Abstract
When mammalian cells experience radiation insult, DNA replication is stalled to prevent erroneous DNA synthesis. UV-irradiation triggers proteolysis of Mcm10, an essential human replication factor, inhibiting the ongoing replication. Here, we report that Mcm10 associates with E3 ubiquitin ligase comprising DNA damage-binding protein, DDB1, cullin, Cul4 and ring finger protein, Roc1. Depletion of DDB1, Roc1 or Cul4 abrogates the UV-triggered Mcm10 proteolysis, implying that Cul4–Roc1–DDB1 ubiquitin ligase mediates Mcm10 downregulation. The purified Cul4–Roc1–DDB1 complex ubiquitinates Mcm10 in vitro, proving that Mcm10 is its substrate. By screening the known DDB1 interacting proteins, we discovered that VprBP is the substrate recognition subunit that targets Mcm10 for degradation. Hence, these results establish that Cul4–DDB1–VprBP ubiquitin ligase mediates the stress-induced proteolysis of replication factor, Mcm10.
INTRODUCTION
Eukaryotic DNA replication initiates with the binding of a six-subunit origin recognition complex (ORC) to multiple sites on the chromosomes called the origins of replication in the M and G1 phases of the cell cycle. Thereafter, Cdc6 and Cdt1 are recruited to the ORC bound origins followed by loading of the replicative helicase, Mcm2-7 (mini chromosome maintenance) complex, assembling the pre-replicative complex (1). Recent work suggests that activation of pre-replicative complex occurs during the late G1 or early S phase when Dbf-dependent kinase (DDK) phosphorylates the Mcm2–7 helicase, promoting the stable recruitment of replication proteins, Sld3 and Cdc45. Activation of S-phase CDKs (Cyclin E/Cdk2 and Cyclin A/Cdk2) leads to phosphorylation of Sld3 promoting its binding to TopBP1 as well as recruitment of GINS complex and DNA polymerase epsilon to the origins of replication. The unwound single strands are stabilized by RPA (the single-stranded DNA binding protein), and the DNA polymerase alpha-primase is next loaded to the unwound origins. The first RNA-primed DNA fragments are synthesized by DNA polymerase alpha thereby beginning DNA synthesis.
The stage of replication initiation at which Mcm10 is recruited to the origins remains debatable and may vary between species (2–7). In Xenopus and humans, the chromatin-bound Mcm2-7 helicase complex is essential for the loading of Mcm10 (6,8). Though previous studies claim that Mcm10 associates with chromatin before DDK and S-CDK are activated, recent studies demonstrate that Mcm10 is recruited after the activation of S-CDKs (7,8). The essential role of Mcm10 in initiation and elongation of DNA replication has been exhibited across species (2–6). Mutations in Mcm10 lead to stalled replication and cell cycle arrest (5,9). It has been shown that Mcm10 physically interacts with and stabilizes DNA polymerase alpha and helps to maintain its association with chromatin (2,3). Controlled proteolysis of the Xenopus laevis Mcm10 revealed N-terminal-, internal (ID)- and C-terminal (CTD)- structured domains. DNA binding activity of Xenopus Mcm10 was mapped to the ID and CTD, both of which bind to single- and double-stranded DNA (10). The ID and CTD domains retain the ability to independently bind to the N-terminus of the p180 subunit of DNA polymerase alpha-primase. Electron microscopic analysis of human Mcm10 reveals a ring-shaped hexamer with large central and smaller lateral channels and a system of inner chambers suggesting a docking module for assembly of the molecular machinery required for eukaryotic DNA replication (11).
The activity of key cell cycle and replication proteins is regulated by ubiquitination, the specificity being dictated by E3 ubiquitin ligases (12). The largest family, ring-finger domain E3 ligases, comprises multimeric cullin-based E3 subfamily (CRL) and monomeric RING finger E3s (13). A prototype CRL consists of a core cullin protein, with a 150 amino acid cullin domain which mediates binding to the ring-finger protein (14,15). The ‘substrate recognition subunit’ which defines the target is linked to the cullin via an ‘adaptor protein’. In humans, there are more than seven different cullin proteins, which associate with distinct adaptors and substrate recognition subunits, thereby generating numerous functional E3 ligases each with a distinct specificity. The role of Cul1-based CRLs comprising Cul1, Rbx1 with Skp1 as an adaptor protein and Skp2 as substrate recognition subunit, has been well established in cell cycle progression (16,17). However, the role of Cul4-based E3 ligase was realized later after the discovery of its function in mediating the radiation triggered degradation of CDK inhibitor, p21 and replication initiator, Cdt1 (18–20). It was discovered that the unperturbed S phase degradation of p21 is carried out redundantly by two E3 ligases: SCFSKP2 as well as Cul4-dependent CRL4CDT2 (20,21). UV-triggered degradation of replication initiator, Cdt1, is dependent on Cul4, Cdt2, PCNA and DDB1, establishing the vitality of the CRL4 complex in stress response (22–26). DDB1 is known to accumulate at the site of DNA damage having a crucial role in nucleotide excision repair (27,28). Further, it was observed that the xeroderma pigmentosum (XP) group E cells lack DDB1 which causes a repair defect seen in these patients (29). The crystal structure of the DDB1 complex which detects UV lesions provides useful insights into the mode of DNA damage recognition (30). The association of DDB1 with Cul4–Roc1 E3 ligase is conserved from yeast to humans to regulate DNA repair and replication (31). We have previously reported that Mcm10, an essential human replication factor, is selectively proteolyzed after UV-irradiation to inactivate the replication machinery (32). In this study, we attempted to identify the ubiquitin ligase that mediates stress-induced Mcm10 proteolysis. We observed that Mcm10 downregulation is independent of the cell cycle regulatory E3 ligase, SCFSkp2, and Cdt2, the substrate recognition subunit essential for Cdt1 ubiquitination. Our study demonstrates that human cells utilize CRL4VprBP ubiquitin ligase to downregulate Mcm10 during stress.
MATERIALS AND METHODS
Cell culture, chemicals, antibodies, cell synchronization and FACS analysis
Cell lines were maintained in DMEM supplemented with fetal bovine serum and antibiotics. Specific chemicals and antibodies used in this study are mentioned in the supplementary data. To evaluate the effect of depletion of target gene on Mcm10 levels, HeLa and 293T cells were exposed to 200 J/m2 UV radiation and were harvested after 4 h. HeLa cells were transfected with specific siRNA oligos consecutively for three days and then blocked with 9 µM RO-3306 for 18 h. For cell cycle analysis, the cells were fixed with 70% ethanol after washing with 1× PBS. After fixing, the cell pellet was resuspended in 1× PBS with 0.1% Triton X-100, 20 µg/ml RNase and 70 µg/ml propidium iodide, and then stained cells were analyzed by flow cytometry. The flow cytometry data were acquired on Becton Dickinson FACS Calibur machine by Cell Quest Pro software. Cell cycle analysis was done by Dean/Jett/Fox method using the FlowJo software.
RNAi silencing and reverse-transcriptase PCR
The siRNA sequences are available upon request. Transfection of siRNA targeting endogenous genes was carried out using Lipofectamine 2000 (Invitrogen). We transfected 40–80 nM siRNA duplexes on three consecutive days, and the cells were harvested 24 h after the last transfection to evaluate the levels of protein and mRNA by immunoblotting and reverse-transcriptase PCR, respectively. For reverse-transcriptase PCR, RNA was extracted using the TRIzol method, and 0.25–1 µg RNA was used for cDNA synthesis. The primers used for PCR are described in Supplementary Data.
Immunoblotting, immunoprecipitation, ubiquitination and interaction studies
Cells of almost equal confluency were lysed in proportionate volumes of Laemmli buffer for immunoblotting. For immunoprecipitation, cell lysate was incubated with specific antibody along with protein A sepharose, and after washing, it was eluted with Laemmli buffer. For in vivo ubiquitination studies, 293T cells were transfected with HA-tagged DDB1, Cul4a and Roc1 (CRL4–DDB1), HA-tagged ubiquitin or myc-tagged Mcm10 followed by immunoprecipitation using myc antibody. To study in vitro ubiquitination of Mcm10, 293T cells were transfected with HA-tagged DDB1, Cul4, Roc1 or VprBP. Endogenous Mcm10 and HA-tagged proteins were immunoprecipitated separately, and the HA tagged proteins were eluted with HA peptide. Immunoprecipitated Mcm10 was incubated with the purified HA proteins in the presence of E1, E2 (UbcH5c), biotinylated-ubiquitin, ATP, MG132 for 1 h at 37°C. To identify the interacting domains of Mcm10 and VprBP, plasmids expressing Mcm10 and VprBP were co-transfected and immunoprecipitation was carried out. To test the binding between purified Mcm10 and VprBP, His-tagged Mcm10 (cloned in pET28a vector) was purified on nickel-NTA column (Qiagen) and eluted with 0.25 M imidazole. Escherichia coli expressing GST-tagged VprBP was lysed in binding buffer (50 mM sodium phosphate buffer, pH 7.4, 450 mM NaCl, 1% Triton X-100), and clarified supernatant was incubated with glutathione agarose column, pre-blocked with 5% BSA. After binding for 3–4 h at 25°C, beads were washed to remove the unbound protein. His-tagged Mcm10 was incubated with the GST-tagged protein bound to glutathione beads for 1.5 h at 25°C, and the co-immunoprecipitated protein was identified by immunoblotting with anti-Mcm10 antibody.
RESULTS
Requirement of factors for UV-triggered Mcm10 degradation
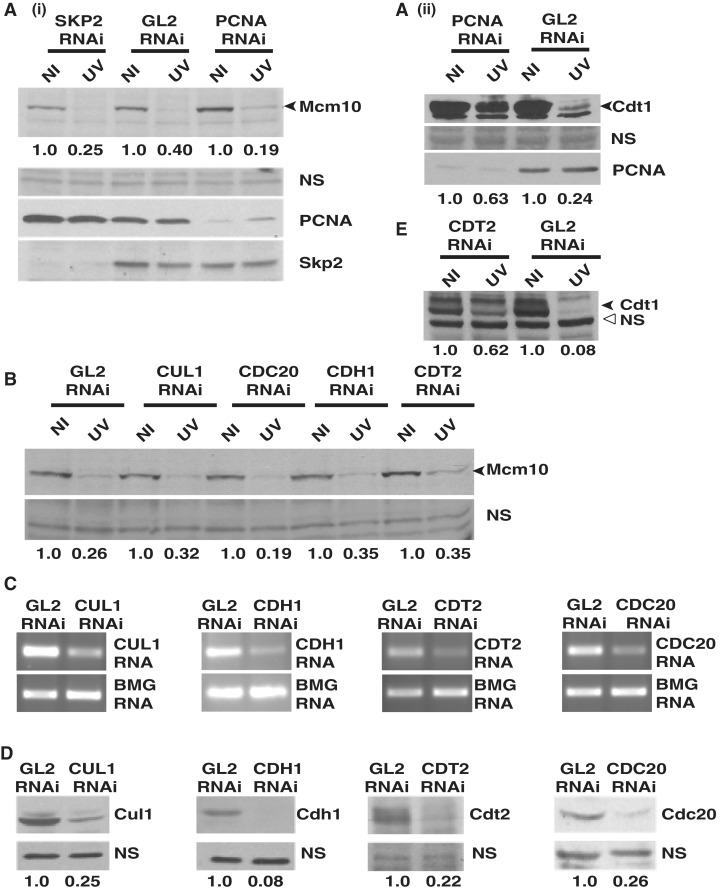
To identify the E3 ligase mediating the UV-triggered degradation of Mcm10, putative E3 ligase components were silenced by RNAi. HeLa cells were transfected with specific siRNA duplexes on three consecutive days, exposed to UV 24 h after the last transfection and harvested 4 h later. Protein bands were quantitated, and the levels of Mcm10 were expressed after normalizing with a non-specific band. We tested the requirement of the substrate-recognition subunits: PCNA and Skp2, which are essential for Cdt1 degradation. In PCNA and SKP2 siRNA-transfected cells, the level of Mcm10 was reduced to 19% and 25%, respectively, after UV-irradiation implying that silencing of these genes do not alter the UV-triggered Mcm10 downregulation (Figure 1A). Silencing of PCNA effectively inhibited its function as evidenced by reduction of UV-triggered Cdt1 proteolysis (Figure 1A, part ii). Skp, Cullin, F-box E3 ubiquitin ligase carries out the timely ubiquitination and subsequent degradation of many key cell cycle regulators, thereby mediating the progression from one phase of the cell cycle to the next. RNAi-targeted silencing of CUL1 reduced its protein to 25% of control levels, but it did not block Mcm10 degradation (Figure 1B–D). Cdt2 functions as a substrate recognition subunit mediating the stress-induced degradation of the replication protein, Cdt1. Though CDT2 silencing by RNAi reduced its protein to 22% of control levels, the proteolysis of Mcm10 remained unaltered (Figure 1B). Silencing of CDT2 alleviated the UV-triggered proteolysis of Cdt1, demonstrating that Cdt2 function was effectively debilitated (Figure 1E). Cdc20, Cdh1, FBXW7 and beta-TRCP function as substrate recognition subunits in mediating ubiquitination of various substrates. Depletion debilitated the function of these E3 ligases as indicated by the decrease of specific mRNA and protein levels as well as stabilization of substrates such as cyclin E (Figure 1C and D, Supplementary Figure S1B–D). However, UV-triggered Mcm10 degradation was not reduced, implying that Mcm10 degradation is independent of these substrate recognition subunits (Figure 1B, Supplementary Figure S1A).
Figure 1.
Mcm10 degradation post-UV-irradiation is independent of PCNA, Skp2, Cul1, Cdc20, Cdh1 and Cdt2. (A) HeLa cells were transfected on three consecutive days with PCNA, SKP2 or control GL2 siRNA, exposed to UV 24 h after the last transfection and harvested 4 h later. The levels of Mcm10 (black arrowhead) and a non-specific band (NS) were quantitated in non-irradiated (NI) and UV-irradiated (UV) cells. Mcm10 levels were expressed after normalizing with the non-specific band to correct for loading differences between different lanes. The numbers in panel (A) indicate the levels of Mcm10 protein in UV-irradiated cells relative to non-irradiated cells after specific siRNA transfections. Each RNAi depletion experiment has been repeated multiple times and the level of Mcm10 protein was quantitated using the Quantity One Software (version 4. 6. 3; Bio-Rad). (B) HeLa cells were transfected with CUL1, CDC20, CDH1, CDT2 or control GL2 siRNA, and as described in part (A) the level of Mcm10 protein was evaluated. The decrease in the level of proteins was confirmed by immunoblotting, whereas the decrease of mRNA was confirmed by RT-PCR. The levels of protein (D) of the genes silenced by RNAi in part (B) were determined and the numbers indicate the levels following specific siRNA depletion relative to control GL2-transfected cells. (E) Suppression of UV-triggered Cdt1 degradation demonstrates effective silencing of CDT2. The numbers indicate the levels of Cdt1 in UV-irradiated cells relative to non-irradiated cells. β-2 microglobulin (BMG) served as the internal RNA loading control.
E3 ligase constituting of Roc1–Cul4–DDB1 mediates the degradation of Mcm10
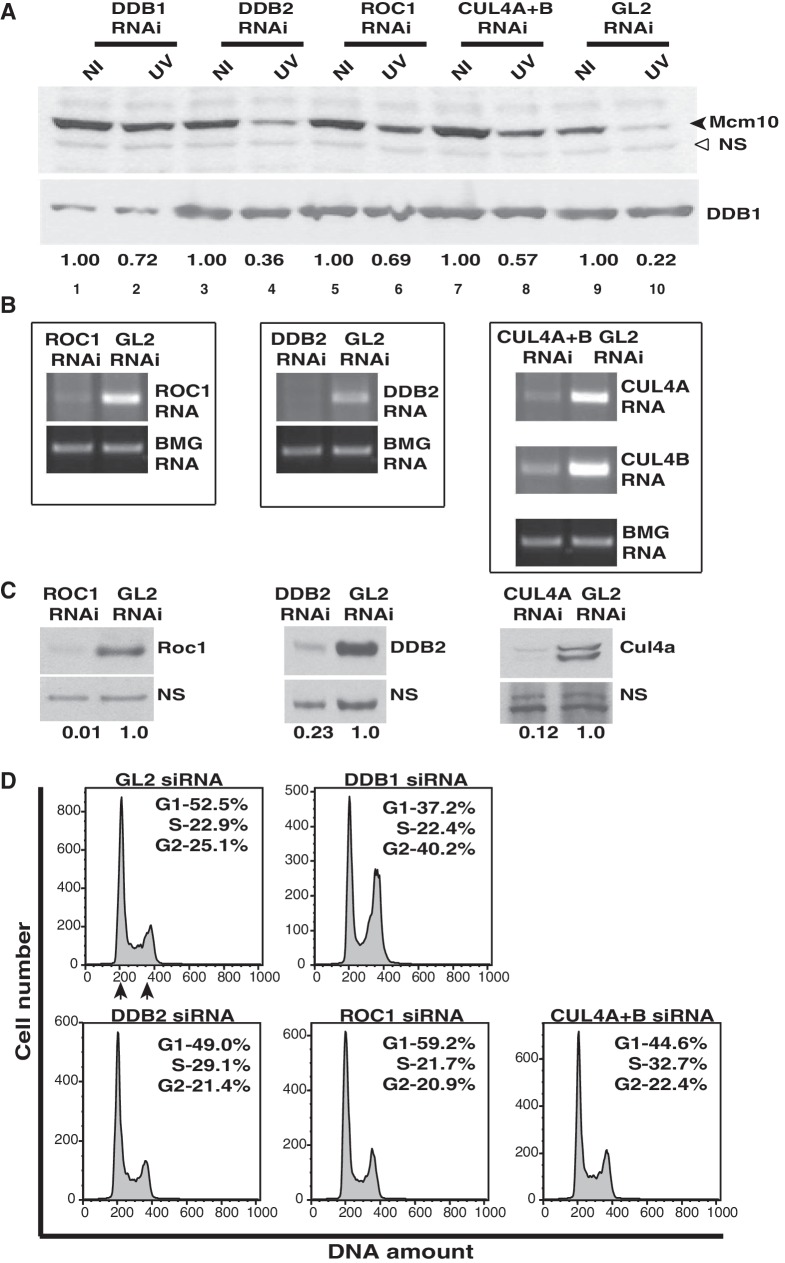
It is known that DDB2 functions as the substrate receptor for the DDB1–CUL4A-based E3 ligase, which ubiquitinates histone H2A in response to UV-irradiation (34). The depletion of DDB2 leads to a minor reduction in the UV-triggered degradation of Mcm10, possibly by an indirect effect on the stability of Mcm10 (Figure 2A and B). The siRNA duplexes designed against DDB1, ROC1 or CUL4A+B significantly reduced the respective mRNA and protein levels (Figure 2A–C). We observed that the Mcm10 protein was decreased to 22% following UV-irradiation. After DDB1 RNAi, Mcm10 decrease post-UV-irradiation was limited to 72%. This demonstrates that the silencing of DDB1 significantly reduces the UV-triggered degradation of Mcm10 (Figure 2A, top panel, compare Lane 2 with Lane 10). We also observed that after silencing the ring-finger protein, ROC1 or CUL4A+B, the Mcm10 protein was decreased to 69% and 57% of non-irradiated levels, respectively (Figure 2A, compare Lanes 6 and 8 with Lane 10). Therefore, Roc1, Cul4 and DDB1 are required for the UV-triggered degradation of Mcm10.
Figure 2.
Mcm10 degradation post-UV-irradiation is mediated by Roc1–Cul4–DDB1 E3 ubiquitin ligase. (A) HeLa cells were transfected on three consecutive days with indicated siRNAs, and 24 h after the last transfection, the cells were exposed to UV, harvested 4 h later and Mcm10 levels were analyzed in non-irradiated (NI) and UV-irradiated (UV) cells. The black and hollow arrowheads point to Mcm10 and non-specific band, respectively. The decrease in the level of DDB1, Roc1, DDB2 and Cul4a proteins was confirmed by immunoblotting (A and C), whereas the decrease of ROC1, DDB2, CUL4A and CUL4B mRNA was confirmed by RT-PCR (B). The levels of protein (C) of the siRNA-targeted genes were determined, and the numbers indicate the levels following specific siRNA depletion relative to control GL2-transfected cells. (D) Cell cycle distribution was determined by flow cytometry of propidium iodide-stained DNA of HeLa cells, as described in (A). The inset shows the percentage of total cells that are present in different phases, and G1 and G2 peaks have been marked by arrows.
Since Mcm10 levels naturally oscillate during the cell cycle, we wanted to confirm if the stabilization of Mcm10 is due to the silencing of specific genes or due to changes in the cell cycle as a result of siRNA depletion of essential proteins (35). Therefore, we simultaneously evaluated the cell cycle distribution after siRNA depletion of each target gene. It is important to note that the genes depleted by RNAi are essential for cell cycle progression, and therefore some variations in cell cycle distribution were observed (Figure 2D). Mcm10 is present in S and G2 phases but is absent from G1 phase, and therefore we determined the ratio of cells in the Mcm10 positive phase (S + G2) to Mcm10 negative phase (G1) after individual siRNA depletions. We hypothesized that if this ratio does not change significantly, then the variations in cell cycle are unlikely to contribute to the changes in Mcm10 levels. We observed that the population of cells present in the Mcm10 positive phase after siRNA transfection of GL2 (control), DDB1, DDB2, ROC1 and CUL4A+B were 48, 63, 51, 43 and 55%, respectively (Figure 2D). Therefore, the fraction of cells present in the Mcm10 positive phase did not vary significantly except after DDB1 silencing where the Mcm10 positive phase increased from 48% to 63%. However, a 15% increase in Mcm10 positive phase cells is unlikely to cause a significant stabilization of Mcm10 after UV-irradiation (compare Lane 2 with Lane 10 of Figure 2A). Since UV triggers Mcm10 proteolysis throughout the cell cycle, accumulation within a particular phase will not affect the UV-mediated downregulation (32).
Mcm10 associates with DDB1, Cul4 and Roc1
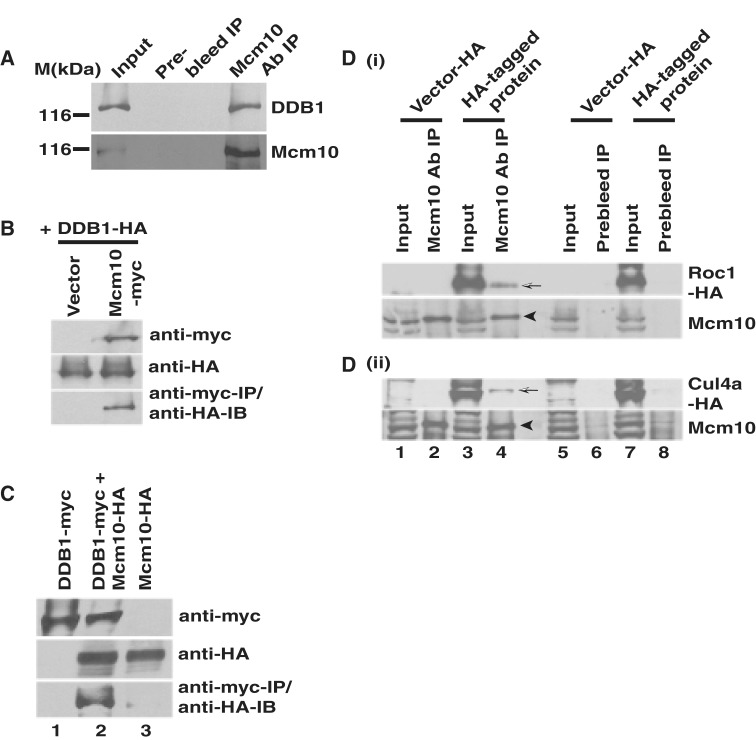
We previously reported that upon UV irradiation, human Mcm10 protein is degraded by the 26S proteasome, and therefore our study revealed a mechanism by which mammalian cells effectively inhibit the replication machinery during stress (32). To identify the E3 ligase that ubiquitinates Mcm10 following UV-irradiation, we evaluated the association of Mcm10 with Cullin 4-based E3 ligase (CRL4), comprising cullin, Cul4, ring-finger protein, Roc1 and adaptor protein, DDB1. Utilizing independent siRNAs, we have previously established the specificity of Mcm10 band detected by the anti-Mcm10 antibody used in the present study (32). Mcm10 was efficiently immunoprecipitated from 293T cell lysates with antibody against Mcm10 (Figure 3A, bottom panel). We resolved the same immunoprecipitate on a separate gel and probed it with anti-DDB1 antibody to identify the associated DDB1 (Figure 3A, top panel). We detected endogenous DDB1 in Mcm10 immunoprecipitate, implying a natural association between the two proteins. Utilizing myc antibody for immunoprecipitation, we observed that myc-tagged Mcm10 associates with HA-tagged DDB1 (Figure 3B, third panel). We also observed that HA-tagged Mcm10 is co-immunoprecipitated with myc-tagged DDB1 (Figure 3C, third panel, Lane 2). Previous interaction studies have relied on overexpressed proteins as it has been difficult to observe an endogenous interaction, and we also observed that buffer extracted endogenous Roc1 and Cul4a were poorly detected (20,33). Therefore to evaluate the interaction of Mcm10 with Roc1 and Cul4a, 293T cells were transfected with plasmids expressing HA-tagged Roc1 or Cul4a and immunoprecipitation was carried out with anti-Mcm10 antibody (Figure 3D). Mcm10 antibody, but not the prebleed, co-immunoprecipitated a fraction of expressed Roc1-HA and Cul4a-HA demonstrating an in vivo association of endogenous Mcm10 with Roc1 and Cul4a (Figure 3D, part i and ii, Lane 4). We also observed that Mcm10 antibody efficiently co-immunoprecipitated myc-tagged Roc1 and Cul4a, further corroborating the physical association (Supplementary Figure S1E, part i and ii, Lane 4). The above observations establish that Mcm10 associates with CRL4.
Figure 3.
Endogenous Mcm10 associates with DDB1 and Roc1. (A) 293T cells were lysed under mild conditions, and the Mcm10 protein was immunoprecipitated with anti-Mcm10 antibody and immunoblotted with anti-DDB1 antibody. Input refers to 10% of the total protein that was immunoprecipitated. (B) Exogenously expressed Mcm10 and DDB1 associate with each other. The 293T cells were co-transfected with plasmids expressing myc-tagged Mcm10 and HA-tagged DDB1 followed by immunoprecipitation of myc-Mcm10 using myc antibody. The expression of myc-tagged Mcm10 and HA-tagged DDB1 has been shown in the first and second panels, respectively, whereas the third panel displays the co-immunoprecipitated HA-tagged DDB1. (C) 293T cells were co-transfected with plasmids expressing myc-tagged DDB1 and HA-tagged Mcm10 followed by immunoprecipitation with myc antibody. The expression of myc-tagged DDB1 and HA-tagged Mcm10 has been shown in the first and second panels, respectively, whereas the third panel displays the co-immunoprecipitated HA-tagged Mcm10. (D) Mcm10 associates with exogenously expressed Roc1 and Cul4a. The 293T cells were transfected with pcDNA3-HA vector, pcDNA3-HA-Roc1 or pcDNA3-HA-Cul4a, lysed under mild conditions, and immunoprecipitation was carried out with either anti-Mcm10 antibody or pre-immune serum. Mcm10 band has been indicated by black arrowhead in the second and fourth panels (Lane 4), whereas co-immunoprecipitation of Roc1-HA (part i) and Cul4a-HA (part ii) was evaluated by anti-HA antibody (indicated by shaded arrow in the first and third panels, Lane 4).
Complementation of Roc1 protein reverses the suppression of UV-triggered Mcm10 degradation
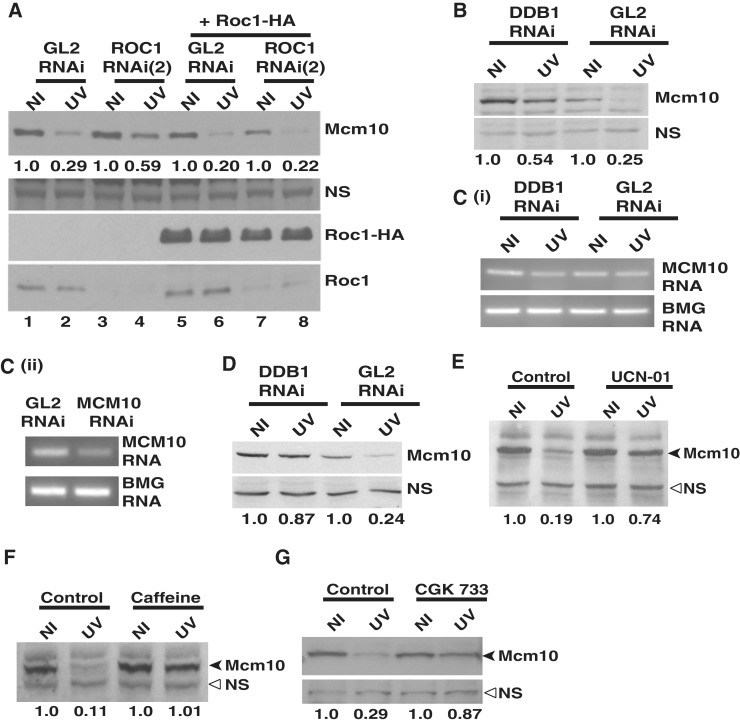
We tested if overexpression of siRNA-resistant ROC1 can reverse the suppression of UV-triggered Mcm10 degradation following Roc1 depletion. ROC1 siRNA (2) is complementary to the 435-455 nucleotide of ROC1 mRNA, which lies within the 3'-untranslated region. ROC1 siRNA (2) targets the endogenous mRNA without depleting the exogenous mRNA, which expresses only the coding sequence of ROC1. 293T cells were co-transfected with either GL2 or ROC1 siRNA (2) along with pcDNA3-HA-Roc1 on three consecutive days as indicated (Figure 4A). Twenty-four hours after the last transfection, the cells were exposed to UV, harvested 4 h later, and levels of Mcm10, endogenous Roc1 and exogenous Roc1-HA were analyzed. ROC1 siRNA (2) depleted the endogenous Roc1 protein, whereas the exogenous Roc1-HA protein was immune to siRNA depletion. The depletion of Roc1 limited the UV-triggered Mcm10 protein decrease to 59% versus 29% in GL2 siRNA-transfected cells (compare Lane 2 with Lane 4). However, on co-expressing the siRNA-resistant Roc1-HA, the Mcm10 levels were almost similar after GL2 and ROC1 siRNA (compare Lane 6 with Lane 8). Therefore, we conclude that complementation of Roc1 protein after UV-irradiation reversed the suppression of UV-triggered Mcm10 degradation due to Roc1 depletion, strongly implicating Roc1 in the Mcm10 turnover.
Figure 4.
DDB1-dependent E3 ubiquitin ligase does not regulate the stability of Mcm10 by altering its mRNA levels. (A) Overexpression of siRNA-resistant Roc1 reverses the suppression of UV-triggered Mcm10 degradation following Roc1 depletion. HeLa cells were transfected on three consecutive days with either GL2 or ROC1 siRNA (2) along with plasmid expressing HA-tagged Roc1 as indicated. 24 h after the last transfection, the cells were exposed to UV, harvested 4 h later and Mcm10 levels were analyzed. The numbers in panel (A) indicate the levels of Mcm10 protein in UV-irradiated cells relative to non-irradiated cells after specific siRNA and plasmid transfection. The expression of HA-tagged Roc1 (third panel) and decrease in the levels of endogenous Roc1 (last panel) was confirmed by immunoblotting. (B) and (C) HeLa cells were transfected with DDB1 siRNA, and the levels of Mcm10 mRNA and protein were analyzed 4 h after UV irradiation. The numbers indicate the levels of Mcm10 protein (B) in UV-irradiated cells relative to non-irradiated cells after GL2 or DDB1 siRNA transfection. Panel C (ii) displays that under similar PCR amplification conditions, reduced levels of Mcm10 mRNA can be detected after RNAi indicating that the PCR amplification was at non-saturating levels. (D) HeLa cells were transfected with DDB1 siRNA, and the stability of Mcm10 was analyzed 2 h after UV irradiation. The numbers indicate the levels of Mcm10 protein in UV-irradiated (UV) cells relative to non-irradiated (NI) cells after GL2 or DDB1 siRNA transfection. (E–G) UV-triggered Mcm10 degradation is blocked in the presence of UCN-01, CGK 733 and caffeine. Cells were treated with 300 nM UCN-01, 25 mM caffeine or 10 µM CGK 733 for 4 h followed by UV exposure, and the cells were harvested 2 h later for analysis of Mcm10 protein. The numbers in panels (E), (F) and (G) indicate the levels of Mcm10 protein in UV-irradiated cells relative to non-irradiated cells after specific treatment. NS points to a non-specific band that displays equal protein loading in different lanes while β-2 microglobulin (BMG) serves as the internal RNA loading control.
To further authenticate that the stabilization of Mcm10 is due to DDB1 depletion and not due to the increased G2 phase, we utilized a Cdk1 inhibitor, RO-3306, which inhibits the activity of Cyclin B1/Cdk1 and reversibly arrests human cells at the G2/M boundary (36). Though the Ki of RO-3306 for Cdk1 is 35 nM and use of 9 µM may inhibit many other kinases, our objective to use RO-3306 was to synchronize GL2 and DDB1 siRNA-transfected cells, so that the effect of DDB1 depletion on Mcm10 levels could be assayed in equivalently distributed cell population. HeLa cells transfected with either GL2 or DDB1 siRNA for three consecutive days were incubated with DMSO or 9 µM RO-3306 for 18 h (Supplementary Figure S3A–B). As observed previously, the combined S and G2 phase increased from 48% to 61% in DDB1-depleted cells. RO-3306-blocked GL2-transfected cells also showed a marked increase in the combined S and G2 phase population from 48% to 77%. Since the treatment with RO-3306 blocked the cells at the G2/M boundary, the depletion of DDB1 did not further increase the S and G2 phases. Though there were negligible differences in cell cycle distribution between GL2 and DDB1 transfected cells, the depletion of DDB1 still led to stabilization of Mcm10 after UV-irradiation (Supplementary Figure S3A, compare Lanes 1 and 2 with Lanes 3 and 4). Therefore, the above result confirms that Mcm10 stabilization results from depletion of DDB1 and not due to variations in the cell cycle distribution. To rule out the effect of off-target silencing by siRNA duplexes in stabilization of Mcm10, we silenced Roc1–Cul4–DDB1 by targeting different regions of the respective mRNA. Knockdown of DDB1, ROC1 or CUL4A+B by non-overlapping siRNA oligos reduced the UV-triggered Mcm10 degradation, confirming their involvement in Mcm10 turnover (Supplementary Figure S2A–E).
Stabilization of Mcm10 following DDB1 depletion is not due to transcriptional regulation
After establishing the role of CRL4 in Mcm10 downregulation, we wanted to establish that the stabilization of Mcm10 is because of decreased proteolysis and not due to upregulated transcription of Mcm10 after DDB1 depletion. DDB1 depletion limited the UV-triggered Mcm10 proteolysis, but its mRNA levels did not increase, confirming that the Mcm10 stabilization is not due to transcriptional upregulation (Figure 4B and C). Though the RT-PCR is not quantitative but under similar PCR amplification conditions, the Mcm10 mRNA levels were reduced after siRNA depletion, suggesting that transcription of Mcm10 was not significantly upregulated after DDB1 depletion (Figure 4C, part i and ii) (32). Moreover, we observed Mcm10 stabilization within 2 h of UV-irradiation, a short time-period for transcription to affect protein levels (Figure 4D). We wanted to ascertain if the canonical DNA damage checkpoint pathway has a role in mediating Mcm10 downregulation. U2OS cells were treated with 300 nM UCN-01 (Chk1 inhibitor) for 4 h followed by UV exposure, and the cells were harvested 2 h later. Incubation with UCN-01 suppressed Mcm10 downregulation following UV-irradiation suggesting involvement of Chk1 in Mcm10 downregulation (Figure 4E). Treatment with 25 mM caffeine or 10 µM CGK 733, which inhibits PI3K kinases, suppressed Mcm10 proteolysis indicating that PI3K kinases may be involved in Mcm10 downregulation (Figure 4F and G).
Overexpression of CRL4–DDB1 generates ubiquitinated forms of Mcm10
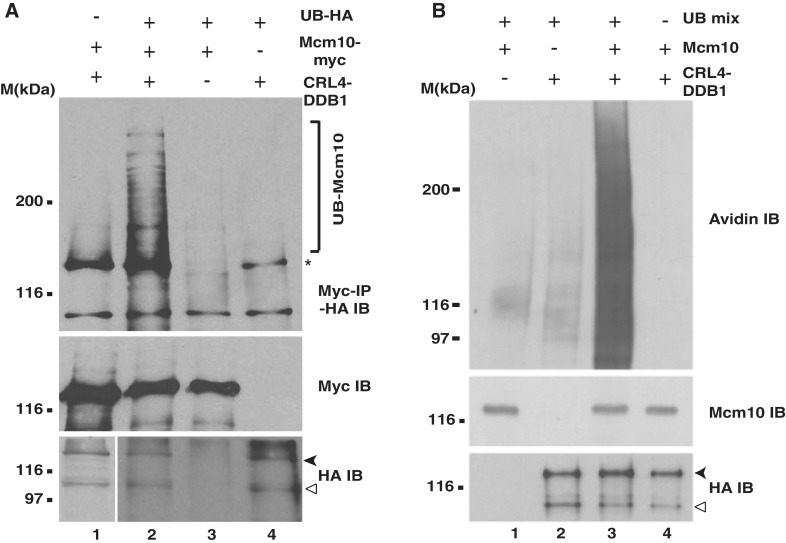
Mcm10 downregulation was blocked by proteasome inhibitors, and therefore, we wanted to test the ubiquitination of Mcm10 by an in vivo ubiquitination assay. On co-expressing HA-tagged ubiquitin, myc-tagged Mcm10 and HA-tagged DDB1, Cul4a and Roc1 (CRL4–DDB1) in 293T cells, we observed a smear of Mcm10 bands (Figure 5A, top panel, Lane 2; depicted by ‘]’). To prove that they are indeed ubiquitinated and not some other modified forms of Mcm10, we removed HA-tagged ubiquitin and observed that the smear completely disappeared, demonstrating that it is dependent on expression of HA-tagged ubiquitin (compare Lane 1 with Lane 2). Similarly, on removing myc-tagged Mcm10, the bands of Mcm10 disappeared confirming that they are the ubiquitinated forms of Mcm10 and not some other ubiquitinated protein detected by HA antibody (compare Lane 2 with Lane 4). The ubiquitinated bands of Mcm10 were dependent on the expression of CRL4–DDB1, establishing that this E3 ligase ubiquitinates Mcm10 (compare Lane 2 with Lane 3).
Figure 5.
DDB1-dependent E3 ligase ubiquitinates Mcm10. (A) 293T cells were transfected with myc-tagged Mcm10, HA-tagged ubiquitin, and HA-tagged DDB1, Cul4a and Roc1 (CRL4–DDB1) as indicated followed by incubation with 25 µM MG132 and exposure to UV-radiation. Subsequently, cells were harvested, and the cell lysates were immunoprecipitated with myc antibody followed by immunoblotting with HA antibody. The bottom panel shows the expression of HA-DDB1 (black arrowhead) and HA-Cul4a (hollow arrowhead), whereas the second panel from top displays the immunoprecipitated myc-Mcm10. The ubiquitinated forms of Mcm10 displayed in the top panel have been normalized to the amount of immunoprecipitated myc-Mcm10. ‘]’ in the top panel highlights the ubiquitinated forms of Mcm10 (UB-Mcm10), whereas asterisk points to a non-specific band. Due to the smear caused due to the expression of HA-ubiquitin, lower exposure has been shown in Lanes 2–4 of the bottom panel. (B) DDB1-dependent E3 ligase ubiquitinates Mcm10 in vitro. 293T cells were transfected with plasmids expressing HA-tagged DDB1, Cul4a and Roc1 (CRL4–DDB1), treated with 25 µM MG132 and UV-irradiated. Purified HA-tagged CRL4 was incubated with immunoprecipitated Mcm10 and ubiquitination mix (UB) as indicated and resolved on a SDS-PAGE gel. The ubiquitinated Mcm10 was displayed by an avidin–HRP immunoblot which detected the biotinylated-ubiquitin linked to Mcm10 (top panel). The bottom panel shows the expression of HA-DDB1 (black arrowhead) and HA-Cul4a (hollow arrowhead), whereas the middle panel displays the immunoprecipitated endogenous Mcm10.
DDB1-dependent E3 ligase ubiquitinates Mcm10 in vitro
To ascertain if the CRL4 complex can ubiquitinate Mcm10 in vitro, 293T cells were transfected with plasmids expressing HA-tagged DDB1, Cul4a and Roc1 (CRL4–DDB1) as indicated in Figure 5B. Two days after the transfection, cells were treated with MG132, UV-irradiated and harvested 4 h later. Anti-Mcm10 and anti-HA antibodies were used to immunoprecipitate endogenous Mcm10 and HA-tagged CRL4, respectively. Immunoprecipitated Mcm10 was incubated with the eluted HA-CRL4 and biotinylated-ubiquitin in the presence of E1, E2 (UbcH5c), ATP, MG132 for 1 h at 37°C for carrying out the ubiquitination reaction whose reaction products were immunoblotted with avidin–HRP. On incubating the immunoprecipitated Mcm10 with HA-CRL4 and ubiquitination mix, we observed a continuum of bands in the avidin–HRP immunoblot (Figure 5B, Lane 3, top panel). Though MG132 would slow the degradation of Mcm10, UV-irradiation would inadvertently lead to generation of fragments of Mcm10 which would be ubiquitinated to varying extents in the in vitro reaction, generating a continuum of bands. The appearance of higher molecular weight bands was dependent on Mcm10 as non-specific proteins were not ubiquitinated. The biotinylated-ubiquitin bands disappeared on excluding either the E1 and E2 enzymes (Lane 4), Mcm10 (Lane 2) or CRL4 complex (Lane 1), proving that these bands are indeed ubiquitinated forms of Mcm10 generated in vitro by the CRL4 complex.
Substrate recognition subunit, Vpr-binding protein (VprBP) is essential for the UV-triggered Mcm10 degradation
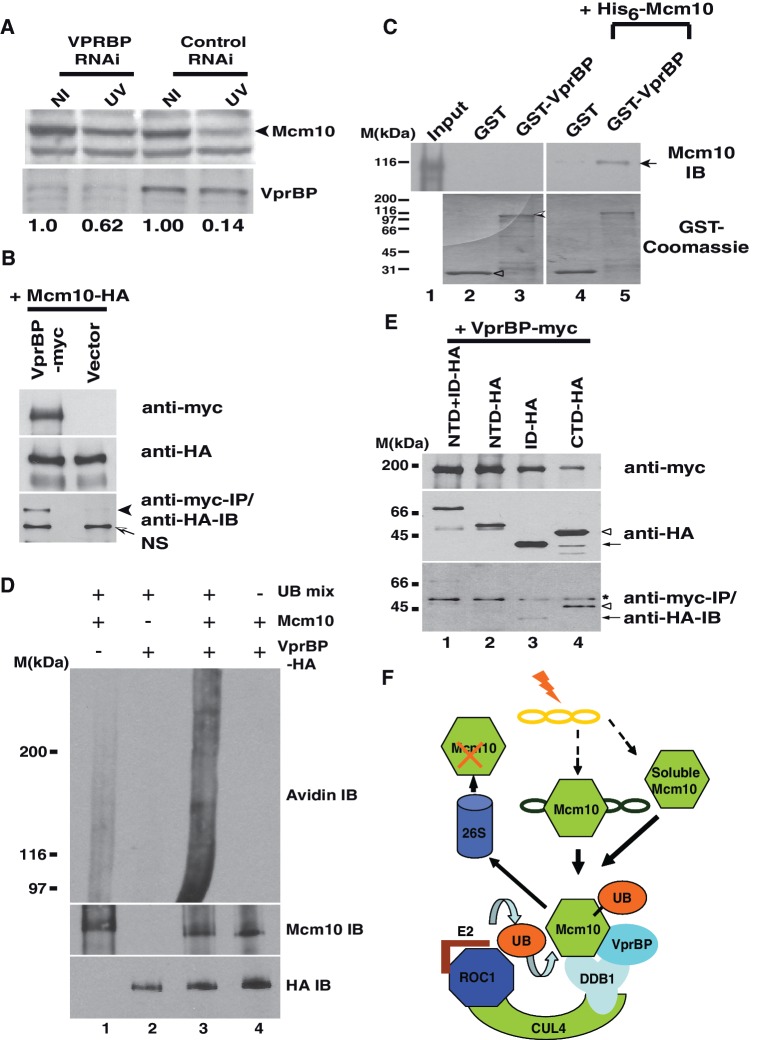
To identify the substrate recognition subunit that targets Mcm10, we depleted the known DDB1 interacting proteins by RNAi and assayed the effect on UV-triggered Mcm10 degradation (Table 1). It is known that human HIV1 arrests host cells at the G2 phase, but the underlying mechanism still remains elusive (37). Proteomic analysis identified a protein, VprBP (HIV Vpr-binding protein) that serves as the substrate recognition subunit for the Cul4A–DDB1 complex (38–40). The siRNA duplex designed to target 965-983 nucleotides of VPRBP mRNA significantly reduced the protein to 14% of control levels. After VPRBP RNAi, Mcm10 decrease post-UV-irradiation was limited to 62%, demonstrating that the depletion of VPRBP significantly reduces the degradation of Mcm10 (Figure 6A). To rule out the effect of off-target silencing by siRNA duplexes on the stabilization of Mcm10, we silenced VPRBP by targeting a different region of its mRNA (1166–1184 nt). We observed that independent siRNA duplexes utilized to silence VPRBP (VPRBP-2) significantly reduced its protein levels and also the UV-triggered Mcm10 degradation, establishing its involvement in the Mcm10 turnover (Supplementary Figure S2F). Evaluation of the cell cycle distribution revealed that the population of cells present in Mcm10 positive phase after siRNA transfection of GL2 (control) and VPRBP were 51 and 55%, respectively (Supplementary Figure S2G). The fraction of cells present in the Mcm10 positive phase did not change significantly indicating that the stabilization of Mcm10 after VprBP depletion is not due to changes in the cell cycle.
Table 1.
DDB1 interacting proteins tested for regulating Mcm10 post-UV-irradiation
| WDTC1 | IQWD1 | WSB1 | WDR5B |
| WDR32 | WDR68 | WDR23 | KATNB1 |
| DCAF11 | DCAF17 | WDR76 | TLE3 |
| WD21A | FBXW5 | WDR82 | NLE1 |
| WDR40A | VprBP | NUP43 | LIS1 |
| WDSOF1 | WDR53 | RBBP5 | IFRG15 |
| PHIP | WDR57 | WDR5 | TRPC4AP |
| CSA | WDR61 | WSB2 | DET1 |
| FBXW8 | RBBP7 | PWP1 | DDB2 |
| DCAF15 | Gβ2 | mβTrCP | CDT2 |
Figure 6.
VprBP is essential for UV triggered Mcm10 degradation. (A) HeLa cells were transfected on three consecutive days with indicated siRNAs, and 24 h after the last transfection, the cells were exposed to UV, harvested 4 h later and Mcm10 levels were analyzed in non-irradiated (NI) and UV-irradiated (UV) cells. The black arrowhead points to Mcm10 band. Immunoblot in the lower panel confirms the decrease in the levels of VprBP. (B) Exogenously expressed Mcm10 and VprBP associate with each other. 293T cells were co-transfected with plasmids expressing myc-tagged VprBP and HA-tagged Mcm10 followed by immunoprecipitation of myc-VprBP using myc antibody. The expression of myc-tagged VprBP and HA-tagged Mcm10 has been shown in the first and second panels, respectively, whereas the third panel displays the co-immunoprecipitated HA-tagged protein. NS points to a non-specific band. (C) Purified His6-Mcm10 binds with GST-VprBP. Purified His6-Mcm10 was incubated with GST-tagged C-terminus VprBP or plain GST bound glutathione-agarose beads as indicated. Bound His6-Mcm10 was detected by immunoblotting with anti-Mcm10 antibody (top panel; black arrow). Lane 1 (Input) shows 1% of His6-Mcm10 added for binding assays. In Lanes 2–3, His6-Mcm10 was not added to rule out any artifacts in detection due to GST-tagged proteins. Coomassie-stained gel (bottom panel) is shown to indicate the levels of the GST fusion proteins used in this binding experiment. Hollow arrowhead and shaded arrowhead point to GST and GST tagged C-terminus VprBP (864-1507), respectively. (D) VprBP-associated complex ubiquitinates Mcm10 in vitro. 293T cells were transfected with plasmids expressing HA-tagged VprBP, treated with 25 µM MG132 and UV-irradiated. Purified HA-tagged VprBP was incubated with immunoprecipitated Mcm10 and ubiquitination mix (UB) as indicated and resolved on a SDS-PAGE gel. The ubiquitinated Mcm10 was displayed by an avidin–HRP immunoblot which detected the biotinylated-ubiquitin linked to Mcm10 (top panel). The bottom panel shows the expression of HA-VprBP, whereas the middle panel displays the immunoprecipitated endogenous Mcm10. (E) VprBP associates with CTD domain of Mcm10. 293T cells were transfected with HA-tagged NTD + ID, NTD, ID or CTD Mcm10 along with myc-tagged VprBP as indicated followed by immunoprecipitation with myc antibody. Top panel displays expression of myc-tagged VprBP, middle panel displays the expression of HA-tagged Mcm10 fragments, whereas the bottom panel shows the co-immunoprecipitated HA-tagged Mcm10. Hollow arrowhead and black arrow point to HA-tagged CTD and ID Mcm10, respectively. Asterisk depicts the IgG heavy chain. (F) A model depicts that CRL4–DDB1 ubiquitin ligase mediates the UV-triggered proteolysis of Mcm10. This E3 ligase comprises a cullin, Cul4, a ring-finger protein, Roc1, an adaptor protein, DDB1, and substrate-recognition subunit (SRS), VprBP to which soluble and chromatin bound Mcm10 would bind for transfer of ubiquitin molecules from E2 conjugating enzyme. Mcm10 would be subsequently degraded by the 26S proteasome.
To study the interaction of Mcm10 with VprBP, myc-tagged VprBP and HA-tagged Mcm10 were expressed in 293 T cells. We observed HA-tagged Mcm10 in anti-VprBP immunoprecipitate, confirming an association between Mcm10 and VprBP (Figure 6B). The observed in vivo interaction of Mcm10 with VprBP could be direct or mediated by other factors. Therefore, to test if Mcm10 directly binds with VprBP, His6-tagged Mcm10 was expressed in bacteria, purified on a Ni-NTA column and was eluted with 0. 25 M imidazole. Since full-length VprBP was insoluble, we expressed C terminus of VprBP (864–1507) tagged to GST in E. coli, and it was bound to glutathione agarose column (Figure 6C, bottom panel). We observed that purified His6-Mcm10 binds to C-terminal VprBP on the glutathione column (Lane 5). However, His6-Mcm10 did not bind to plain GST which was heavily expressed, ruling out non-specific sticking to the glutathione-agarose column (compare Lane 4 with Lane 5). These results provide strong evidence that VprBP serves as a substrate recognition subunit targeting Mcm10 to CRL4 ubiquitin ligase.
VprBP-associated complex ubiquitinates Mcm10 in vitro
293T cells were transfected with plasmid expressing HA-tagged VprBP, and two days after the transfection cells were treated with MG132, UV-irradiated and harvested 4 h later. Anti-Mcm10 and anti-HA antibodies were used to immunoprecipitate endogenous Mcm10 and HA-tagged VprBP, respectively. Immunoprecipitated Mcm10 was incubated with HA-peptide eluted HA-VprBP in the presence of ubiquitination mix, and the ubiquitinated proteins were detected by avidin–HRP which detects biotinylated-ubiquitin linked to Mcm10 at a higher efficiency than the anti-Mcm10 antibody. We observed a continuum of bands demonstrating the in vitro ubiquitination of Mcm10 by VprBP-associated complex (Figure 6D, Lane 3, top panel). If Mcm10 was excluded from the reaction, the observed bands disappeared corroborating that they are ubiquitinated forms of Mcm10 (compare Lanes 2 and 3). The biotinylated-ubiquitin bands also disappeared on excluding the E1 and E2 enzymes from the reaction proving that these bands are indeed ubiquitinated forms generated in the in vitro reaction (Lane 4). Further, if the immunoprecipitated HA-VprBP was excluded from the reaction, these bands disappeared demonstrating that Mcm10 is directly ubiquitinated by the VprBP-associated complex in vitro (Lane 1).
VprBP binds to the C-terminal domain of Mcm10
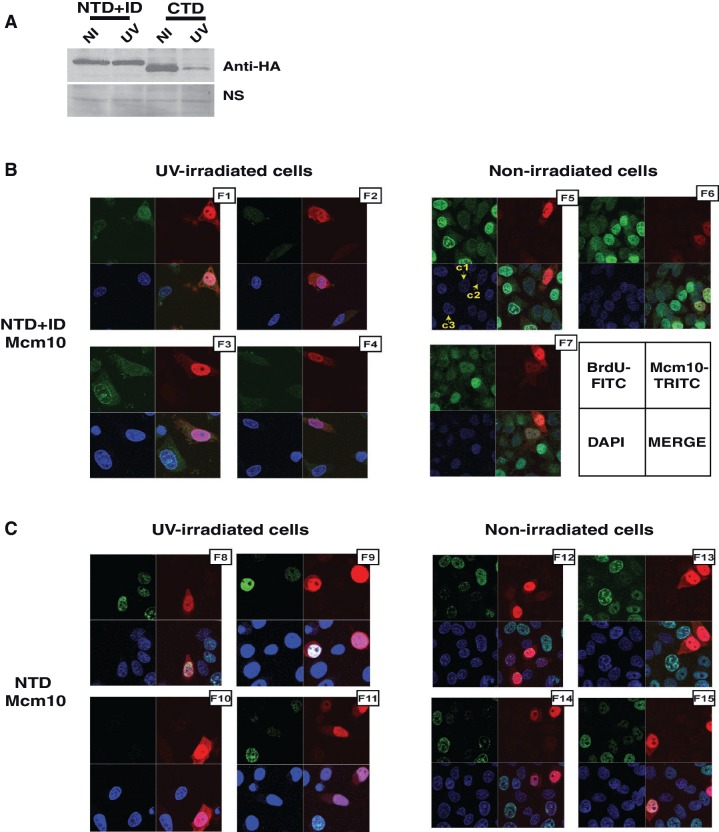
To identify the domains of Mcm10 essential for forming a complex with VprBP, myc-tagged VprBP was expressed in 293T cells along with different regions of Mcm10 fused to HA-tag. The structural architecture of vertebrate Mcm10 has been determined, and the limited proteolytic digestion reveals an independently folded globular N-terminal domain (NTD) and inner domain (ID) connected to the C-terminal domain (CTD) via a linker region (10,11). We observed that the CTD domain, which is rapidly degraded after UV-irradiation, displays a robust association with myc-VprBP (Figures 6E and 7A). On the other hand when we expressed ID domain itself or in combination with NTD domain, a weak physical interaction with VprBP was observed. NTD and ID domains are resistant to UV-triggered degradation, suggesting that the binding may not be sufficient for UV-triggered degradation. Therefore, we conclude that VprBP binds to the CTD domain of Mcm10, causing its degradation.
Figure 7.
Expression of NTD and ID domains of Mcm10 does not rescue the block of replication in UV checkpoint arrest. (A) NTD + ID domain is stable, whereas CTD is degraded after UV irradiation. U2OS cells expressing different domains of Mcm10 were exposed to UV radiation and harvested 2 h later for evaluating the levels of the respective domains by anti-HA immunoblotting. (B) HeLa cells were transfected with plasmid expressing NTD + ID domain of Mcm10 fused to a nuclear localization signal, and BrdU incorporation was measured 16–24 h after exposure to 40 J/m2 UV radiation (F1–F4). BrdU incorporation was also measured without exposure to UV radiation (F5–F7). The figure displays a few randomly selected 75 × 75 µm fields from each sample. HeLa cells were visualized for DNA staining, BrdU incorporation and Mcm10 overexpression by DAPI, FITC and TRITC immunofluorescence, respectively. The arrangement of immunofluorescence images obtained from a single field has been illustrated in the bottom-right panel. Cells that were positive for BrdU signal were counted to calculate the % BrdU incorporation in Mcm10 overexpressing and non-Mcm10 overexpressing cells. Due to cell death resulting from UV-irradiation, the confluency is lower than non-irradiated cells. To rule out bleed-through of fluorescent signals, cells that have different status of DAPI, FITC and TRITC signals have been marked: Cell marked as c1 is positive for DAPI (nuclei staining) but negative for FITC (BrdU) and TRITC (Mcm10) signal, whereas c2 is positive for TRITC but negative for FITC signal and c3 is positive for FITC but negative for TRITC signal. The effect of full-length Mcm10 overexpression on BrdU incorporation has been described previously (27). (C) HeLa cells were transfected with plasmid expressing NTD domain of Mcm10 fused to a nuclear localization signal, and BrdU incorporation was measured with and without exposure to UV radiation as described in panel B.
Overexpression of NTD and ID domains of Mcm10 do not complement DNA replication post-UV irradiation
In the present study, we have expressed the UV-resistant NTD + ID domain, which does not interact with VprBP, and evaluated its effect on the rate of DNA replication following UV irradiation (Figure 7). HeLa cells were transfected with plasmid expressing the NTD + ID domain, and BrdU incorporation was measured with and without exposure to UV radiation in NTD + ID overexpressing and non-overexpressing cells (Figure 7B, F1–F7). First, we observed that overexpression of NTD + ID fragment itself did not affect the rate of DNA replication significantly in non-irradiated cells (44.17% versus 57.44%) (Figure 7B and Table 2). As expected, UV-irradiation decreased the percentage of cells incorporating BrdU (16.77 and 44.17% with and without UV-irradiation in control non-Mcm10 overexpressing cells). The overexpression of NTD + ID fragment could not significantly complement the DNA replication following UV irradiation (21.42% in Mcm10 overexpressing cells versus 16.77% in non-Mcm10 overexpressing cells). Similarly, overexpression of NTD fragment could not rescue DNA replication following UV irradiation (Figure 7C, F8–F15 and Table 2). In contrast, we have previously observed that the BrdU incorporation was higher in full-length Mcm10-overexpressing cells (43.73%) as compared with non-Mcm10 expressing cells (20.56%) after UV-irradiation (32). Though the full-length Mcm10 has the requisite degradation signals, Mcm10 levels were sustained post-irradiation by saturating the degradation machinery by overexpression. It should be noted that the C-terminus of Mcm10, which is absent in the NTD + ID fragment used for complementation, provides vital function. CTD contains two Zn binding motifs and a winged helix domain which binds to both ss- and ds-DNA as well as the catalytic subunit of DNA polymerase alpha. It is also observed that Mcm10 deleted at the C-terminus is unable to interact with MCM complex or support cell growth (41). Therefore, we conclude that the NTD and ID domains of Mcm10 are not able to attenuate the inhibition of DNA replication post-UV irradiation.
Table 2.
BrdU incorporation after expression of different domains of Mcm10
| Mcm10 domain overexpressed | Mcm10 overexpressing cells |
Mcm10 non-overexpressing cells |
||
|---|---|---|---|---|
| Non- irradiated | UV- irradiated | Non- irradiated | UV- irradiated | |
| NTD + ID | 57.44 | 21.42 | 44.17 | 16.77 |
| NTD | 31.11 | 20.31 | 44.92 | 20.10 |
DISCUSSION
Since binding of Mcm10 to the origins of DNA replication causes activation of the pre-replicative complexes to initiate replication, we believe that during stress, this crucial step is targeted by ubiquitination machinery to arrest replication. Work in the last few years has established the essential role of Cul4 and DDB1 in the UV triggered degradation of replication initiator, Cdt1 and cyclin-dependent kinase inhibitor p21 (18–21,42). We demonstrate that Mcm10 downregulation requires the Cul4–DDB1 complex but is independent of Skp2 or PCNA, the cofactors required for Cdt1 degradation (25,26). There are around 60 proteins predicted to interact with Cul4 or DDB1 (termed as DCAF: DDB1- and CUL4-associated factors) that serve as substrate recognition subunits (SRS). We depleted the known DDB1 interacting proteins by RNAi and assayed its effect on UV-triggered Mcm10 degradation. Though the efficiency of RNAi-mediated silencing of different target genes was variable, our screen identified VprBP as the substrate recognition subunit that targets Mcm10 (Table 1). It is well accepted that HIV1-encoded protein, Vpr, is essential for inducing host cell cycle arrest (43,44). It was subsequently reported that Vpr mediates G2 arrest by engaging an ubiquitin ligase complex containing DDB1 and Cul4. It was also reported that Vpr binding partner, VprBP, serves as a substrate recognition subunit in the degradation of unidentified factors that are required for cell cycle progression from G2 to mitosis (38–40,45–47). We have discovered a target of Cul4–DDB1–VprBP ubiquitin ligase, and therefore it is quite likely that replication factors like Mcm10 may be targeted during HIV1 infection for accomplishing a cell cycle arrest. If that turns out to be true, it would explain how mammalian replication and cell cycle progression is subverted by HIV1. Our work alludes to the possibility that VprBP may be a key regulator of replication machinery, and its role in the stability of other replication factors should be investigated. It has also been demonstrated recently that VprBP-associated E3 ligase regulates the stability of uracil DNA glycosylase (48). Significant research work has demonstrated the functions of VprBP-dependent ubiquitin ligase including the effect on cell cycle and the flexibility of VprBP to associate with different ubiquitin ligases for targeting substrates (49–53). The VprBP–DDB1 complex is known to associate with ring-finger-based ubiquitin ligases targeting tumor suppressor, Merlin (54,55). The loading of Merlin to Roc1–Cullin4A–DDB1 ubiquitin ligase requires VprBP. We have previously reported that Mcm10 levels were not altered after exposure to gamma radiation or DNA damaging chemicals, and therefore we conclude that the downregulation of Mcm10 occurs by a pathway that is specifically induced by UV-mediated DNA damage (32). Inhibitors of canonical checkpoint pathways were able to attenuate UV-triggered Mcm10 degradation, suggesting their role in Mcm10 downregulation (Figure 4E–G).
We have demonstrated that the complementation of Mcm10 partially obviates the UV-triggered inhibition of replication, and therefore downregulation of Mcm10 is pivotal in inhibiting replication upon UV-irradiation (32). It is well documented that unwarranted activity of replication genes leads to incorporation of genomic lesions, and our study suggests that CRL4VprBP ubiquitin ligase may prevent such aberrations (22,56,57). The downregulation of Mcm10 occurs at higher doses of UV radiation and is temporally separated from instantaneous Cdt1 downregulation displaying the cellular resilience in responding to stress (58). Our study suggests that CRL4VprBP ubiquitin ligase may be involved in regulating the levels of other replication proteins and their identification would help us in completely understanding the role of this E3 ligase in cellular physiology. Mcm10 is a naturally oscillating replication protein which is proteolyzed during mitosis, and in a separate study, we have observed that Mcm10 proteolysis during mitosis is mediated by CRL4VprBP(59). In the present study, we have observed that p53-negative HeLa cells degrade Mcm10 post-UV-irradiation. Treatment with inhibitors of ATM/ATR kinases, CGK 733 and caffeine, reduced UV-triggered Mcm10 degradation. Since UCN01 also blocks Mcm10 downregulation, it is likely that UV-induced lesions activate the ATR and CHK1 kinases that lead to Mcm10 degradation. However, it has been observed that radiation triggered Cdt1 degradation was impervious to treatment with caffeine (19). The authors also noted that siRNA depletion of Chk1, Chk2, ATM and ATR, singly or in combination, failed to block Cdt1 degradation. In contrast, UV-triggered degradation of p12 subunit of polymerase delta did not occur in conditional ATR−/− cells (60). Therefore, the involvement of the canonical checkpoint pathway in the downregulation of replication proteins needs a detailed investigation in different genetic backgrounds. In conclusion, we establish that E3 ubiquitin ligase composed of Cul4, Roc1, DDB1 and VprBP imparts a vital role in regulating replication by mediating the stress-induced turnover of replication factor, Mcm10 (our model is described in Figure 6F).
SUPPLEMENTARY DATA
The Supplementary Data are available at NAR Online: Supplementary Figures 1–3 and Supplementary Materials and Methods.
FUNDING
Department of Biotechnology, Government of India and NET fellowship (to M.K. and A.S). Funding for open access charge: National Institute of Immunology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sunder Bisht, Md. Sadique Ali, Kanika Jain, Aishana Singh, Nirmal, Subha, Krishan and Swarnabh for helping in various parts of this paper.
REFERENCES
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Ricke RM, Bielinsky AK. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol. Cell. 2004;16:173–185. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Ricke RM, Bielinsky AK. A conserved Hsp10-like domain in Mcm10 is required to stabilize the catalytic subunit of DNA polymerase-alpha in budding yeast. J. Biol. Chem. 2006;281:18414–18425. doi: 10.1074/jbc.M513551200. [DOI] [PubMed] [Google Scholar]
- 4.Das-Bradoo S, Ricke RM, Bielinsky AK. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol. Cell. Biol. 2006;26:4806–4817. doi: 10.1128/MCB.02062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merchant AM, Kawasaki Y, Chen Y, Lei M, Tye BK. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:3261–3271. doi: 10.1128/mcb.17.6.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye BK. Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 2000;14:913–926. [PMC free article] [PubMed] [Google Scholar]
- 7.Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol. Cell. 2002;9:233–240. doi: 10.1016/s1097-2765(02)00456-2. [DOI] [PubMed] [Google Scholar]
- 9.Kawasaki Y, Hiraga S, Sugino A. Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae. Genes Cells. 2000;5:975–989. doi: 10.1046/j.1365-2443.2000.00387.x. [DOI] [PubMed] [Google Scholar]
- 10.Robertson PD, Warren EM, Zhang H, Friedman DB, Lary JW, Cole JL, Tutter AV, Walter JC, Fanning E, Eichman BF. Domain architecture and biochemical characterization of vertebrate Mcm10. J. Biol. Chem. 2008;283:3338–3348. doi: 10.1074/jbc.M706267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okorokov AL, Waugh A, Hodgkinson J, Murthy A, Hong HK, Leo E, Sherman MB, Stoeber K, Orlova EV, Williams GH. Hexameric ring structure of human MCM10 DNA replication factor. EMBO Rep. 2007;8:925–930. doi: 10.1038/sj.embor.7401064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koepp DM, Harper JW, Elledge SJ. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 14.Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 15.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, Shirane M, Tsunematsu R, Tsukiyama T, Ishida N, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 2000;19:2069–2081. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol. Cell. 2007;27:462–473. doi: 10.1016/j.molcel.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higa LA, Mihaylov IS, Banks DP, Zheng J, Zhang H. Radiation-mediated proteolysis of CDT1 by CUL4–ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 2003;5:1008–1015. doi: 10.1038/ncb1061. [DOI] [PubMed] [Google Scholar]
- 19.Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1–CUL4A–ROC1 ligase in response to DNA damage. Nat. Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 20.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22:2507–2519. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423:885–889. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- 23.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4–Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Sansam CL, Shepard JL, Lai K, Ianari A, Danielian PS, Amsterdam A, Hopkins N, Lees JA. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 2006;20:3117–3129. doi: 10.1101/gad.1482106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 2006;8:84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- 26.Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 2006;281:6246–6252. doi: 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- 27.Wakasugi M, Kawashima A, Morioka H, Linn S, Sancar A, Mori T, Nikaido O, Matsunaga T. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 2002;277:1637–1640. doi: 10.1074/jbc.C100610200. [DOI] [PubMed] [Google Scholar]
- 28.Seki N, Hayashi A, Hattori A, Kozuma S, Sasaki M, Suzuki Y, Sugano S, Muramatsu M, Saito T. cDNA cloning, tissue expression, and chromosomal assignment of a mouse gene, encoding a 127 kDa UV-damaged DNA binding protein which is defective in XPE cells. DNA Res. 1999;6:319–322. doi: 10.1093/dnares/6.5.319. [DOI] [PubMed] [Google Scholar]
- 29.Hwang BJ, Toering S, Francke U, Chu G. p48 Activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol. Cell Biol. 1998;18:4391–4399. doi: 10.1128/mcb.18.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH. Structural basis of UV DNA-damage recognition by the DDB1–DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 32.Sharma A, Kaur M, Kar A, Ranade SM, Saxena S. Ultraviolet radiation stress triggers the down-regulation of essential replication factor Mcm10. J. Biol. Chem. 2010;285:8352–8362. doi: 10.1074/jbc.M109.041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondo T, Kobayashi M, Tanaka J, Yokoyama A, Suzuki S, Kato N, Onozawa M, Chiba K, Hashino S, Imamura M, et al. Rapid degradation of Cdt1 upon UV-induced DNA damage is mediated by SCFSkp2 complex. J. Biol. Chem. 2004;279:27315–27319. doi: 10.1074/jbc.M314023200. [DOI] [PubMed] [Google Scholar]
- 34.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS. The DDB1–CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl Acad. Sci. USA. 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izumi M, Yatagai F, Hanaoka F. Cell cycle-dependent proteolysis and phosphorylation of human Mcm10. J. Biol. Chem. 2001;276:48526–48531. doi: 10.1074/jbc.M107190200. [DOI] [PubMed] [Google Scholar]
- 36.Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl Acad. Sci. USA. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui J, Tungaturthi PK, Ayyavoo V, Ghafouri M, Ariga H, Khalili K, Srinivasan A, Amini S, Sawaya BE. The role of Vpr in the regulation of HIV-1 gene expression. Cell Cycle. 2006;5:2626–2638. doi: 10.4161/cc.5.22.3442. [DOI] [PubMed] [Google Scholar]
- 38.Wen X, Duus KM, Friedrich TD, de Noronha CM. The HIV1 protein Vpr acts to promote G2 cell cycle arrest by engaging a DDB1 and Cullin4A-containing ubiquitin ligase complex using VprBP/DCAF1 as an adaptor. J. Biol. Chem. 2007;282:27046–27057. doi: 10.1074/jbc.M703955200. [DOI] [PubMed] [Google Scholar]
- 39.Le Rouzic E, Belaidouni N, Estrabaud E, Morel M, Rain JC, Transy C, Margottin-Goguet F. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4–DDB1 ubiquitin ligase. Cell Cycle. 2007;6:182–188. doi: 10.4161/cc.6.2.3732. [DOI] [PubMed] [Google Scholar]
- 40.Hrecka K, Gierszewska M, Srivastava S, Kozaczkiewicz L, Swanson SK, Florens L, Washburn MP, Skowronski J. Lentiviral Vpr usurps Cul4–DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl Acad. Sci. USA. 2007;104:11778–11783. doi: 10.1073/pnas.0702102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JK, Seo YS, Hurwitz J. The Cdc23 (Mcm10) protein is required for the phosphorylation of minichromosome maintenance complex by the Dfp1-Hsk1 kinase. Proc. Natl Acad. Sci. USA. 2003;100:2334–2339. doi: 10.1073/pnas.0237384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T. CDK inhibitor p21 is degraded by a PCNA coupled Cul4–DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 2008;283:29045–29052. doi: 10.1074/jbc.M806045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jowett JB, Planelles V, Poon B, Shah NP, Chen ML, Chen IS. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J. Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogel ME, Wu LI, Emerman M. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J. Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan L, Ehrlich E, Yu XF. DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J. Virol. 2007;81:10822–10830. doi: 10.1128/JVI.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belzile JP, Duisit G, Rougeau N, Mercier J, Finzi A, Cohen EA. HIV-1 Vpr-mediated G2 arrest involves the DDB1–CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog. 2007;3:e85. doi: 10.1371/journal.ppat.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCall CM, Miliani de Marval PL, Chastain PD, 2nd, Jackson SC, He YJ, Kotake Y, Cook JG, Xiong Y. Human immunodeficiency virus type 1 Vpr-binding protein VprBP, a WD40 protein associated with the DDB1–CUL4 E3 ubiquitin ligase, is essential for DNA replication and embryonic development. Mol. Cell. Biol. 2008;28:5621–5633. doi: 10.1128/MCB.00232-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahn J, Vu T, Novince Z, Guerrero-Santoro J, Rapic-Otrin V, Gronenborn AM. HIV-1 Vpr loads uracil DNA glycosylase-2 onto DCAF1, a substrate recognition subunit of a cullin 4A-ring E3 ubiquitin ligase for proteasome-dependent degradation. J. Biol. Chem. 2010;285:37333–37341. doi: 10.1074/jbc.M110.133181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson S, Xiong Y. Targeting protein ubiquitylation: DDB1 takes its RING off. Nat. Cell Biol. 2009;11:379–381. doi: 10.1038/ncb0409-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maddika S, Chen J. Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat. Cell Biol. 2009;11:409–419. doi: 10.1038/ncb1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casey L, Wen X, de Noronha CM. The functions of the HIV1 protein Vpr and its action through the DCAF1.DDB1.Cullin4 ubiquitin ligase. Cytokine. 2010;51:1–9. doi: 10.1016/j.cyto.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Transy C, Margottin-Goguet F. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4–DDB1 ubiquitin ligase. Cell Cycle. 2009;8:2489–2490. doi: 10.4161/cc.8.16.9129. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Feng S, Chen F, Chen H, Wang J, McCall C, Xiong Y, Deng XW. Arabidopsis DDB1–CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell. 2008;20:1437–1455. doi: 10.1105/tpc.108.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang J, Chen J. VprBP targets Merlin to the Roc1–Cul4A–DDB1 E3 ligase complex for degradation. Oncogene. 2008;27:4056–4064. doi: 10.1038/onc.2008.44. [DOI] [PubMed] [Google Scholar]
- 55.Li W, You L, Cooper J, Schiavon G, Pepe-Caprio A, Zhou L, Ishii R, Giovannini M, Hanemann CO, Long SB, et al. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell. 2010;140:477–490. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gopalakrishnan V, Simancek P, Houchens C, Snaith HA, Frattini MG, Sazer S, Kelly TJ. Redundant control of rereplication in fission yeast. Proc. Natl Acad. Sci. USA. 2001;98:13114–13119. doi: 10.1073/pnas.221467598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yanow SK, Lygerou Z, Nurse P. Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J. 2001;20:4648–4656. doi: 10.1093/emboj/20.17.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma A, Kar A, Kaur M, Ranade SM, Sankaran A, Misra S, Rawat K, Saxena S. Specific replication factors are targeted by different genotoxic agents to inhibit replication. IUBMB Life. 2010;62:764–775. doi: 10.1002/iub.380. [DOI] [PubMed] [Google Scholar]
- 59.Kaur M, Sharma A, Khan M, Kar A, Saxena S. Mcm10 proteolysis initiates before the onset of M-phase. BMC Cell Biol. 2010;11:84. doi: 10.1186/1471-2121-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang S, Zhou Y, Trusa S, Meng X, Lee EY, Lee MY. A novel DNA damage response: rapid degradation of the p12 subunit of DNA polymerase delta. J. Biol. Chem. 2007;282:15330–15340. doi: 10.1074/jbc.M610356200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.