Abstract
The RNA-binding protein Lin28 regulates the processing of a developmentally important group of microRNAs, the let-7 family. Lin28 blocks the biogenesis of let-7 in embryonic stem cells and thereby prevents differentiation. It was shown that both RNA-binding domains (RBDs) of this protein, the cold-shock domain (CSD) and the zinc-knuckle domain (ZKD) are indispensable for pri- or pre-let-7 binding and blocking its maturation. Here, we systematically examined the nucleic acid-binding preferences of the Lin28 RBDs and determined the crystal structure of the Lin28 CSD in the absence and presence of nucleic acids. Both RNA-binding domains bind to single-stranded nucleic acids with the ZKD mediating specific binding to a conserved GGAG motif and the CSD showing only limited sequence specificity. However, only the isolated Lin28 CSD, but not the ZKD, can bind with a reasonable affinity to pre-let-7 and thus is able to remodel the terminal loop of pre-let-7 including the Dicer cleavage site. Further mutagenesis studies reveal that the Lin28 CSD induces a conformational change in the terminal loop of pre-let-7 and thereby facilitates a subsequent specific binding of the Lin28 ZKD to the conserved GGAG motif.
INTRODUCTION
MicroRNAs (miRNAs) are small regulatory RNAs, typically ∼22 nt in length, which regulate target mRNAs through complementary base pairing. The biogenesis of miRNAs can be divided into two major steps. First, the nuclear localized ribonuclease III Drosha, in a complex with DGCR8 (Pasha in invertebrates), cleaves a long primary miRNA transcript (pri-miRNA), creating a characteristic 60–80 nt hairpin structure with a 2-nt 3′ overhang (1,2). After export of this precursor-miRNA (pre-miRNA) by exportin-5:RanGTP (3,4), it is further cleaved by Dicer to yield the mature miRNA that functions in the RNA-induced silencing complex (RISC) (5–7).
The highly conserved let-7 miRNA is known to regulate developmental timing and proliferation (8–10). Mature let-7 is absent or only present in low amounts in various stem and progenitor cells but strongly induced upon tissue differentiation. Multiple oncogenes, such as RAS, HMGA2, c-MYC, Cdc25a and Cdk6 are targets of let-7, and it has been reported that let-7 can act as a tumor suppressor in lung cancers (11–13). Interestingly, mature let-7 is downregulated in various cancer and stem cells while levels of pri- and pre-let-7 are not affected, suggesting a regulation of let-7 maturation rather than at transcription levels (14).
The RNA-binding protein Lin28 (from cell lineage abnormal 28) was shown to regulate the maturation of let-7 and is involved in the regulation of pluripotency, development and differentiation (15–18). In Caenorhabditis elegans, mutations in the LIN28 gene disturbed the developmental progression of the worm (19). Further experiments revealed that Lin28 is abundant early in nematode development but is downregulated during larval stages L1–L3 (20,21), whereas the amount of mature let-7 shows an opposite pattern (11,22). This opposing expression pattern is also valid for mammalian cells, in which the corresponding homologs Lin28a and Lin28b are mainly expressed in undifferentiated embryonic stem cells (18,23,24). Strikingly, mature let-7 itself is involved in the post-transcriptional repression of LIN28 upon differentiation. It can bind to the 3′-untranslated region (UTR) of the Lin28 mRNA and thereby enables a double-negative feedback loop (17,25). Reciprocally, Lin28 was used to reprogram adult human fibroblasts to induced pluripotent stem cells in cooperation with Oct4, Sox2 and Nanog (26). These observations, in conjunction with the fact that LIN28b and less frequently LIN28a are reactivated in many tumor cells, indicate the functional relevance of Lin28 in maintaining and reconstituting pluripotency (27).
Recent studies revealed the molecular basis for the selective inhibition of let-7 biogenesis by Lin28. According to these, Lin28 can inhibit the maturation of let-7 in two different ways. First, Lin28 binds specifically the terminal loop of pri- and pre-let-7, thereby interfering with cleavage by Drosha and Dicer, respectively (16–18,28). Second, Lin28 is thought to promote terminal uridylation of the 3′-end of pre-let-7 by recruiting a terminal uridyl-transferase (TUT4/Zcchc11 in mammals, PUP-2 in C. elegans) (15,29,30). The uridylated pre-let-7 is resistant to cleavage by Dicer and thus serves as a signal for degradation. Furthermore, it was shown that a conserved GGAG motif within the terminal loop (pre-element, preE; Figure 1) of pre-let-7 is essential for efficient binding and uridylation (31). Recent structural studies showed that this sequence-specific interaction is mediated by the two CCHC zinc knuckles forming the zinc-knuckle domain (ZKD) of Lin28 (32,33). However, because the observed ZKD-binding site is inaccessible within a double-helical stem region in most pre-let-7 (Figure 1B), binding of Lin28 requires an extensive remodeling of preE. The N-terminal cold-shock domain (CSD) may mediate this remodeling, as CSDs are known to bind to a wide range of single-stranded nucleic acids and can act as RNA chaperones (34–37).
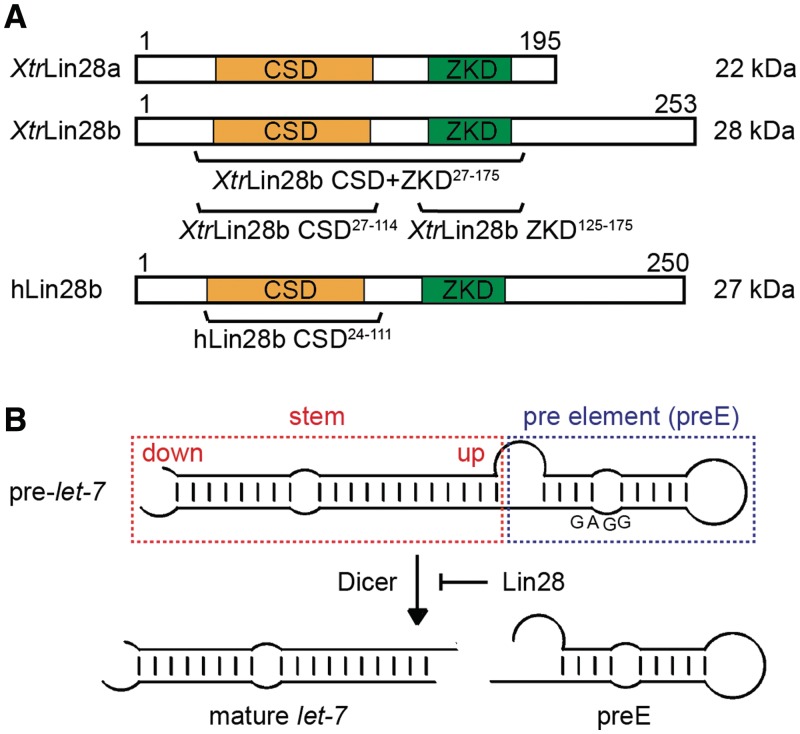
Figure 1.
Schematic representation of the domain structure of Lin28b and the processing of pre-let-7 by Dicer. (A) Domain structure of XtrLin28a and b and human (h) Lin28b with the molecular weight and amino acid positions indicated. The truncated Lin28 variants used in this study are shown below the corresponding domain representation. (B) Lin28 inhibits the Dicer-mediated pre-let-7 processing by binding to the pre-element (preE).
To obtain functional and mechanistic insights into Lin28-mediated inhibition of let-7 maturation, we undertook a detailed structural and biochemical analyses of the Lin28 nucleic acid interactions. The data presented here demonstrate how Lin28 can associate with a wide range of different preE RNAs with diverse secondary structure and specifically inhibit the maturation of pre-let-7. We show that Lin28 binds in a stepwise manner to pre-let-7, a process that can be subdivided into an initial binding of the CSD followed by an extensive remodeling of preE and subsequent binding of the ZKD. Further mutagenesis experiments in context of the isolated Lin28b RNA-binding domains (RBDs) and full-length Lin28b support our conclusion that the CSD is responsible for remodeling pre-let-7, while the ZKD ensures a specific and directional binding.
MATERIALS AND METHODS
Cloning and protein overexpression
The cDNAs for human Lin28b and Xenopus tropicalis (Xtr) Lin28 homologs A and B (hLin28b, XtrLin28a and XtrLin28b) were obtained from Source Bioscience (Nottingham) and used for designing the following constructs: full-length protein (XtrLin28b2−253), an N- and C-terminally truncated protein containing the CSD and ZKD (XtrLin28b CSD+ZKD27−175), CSD-only constructs (XtrLin28b CSD27−114 and hLin28b CSD24−111) and a ZKD-only construct (XtrLin28b ZKD125−175). The coding sequence of the respective constructs were polymerase chain reaction (PCR)-amplified and cloned into the bacterial expression vector pQLinkH (38) via BamHI and NotI restriction sites. Mutants of Xtrlin28b and its isolated domains were generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according the manufacturer’s protocol. The N-terminally His7-tagged proteins were overexpressed in Escherichia coli Rosetta (DE3) cells at 293 K overnight. Cells were grown in Terrific Broth medium and induced at an OD600 of 1.5 with 0.5 mM isopropyl β-d-1-thiogalactopyranoside. After harvesting cells by centrifugation, pellets were either directly used for purification or stored at 193 K.
Protein purification
The purification procedure comprised the following steps: (i) Ni/Zn affinity chromatography on a 5 ml HisTrap FF crude column (GE Healthcare); (ii) cleavage of the His7 tag by tobacco etch virus (TEV) protease; (iii) reapplication of the cleaved protein on the Ni/Zn affinity column; (iv) chromatography using a 5 ml heparin column (GE Healthcare); and (v) size-exclusion chromatography on a Superdex 75 prep-grade column (16 × 60 cm or 26 × 60 cm, GE Healthcare). All purification steps were carried out on ice or at 277 K. For constructs containing the ZKD, Ni2+ HisTrap columns were charged with Zn2+ according to the manufacturer’s protocol to avoid a Zn2+ to Ni2+ exchange within the zinc finger. In addition, purification buffers were supplemented with 10 μM ZnSO4 for these proteins.
Escherichia coli cells overexpressing the protein of interest were resuspended in phosphate-buffered saline (PBS) lysis buffer [1× PBS, pH 7.4, 0.5 M NaCl, 5% (v/v) glycerol and 1 mM dithiothreitol (DTT)], supplemented with 1 μg/ml DNase I (Roche), 1 U/ml Benzonase (Novagen) and one tablet of EDTA-free Complete Protease Inhibitor (Roche). After mechanical lysis of the bacterial suspension using a microfluidizer (Microfluidics) or sonication (SONOPULS HD 2200, Bandelin), the soluble fraction of the lysate was prepared by centrifugation at 50 000g for 45 min. The supernatant was filtered through a 0.2 µm pore size filter and applied on a 5 ml HisTrap column, equilibrated with buffer I (20 mM Tris/HCl, pH 8.0, 0.5 M NaCl, 1 mM DTT and 5 mM imidazole). After washing with at least 10 column volumes of buffer I supplemented with 25 mM imidazole, the protein was eluted using a linear imidazole gradient (25–250 mM). The eluate was then dialyzed overnight against buffer I in the presence of 1 mg TEV protease per 20 mg protein and reapplied on a 5 ml HisTrap FF column. The flow-through was collected, dialyzed for at least 4 h against buffer II (20 mM Bis–Tris, pH 6.5, 1 mM DTT) and applied on a 5 ml heparin column equilibrated with buffer II. The target protein was eluted using a linear gradient of 0–1 M NaCl, dialyzed against buffer III (20 mM Tris/HCl, pH 7.5, 1 mM DTT and 100 mM KCl/360 mM NaCl, respectively), concentrated to a volume of 5–13 ml and applied to the final size-exclusion chromatography using buffer III as running buffer. The peak fractions were pooled and concentrated to about 10 mg/ml. After flash-freezing in liquid nitrogen, the proteins were stored at 193 K.
Protein crystallization
The purified XtrLin28b CSD was crystallized either alone or in complex with the ssDNA oligonucleotides dT7 (TTT TTT T) or dT6 (TTT TTT) or the ssRNA oligonucleotide rU6 (UUU UUU). ssDNA oligonucleotides were obtained from Biotez (Berlin) and ssRNA oligonucleotides from Biomers (Ulm). All crystallization trials were performed using the vapor diffusion method by mixing equal volumes of protein or protein–oligonucleotide complex solution and reservoir buffer at either 277 K for the XtrLin28b CSD–oligonucleotide complexes and hLin28b CSD apo protein or at 293 K for the XtrLin28b CSD apo protein. Crystals of the XtrLin28b CSD apo protein could be obtained with 10 mg/ml protein (in 20 mM Tris/HCl, pH 8.0, 360 mM NaCl and 5 mM DTT) and 2.5 M sodium acetate, 0.1 M HEPES, pH 7.0 as reservoir solution. The hLin28b CSD apo protein was crystallized at 11 mg/ml (in 20 mM Tris/HCl, pH 8.0, 340 mM NaCl and 5 mM DTT) using 2 M ammonium sulfate, 0.2 M NaCl and 0.1 M MES, pH 6.5 as reservoir solution. For co-crystallization of XtrLin28b CSD with DNA or RNA oligonucleotides, the protein was mixed with a 1.2-fold molar excess of the corresponding oligonucleotide, followed by gel filtration on a Superose S12 10–300 GL column (GE Healthcare) to separate the complex from unbound oligonucleotides. Crystals for XtrLin28b CSD in complex with dT6 were grown using 0.75 mM protein–DNA complex (in 20 mM Tris/HCl, pH 7.5, 100 mM KCl, 2 mM MgCl2 and 1 mM DTT) and 0.1 mM Bis–Tris, pH 5.5, 15% (w/v) PEG 3350 and 0.1 M sodium thiocyanate as reservoir buffer. The XtrLin28b CSD/dT7 complex (1 mM in 20 mM Tris, pH 7.5, 100 mM KCl and 1 mM DTT) was crystallized using 17% (w/v) PEG 3350, 0.2 M sodium thiocyanate as reservoir buffer, whereas the crystallization of XtrLin28b CSD bound to rU6 could be achieved under similar conditions but using a reservoir buffer containing 20% (w/v) PEG 3350 and 0.2 M potassium thiocyanate. After soaking in a cryoprotectant consisting of reservoir solution supplemented with 25–30% (v/v) glycerol, the crystals were flash-frozen in liquid nitrogen.
Data collection, structure determination and refinement
X-ray diffraction data were collected at 100 K and a wavelength of 0.9184 Å at beamline BL 14.1 or BL 14.2 of the Helmholtz-Zentrum Berlin für Materialien und Energie and Freie Universität Berlin at BESSY (39). Initial indexing and determination of an optimal data collecting strategy was performed using iMOSFLM (40). The recorded diffraction data for the XtrLin28b CSD crystals were indexed, integrated and scaled with X-ray detector software (XDS) (41) and for the hLin28b CSD crystals with HKL2000 (42). For the native protein, the phase problem could be solved by molecular replacement using Phaser and Salmonella thyphimurium cold-shock protein (3I2Z) as a template. The structures of hlin28b CSD and the RNA- or DNA-bound XtrLin28b CSDs were solved similarly but using the apo XtrLin28b CSD as a template. After initial model building by ARP/wARP (43), the structure was refined using REFMAC 5.5 (44) in multiple rounds, while the DNA or RNA oligonucleotides were placed manually into the density using COOT (45). Statistics on data collection and refinement are summarized in Supplementary Tables S4 and S5. The graphics program COOT was used for model building and structure analysis. Molecular drawings and root-mean-square deviation (RMSD) values were created or calculated with PyMOL Molecular Graphics System (Version 1.3, Schrödinger, LLC). Hydrogen bonding was analyzed using the University of California at San Francisco Chimera program (46). The resulting structures of the XtrLin28b CSD alone or bound to dT6, dT7 or rU6 as well as the apo hLin28b CSD structure were deposited in the RCSB Protein Data Bank (http://www.pdb.org).
Fluorescence titration of Lin28 CSD with oligonucleotides
The intrinsic fluorescence of Trp39 of XtrLin28b CSD is quenched upon binding of nucleic acids and was therefore used as a probe to examine the DNA-binding preferences of the protein. For determination of the equilibrium dissociation constants, 50 nM XtrLin28b CSD was titrated with increasing amounts of oligonucleotide depending on dissociation constants in preliminary experiments in a total volume of 1 ml. After addition of DNA oligonucleotides from a highly concentrated stock solution, the mixture was equilibrated for 1 min under constant stirring. Finally, the fluorescence of Trp39 was measured at 350 nm upon excitation at 280 nm. All experiments were performed using a Cary-Eclipse fluorescence spectrofluorimeter at 293 K in 20 mM Tris/HCl, pH 7.5, 60 mM KCl, supplemented with 40 μM N-acetyltryptophanamide. The fluorescence was corrected for inner filter effects, buffer and oligonucleotide fluorescence and dilution. The changes of fluorescence were analyzed according to the following binding equation (47,48):
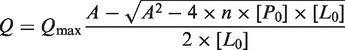 |
with A = KD + [P0] + [L0], where Q represents the quenching of the intrinsic Trp39 fluorescence after each addition of oligonucleotide, Qmax the maximum quenching upon saturation of the protein with ligand, n the stoichiometric ratio of the protein–ligand complex, KD the equilibrium dissociation constant and [P0] and [L0] the overall concentrations of either XtrLin28b CSD and the respective oligonucleotide.
Isothermal titration calorimetry
All isothermal titration calorimetry (ITC) experiments were carried out at 288 K in a VP-ITC (Microcal) in 20 mM Tris/HCl, pH 8.0 and 60 mM KCl. Prior to the measurement, all protein constructs and synthetic oligonucleotides were dialyzed against this buffer. Depending on the construct and the respective oligonucleotide, protein concentrations between 4 and 15 μM were used. All binding isotherms and thermodynamic parameters were fitted and calculated using the MICROCAL ORIGIN software.
Electrophoretic mobility shift assay
All RNA constructs used for electrophoretic mobility shift assay (EMSA) were in vitro transcribed using the T7 Transcription Kit from Roboklon (Berlin) according to the manufacturer’s protocol. To ensure optimal transcription, slightly shortened pre-let-7 templates with a G at the 5′-end were designed. However, this change in sequence had no effect on pre-let-7 secondary structure according to Mfold (49). All templates were cloned and fused to a T7 RNA-polymerase promoter (TAATA CGACT CACTA TAG) and amplified via PCR. Radiolabeling was performed during in vitro transcription using α-32P-ATP (Perkin Elmer) or afterward using γ-32P UTP (Perkin Elmer) for 5′-end labeling.
Binding reactions were conducted using 1 nM of α-32P-ATP (Perkin Elmer) labeled pre-miRNA as a probe in a total volume of 20 μl together with 10 μg of yeast tRNA and the indicated amounts of recombinant Lin28 protein. The labeled RNA was first heated to 368 K for 2 min and immediately cooled on ice for 3 min. Binding buffer contained 20 mM Tris/HCl, pH 7.5, 60 mM KCl, 10 U of Ribonuclease Inhibitor (Roboklon) and 1 mM DTT. After 30 min of incubation at room temperature, the EMSA sample was mixed in a 1:1 ratio with loading buffer (similar to binding buffer but supplemented with 50% (v/v) glycerol, 0.1% (w/v) bromophenol blue and 0.1% (w/v) xylene cyanol) and resolved on a native 10% (w/v) polyacrylamide gel. RNA was visualized by phosphorimaging, and band intensities of scanned gels were quantified using ImageQuant (GE Healthcare). The total amount of probe in each binding reaction was normalized against the unbound probe (in the absence of recombinant protein). The data were fitted to a one-site specific binding function with Hill slope, thereby taking a possible cooperative binding into account. The following binding equation of the nonlinear curve fitting method of GraphPad Prism (version 5.0a for Mac, GraphPad Software, La Jolla, CA, USA) was used:
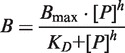 |
where B represents the amount of complex as a function of the protein concentration [P], Bmax the observed maximum fraction of complex, h the Hill coefficient and KD the dissociation constant.
In vitro pre-miRNA processing assay
The 10 000 cpm in vitro transcribed and 32P 5′-end labeled pre-miRNA was incubated with the indicated amounts of the corresponding Lin28 variant for 30 min on ice in 20 mM Tris/HCl, pH 7.5, 60 mM KCl, 10 mM MgCl2 and 1 mM DTT. Two units of recombinant human Dicer (Genlantis) were added to each sample and incubated for 2 h at 310 K. The reaction was stopped by mixing 4 µl Dicer Stop Solution (Genlantis) with 20 µl sample and heated to 368 K for 5 min. All samples were resolved on 10% (w/v) TBE–polyacrylamide gel supplemented with 8 M urea and visualized by autoradiography.
RNA remodeling assay
A truncated Xtr-pre-let-7 g* was synthesized by Thermo Scientific. This construct contained a 5′-dabcyl and a 3′-fluorescein label that are in close proximity to each other according to secondary structure predictions using Mfold (Figure 5A). For determination of the remodeling activity of the used Lin28 constructs, 200 nM of Xtr-pre-let-7 g* was titrated with increasing amounts of protein in a total volume of 1 ml. After addition of protein from a highly concentrated stock solution, the mixture was equilibrated for 1 min under constant stirring. Finally, the fluorescence of fluorescein was measured at 522 nm upon excitation at 495 nm. All experiments were performed using a Cary-Eclipse fluorescence spectrofluorimeter at 293 K in 20 mM Tris/HCl, pH 7.5, 60 mM KCl, 10 mM MgCl2 and 1 mM DTT. The fluorescence was corrected for inner filter effects, buffer and dilution.
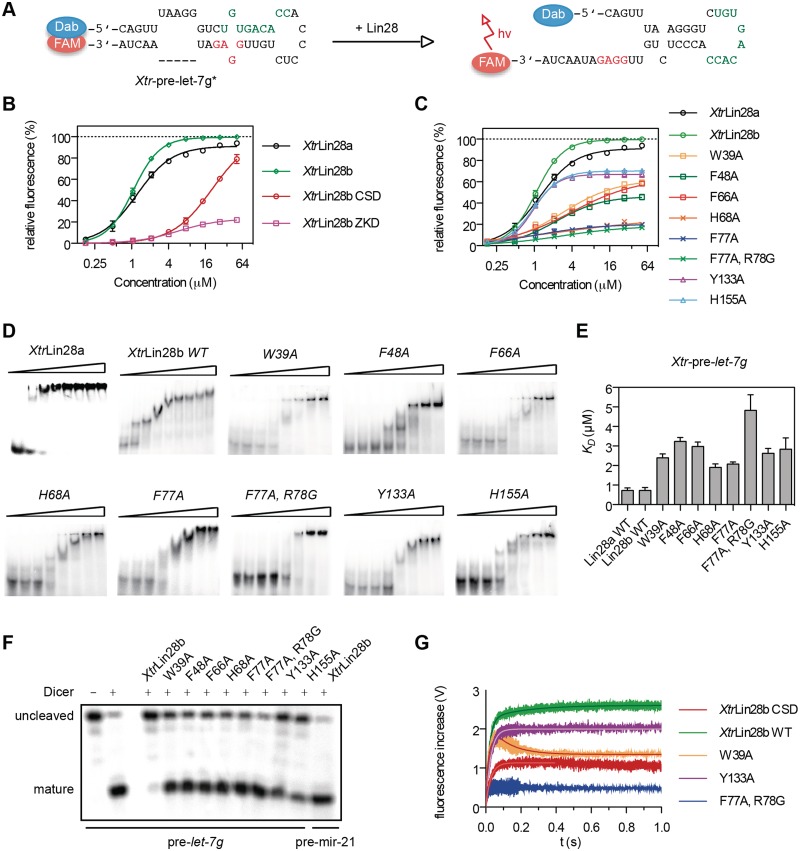
Figure 5.
The Lin28 CSD remodels the terminal loop of pre-let-7 and facilitates a subsequent binding of the ZKD to the conserved GGAG motif. (A–C) RNA remodeling assay. A truncated Xtr-pre-let-7g that contains only the preE sequence and 5 bp of the upper stem region (Xtr-pre-let-7g*) was incubated with increasing concentrations of the corresponding Lin28 protein. The 5′-end of the RNA was modified with the quencher dabcyl (Dab), and the adjacent 3′-end harbored a fluorescein (FAM) label. The sequence and secondary structure of the RNA are indicated and the known ZKD- and CSD-binding sites are marked in red and green, respectively. The increase of fluorescence was plotted as a function of titrated Lin28 protein. All experiments were performed using a Cary-Eclipse fluorescence spectrometer at 293 K in 20 mM Tris (pH 7.5), 60 mM KCl, 10 mM MgCl2 and 1 mM DTT. (D) EMSAs with Xtr-pre-let-7g as a probe, mixed with increasing concentrations of XtrLin28a and the indicated XtrLin28b variants (0, 0.4, 0.8, 1.6, 3.2, 6.4, 13, 26 µM). All EMSAs were performed using 1 nM α-32P-ATP-labeled RNA. (E) Apparent dissociation constants from EMSAs shown in (D). Band intensities were quantitated from three independent experiments and used to generate the binding data. (F) In vitro pre-miRNA processing reaction on Xtr-pre-let-7g. 10 µM of the indicated XtrLin28b variant was added to 1 nM 32P 5′-end-labeled Xtr-pre-let-7g in the presence or absence of human Dicer. The samples were resolved on a 10% (w/v) denaturing PAGE and visualized by autoradiography. (G) Pre-steady-state kinetics of Lin28-mediated binding and remodeling of Xtr-pre-let-7g*. After rapid mixing of Xtr-pre-let-7g* with 15 µM (final concentration) of the indicated XtrLin28b variant, the change of FAM fluorescence was monitored for 1 s using a Chirascan stopped-flow instrument. Traces of at least eight replicates were fitted to a mono- (Y133A) or biexponential curve (Xtrlin28b), respectively (solid lines). For W39A, the time courses of the first 80 ms were fitted to a single exponential association curve, while traces from 90 ms to 1 s were fitted to a one-phase decay curve (see Supplementary Figure S11).
Kinetic measurements of RNA remodeling
The kinetics of Lin28-mediated Xtr-pre-let-7 g* remodeling were measured using 15 µM Lin28 variant and 100 nM Xtr-pre-let-7g* (final concentration for each) on a Chirascan SF.3 spectrometer (Applied Photophysics). The path length of the observation chamber was 10 mm. The kinetic was followed by fluorescence above 515 nm after excitation at 490 nm (5 nm bandwidth). The experiments were performed at 293 K in 20 mM Tris/HCl, pH 7.5, 60 mM KCl, 10 mM MgCl2 and 1 mM DTT. The fluorescence was corrected for buffer and dilution. Kinetic curves were measured at least 12 times under identical conditions and fitted either as mono- or biexponential curves, respectively, and analyzed assuming a 1:1 stoichiometry.
RESULTS
The Lin28 ZKD binds with high specificity to single-stranded GGAG or GGUG sequences
To understand the binding preferences of Lin28 to DNA and RNA, we first examined the binding of the isolated RBDs of XtrLin28b (Figure 1) to single-stranded oligonucleotides. It has been reported that Lin28 variants with mutations in the ZKD were impaired in pre-let-7 binding (28,31). Further experiments using electrophoretic mobility shift (EMSA) and in vitro uridylation assays showed that the conserved GGAG or GGUG motif within preE is critical for Lin28 binding and Lin28-mediated uridylation by TUT4 (31).
To confirm that the Lin28 ZKD mediates this binding preference, we performed isothermal titration calorimetry (ITC) experiments with the isolated ZKD and small ssRNA oligonucleotides. The data show that XtrLin28b ZKD binds tightly to GGAG- and GGUG-containing oligonucleotides with dissociation constants (KD) of 45 or 32 nM, respectively (Table 1 and Supplementary Figure S1). A single G-to-A mutation at the first or second position within the GGAG motif impaired binding only slightly (rZ3, rZ4), while mutating both Gs led to a >250-fold decrease in binding affinity (rZ5). A mutation of the fourth position strongly impaired the binding of the ZKD (rZ6, rZ7). This is consistent with previously observed NMR structures of the ZKD of the nucleocapsid protein of HIV type 1 (HIV-1 NC), in which each zinc knuckle mediates a specific contact to G-2 or G-4 of the GGAG/GGUG motif, respectively (50,51). On the protein level, key residues for a specific interaction within the GGAG motif are Phe133 in the first and His155 in the second zinc knuckle, because a mutation toward A caused a severe loss of binding affinity. Both residues were shown to interact with each other and at least one G of the GGAG motif, thereby establishing a kinked conformation in the RNA backbone (32,33).
Table 1.
Equilibrium dissociation constants of XtrLin28b ZKD binding to ssRNA oligonucleotides derived from ITC experimentsa
| ZKD | ||
|---|---|---|
| Ligand | Sequence | KD (nM) |
| rZ1 | AA GGAG AA | 45 ± 4 |
| rZ2 | AA GGUG AA | 32 ± 2 |
| rZ3 | AA AGAG AA | 200 ± 30 |
| rZ4 | AA GAAG AA | 600 ± 170 |
| rZ5 | AA AAAG AA | 11 900 ± 2200 |
| rZ6 | AA GGAA AA | n.d.b |
| rZ7 | AA AGAA AA | 6800 ± 2100 |
| rZ8 | AA GGGG AA | n.d.b |
| rA8 | AA AAAA AA | n.d.b |
| rU7 | UU UUUU U | n.d.b |
| ZKD variants | ||
|---|---|---|
| Mutation | Sequence | KD (nM) |
| Y133A | AA GGAG AA | 2200 ± 700 |
| H155A | AA GGAG AA | 2600 ± 600 |
Additionally calculated thermodynamic parameters are listed in the supporting information (Supplementary Table S1).
aAll experiments were performed at 288 K in 20 mM Tris/HCl, pH 8.0 and 60 mM KCl.
bNot determined due to insufficient affinity.
Remarkably, no binding was observable for adenosine or uridine oligomers (rA8, rU7), and XtrLin28b ZKD did not bind to a GGGG-containing oligonucleotide, indicating that G itself cannot act as a spacing residue (rZ8). Taken together, the data show that two Gs spaced by at least one nucleotide other than G are specifically recognized by the Lin28 ZKD.
The Lin28 CSD preferentially binds to pyrimidine-rich heptanucleotides with high affinity but limited sequence specificity
We and others demonstrated previously that bacterial cold-shock proteins (Csps) and eukaryotic CSDs show a preference for pyrimidine-rich ssDNA/ssRNA oligonucleotides with KD values in the low nanomolar range (34–37). To test whether XtrLin28b CSD shows a similar sequence selectivity and affinity, we systematically analyzed the binding preferences of XtrLin28b CSD to ssDNA oligonucleotides using fluorescence titration experiments.
First, we addressed the question how many nucleotides are bound by XtrLin28b CSD. Therefore, we used oligothymidines in the range from 5 to 9 nt and analyzed the respective KD values. The binding affinity constantly increases from 5 - to 7-mers with KD values ranging from 680 to 58 nM at a 1:1 stoichiometric ratio. No further substantial increase in binding strength could be obtained for longer ssDNA oligonucleotides, indicating that XtrLin28b CSD preferentially binds hepta- or octanucleotides (Supplementary Figure S2 and Table 2). However, XtrLin28b CSD shows lower affinity toward ssDNA oligonucleotides than bacterial Csps. In the case of Bacillus subtilis CspB or Bacillus caldolyticus Csp, dT7 was bound 25 - to 50-fold tighter (35,37).
Table 2.
Equilibrium dissociation constants of XtrLin28b CSD:ssDNA complexes derived from fluorescence titration experiments assuming a 1:1 stoichiometry
| Ligand | Sequence | KD (nM) |
|---|---|---|
| dT5 | TTT TT | 680 ± 40 |
| dT6 | TTT TTT | 169 ± 30 |
| dT7 | TTT TTT T | 59 ± 5 |
| dT8 | TTT TTT TT | 32 ± 1 |
| dT9 | TTT TTT TTT | 44 ± 3 |
| dA1 | ATT TTT T | 84 ± 5 |
| dA2 | TAT TTT T | 311 ± 24 |
| dA3 | TTA TTT T | 137 ± 10 |
| dA4 | TTT ATT T | 124 ± 12 |
| dA5 | TTT TAT T | 176 ± 8 |
| dA6 | TTT TTA T | 147 ± 15 |
| dA7 | TTT TTT A | 113 ± 15 |
| dA7 | AAA AAA A | – |
| dG1 | GTT TTT T | 12 ± 1 |
| dG2 | TGT TTT T | 39 ± 4 |
| dG3 | TTG TTT T | 139 ± 8 |
| dG4 | TTT GTT T | 119 ± 16 |
| dG5 | TTT TGT T | 45 ± 4 |
| dG6 | TTT TTG T | 81 ± 10 |
| dG7 | TTT TTT G | 80 ± 10 |
| dG7 | GGG GGG G | – |
| dC1 | CTT TTT T | 45 ± 4 |
| dC2 | TCT TTT T | 137 ± 22 |
| dC3 | TTC TTT T | 71 ± 3 |
| dC4 | TTT CTT T | 73 ± 6 |
| dC5 | TTT TCT T | 154 ± 26 |
| dC6 | TTT TTC T | 87 ± 14 |
| dC7 | TTT TTT C | 59 ± 4 |
All experiments were performed at 293 K in 20 mM Tris/HCl, pH 7.5, 50 mM KCl and 40 µM N-acetyltryptophanamide.
Next, the oligonucleotide sequence was altered progressively using heptathymidine (dT7) as a reference. The binding analysis showed that XtrLin28b CSD has the highest affinity toward oligonucleotides containing a G at position 1 (dG1; Table 2). This nucleotide exchange led to a 5-fold decrease in the KD. Furthermore, at positions 2 and 5, a T is preferred over A or C (dT7 versus dA2/5 and dC2/5). On the other hand, mutations at positions 3, 4, 6 and 7 caused only subtle changes in the binding affinities with KD values ranging from 39 to 147 nM, indicating that XtrLin28b CSD has only limited sequence specificity. However, as purine heptanucleotides did not bind to XtrLin28b CSD, the protein seems to have a slight preference for pyrimidine-rich ssDNA (dT7 versus dG7 and dA7).
To verify the KD values obtained earlier by an independent method, we performed ITC measurements with ssDNA and ssRNA oligonucleotides. As observed before, XtrLin28b CSD displayed limited sequence specificity except for a preferential binding of G at position 1 and T at position 2 (dG1, dG1b; Supplementary Table S2). On the RNA level, binding of the oligouridines rU6 and rU7 was significantly impaired with an associated 10 - to 15-fold increase in KD with respect to dT6 and dT7 (Table 3 and Supplementary Table S3). A U-to-G exchange at position 1, however, led to similar KD values as for the corresponding DNA oligonucleotide (rG1; Table 3, versus dG1, Supplementary Table S2), showing that the differences between rU6/rU7 and dT6/dT7 can be traced back to this particular binding subsite. Interestingly, a heptameric ssRNA sequence derived from a single-stranded part of the Xtr-pre-let-7f preE showed the highest affinity with a KD of 13 nM (rlet-7f1; Table 3 and Supplementary Figure S3A). This short and rather pyrimidine-rich sequence motif contains the relevant G at position 1 and U at positions 2 and 5. Upon mutation of these nucleotides against A, the binding of XtrLin28b CSD is dramatically impaired (rlet-7f2,3).
Table 3.
Equilibrium dissociation constants of the binding of XtrLin28b CSD and XtrLin28b CSD variants to ssRNA oligonucleotides derived from ITC experimentsa
| CSD | ||
|---|---|---|
| Ligand | Sequence | KD (nM) |
| rU6 | UUU UUU | 2760 ± 790 |
| rU7 | U UUU UUU | 530 ± 65 |
| rG1 | G UUU UUU | 35 ± 2 |
| rG1b | G UCU UUU | 27 ± 3 |
| rGA6 | G AAA AAA | n.d.b |
| rlet-7f | G UCA CAC | 46 ± 3 |
| rlet-7f1 | G UCA UAC | 13 ± 2 |
| rlet-7f2 | G UCA AAC | 54 ± 9 |
| rlet-7f3 | G ACA AAC | 1360 ± 600 |
| rlet-7f4 | GG UCA CAC | 66 ± 9 |
| rlet-7f5 | G UCA CAC C | 18 ± 2 |
| rlet-7f6 | GG UCA CAC C | 21 ± 4 |
| CSD variants | ||
|---|---|---|
| Mutation | Sequence | KD (nM) |
| W39A | G UCA CAC C | 1130 ± 110 |
| F48A | G UCA CAC C | 1610 ± 340 |
| F66A | G UCA CAC C | 1440 ± 200 |
| H68A | G UCA CAC C | 540 ± 50 |
| F77A | G UCA CAC C | 1180 ± 60 |
Additionally calculated thermodynamic parameters are listed in the supporting information (Supplementary Table S3).
aAll experiments were performed at 288 K in 20 mM Tris/HCl, pH 8.0 and 60 mM KCl.
bNot determined due to insufficient affinity.
Next, we analyzed the optimal RNA strand length for CSD binding using the natural preE-let-7f as a template. While an additional G at the 5′-end slightly impaired binding, an extra C at the 3′-end (position 8) increased the binding affinity considerably (rlet-7f, rlet-74 and rlet-75). Adding both nucleotides led to a similar KD as seen for rlet-75 (Table 3 and Supplementary Figure S3B).
Taken together, XtrLin28b CSD preferentially binds single-stranded pyrimidine-rich oligonucleotides with up to eight bases. However, it only has moderate sequence specificity, as sequence-specific binding is only mediated at positions 1, 2 and, partially, 5.
The structure of the Lin28 CSD reveals a preformed nucleic acid-binding platform
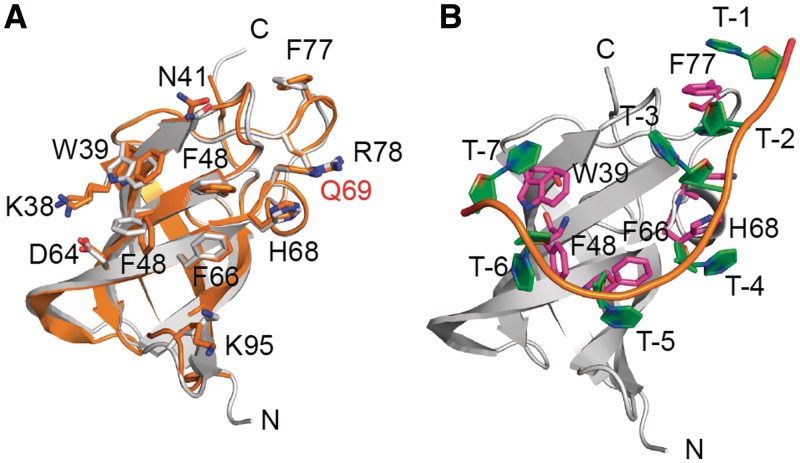
To investigate the structural basis for the observed sequence preferences, we crystallized human hLin28b CSD in the apo and XtrLin28b CSD in the apo and in nucleotide-bound forms. First, we succeeded in determining the crystal structure of apo XtrLin28b CSD and hlin28b CSD at 1.06 and 1.95 Å resolution, respectively (Supplementary Tables S4 and S5). Both structures are remarkably similar with an RMSD of the backbone Cα atoms of 0.2 Å (Figure 2A). The monomeric Lin28b CSDs reveal the CSD-typical oligosaccharide/oligonucleotide-binding fold (52–56), which consists of a β-barrel composed of five anti-parallel β-strands (Supplementary Figure S4). In general, the overall architecture is quite similar to those of other CSDs; however, both Lin28b CSDs show a higher structural similarity to bacterial Csps than to eukaryotic CSD-containing proteins.
Figure 2.
Comparison between apo Lin28 CSDs and oligonucleotide-bound Lin28 CSD. (A) Superimposition of apo XtrLin28b CSD (gray) and hLin28b CSD (orange). Highly conserved residues that are involved in nucleic acid binding are indicated in stick representation and marked according to their position within the XtrLin28b CSD. Gln69, which corresponds to a highly conserved Phe in bacterial CSPs, is indicated in red. (B) Cartoon representation of dT7 bound to XtrLin28b CSD. The aromatic side chains that are involved in direct stacking interactions with the dT7 bases are shown in stick representation.
Consistent with this, the globular units of both Lin28b CSDs resemble their bacterial homologs in having a strong amphipathic character. While one side of the protein shows a rather negative surface potential, the opposite surface forms a hydrophobic platform interspersed with highly conserved exposed aromatic residues that are surrounded by basic and polar residues (Figure 2A, Supplementary Figure S4A and B). The latter platform can associate with single-stranded nucleic acids and thus accommodates the majority of the highly conserved residues (Supplementary Figure S5). Most of these residues are known to interact with the nucleic acid ligand via hydrophobic and hydrogen bond contacts and are therefore highly conserved both in sequence and structure (35,55).
Compared with their bacterial homologs, Lin28b CSDs display three structural differences (Supplementary Figure S5). First, β-strand 2 and the following loop are extended by eight amino acid insertion. Second, two surface-exposed Lys within β-strand 4 create a rather basic surface next to the hydrophobic platform. Finally, a conserved Phe that was shown to be involved in base-stacking interactions with nucleic acid bases in bacterial Csps (34,37,57) is replaced by Gln (Q69; Figure 2A).
The binding of ssDNA and ssRNA to the Lin28b CSD is dominated by base-stacking interactions
We next co-crystallized XtrLin28b CSD in complex with hexathymidine (dT6) or heptathymidine (dT7), respectively (Supplementary Table S4). Both dT6 and dT7 bind in an identical curved conformation to the hydrophobic platform of XtrLin28b CSD (Supplementary Figure S6A). The bases point to the protein surface and interact mainly via stacking with the surface exposed Phe77, Phe66, Phe48 and Trp39 as well as His68 (Figure 2B). A substitution of these individual side chains by Ala resulted in a dramatic decrease in binding affinity with respect to the preE-derived rlet-f5 oligonucleotide (see Table 3 and Supplementary Figure S3C). Moreover, a replacement of the aromatic residues impaired binding more strongly than the His68Ala mutation. This underlines the dominance of base-stacking interactions in the RNA:protein contacts. The observed binding site is already preformed in the apo protein, and therefore only subtle changes are recognizable upon nucleic acid binding. From the four conserved aromatic residues that are directly involved in base-stacking interactions, only the side chain of Phe48 performs a conformational change upon nucleotide binding (see Supplementary Figure S4D).
In comparison to the dT6-bound structure, the dT7:XtrLin28b CSD complex shows an additional nucleotide (T-1) at the 5′-end that stacks on Phe77 (Supplementary Figure S6). Interestingly, nucleoside binding to this site was not observed in previous crystal structures of CSDs with bound oligonucleotides, probably due to the lack of specific hydrogen-bonded contacts with the nucleoside. However, the formation of a three-membered stack between T-1, Phe77 and T-2 (Figure 3A) is consistent with the observed increase in binding affinity from 6- to 7-mers. This continuous stack also explains the favored G at binding subsite 1, as the purine base dramatically improves the stacking interactions with Phe77 and T-2 as judged by modeling. In the case of a G, the base is likely further recognized by specific hydrogen bonds, as an A at position 1 impaired binding.
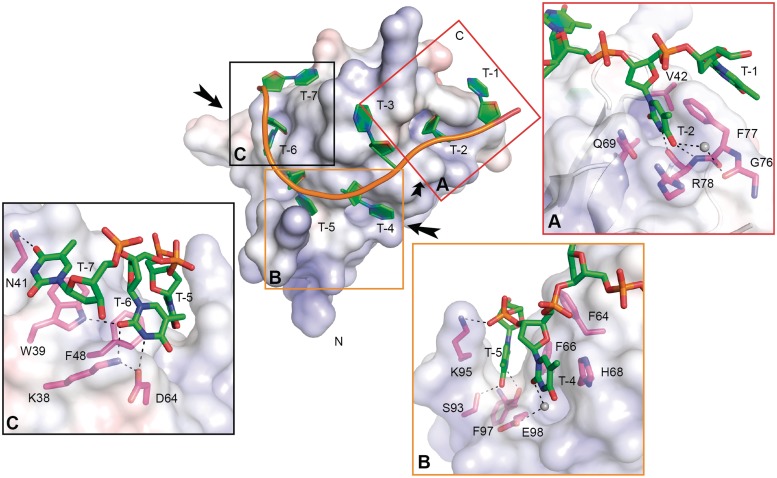
Figure 3.
Molecular interactions that promote binding of dT7 to XtrLin28b CSD. The electrostatic surface potential of XtrLin28b CSD upon binding to dT7 was calculated with APBS for pH 7 with a range from −10 (red) to +10 kT (blue). The bound oligonucleotide is shown in cartoon representation. Water molecules are shown as gray spheres. (A) Detailed view of T-2 bound to the hydrophobic pocket at binding subsite 2. Besides hydrophobic contacts including a three-membered stack with Phe77 and T-1, T/U-specific contacts are mediated via the backbone atoms of Arg78 and Phe77. (B) Detailed view of T-5 bound in the hydrophobic cleft created by Lys95 and Phe66. Ser93 and Phe97 form hydrogen bonds with O4 and HN3 of T-5. Binding of T-4 is only mediated via stacking interactions with His68 and a water molecule-mediated hydrogen bond. (C) Detailed view of binding subsites 6 and 7. T-6 and T-7 are mainly bound via stacking interactions with Phe48 and Trp39, respectively.
T-2 is bound in a hydrophobic pocket mainly formed by Phe77, Gln69, Val42, Phe40 and the main chain of Arg78. Apart from these hydrophobic interactions, the T-2 base is specifically bound by three hydrogen bonds to the backbone of Arg78 and, via a bridging water molecule, to the backbone of Gly76 (Figure 3A). A similar sequence-specific interaction mode was reported previously for bacterial Csps (34) and thus seems to explain the conserved preferential binding of U/T at this site. Further base-specific contacts are made by the backbones of Phe97 and Ser93 through groups of T-5 that are normally involved in Watson–Crick TA base pairing (Figure 3B and C). At binding subsite 6, Lys38 and Trp39 contact O2 of T-6, while Asp64 is connected to HN3 of the same base and in addition forms a salt bridge with Lys38 (Figure 3C). While T-4 and T-7 are mainly bound through stacking interactions with the bases, T-3 is not specifically bound but contacts another protein molecule and thus occupies binding subsite 2 within another monomer of the asymmetric unit (Supplementary Figure S7).
To examine whether ssRNA binds in the same way as ssDNA, we co-crystallized XtrLin28b CSD in the presence of various small oligoribonucleotides and finally succeeded in obtaining the structure of XtrLin28b CSD bound to rU6 (Supplementary Table S4). Interestingly, the binding mode is different from that observed in the ssDNA-bound complexes as only one ssRNA molecule binds to four different protein molecules, two belonging to the same asymmetric unit (Supplementary Figure S8). The uridine is bound in the T/U-specific hydrophobic pocket mainly formed by Phe77, Gln69 and Val42. However, the uracil base adopts a different orientation and is bound through hydrogen bonds to O2 and HN3 via the backbone of Arg78. Probably, the unique binding geometry observed in this structure is a crystallization artefact, because a 4:1 stoichiometry between RNA and Lin28 was not observed in any of the solution binding experiments, which are completely in agreement with the binding mode observed in dT7-bound XtrLin28b CSD.
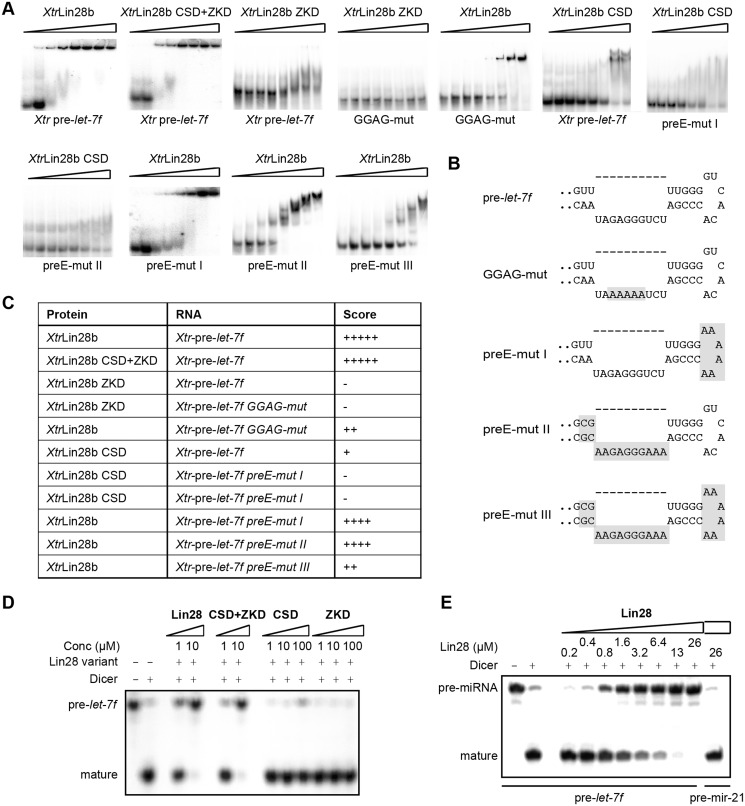
Determinants of Lin28:pre-let-7 interactions
To understand how the observed binding preferences of the individual Lin28 RBDs result in a specific recognition of the natural substrate, we performed EMSAs and Dicer in vitro processing assays with Xtr-pre-let-7f. First, we tested if other parts than the Lin28b RBDs contribute to binding of pre-let-7f. Therefore, we created a variant lacking the C-terminal 27 and the N-terminal 78 residues of the protein (XtrLin28b CSD+ZKD; see Figure 1). This variant shows a similar binding affinity as the full-length protein with a KD of 1.6 ± 0.1 µM for each (Figure 4A and C). Moreover, binding of both proteins is highly cooperative, as the binding data could best be fitted to a Hill equation with a Hill coefficient higher than 2 (see Supplementary Figure S10). Unlike the isolated domains, XtrLin28b CSD+ZKD is able to inhibit the processing of pre-let-7 by Dicer to a similar extent (Figure 4D) as the full-length protein. Moreover, the degree of inhibition strongly depends on the Lin28 concentration and thus reflects the competitive nature between Dicer and Lin28 with respect to the accessibility to the Dicer cleavage site within Xtr-pre-let-7 (Figure 4E).
Figure 4.
Determinants of Lin28:pre-let-7f interactions. (A) EMSAs with Xtr-pre-let-7f as a probe, mixed with increasing concentrations of protein (for XtrLin28b and XtrLin28b CSD+ZKD: 0, 0.4, 0.8, 1.6, 3.2, 6.4, 13, 26 µM; for XtrLin28b CSD and ZKD: 0, 0.8, 1.6, 3.2, 6.4, 13, 26, 51 µM). All EMSAs were performed using 1 nM α-32P-ATP-labeled RNA. (B) The terminal loop sequence and secondary structure of the Xtr-pre-let-7f mutants as predicted by Mfold (49). Regions that were mutated are shaded. (C) Results from EMSA experiments shown in (A). The binding affinity was scored according to the following dissociation constant ranges: +++++, 0.8–1.6 µM; ++++, 1.6–3.2 µM; +++, 3.2–6.4 µM; ++, 6.4–12.8 µM; +, 12.8–25.6 µM; −, >25.6 µM. (D and E) In vitro pre-miRNA processing reaction on 32P 5′-end-labeled Xtr-pre-let-7f. The indicated concentrations (in micromolar) of XtrLin28b, XtrLin28b RBDs, XtrLin28b CSD and XtrLin28b ZKD were added to 1 nM Xtr-pre-let-7f in the presence or absence of human Dicer. The samples were resolved on a 10% (w/v) denaturing PAGE and visualized by autoradiography.
Next, we checked the binding of the isolated ZKD to Xtr-pre-let-7f. Surprisingly, hardly any binding is observable for this variant, even though, according to the lowest energy structure by Mfold (49), the GGAG motif should be single stranded (Figure 4B). Thus, the data imply that the bases of the conserved GGAG motif are not freely accessible for the ZKD, since the presence of the GGAG motif is essential for a proper binding of the full-length protein (Figure 4A).
The isolated XtrLin28b CSD, on the other hand, binds to Xtr-pre-let-7f with about nine times lower affinity compared to the full-length protein. Moreover, up to two differently migrating complexes are visible, indicating that the CSD can bind to more than one site. Considering the binding preference of XtrLin28 CSD for single-stranded pyrimidine-rich sequences, we mutated two potential CSD-binding sites within preE of Xtr-pre-let-7f: one in the terminal hairpin loop, harboring the rlet-7f5 sequence (preE-mut I), and one in a bulge close to the upper stem region and adjacent to the Dicer cleavage site (preE-mut II). Interestingly, both mutations impair CSD binding, as hardly any complex band is visible in both cases. Consistent with this, these mutations cause a comparable loss of binding affinity in context of the wild-type (WT) protein, even though only a combination of both mutations (preE-mut III) has a similar effect as the GGAG-mut. Nam et al. (32) showed that the CSD domain of mLin28 indeed binds to a sequence corresponding to preE-mut I. However, as preE-mut II also had a considerable influence on binding, we suggest that additional elements within preE have an influence on the binding reaction of both the CSD and full-length Lin28.
The Lin28 CSD remodels the terminal loop and the upper stem of pre-let-7 while the ZKD ensures a stable binding and thus efficient blocking of pre-let-7 processing
The observation that the isolated CSD, but not the ZKD, binds to Xtr-pre-let-7f with a reasonable affinity suggests a multistep binding reaction in which the CSD first binds and imposes a structural change within preE and thereby facilitates a subsequent binding of the ZKD. To confirm this hypothesis, we performed an RNA remodeling assay using a truncated Xtr-pre-let-7g* that contains the entire terminal loop and five bases of the double-stranded upper stem region. The blunt end of Xtr-pre-let-7g* was modified with a fluorescence quencher pair to monitor a potential RNA unwinding of the double-stranded upper stem region upon protein binding (Figure 5A). Both full-length XtrLin28 isoforms and XtrLin28b CSD show a clear increase in fluorescence and thus an RNA remodeling activity, while for the ZKD only a slight increase in fluorescence is observable (Figure 5B). The increase in fluorescence is again cooperative for WT proteins and the isolated CSD and reflects their different binding affinities to Xtr-pre-let-7g (Figure 5B, D and E).
The ability of the isolated Lin28 CSD to remodel RNA implies that mutations in the CSD that impair the observed RNA interactions surface should lead to a stronger loss of remodeling activity than mutations in the Lin28 ZKD. Indeed, this is the case for all examined CSD mutations even though the KD values of the analyzed variants with respect to Xtr-pre-let-7g vary only slightly (Figure 5C–E). Upon closer inspection, the Lin28 mutations can be clustered into three classes. First, the ZKD variants XtrLin28b Tyr133Ala and His155Ala have a similar remodeling activity as the WT protein at low concentrations but only reach up to 70% of the WT’s activity at saturation. Second, variations in the CSD affecting binding subsites 5–7 (XtrLin28b Phe66Ala, Phe48Ala and Trp39Ala) still show modest remodeling activity that almost reaches the level of the ZKD variants at high concentrations. Finally, XtrLin28b His68Ala (binding subsite 4), Phe77Ala and the double variant XtrLin28b Phe77Ala/Arg78Gly (binding subsites 1 and 2) show hardly any remodeling activity (Figure 5C). This observation is highly interesting, as in the corresponding EMSAs all analyzed variants still shifted Xtr-pre-let-7g completely at high concentrations (Figure 5D) even though the CSD-binding site within pre-let-7g (32) is inaccessible according to the predicted secondary structure (Figure 5A). Consequently, XtrLin28b Phe77Ala and His68Ala might bind in an alternative mode to Xtr-pre-let-7g. Nevertheless, all variants are impaired in binding to Xtr-pre-let-7g and consequently lead to an increased pre-let-7 processing by Dicer (Figure 5D–F).
To gain further insights into the observed remodeling reaction, we measured the kinetics of remodeling by stopped-flow mixing experiments performed with the WT protein, Lin28 variants of each class and the isolated CSD under saturation conditions. Mixing of WT with Xtr-pre-let-7g* resulted in an association reaction with two phases (Figure 5G and Supplementary Figure S11). A major fast reaction (85% of total amplitude; time constant of the reaction τ1 = 20 ms) is followed by a minor slow phase (τ2 = 210 ms). The time course of the XtrLin28b Tyr133Ala remodeling activity could be fitted to either a monoexponential (τ = 28 ms) or a biexponential equation (τ1 = 26 ms, τ2 = 510 ms) with a very weak second time constant making up only 3.7% of the total amplitude. This indicates that the second rate observed in the WT can be traced back to the binding of the ZKD. Considering the flexible linker between both RBDs and the structural difference between apo Lin28 ZKD and RNA-bound Lin28 ZKD (32,33), the slow second time constant may reflect the necessary structural rearrangement in these regions to facilitate binding. The isolated CSD shows a one-phase association reaction with a similar time constant as observed for XtrLin28b Tyr133Ala (τ1 = 29 ms), even though the amplitude is much lower. For XtrLin28b Trp39Ala, after a fast association reaction (τ1 = 15 ms) that is completed in 80 ms, a slow dissociation reaction is followed (τ2 = 110 ms). The decrease of fluorescence might reflect the impaired binding of this construct and thus a higher dissociation rate compared to WT protein, as the fast remodeling rate is not impaired. For XtrLin28b Phe77Ala also the fast rate was considerably impaired, indicating that the corresponding residues are crucial for the remodeling reaction.
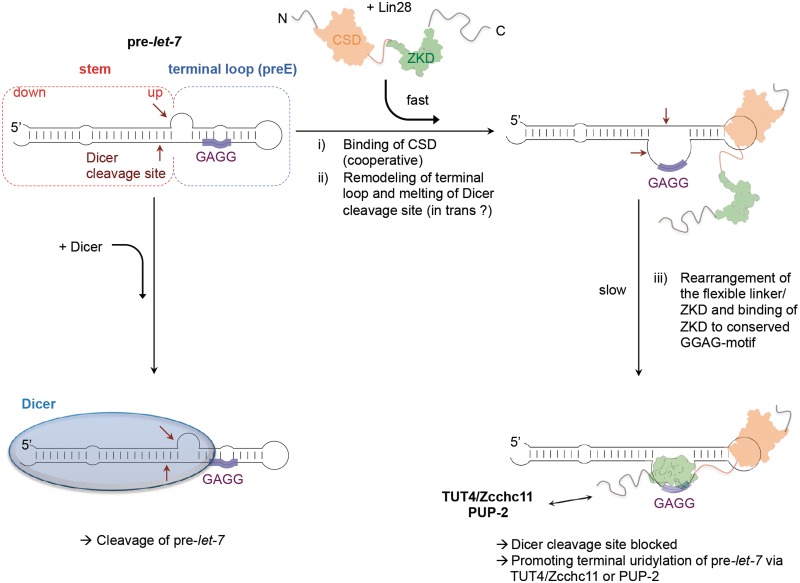
Taken together, the data clearly demonstrate that the XtrLin28b CSD imposes a structural change within preE of Xtr-pre-let-7g, which facilitates a subsequent binding of the ZKD. However, as the isolated CSD is unable to block the processing of Xtr-pre-let-7g alone, the ZKD probably serves as an anchor for directional binding and thus ensures a constant opening of the Dicer cleavage site (Figure 6).
Figure 6.
Schematic model of Lin28-mediated binding and inhibition of pre-let-7 maturation. In the absence of Lin28, the PAZ domain of Dicer recognizes the 2 nt 3′-overhang of pre-let-7 and cleaves the substrate about 22 nt from the end (Dicer cleavage site, indicated by dark red arrows). In the presence of Lin28, the CSD first binds with its preformed nucleic acid-binding platform to pyrimidine-containing single-stranded parts in the terminal loop (preE) of pre-let-7. In a fast reaction, the CSD then remodels the preE region and melts a part of the upper stem region including the Dicer cleavage site. As the binding of Lin28 to pre-let-7 is highly cooperative, the remodeling of pre-let-7 may be facilitated in trans by another Lin28 molecule. Once the conserved GGAG is freely accessible, the Lin28 ZKD mediates a specific binding to this motif and anchors Lin28 in this position. This second reaction is rather slow, as both the ZKD and the flexible basic linker between the domains have to perform larger conformational changes as judged from apo and nucleotide-bound structures. As a consequence, the Dicer cleavage site remains constantly open, and Lin28 inhibits the cleavage by Dicer. The sequence-specific interaction via the ZKD thus ensures a directional positioning of Lin28 to pre-let-7, which may allow promoting the terminal uridylation of pre-let-7 by TUT4/Zcchc11 or PUP-2.
DISCUSSION
We found that the Lin28 CSDs bind with high affinity to single-stranded nucleic acids via a conserved nucleic acid-binding platform mainly formed of exposed side chains that are surrounded by basic surface residues. This binding platform is already preformed in the apo protein, and consequently only subtle changes are observed upon nucleic acid binding. The crystal structure of the XtrLin28b CSD bound to dT7 reveals the binding of up to seven nucleotides. This observation is in good agreement with the binding data, as the CSD showed the highest affinity for 7- to 9-mers. Such binding preferences have also been described for bacterial Csps (34–37,58–60) and recently also for mLin28a in complex with more complete preE structures (32). The presence of a closed RNA loop as well as elements of the basic linker regions in these structures probably triggers the binding of an additional eighth and ninth nucleotide.
Regarding sequence specificity, our structural data reveal sequence-specific binding at positions 2 and 5 only, because at these binding sites only T/U-specific hydrogen bond contacts are mediated with the base. Moreover, at position 6, the presence of a salt bridge between Lys38 and Asp64, which is important for the formation of hydrogen bonds to T-6, limits the flexibility and consequently the size of this binding pocket. However, as hardly any contacts are mediated with the sugar–phosphate backbone, the DNA/RNA backbone is rather flexible and can adopt its conformation to bind a wide range of different RNA sequences. For example, protein-binding subsite 2 can also harbor a G as seen in the mLin28:preE-let-7f structure, as here the sugar–phosphate backbone moves away from the binding pocket and thereby enables the larger purine to bind into the pocket without disrupting the hydrogen bond network (Supplementary Figure S12). This flexibility is also valid for the other binding sites and reflects the dominance of stacking interactions in the binding interface.
Nevertheless, XtrLin28b CSD resembles its bacterial homologs in having a clear preference for pyrimidine-rich sequences even though the overall specificity is rather low. The lower affinity toward ssDNA oligonucleotides compared to bacterial Csps can be explained with the presence of Gln69 instead of a Phe that otherwise contributes to base-stacking interactions in bacterial Csps. Except for the polyuridines rU6 and rU7, no substantial difference in binding between ssRNA and ssDNA oligonucleotides was detected. In general, we always observe high binding affinities for oligonucleotides containing a G1UNNUNN7 or G1TNNTNN7 motif. This motif is slightly different from the recently proposed CSD-binding motif [N1GNGAYN7NN (32)] especially with respect to the corresponding protein-binding subsite. The discrepancy might be explained by the use of small oligonucleotides in our binding experiments which might shift in register. However, based on our structural and biochemical studies, a three-membered stack between the first base, Phe77 and the second base seems to be more favorable than the interaction at the eighth or ninth binding site observed in the mLin28:preE-let7 structures (32). Moreover, in the Xtrlin28b CSD:rU6 structure, U was bound at binding subsite 2, indicating that this particular binding subsite has a high affinity toward U. Therefore, it is likely that the oligonucleotides used for solution binding experiments in our study indeed bound as seen in the Xtrlin28b CSD:dT7 structure.
The broad spectrum of potential CSD-binding sequences probably reflects the low sequence conservation within its natural substrate, the preE of pre-let-7 (see Supplementary Figure S9). Consequently, sequence-specific binding of Lin28 is likely mediated through the ZKD. Structural studies with homologous ZKDs revealed a specific binding of a GGAG- or GGAG-like motif (50,51). Interestingly, this motif is highly conserved within preEs of pre-let-7. However, we could show that the association of Lin28 with pre-let-7 is also strongly dependent on the Lin28 CSD, as the isolated XtrLin28b ZKD could hardly bind to Xtr-pre-let-7f in vitro. Thus, a mutation of the GGAG motif to AAAA did not disrupt binding of WT XtrLin28b completely. Moreover, our biochemical analysis of XtrLin28b ZKD showed that it binds with high affinity to motifs containing two Gs, which are preferentially spaced by one nucleotide other than G. This reflects the observation that the GGAG motif is present in variable forms in pre-let-7 such as GGUG, GGCG, GGAG and UGXG. It seems likely that the ZKD specifically recognizes this variable motifs as well and thereby contributes to a stable binding of pre-let-7 and facilitates a subsequent TUT4-mediated uridylation of pre-let-7, as this process was shown to be dependent on the GGAG motif (31).
A recent study proposed that Lin28 induces a structural change within preE of pre-let-7g and thus directly prevents the processing of pre-let-7g by Dicer (61). Using enzymatic footprinting, the authors reported that upon Lin28 binding a part of the upper stem of pre-let-7 becomes more susceptible to cleavage by single-strand-specific ribonucleases. We now could demonstrate that the structural change within pre-let-7g is mediated through the action of the CSD, as only mutations within this domain impaired the remodeling activity significantly. The function of CSDs as protein domains that can induce local secondary structural changes within RNA was already proposed for bacterial Csps in the process of transcription anti-termination (62–64). In the case of E. coli CspE, a melting pathway was suggested, in which two conserved Phe and one His, resembling Phe46 and Phe66 (binding subsite 5) and His68 (binding subsite 4) in XtrLin28b CSD, intercalate between bases of the stem. Indeed, His68 and Phe77 (binding subsites 1 and 2) are essential for the remodeling activity of Lin28, and it is likely that a similar melting pathway exists for the Lin28 CSD.
Keeping in mind that most preE secondary structures of pre-let-7 differ considerably from the structure observed in the mLin28:preE complexes, the CSD-dependent remodeling seems to be essential for a directional binding as proposed by Nam et al. (32). According to Mfold, in 6 (pre-let-7 a-1, a-2, b, c, g, f-2) out of 11 pre-let-7 miRNAs, the GGAG motif is inaccessible. Moreover, often multiple secondary structures with almost the same energy exist. Based on our data, we propose a multi-step binding model according to which Lin28 CSD first binds a pyrimidine-rich single-stranded sequence within preE and induces a conformational change that facilitates a subsequent binding of the ZKD to the conserved GGAG motif. As the binding of XtrLin28b and the isolated XtrLin28b CSD to Xtr-pre-let-7f is highly cooperative [Supplementary Figure S10 (65)], it is possible that the remodeling is performed in trans by another Lin28 molecule. Moreover, the action of the Lin28 CSD also leads to a melting of the upper stem region including the Dicer cleavage site (32,61). The sequence-specific binding of the ZKD to the GGAG motif adjacent to this site may ensure a constant opening of the corresponding site and thereby directly inhibit the processing by Dicer. This would also explain why both intact RBDs are required for an efficient inhibition of pre-let-7 processing by Dicer even though the isolated CSD and most of the analyzed variants are able to bind and remodel pre-let-7.
Apart from inhibiting let-7 biogenesis, Lin28 also exhibits let-7-independent functions. Multiple studies showed that Lin28 binds to a variety of mRNAs and stimulates the translation of factors associated with cell growth and survival (66–70). This let-7-independent function of Lin28 is mediated via protein interactions. As an N- and C-terminally truncated construct of XtrLin28b, harboring the two RBDs only, was still able to bind to pre-let-7 with similar affinity as the full-length protein, we conclude that regions outside the RBDs have other functions than RNA binding and may be involved in protein–protein interactions.
ACCESSION NUMBERS
PDB 3ulj, 4a75, 4a76, 4alp and 4a4i.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 1–12.
FUNDING
Funding for open access charge: Max-Delbrück-Centrum für Molekulare Medizin Berlin-Buch.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tracy Dornblut, Janett Tischer, Ingrid Berger and Michaela Naschke for excellent technical assistance. We are also thankful to Yvette Roske, who assisted in diffraction data collection and processing, and acknowledge Uwe Müller and the beamline support by the staff of the Helmholtz-Zentrum Berlin für Materialien und Energie at BESSY. The Protein Sample Production Facility at the Max Delbrück Center is funded by the Helmholtz Association of German Research Centres. U.H., A.S. and F.M. conceived and coordinated the study. F.M., A.S. and N.D. performed the experiments. F.M. and A.S. analyzed the data. F.M. wrote the manuscript. All authors read and approved the manuscript.
REFERENCES
- 1.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 6.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev. Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 9.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 10.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 11.Büssing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Johnson S, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert K, Brown D, Slack F. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wulczyn F, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- 15.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl E, Nitsch R, Wulczyn F. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 18.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambros V, Horvitz H. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 20.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 21.Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- 22.Reinhart B, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz H, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 23.Richards M, Tan S-P, Tan J-H, Chan W-K, Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- 24.Yang DH, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr. Patterns. 2003;3:719–726. doi: 10.1016/s1567-133x(03)00140-6. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Lin X, Zhong X, Kaur S, Li N, Liang S, Lassus H, Wang L, Katsaros D, Montone K, et al. Double-negative feedback loop between reprogramming factor LIN28 and microRNA let-7 regulates aldehyde dehydrogenase 1-positive cancer stem cells. Cancer Res. 2010;70:9463–9472. doi: 10.1158/0008-5472.CAN-10-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, Vodyanik M, Smuga-Otto K, Antosiewicz-Bourget J, Frane J, Tian S, Nie J, Jonsdottir G, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 27.Viswanathan S, Powers J, Einhorn W, Hoshida Y, Ng T, Toffanin S, O’Sullivan M, Lu J, Phillips L, Lockhart V, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 29.Hagan J, Piskounova E, Gregory R. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heo I, Joo C, Kim Y-K, Ha M, Yoon M-J, Cho J, Yeom K-H, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loughlin FE, Gebert LF, Towbin H, Brunschweiger A, Hall J, Allain FH. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat. Struct. Mol. Biol. 2012;19:84–89. doi: 10.1038/nsmb.2202. [DOI] [PubMed] [Google Scholar]
- 34.Max KEA, Zeeb M, Bienert R, Balbach J, Heinemann U. T-rich DNA single strands bind to a preformed site on the bacterial cold shock protein Bs-CspB. J. Mol. Biol. 2006;360:702–714. doi: 10.1016/j.jmb.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 35.Max KEA, Zeeb M, Bienert R, Balbach J, Heinemann U. Common mode of DNA binding to cold shock domains. Crystal structure of hexathymidine bound to the domain-swapped form of a major cold shock protein from Bacillus caldolyticus. FEBS J. 2007;274:1265–1279. doi: 10.1111/j.1742-4658.2007.05672.x. [DOI] [PubMed] [Google Scholar]
- 36.Zeeb M, Balbach J. Single-stranded DNA binding of the cold-shock protein CspB from Bacillus subtilis: NMR mapping and mutational characterization. Protein Sci. 2003;12:112–123. doi: 10.1110/ps.0219703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeeb M, Max KEA, Weininger U, Löw C, Sticht H, Balbach J. Recognition of T-rich single-stranded DNA by the cold shock protein Bs-CspB in solution. Nucleic Acids Res. 2006;34:4561–4571. doi: 10.1093/nar/gkl376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheich C, Kümmel D, Soumailakakis D, Heinemann U, Büssow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35:e43. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinemann U, Büssow K, Mueller U, Umbach P. Facilities and methods for the high-throughput crystal structural analysis of human proteins. Acc. Chem. Res. 2003;36:157–163. doi: 10.1021/ar010129t. [DOI] [PubMed] [Google Scholar]
- 40.Leslie A. 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. No. 26. [Google Scholar]
- 41.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Perrakis A, Langer G, Cohen SX, Lamzin VS. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Ferrin TE, Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 47.Eftink MR. Fluorescence methods for studying equilibrium macromolecule-ligand interactions. Methods Enzymol. 1997;278:221–257. doi: 10.1016/s0076-6879(97)78013-3. [DOI] [PubMed] [Google Scholar]
- 48.Lohman TM, Bujalowski W. Thermodynamic methods for model-independent determination of equilibrium binding isotherms for protein-DNA interactions—spectroscopic approaches to monitor binding. Methods Enzymol. 1991;208:258–290. doi: 10.1016/0076-6879(91)08017-c. [DOI] [PubMed] [Google Scholar]
- 49.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 51.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 52.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Bläsi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 53.Schindelin H, Jiang W, Inouye M, Heinemann U. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1994;91:5119–5123. doi: 10.1073/pnas.91.11.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaikam V, Karlson DT. Comparison of structure, function and regulation of plant cold shock domain proteins to bacterial and animal cold shock domain proteins. BMB Rep. 2010;43:1–8. doi: 10.5483/bmbrep.2010.43.1.001. [DOI] [PubMed] [Google Scholar]
- 55.Horn G, Hofweber R, Kremer W, Kalbitzer HR. Structure and function of bacterial cold shock proteins. Cell. Mol. Life Sci. 2007;64:1457–1470. doi: 10.1007/s00018-007-6388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mihailovich M, Militti C, Gabaldón T, Gebauer F. Eukaryotic cold shock domain proteins: highly versatile regulators of gene expression. Bioessays. 2010;32:109–118. doi: 10.1002/bies.200900122. [DOI] [PubMed] [Google Scholar]
- 57.Max KEA, Wunderlich M, Roske Y, Schmid FX, Heinemann U. Optimized variants of the cold shock protein from in vitro selection: structural basis of their high thermostability. J. Mol. Biol. 2007;369:1087–1097. doi: 10.1016/j.jmb.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 58.Bienert R, Zeeb M, Dostál L, Feske A, Magg C, Max K, Welfle H, Balbach J, Heinemann U. Single-stranded DNA bound to bacterial cold-shock proteins: preliminary crystallographic and Raman analysis. Acta Crystallogr. D Biol. Crystallogr. 2004;60:755–757. doi: 10.1107/S0907444904002422. [DOI] [PubMed] [Google Scholar]
- 59.Lopez MM, Makhatadze GI. Major cold shock proteins, CspA from Escherichia coli and CspB from Bacillus subtilis, interact differently with single-stranded DNA templates. Biochim. Biophys. Acta. 2000;1479:196–202. doi: 10.1016/s0167-4838(00)00048-0. [DOI] [PubMed] [Google Scholar]
- 60.Lopez MM, Yutani K, Makhatadze GI. Interactions of the cold shock protein CspB from Bacillus subtilis with single-stranded DNA. Importance of the T base content and position within the template. J. Biol. Chem. 2001;276:15511–15518. doi: 10.1074/jbc.M010474200. [DOI] [PubMed] [Google Scholar]
- 61.Lightfoot HL, Bugaut A, Armisen J, Lehrbach NJ, Miska EA, Balasubramanian S. A LIN28-dependent structural change in pre-let-7g directly inhibits dicer processing. Biochemistry. 2011;50:7514–7521. doi: 10.1021/bi200851d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phadtare S, Inouye M, Severinov K. The mechanism of nucleic acid melting by a CspA family protein. J. Mol. Biol. 2004;337:147–155. doi: 10.1016/j.jmb.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 63.Phadtare S. Unwinding activity of cold shock proteins and RNA metabolism. RNA Biol. 2011;8:394–397. doi: 10.4161/rna.8.3.14823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phadtare S, Severinov K. Nucleic acid melting by Escherichia coli CspE. Nucleic Acids Res. 2005;33:5583–5590. doi: 10.1093/nar/gki859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Desjardins A, Yang A, Bouvette J, Omichinski JG, Legault P. Importance of the NCp7-like domain in the recognition of pre-let-7g by the pluripotency factor Lin28. Nucleic Acids Res. 2012;40:1767–1777. doi: 10.1093/nar/gkr808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balzer E, Heine C, Jiang Q, Lee V, Moss E. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137:891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- 67.Peng S, Chen L-L, Lei X-X, Yang L, Lin H, Carmichael GG, Huang Y. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29:496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- 68.Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–1248. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu B, Huang Y. Histone H2a mRNA interacts with Lin28 and contains a Lin28-dependent posttranscriptional regulatory element. Nucleic Acids Res. 2009;37:4256–4263. doi: 10.1093/nar/gkp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.