Abstract
Dynamic acetylation of all lysine-4-trimethylated histone H3 is a complex phenomenon involved in Immediate-early gene induction in metazoan eukaryotes. Higher eukaryotes express repeated copies of three closely related H3 variants, inaccessible to genetic analysis. We demonstrate conservation of these phenomena in Dictyostelium which has three single-copy H3 variant genes. Although dynamic acetylation is targeted to two H3 variants which are K4-trimethylated, K9-acetylation is preferentially targeted to one. In cells lacking Set1 methyltransferase and any detectable K4-trimethylation, dynamic acetylation is lost demonstrating a direct link between the two. Gene replacement to express mutated H3 variants reveals a novel interaction between K4-trimethylation on different variants. Cells expressing only one variant show defects in growth, and in induction of a UV-inducible gene, demonstrating the functional importance of variant expression. These studies confirm that dynamic acetylation targeted to H3K4me3 arose early in evolution and reveal a very high level of specificity of histone variant utilization in a simple multicellular eukaryote.
INTRODUCTION
The control of inducible genes in higher eukaryotes is a complex process requiring activation of signalling cascades directed to specific transcription factors and co-activators, polymerase II engagement and a host of transient, region-specific histone modifications across the gene (1–3). Our recent work, using the inducible genes c-fos and c-jun as a model, has established four layers of histone modification which are experimentally separable into different phases corresponding to stages of transcriptional activation (2). First, in quiescent unstimulated cells, histone 3 lysine-4 trimethylation (H3K4me3) and H3 acetylation can be found across the promoter and start-site of these genes, and H3K79 and H3K36 methylation across the body and at the 3′-end respectively (2,4). A second layer, also observed in quiescent cells, is highly dynamic turnover of H3 acetylation which is specifically targeted to K4-trimethylated H3 tails across the promoter and start-site (5,6). The third layer, dependent upon signalling cascades but not on transcriptional activation, arises upon stimulating these cells with various agonists; this includes phosphorylation of histone H3 on serine 10 (7,8) mediated via MAP kinase cascades (9,10), and enhanced H3 acetylation across promoter and start-site (4,9,10). Finally, there is a set of transcription-dependent modifications including further H3K4 and K36 methylation, which may be laid down during the passage of RNA pol II across these genes by enzymes associated with the polymerase (4,6). Specific enzymes that mediate this complex process, such as Setd2 for H3K36 trimethylation in layers 1 and 4 (4), p300/CBP for dynamic turnover of acetylation in layer 2 (6,11) and MSK1/2 for H3S10 phosphorylation in layer 3 (10), have been identified.
The importance of histone H3 variants in dynamic modification and regulation of gene induction (12,13) is difficult to address in higher eukaryotes in which H3 genes are highly repeated. In mammalian cells, three major H3 variants have been described and other minor variants have been identified more recently(14), adding the potential for an extra layer of complexity to the role of histone modifications in nuclear processes. H3.1 and H3.2 are replication-dependent histones, repopulating the genome during DNA replication [reviewed in (13)]. H3.3 is a replication-independent variant which replaces H3.2 and H3.1 outside S phase, and is enriched at sites of active transcription in some cases [reviewed in (13)]. These variants differ by only around four amino acids making biochemical analysis of variants difficult. Furthermore, genetic manipulation of histone genes is not possible in higher eukaryotes due to massive duplication and presence of high copy number arrays of genes encoding H3.1 and H3.2. An additional important issue is the growing realization that some of the complexity in histone proteins and their modification may have arisen during evolution specifically for metazoan multicellular organisms, where processes such as embryological development, morphogenesis and pattern formation may place more intricate and subtle demands on the basic mechanisms of gene induction [reviewed in (1)]. These considerations prompt a search for simpler multicellular organisms in which some or all of these complex mechanisms are conserved, but have single copy histone genes amenable to genetic manipulation.
All these considerations are uniquely met in the social amoeba Dictyostelium discoideum. Feeding Dictyostelium cells grow as individual amoebae, but starvation triggers a developmental life cycle in which cells aggregate to form a multicellular organism. After 24 h, a fruiting body is formed consisting of two major cell types, spore and stalk cells (15). As it contains both unicellular amoeboid and multicellular developmental phases, Dictyostelium is a potentially valuable model organism for studying the role of epigenetic and/or histone modifications in a developmental programme associated with the evolution of multicellularity. The complete genome sequence is available and molecular genetics of this haploid organism facilitate targeted gene disruption and replacement experiments (16).
The Dictyostelium genome contains genes encoding three histone H3 variants, each a single copy, in addition to two divergent variants one of which is a centromeric form (17,18). Virtually every known site of H3 modification is conserved in these variants and some, including methylation of H3K4 and K79, have been proven using modification-specific antisera (19,20). Furthermore, single enzymes are responsible for these modifications, as disruption of genes encoding Set1 or Dot1 leads to complete loss of methylation of H3K4 or H3K79 respectively (19,20). Acetylation of H3 has also been described and disruption of a gene encoding one of two potential histone deacetylases leads to increased levels of acetylation (21). The presence of H3K9me2 and low levels of DNA methylation demonstrate that Dictyostelium contains chromatin marks associated with heterochromatic gene silencing (22,23).
Here, we report the expression and modifications associated with histone H3 variants in Dictyostelium. We show that variants are selectively modified and that key features of the complexity seen in higher eukaryotes, such as the preferential dynamic acetylation of K4-trimethylated H3, are observed. Ablation of all K4-methylation by deletion of set1 proves that Set1-mediated K4-methylation is essential for targeted dynamic histone H3 acetylation. These studies show that dynamic histone acetylation targeted to K4-trimethylated histone H3 is conserved and also reveal a novel interaction between modifications on variants as mutation of a modification site on one variant alters modification of the same site on another. Disruption of genes encoding two H3 variants, leaving an organism expressing a single H3 variant, leads to selective impairment of gene induction and defects in growth and development.
MATERIALS AND METHODS
Materials
All chemicals and reagents were purchased from Sigma-Aldrich or Merck unless otherwise stated. Molecular biology products were purchased from Sigma-Aldrich, Promega, Boehringer Ingelheim, Invitrogen, BIOLINE, and New England Biolabs. Histone deacetylase inhibitor trichostatin A (TSA) was from Errant Gene Therapeutics, LLC.
Growth and development of Dictyostelium
Dictyostelium cells were grown axenically at 22°C and developed on Millipore filters as described (24). Exponential vegetative growth stage cells (2–6 × 106 cells/ml) were used for all experiments. Cells were also grown on SM–Agar plates in association with K. aerogenes lawns or heat-killed K. aerogenes at indicated density in KK2 buffer (19 mM KH2PO4, 3.6 mM K2HPO4) in shaking suspension at 22°C.
Generation of Dictyostelium H3b, H3c and H3b/c−/− disruption and H3bK4A strains
The strains set1−, H3aK4A and H3aK4K are previously described (19,25). To generate H3b/c double disruption cells, two fragments −154 to −1010 of H3b genomic and 1457–2491 of H3c genomic (see dictyBase.org) were cloned either side of a blasticidin resistance cassette (bsr), deleting all the H3b sequence except the start codon and all but the extreme 3′-end of H3c. To generate H3b disruption cells, the targeting fragment was 215–2250 of the H3b locus, with the bsr cassette inserted into the EcoRI site of H3b. For H3c disruption, the targeting fragment spans from the HindIII site of H3b to 2491 of H3c and the bsr cassette was put in a linkered PacI site in H3c. In H3bK4A cells, GCA replaces AAA in a fragment containing from 388 to 2300 of the H3b locus and the bsr cassette was put in the EcoRV site downstream of the STOP codon. Linearized constructs were electroporated in Ax2 cells and Bsr resistant clones screened for homologous recombination events.
Protein extraction and gel electrophoresis
Histones from C3H 10T1/2 mouse fibroblasts, Drosophila S2 and Dictyostelium cells were prepared as described previously (26,27). Histones were separated by 18% SDS, or 15% acid-urea (AU) polyacrylamide gel electrophoresis and analysed by western blotting as described previously (4). For mass spectrometry (LC–ESI/MS/MS, collaboration with B Thomas) HPLC grade reagents were used. Antibodies were used at the following dilutions: total H3 (1:5000; ab9050; Abcam); H3K14ac (1:5000; 07-353; Millipore); H3R2me2a (1:2500; ab80075; Abcam); Rabbit anti-H3K4me3 (1:50 000) and anti-H3K9ac (1:5000) antibodies were generated in this laboratory (4). Western blots were quantified using ImageJ having established the linear range, and corrected against histone H3 signal as a loading control. Results shown are the average of at least three independent experiments.
RNA extraction and northern blotting
Total cellular RNA was extracted from 2 × 107 cells by TRI RNA Isolation Reagent (Sigma-Aldrich). The DNA probes for the detection of the rnrB, repB, repD were PCR amplified according to previous reports (28,29).
Mixing experiments
Exponential suspension growth cells were harvested and washed twice with KK2 buffer. Cells were re-suspended in KK2 buffer at 1 × 107 cells/ml and then incubated with pre-warmed CellTracker™ Green CMFDA (5-chloromethylfluorescein diacetate) (C2925, Invitrogen) at 10 μM or equivalent volume of DMSO on a shaker at 220 rpm at 22°C for 45 min. The cells were washed twice with KK2 buffer and resuspended at 1 × 107 cells/ml. Labelled and unlabelled cells were mixed at different ratios and developed on Millipore filters on an LPS soaked pad as described previously. The spores were harvested after 2 days and visualized by fluorescence microscopy after staining all spores with DAPI (H-1200, Vector). The percentage of spores labelled with CellTracker™ Green was determined.
RESULTS
Expression of histone H3 variant proteins in Dictyostelium
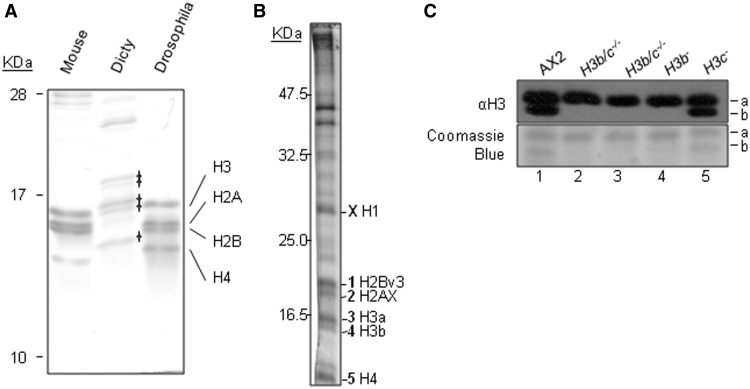
An optimized acid-extraction and electrophoresis protocol for histones from vegetatively growing Dictyostelium cells (27) generated preparations enriched in five major coomassie-stainable protein bands in the region where histones normally migrate on SDS–polyacrylamide gels (Figure 1A). These bands were subject to mass spectrometry (Figure 1B) and we identified histone H2AX [Band 2 (27)], H2Bv3 (Band 1) and H4 (Band 5), as previously reported (17) and also histone H1 [Band X; (30,31)]. The two genes encoding H4 encode proteins of identical amino acid sequence, so we have not been able to determine if both are expressed by this analysis. Of the three predicted H3 proteins, we found evidence for expression of H3a (Band 3) and H3b (Band 4) as peptides corresponding to regions that distinguished the two proteins were present. The observation of H3a in the upper of the two H3 bands is consistent with the presence of a three amino acid insertion in its amino terminal tail (see sequence comparison in Supplementary Figure S1). Differences in the length of the H3 N-terminal tail are highly unusual but not unique, as a number of predicted plant histone H3 proteins also contain small variations. The three extra amino acids in Dictyostelium H3a are all serines and phosphorylation in this region has been detected by mass spectrometry (17). No H3c-specific peptides were identified, although some of the H3b peptides identified would also be present in H3c.
Figure 1.
Histone H3 variants. (A) An optimized histone preparation by acid extraction of nuclei from exponentially growing Ax2 cells was analysed by 18% SDS–PAGE alongside with mouse and Drosophila preparations, visualized by Coomassie blue staining. Five major bands were apparent in the Dictyostelium sample in the area where histones are expected to run. The positions of mouse and Drosophila histones are marked. (B) Histone proteins from vegetative Dictyostelium cells were extracted by the optimized method and bands excised from the gel for analysis by mass spectrometry. Peptides derived from a number of histone proteins were identified, as indicated. (C) Expression of histone H3 variants in vegetative stage Dictyostelium were confirmed by western blot analysis of acid extracted histones from H3 variant deficient strains, H3b/c−/−, H3b− and H3c− using anti-histone H3 polyclonal antibody. Equal loading was confirmed by staining with coomassie blue. The second H3b/c−/− sample is from cells in which the Bsr cassette has been removed using cre recombinase.
To confirm the identity of H3 bands, we generated strains in which genes encoding individual H3 variants were deleted (Figure 1C), which is possible in this organism due to their existence as single copy genes. We were unable to generate strains carrying H3a gene deletions. This is unlikely to be due to any technical problem with targeting this gene because we were able to carry out gene replacements at this locus. The targeting fragment for disruption did hit the locus several times, but these were incomplete targeting events leaving the coding region intact; this shows that targeted recombination events do occur here. It seems likely therefore that H3a is an essential protein and its deletion results in inviability.
Western blotting analysis of histones from cells in which H3b, H3c or both were deleted, showed that antisera against H3 normally detect two bands but the lower H3 band is missing in strains in which the gene encoding H3b was deleted, either on its own, or with simultaneous deletion of that encoding H3c (Figure 1C). Histones extracted from the strain with only the H3c gene deleted were indistinguishable from parental cells (Figure 1C). Together with the mass spectrometry data above and consistent with the three amino acids insertion, this proves conclusively that the upper of the two bands contains H3a and that the lower band is predominantly H3b. The H3c protein, predicted to have the same molecular weight as H3b, is either expressed at levels too low to be detectable or not expressed at all in these cells. We were able to detect mRNA encoding H3c by reverse transcriptase PCR (data not shown). Low level of expression of this gene is consistent with RNA sequencing data which suggests far fewer transcripts from the gene encoding H3c than from those encoding H3a or H3b (available at dictybase.org). Quantification of the staining intensities of the H3a and H3b bands by Coomassie blue reveals that the lower band is ∼32 ± 3.9% (SDM, n = 6) of the total H3 protein.
Selective acetylation and methylation of histone H3 variants in Dictyostelium
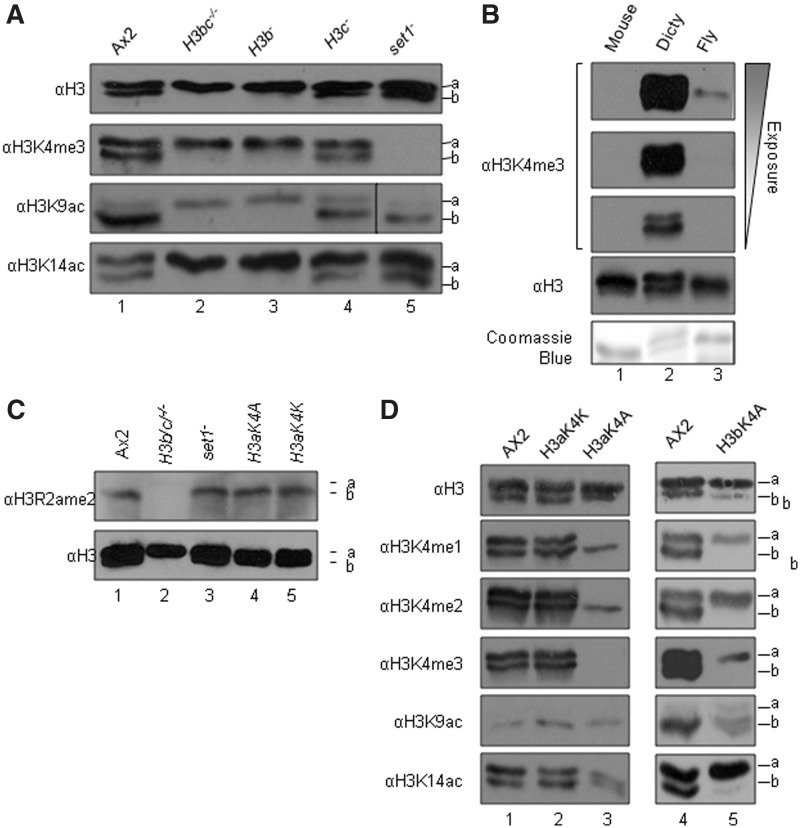
In Dictyostelium, methylation of histone H3 on lysines 4, 9 and 79 is known to be conserved, as is acetylation (17,19–21,32). The ability to separate H3a and H3b proteins by electrophoresis provides a unique opportunity to investigate differential modification of these two variants by western blot using well-characterized antisera recognising H3K9Ac and H3K4me3 (raised against peptides spanning N-terminal H3 residues 1–20 and 1–10) (5) (Figure 2A). Importantly, H3a, H3b and H3c are all identical in amino acid sequence from residue 1–19, so antisera against modifications in this region react equally well to all three variants. In parental Ax2 cells, equal amounts of K4me3 are detected in the upper and lower bands. As H3a is present at around twice the level of H3b, this suggests that H3b preferentially carries this mark. An important feature of H3K4 trimethylation in Dictyostelium is its presence in extremely high amounts compared to higher eukaryotes. Drosophila H3 yields an H3K4me3 immunostaining band approximately 11 times stronger than that seen in mammalian cells (6). However, neither mouse nor Drosophila H3K4me3 is even detectable at short exposures of blots when Dictyostelium H3K4me3 provides an optimal signal, despite containing equivalent amounts of H3 protein (Figure 2B). This shows that the relative level of Dictyostelium H3K4me3 massively exceeds that found in Drosophila or mouse. As this mark is associated with the 5′-ends of genes, this may reflect the relative gene density, there being much less non-coding DNA, <40% of the genome in Dictyostelium, compared to Drosophila or mouse (16). This is consistent with previous reports regarding high levels of this modification in Tetrahymena and yeast (33). Alternatively, it is possible that this mark is more widespread with an alternative role in Dictyostelium. All H3K4me3 disappears in cells in which the lysine methyltransferase Set1 is deleted (Figure 2A).
Figure 2.
Histone modifications on H3 variants. Acid extracts from nuclei of exponentially growing Dictyostelium cells (A, B, C) and mouse, Drosophila (fly) (B) were resolved by 18% SDS–PAGE and western blotted using antisera specific for H3, H3K4me3, H3K9ac, H3K14ac, H3R2ame2, H3K4me1 or H3K4me2 as indicated. The relative position of H3a (a) and H3b (b) are shown. (B) Different exposure times of the same blot are shown. (D) Six independent clones for H3aK4A and two for H3aK4K and H3bK4A were tested. Coomassie blue staining or blotting with anti-H3 was used to demonstrate equal loading of H3.
Selective variant modification is very obvious for H3K9 acetylation, where the majority (90 ± 2%) of this modification is found in H3b with relatively little detected on H3a (Figure 2A). In cells in which the gene encoding H3b is deleted (H3b/c−/− and H3b−), a low level of K9 acetylation remains detectable on endogenous H3a, and although there is an increase on H3a relative to parental Ax2 cells, the level of H3aK9Ac does not reach that seen on H3b in wild-type cells, so there is an overall reduction in the level of H3K9Ac (Figure 2A). By comparison, the distribution between variants is different for K14 acetylation, which is equally apparent on both variants and upon deletion of H3b, increases on H3a. No major change in the levels of K9 acetylation (Figure 2A) was seen in set1− cells which lack all K4 methylation (20). Analysis of three independent experiments revealed the level of K9Ac in set1−cells to be 0.97 ± 0.21 relative to a level of 1 in parental Ax2 cells.
Selective variant modification is also apparent for asymmetric dimethylation of arginine 2 (H3R2ame2), which is restricted to H3b and not re-distributed to H3a upon deletion of H3b (Figure 2C). H3R2me2a in the histone H3 tail has been reported to manage deposition of H3K4me3 in both mammals and yeast (34,35) with the presence of R2ame2 leading to loss of K4 trimethylation. Thus, H3 variants in Dictyostelium are clearly differentially utilized for different histone modifications.
Interaction between variants: K4A substitution on H3a results in loss of lysine 4-trimethylation on H3b
Muramoto et al. (2010) previously reported that replacing the gene encoding H3a with a mutated version in which K4 is replaced with alanine (K4A) led to complete loss of K4me3 in Dictyostelium (25) which suggested that only H3a was trimethylated at K4. This previous analysis was performed by western blot but under gel conditions which did not separate the H3a and H3b variants. In our data, we clearly see K4-trimethylation of both H3a and H3b (Figure 2A). To resolve the discrepancy, we analysed H3 variant methylation in H3K4A gene replacement strains (Figure 2D). As expected, we could detect no mono, di or trimethylation of the mutant H3aK4A. Surprisingly, however, expression of H3aK4A led to complete loss of detectable trimethylation of K4 on H3b and a reduction in the levels of mono and di methylation of this residue, though these were still detectable (Figure 2D). As a control for this unexpected and novel observation, the endogenous gene was also replaced with an H3K4K version; this behaved identically with wild-type, indicating that this is not an artefact of genetic manipulation procedures. This confirms the previous observation that replacement of H3a with a mutant containing non-modifiable alanine at K4 leads to complete loss of K4 trimethylation, but raises the new and intriguing question of why trimethylation of K4 on H3b is also lost. To determine if a reverse inhibition would occur if H3b was mutated we replaced the endogenous H3b gene with a version in which K4 was mutated to alanine. Interestingly in this case, trimethylation of K4 on H3a was still apparent, although somewhat reduced (Figure 2D). H3bK4A also shows a reduced level of K9 and K14 acetylation, but it should be noted that the total amount of H3bK4A is lower in these cells compared to usual levels of H3b. These experiments show that there is variant specificity in the ability of H3K4A expression to eliminate trimethylation of the other H3 variant.
Dynamic acetylation of Dictyostelium H3
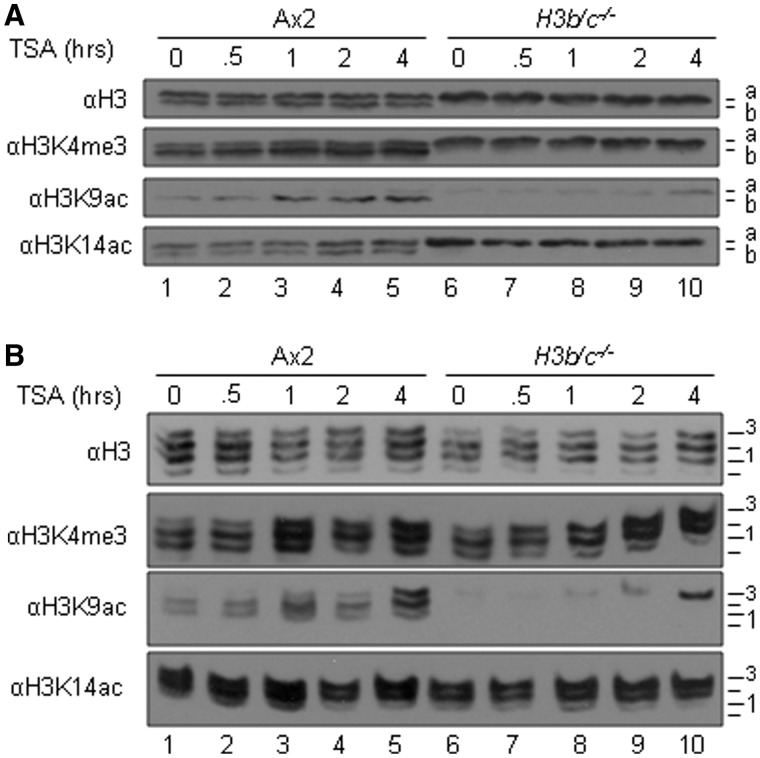
Conservation of the majority of known modification sites in H3a and H3b suggests conservation of the pathways and enzymes regulating these events. We have established that rapid dynamic turnover of acetylation of H3 targeted towards tails that are already modified by K4me3 is a common phenotype observed in human, mouse and Drosophila cells (5,6). This characteristic is visualized by treatment of cells with the histone deacetylase (HDAC) inhibitor Trichostatin A (TSA) and monitoring the rapid acetylation that follows by western blotting on SDS–PAGE (Figure 3A) and on AU gels (Figure 3B) where histone H3 is resolved into a ladder of bands corresponding to increasing modification. In these experiments, therefore, the upward shift of H3 bands upon TSA treatment indicates increasing acetylation. The evolutionary conservation of histone H3 modifications observed in Dictyostelium led us to investigate whether the same was true in this protist, and extend this to asking whether there was variant-selectivity in this phenotype.
Figure 3.
Dynamic acetylation of histone H3 is conserved in Dictyostelium and preferentially found on H3b. (A–B) Histone extracts from TSA (1 μg/ml, 3.3 μM) treated exponentially growing Ax2 and H3b/c−/− cells were isolated at various times of treatment up to 4 h and analysed by (A) 18% SDS–PAGE or (B) 15% acetic AU (AU) gel electrophoresis and western blots were performed with anti-H3K4me3, H3K9ac, -H3K14ac and -H3 polyclonal antibodies to show the induction level and distribution. A. Positions of histone H3 variants are shown on the right of each panel. (B) Position of differentially charged H3 species are labelled 0 = presumed unmodified to 3.
In the parental Ax2 cells, the distribution of modifications of histone H3 is exactly as described above, with H3K4me3 biased towards H3b and H3K9ac almost completely restricted to H3b. Upon TSA treatment, H3K9ac is clearly increased but remains predominantly on H3b, with only a slight increase seen in H3a. H3K14 acetylation, by contrast, is distributed between both H3a and H3b and only increases very slightly on both upon TSA treatment. In the absence of H3b, there is slightly more H3K9 acetylation on the H3a variant; TSA treatment causes a further increase but this does not reach the same total level as that seen in parental cells (Figure 3A and B, quantified in middle panel, Figure 4C). Acetylation at H3K14 was not increased by TSA treatment in the H3b/c−/− mutant (Figure 3A and B; middle panel, Figure 4C).
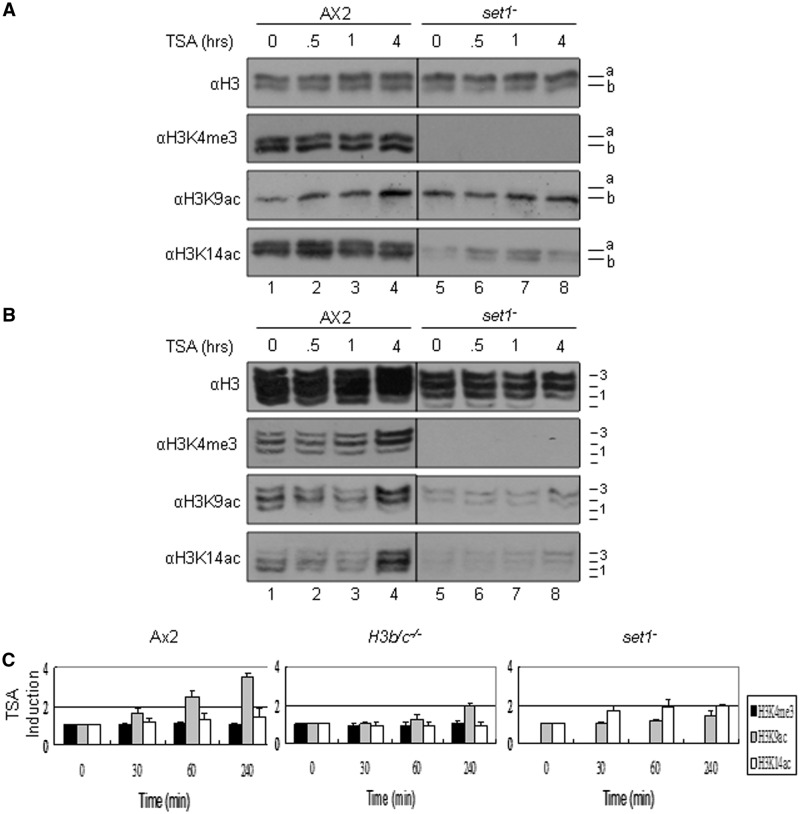
Figure 4.
Histone lysine 4 trimethylation is crucial for dynamic acetylation of histone H3 in Dictyostelium. (A–B) Histone extracts from TSA (1 μg/ml, 3.3μM) treated exponentially growing Ax2 and set1− cells were isolated at various times of treatment up to 4 h and analysed by (A) 18% SDS–PAGE or (B) 15% acetic AU (AU) gel electrophoresis and western blots were performed with anti-H3K4me3, H3K9ac, -H3K14ac and -H3 polyclonal antibodies to show the induction level and distribution. A. Positions of histone H3 variants are shown on the right of each panel. B. Position of differentially charged H3 species are labelled 0 = presumed unmodified to 3. (C) Quantification of changes in histone modification following TSA treatment of Ax2, H3b/c−/− and set1− cells. The western blots from SDS–PAGE analysis of three independent experiments as described in Figures 3A and 4A were used to quantify the modification levels, normalized to the level of total H3 extracted. In all cases the starting signal prior to TSA treatment is denoted as 1. Values are mean ± SD (n = 3).
To investigate whether TSA-stimulated acetylation is preferentially targeted to K4-trimethylated histone H3, these histones were also analysed on AU gels, where the extent of acetylation can be monitored by its position on the H3 ladder (Figure 3B). By blotting these gels with an anti-H3K4me3 antibody, any preferential acetylation similar to that seen in higher eukaryotes is apparent from its upward shift through the H3 ladder, relative to total H3. In both wild-type and H3b/c−/− cells, four clear bands (labelled 0 = unmodified to 3 in the figure) are apparent on the anti-H3 blots. Comparing the total H3 bands with H3K4me3, it is very clear that there is preferential acetylation of the latter, indicated by their shift towards the highest positions over this period, as the highest positions represent the most acetylated forms. This is especially clear in H3b/c−/− cells that are only expressing H3a, the remaining K4-trimethylated H3a is very clearly highly acetylated to occupy the highest positions on the ladder. This loss of signal at the lowest positions demonstrates that all trimethylated H3 of both variants is subject to dynamic acetylation. K14 acetylated bands occupy the highest positions on the ladder in untreated cells and are not significantly shifted further on TSA treatment.
These results demonstrate that H3K4me3 is clearly targeted for preferential acetylation in Dictyostelium and this link between modifications has arisen early in evolution. The response is slower than that in mouse and Drosophila as the shift is not apparent until 60 min of treatment whereas it is apparent after 15 min in mouse. Possible explanations include that Dictyostelium may be more resistant to TSA due to poor uptake or increased metabolism, or because of the extremely high level of K4methylated H3 in Dictyostelium.
TSA-stimulated acetylation is lost in set1− cells lacking H3K4me3
The above data indicates that the link between K4-trimethylated H3 and dynamic acetylation is observed in Dictyostelium and has arisen early in evolution. In contrast to mammalian and fly cells where multiple enzymes are responsible for modification of histones, it is possible in Dictyostelium to ask if there is a direct link between K4-trimethylation and dynamic acetylation by monitoring TSA-stimulated histone H3 acetylation in set1− cells that lack K4-methylation(19). TSA-stimulated histone H3 acetylation in these cells was analysed on SDS (Figure 4A) and AU (Figure 4B) gels. As shown previously, there is no H3K4 mono, di or trimethylation detectable in set1− cells (19) and we confirmed this using our antibodies (data not shown). The basal level of K14 acetylation was low in the set1− cells compared to wild-type, suggesting a link between acetylation and K4-methylation. Upon TSA treatment, the increase in H3K9 acetylation that is seen in the Ax2 cells over a 4-h time course is not observed in the set1− cells (Figure 4A and C). By contrast, similar analysis in cells containing disruption of the set2 gene which lack detectable methylation of H3K36 shows levels of TSA-induced K9 acetylation comparable to parental cells (Supplementary Figure S2), consistent with the loss of induction in set1− cells being specific. Therefore Set1 is essential for targeting rapid TSA-stimulated acetylation to histone H3, although it remains unclear if this is due to Set1 participating in protein complexes that mediate targeted methylation and acetylation of these tails, or if K4-methylation mediated by Set1 is an essential pre-requisite for its subsequent dynamic acetylation.
Genes encoding H3b and H3c confer an advantage to cells
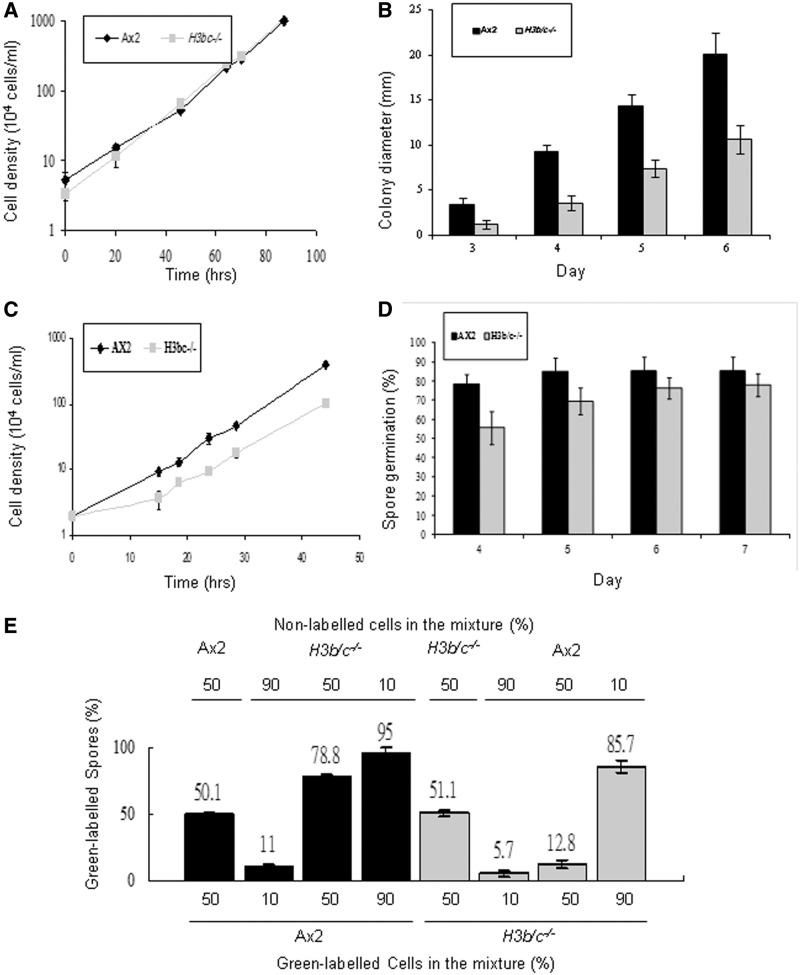
It is clear that H3a shows differential modification relative to H3b but the viability of H3b/c−/−cells reveals that H3b and H3c are not essential. Is there, therefore, any biological advantage to the cell in having these histone variants which would explain their conservation across evolution? To address this question we examined the growth rate of the H3b/c−/− cells in comparison to parental Ax2 cells. When grown in axenic media, we could detect no difference in growth rate between the strains (Figure 5A). However, in the wild these amoebae live in the soil and feed upon bacteria, so their growth rate on bacteria was also determined. When grown on a lawn of Klebsiella aerogenes, the colonies generated by the H3b/c−/− cells were consistently smaller than those from wild-type cells (Figure 5B). There are a number of reasons for reduced colony size, including changes in cell motility as well as growth rate, so we determined the growth rate of cells grown in a suspension of heat-killed bacteria. The H3b/c−/− cells showed a significantly reduced growth rate with a bacterial food source (Figure 5C).
Figure 5.
Dictyostelium H3b/c−/− cells show growth and developmental defects. (A) Axenic growth. Ax2 and H3b/c−/− cells were diluted to a density of 4 × 104 cells/ml and grown in shaken suspension in HL5 media at 22°C and cell numbers were determined at the indicated intervals. Values are mean ± SD (n = 4). (B) Exponentially growing Ax2 and H3b/c−/− cells were diluted and plated at low density on SM agar plates in association with Klebsiella aerogenes (Ka). Diameter of colonies derived from single cells was measured every day from the third to the sixth day. More than 30 random colonies were measured in each experiment and mean values of three independent experiments are shown ± SD. (C) Ax2 and H3b/c−/− cells were diluted to a density of 2 × 104 cells/ml and grown in shaking suspension in association with heat killed Ka (109 Ka/ml) at 22°C and cell numbers were determined at the indicated intervals. Values are mean ± SD (n = 4). (D) Three hundred Ax2 and H3b/c−/− spores were isolated from 2-day-old fruiting bodies and plated on a Ka lawn on SM agar plates. The number of visible colonies was counted each day for up to 7 days. Results are expressed as the percentage of spores plated. Values are mean ± SD (n = 3). (E) Exponentially growing Ax2 or H3b/c−/− cells were labeled with Green Cell Tracker Dye and mixed with unlabelled cells of the other genotype in the percentages indicated and the mixtures developed on Millipore filters on an LPS soaked pad for 48 h. In each case over 2000 spores were counted and the percentage of labelled spores is shown. In all cases the experiments were performed labelling the wild-type cells mixed with unlabelled H3b/c−/− cells and with labelled H3b/c−/− cells mixed with unlabelled parental cells to ensure there was no bias due to the labelling procedure. Controls were carried out in which labelled cells were mixed with unlabelled cells of the same genotype to confirm that the dye loading did not bias cell fate. Values are mean ± SD (n = 4).
The asexual life cycle of Dictyostelium is triggered by starvation and leads to generation of a fruiting body in which ∼80% of cells differentiate into spore cells with the potential to germinate and re-enter the proliferative cycle. The remaining 20% of cells terminally differentiate into stalk cells which will never re-enter the proliferative phase and so are clearly at a disadvantage. H3b/c−/− cells were able to generate fruiting bodies (data not shown). Development of this null strain was consistently somewhat prolonged compared to parental cells, taking up to 4 h longer to generate mature fruiting bodies, although the precise extent of the delay was somewhat variable (data not shown). It is clear that H3b and H3c are not necessary for the developmental process and differentiation, consistent with Drosophila where flies lacking expression of H3.3 are phenotypically normal (36,37). The fruiting bodies obtained from the H3b/c−/− cells contained similar numbers of spores and these spores germinated with efficiency indistinguishable from parental cells (Figure 5D).
A special feature of the Dictyostelium developmental process is that it is not necessarily clonal. The initial stages involve around 105 cells aggregating together and this initial aggregate can contain cells of different genetic backgrounds. In theory the proportion of cells of different backgrounds in the spore head should reflect the proportion entering the aggregate. However, it has become apparent that certain genetic backgrounds are able to form a higher proportion of spores than expected (38), clearly putting the ‘cheater’ cells at an advantage as they are overrepresented following spore germination. In order to determine the propensity of H3b/c−/− cells to form spores in chimaeric fruiting bodies, mixtures were generated with different proportions of parental and H3b/c−/− cells (Figure 5E). One of the two cell types was labelled prior to mixing and the proportion of spores generated by the labelled cells determined by counting. In these mixed fruiting bodies, the H3b/c−/− cells were less able to form spores than the parental cells containing all three H3 genes (Figure 5E).
The reduced growth rate when feeding on bacteria and the reduced proportion of spores formed in chimaeric fruiting bodies, suggests that the lack of the genes encoding H3b and H3c would be a selective disadvantage to cells outside the laboratory environment.
Gene expression in the absence of H3b and H3c
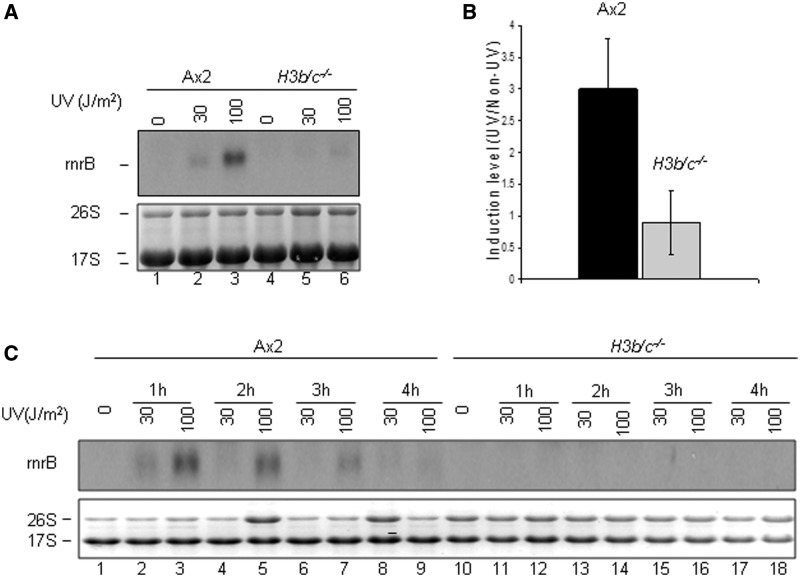
The ability of H3b/c−/− cells to grow and develop reveals that the majority of gene expression must be able to take place, with no catastrophic failures of gene regulation. This is consistent with data from Drosophila lacking one variant (37) and with our observations on the distribution of histone modifications associated with active gene transcription (4). However, this does not rule out the possibility that there are more subtle changes in the kinetics or extent of regulated gene expression. Relatively few genes have been identified in Dictyostelium whose expression is induced very rapidly on receipt of signal and known to be independent of protein translation, a so-called immediate early gene. One gene that does fall into that category is the rnrB gene which is induced in response to a number of agents which damage DNA such as UV light, even in the presence of cycloheximide (28). The rnrB gene was induced in a dose-dependent manner in Ax2 cells but induction was significantly reduced in H3b/c−/− cells (Figure 6A and B). Extending the analysis to 4 h following UV treatment demonstrated that induction was not simply delayed (Figure 6C). However, not all UV-inducible genes showed this dependence on H3b expression as repB and repD induction was comparable in the H3b/c−/− and Ax2 cells (Supplementary Figure S3).
Figure 6.
UV induction of rnrB gene in AX2 and H3b/c−/− cells. (A) Exponentially growing Ax2 and H3b/c−/− cells were exposed to UV light (30 or 100 J/m2), RNA was extracted after 1 h of recovery and expression of rnrB analysed by northern blot. Staining of ribosomal RNA was used to demonstrate equal loading of RNA. (B) The autoradiographs from six independent experiments as described in A were quantified. Bars represent mean ± SDM (C) Exponentially growing Ax2 and H3b/c−/− cells were exposed to UV light (30 or 100 J/m2). RNA was extracted after various times of recovery and analysed in A.
These results show that the absence of histone variants leads to subtle changes in gene expression that have the potential to alter cell behaviour and suggests that the evolution of histone variants has led to an important increase in complexity of nucleosome modification systems with functional consequences.
DISCUSSION
Two major features of the evolution of histone H3 genes in higher eukaryotes are first, the rise of variants, particularly their divergence into replication-dependent forms, such as H3.1 and H3.2 in mammals and replication-independent or ‘replacement’ forms such as H3.3, and secondly, the massive duplication of the genes encoding H3.1 and H3.2 to form highly repeated arrays (12,13). The three genes encoding H3 variants in Dictyostelium do not fall into the classic pattern for replication-dependent and -independent genes in higher eukaryotes. All three are single copy genes, although H3b and H3c form a tandem repeat, and all three have an intron towards the 3′-end (dictyBase.org) (Supplementary Figure S4). Protein sequence comparison reveals that all three Dictyostelium H3 variants contain features characteristic of H3.3 proteins and phylogenetic analysis is consistent with the divergence of the Dictyostelium variants prior to the split between H3.1/2 and H3.3 in mammals (Supplementary Figure S4).
The ability to separate H3a and H3b by electrophoresis has facilitated analysis of post-translational modification of variants using western blots. In other systems determining the modification status of histone variants relies either on expressing tagged versions of variants, purification of the variants and/or mass spectrometric analysis of long peptides which contain both the modified site and one or more variant-specific residues (13,39). All of these are problematic when compared to the western analysis of endogenous H3 described here. Some marks associated with promoters and active genes (H3K4me3 and K14Ac) are relatively abundant on both variants, whereas others show marked variant preference with K9Ac and asymmetric R2dimethylation both preferentially present on H3b. In the absence of H3b, an increased level of K9Ac, but no trace of aR2dimethylation, appears on H3a.
Cells in which endogenous H3a has been mutated to H3K4A show loss of K4me3 on H3b with reduced, though still detectable, levels of mono and dimethylation of H3b. The reason for this is not clear. When the equivalent substitution was introduced into the endogenous H3b, K4me3 on H3a was still detectable albeit at a reduced level. We have not detected any difference in the mRNA expression levels of a number of subunits predicted to encode known H3K4 methylases and demethylases (unpublished observations). H2b monoubiquitination is proposed to target H3 for K4 methylation, but we detected no evidence for difference in monoubiquitination levels and H3 aR2methylation levels were unchanged in H3K4A cells (unpublished observations). It is possible that this loss of H3bK4me3 reveals a hitherto unknown level of crosstalk between modifications on histone variants. In Drosophila, H3K4me3 levels are normal in flies lacking H3.3 compared to wild-type but dramatically reduced in flies expressing an H3.3K4A transgene (37). These results were interpreted to mean that this mark is normally carried by H3.3, H3.2 only becoming K4-trimethylated in the absence of H3.3. However our results showing crosstalk in K4-trimethylation between H3 variants in Dictyostelium suggest the possibility of an alternative interpretation.
This is the first demonstration in a protist of conservation of dynamic H3 acetylation and targeting of this modification to histone tails already modified by K4-trimethylation. This suggests that this pathway arose early in evolution and also offers the opportunity to study it by genetic means. Although both variants contain K4me3 and show TSA-induced acetylation, H3b is preferentially targeted for rapid acetylation of K9. Even in the absence of H3b, H3a shows little K9 acetylation. K9 acetylation may also be distinguished from K14 acetylation which is more equally distributed between the variants and which shows little increase on treatment with TSA. Mass spectrometric analysis in Arabidopsis revealed very few histone tails which were acetylated on both residues, consistent with these modifications not being commonly present on the same histone tails (40).
In Drosophila, flies lacking H3.3 develop phenotypically normally, although both sexes are sterile (36,37). The phenotypic defects we report here for H3b/c−/− cells are found during the asexual stages of the Dictyostelium life cycle, demonstrating that variants can play an important role outside the germ cells. This is consistent with the situation in mouse where loss of the H3.3-specific chaperone HIRA leads to early embryonic lethality, and developmental defects in an H3.3 hypomorph suggest a more important role for H3.3 in mammalian development (41,42). The reduced induction of the rnrB gene in response to UV would suggest that some genes are particularly sensitive to loss of the H3b and/or H3c variant and the phenotype would suggest that this includes one or more genes required for efficient growth on bacteria and for development in chimaeras.
In summary, we have demonstrated evolutionary conservation of the targeting of dynamic H3 acetylation to H3 tails containing trimethylated K4 in a protozoan. Loss of the H3b variant and general reduction in this targeted acetylation can be tolerated by cells and apart from the UV-inducible rnrB gene, the majority of gene expression is not compromised. Furthermore, cells expressing only one variant show reduced growth on bacteria and reduced capacity to generate spores in mixed populations revealing a potential role for H3 variants in the natural environment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
The Wellcome Trust [no. 081548/Z/06/Z], with some additional support from the EPA Cephalosporin Fund. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to all members of the Pears, Mahadevan and Chubb labs for their input, to Po-Hsien Liu for early experimental work and to B. Thomas for mass spectrometry.
REFERENCES
- 1.Hazzalin CA, Mahadevan LC. MAPK-regulated transcription: a continuously variable gene switch? Nat. Rev. Mol. Cell. Biol. 2002;3:30–40. doi: 10.1038/nrm715. [DOI] [PubMed] [Google Scholar]
- 2.Lee BM, Mahadevan LC. Stability of histone modifications across mammalian genomes: implications for 'epigenetic' marking. J. Cell Biochem. 2009;108:22–34. doi: 10.1002/jcb.22250. [DOI] [PubMed] [Google Scholar]
- 3.Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 4.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hazzalin CA, Mahadevan LC. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol. 2005;3:e393. doi: 10.1371/journal.pbio.0030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crump NT, Hazzalin CA, Bowers EM, Alani RM, Cole PA, Mahadevan LC. Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc. Natl Acad. Sci. USA. 2011;108:7814–7819. doi: 10.1073/pnas.1100099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahadevan LC, Willis AC, Barratt MJ. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell. 1991;65:775–783. doi: 10.1016/0092-8674(91)90385-c. [DOI] [PubMed] [Google Scholar]
- 8.Davie JR. MSK1 and MSK2 mediate mitogen- and stress-induced phosphorylation of histone H3: a controversy resolved. Sci. STKE. 2003 doi: 10.1126/stke.2003.195.pe33. 2003, PE33. [DOI] [PubMed] [Google Scholar]
- 9.Thomson S, Hollis A, Hazzalin CA, Mahadevan LC. Distinct stimulus-specific histone modifications at hsp70 chromatin targeted by the transcription factor heat shock factor-1. Mol. Cell. 2004;15:585–594. doi: 10.1016/j.molcel.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem. Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the "H3 barcode hypothesis". Proc. Natl Acad. Sci. USA. 2006;103:6428–6435. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: no substitute for histone H3.3. Curr. Opin. Genet. Dev. 2010;20:110–117. doi: 10.1016/j.gde.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiedemann SM, Mildner SN, Bonisch C, Israel L, Maiser A, Matheisl S, Straub T, Merkl R, Leonhardt H, Kremmer E, et al. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J. Cell Biol. 2010;190:777–791. doi: 10.1083/jcb.201002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strmecki L, Greene DM, Pears CJ. Developmental decisions in Dictyostelium discoideum. Dev. Biol. 2005;284:25–36. doi: 10.1016/j.ydbio.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevense M, Chubb JR, Muramoto T. Nuclear organization and transcriptional dynamics in Dictyostelium. Dev. Growth Diff. 2011;53:576–586. doi: 10.1111/j.1440-169X.2011.01271.x. [DOI] [PubMed] [Google Scholar]
- 18.Dubin M, Fuchs J, Graf R, Schubert I, Nellen W. Dynamics of a novel centromeric histone variant CenH3 reveals the evolutionary ancestral timing of centromere biogenesis. Nucleic Acids Res. 2010;38:7526–7537. doi: 10.1093/nar/gkq664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chubb JR, Bloomfield G, Xu Q, Kaller M, Ivens A, Skelton J, Turner BM, Nellen W, Shaulsky G, Kay RR, et al. Developmental timing in Dictyostelium is regulated by the Set1 histone methyltransferase. Dev. Biol. 2006;292:519–532. doi: 10.1016/j.ydbio.2005.12.054. [DOI] [PubMed] [Google Scholar]
- 20.Muller-Taubenberger A, Bonisch C, Furbringer M, Wittek F, Hake SB. The histone methyltransferase Dot1 is required for DNA damage repair and proper development in Dictyostelium. Biochem. Biophys. Res. Commun. 2011;404:1016–1022. doi: 10.1016/j.bbrc.2010.12.101. [DOI] [PubMed] [Google Scholar]
- 21.Sawarkar R, Visweswariah SS, Nellen W, Nanjundiah V. Histone deacetylases regulate multicellular development in the social amoeba Dictyostelium discoideum. J. Mol. Biol. 2009;391:833–848. doi: 10.1016/j.jmb.2009.06.067. [DOI] [PubMed] [Google Scholar]
- 22.Kuhlmann M, Borisova BE, Kaller M, Larsson P, Stach D, Na J, Eichinger L, Lyko F, Ambros V, Soderbom F, et al. Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi. Nucleic Acids Res. 2005;33:6405–6417. doi: 10.1093/nar/gki952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh M, Curk T, Xu Q, Zupan B, Kuspa A, Shaulsky G. Developmentally regulated DNA methylation in Dictyostelium discoideum. Eukar. Cell. 2006;5:18–25. doi: 10.1128/EC.5.1.18-25.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greene DM, Hsu DW, Pears CJ. Control of cyclin C levels during development of dictyostelium. PLoS One. 2010;5:e10543. doi: 10.1371/journal.pone.0010543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muramoto T, Muller I, Thomas G, Melvin A, Chubb JR. Methylation of H3K4 Is required for inheritance of active transcriptional states. Curr. Biol. 2010;20:397–406. doi: 10.1016/j.cub.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Clayton AL, Rose S, Barratt MJ, Mahadevan LC. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 2000;19:3714–3726. doi: 10.1093/emboj/19.14.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hudson JJ, Hsu DW, Guo K, Zhukovskaya N, Liu PH, Williams JG, Pears CJ, Lakin ND. DNA-PKcs-dependent signaling of DNA damage in Dictyostelium discoideum. Curr. Biol. 2005;15:1880–1885. doi: 10.1016/j.cub.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 28.Gaudet P, Tsang A. Regulation of the ribonucleotide reductase small subunit gene by DNA-damaging agents in Dictyostelium discoideum. Nucleic Acids Res. 1999;27:3042–3048. doi: 10.1093/nar/27.15.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SK, Yu SL, Garcia MX, Alexander H, Alexander S. Differential developmental expression of the rep B and rep D xeroderma pigmentosum related DNA helicase genes from Dictyostelium discoideum. Nucleic Acids Res. 1997;25:2365–2374. doi: 10.1093/nar/25.12.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parish RW, Schmidlin S. A lysine-rich protein functions as an H1 histone in Dictyostelium discoideum chromatin. Nucleic Acids Res. 1985;13:15–30. doi: 10.1093/nar/13.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hauser LJ, Dhar MS, Olins DE. Dictyostelium discoideum contains a single-copy gene encoding a unique subtype of histone H1. Gene. 1995;154:119–122. doi: 10.1016/0378-1119(94)00904-7. [DOI] [PubMed] [Google Scholar]
- 32.Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr. Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, et al. Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 34.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 35.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hodl M, Basler K. Transcription in the absence of histone H3.3. Curr. Biol. 2009;19:1221–1226. doi: 10.1016/j.cub.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 37.Sakai A, Schwartz BE, Goldstein S, Ahmad K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr. Biol. 2009;19:1816–1820. doi: 10.1016/j.cub.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santorelli LA, Thompson CR, Villegas E, Svetz J, Dinh C, Parikh A, Sucgang R, Kuspa A, Strassmann JE, Queller DC, et al. Facultative cheater mutants reveal the genetic complexity of cooperation in social amoebae. Nature. 2008;451:1107–1110. doi: 10.1038/nature06558. [DOI] [PubMed] [Google Scholar]
- 39.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 40.Johnson L, Mollah S, Garcia BA, Muratore TL, Shabanowitz J, Hunt DF, Jacobsen SE. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 2004;32:6511–6518. doi: 10.1093/nar/gkh992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol. Cell, Biol. 2002;22:2318–2328. doi: 10.1128/MCB.22.7.2318-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Couldrey C, Carlton MB, Nolan PM, Colledge WH, Evans MJ. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum. Mol. Genet. 1999;8:2489–2495. doi: 10.1093/hmg/8.13.2489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.