Abstract
In most bacteria, the timing and synchrony of initiation of chromosomal replication are determined by the binding of the AAA+ protein DnaA to a set of high- and low-affinity sites found within the origin of chromosomal replication (oriC). Despite the large amount of information on the role and regulation of DnaA, the actual structure of the DnaA–oriC complex and the mechanism by which it primes the origin for the initiation of replication remain unclear. In this study, we have performed magnetic tweezers experiments to investigate the structural properties of the DnaA–oriC complex. We show that the DnaA-ATP–oriC complex adopts a right-handed helical conformation involving a variable amount of DNA and protein whose features fit qualitatively as well as quantitatively with an existing model based on the crystal structure of a truncated DnaA tetramer obtained in the absence of DNA. We also investigate the topological effect of oriC’s DNA unwinding element.
INTRODUCTION
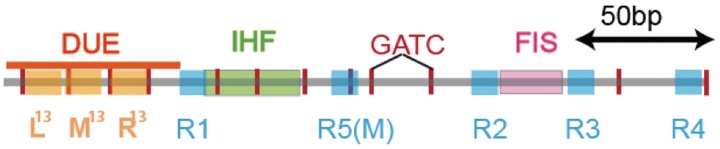
Initiators are trans-acting proteins that remodel DNA structure at the origin of replication where the replisome is recruited. Origin Recognition Complex (ORC) proteins are the initiators in eukaryotes and archaea while the DnaA protein is the initiator in most prokaryotes. Initiators of the different kingdoms share numerous features in their structure, for instance, the presence of AAA+ domains (1), as well as in their mechanisms of action (2–7). The DnaA-dependent initiation sequence in Escherichia coli is the most intensively studied system of initiation and the best understood (5,8): the ATP-bound form of DnaA binds the 9-mer DnaA-binding sites within oriC in a cooperative fashion followed by binding to the lower affinity, DnaA-ATP-specific sites (Figure 1). DnaA-ATP then invades the adjacent 30 bp A+T-rich DNA unwinding element (DUE), which contains additional 13-mer binding sites recognized by DnaA-ATP (Figure 1). Structural and biochemical assays suggest that this invasion stabilizes the single-stranded form of the DUE where the replication machinery is eventually recruited (8–11). The initiation of a round of replication stimulates the inactivation of oriC-bound DnaA by enhancing the rate of hydrolysis of the ATP bound to DnaA by the regulatory inactivation of DnaA (RIDA) process mediated by the Hda protein and the β-clamp (12). The hydrolysis-driven switch from ATP-bound DnaA to the ADP form is one of the most important processes that contribute to the inhibition of re-initiation of replication within the same cell cycle (13). The ADP-bound form of DnaA is able to bind to 9-mer DnaA-binding sites (R in Figure 1), with similar affinity compared to DnaA-ATP (14); however, it is unable to form multimeric structures characteristic of DnaA-ATP (8,15) and is inactive for initiation (16).
Figure 1.
The E. coli origin of replication bears five 9-mer DnaA-binding sites (R1, R2, R3, R4 and R5) as well as three 13-mer binding sites included in an A+T-rich DNA unwinding element (DUE). Although the 9-mers show no differential specificity between DnaA-ATP and DnaA-ADP, the 13-mers specifically recruit DnaA-ATP. In addition to DnaA-binding sites, oriC hosts-specific binding sites for IHF, SeqA and FIS, three proteins that regulate the activity of DnaA.
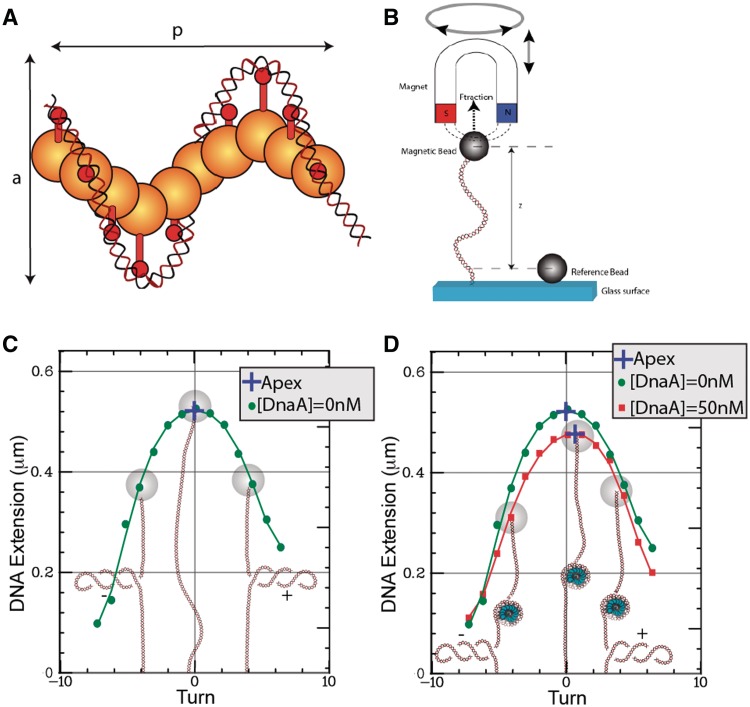
The oligomeric structure of DnaA is still a matter of debate. The four structural domains of the protein were first identified by sequence alignment, with domains I and III involved in DnaA–DnaA interaction and domain IV in DNA binding (17,18). Domain III contains the AAA+ motif that mediates ATP binding and hydrolysis. Early electron microscopy studies and bulk biochemical assays suggested that DNA wraps around a core of multiple DnaA proteins in a left-handed structure equivalent to negative supercoiling (6,9,19,20). More recently, crystallographic studies resolved the structural properties of DnaA, namely domain IV of DnaA from E. coli bound to DNA, as well as an oligomeric form of domains III and IV of DnaA from Aquifex aeolicus in the absence of DNA (21,22). Based on these structures, it has been proposed that DnaA could form an oligomeric right-handed helix with domain IV facing outward so that oriC would wrap around this protein scaffold (Figure 2A). However, when the DNA was modeled onto the crystal structure of the oligomeric form, domains III and IV exhibited steric clash with each other, requiring further investigation of the structure in the presence of DNA (15). Here, we show that magnetic tweezers can be used to assess the structural feature of the complex formed between wild-type DnaA and oriC from E. coli, in solution and at the single-molecule level.
Figure 2.
Single DNA micromanipulations with magnetic tweezers. (A) The DnaA-ATP–oriC is modeled as a right-handed helical structure with DNA-binding domain (red stick) facing outward. The structural properties of the helix are characterized by its pitch, p, and its diameter, a. (B) Sketch of the experimental setup: a single ∼2 kb DNA is attached at one end to a magnetic bead and at the other to a functionalized glass surface. A pair of permanent magnets located above the sample generates a vertical extending force, F, on the DNA and constrains the rotation of the bead and hence DNA’s supercoiling. In all experiments presented here, the force F = 0.2 pN. The end-to-end extension, z, of the DNA is measured in real time (30 Hz) by monitoring the bead position with ∼5 nm accuracy. To account for microscope drift, the position of a second bead fixed to the surface (reference bead) is simultaneously monitored. (C) Typical rotation–extension curve with, superimposed, the corresponding DNA conformation and polynomial fit. The apical point of the curve is computed by polynomial fitting and is indicated by a blue cross on the curve. (D) Rotation–extension curves, and their fits, performed on the same single DNA molecule containing the oriC sequence in the absence of protein (green circles) and in the presence of 50 nM of DnaA-ATP (red squares). As shown on the superimposed sketch, the change in the DNA conformation imposed by the formation of a nucleoprotein complex (blue structure) results in a shift in the coordinates of the apex. For this particular experiment, the shifts in apex coordinates are Δz = −45 nm and Δr = 0.54 turns.
Due to their ability to directly monitor at high-resolution protein–DNA interactions (down to subpiconewton forces and with nanometre-scale sensitivity), single-molecule nano-manipulation techniques (e.g. optical tweezers and magnetic tweezers) have been extensively used to determine the mechanical and structural properties of DNA both with and without DNA-binding proteins (23–27). Using a magnetic tweezers assay (Figure 2B), we were able to probe the structural properties of the DnaA–oriC complex and observe the formation of a highly stable positively supercoiled structure. These results are in agreement with the model introduced by Erzberger et al. (11,22) but in contradiction to the left-handed topology of the structure proposed by Bramhill and Kornberg (6).
MATERIALS AND METHODS
Protein expression and purification
The DnaA protein was over-expressed in E. coli strain WM2287 from the pdnaA116 plasmid. Protein over-expression was induced at OD600 = 0.5 for 3 h at 37°C by addition of 0.3 mM IPTG. The purification was performed as described in (28) with the modifications described below. After precipitation with 0.28 g/ml (NH4)2SO4, the pellet containing DnaA was re-suspended and desalted (Illustra NAP-25, GE Healthcare). The protein solution was then applied to a cation exchange resin (MonoS 5/50 GL, GE Healthcare) and fractionated against a 0.1–1 M gradient of potassium glutamate. Fractions containing DnaA were pooled and loaded onto a gel filtration column (Superdex 75 HR 10/300, GE healthcare) previously equilibrated in LG-buffer (45 mM Hepes/KOH, pH 7.6, 600 mM potassium glutamate, 1 mM DTT, 10 mM magnesium acetate, 0.5 mM EDTA and 10% glycerol). Purified fractions were pooled, snap-frozen and stored in individual aliquots at −80°C. Protein concentration was ∼5 μM as measured using a standard Bradford assay.
Single-molecule assay
Individual, linear, double-stranded DNA molecules 2 kb in length and containing oriC at their center were attached at one end to a 1 µm magnetic bead (MyOne Dynabeads, Dynal) and at the other end to a functionalized glass surface (Figure 2B) according to (29). To accomplish this, the 2 kb DNA was ligated at one end to a 1 kb DNA fragment multiply labeled with digoxigenin, and at the other end to a 1 kb DNA fragment multiply labeled with biotin. The DNA was then allowed to bind to streptavidin-coated magnetic beads. The bead–DNA solution was then deposited on an anti-digoxigenin-coated glass surface, and, when the DNA was bound to the glass surface, a tethered DNA–bead system formed. The sample was placed on an inverted microscope through which bead images can be viewed and tracked by computer-aided videomicroscopy (PicoTwist). To compensate for microscope drift, the position of a second bead, directly fixed to the surface of the capillary, was simultaneously monitored (Figure 2B). From the difference in z positions between the two beads (the magnetic bead and the bead attached to the surface), the end-to-end extension, z, of the DNA was determined. A pair of permanent rare-earth magnets located above the sample is used to generate a magnetic field with which the bead, and hence the DNA, is manipulated. The magnets are mounted on computer-controlled translation and rotation stages. The vertical distance between the magnets and the glass surface determines the vertical extending force applied to the DNA via the bead [for these experiments, the force is set to a very weak value, namely 0.2 pN (1 pN = 10−12 Newtons)], while the angular position of the magnets determines the angular position of the bead and hence quantitatively sets the supercoiling of the DNA. The extension of the DNA can then be measured as a function of its mechanical constraints; in the experiments described here we vary supercoiling of the DNA by rotating the magnets, while keeping the extending force constant by holding constant the position of the magnets above the sample (see below).
Unless specified otherwise, the standard conditions under which experiments were performed were at 34°C with 50 nM DnaA in buffer consisting of 25 mM Hepes/KOH, pH 7.6, 100 mM potassium acetate, 5 mM magnesium acetate, 0.1% Tween-20, 0.5 mg/ml BSA, 300 µM Mg-ATP and 170 pM of 2 kb competitor DNA. These conditions are referred to in the text as the standard conditions. Prior to injection into the reaction chamber, DnaA was diluted to 50 nM in the reaction buffer described above and pre-incubated at room temperature for 30 min. Experiments on oriCΔDUE were conducted at both 50 and 75 nM of DnaA.
Rotation–extension curves
As mentioned above, the field generated by a pair of permanent magnets constrains the rotation of the magnetic bead and applies an extending force of 0.2 pN to the DNA [as calibrated from the thermal fluctuations in the bead’s position (30)]. Rotation of the magnets by one turn induces rotation of the magnetic bead by one turn and results in a change in the topology of the DNA [quantified by the linking number, Lk (31)] of +1 for counterclockwise rotation and −1 for clockwise rotation (as viewed from above the bead). Under the weak extending force employed here, supercoiled DNA adopts a plectonemic conformation where the DNA double helix is intertwined upon itself (Figure 2C). As the DNA in the plectonemic phase does not participate in the vertical extension of the DNA tether, the growth of the plectonemic phase decreases the distance between the bead and the surface. Figure 2C shows the vertical extension of a single DNA molecule as a function of the rotation of the magnets (rotation–extension curve). Under our physico-chemical conditions, the rotation–extension curve is symmetric for naked DNA, with an apex representing the torsionally relaxed state of the DNA. This apical point of rotation–extension curves can be computed with subrotational and nanometric accuracy by fitting the rotation–extension curves with a polynomial function of the sixth degree.
Single-molecule detection of a DnaA–oriC complex
So as to characterize the DNA structure stabilized by DnaA, we proceed as follows: we first measure a rotation–extension curve of the tethered DNA molecule in the absence of protein (Figure 2C). Then, the protein is added to the sample chamber and incubated for 15 min before measuring a second rotation–extension curve (Figure 2D). The shift in the position of the apex provides structural information on the complex formed on the DNA. Indeed, the shift in the position of the apex along the rotation axis (Δr) relates to the topology of the nucleoprotein complex (see Supplementary Data). Since the fraction of DNA involved in the complex no longer participates in the total vertical extension of the DNA (see Supplementary Data), a reduction of the extension at the apex (Δz) is also observed. The measure of (Δz, Δr) on several single molecules can then be reported on a 2D plot (Figure 3) to infer the structural properties of the underlying DnaA–oriC complex.
Figure 3.
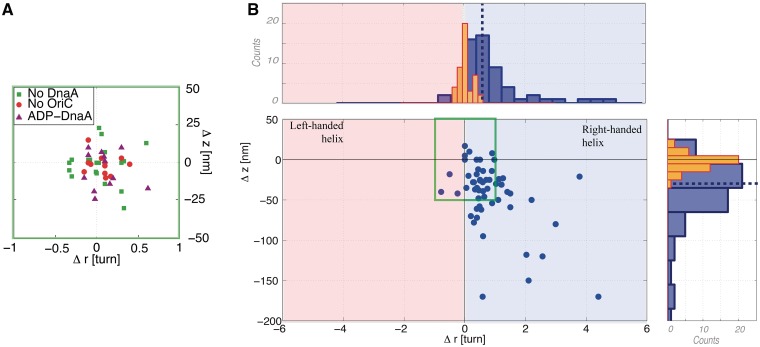
Shifts in apex coordinates (Δr, Δz) obtained after incubating nanomanipulated DNA with DnaA under various conditions and for a fixed extending force F = 0.2 pN. (A) Shift in apex coordinates for: (i) no DnaA present in the injected solution (green squares, n = 20), 〈Δr〉 = 0.04 ± 0.06 turns (SD = 0.25 turns) and 〈Δz〉 = 1 ± 3 nm (SD = 12 nm), (ii) DNA lacking oriC sequence (red circles, n = 13): 〈Δr〉 = 0.08 ± 0.04 turns (SD = 0.16 turns) and 〈Δz〉 = 1 ± 2 nm (SD = 6 nm) and (iii) when ADP is substituted to ATP in the reaction mix (purple triangles, n = 12): 〈Δr〉 = 0.09 ± 0.06 turns (SD = 0.21 turns) and 〈Δz〉 = −5 ± 3 nm (SD = 12 nm). (B) Shifts in apex coordinates under standard conditions: DNA bearing the oriC sequence, DnaA-ATP (blue circles, n = 57). Pink is the predicted area for a left-handed helical complex and the light-blue region is the predicted area for a right-handed helical complex. The corresponding projection on both axes is depicted in blue on both the upper histogram (projection along the axis of rotation) and right histogram (projection along the axis of extension). The orange bars correspond to the distribution of the combined controls from panel A. The dotted line corresponds to the maximum of likelihood for the Gaussian fit applied to the clustered points along the two axes as calculated in Supplementary Figure S3 (see main text for details).
RESULTS
Method resolution
Our approach holds intrinsic limitations on the amount of data points that can be acquired within one experimental session and the accuracy of the measured parameters. Indeed, each experimental point relating apex coordinates shifts is based on a measurement performed on a single DNA molecule, which cannot be recycled for a new, independent measurement due to the very high stability of the nucleoprotein complex (see below and Supplementary Data). In order to test the intrinsic resolution of the method, we first performed experiments in the absence of DnaA. Thus, after measuring the first rotation–extension curve, we performed a blank injection of reaction buffer containing no DnaA, and then re-measured the rotation–extension curve. Analysis of these experiments shows that the values for (Δr, Δz) measured on individual molecules are centered about zero with a mean change in rotation Δr = 0.04 ± 0.06 turns (SD = 0.25 turns, n = 20) and a mean change in extension Δz = 1 ± 3 nm (SD = 12 nm, n = 20) (Figure 3A, green squares). Since no structural change is induced on the DNA in the absence of protein, this first control is a direct evaluation of the intrinsic resolution of our approach.
Structural properties of the DnaA–oriC complex, specificity and requirement for ATP
In order to study the structural properties of the DnaA–oriC complex, we performed experiments in which DnaA-ATP is injected into the reaction cell and incubated for 15 min prior to measuring the second rotation–extension curve to determine shifts in apex coordinates (Δr, Δz) (Figure 3B, blue circles).
From a qualitative standpoint, the data indicate that the vast majority (80%) of complexes induce a positive shift in DNA topology accompanied by a decrease in DNA extension. Comparison of these data with values obtained after blank injections shows that the shifts in apex coordinates after incubation with DnaA-ATP are statistically significant as they lie above the resolution limit as determined from the control experiments (Figure 3A and histograms in Figure 3B). We thus conclude that these shifts in apex coordinates reflect a structural change induced by DnaA-ATP on the DNA. From a more quantitative standpoint, 85% of data points cluster in a Gaussian fashion with mean shift in extension Δz = −32 ± 5 nm (SD = 28 nm, n = 48) and mean shift in rotation Δr = 0.60 ± 0.05 turns (SD = 0.37 turns; n = 48) (Supplementary Figure S3). While most data points fall into this cluster, roughly 15% appear as outliers with either |Δz| > 100 nm or Δr > 2 turns, giving mean values Δz = −129 ± 26 nm (SD = 77 nm, n = 9) and Δr = 2.8 ± 0.5 turns (SD = 1.4 turns; n = 9). We note that the ratio Δz/Δr is similar for points in the cluster and the outliers (respectively, −53 ± 9 nm/turn and −46 ± 12 nm/turn). Taken all together, the data gave a mean shift in rotation Δr = 0.91 ± 0.14 turns (SD = 1.08 turns, n = 57) and a mean shift in extension Δz = −47 ± 7 nm (SD = 51 nm, n = 57; Δz/Δr = −52 ± 9 nm/turn).
Whereas the standard deviation of experimental points measured in the control experiments is due to the intrinsic error in determining the position of the apex, when DnaA is present the standard deviation increases ∼2-fold (for data points that distribute in a Gaussian fashion). This significant increase in variance is not unusual for protein–DNA interactions (23) and can be a sign of molecular heterogeneity, in other words of variability between different DnaA–oriC complexes. Conformations and localization of monomers may vary slightly from filament to filament, for instance, or the number of DnaA oligomers involved in the complex, and thus its extent, could vary from complex to complex leading to additional variance in the observed shifts. As the observed standard deviation is the sum in quadrature of the ‘intrinsic’ standard deviation (related to measurement error) and the ‘extrinsic’ standard deviation (DnaA-related variability), we estimate that DnaA-related variability contributes 0.27 turns and 25 nm to the observed experimental standard deviation.
To determine the specificity of formation of this complex, we investigated the response of DNA molecules lacking the oriC sequence to incubation with DnaA-ATP and competitor DNA at the same concentrations as in the previous experiments. With DNA substrates lacking oriC, no significant shifts in the apex’s position were observed in the presence of DnaA-ATP (Figure 3A, red circles), essentially identical to control results obtained in the absence of DnaA. This shows that the formation of the complex, as observed in our assay, is dependent on the presence of the oriC sequence.
To determine the role of the available nucleotide in formation of the complex, we replaced ATP by ADP in the reaction mix (ADP was also used instead of ATP in the pre-incubation step, see ‘Materials and Methods’ section). In these conditions, no significant shifts in apex coordinates were observed (Figure 3A, purple triangles). This confirms that the binding of DnaA-ADP to oriC does not result in any structural changes detectable by our assay. Since it has been shown that DnaA-ADP is unable to form an oligomeric structure this implies that the signal observed in the presence of DnaA-ATP results from an oligomerization of the protein dependent on the presence of the DnaA sites at the origin.
In summary, the above experiments demonstrate that the DNA present in the DnaA–oriC complex possesses a preferred topology and level of compaction, and furthermore that the nucleoprotein complex we observe is both sequence- and nucleotide specific.
Characterization of oriC mutants lacking DUE
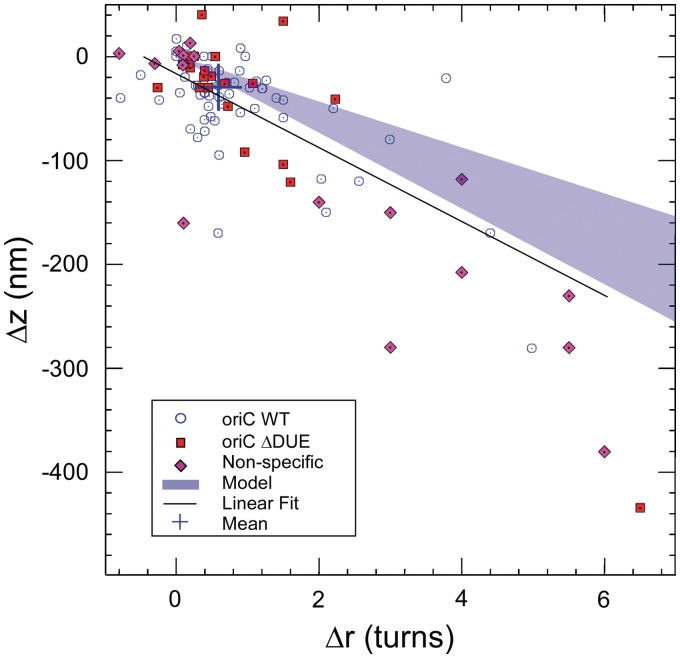
The DUE plays a key role during DnaA-mediated initiation of replication at oriC. This A+T-rich region of oriC has traditionally been believed to undergo melting over several turns of the double helix during formation of the DnaA–oriC complex (9,32), although recent structural results (see (11), discussed below) may lead to a re-examination of this model. Nevertheless, simple unwinding of the DUE would be predicted to have a topological effect that could be directly investigated with our approach (23) (see Supplementary Data). We thus conducted experiments on a mutant of oriC lacking the DUE segment (oriCΔDUE; Figure 4, red squares). Under those conditions, the shifts in apex coordinates induced by the formation of the initiation complex fall in the same range as the ones resulting from standard conditions (Figure 4, blue circles). We conclude that the complexes we detect formed on oriC wt or on oriCΔDUE share the same structural properties, and thus that in this topological assay, the DUE does not significantly affect the overall topology of the DnaA–oriC complex. This somewhat surprising result is further discussed below, in particular in light of recent structural data pertaining to the structure of a DnaA–ssDNA cocrystal.
Figure 4.
Comparison with model for DnaA-ATP–oriC nucleoprotein filament. The shaded area reflects the experimental area accounted for by the structural model proposed by Erzberger, Mott et al. using the mechanical model introduced by Hegner et al. (22,33). In this case, the radius and pitch of the helix are as discussed in the Supplementary Data, and the shaded area is delimited at bottom by the case for a flexible helical filament (Ξ = 30 nm), and at top by the case for a rigid helical filament (Ξ = 180 nm). Empty blue circles: standard conditions (from Figure 3). Red squares: DNA bearing a mutated version of oriC lacking the DUE (oriCΔDUE, n = 15). Purple diamonds: DNA lacking oriC and incubated with DnaA in the absence of bulk competitor DNA (n = 16). The complete data set collected under standard condition are linearly fitted (solid line, y = Ax + B with A = −31 nm/turn and B = −20 nm). The blue cross represents the mean for points which distribute in a Gaussian fashion.
Formation of a non-specific complex
All the above experiments were carried out in the presence of bulk, non-specific, 2 kb competitor DNA (170 pM) that does not contain the oriC sequence. This is required in order to distinguish specific from non-specific effects of DnaA binding. Indeed, results obtained using the control DNA substrates lacking oriC, but this time in the absence of competitor DNA (Figure 4, purple diamonds) show that the presence of a large excess of DnaA-ATP compared to the amount of tethered DNA molecules within the capillary (on the order of attomolar) can result in the formation of oligomeric complexes independently of the presence of specific binding sites.
Stability of the DnaA-ATP–oriC complex
Washing the sample with reaction buffer does not result in a significant dissociation of the protein over a period of 1 h. In addition, the change in bead rotation to create a negatively supercoiled DNA does not subsequently affect the stability of the nucleoprotein complex (Supplementary Figure S1).
A model for the mechanical and structural properties of the DnaA–oriC complex
The crystal structure of the oligomeric form of DnaA (PDB entry 2HCB) provides geometrical information which can serve as a starting point for predicting the shift in the apex position induced by formation of the nucleoprotein complex (see Supplementary Figure S2 and Figure 4). Erzberger et al. propose that DNA tracks the outside edge of the DnaA helix, leading us to model the nucleoprotein helix as having the same pitch as the protein helix P = 17.8 nm, but with a radius R = 7.1 ±1 nm (R = Rhelix + rDnaA + rDNA, with the protein helix radius Rhelix ∼ 4.5 ±1 nm, the protein monomer radius rDnaA ∼ 1.6 ± 0.3 nm, and the DNA radius rDNA = 1 nm as determined from the respective crystal structures).
Our observation that complex formation causes the apex position to shift to more positive values of supercoiling, while also decreasing in extension, is qualitatively consistent with this model. Indeed, sequestering a certain contour length of DNA along the outer surface of such a wide, right-handed, protein-determined suprahelix is expected to both decrease the extension of the DNA and ‘lock in’ positive DNA topology, causing one to find the topologically relaxed state of the DNA for more positive values of rotation than in the absence of such a structure. Thus formation of one helical pitch of right-handed nucleoprotein filament with the dimensions cited above has a topological ‘cost’ of Δr = 0.63 turns (see Supplementary Data).
Next, to quantitatively relate the shift in DNA rotation to the shift in DNA extension one must have information on how the complex is extended by the applied force, which depends on the mechanical bending rigidity (or persistence length) of the complex. However, such information is not directly provided by the crystal structure. In addition, it cannot be derived experimentally with our approach because only a very small portion of the total DNA, on the order of 10%, is involved in the complex. Nevertheless, Hegner et al. (32) developed an analytical model to compute the mechanical properties of a helical protein complex from the pitch and radius of the helix and the thickness of the protein monomer as measured on the crystal structure. This model successfully predicted the bending rigidity of a RecA–DNA complex as measured experimentally with optical tweezers (33). DnaA and RecA are closely related, with similar helical structures when bound to both dsDNA and ssDNA (11). In order to estimate the mechanical properties of the DnaA–oriC filament, we have applied this analytical model to the structural model introduced by Erzberger et al. and computed the bending rigidity of the complex (the DNA does not affect the bending modulus of the helical complex, see Supplementary Data). The predicted persistence length of the complex is ∼60 nm, with values ranging from 30 to 180 nm given the errors in estimating the radius of the protein helix and the DnaA monomer.
From the structural model and the estimates of the filament’s persistence length, we have computed the range of possible ratios Δz/Δr (see Supplementary Data). By considering a single pitch of helical DnaA–oriC nucleoprotein filament with small persistence length (30 nm) we find Δz/Δr = −36 nm/turn, while for a single pitch nucleoprotein filament with large persistence length (180 nm), we estimate Δz/Δr = −22 nm/turn. These delimiting boundaries are shaded on the (Δz, Δr) plot shown in Figure 4, with the mean measurement of experimentally observed shifts lying just outside of this zone (with Δz/Δr = −53 ± 9 nm/turn). Thus, the closest fit to the model would take place for the shortest persistence length, which would predict a value slightly smaller than what we observe after taking into account measurement uncertainty. For the model to better include our mean measurement would require an increase in the radius of the nucleoprotein helix on the order of a few nanometers, or a further decrease of the persistence length of the nucleoprotein helix, or a combination of the two (see Supplementary Table S1). Both of these are plausible although the model is more sensitive to helix radius than persistence length. For instance, in the nucleoprotein filament model proposed by Erzberger et al., domain IV requires a rotation away from its position as seen in the crystal structure in order to bind the DNA. The range of angles which are compatible with binding to DNA suggest an increase (on the order of 10%) of the radius of the nucleoprotein helix formed. Furthermore, crystal packing forces would also be expected to act in a predominantly lateral fashion on the protein helix and tend to reduce its radius relative to its solution state. Both of these effects would lead to better inclusion of our mean measurement in the bounds of the model (see Supplementary Table S1). Finally, we note that an increase in helix radius couples to a decrease in helix persistence length in the Hegner model, underscoring the internal consistency of selecting the lower bounds for the persistence length while simultaneously proposing that the nucleoprotein helix may in fact be slightly wider than proposed by Erzberger et al.
With a mean change in rotation Δr = 0.6 for points in the cluster, the structural model suggests that roughly one pitch of nucleoprotein helix is formed (see Supplementary Data). With ∼50 nm (∼170 bp) of DNA sequestered in one pitch of the complex, we estimate that this is the mean amount of DNA involved in the cluster of complexes we observe. Based on the crystal structure, this would involve on average approximately 8 molecules of DnaA, with fluctuations on the order to 3–4 monomers based on our estimate of DnaA-related variability.
Clearly, this amount can vary quite widely, as the significant presence of outliers beyond the Gaussian cluster attests, and for the entire dataset the mean amount of DNA engaged in the complex approaches 260 bp. These values are consistent with the spatial extent of oriC (245 bp) excluding the DUE. Thus, overall, a geometrical model accounting for polymer elasticity captures the main features of the experimentally derived change in DNA conformation upon DnaA binding to oriC.
DISCUSSION
We report a structural study of the DnaA–oriC complex using single-molecule magnetic tweezers. Control experiments where either DNA or DnaA is inactive for initiation (DNA lacking oriC or DnaA-ADP) demonstrate that the magnetic tweezers assay allows for characterization of the effect of the specific binding of DnaA-ATP to oriC and the formation of a complex at the single-molecule level.
As shown by the slow dissociation measured here upon washing the sample with buffer, the DnaA-ATP–oriC oligomeric complex appears to be very stable (see Supplementary Figure S1). Previous work by Messer and coworkers (8,34) measured the dissociation constant from short DNA fragments containing one or two DnaA sites but not from the complete origin sequence. This new result is consistent with recent work showing that the binding of DnaA to the origin inhibits re-methylation of the newly synthesized DNA strand for a significant amount of time after initiation has taken place (35). This emphasizes the role of auxiliary proteins for the regulation of the initiation of replication. In addition, the presence of DnaA-binding sites that do not discriminate between DnaA-ATP and DnaA-ADP (R boxes in Figure 1) have been shown to play an important regulatory role in the activity of the origin (36), indicating that the binding of DnaA-ADP could also play a role in the regulation of the formation of the DnaA-ATP oligomeric complex at oriC. Furthermore, systems such as RIDA appear to be essential to undo the complex, and to render oriC available for the next replication round (37).
The distribution of the shifts in the position of the apex resulting from the formation of a DnaA-ATP–oriC complex (Figure 4, blue circles) displays numerous features that can be linked to the underlying structure of the complex. First, Δr is nearly systematically positive and Δz is nearly systematically negative. These points imply, respectively, that the topology of the DNA within the complex is equivalent to positive supercoiling, and that DNA contour length is sequestered by the nucleoprotein complex. Second, Δz and Δr are strongly correlated for each molecule (with a P-value < 10−3 according to Pearson’s test). This implies that the relationship between contour length of DNA sequestered in the complex and topology of that DNA is non-random and instead reflects the structure of the complex. This is supported by the fact that both points in the cluster and outliers have the same ratio <Δz>/<Δr>. Finally, the values for the shift in apex position in rotation and extension are widely spread, with Δr of up to five turns and Δz reaching −280 nm. In summary, our results in standard conditions support a structural model for a nucleoprotein filament which is a (i) topologically right-handed (ii) helix with (iii) variable extent:
(a) While Bramhil et al. have suggested a left-handed complex, the crystal structure described by Erzberger et al. supports a right-handed helix (6,22). Our observation that Δr is nearly systematically positive strongly supports the right-handed helical model and excludes the left-handed model.
(b) DnaA is a member of the AAA+ family of proteins that includes numerous proteins that form oligomers (usually 6–7) with a donut-like shape, such as MCM (38) and ClpX (39) or a helical shape such as RecA (40,41). Recent structural studies show DnaA forms a right-handed helix (11,21) and our observation of coupled changes Δz and Δr supports such helical conformations. The structural parameters proposed by Erzberger et al. (pitch and radius of the complex) predict the coupled changes (Δr, Δz) represented by the shaded area in Figure 4, which captures the essence of our experimental observations. This suggests that binding of DnaA to oriC does not significantly affect the oligomeric form of the protein observed by crystallography in the absence of DNA.
(c) As suggested by Leonard and Grimwade (42) in a recent review, we observe that DnaA is prone to oligomerize beyond oriC and vary in the extent of the complex formed. In Leonard’s model, the formation of DnaA–oriC complex nucleates at high-affinity sites and propagates to lower affinity or cryptic sites, by ‘filling the gap’ (42). Tolerance for variability in helix formation from one DNA molecule to the next could be a consequence of the success of DnaA in driving initiation of replication for numerous different oriC organizations (43). It could also reflect the fact that the filament is structurally heterogeneous, with different monomers engaged on the DNA to different extents. Furthermore, the numerous regulatory proteins that target the formation of the DnaA–oriC complex to fine-tune the rate of initiation would act by changing the availability of the gap between DnaA-binding sites. For instance, following initiation, SeqA binds to hemimethylated oriC, preventing DnaA from forming a new active complex at oriC and inhibiting reinitiation during the same cycle (44). In addition, more recently SeqA has also been shown to direct the correct sequential binding of DnaA at the origin (45).
Effect of the DUE
One of the major transitions during the initiation process is the DnaA-dependent denaturation of the DUE region where the replisome assembles starting with DnaB and DnaC (9,46). The form taken by DNA in the DUE has long been a topic of controversy. On one hand, the susceptibility to chemical attack shown by this A+T-rich region in the presence of DnaA led to the suggestion that it became topologically unwound by DnaA (9,47). More recently, the structure of the DnaA filament in complex with ssDNA indicates that the nucleic acid is highly extended in a manner similar to that seen in the RecA–ssDNA filament (11). The structure does not provide information, however, on the topology of the two strands of DNA, which is what is detected in our assay. Thus, our method is either unable to detect DnaA-driven unwinding of the DUE, for instance, if it occurs with both DNA strands sequestered on the surface of the protein filament, or alternatively additional protein components such as IHF and/or SSB will be required to form and/or capture a DUE transiently unwound by DnaA.
CONCLUSIONS
Using magnetic tweezers, we have obtained topological information on the nucleoprotein filament formed on the origin of replication by DnaA-ATP, the protein responsible for initiating DNA replication. We have observed that the oriC sequence involved in the complex adopts a right-handed helical structure. These features do not fit the prediction for the structure of DnaA–oriC proposed by Bramhill and Kornberg (6), but they do fit, both qualitatively and quantitatively, the one introduced by Erzberger et al. (22). Although we were unable to unequivocally detect melting of the DUE, recent data on the DnaA–ssDNA structure suggest that such melting could be more complex than simply topologically driven, i.e. compensatory melting due to formation of the adjacent, positively supercoiled nucleoprotein helix. This helical structure is very similar to the hexameric structure of the initiator ORC proteins from archaea and eukarya, two members of the AAA+ superfamily and emphasizes the universality of the initiators’ structure (7,15,22). In addition, this helical structure and the observed propensity to vary in extent of the complex formed echoes the model for regulation of initiation introduced by Leonard and Grimwade (42). By providing a way to detect binding of DnaA to oriC in a single-molecule assay, these experiments with DnaA may allow for progressive reconstitution of replication initiation, for instance by forming a platform from which to detect DNA unwinding by DnaB helicase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–3 and Supplementary Data.
FUNDING
The authors wish to acknowledge funding support from the CNRS ATIP Program and European Science Foundation EURYI Program (to T.R.S.), as well as from the C'NANO IDF and Association pour la Recherche sur le Cancer programs (to S.Z.). Core funding was also provided by the CNRS and the University of Paris Diderot. Funding for open access charge: CNRS core funding.
Conflicts of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Masamichi Kohiyama and John Marko for useful discussions as well as Rob Phillips for comments on the manuscript.
REFERENCES
- 1.Giraldo R. Common domains in the initiators of DNA replication in Bacteria, Archaea and Eukarya: combined structural, functional and phylogenetic perspectives. FEMS Microbiol. Rev. 2003;26:533–554. doi: 10.1111/j.1574-6976.2003.tb00629.x. [DOI] [PubMed] [Google Scholar]
- 2.Georgescu RE, O'Donnell M. Structural biology. Getting DNA to unwind. Science. 2007;317:1181–1182. doi: 10.1126/science.1147795. [DOI] [PubMed] [Google Scholar]
- 3.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 4.Dueber ELC, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 5.Kornberg A, Baker T. DNA Replication. 2nd edn. New York: W.H. Freeman and Company; 1991. [Google Scholar]
- 6.Bramhill D, Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988;54:915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- 7.Kawakami H, Katayama T. DnaA, ORC, and Cdc6: similarity beyond the domains of life and diversity. Biochem. Cell Biol. 2010;88:49–62. doi: 10.1139/o09-154. [DOI] [PubMed] [Google Scholar]
- 8.Speck C, Messer W. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 2001;20:1469–1476. doi: 10.1093/emboj/20.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gille H, Messer W. Localized DNA melting and structural pertubations in the origin of replication, oriC, of Escherichia coli in vitro and in vivo. EMBO J. 1991;10:1579–1584. doi: 10.1002/j.1460-2075.1991.tb07678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S, Farhana A, Hasnain SE. In-vitro helix opening of M. tuberculosis oriC by DnaA occurs at precise location and is inhibited by IciA like protein. PLoS One. 2009;4:e4139. doi: 10.1371/journal.pone.0004139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duderstadt KE, Chuang K, Berger JM. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478:209–213. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato J, Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimitsu K, Su'etsugu M, Yamaguchi Y, Mazda K, Fu N, Kawakami H, Katayama T. Modes of overinitiation, dnaA gene expression, and inhibition of cell division in a novel cold-sensitive hda mutant of Escherichia coli. J. Bacteriol. 2008;198:5368–5381. doi: 10.1128/JB.00044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGarry KC, Ryan VT, Grimwade JE, Leonard AC. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl Acad. Sci. USA. 2004;101:2811–2816. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duderstadt KE, Mott ML, Crisona NJ, Chuang K, Yang H, Berger JM. Origin remodeling and opening in bacteria relies on distinct assembly states of the DnaA initiator. J. Biol. Chem. 2010;285:28229–28239. doi: 10.1074/jbc.M110.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speck C, Weigel C, Messer W. ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 1999;18:6169–6176. doi: 10.1093/emboj/18.21.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutton MD, Kaguni JM. The Escherichia coli dnaA gene: four functional domains. J. Mol. Biol. 1997;274:546–561. doi: 10.1006/jmbi.1997.1425. [DOI] [PubMed] [Google Scholar]
- 18.Messer W, Blaesing F, Majka J, Nardmann J, Schaper S, Schmidt A, Seitz H, Speck C, Tüngler D, Wegrzyn G, et al. Functional domains of DnaA proteins. Biochimie. 1999;81:819–825. doi: 10.1016/s0300-9084(99)00215-1. [DOI] [PubMed] [Google Scholar]
- 19.Crooke E, Thresher R, Hwang DS, Griffith J, Kornberg A. Replicatively active complexes of DnaA protein and the Escherichia coli chromosomal origin observed in the electron microscope. J. Mol. Biol. 1993;5:16–24. doi: 10.1006/jmbi.1993.1481. [DOI] [PubMed] [Google Scholar]
- 20.Funnell BE, Baker TA, Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem. 1987;25:10327–10334. [PubMed] [Google Scholar]
- 21.Fujikawa N, Kurumizaka H, Nureki O, Terada T, Shirouzu M, Katayama T, Yokoyama S. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003;31:2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 23.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 25.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 26.Bancaud A, Wagner G, Silva NCE, Lavelle C, Wong H, Mozziconacci J, Barbi M, Sivolob A, Le Cam E, Mouawad L, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol. Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 27.Lipfert J, Wiggin M, Kerssemakers JW, Pedaci F, Dekker NH. Freely orbiting magnetic tweezers to directly monitor changes in the twist of nucleic acids. Nat. Commun. 2011;2:439. doi: 10.1038/ncomms1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olliver A, Saggioro C, Herrick J, Sclavi B. DnaA-ATP acts as a molecular switch to control levels of ribonucleotide reductase expression in Escherichia coli. Mol. Microbiol. 2010;76:1555–1571. doi: 10.1111/j.1365-2958.2010.07185.x. [DOI] [PubMed] [Google Scholar]
- 29.Revyakin A, Ebright RH, Strick TR. Single-molecule DNA nanomanipulation: improved resolution through use of shorter DNA fragments. Nat. Methods. 2005;2:127–138. doi: 10.1038/nmeth0205-127. [DOI] [PubMed] [Google Scholar]
- 30.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 31.Fuller FB. The writhing number of a space curve. Proc. Natl Acad. Sci. USA. 1971;68:815–819. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowalski D, Eddy MJ. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989;8:4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hegner M, Smith SB, Bustamante C. Polymerization and mechanical properties of single RecA-DNA filaments. Proc. Natl Acad. Sci. USA. 1999;96:10109–10114. doi: 10.1073/pnas.96.18.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaper S, Messer W. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 1995;270:17622–17626. doi: 10.1074/jbc.270.29.17622. [DOI] [PubMed] [Google Scholar]
- 35.Bach T, Morigen, Skarstad K. The initiator protein DnaA contributes to keeping new origins inactivated by promoting the presence of hemimethylated DNA. J. Mol. Biol. 2008;384:1076–1085. doi: 10.1016/j.jmb.2008.09.042. [DOI] [PubMed] [Google Scholar]
- 36.Miller DT, Grimwade JE, Betteridge T, Rozgaja T, Torgue JJ-C, Leonard AC. Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc. Natl Acad. Sci. USA. 2009;106:18479–18484. doi: 10.1073/pnas.0909472106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leonard AC, Grimwade JE. Initiating chromosome replication in E. coli: it makes sense to recycle. Genes Dev. 2009;23:1145–1150. doi: 10.1101/gad.1809909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadia M, Klein MG, Chen XS. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc. Natl Acad. Sci. USA. 2008;105:20191–20196. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 41.Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 42.Leonard AC, Grimwade JE. Regulating DnaA complex assembly: it is time to fill the gaps. Curr. Opin. Microbiol. 2010;13:766–772. doi: 10.1016/j.mib.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao F, Zhang C-T. DoriC: a database of oriC regions in bacterial genomes. Bioinformatics. 2007;23:1866–1867. doi: 10.1093/bioinformatics/btm255. [DOI] [PubMed] [Google Scholar]
- 44.Nievera C, Torgue JJ, Grimwade JE, Leonard AC. SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol. Cell. 2006;24:581–592. doi: 10.1016/j.molcel.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waldminghaus T, Skarstad K. The Escherichia coli SeqA protein. Plasmid. 2009;61:141–150. doi: 10.1016/j.plasmid.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Hwang DS, Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 1992;267:23083–23086. [PubMed] [Google Scholar]
- 47.Landoulsi A, Kohiyama M. DnaA protein dependent denaturation of negative supercoiled oriC DNA minicircles. Biochimie. 2001;83:33–39. doi: 10.1016/s0300-9084(00)01223-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.