULTS
Identification of a compound that inactivates RAD51 in vitro and inhibits HR in cells
To identify small molecules that modify the DNA binding properties of HsRAD51, we used a fluorescence polarization (FP) based method that involves incubation of HsRAD51 protein with oligonucleotides that are end-labeled with a fluorescent tag. The binding of HsRAD51 to this substrate ssDNA generates an increase in the polarization of light emitted by the fluorescent tag. A naïve 10 000 compound library (Chembridge DIVERSetTM) was screened, and the details were previously described (19). This primary screen identified 76 small molecules that reduced FP signal by ≥50%.
These 76 ‘hit’ compounds were subjected to secondary screening that evaluates their ability to inhibit HR efficiency in cells. This assay makes use of human cells containing a stably-transfected reporter construct that carries two non-functional copies of green fluorescence protein (GFP), one of which is interrupted by an I-SceI endonuclease site. Induction of a DSB at the I-SceI site can lead to repair by homologous gene conversion that generates a functional copy of GFP (22). Cells were transfected with an I-SceI expressing plasmid and exposed to candidate compounds for 24 h thereafter. Fluorescence assisted cell sorting (FACS) analysis revealed eight compounds that were capable of inhibiting GFP gene conversion frequency by >50%, at compound concentrations that still allowed cell viability of ≥90%.
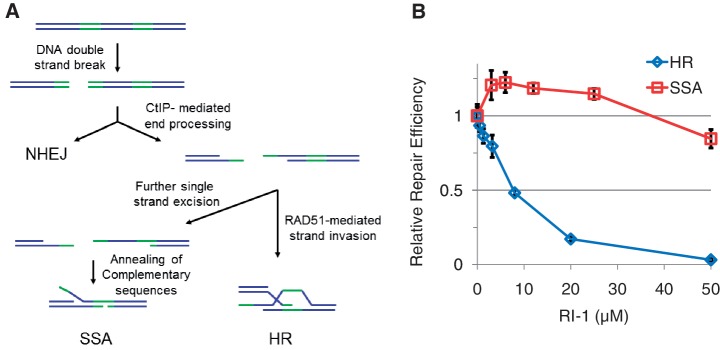
A tertiary screen was employed to determine which of these eight compounds were disrupting HR by specifically inhibiting RAD51. This was performed using cells containing a reporter construct that measures the efficiency of single-strand annealing (SSA). SSA is a pathway that repairs DSBs occurring between a pair of tandem-repeated sequences (Figure 1A), such that single strand degradation from the site of the DSB creates a pair of ssDNA tracts with extensive complementary sequence (25). The two ssDNA tails then anneal to heal the DSB, and the DNA originally located between the repeats is thereby deleted. SSA does not involve the NHEJ pathway, nor does it require most HR-related proteins (26). Interestingly, SSA efficiency is known to be elevated in situations where RAD51 function has been disrupted by a variety of means, including RNAi knockdown, expression of a dominant negative RAD51 mutant, or expression of the BRCA2-derived BRC3 peptide (23,27,28). We exploited this effect to better characterize the eight HR-inhibitory compounds. Seven of the eight compounds exerted no significant impact on SSA or generated an inhibitory effect. However, one compound (hereafter termed RI-1 for RAD51 inhibitor #1) significantly stimulated SSA, in the same compound concentration range (∼5–30 µM) that was required to inhibit HR efficiency (Figure 1B). Some inhibition of SSA could be generated with RI-1 concentrations above this range (>30 µM), but this was likely related to compound-induced cytotoxicity.
Figure 1.
RI-1 specifically inhibits HR in cells. (A) Pathways involved in the repair of DNA breaks include non-homologous end joining (NHEJ), single strand annealing (SSA) and homologous recombination (HR). A simplified schematic of DNA repair reporters is represented in cartoons, with green portions representing repeats of a homologous DNA sequence. (B) RI-1 modulates the DNA repair efficiency in U2OS cells containing an HR reporter and HEK293 cells containing an SSA reporter. Cells were electroporated with an I-SceI endonuclease-expressing plasmid, incubated with RI-1, and subjected to FACS analysis. Relative repair efficiencies were measured as described in ‘Materials and Methods’ section.
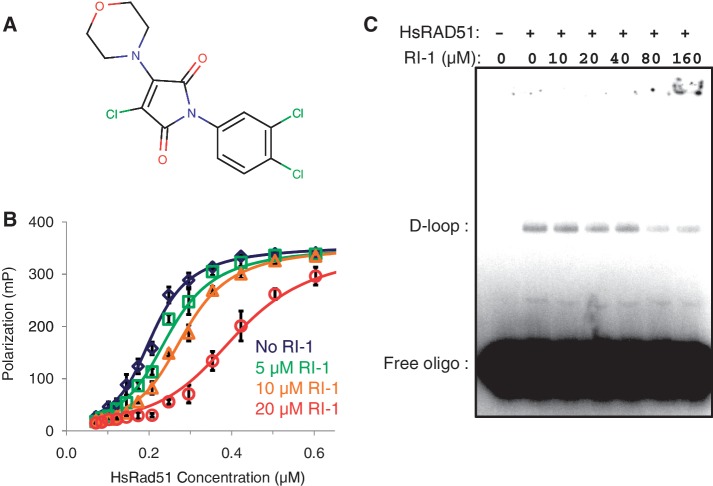
The chemical structure of RI-1 (3-chloro-1-(3,4-dichlorophenyl)-4-(4-morpholinyl)-1H-pyrrole-2,5-dione) is shown in Figure 2A. Physiochemical properties of RI-1 include a molecular weight of 362 g/mol and a CLogP of 4.25. It contains five H-bond acceptors and no H-bond donors. Therefore, RI-1 satisfies all four ‘Lipinski rules’ for predicting drug likeness (29).
Figure 2.
RI-1 inhibits human RAD51 in biochemical assays. (A) Chemical structure of RI-1 is displayed. (B) Human RAD51 protein was allowed to bind fluorescently-tagged ssDNA following a pre-incubation in various concentrations of RI-1. DNA binding is reported as a function of fluorescence polarization as described in the ‘Materials and Methods’ section. (C) HsRAD51 was allowed to generate joint molecules (‘D-loops’) in the presence of RI-1. A representative phosphorimage of an agarose gel is displayed, and the positions of free oligo and oligo associated with D-loops are indicated.
The inhibitory activity of RI-1 was quantified in the above described FP assay, which measures the binding of HsRAD51 to ssDNA. RI-1 generated a concentration-dependent inhibition of HsRAD51 (Figure 2B). The IC50 of RI-1 was generally in the 5–30 µM range, though this depended on the concentration of protein included in the assay. RI-1 induced no measurable effect on FP signal in the absence of RAD51, suggesting that the compound interacts with RAD51 protein rather than the substrate DNA.
Strand assimilation assays were also performed to assess the effect of RI-1 on RAD51’s recombination function in vitro. In this assay, HsRAD51 was first allowed to assemble into filaments on a 32P-labeled ssDNA oligonucleotide in the presence or absence of RI-1. A homology-containing super-coiled target plasmid was then added, and the two were allowed to generate a homology-dependent joint molecule referred to as a ‘D-loop’. RI-1 was found to inhibit the formation of D-loops in a concentration-dependent manner (Figure 2C).
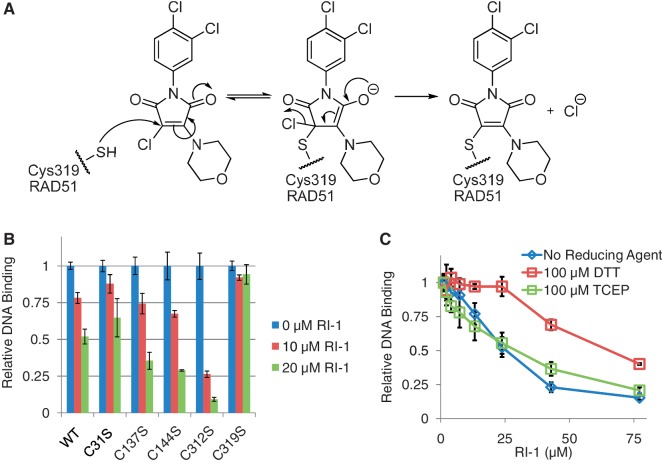
RI-1 directly binds to human RAD51 at cysteine-319
RI-1 is chemically composed of three rings, one of which is a chloromaleimide group that is expected to act as a Michael acceptor and react with RAD51 according to a conjugate addition–elimination mechanism (Figure 3A). Maleimides are frequently used to modify cysteine residues and halomaleimides have previously been shown to selectively react with the sulfhydryl side chain of cysteine residues present in intact proteins (30). Therefore, it is likely that RI-1 reacts with RAD51 in a similar fashion. To test this hypothesis, site-directed mutagenesis was performed to generate cysteine to serine alterations at each of the five cysteines that exist in HsRAD51. All five mutant proteins were purified and found to exhibit comparable binding activity to ssDNA relative to the wild-type (WT) protein, though the C312S was slightly less active. Four of these five C-to-S mutants could be significantly inhibited by RI-1, using compound concentrations in the 10–20 µM range. However, the C319S mutant was significantly less susceptible to inhibition by RI-1, suggesting that C319 is the binding target of RI-1 (Figure 3B).
Figure 3.
RI-1 directly binds to human RAD51 at cysteine-319. (A) A chemical mechanism is proposed, by which RI-1 binds a cysteine residue on HsRAD51. (B) Purified HsRAD51 proteins (at 0.2 µM) containing cysteine to serine mutations were tested for ability to bind ssDNA, in the presence or absence of RI-1. Relative DNA binding is reported as a function of fluorescence polarization as described in the ‘Materials and Methods’ section, normalized to the 0 µM RI-1 condition of each protein. (C) The ability of RI-1 to inhibit the ssDNA binding activity of WT HsRAD51 protein (0.25 µM) was evaluated in the presence of different reducing agents, including dithiothreitol (DTT) and tris(2-carboxyethyl)phosphine (TCEP).
This proposed mechanism of direct covalent attachment between RI-1 and RAD51 was tested further using mass spectrometry. In this assay, purified HsRAD51 was incubated with RI-1 and then digested with trypsin. The resulting peptide fragments were analyzed via LC–MS/MS (tandem mass spectrometry). The inclusion of RI-1 at 10 µM in the reaction induced an increase in the mass of IYDSPCLPEAEAMFAINADGVGDAKD, the peptide that comprises residues 314–339 of HsRAD51. Subsequent MS/MS analysis of this peptide identified a C8H6Cl2NO adduct at residue C319 (Supplementary Figure S1). It should be noted that the molecular weight (MW) of this observed adduct was smaller than the expected 324 Da, which is likely an artifact that occurred during trypsin digest or MS ionization/fragmentation. A proposed structure of this adduct is displayed in Supplementary Figure S2, which is based on published work related to halomaleimide compounds (31).
This mode of RI-1 binding was tested further with additional experiments using different reducing agents. Since dithiothreitol (DTT) is a strong reducing agent with two thiol groups, DTT would be expected to compete with cysteine residues for the reaction with RI-1. As expected, experiments showed that 100 µM DTT increased the IC50 of RI-1 by ∼3-fold, relative to when no reducing agent was present (Figure 3C). This antagonistic effect of DTT appears related to its nucleophilic potential rather than its reducing capability, given that no significant antagonism was observed with tris(2-carboxyethyl)phosphine (TCEP), which is a reducing agent that lacks these nucleophilic thiol groups.
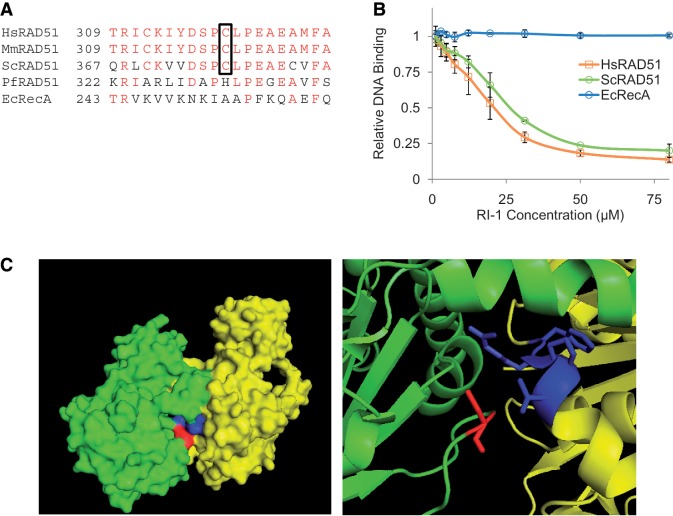
RI-1 binds near a protein–protein interface formed between subunits of RAD51 filaments
The functional significance of the C319 residue of HsRAD51 was further investigated to better elucidate the mechanism of inhibition by RI-1. C319 lies within a portion of sequence that is highly conserved among different eukaryotic species, but far less so in the homologous recombinase proteins of archaeal and prokaryotic species (Figure 4A). Consistent with this observation, RI-1 was capable of inhibiting Saccharomyces cerevisiae RAD51, using compound concentrations comparable with those needed to inhibit HsRAD51 (Figure 4B). In contrast, the E. coli homolog RecA was insensitive to inhibition by RI-1 in the same range of RI-1 concentrations.
Figure 4.
The cysteine binding target of RI-1 is highly conserved among eukaryotic RAD51 proteins and is located in an interface used for protein–protein interactions. (A) A sequence alignment is shown for homologous recombinase proteins of various species. Amino acids with identity are displayed in red, and the cysteine corresponding to C319 in HsRAD51 is boxed. (B) The ability of RI-1 to inhibit the ssDNA binding activity of recombinase proteins from various species (0.35 µM HsRAD51, 0.42 µM ScRAD51, 0.30 µM EcRecA) was evaluated. Relative DNA binding is reported as a function of fluorescence polarization as described in the ‘Materials and Methods’ section. (C) The crystal structure (previously solved and described by Conway et al.) is shown for two adjacent monomers (one monomer displayed in green and one in yellow) of ScRAD51 as a surface rendering (left) and a magnified cartoon (right). The cysteine binding target of RI-1 is displayed in red (within the green monomer) and the α9 chain of the opposing monomer is displayed in blue (within the yellow monomer).
Recombinase protein filaments are known to utilize conserved structural motifs to govern interactions between adjacent protein protomers (20,32). The functional relevance of C319 was examined by viewing its location within the yeast RAD51 filament crystal structure (20). Interestingly, the cysteine target of RI-1 binding (C319 of HsRAD51 corresponds to C377 in ScRAD51) contributes directly to a protomer interface. On closer inspection of this interface, the cysteine residue of one protomer lies in close proximity to the α9 chain of an adjacent protomer (Figure 4C). Furthermore, this cysteine resides within a loop of protein that serves as an ATP cap, which overlays the inter-subunit ATP binding site (33,34). Based on these observations, it is very likely that binding of RI-1 to RAD51 destabilizes a protein–protein interface that is required for filament formation and subsequent recombinase activity, possibly by modulating RAD51’s interactions with ATP.
RI-1 inhibits the formation of RAD51 foci in human cells
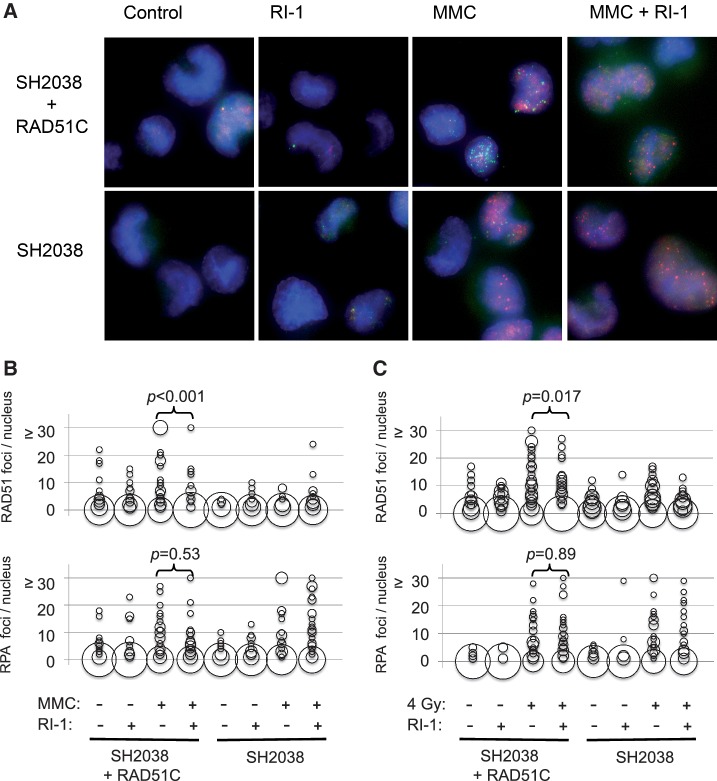
Since RAD51 filaments can be imaged as sub-nuclear foci (35), immunofluorescence microscopy was used to directly visualize the effect of RI-1 on HR in cells. These assays were performed using a previously described isogenic cell line pair that differs in RAD51C expression (21). These cells were selected because RAD51C is a component of RAD51 paralog complexes that are known to be required for coordinated assembly of RAD51 at sites of RPA-coated DNA (36). Microscopic images were quantified, and differences between conditions were tested via the Mann–Whitney rank sum test. As expected, the two cell lines exhibited comparable RPA focus formation (P = 0.46) following treatment with the inter-strand DNA cross-linking drug MMC, however the RAD51C mutant cells had significantly impaired RAD51 focus formation (P < 0.001). When RI-1 was added to media containing MMC, the RAD51C-complemented cells exhibited staining that mimicked the RAD51C mutant phenotype (Figure 5A and B). Specifically, incubation of RI-1 with RAD51C-complemented cells inhibited MMC-induced RAD51 focus formation (P < 0.001), while not significantly affecting RPA focus formation (P = 0.53). The lack of RPA focus inhibition is important, because it supports the claim that RI-1 specifically inhibits RAD51 rather than upstream components of HR or more global cellular processes.
Figure 5.
RI-1 disrupts the formation of RAD51 foci after DNA damage. (A) Immortalized human fibroblasts (SH2038 ± RAD51C) were incubated for 8 h in media containing 150 nM mitomycin C (MMC) and/or 20 µM RI-1. Cells were subsequently harvested and indirectly immunostained. Representative images are displayed with HsRAD51 foci displayed in green and RPA foci displayed in red. (B) In total, 50 randomly selected nuclei per treatment group were examined and quantified. The size of each ‘bubble’ represents the number of cells having a given number of foci/nucleus. (C) The above described experiments were repeated using 4 Gy in place of MMC, and cells were similarly processed 8 h after irradiation.
Of note, these experiments were performed using the same compound concentration range that inhibited HsRAD51 in the aforementioned biochemical assays. The effect was not due to altered HsRAD51 protein levels, given that western blotting showed cellular HsRAD51 levels to be unaffected by RI-1 at these concentrations and incubation lengths (Supplementary Figure S3). These results were replicated using ionizing radiation in place of MMC (Figure 5C), which minimizes the possibility that RI-1 might have directly interacted with MMC and inactivated its cross-linking activity.
RAD51 foci are known to appear in S/G2 portions of the cell cycle. Therefore, additional experiments were performed to ensure that RI-1 inhibited the assembly of RAD51 foci, as opposed to simply preventing progression of cells into S/G2. The MMC-based cytology experiments were repeated, such that S-phase cells were allowed to incorporate the thymidine analog EdU for 3 h prior to harvest. Analysis of the resulting EdU-positive cells demonstrated that RI-1 inhibited the appearance of MMC-induced RAD51 foci (P = 0.007) in these S/G2-selected cells (Supplementary Figure S4). Taken together, these results suggest that RI-1 can gain intracellular access to human cells and can specifically interfere with RAD51 functions in cells following genotoxic injury.
RI-1 sensitizes human cancer cells to cross-linking chemotherapy
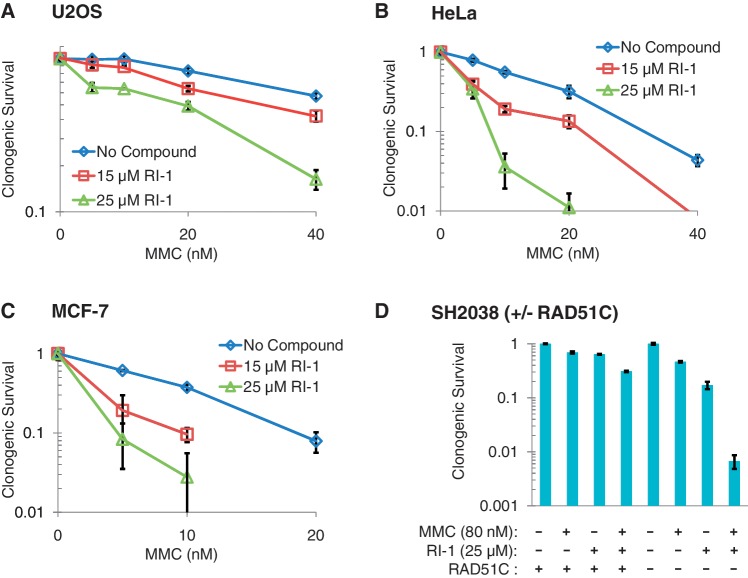
As discussed earlier, cells that harbor defects in HR genes are hypersensitive to various chemotherapy drugs, especially to drugs that induce inter-strand DNA cross-links. Therefore, we hypothesized that RI-1 would sensitize human cancer cells to MMC-induced lethality. Three common cancer cell lines (HeLa, MCF-7 and U2OS) were selected for these experiments, because they represent a wide range of human cancer types. Incubation of cells in RI-1 alone for 24 h generated some degree of single-agent toxicity in all three cancer cell lines, with LD50 values in the 20–40 µM range (Supplementary Figure S5).
To test for sensitization, cells were treated sequentially for 24 h with MMC followed by 24 h of RI-1. This experimental design was selected to eliminate possible artifacts that could potentially occur if MMC and RI-1 were to interact directly with one another. In all three cell types, RI-1 significantly sensitized cells to MMC (Figure 6A–C). The magnitude of sensitization was calculated as a function of the concentration of MMC required to kill 50% of cells. The magnitude of sensitization achieved with 25 µM RI-1 was 2.5–3 fold, depending on the cell type. These data suggest that RI-1 can specifically interfere with RAD51 functions in human cancer cells, thereby rendering them more susceptible to the lethal effects of oncologic treatment.
Figure 6.
RI-1 sensitizes human cell lines to DNA cross-linking chemotherapy. Five human cell lines were sequentially incubated for 24 h in media containing varying concentrations of mitomycin C (MMC), followed by 24 h in media containing RI-1. Cells were then allowed to grow in drug-free media for an additional 7–9 days. (A–C) Average survival for each condition is normalized to the MMC-free control of that condition. (D) For each cell line (RAD51C + or −), survival is normalized to cells receiving no treatment.
Additional survival assays were performed to confirm the specificity of RI-1 to RAD51 in a genetic model. Since mammalian cells require at least some RAD51 function for survival, it is difficult to evaluate RI-1 in human cells that have undergone complete RAD51 protein depletion or carry RAD51 null mutations. As an alternative, we tested the effects of RI-1 on an isogenic pair of immortalized human fibroblasts that differs in RAD51C (21). As discussed previously, the RAD51C mutant cells are known to exhibit reduced assembly of RAD51 at sites of DNA damage. As expected, the RAD51C mutant cells were relatively sensitive to MMC, relative to the RAD51C complemented cells. Interestingly, RI-1 plus MMC generated synergistic cell kill in RAD51C mutant cells that far exceeded the cell kill observed in RAD51 complemented cells, suggesting that cells with pre-existing defects in RAD51 assembly are specifically susceptible to RI-1 (Figure 6D). Taken together with the cytology-based results, these data provide evidence that RI-1 acts specifically to reduce RAD51 assembly in cells.
DISCUSSION
RI-1 sensitizes cells to DNA damage by directly and specifically disrupting HsRAD51. Our results demonstrate that this compound covalently attaches to HsRAD51 protein, thereby inhibiting the ability of RAD51 to form filaments on ssDNA. We also show that RI-1 can inhibit sub-nuclear accumulation of HsRAD51 protein at sites of DNA damage, and this inhibitory activity sensitizes various cancer cell types to cross-linking chemotherapy.
The covalent attachment that is formed has allowed us to map the RI-1 binding site on the surface of HsRAD51 protein. Analysis of ScRAD51 filament crystal structures suggests that RI-1 likely interferes with an interface used by protein subunits as they oligomerize into helical filaments around ssDNA (20). This surface of RAD51 has previously been implicated as an interaction interface based on studies of the Y315 residue (37), which is located only four residues away from C319 of HsRAD51. Furthermore, this cysteine resides within a loop of protein that serves as an ATP cap, which overlays the inter-subunit ATP binding site (33,34). This observation raises the possibility that RI-1 may inhibit RAD51 oligomerization by modulating its interactions with ATP. In addition to the filament-promoting role of this RAD51 protein interface, this protein surface might also serve other roles in HR. Specifically, it might coordinate the exchange of other HR proteins, since a C377Y mutation in budding yeast RAD51 (i.e. a mutation in the residue orthologous to HsRAD51 C319) was previously shown to modulate interactions between ScRAD51 and other HR-related proteins (RAD54 and RAD52) in a two-hybrid system (38). Thus, RI-1 binding to RAD51 could possibly inhibit cellular HR via additional mechanisms independent of its effects on RAD51 oligomerization.
Several other agents and potential strategies have been previously proposed for inhibiting HR. For example, mirin is a chemical inhibitor of the MRN (Mre11-Rad50-Nbs1) complex, which is capable of inhibiting HR in cells (39). However, consistent with the upstream role of MRN in sensing and signaling DNA damage, mirin generates a far broader range of cellular effects than RI-1, including inhibition of Ataxia telangiectasia mutated (ATM) activation, loss of G2/M cell cycle checkpoint and down-regulation of NHEJ repair efficiency (39,40). Additionally, the Kurumizaka and Mazin labs have identified several compounds that inhibit HsRAD51 in purified biochemical systems (41–44). Of these, B02 is the only compound shown to sensitize cells to DNA damage; it appears to do so by depleting RAD51 protein, which requires relatively long (10–12 h) incubations of cells with B02. Several groups have proposed utilizing peptide-based antagonists of either RAD51 or RAD51 paralog proteins to inhibit HR in cells (45–47). However, although these highly specific strategies are appealing in principle, their potential uses are limited by practical difficulties of transporting peptide-based agents across cell membranes and/or of administering them pharmacologically. Likewise, similar pharmacologic barriers will also probably limit the development of DNA aptamers as HR inhibitors in cells, even though these agents can specifically dissociate RAD51–DNA complexes in vitro (48).
As an alternative to directly inactivating RAD51 protein, some have proposed inhibiting cellular HR by down-regulating RAD51 at the transcriptional level. These strategies also have limitations that distinguish them from RI-1. For example, RAD51 protein levels have been successfully knocked down in cell culture experiments with ribozymes, siRNA and antisense oligonucleotides (9,18,49,50), but extension of any of these techniques into a drug development setting would presumably be challenging. Several existing drugs (including inhibitors of tyrosine kinases, chk1 and Poly (ADP-ribose) polymerase (PARP)) have also been noted to generate unanticipated reductions in RAD51 expression (17,51–54). Although it may be possible to exploit this RAD51 down-regulation clinically, these drug effects will be challenging to study because they occur in the background of other intended and unintended drug effects.
Our knowledge of the RI-1 binding site on RAD51 creates an important opportunity for chemical optimization of the compound. Preliminary structure–activity relationship experiments suggest that RI-1 has affinity for a pocket on the RAD51 protein surface, and that this reversible interaction becomes permanent when C319 reacts with the Michael acceptor of RI-1. If medicinal chemistry optimization of RI-1 can further exploit the reversible aspects of this affinity, then the covalent mode of attachment may prove to be dispensable in second generation chemical analogs. Alternatively, second generation analogs might continue to require covalent modification for activity, however this would not necessarily impede their development as drugs. Several commonly used drugs, including the anticancer agents MMC and sunitinib, contain Michael acceptors and still have tolerable toxicity profiles in patients.
RI-1 represents a powerful tool for future investigations on mechanisms of DNA repair. Presently, it is technically challenging to down-regulate RAD51 in living mammalian cells, since RAD51 is an essential gene. Although some methods have allowed for limited reductions of RAD51 activity in cells, including RNAi knockdown or expression of dominant negative proteins (23,27,28), these methods are slow and/or require new protein synthesis or protein turnover. RI-1 is expected to inhibit quickly, making novel analyses of RAD51 function possible. By disrupting filaments at intermediate stages, for example, it should be possible to test whether RAD51 filaments contribute to more than one step of DSB repair. Alternatively, it may help characterize the factors involved in disassembly of RAD51 filaments during repair. For example, RI-1 may be able to bypass the filament disassembly functions of the multi-functional recombination proteins RAD54 and RAD54B. These and related approaches may prove invaluable in future efforts to dissect the network of reaction pathways that contribute to DNA repair in cells.
Pharmacologic evaluation and testing in animal tumor models will be necessary to define the full potential of RI-1 and related second generation analogs as drug candidates. The ability of RI-1 to sensitize cells to DNA damage by directly and specifically disrupting HR repair provides considerable support for this therapeutic strategy. As such, this work represents an important advance in oncology drug development.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5 and Supplementary Methods.
FUNDING
The National Institutes of Health (NIH) grant [CA142642-02 2010-2015 to P.P.C., D.K.B., A.P.K., 2T32CA009594 to B.B.]. Funding for open access charge: NIH grant [CA142642-02].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Geoff Greene, Helmut Hanenberg, Maria Jasin, Phoebe Rice and Kathryn Stone for helpful conversations and/or materials. We also thank Jennifer Grubb for help with optimizing microscopy methods.
REFERENCES
- 1.Tebbs RS, Zhao Y, Tucker JD, Scheerer JB, Siciliano MJ, Hwang M, Liu N, Legerski RJ, Thompson LH. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc. Natl Acad. Sci. USA. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, et al. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 3.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 2001;477:131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 5.Martin RW, Orelli BJ, Yamazoe M, Minn AJ, Takeda S, Bishop DK. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res. 2007;67:9658–9665. doi: 10.1158/0008-5472.CAN-07-0290. [DOI] [PubMed] [Google Scholar]
- 6.Vispe S, Cazaux C, Lesca C, Defais M. Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res. 1998;26:2859–2864. doi: 10.1093/nar/26.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, Pierce AJ, Fishel R, Skorski T. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol. Cell. 2001;8:795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 8.Bello VE, Aloyz RS, Christodoulopoulos G, Panasci LC. Homologous recombinational repair vis-a-vis chlorambucil resistance in chronic lymphocytic leukemia. Biochem. Pharmacol. 2002;63:1585–1588. doi: 10.1016/s0006-2952(02)00954-1. [DOI] [PubMed] [Google Scholar]
- 9.Hansen LT, Lundin C, Spang-Thomsen M, Petersen LN, Helleday T. The role of RAD51 in etoposide (VP16) resistance in small cell lung cancer. Int. J. Cancer. 2003;105:472–479. doi: 10.1002/ijc.11106. [DOI] [PubMed] [Google Scholar]
- 10.Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair. 2008;7:686–693. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hine CM, Seluanov A, Gorbunova V. Use of the Rad51 promoter for targeted anti-cancer therapy. Proc. Natl Acad. Sci. USA. 2008;105:20810–20815. doi: 10.1073/pnas.0807990106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maacke H, Opitz S, Jost K, Hamdorf W, Henning W, Kruger S, Feller AC, Lopens A, Diedrich K, Schwinger E, et al. Over-expression of wild-type Rad51 correlates with histological grading of invasive ductal breast cancer. Int. J. Cancer. 2000;88:907–913. doi: 10.1002/1097-0215(20001215)88:6<907::aid-ijc11>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 13.Mitra A, Jameson C, Barbachano Y, Sanchez L, Kote-Jarai Z, Peock S, Sodha N, Bancroft E, Fletcher A, Cooper C, et al. Overexpression of RAD51 occurs in aggressive prostatic cancer. Histopathology. 2009;55:696–704. doi: 10.1111/j.1365-2559.2009.03448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connell PP, Jayathilaka K, Haraf DJ, Weichselbaum RR, Vokes EE, Lingen MW. Pilot study examining tumor expression of RAD51 and clinical outcomes in human head cancers. Int. J. Oncol. 2006;28:1113–1119. [PubMed] [Google Scholar]
- 15.Qiao GB, Wu YL, Yang XN, Zhong WZ, Xie D, Guan XY, Fischer D, Kolberg HC, Kruger S, Stuerzbecher HW. High-level expression of Rad51 is an independent prognostic marker of survival in non-small-cell lung cancer patients. Br. J. Cancer. 2005;93:137–143. doi: 10.1038/sj.bjc.6602665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takenaka T, Yoshino I, Kouso H, Ohba T, Yohena T, Osoegawa A, Shoji F, Maehara Y. Combined evaluation of Rad51 and ERCC1 expressions for sensitivity to platinum agents in non-small cell lung cancer. Int. J. Cancer. 2007;121:895–900. doi: 10.1002/ijc.22738. [DOI] [PubMed] [Google Scholar]
- 17.Russell JS, Brady K, Burgan WE, Cerra MA, Oswald KA, Camphausen K, Tofilon PJ. Gleevec-mediated inhibition of Rad51 expression and enhancement of tumor cell radiosensitivity. Cancer Res. 2003;63:7377–7383. [PubMed] [Google Scholar]
- 18.Ito M, Yamamoto S, Nimura K, Hiraoka K, Tamai K, Kaneda Y. Rad51 siRNA delivered by HVJ envelope vector enhances the anti-cancer effect of cisplatin. J. Gene Med. 2005;7:1044–1052. doi: 10.1002/jgm.753. [DOI] [PubMed] [Google Scholar]
- 19.Jayathilaka K, Sheridan SD, Bold TD, Bochenska K, Logan HL, Weichselbaum RR, Bishop DK, Connell PP. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc. Natl Acad. Sci. USA. 2008;105:15848–15853. doi: 10.1073/pnas.0808046105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 21.Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 22.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowalczykowski SC, Paul LS, Lonberg N, Newport JW, McSwiggen JA, von Hippel PH. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986;25:1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- 25.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansour WY, Schumacher S, Rosskopf R, Rhein T, Schmidt-Petersen F, Gatzemeier F, Haag F, Borgmann K, Willers H, Dahm-Daphi J. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res. 2008;36:4088–4098. doi: 10.1093/nar/gkn347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 30.Smith ME, Schumacher FF, Ryan CP, Tedaldi LM, Papaioannou D, Waksman G, Caddick S, Baker JR. Protein modification, bioconjugation, and disulfide bridging using bromomaleimides. J. Am. Chem. Soc. 2010;132:1960–1965. doi: 10.1021/ja908610s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan CP, Smith ME, Schumacher FF, Grohmann D, Papaioannou D, Waksman G, Werner F, Baker JR, Caddick S. Tunable reagents for multi-functional bioconjugation: reversible or permanent chemical modification of proteins and peptides by control of maleimide hydrolysis. Chem. Commun. 2011;47:5452–5454. doi: 10.1039/c1cc11114k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 33.Wu Y, He Y, Moya IA, Qian X, Luo Y. Crystal structure of archaeal recombinase RADA: a snapshot of its extended conformation. Mol. Cell. 2004;15:423–435. doi: 10.1016/j.molcel.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 34.Wu Y, Qian X, He Y, Moya IA, Luo Y. Crystal structure of an ATPase-active form of Rad51 homolog from Methanococcus voltae. Insights into potassium dependence. J. Biol. Chem. 2005;280:722–728. doi: 10.1074/jbc.M411093200. [DOI] [PubMed] [Google Scholar]
- 35.Bishop DK. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 36.Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA- catalyzed DNA strand exchange. Genes Dev. 2001;15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conilleau S, Takizawa Y, Tachiwana H, Fleury F, Kurumizaka H, Takahashi M. Location of tyrosine 315, a target for phosphorylation by cAbl tyrosine kinase, at the edge of the subunit-subunit interface of the human Rad51 filament. J. Mol. Biol. 2004;339:797–804. doi: 10.1016/j.jmb.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 38.Krejci L, Damborsky J, Thomsen B, Duno M, Bendixen C. Molecular dissection of interactions between Rad51 and members of the recombination-repair group. Mol. Cell Biol. 2001;21:966–976. doi: 10.1128/MCB.21.3.966-976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat. Chem. Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 41.Ishida T, Takizawa Y, Kainuma T, Inoue J, Mikawa T, Shibata T, Suzuki H, Tashiro S, Kurumizaka H. DIDS, a chemical compound that inhibits RAD51-mediated homologous pairing and strand exchange. Nucleic Acids Res. 2009;37:3367–3376. doi: 10.1093/nar/gkp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takaku M, Kainuma T, Ishida-Takaku T, Ishigami S, Suzuki H, Tashiro S, van Soest RW, Nakao Y, Kurumizaka H. Halenaquinone, a chemical compound that specifically inhibits the secondary DNA binding of RAD51. Genes Cells. 2011;16:427–436. doi: 10.1111/j.1365-2443.2011.01494.x. [DOI] [PubMed] [Google Scholar]
- 43.Huang F, Motlekar NA, Burgwin CM, Napper AD, Diamond SL, Mazin AV. Identification of specific inhibitors of human RAD51 recombinase using high-throughput screening. ACS Chem. Biol. 2011;6:628–635. doi: 10.1021/cb100428c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang F, Mazin OM, Zentner IJ, Cocklin S, Mazin AV. Inhibition of homologous recombination in human cells by targeting RAD51 recombinase. J. Med. Chem. 2012;55:3011–3020. doi: 10.1021/jm201173g. [DOI] [PubMed] [Google Scholar]
- 45.Connell PP, Siddiqui N, Hoffman S, Kuang A, Khatipov EA, Weichselbaum RR, Bishop DK. A hot spot for RAD51C interactions revealed by a peptide that sensitizes cells to cisplatin. Cancer Res. 2004;64:3002–3005. doi: 10.1158/0008-5472.can-03-3608. [DOI] [PubMed] [Google Scholar]
- 46.Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH. Expression of BRC repeats in breast cancer cells disrupts the BRCA2- Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J. Biol. Chem. 1999;274:32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- 47.Nomme J, Renodon-Corniere A, Asanomi Y, Sakaguchi K, Stasiak AZ, Stasiak A, Norden B, Tran V, Takahashi M. Design of potent inhibitors of human RAD51 recombinase based on BRC motifs of BRCA2 protein: modeling and experimental validation of a chimera peptide. J. Med. Chem. 2010;53:5782–5791. doi: 10.1021/jm1002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez SF, Renodon-Corniere A, Nomme J, Eveillard D, Fleury F, Takahashi M, Weigel P. Targeting human Rad51 by specific DNA aptamers induces inhibition of homologous recombination. Biochimie. 2010;92:1832–1838. doi: 10.1016/j.biochi.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 49.Ohnishi T, Taki T, Hiraga S, Arita N, Morita T. In vitro and in vivo potentiation of radiosensitivity of malignant gliomas by antisense inhibition of the RAD51 gene. Biochem. Biophys. Res. Commun. 1998;245:319–324. doi: 10.1006/bbrc.1998.8440. [DOI] [PubMed] [Google Scholar]
- 50.Collis SJ, Tighe A, Scott SD, Roberts SA, Hendry JH, Margison GP. Ribozyme minigene-mediated RAD51 down-regulation increases radiosensitivity of human prostate cancer cells. Nucleic Acids Res. 2001;29:1534–1538. doi: 10.1093/nar/29.7.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hegan DC, Lu Y, Stachelek GC, Crosby ME, Bindra RS, Glazer PM. Inhibition of poly(ADP-ribose) polymerase down-regulates BRCA1 and RAD51 in a pathway mediated by E2F4 and p130. Proc. Natl Acad. Sci. USA. 2010;107:2201–2206. doi: 10.1073/pnas.0904783107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ko JC, Tsai MS, Kuo YH, Chiu YF, Weng SH, Su YC, Lin YW. Modulation of Rad51, ERCC1, and thymidine phosphorylase by emodin result in synergistic cytotoxic effect in combination with capecitabine. Biochem. Pharmacol. 2011;81:680–690. doi: 10.1016/j.bcp.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Welsh JW, Mahadevan D, Ellsworth R, Cooke L, Bearss D, Stea B. The c-Met receptor tyrosine kinase inhibitor MP470 radiosensitizes glioblastoma cells. Radiat. Oncol. 2009;4:69. doi: 10.1186/1748-717X-4-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parsels LA, Morgan MA, Tanska DM, Parsels JD, Palmer BD, Booth RJ, Denny WA, Canman CE, Kraker AJ, Lawrence TS, et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol. Cancer Ther. 2009;8:45–54. doi: 10.1158/1535-7163.MCT-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.