Abstract
The yeast IMD2 gene encodes an enzyme involved in GTP synthesis. Its expression is controlled by guanine nucleotides through a set of alternate start sites and an intervening transcriptional terminator. In the off state, transcription results in a short non-coding RNA that starts upstream of the gene. Transcription terminates via the Nrd1-Nab3-Sen1 complex and is degraded by the nuclear exosome. Using a sensitive terminator read-through assay, we identified trans-acting Terminator Override (TOV) genes that operate this terminator. Four genes were identified: the RNA polymerase II phosphatase SSU72, the RNA polymerase II binding protein PCF11, the TRAMP subunit TRF4 and the hnRNP-like, NAB3. The TOV phenotype can be explained by the loss of function of these gene products as described in models in which termination and RNA degradation are coupled to the phosphorylation state of RNA polymerase II's repeat domain. The most interesting mutations were those found in NAB3, which led to the finding that the removal of merely three carboxy-terminal amino acids compromised Nab3's function. This region of previously unknown function is distant from the protein's well-known RNA binding and Nrd1 binding domains. Structural homology modeling suggests this Nab3 ‘tail’ forms an α-helical multimerization domain that helps assemble it onto an RNA substrate.
INTRODUCTION
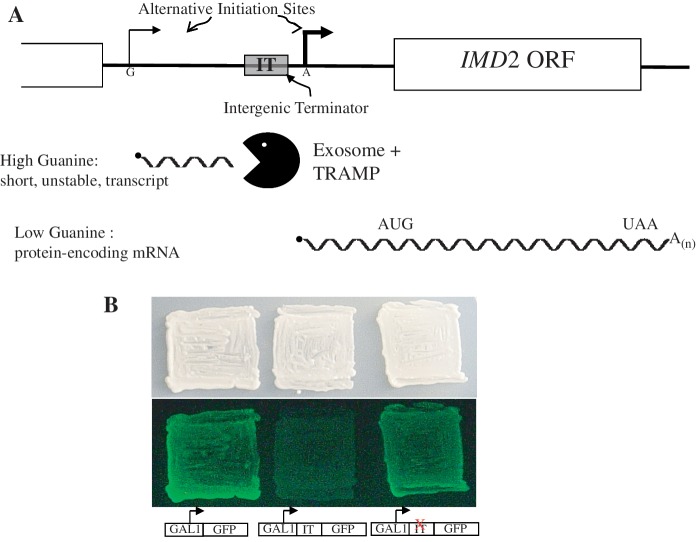
An interesting role of transcription termination in controlling nucleotide-regulated genes has recently been brought to light in Saccharomyces cerevisiae (1,2). IMD2 encodes IMP dehydrogenase, an important enzyme in the de novo guanine nucleotide biosynthetic pathway. Its transcription is regulated by cellular guanine nucleotides through alternative start sites and an intergenic terminator (IT) (Figure 1A). When GTP is abundant, transcripts initiate at an upstream G-residue. Due to the strong terminator, this initiation choice results in a short, non-coding transcript that is degraded. In this manner, IMD2 attains the ‘off’ state when IMP dehydrogenase activity is not needed (high GTP levels). Termination employs the RNA binding heterodimer Nrd1–Nab3 and the putative helicase Sen1, all encoded by essential genes (3–5). This system operates to terminate a number of other small RNAs, including snRNAs, snoRNAs and cryptic unstable transcripts (CUTs) (6). The CUT upstream of IMD2 is degraded by the nuclear exosome (3,7). When IMD2 expression is needed at times of GTP scarcity, RNA polymerase II (pol II) initiates at an adenine downstream of the terminator due to the unavailability of GTP needed for initiation at the upstream site (Figure 1A) (4). As a result, full length IMD2 mRNA is produced. Hence, pol II is the proximal sensor of intracellular GTP abundance, the use of which governs start site choice and whether transcription is terminated prematurely or allowed to go through the open reading frame (4).
Figure 1.
(A) Model for the regulation of IMD2 by GTP. The choice of start sites for IMD2 transcription varies as a function of GTP levels (4). Under GTP-replete conditions, initiation starts upstream (thin bent arrow) and terminates at the IT. The resulting non-coding RNA (short wavy line) is degraded by the exosome and IMD2 mRNA is not produced. When GTP is scarce, nucleotide-starved pol II bypasses the upstream start in favor of an adenine-initiated downstream start (bold bent arrow). The IT is bypassed, full-length mRNA is made (long wavy line) and expression of IMD2 results. (B) IMD2’s terminator quenches GFP reporter expression in yeast. Yeast strains harboring a plasmid that contains (DY1514; center patch), or lacks (DY1513; left patch) the IMD2 intergenic terminator between the GAL1 promoter and GFP reading frame, were patched onto galactose-containing medium and photographed under white light (top) or blue light to monitor GFP fluorescence (bottom). A strain with a plasmid containing a previously described point mutation in the IMD2 terminator (36) was tested in the patch on the right.
NAB3 encodes a protein with an RNA recognition motif (RRM), which is important for cell growth and allows the protein to interact with the sequence UCUU in an RNA substrate (8–11). This central core of Nab3 has been expressed in Escherichia coli and dimerizes with Nrd1. The resulting heterodimer binds RNA with high affinity (12). The protein is peculiar in that it has an amino-terminal region very rich in aspartate (D) and glutamate (E) and a carboxy-terminal region rich in proline (P) and glutamine (Q), with the respective regions showing long runs of D, E, P and Q homopolymers. The function of these repeat sequence domains is not known. The amino-terminal D/E-rich region is dispensable for cell viability, whereas the carboxy terminal ≈30% of the protein, including the P/Q region, is not (12). The Nab3–Nrd1 complex is thought to recruit the Sen1 helicase to appropriate elongation complexes to provoke termination (13–15). Nrd1 possesses a pol II carboxy-terminal domain (CTD) interaction region that preferentially binds to the pol II CTD phosphorylated on ser5 of its repeating heptapeptide unit (16). This interaction, along with Nab3's recognition of its binding site in RNA, is thought to contribute to the specificity of the system in enabling the termination complex to recognize only some promoter proximal elongation complexes, particularly those that generate short RNAs (12,14–20). In addition, Nrd1 possesses an RRM and recognizes RNA containing the GUA[A/G] sequence (8,21,22). Association of the two RRM-containing proteins Nab3 and Nrd1 yields cooperative RNA binding (12). A number of proteome-wide yeast protein–protein interaction maps have documented the physical interactions between Nrd1, Nab3 and Sen1 (23,24). The phosphorylation pattern of the repeat domain of the large subunit of pol II appears to be a marker for promoter proximal Nab3–Nrd1-dependent termination of short RNAs and may explain some of the specificity of termination for short RNAs (16,25).
In addition to participating in this alternative form of transcription termination, the Nrd1–Nab3–Sen1 complex also recruits the nuclear exosome's ribonuclease activity to degrade or process many of the resulting small RNAs (26). The coupling of termination and RNA hydrolysis employs the TRAMP complex, which is composed of a copy of the non-canonical polyA polymerase Trf4 or Trf5, a copy of the zinc knuckle protein Air1 or Air2 and the Mtr4 RNA helicase (19,20,27,28). The TRAMP complex interacts with Nrd1 as well as the nuclear exosome and stimulates the latter's ribonuclease function (15,19,28,29). Through this coupling role, TRF4 is involved in the metabolism of CUTs and the processing of small non-coding RNAs (30).
Here we use a genetic screen in Saccharomyces cerevisiae to identify TOV (‘terminator override’) genes. Based upon an analysis of RNA metabolism, two classes of mutants could be identified. One group appeared to be primarily RNA degradation deficient and two independent isolates were shown to contain mutations in the TRAMP component, TRF4. The other class displayed terminator read-through defects. Two of the mutant strains had alterations in SSU72. One had a mutation in PCF11. Another two mutants in this class fell within the NAB3 gene. The tov changes represent new mutant alleles for each of these termination-related genes. The nab3 alleles were particularly interesting since they revealed a new portion of this hnRNP-like protein in a short region of its carboxy-terminal amino acids. Structural homology suggests that this region is functionally akin to an α-helix in hnRNP C, which serves to multimerize that RNA binding protein on substrate RNAs. These findings show the effectiveness of sorting live fluorescent yeast as a genetic screen and of identifying mutations by whole genome sequencing. They also reveal a new degree of complexity in the Nab3 protein.
MATERIALS AND METHODS
Cell growth and media
Yeast strains used in this study are presented in Table 1. Cells were grown in rich media (YPD) or standard selective ‘drop-out’ media (SC-ura, SC-leu or SC-ura-leu) at 30°C unless otherwise indicated. For galactose induction, cells were grown in SC-ura in which glucose was replaced by raffinose as a carbon source and 2% galactose was added as indicated or cells were grown continuously in galactose. Plasmids were introduced into yeast strains using the lithium acetate procedure (31).
Table 1.
Yeast strains used in this study
| 1A1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [pREF-GFP kan (URA3)] tov1 | This study |
| 1A1F | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov1 | This study |
| 2A1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [pREF-GFP kan (URA3)] tov2 | This study |
| 2A1F | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov2 | This study |
| 2B1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [pREF-GFP kan (URA3)] tov5 | This study |
| 2B1F | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov5 | This study |
| 2C1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [pREF-GFP kan (URA3)] tov6 | This study |
| 2C1F | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov6 | This study |
| 2D1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [pREF-GFP kan (URA3)] tov7 | This study |
| 2D1F | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov7 | This study |
| 3A1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [pREF-GFP kan (URA3)] tov3 | This study |
| 3A1F | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov3 | This study |
| 4A1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [pREF-GFP kan (URA3)] tov4 | This study |
| 4A1F | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov4 | This study |
| ACY1223 | MATa ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab3-11 | A. Corbett-Emory U. |
| DY1513 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [pGAL-GFP kan (URA3)] | This study |
| DY1514 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [pREF-GFP kan (URA3)] | This study |
| DY1514F | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | This study |
| DY2043 | MATa lys2-A14 ade5-1 his7-2 leu2-3,112 trp1-289 ura3-52 [pREF-GFP kan (URA3)] | This study |
| DY2044 | MATa lys2-A14 ade5-1 his7-2 leu2-3,112 trf4::KANMX trp1-289 ura3-52 [pREF-GFP kan (URA3)] | This study |
| DY2222 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 met15Δ0/MET15 NAB3/nab3::kanMX ura3Δ0/ura3Δ0 [pREFGFP kan (URA3)] | This study |
| DY2233 | MATα his3Δ1 leu2Δ0 (lys2, met15 status unknown) nab3::kanMX ura3Δ0 [pRS315-nab3-Q784X (LEU2)] | This study |
| DY2240 | MATα his3Δ1 leu2Δ0 (lys2, met15 status unknown) nab3::kanMX ura3Δ0 [pRS315-nab3-Q784X (LEU2)] [pREFGFP kan (URA3)] | This study |
| DY30009 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 chr. XII::GAL-GFP-kan | This study |
| DY3001 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 chr. XII::REF-GFP-kan | This study |
| DY3003 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov1 chr. XII::REF-GFP-kan | This study |
| DY30059 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov2 chr. XII::REF-GFP-kan | This study |
| DY30079 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov3 chr. XII::REF-GFP-kan | This study |
| DY3009 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov4 chr. XII::REF-GFP-kan | This study |
| DY30109 | MATa ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab3-11 [pNAB3] | This study |
| DY3011 | MATa ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab3-11 [pRS315] | This study |
| DY3012 | MATa ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab3-11 [pRS315-nab3-Q784X] | This study |
| DY3013 | MATa ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab3-11 [pnab3-11] | This study |
| DY3018 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov1 [pREF-GFP kan (URA3)] [pRS315 (LEU2)] | This study |
| DY3019 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov1 [pREF-GFP kan (URA3)] [pRS315-NAB3 (LEU2)] | This study |
| DY30229 | MATα his3Δ1 leu2Δ0 (lys2, met15 status unknown) nab3::kanMX ura3Δ0 [pRS316-nab3-11 (URA3)] | This study |
| DY3029 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov2 [pREF-GFP-kan (URA3)] [pTRF4 (LEU2)] | This study |
| DY3036 | MATα his3Δ1 leu2Δ0 (lys2, met15 status unknown) nab3::kanMX ura3Δ0 [pRS315 nab3FL (LEU2)] [pREFGFP kan (URA3)] | This study |
| DY3038 | MATα his3Δ1 leu2Δ0 (lys2 and met15 status unknown) nab3::kanMX ura3Δ0 [pRS315 nab3Δ3 (LEU2)] [pREFGFP kan (URA3)] | This study |
| DY3060 | MATα his3Δ1 leu2Δ0 (lys2, met15 status unknown) nab3::kanMX [pRS315 nab3CΔ1 (LEU2)] [pREFGFP kan (URA3)] | This study |
| DY3061 | MATα his3Δ1 leu2Δ0 (lys2, met15 status unknown) nab3::kanMX [pRS315 nab3CΔ2 (LEU2)] [pREFGFP kan (URA3)] | This study |
| DY3078 | MATa/α lys2-A14/lys2-A14 ade5-1/ade5-1 his7-2/his7-2 leu2-3,112/leu2-3,112 trf4::KANMX/TRF4 trp1-289/trp1-289 ura3-52/ura3-52 | This study |
| DY3082 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov6 pREF-GFP-kan [pTRF4 (LEU2)] | This study |
| DY3083 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov6 [pREF-GFP-kan (URA3)] [ptrf4-13A (LEU2)] | This study |
| DY3084 | MATa lys2-A14 ade5-1 his7-2 leu2-3,112 trp1-289 ura3-52 | This study |
| DY3085 | MATa lys2-A14 ade5-1 his7-2 leu2-3,112 trf4::KANMX trp1-289 ura3-52 | This study |
| DY3088 | MATa lys2-A14 ade5-1 his7-2 leu2-3,112 trf4::KANMX trp1-289 ura3-52 [pREF-GFP kan (URA3)] [pTRF4 (LEU2)] | This study |
| DY3092 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov2 pREF-GFP-kan-URA3 [ptrf4-S430stop (LEU2)] | This study |
| DY3114 | MATa ura3-52 trp1Δ63 leu2Δ1 [pREF-GFP kan (URA3)] | This study |
| DY3115 | MATa ura3-52 trp1Δ63 leu2Δ1 Ub-DHFRts-HA-SSU72-URA3 [pREF-GFP kan (URA3)] | This study |
| DY3625 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [pREF-GFP kan (URA3)] tov7 [pRS315-SSU72 (LEU2)] | This study |
| DY3626 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [pREF-GFP kan (URA3)] tov7 [pRS315-ssu72-2D (LEU2)] | This study |
| DY3627 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov5 [pREF-GFP kan (URA3)] [pRS315 (LEU2)] | This study |
| DY3628 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tov5 [pREF-GFP kan (URA3)] [pRS315-PCF11 (LEU2)] | This study |
| H291 | MATa ura3-52 trp1Δ63 leu2Δ1 Ub-DHFRts-HA-SSU72-URA3 | (32) |
| H98 | MATa ura3-52 trp1Δ63 leu2Δ1 | (32) |
| YH991 | MATa/α lys2-A14/lys2-A14 ade5-1/ade5-1 his7-2/his7-2 leu2-3,11/2 leu2-3,11 trp1-289/trp1-289 ura3-52/ura3-52 | N. Degtyareva P. Doetsch-Emory U. |
| YSC1021-669423 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 met15Δ0/MET15 NAB3/nab3::kanMX ura3Δ0/ura3Δ0 | Open Biosystems |
Plasmid/strain construction and genomic integration
Oligonucleotides used in this study are listed in Table 2. The plasmid pREF-GFP was assembled by a three-way ligation of a PCR product encoding the yeast IMD2 IT (HindIII-PstI cut product made with primers ‘oPur5P-F170’ and ‘oR90-PstI’) into the plasmid pYES2 (Invitrogen; cut with HindIII and XhoI) along with a PCR product encoding the GFP open reading frame (PstI-XhoI cut PCR product made with primers ‘oGFP-atg’ and ‘oGFP-stop’). The kanamycin resistance gene from pFA6a-KanMX4 (33) was excised by digestion with EcoRI and BamHI, filled in with DNA polymerase and inserted into the blunted BglI site of the ampicillin resistance gene in pREF-GFP to yield pREF-GFP-Kan.
Table 2.
Oligonucleotides used in this study
| SNR33up | 5′-cggaacggtacataagaatagaagag-3′ |
| SNR33down | 5′-taaagaaaacgataagaactaacc-3′ |
| oIMD2P-F500B | 5′-gactagtgcggccgcatcggttgagcgcgatatta-3′ |
| oPur5P-R 00 | 5′-ctgatcaggatccggccattgcttttgctactt-3′ |
| oPur5P-F170 | 5′-ggggtaccaagcttttccgtattctattctattccttgc-3′ |
| oR90-PstI | 5′-cgcctgcagaacaaaatgcgtttatgacagtt-3′ |
| oGFP-atg | 5′-tctagactgcagatggctagcaaaggagaagaact-3′ |
| oGFP-stop | 5′-tgaattctcgagttagcagccggatcctttgtata-3′ |
| Nab3Prom-UP-BamHI | 5′-cacgggatccagtgtaaccctgaattgtgaagag-3′ |
| Nab3cds-Dwn-XhoI | 5′-atatctcgagcagaggaaacaaatgaagaggtgcg-3′ |
| 5-GalGFP(C) | 5′-gttaaatgctacgactcggcatatactgtgctcgtttttagcgtggggatgatccactagt-3′ |
| 3-GalGFP(C) | 5′-ttcgttaaacgtctgtagaggtgccgaacaatttttgtctgcaatgccaacaacgttgcg-3′ |
| Nab3 Δ1 tail Fwd | 5′-aataatgttcaaagtctattagatagtttagcaaaactacaatagc-3′ |
| Nab3 Δ1 tail REV | 5′-tcgagctattgtagttttgctaaactatctaatagactttgaacat-3′ |
| Nab3 Δ2 tail Fwd | 5′-aataatgttcaaagtctattagatagtttagcaaaactatagc-3′ |
| Nab3 Δ2 tail REV | 5′-tcgagctatagttttgctaaactatctaatagactttgaacat-3′ |
| Nab3-15aa-tail Fw | 5′-aataatgttcaaagtctattagatagtttagcaaaatagc-3′ |
| Nab3-15aa-tail Re | 5′-tcgagctattttgctaaactatctaatagactttgaacat-3′ |
| Nab3-18aa-tail Fw | 5′-aataatgttcaaagtctattagatagtttagcaaaactacaaaaatagc-3′ |
| Nab3-18aa-tail Re | 5′-tcgagctatttttgtagttttgctaaactatctaatagactttgaacat-3′ |
| Trf4-5′ UTR-MX4-FWD | 5′-gaatatacgaccaaaaacgttacgctttcataaagtgtgaataagcaagggaactatacttgaaatacatggaggcccagaataccctcc |
| Trf4-3′ UTR-MX4-REV | 5′-aacacacattctatccaggtacacagtgatgtacagttcagtgcatcatttaaacaaaaaggcacatacagtatagcgaccagcattcac |
| AC4037-SacI-Trf4-Fwd | 5′-atatgagctctacgtatacatatctatataatatgccttgtactttacgc-3′ |
| AC4040-XhoI-Trf4-Rev | 5′-atatctcgagttaaatcaatttattttacgttacgatagagcc-3′ |
| SSU72-Bam-For | 5′-tataggatcctcgtttgatcatcctttacg-3′ |
| SSU72-Xho-Rev | 5′-tatactcgagcagtcctatatctggtgatgtg-3′ |
| Pcf11BamHIFor | 5′-tataggatccagcggcgaagaagtctagc-3′ |
| Pcf11SalIRev | 5′-tatagtcgactgggcttttggctacatatg-3′ |
The plasmid pRS315-nab3Q784X was made by generating a PCR product from the genome of the mutant strain 1A1 using the oligonucleotides ‘Nab3Prom-UP-BamHI’ and ‘Nab3cds-Dwn-XhoI’. The product was cut with XhoI and BamHI and inserted into similarly cut pRS315. The nab3-11 allele was amplified by PCR using the same primers and genomic DNA from strain ACY1223. It was digested with XhoI and BamHI and inserted into pRS315, to yield plasmid pnab3-11. A PCR product of wild-type NAB3 was generated from BY4742 genomic DNA using the same primers and inserted into pRS316 cut with BamHI and XhoI to yield pRS316-NAB3.
The Nab3 tail deletion series was made by subcloning a 1.2 kbp HindIII-XhoI fragment encoding the 3′-end of the coding sequence and untranslated region into similarly cut pBluescript II KS(−) (Stratagene, Inc.). This plasmid was cut with BspMI and XhoI into which was inserted a series of oligonucleotide duplexes with compatible overhangs and which encoded portions of the Nab3 tail (‘Δ1’: ‘Nab3Δ1 tail Fwd’ and ‘Nab3Δ1 tail REV’; ‘Δ2’: ‘Nab3Δ2 tail Fwd’ and ‘Nab3Δ2 tail REV’; ‘Δ3’: ‘Nab3-15aa-tail Fw’ and ‘Nab3-15aa-tail Re’ and full length (‘FL’): ‘Nab3-18aa-tail Fw’ and ‘Nab3-18aa-tail Re’). These constructs generated a series of intermediate plasmids from which fragments were removed to build the deletion derivatives of Nab3. Restriction fragments encoding these rebuilt 3′-ends were individually removed from their respective Bluescript vectors by digestion with HindIII and XhoI and inserted into similarly cut pRS315-NAB3 from which the natural sequence was removed. This resulted in the final plasmids pRS315-nab3CΔ1, pRS315-nab3CΔ2, pRS315-nab3CΔ3 and pRS315-nab3FL.
pTRF4 was obtained from Dr A. Corbett (Emory U.) and contained the wild-type copy of TRF4 inserted into pRS315. The plasmid ptrf4-13A was assembled from a XhoI and SacI digested PCR product made from genomic DNA of strain 2A1 (tov2) using oligonucleotides ‘AC4037-SacI-Trf4-Fwd’ and ‘AC4040-XhoI-Trf4-Rev’ and inserted into similarly cut pRS315. Similarly, ptrf4S430stop was made by amplifying the TRF4 allele in genomic DNA from 2C1 (tov6) and inserting it into pRS315 as was done for ptrf4-13A. pRS315-SSU72 and pRS315-ssu72-2D were made by inserting a PCR product made from wild-type or 2D1 genomic DNA, respectively (using primers SSU72-Bam-For and SSU72-Xho-Rev), into BamHI and XhoI cut pRS315. pRS315-PCF11 was made by amplifying from genomic DNA using the oligonucleotide ‘Pcf11BamHIFor’ and ‘Pcf11SalIRev’, cutting the product with BamHI and SalI and inserting it into pRS315.
Strains DY1513 and DY1514 were made by transforming BY4742 (Open Biosystems) with the plasmids pGAL-GFP-kan and pREF-GFP-kan, respectively. Isolates that were selected by FACS were named 1A1, 2A1, 3A1, 4A1, 2B1, 2C1 and 2D1 and assigned tov numbers one through seven, respectively. These strains (and their parent strain, DY1514) were cured of the reporter plasmid by growth on 5-flouro-orotic acid (FOA) and named 1A1F, 2A1F, 3A1F, 4A1F, 2B1F, 2C1F, 2D1F and DY1514F, respectively. Strain 2A1 was transformed with pTRF4 or ptrf4S430stop to generate DY3029 and DY3092, respectively. Strain 2B1 was transformed with pREF-GFP-kan and either pRS315-PCF11 (DY3628) or pRS315 (DY3627).
To make a family of strains lacking chromosomal NAB3, a diploid heterozygous for a kanMX disruption of the gene (Open Biosystems YSC1021-669423) was transformed with pRS315-nab3Q784X to yield DY2222. This strain was sporulated and a resulting haploid (DY2233) lacking chromosomal NAB3 and containing the plasmid was isolated. This strain was transformed with pREF-GFP-kan to yield DY2240. DY2233 was transformed with URA3-marked pnab3-11 and screened for the loss of LEU2-marked pRS315-nab3Q784X to generate DY30229. DY30229 was independently transformed with pRS315-nab3CΔ1, pRS315-nab3CΔ2, pRS315-nab3CΔ3 or pRS315-nab3FL and each was grown on FOA to select for loss of pnab3-11. The resulting strains were transformed with pREF-GFP-kan to generate DY3060, DY3061, DY3038 and DY3036, respectively.
In some strains, a genomic copy of the GFP reporter transcription unit was integrated into a gene-poor region of chromosome XII. The oligonucleotides ‘5-GalGFP(C)’ and ‘3-GalGFP(C)’ were used to amplify the integration cassette from plasmids pREF-GFP-kan or pGal-GFP-kan. The former plasmid contains IMD2's IT sequence between the GAL1 promoter and the GFP reading frame, the latter does not. The PCR product from pREF-GFP-kan was transformed into DY1514F, 1A1F, 2A1F, 3A1F and 4A1F using high-efficiency lithium acetate transformation and G418 resistant isolates were selected and named DY3001, DY3003, DY30059, DY30079 and DY3009. Similarly, the product from pGal-GFP-kan was integrated into DY1514F to yield DY30009. Accuracy of the integration events was confirmed by PCR from genomic DNA.
TRF4 was deleted by homologous recombination in the diploid yeast strain YH991 using a PCR product made with the oligonucleotides Trf4-5′UTR-MX4-FWD and Trf4-3′UTR-MX4-REV. Integration was confirmed by PCR from genomic DNA of this diploid, DY3078. The diploid was sporulated and genomic DNA was prepared from each spore dissected from a tetrad to again confirm integration and kanamycin resistance that segregated 2:2. Sister spores with either an intact TRF4 (DY3084) or deleted TRF4 (DY3085) were transformed with pREF-GFP-kan to generate DY2043 and DY2044, respectively. DY2044 was transformed with pTRF4 to generate DY3088.
Strain DY3625 and DY3626 were made following transformation of 2D1 with pRS315-SSU72 or pRS315-ssu72-2D, respectively. DY3114 and DY3115 were made by transforming pREF-GFP-kan into H98 or H291, respectively.
Northern blots
Cultures were grown to an optical density (600 nm) of 0.4–0.6, washed in water and frozen at −80°C. Where indicated, 0.5 mM guanine for 30 min or 15 µg/ml mycophenolic acid for 2 hr was added to the media. RNA was extracted and separated on 1% agarose gels for transfer to Zeta-Probe GT Genomic filters as described (34–36). Probes were generated by PCR from plasmid DNA (for GFP) or yeast genomic DNA using the primer pairs ‘SNR33up/SNR33down’, ‘oIMD2P-F500B’/‘oPur5-R00’ and ‘oGFP-atg’/‘oGFP-stop’ for SNR33, IMD2 and GFP, respectively. Probes were labeled using random primers (Invitrogen), Klenow DNA polymerase and α-32P-dATP, (Perkin Elmer). Filters were hybridized at 42°C overnight, washed and exposed to HyBlot CL film (Denville Scientific Inc.) with an intensifying screen.
Fluorescence-activated cell sorting and analysis
Parental strain DY1514 was grown overnight at 30°C in SC-ura (raffinose) with 2% galactose. Cells were resuspended to a density of 1.33 OD/ml in 1.5 ml and were sorted at 488 nm using a FACSAria II (Becton–Dickinson) and FACSDiva software. Selected cells were plated onto SC-ura plates incubated at 30°C and replica plated onto SC-ura + galactose. Fluorescent colonies were identified by illumination with a Dark Reader® lamp and filter glasses (Clare Chemical), cultured and named. Flow cytometry analysis employed the LSRII machine (Becton–Dickinson) at 488 nm excitation wavelength.
Whole genome sequencing
Genomic DNA was purified and subjected to high-throughput sequencing by HiSeq2000 (Illumina Inc.). In brief, the purified DNA was sheared to an average fragment size of 300 bp using acoustic focusing. The SPRI works platform was used to create libraries, which were sequenced on an Illumina HiSeq2000 with a read length of 50 bases plus 7 bases for the multiplex tags. The resulting sequence was extracted and de-multiplexed using Illumina's SCS2.8 software. The data were then analyzed using the CLC Genomics Workbench package. Imported reads were trimmed to remove low-quality sequence as well as any reads of <36 bases in length. Reads were mapped to the published yeast genome obtained from the Saccharomyces Genome Database, yielding an average coverage of 95× across the genome. Variations (SNPs and InDels) were called using a minimum Q score of 20 and a coverage minimum of 25×. This yielded over 500 SNPs and 350 InDels per strain. We focused on SNPs in protein-encoding sequences to look for differences between strains. Using data from other similar strains as well as gene annotation, variations that were common or unlikely to be related to the phenotype were removed from consideration. All variations in the reduced subset were manually checked to ensure that the variation call was accurate.
Western blotting
Yeast cells were collected, washed with water and lysed with SDS loading buffer before being resolved on a 5% polyacrylamide gel. Proteins were transferred to nitrocellulose electrophoretically and probed with monoclonal antibodies against Pgk1 (2C12; Invitrogen, Inc.) or Nab3 (2F12-2H2; Dr M. Swanson, U. Fla.). Signal was detected using horse radish peroxidase-conjugated anti-mouse IgG (Amersham-General Electric, Co.), chemiluminescent substrate (Pierce, Inc.) and XOMAT film (Kodak).
RESULTS
A genetic screen for terminator function
We previously characterized IMD2's IT, an ≈100 bp element that blocks elongation and provokes exosome-mediated degradation of the resulting short RNAs (3,37). When placed downstream of the GAL1 promoter, the IT is a potent terminator. cis-Acting mutations were identified in the IT that inactivated its activity allowing full length RNA synthesis (4,37). We reasoned that including the terminator between an inducible promoter and the GFP reading frame could form the basis of a sensitive genetic screen for trans-acting mutations in genes that encode proteins needed for IT function. Loss of terminator activity would be observed as transcriptional read-through yielding fluorescent cells that could be captured by fluorescence-activated cell sorting (FACS) and clonally isolated.
As a proof of principle, we introduced into yeast a plasmid with the IT inserted between the GAL1 promoter and GFP encoding sequences and plated them on galactose-containing medium. The presence of the IT effectively prevented what was otherwise strong GFP fluorescence (Figure 1, center versus left patches). When we included the version of the IT bearing a point mutation known to give transcriptional read-through (4,37), fluorescence was restored (right patch). Thus, the IT functions as a potent switch for interrupting GFP expression in live yeast cells.
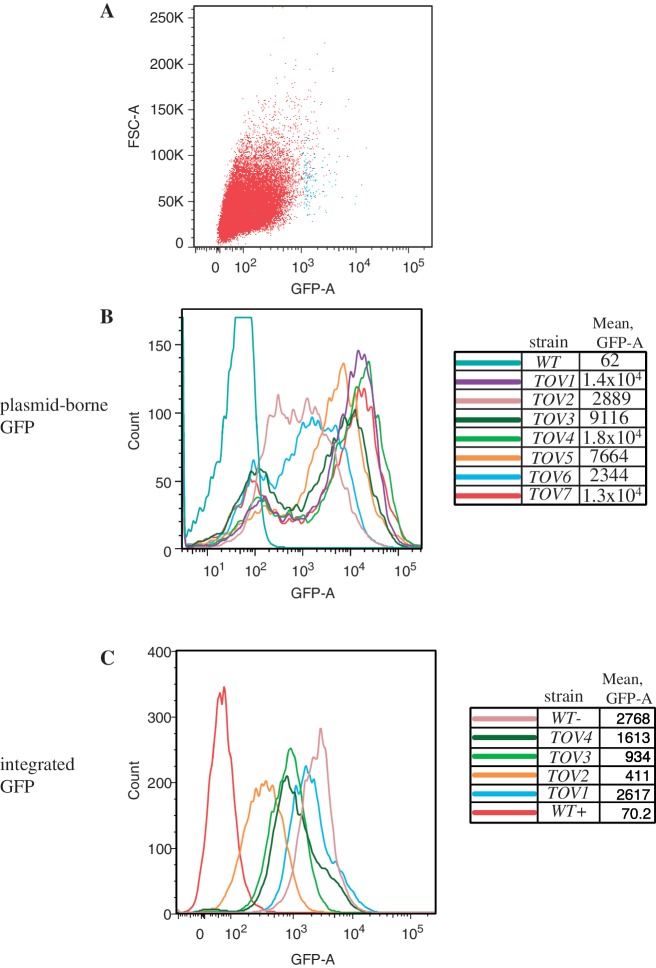
Cells transformed with the GAL-IT-GFP reporter (Figure 1, center patch) were induced with galactose and subjected to FACS sorting. We expected that cells with spontaneous, trans-acting mutations that reduce IT function, would yield mutants that score positive for fluorescence. From 107 starting cells, we collected ≈0.1% of those with the strongest fluorescent signal. Figure 2A shows the profile of the presorted (red) and post-sorted (blue) populations. The latter group of cells was plated onto galactose-containing media and seven of the strongest fluorescent colonies were pursued (tov1–tov7). The fluorescence intensity of a population grown from each founder colony was quantified by flow cytometry (Figure 2B). For most isolates, a bimodal pattern was obtained in which a peak at ≈102 fluorescent units was seen along with a strong fluorescent signal at ≈103–104 units. The former represents the background of endogenous fluorescence from live yeast cells without a GFP-containing plasmid (data not shown). This peak is also the signal from cells of the starting strain that contains the terminator-repressed GFP plasmid, demonstrating the efficiency of the IT (Figure 2B, ‘WT’, blue-green line and 2C, ‘WT+’ red line). The tov mutants clearly gained GFP fluorescence relative to the wild-type population. A small peak of cells with a fluorescence value of ≈102 in the tov mutant populations are non-expressers. This subset results from reporter plasmid loss that has been described previously (38,39). Even when this group is included in the calculation, mean fluorescence from each of the tov populations was substantially increased over the parent strain, with the maxima of positive peaks showing values of >103 units for tov2 and tov6 and ≥104 fluorescent units for the remaining strains (Figure 2B). These latter values closely approached the level of expression of a wild-type cell with a reporter plasmid that has no terminator placed between the GAL1 promoter and GFP (data not shown). To ensure that the genetic changes observed were not linked to the reporter plasmid, strains were cured of their URA3-marked plasmids by growth on FOA and retransformed with fresh plasmid. All tov strains fluoresced upon retransformation (data not shown).
Figure 2.
Flow cytometry analysis of candidate yeast mutants expressing GFP from a reporter. (A) Plot of forward scatter area (particles) versus GFP fluorescence area for yeast strain DY1514, the parent strain to the tov mutants, prior to sorting (red, 50 000 cells) and post-sorting (blue, 150 cells). (B) Particle count versus GFP area is plotted for yeast strains designated tov1 through tov7 bearing the GAL-IT-GFP reporter plasmid. Flow cytometry was independently performed on 10 000 cells each of the wild-type and tov strains at an excitation wavelength of 488 nm using a LSRII (Becton–Dickson) digital analyzer. Bimodality results from plasmid loss (see text). (C) Particle count versus GFP area intensity plot for wild-type and tov1 through tov4 in which a copy of the GAL-IT-GFP reporter transcription unit was integrated. Quantification of the fluorescence of 10 000 cells from each strain was plotted as in part B. The wild-type cells with an integrated GFP reporter that contained (WT+) or lacked (WT−) the IT were also analyzed as negative and positive controls for GFP expression, respectively.
To obtain a more accurate measure of mean fluorescence intensity differences, we integrated the GAL-IT-GFP reporter into a relatively gene-free region of chromosome XII for four of the tov strains, tov1, tov2, tov3 and tov4. This obviated the need for an extrachromosomal reporter plasmid and eliminated the problem of plasmid loss. As a result, bimodality disappeared and single peaks were obtained from populations that also displayed lower fluorescent intensities due to the strictly single copy nature of the integrated transcription units (Figure 2C). These four mutant strains displayed mean intensities 6 (tov2)–37 (tov1)-fold higher than the parental wild-type strain. All four tov strains retained full expression of GFP in the absence of the IT, ruling out an indirect cause for fluorescent changes, such as a defective galactose response (data not shown).
None of the isolates displayed a temperature-sensitive growth phenotype at 37°C (data not shown). When the tov1, tov2, tov3 and tov4 mutant haploid strains were mated to an isogenic wild-type strain, GFP fluorescence was lost in the presence of galactose, demonstrating that the mutations were recessive (data not shown).
Termination and/or RNA degradation is defective in tov mutants
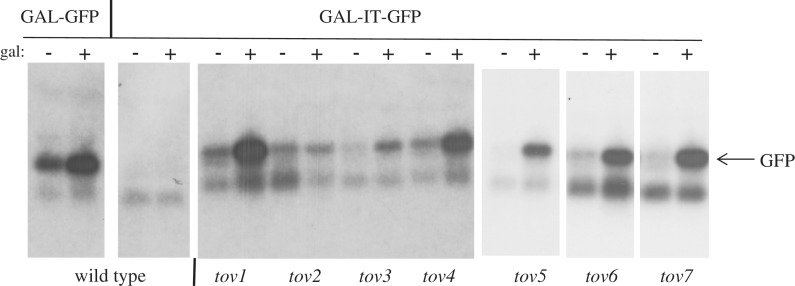
To test if pol II in these GFP-positive mutants bypassed the IT, we analyzed RNA derived from the reporter plasmid by northern blotting with a GFP probe. In wild-type cells, placement of the IT between the GAL1 promoter and GFP effectively turned off mRNA production (Figure 3, left two panels GAL-GFP versus GAL-IT-GFP), whereas in all the tov isolates, expression was restored following galactose induction, albeit to varying levels. These results are consistent with the fluorescence data suggesting that the mutants suffered from terminator override at the level of transcription.
Figure 3.
Northern blot of GFP mRNA. Cells were grown in the presence or absence of galactose as indicated. Total RNA was isolated from the indicated primary tov strains, the isogenic wild-type strain containing the GAL-IT-GFP plasmid (DY1514) or isogenic wild-type strain with a GAL-GFP reporter plasmid lacking the IT (DY1513). The agarose gel was blotted to a membrane and the filter was probed with a radio-labeled PCR product containing the GFP reading frame. The 12 lanes on the left were from a single gel and filter. The six lanes on the right were from a different single gel and filter (the origin of the higher mobility, cross-hybridizing band seen in all lanes is unknown).
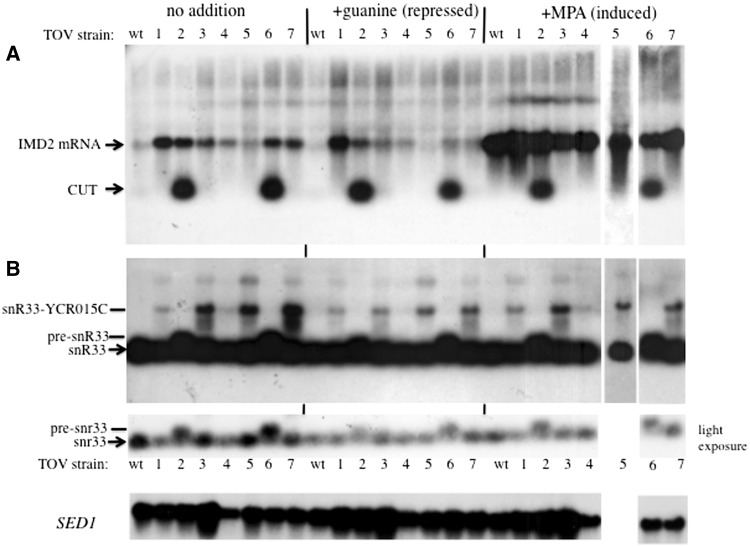
To test if the mutations resulted in changes in expression of endogenous IMD2, northern blots were probed to detect the short upstream CUT (Figure 1A), as well as full length IMD2 mRNA, as described previously (3). RNA was prepared from untreated cells and from cells grown with 0.5 mM guanine (the repressed state in which the short upstream transcript is made but normally degraded). Cells treated with 15 µg/ml mycophenolic acid, a drug that inhibits IMP dehydrogenase and reduces GTP levels (the induced state), were also analyzed (Figure 4A). For tov2 and tov6 cells, IMD2's short upstream RNA was strongly stabilized, similar to what was seen previously for an RRP6-deletion strain (3). This result suggested that tov2 and tov6 harbored mutations in exosomal subunits or proteins that couple exosomal degradation to termination. For most of the other tov strains, a readily detectable increase in the level of full length IMD2 mRNA over wild-type was observed during standard vegetative growth (‘no addition’) and repressed (‘+guanine’) conditions (Figure 4A), consistent with the idea that pol II reads through the endogenous IMD2 terminator in these mutants.
Figure 4.
Northern blot of IMD2 and SNR33 transcripts. (A) RNA was isolated from the tov1–tov7 strains 1A1F, 2A1F, 3A1F, 4A1F, 2B1F, 2C1F and 2D1F and the otherwise wild-type strain DY1514F and analyzed by northern blotting using a radio-labeled IMD2 probe that detects both the short, intergenic CUT and mature IMD2 mRNA. (B) The filter was stripped and rehybridized to radio-labeled snR33 DNA (middle panel) that detects the mature snoRNA, as well as the precursor and read-through versions of the transcript (pre-snR33 and snR33-YCR015C, respectively). A lighter exposure of the SNR33-hybridized filter is shown at the bottom to help resolve snR33 from pre-snR33. All lanes are derived from a single gel and filter except for the +MPA tov5 lane. As a loading control, the filter was stripped and reprobed for SED1 mRNA.
To test the generality of these observations, we stripped and re-probed the northern filter to examine a small non-coding RNA, snR33, whose biogenesis is known to involve the Nrd1-Nab3-Sen1 machinery (14,16,17). The resulting terminated RNA is a precursor that is trimmed to its mature length by the exosome. Defects in snR33 termination have been observed as read-through into the downstream ORF (17). Strains tov2 and tov6 accumulated the pre-snR33 transcript that was otherwise absent from wild-type cells (Figure 4B, see light exposure). This phenotype is similar to that seen when exosomal subunits are mutated (7,17,20,29,40). Strains with tov1, 3, 4, 5 and 7 showed an accumulation of aberrant SNR33–YCR015C fusion RNAs that were longer than the mature transcript and its precursor (Figure 4B). This is a similar phenotype to that seen previously for sen1 mutants (17). It is notable that there were differences in the magnitude of the changes in aberrant snoRNA versus IMD2 RNA depending upon the mutant strain. For example, tov5 had little effect on full length IMD2 mRNA but a strong effect upon snoRNA read-through (Figure 4). Taken together, these findings are an important validation of the GFP/FACS screen, showing it can reveal trans-acting factors involved in terminator function.
tov2 and tov6 are allelic to TRF4
To identify tov mutations, a subset of strains were subjected to whole genome sequencing using high-throughput Illumina technology. We focused on single nucleotide polymorphisms that differed between the starting strain DY1514 and the tov strains. Sequencing revealed that, relative to its parent strain or the Saccharomyces Genome Database reference sequence, tov2 and tov6 have alterations in the coding sequence of TRF4, a subunit of the TRAMP complex. Tov2 had an additional adenine in a tract of 12 A-residues in TRF4, which produces a frame shift resulting in a Trf4 protein truncated after its first 264 amino acids. This truncated protein would lack its nucleotide binding domain and one of the three aspartic acids of the catalytic triad needed for polyadenylation activity (41,42). The tov6 strain had a C–G substitution in TRF4 that changed Ser430 to a stop codon. This truncates the protein further downstream than tov2's frame shift in the domain called the ‘central’ region that is conserved between canonical polyA polymerases (41). Both changes were confirmed after amplifying the region and sequencing the products by conventional Sanger methods.
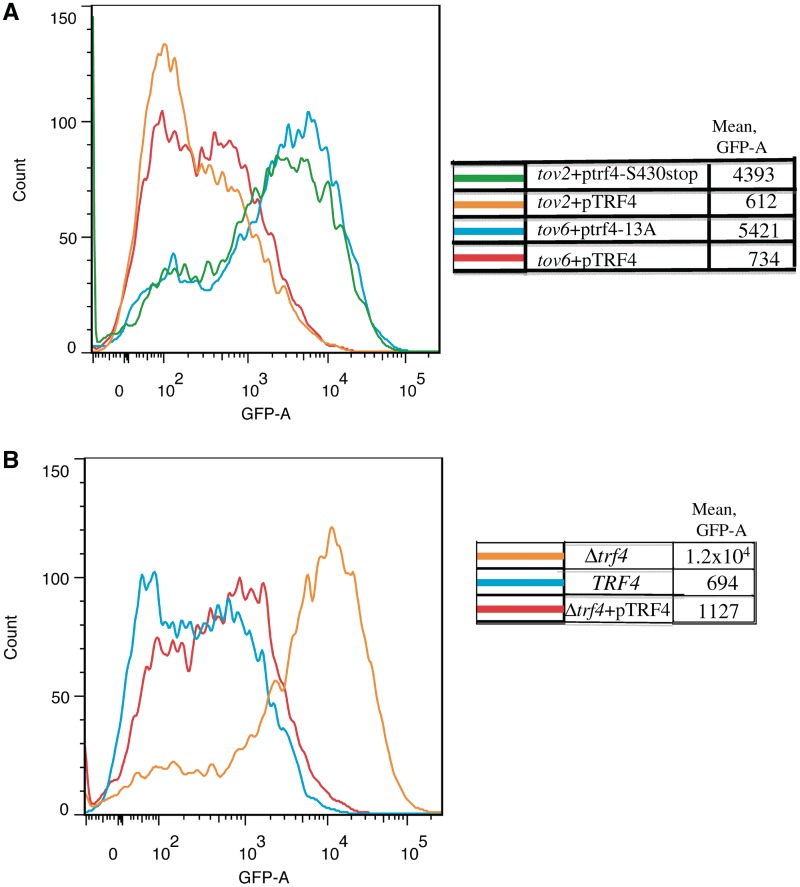
If these mutations were causal for the terminator override phenotype, then a copy of wild-type TRF4 on a plasmid should extinguish the fluorescence of the tov2 and tov6 strains containing the GAL-IT-GFP reporter plasmid. Indeed, the wild-type TRF4 restored terminator function to both strains (orange and red lines, Figure 5A). The mutant trf4 alleles were amplified from tov2 and tov6, inserted into plasmids and tested for their abilities to cross-complement the tov6 and tov2 strains, respectively. Neither mutant allele could rescue the GFP-expressing phenotype of the other (blue and green lines, Figure 5A), yielding cells with strong fluorescence levels similar to that of cells with empty plasmids (data not shown). These results provide evidence that the tov2 and tov6 mutations are two different, non-complementing, loss-of-function, mutations in TRF4.
Figure 5.
Rescue of tov2, tov6 and Δtrf4 strains with TRF4 derivatives. (A) Flow cytometry of the tov2 and tov6 strains containing the GAL-IT-GFP reporter plasmid and a plasmid expressing the indicated TRF4 alleles. Strains DY3029 (‘tov2 + pTRF4’), DY3092 (‘tov2 + ptrf4-S430stop’), DY3082 (‘tov6 + pTRF4’) and DY3083 (‘tov6 + ptrf4-13A’) were grown on galactose, subjected to flow cytometry and the GFP intensities were plotted versus cell count. (B) Flow cytometry of galactose-induced cells deleted for chromosomal TRF4 (DY2044) or with intact TRF4 (DY2043) and transformed with the GAL-IT-GFP reporter. DY3088 (Δtrf4 + pTRF4) lacked chromosomal TRF4 but contained wild-type TRF4 on a plasmid.
If tov2 and tov6's phenotypes were due to their respective mutations in TRF4, then a complete deletion of TRF4 should yield GFP-positive cells by flow cytometry after transformation with the GAL-IT-GFP reporter plasmid. This was indeed the case (orange versus blue, Figure 5B). Wild-type TRF4 on a plasmid restored substantial IT function (red). This provided independent evidence that the nonsense mutations in TRF4 found in tov2 and tov6 were causally involved in their terminator read-through phenotype.
TOV genes and pol II's CTD: tov3 and tov7 are allelic to SSU72 and tov5 is allelic to PCF11
Whole genome sequencing also revealed that the tov3 and tov7 strains both harbored the same nucleotide substitution in SSU72, an RNA polymerase II phosphatase previously implicated in termination. The A–T change was confirmed by Sanger sequencing of a PCR product from tov3 and tov7 genomic DNA and was unique to these two strains. The resulting change converts leucine84 to a phenylalanine.
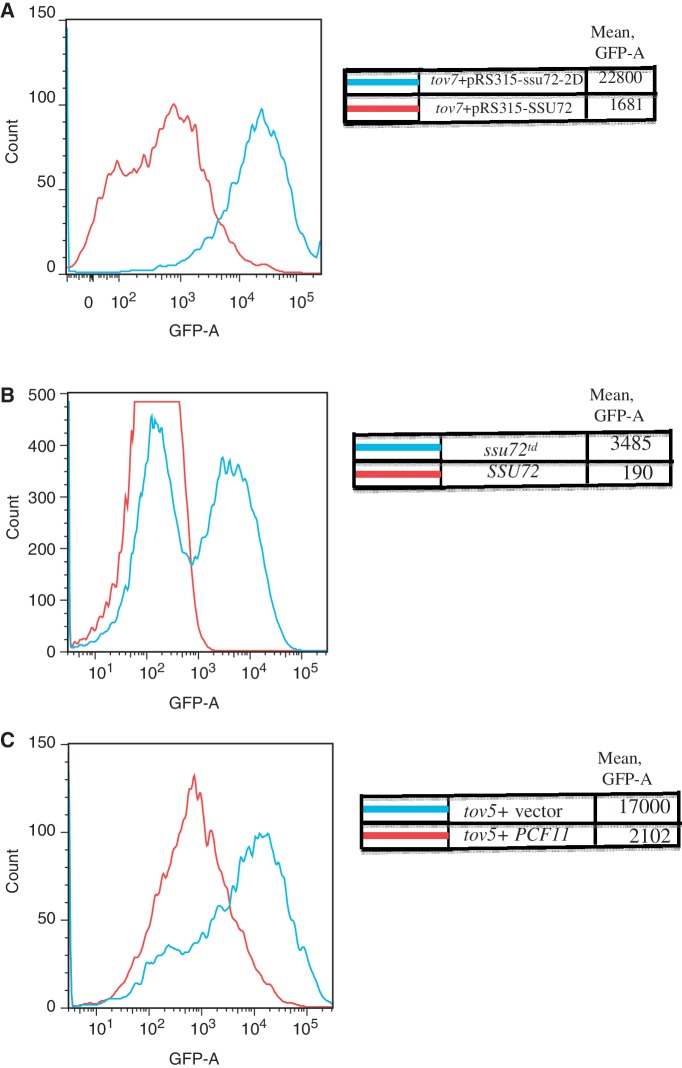
To prove that the ssu72 mutation was causal, we introduced plasmids containing either the wild-type or tov3 (= tov7) allele of SSU72 into the tov7 strain with the GAL-IT-GFP reporter plasmid. Strong termination was restored to the tov strain by the wild-type, but not the mutant, copy of SSU72 (Figure 6A, red versus blue). Similar results were observed for the tov3 strain containing an integrated copy of the GAL-IT-GFP reporter (data not shown).
Figure 6.
Terminator override in ssu72 (tov7) and pcf11 (tov5) mutants. (A) A tov7 strain transformed with the GAL-IT-GFP reporter plasmid and either a plasmid with a wild-type copy of SSU72 (DY3625) or its L84F allele (‘ssu72-2D’; DY3626), were induced with galactose and subjected to flow cytometry. (B) Yeast strains containing an intact (DY3114) or degron derivative (DY3115) of SSU72 were transformed with the GAL-IT-GFP reporter plasmid, induced with galactose and analyzed by flow cytometry. Each curve is the result of averaging five runs of 10 000 cells each. (C) A tov5 strain was transformed with the GAL-IT-GFP reporter plasmid and either a plasmid with a wild-type copy of PCF11 (DY3628) or the empty pRS315 plasmid (DY3627) was induced with galactose and analyzed by flow cytometry.
We also tested if an independently generated, conditional allele of SSU72 (degron strain H291; courtesy of M. Hampsey) (Table 1), showed read-through of the IMD2 IT. Following a shift to the non-permissive temperature, cells containing the GAL-IT-GFP reporter were GFP positive relative to control cells (6B). Together the experiments of Figure 6 show that Ssu72 functions at IMD2 and is the causal basis for terminator override in tov3 and tov7.
Similarly, genome sequencing revealed that the tov5 strain contained a point mutation in PCF11 gene that was not found in any other tov strains. PCF11 is an essential gene that encodes a termination-related protein known to associate with the pol II large subunit's CTD. Conventional Sanger sequencing confirmed this change in tov5, which changes tyrosine94 to serine. A wild-type copy of PCF11 on a plasmid was effective at reversing GFP reporter expression when transformed into a tov5 strain (Figure 6C). Both PCF11 and SSU72 strongly implicate the phosphorylation state of the CTD in IMD2's regulation via the IT.
tov1 and tov4 are allelic to NAB3 and reveal an important function for the protein's extreme carboxy-terminal residues
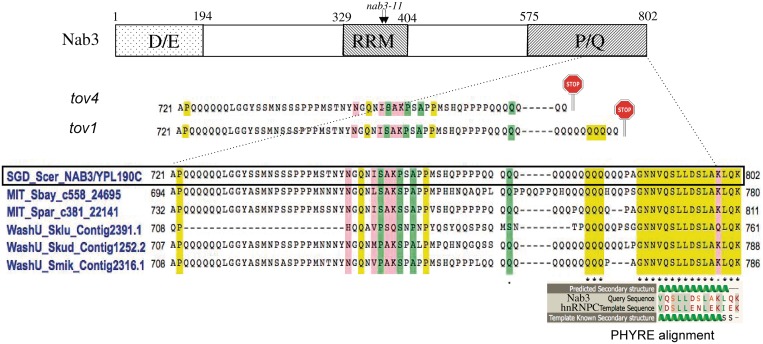
Early in the process of characterizing the tov genes, we explored the possibility that mutations fell in genes previously known to control termination and processing of small RNAs such as CUTs and snoRNAs. We selected SEN1, RRP6, NAB3 and NRD1 as candidate open reading frames for direct conventional sequencing using individually amplified regions from the genomic DNA of some of the tov strains. All strains were wild-type for these except tov1 and tov4 that showed novel TAA nonsense mutations in NAB3. Both fell within a stretch of CAA codons, encoding a tract of 16 glutamine residues near the carboxy terminus. This resulted in the loss of 19 and 27 amino acids from the end of carboxy-terminal end of Nab3 (Figure 7). Thus, it appeared that there was a short region of Nab3's carboxyl-terminal tail that was functionally important for termination. Interestingly, 15 of the 16 carboxy-terminal residues are completely conserved among six Saccharomyces species (asterisks), even though the adjacent sequence, including the glutamine repeat tract, is somewhat variable (Figure 7).
Figure 7.
Map of Nab3 with expansion of the carboxy-terminal sequence and its alignments. A schematic map (not to scale) of the domains of S. cerevisiae Nab3 is shown at the top with the approximate positions of the D/E- and P/Q-rich regions. The RRM piece analyzed by X-ray crystallography (10) is delineated as are the positions of the missense changes in the nab3-11 mutant. A CLUSTALW alignment of the indicated regions of Nab3 from S. cerevisiae (boxed), S. bayanus, S. paradoxus, S. kluyveri, S. kudriavzevii and S. mikatae, is shown in the centre (43,44). Shading designates identities (yellow), strong similarities (pink) and weak similarities (green). The positions of nonsense mutations (stop signs) for the tov1 and tov4 isolates are shown above the alignment. The structural homology alignment generated by the PHYRE algorithm is shown for the known structure of a portion of hnRNP C (template) and the Nab3 tail (query).
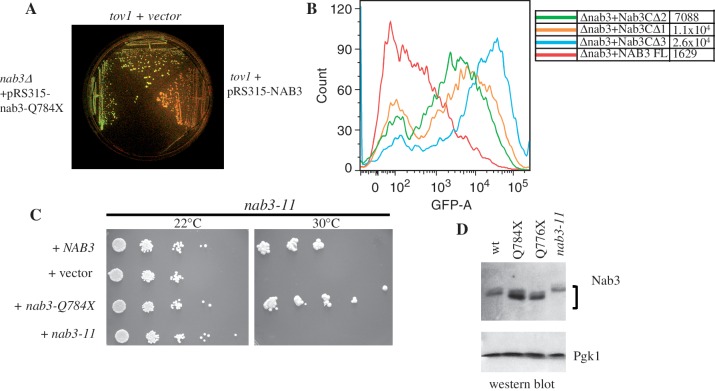
To confirm that the terminator override phenotype was due to the tov1 mutation resulting in the loss of 19 carboxy-terminal amino acids (henceforth referred to as nab3-Q784X), we transformed a diploid heterozygous for a deletion of NAB3, with a plasmid encoding the nab3-Q784X allele expressed from its own promoter. The diploid was sporulated and a haploid was selected that bore the deletion of chromosomal NAB3 and contained the plasmid expressing nab3-Q784X. When this strain was transformed with the GAL-IT-GFP reporter plasmid to test for terminator function, GFP was expressed at levels comparable to the original mutant strain, providing strong evidence that this genetic change alone was sufficient to yield the terminator override phenotype (Figure 8A, left sector). In addition, when wild-type NAB3 was introduced on a plasmid, terminator function (loss of GFP) was restored to both the tov1 (Figure 8A, right sector) and tov4 (data not shown) strains. Genetic complementation was also observed when tov1 cells were mated to a tov2 strain in which NAB3 was wild-type (data not shown).
Figure 8.
Testing of wild-type and mutant NAB3 function. (A) tov1 strains containing the GAL-IT-GFP reporter plasmid and either an empty control vector control (DY3018; top sector) or a plasmid with the NAB3 gene (DY3019; right sector), were grown on galactose along with a strain (DY2240; left sector) deleted for chromosomal NAB3 and containing a plasmid expressing the tov1 mutation of NAB3 (nab3-Q784X). The plate was photographed under blue light to detect GFP fluorescence. (B) A yeast strain lacking its chromosomal copy of NAB3 and containing the GAL-IT-GFP reporter plasmid was transformed with a second plasmid containing the wild-type (‘NAB3FL’; DY3036), the Δ3 (‘nab3CΔ3’; DY3038), the Δ2 (‘nab3CΔ2’; DY3060) or the Δ1 (‘nab3CΔ1’; DY3061) derivatives of NAB3, as indicated. Cells were grown in the presence of galactose and analyzed by flow cytometry. (C) A strain with the nab3-11 allele was transformed with a plasmid containing either the wild-type NAB3 (DY30109), the tov1 allele nab3-Q784X (DY3012), the nab3-11 allele (DY3013) or an empty vector (DY3011). Cultures were serially diluted 10-fold and the dilutions were spotted on selective medium for growth at 22°C or 30°C, as indicated. (D) Western blot for Nab3 in wild-type (DY1514F) and the indicated mutant strains (1A1F = ‘nab3-Q784X’, 4A1F = ‘nab3-Q776X’, DY30229 = ‘nab3-11’). Pgk1 served as a loading control. The nab3-11 strain was grown at the permissive temperature of 22°C.
To define how much of this carboxy-terminal Nab3 ‘tail’ is required for termination, we constructed a nested set of plasmids that deleted the last 3, 6, 9, 12 or 15 residues of the protein. The tov1 strain was transformed with each of these and tested for GFP expression. None could rescue termination function; all constructs yielded strongly fluorescent cells (data not shown). This suggests that the loss of as few as three of Nab3's carboxy-terminal amino acids sufficed to generate the tov phenotype. This was independently confirmed for the construct lacking the carboxy-terminal three amino acids when it was expressed in a strain deleted for the chromosomal copy of NAB3 (Figure 8B, blue versus red). To refine further the minimum portion of the carboxy-terminal tail that supports efficient termination, we generated Nab3 expressing constructs lacking the last one or two amino acids (Figure 8B, orange and green). These versions of Nab3 were partially defective for termination as reflected by their fluorescence profile. This result indicates that the last amino acid in the Nab3 polypeptide is functionally important and the final three amino acids are critical for Nab3 to function properly in termination.
Previous studies of Nab3's function exploited the temperature-sensitive allele nab3-11, which has two amino acid substitutions (F371L and P374T) in the protein's RNA binding domain (Figure 7, top) (4,8,19). To test if the nab3-Q784X mutation could be distinguished from this previously characterized mutation, we transformed a nab3-11 strain with the plasmid expressing nab3-Q784X. Plasmids containing wild-type NAB3 or nab3-Q784X rescued the temperature sensitivity seen in the parent nab3-11 strain (Figure 8C). This result demonstrates that the nab3-Q784X protein is thermostable and has a functional deficit distinct from that of the well-studied nab3-11 mutant that has an altered RRM.
To learn if the truncated proteins encoded by the tov1 and tov4 alleles of NAB3 were stable proteins (nab3-Q784X and nab3-Q776X, respectively), we carried out western blotting using a previously described monoclonal antibody against Nab3 (11). Both truncated Nab3 derivatives (tov1 and tov4) were at least as abundant as wild-type Nab3 (Figure 8D). The mutant proteins also migrated faster than wild-type Nab3 consistent with their slightly shorter lengths.
DISCUSSION
IMD2 expression is regulated by intracellular guanine nucleotides through an IT that can be considered a eukaryotic attenuator (1–4). In a set of proof-of-principle experiments, we have exploited a genetic screen to identify alleles of genes encoding trans-acting factors that operate this terminator. Prior studies by us and others have exploited GFP fluorescence and copper resistance to identify cis-acting sequences involved in termination, as well as start site choice at IMD2 (4,37). Here, we observed that the fluorescence sorting approach was sensitive enough to identify spontaneous mutations in at least four trans-acting genes known to participate in small RNA biogenesis and metabolism, TRF4, SSU72, PCF11 and NAB3. This set of base and amino acid changes are summarized in Table 3. It is doubtful that this screen has been saturated and it is likely that mutagenesis of our starting strain could provide additional genes and alleles that can add to our understanding of the function of this, or even other, terminators. In addition, high-throughput sequencing of genomes from a number of mutants was effective at identifying causal mutations. Whole genome sequencing reveals numerous strain to strain differences, hence identification of genes expected to affect the process under study is straightforward, as observed here through complementation with plasmid-borne wild-type candidates. Finding genes whose role may be unforeseen would be more difficult, requiring, for example, complementation with a genomic library. The four tov genes identified here represent the following components of the transcription termination machinery: (i) Trf4, part of the TRAMP complex, (ii) Ssu72, an enzyme that modulates RNA polymerase II's CTD's phosphorylation state, (iii) Pcf11, a protein that binds to phospho-CTD and (iv) Nab3, an RNA binding, hnRNP-like protein. Further study of the individual resulting mutant proteins should be of mechanistic value in understanding the distinct regulated termination event at IMD2.
Table 3.
tov Gene identification
| tov isolate | Candidate gene | Base change | Amino acid change |
|---|---|---|---|
| tov1 | NAB3 | C2350T | Q784X |
| tov2 | TRF4 | A795ins | K264fs |
| tov3 | SSU72 | A252T | L84F |
| tov4 | NAB3 | C2326T | Q776X |
| tov5 | PCF11 | A281C | Y94S |
| tov6 | TRF4 | C1289G | S430X |
| tov7 | SSU72 | A252T | L84F |
The TRAMP complex links the termination and ribonuclease machineries by directing terminated RNAs for degradation via the unconventional polyadenylation activity of the Trf4 polyA polymerase (29). It facilitates termination and exosomal hydrolysis of transcripts through a physical association with both the Nrd1–Nab3 and exosomal protein complexes (30). Deletion of TRF4 stabilizes a number of CUTs (7,29,30). Our results confirm that the loss of TRF4 takes the IMD2 CUT out of the degradation pathway (Figure 4). In addition, termination read-through was seen in the trf4 mutants examined here, giving the impression that TRF4 influences both the efficiency of CUT degradation and the tightness of termination. The appearance of this read-through transcript could be due to (i) the sparing of a constitutively produced, extended (non-terminated) RNA that would otherwise be degraded if TRAMP was intact and/or (ii) loss of termination efficiency because TRAMP modulates the termination reaction, perhaps through its interaction with Nab3-Nrd1. A less likely possibility is that the trf4 mutation shifts initiation from the upstream to the downstream start site as an alternative means of bypassing the IT.
The Ssu72 phosphatase and the phosphorylation state of the CTD of the large subunit of pol II, are implicated in transcription termination at a number of genes encoding small non-coding RNAs and mRNAs (13,16,17,25,45–49). While a number of SSU72 mutations have been identified by genetic screens, none have yielded the change we found here (L84F). The atomic level structure of Drosophila Ssu72 shows that the cognate leucine (leu82) lines the groove that accommodates the pol II CTD substrate (49). When site-direct mutagenesis was used to probe the importance of the groove residues by changing them to alanine, the substitution of leu82 did not impact phosphatase activity toward a CTD peptide. We suggest that a change to the larger phenylalanine residue obtained in the screen reported here, would influence peptide binding and phosphatase activity. In any case, this report provides biological evidence in favor of the importance of this substrate binding site residue. Our finding contrasts with an earlier study using a copper resistance assay in which a mutation in SSU72 known to affect termination did not strongly perturb the IMD2 CUT terminator's function (4). A higher sensitivity of the fluorescent assay presented here could be part of the basis for this difference. One hypothesis for the L84F mutation's impact upon termination is that the accumulation of ser5-CTD phosphorylation when phosphatase activity is impaired, relaxes the coupling of Nab3–Nrd1's association with the IMD2 CUT in preparation for termination, resulting in higher read-through of the terminator.
The structure of Pcf11 and the details of its binding to pol II's CTD have also been studied at the atomic level (50,51). The CTD-interaction domain of Pcf11 contains an eight helix hydrophobic core. The tov5 mutation of PCF11 is a new allele that changes a tyrosine in a conserved position important for this helical bundle's packing, to a serine. The protein's folding is likely to be changed; whether this results in a reduction of Pcf11 levels and/or interferes with CTD binding, remains to be determined. Pcf11 is known to play a role in transcription termination for mRNA and non-coding RNAs (17,52–55). Recent work has shown that it associates with IMD2 and the A66D allele (Pcf11-9) disrupts termination at the IMD2 CUT (48). The two phospho-CTD binding proteins, Nrd1 and Pcf11 influence termination in a complex manner in response to a gene-specific CTD modification code (48). Further work is needed to learn if Nrd1 and Pcf11 cooperate at this terminator.
The most interesting of the tov mutations was that found in Nab3, a protein whose study has been hampered by the inability to produce a full length recombinant protein (12). NAB3 was previously demonstrated to be involved in termination at IMD2's IT using the well-studied nab3-11 temperature-sensitive allele (4). However, data presented herein draw attention to a distinct and very small part of Nab3 that has not previously been recognized as important, but which is required for termination at IMD2. The critical RRM of Nab3 and its Nrd1 interaction region are located approximately centrally in the 802 amino acid protein (Figure 7) (8). They lie in the most evolutionarily conserved part of the protein in which lethal mutations have been described (8,9). Biochemical and structural analyses have shown that the RNA binding domain forms a canonical βαββαβ RRM sufficient for high-affinity binding to RNA when paired with a corresponding RRM from Nrd1 (9,10,12,18). Difficulty in expressing full length recombinant Nab3, probably due to its extensive polyQ and polyD/E tracts, has meant there has been little, if any, biochemical analysis of either portion of the protein flanking the RRM, much less the complete protein (12). Hence, the function of the bulk of the Nab3 protein remains mysterious. The spontaneous tov1 and tov4 mutations identified here are not temperature sensitive and they do not result in degradation of Nab3 due to misfolding. These alleles rescue the nab3-11ts allele and thus, appear to harbor a defect distinct from the RRM class of mutations. They would not have been picked up in prior ts screens for NAB3 mutants, nor during site-directed mutagenesis of the RNA binding domain (8,9), thereby endorsing the approach used here as an alternative means of mutant selection.
Nrd1 and Nab3 have been considered to be yeast orthologs of mammalian hnRNP proteins. Mammalian correlates of Nrd1 can be identified that contain a recognizable RNA polymerase II CTD interaction domain, an SE/SR-rich region and an RRM (11,21). In contrast, a Nab3 ortholog is much less discernible in higher eukaryotic genomes, although proteins with a small region of homology to the RRM can be detected using the BLAST algorithm (11). Although the carboxy-terminal piece of Nab3 that was lost to mutation in the tov1 and tov4 strains is not required for cell viability, it is important for the termination reaction and optimal cell proliferation (data not shown). Precisely what the role of this tail region is remains to be learned. It is unlikely to bind RNA (9). A clue to its function is offered by the result of computational analyses using the PHYRE and HHpred algorithms, which assess structural relatedness (56,57). Both analyses predict an α-helical structure for the Nab3 tail and the best fit was to a similar region conserved in the hnRNP C family of proteins (Figure 7 and data not shown). Evidence from NMR and biophysical studies indicates that this element of the human hnRNP C protein forms a tetrameric α-helical bundle (58). This suggests that the cognate region of Nab3 is a protein–protein interaction domain, perhaps one that enables Nab3 to multimerize with itself much the way hnRNP C can (59,60). It is intriguing to consider this possibility given that substrate RNAs often contain multiple tandem Nrd1 and Nab3 recognition sites (18). The organization of such sites is variable between different RNA substrates and the sites often appear redundant by mutational analysis (18). Perhaps the degree of multimerization of the Nab3–Nrd1 complex on RNA accounts for some of the variability between transcripts in their dependence upon Nrd1 and Nab3 for termination or processing. The concept of polymerization of Nrd1–Nab3 dimers on RNA was presented previously based upon (i) the multiplicity of RNA binding sites per transcript and (ii) the repeats in the CTD that may bind several copies of Nrd1 in the vicinity of the nascent transcript (12). Self association of the Nab3 tail could offer an additional source of scaffolding through which an array of Nrd1–Nab3 dimers assemble onto RNA, potentially forming a higher order packaging of this yeast ribonucleoprotein.
Regulation of IMD2 expression has proven useful as a model to study the Nrd1–Nab3–Sen1 termination system, CTD phosphorylation and the degradation of CUTs. Additional work on the mutations reported here should shed light on this complex mechanism.
FUNDING
Funding for open access charge: National Institutes of Health [GM46331 to D.R.].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank R. Karaffa and S. Durham of the Emory FACS Core Facility for cytometry services, the Emory GRA Genome Center for sequencing services, Drs M. Swanson, M. Hampsey, M. Fasken, A. Corbett and P. Doetsch for materials, Dr N. Degtyareva for valuable advice and tetrad dissection and Drs R. Kahn, A. Corbett, X. Cheng and G. Crouse for helpful discussions.
REFERENCES
- 1.Dichtl B. Transcriptional ShortCUTs. Mol. Cell. 2008;31:617–618. doi: 10.1016/j.molcel.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Corden JL. Yeast Pol II start-site selection: the long and the short of it. EMBO Rep. 2008;9:1084–1086. doi: 10.1038/embor.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenks MH, O’Rourke TW, Reines D. Properties of an intergenic terminator and start site switch that regulate IMD2 transcription in yeast. Mol. Cell Biol. 2008;28:3883–3893. doi: 10.1128/MCB.00380-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuehner JN, Brow DA. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell. 2008;31:201–211. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 5.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 6.Lykke-Andersen S, Jensen TH. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie. 2007;89:1177–1182. doi: 10.1016/j.biochi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Davis CA, Ares M., Jr Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad NK, Wilson SM, Steinmetz EJ, Patturajan M, Brow DA, Swanson MS, Corden JL. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics. 2000;154:557–571. doi: 10.1093/genetics/154.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobor F, Pergoli R, Kubicek K, Hrossova D, Bacikova V, Zimmermann M, Pasulka J, Hofr C, Vanacova S, Stefl R. Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein. J. Biol. Chem. 2011;286:3645–3657. doi: 10.1074/jbc.M110.158774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunde BM, Horner M, Meinhart A. Structural insights into cis element recognition of non-polyadenylated RNAs by the Nab3-RRM. Nucleic Acids Res. 2011;39:337–346. doi: 10.1093/nar/gkq751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson SM, Datar KV, Paddy MR, Swedlow JR, Swanson MS. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J. Cell Biol. 1994;127:1173–1184. doi: 10.1083/jcb.127.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll KL, Ghirlando R, Ames JM, Corden JL. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA. 2007;13:361–373. doi: 10.1261/rna.338407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nedea E, Nalbant D, Xia D, Theoharis NT, Suter B, Richardson CJ, Tatchell K, Kislinger T, Greenblatt JF, Nagy PL. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell. 2008;29:577–587. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 14.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 15.Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 16.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol. Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol. Cell. Biol. 2004;24:6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 20.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 21.Steinmetz EJ, Brow DA. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinmetz EJ, Brow DA. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl Acad. Sci. USA. 1998;95:6699–6704. doi: 10.1073/pnas.95.12.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 24.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 25.Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat. Struct. Mol. Biol. 2008;15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- 26.Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat. Rev. Mol. Cell Biol. 2011;12:283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grzechnik P, Kufel J. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol. Cell. 2008;32:247–258. doi: 10.1016/j.molcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia H, Wang X, Liu F, Guenther UP, Srinivasan S, Anderson JT, Jankowsky E. The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell. 2011;145:890–901. doi: 10.1016/j.cell.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 30.Anderson JT, Wang X. Nuclear RNA surveillance: no sign of substrates tailing off. Crit. Rev. Biochem. Mol. Biol. 2009;44:16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- 31.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucl. Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He X, Khan AU, Cheng H, Pappas DL, Jr, Hampsey M, Moore CL. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003;17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 34.Riles L, Shaw RJ, Johnston M, Reines D. Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast. 2004;21:241–248. doi: 10.1002/yea.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw RJ, Wilson JL, Smith KT, Reines D. Regulation of an IMP dehydrogenase gene and its overexpression in drug-sensitive transcription elongation mutants of yeast. J. Biol. Chem. 2001;276:32905–32916. doi: 10.1074/jbc.M105075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyle JW, Shaw RJ, Reines D. Functional distinctions between IMP dehydrogenase genes in providing mycophenolate resistance and guanine prototrophy to yeast. J. Biol. Chem. 2003;278:28470–28478. doi: 10.1074/jbc.M303736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopcewicz KA, O’Rourke TW, Reines D. Metabolic regulation of IMD2 transcription and an unusual DNA element that generates short transcripts. Mol. Cell. Biol. 2007;27:2821–2829. doi: 10.1128/MCB.02159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hegemann JH, Klein S, Heck S, Guldener U, Niedenthal RK, Fleig U. A fast method to diagnose chromosome and plasmid loss in Saccharomyces cerevisiae strains. Yeast. 1999;15:1009–1019. doi: 10.1002/(SICI)1097-0061(199907)15:10B<1009::AID-YEA396>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 39.Ishii J, Izawa K, Matsumura S, Wakamura K, Tanino T, Tanaka T, Ogino C, Fukuda H, Kondo A. A simple and immediate method for simultaneously evaluating expression level and plasmid maintenance in yeast. J. Biochem. 2009;145:701–708. doi: 10.1093/jb/mvp028. [DOI] [PubMed] [Google Scholar]
- 40.Houalla R, Devaux F, Fatica A, Kufel J, Barrass D, Torchet C, Tollervey D. Microarray detection of novel nuclear RNA substrates for the exosome. Yeast. 2006;23:439–454. doi: 10.1002/yea.1369. [DOI] [PubMed] [Google Scholar]
- 41.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamill S, Wolin SL, Reinisch KM. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc. Natl Acad. Sci. USA. 2010;107:15045–15050. doi: 10.1073/pnas.1003505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 44.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 45.Dichtl B, Blank D, Ohnacker M, Friedlein A, Roeder D, Langen H, Keller W. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell. 2002;10:1139–1150. doi: 10.1016/s1097-2765(02)00707-4. [DOI] [PubMed] [Google Scholar]
- 46.Ganem C, Devaux F, Torchet C, Jacq C, Quevillon-Cheruel S, Labesse G, Facca C, Faye G. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 2003;22:1588–1598. doi: 10.1093/emboj/cdg141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinmetz EJ, Brow DA. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 2003;23:6339–6349. doi: 10.1128/MCB.23.18.6339-6349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Zhang M, Zhang Y. Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue. Biochem. J. 2011;434:435–444. doi: 10.1042/BJ20101471. [DOI] [PubMed] [Google Scholar]
- 50.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 51.Noble CG, Hollingworth D, Martin SR, Ennis-Adeniran V, Smerdon SJ, Kelly G, Taylor IA, Ramos A. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat. Struct. Mol. Biol. 2005;12:144–151. doi: 10.1038/nsmb887. [DOI] [PubMed] [Google Scholar]
- 52.Barilla D, Lee BA, Proudfoot NJ. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2001;98:445–450. doi: 10.1073/pnas.98.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadowski M, Dichtl B, Hubner W, Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammell CM, Gross S, Zenklusen D, Heath CV, Stutz F, Moore C, Cole CN. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 2002;22:6441–6457. doi: 10.1128/MCB.22.18.6441-6457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 56.Soding J. Protein homology detection by HMM–HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 57.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 58.Whitson SR, LeStourgeon WM, Krezel AM. Solution structure of the symmetric coiled coil tetramer formed by the oligomerization domain of hnRNP C: implications for biological function. J. Mol. Biol. 2005;350:319–337. doi: 10.1016/j.jmb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 59.Huang M, Rech JE, Northington SJ, Flicker PF, Mayeda A, Krainer AR, LeStourgeon WM. The C-protein tetramer binds 230 to 240 nucleotides of pre-mRNA and nucleates the assembly of 40S heterogeneous nuclear ribonucleoprotein particles. Mol. Cell. Biol. 1994;14:518–533. doi: 10.1128/mcb.14.1.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conway G, Wooley J, Bibring T, LeStourgeon WM. Ribonucleoproteins package 700 nucleotides of pre-mRNA into a repeating array of regular particles. Mol. Cell. Biol. 1988;8:2884–2895. doi: 10.1128/mcb.8.7.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]