Abstract
Multisubunit RNA polymerase (RNAP) is the central information-processing enzyme in all cellular life forms, yet its mechanism of translocation along the DNA molecule remains conjectural. Here, we report direct monitoring of bacterial RNAP translocation following the addition of a single nucleotide. Time-resolved measurements demonstrated that translocation is delayed relative to nucleotide incorporation and occurs shortly after or concurrently with pyrophosphate release. An investigation of translocation equilibrium suggested that the strength of interactions between RNA 3′ nucleotide and nucleophilic and substrate sites determines the translocation state of transcription elongation complexes, whereas active site opening and closure modulate the affinity of the substrate site, thereby favoring the post- and pre-translocated states, respectively. The RNAP translocation mechanism is exploited by the antibiotic tagetitoxin, which mimics pyrophosphate and induces backward translocation by closing the active site.
INTRODUCTION
RNA polymerase (RNAP) mediates the synthesis of an RNA copy of template DNA—an essential step in gene expression. All cellular RNAPs are multisubunit enzymes that share homologous approximately 2500 amino acid catalytic cores (Figure 1). Transcribing RNAPs do not displace the non-template strand, but rather form a transient 9–10 bp RNA–DNA hybrid within the 11–12 bp melted DNA bubble. RNAPs cannot re-engage a prematurely released transcript and therefore must synthesize the entire RNA without dissociating from the nascent RNA–DNA scaffold. Nucleotide addition occurs through the Mg2+-dependent, SN2 nucleophilic attack of the RNA 3′-OH on the α-phosphate of the incoming nucleoside triphosphate (NTP) (1). Catalysis involves near obligatory closure of the active site by a mobile RNAP element called the trigger loop (TL), which orients substrate NTPs for efficient catalysis (2–3). The 3′ terminal nucleotide of the nascent RNA can occupy two distinct positions within the RNAP active site: the nucleophilic site (a.k.a. i site), where the RNA 3′-OH group is activated for attack on substrate NTPs, and the substrate site (a.k.a. i + 1 site), where the new RNA 3′ end is generated after the substrate is joined to the nascent RNA. The former corresponds to the post-translocated state and the latter to the pre-translocated state. Each nucleotide-incorporation event must therefore be followed by translocation of RNAP along the DNA. Upon translocation, the RNA 3′ nucleotide migrates into the nucleophilic site, whereas the substrate site captures a new DNA-template position. RNAP movement is ultimately powered by the free energy liberated as NTPs are condensed into the nascent RNA and pyrophosphate (PPi) is released (4). This energy is presumably used indirectly. According to a proposed Brownian ratchet model, the transcription elongation complex (TEC) oscillates between the pre- and post-translocated states (the RNA 3′ nucleotide alternates between substrate and nucleophilic sites) driven by thermal fluctuations, whereas the growing RNA and incoming NTP substrates bias the assembly towards forward translocation (5–7). Despite the apparent lack of direct coupling between catalysis and translocation, Escherichia coli RNAP is a powerful molecular motor that progresses along the DNA at speeds of ∼20–90 nt s−1 and generates a force of up to 25 pN (6,8). Although the mechanism of RNAP translocation has been discussed extensively for over a decade (9–12) and conceivable models have been proposed based on structures of the TECs (13–14), exonuclease foot-printing analyses (5) and single-molecule experiments (6–7), the translocational motion of RNAP during a single nucleotide addition cycle have not yet been resolved. In this study, we developed an assay for monitoring RNAP translocation over a single base pair in real-time using fluorescent base analogs incorporated into template DNA. Our kinetic and equilibrium studies using this method converge on the identification of active site opening and closure as the central events governing multisubunit RNAP translocation. We also explain how the antibiotic tagetitoxin (TGT) hijacks RNAP′s translocation mechanism, and further propose that PPi mimicry is a common theme among inhibitors of nucleic acid polymerases that target the translocation step.
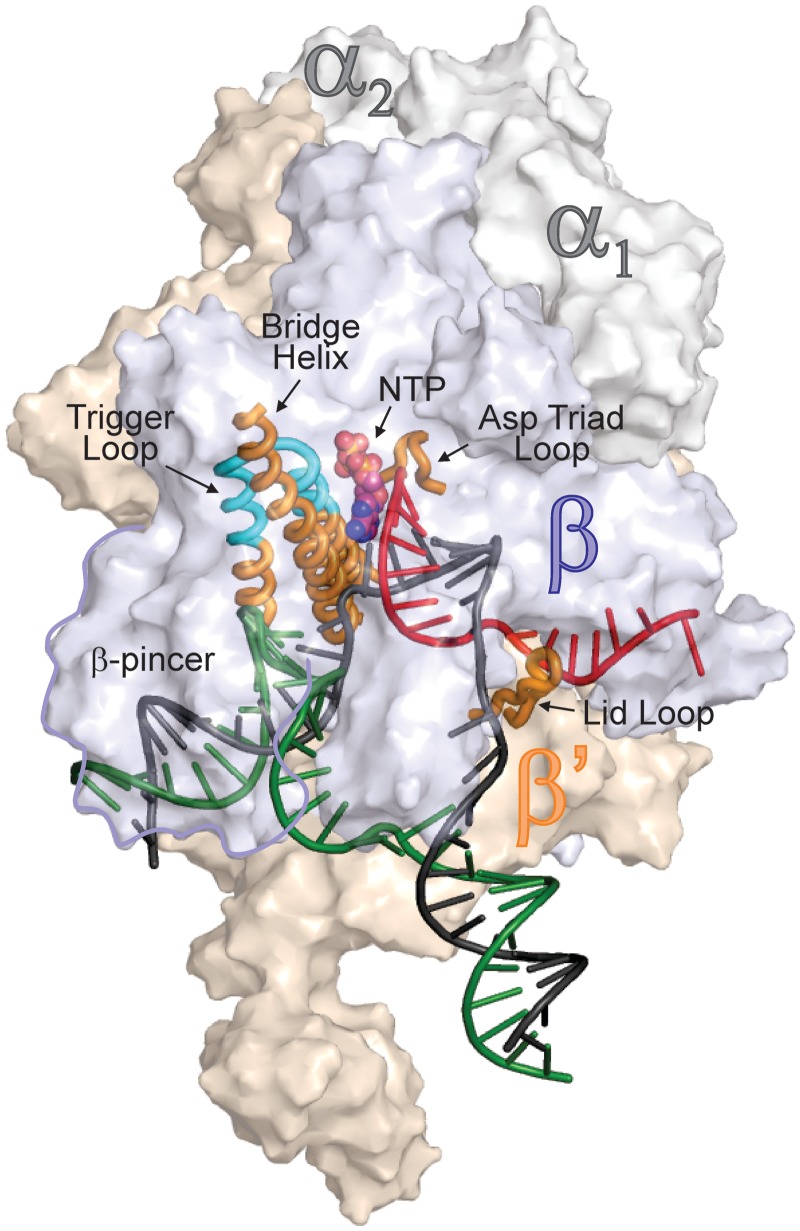
Figure 1.
An overview of bacterial TEC structure. β (light blue), β′ (wheat) and two (gray) subunits are depicted as transparent surfaces; the non-essential ω subunit is obstructed by β and β′ subunits. Template DNA, non-template DNA and RNA are depicted in black, green and red, respectively. The β′ subunit elements discussed throughout the manuscript are depicted as cartoons: the folded TL is colored cyan; the bridge helix (BH), lid loop, TL base and catalytic aspartate triad loop are colored orange. The β-pincer domain is outlined. Substrate NTP atoms are shown as spheres. RNAP is drawn using coordinates of Thermus thermophilus TEC PDB ID 2O5J, and nucleic acids are taken from the E. coli TEC model (15).
MATERIALS AND METHODS
Proteins and reagents
DNA and RNA oligonucleotides were purchased from IBA Biotech (Göttingen, Germany) and Fidelity Systems (Gaithersburg, MD, USA). TGT was from Epicentre (Madison, WI, USA). RNAPs and yeast inorganic pyrophosphatase (PPase) were expressed and purified as described previously (16,17). Escherichia coli phosphate binding protein (PBP) was expressed, purified and labeled with 7-diethylamino-3-[N-(2-maleimidoethyl)carbamoyl]coumarin (MDCC) as described previously (18). Plasmids are listed in Supplementary Table S3.
TEC assembly
TECs (1 µM) were assembled by a procedure developed by Komissarova et al. (19). An RNA primer labeled with Atto680 fluorescent dye at the 5′ end was annealed to template DNA, and incubated with 1.5 µM RNAP for 10 min at 25°C in TB10 buffer (10 mM MgCl2, 40 mM HEPES–KOH, pH 7.5, 80 mM KCl, 5% glycerol, 0.1 mM EDTA and 0.1 mM DTT) and with 2 µM of the non-template DNA for 20 min at 25°C. For TECs used in nucleotide addition measurements, RNA was the limiting component at 1 µM, and the template strand was used at 1.4 µM, whereas for TECs used in translocation and PPi release measurements, the template strand was limiting at 1 µM, and RNA was added at 1.4 µM. Template strand oligonucleotides are listed in Supplementary Table S4.
Nucleotide addition measurements
To determine the incorporation efficiency of NTP, 2′ and 3′ dNTP substrates, 1 µM TEC in a total volume of 20 µl of TB10 buffer was incubated for 10 min (1.5 h for ΔTL RNAP) with 5 µM substrates at 25°C and quenched by adding 80 µl of loading buffer (94% formamide, 13 mM Li4-EDTA and 0.2% Orange G). Time-resolved measurements were performed in an RQF 3 quench-flow instrument (KinTek Corporation, Austin, TX, USA). The reaction was initiated by rapid mixing of 14 µl of 0.5 µM TEC with 14 µl of 400 µM NTP (both solutions in TB10 buffer). The reaction was allowed to proceed for 0.004–10 s at 25°C and quenched with 86 µl of 0.5 M HCl and immediately neutralized by adding 171 µl of loading buffer (290 mM Tris base, 13 mM EDTA, 0.2% Orange G and 94% formamide). Product RNA were separated on 16% denaturing polyacrylamide gels and RNA species were visualized with Odyssey Infrared Imager (Li-Cor Biosciences, Lincoln, NE, USA); band intensities were quantified using ImageJ software (20).
Translocation measurements
Translocation of RNAP along the DNA was assayed by monitoring changes in fluorescence of base analogs incorporated into template DNA. Equilibrium levels of fluorescence of base analogs were determined by recording emission spectra of 6-methyl-isoxanthopterin (6-MI; 0.1–0.2 µM TEC; excitation at 340 nm) and 2-aminopurine (2-AP; 0.5–1 µM TEC; excitation at 330 nm) with an LS-55 spectrofluorometer (PerkinElmer, Waltham, MA, USA). The fluorescence at peak emission wavelength (420 and 375 nm for 6-MI and 2-AP, respectively) was used for data analysis and representation. Prior to spectral measurements, TEC was incubated for 10 min (1.5 h for ΔTL RNAP) with 5 µM substrates and 0.2 nM yeast PPase in 100 µl of TB10 buffer at 25°C. Where indicated, TGT and cytidine-5′-[(α,β)-methyleno]triphosphate (CMPcPP) were added post-catalytically and incubated for an additional 5 min. Time-resolved measurements were performed in an Applied Photophysics (Leatherhead, UK) SX.18MV stopped-flow instrument at 25°C. The reaction was initiated by mixing 60 µl of 0.4 µM TEC with 60 µl of 400 µM NTP (both solutions in TB10 buffer). The NTP solution was supplemented with 10 µM TGT or 400 µM CMPcPP, where indicated. 6-MI and 2-AP fluorophores were excited at 340 and 320 nm, respectively, and emitted light was collected through 400- or 375-nm long pass filters, respectively. At least three individual traces were averaged for each reported curve.
PPi release measurements
The release of PPi from the active site was monitored using a coupled assay involving PPase-mediated PPi hydrolysis and Pi detection with an MDCC fluorophore-labeled PBP (MDCC-PBP), which shows strong fluorescence enhancement upon binding to Pi (21,22). The assay was performed in an Applied Photophysics SX.18MV stopped-flow instrument at 25°C. The reaction was initiated by mixing 60 µl of a 400 µM NTP solution containing 5 µM MDCC-PBP and 100 pM yeast PPase with 60 µl of a 0.4 µM TEC solution containing 5 µM MDCC-PBP and 20 µM yeast PPase (both solutions in TB10 buffer). MDCC-PBP fluorescence was excited at 430 nm, and emitted light was collected through a 455-nm long pass filter. The reported curves were averaged from 6 to 7 individual measurements. The sample syringes and lines of the stopped-flow system, as well as NTP stocks, were treated with Pi-mop (100 µM 7-methyl guanosine, 0.1 U ml−1 purine nucleoside phosphorylase) to reduce Pi contamination (21).
Data analyses
Kinetic data were simultaneously fit to one- and two-step models
defined by a set of differential equations, using the numerical integration capabilities of Scientist 2.01 software (Micromath, St Louis, MO, USA). The heterogeneity of the initial TEC (10–20% of 10- to 30-fold slower TEC) was taken into account by invoking two rate constants for the nucleotide addition step and applying a parameter defining the fraction of the correct (fast) TEC. The rates of PPi release determined by fitting the data to the two-step model were further corrected for the 5.8-ms delay of the fluorescent signal (signal generation rate, 120 s−1) attributed to PPi hydrolysis by PPase and Pi binding to MDCC-PBP. TGT titration data were fit to the dissociation equilibrium equations that accounted for changes in TEC, TGT and CMPcPP concentration upon formation of TEC–TGT and TEC–CMPcPP complexes using Scientist 2.01 software. A detailed description of the models used to fit the data is presented in Supplementary Methods.
RESULTS
Monitoring RNAP translocation using fluorescent base analogs
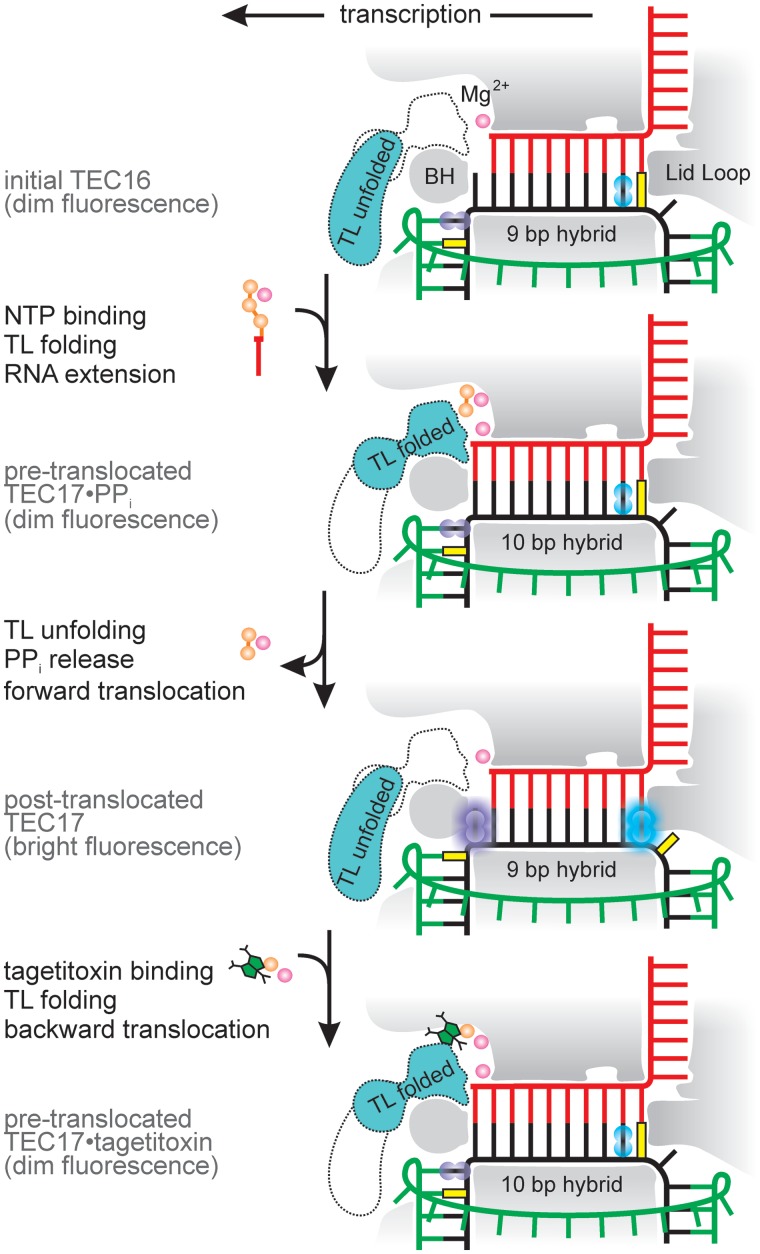
To monitor RNAP translocation over a single base pair step (3.7 Å) in a real time, we developed a beacon-like setup utilizing base analog fluorophores (23–26) and the natural base, guanine, as a quencher. The initial TECs were assembled with E. coli RNAP on a chemically synthesized nucleic acid scaffold containing 16 nt RNAs forming a 9-bp RNA–DNA hybrid and fully complementary template and non-template strands (19). The base analog fluorophore was incorporated into the template strand in positions i + 2 or i − 7, with a neighboring guanine nucleotide (quencher) positioned downstream and upstream, respectively (Figure 2 and Supplementary Figure S1). In TECs that incorporated a nucleotide (extended TECs) and translocated, the fluorophore migrated into i + 1 or i − 8 positions and became separated from the guanine quencher by the β′ subunit bridge helix and the lid loop, respectively, with a concomitant 2- to 5-fold increase in fluorescence.
Figure 2.
Overview of the experimental setup used in this study. RNAP is shaded gray. Two alternative conformations of the TL domain are indicated, with the predominant conformation shaded in turquoise (see ‘Discussion’ section). Template DNA, non-template DNA and RNA are depicted in black, green and red, respectively. Although presented as a bifluorophore for compact representation, each TEC in our experiments possessed only one of the indicated fluorophores: either guanine analog 6-MI (cyan) at i − 7 or adenine analog 2-AP (violet) at i + 2 (for the initial TECs). Guanine, serving as a quencher, is depicted in yellow.
We found that either upstream or downstream placement of fluorophore is suitable for monitoring translocation. However, the upstream placement offers several advantages: (i) the fluorophore is positioned far away from the active site and thus is less likely to respond to translocation-unrelated motions, such as TL folding and substrate NTP binding, (ii) 6-MI, a brighter fluorophore but one more divergent in structure from natural bases, can be employed in place of the 2-AP used in the downstream system. We suggest that the nature of the base is of lesser importance at the upstream side of the RNA–DNA hybrid because i − 8 base contacts with the flexible lid loop are weak, in contrast to intimate and potentially specific interactions of the i + 1 base with the bridge helix and (iii) the transcribed sequence can be chosen freely. Accordingly, most of the data presented here were obtained using upstream placement of the fluorophore, though a limited set of experiments was also performed using downstream placement and produced quantitatively similar results.
Translocation is delayed relative to nucleotide incorporation and occurs shortly after or concurrently with pyrophosphate release
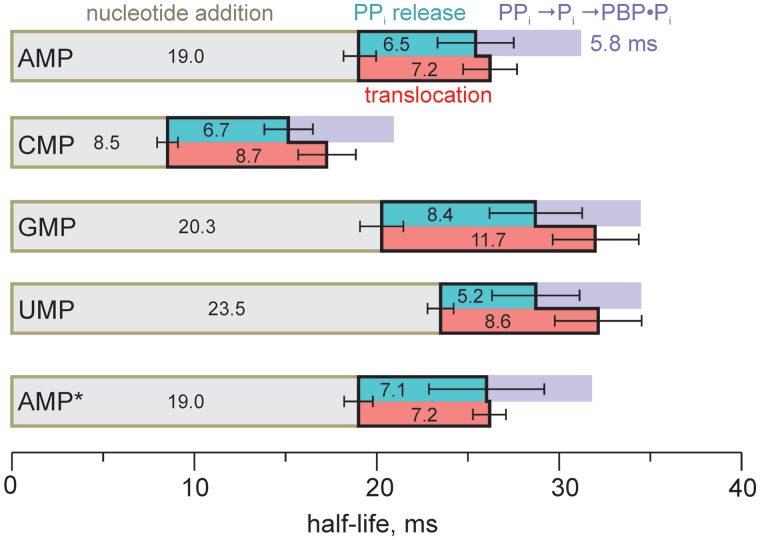
We performed parallel, time-resolved measurements of three individual steps in the AMP, CMP, GMP and UMP addition reactions. RNA extension was monitored utilizing a rapid chemical quench-flow method (27). PPi release from the active site was monitored with a coupled assay involving PPase-mediated PPi hydrolysis and Pi detection with a MDCC-PBP (21). Translocation of RNAP along the DNA was monitored with fluorescent base analogs, as described above. For each of the four incorporated nucleotides, the translocation and PPi release curves were delayed relative to the nucleotide-addition curves (Figures 3 and 4 and Supplementary Figure S2). Fitting the data to a two-step model (Supplementary Methods) revealed that the rates of translocation (t1/2 7–12 ms) and PPi release (t1/2 6–9 ms) were uncorrelated with the rate of nucleotide addition and were largely independent of the nature of the incorporated nucleotide (Figure 4 and Supplementary Table S1). Accordingly, RNAP spends from half (for fast-incorporating CMP; RNA extension t1/2 ∼8 ms) to one-third (for slower incorporating AMP, GMP and UMP; RNA extension t1/2 ∼20 ms) of the total reaction time on post-catalytic steps involving translocation and PPi release. However, the values determined should only be considered in context as the first such estimates ever obtained; large variations may exist depending on the sequence of the RNA–DNA hybrid (94 sequence combinations). Consistent with occlusion of the substrate site in the pre-translocated TEC by the nascent RNA and PPi, a non-hydrolyzable analog of the next substrate NTP did not measurably affect translocation traces (Supplementary Figure S2, inset in GMP addition graph).
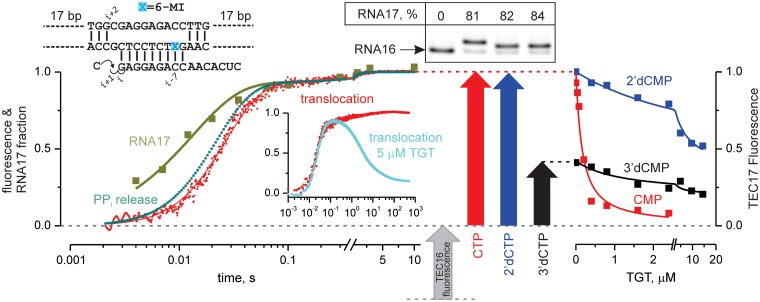
Figure 3.
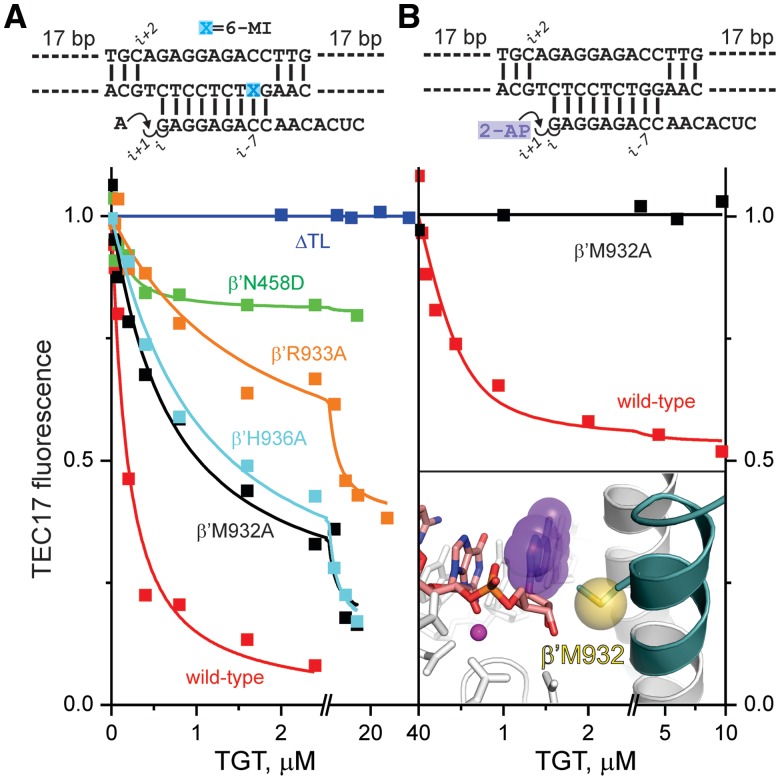
Kinetic and equilibrium analyses of translocation following CMP incorporation. Top: The initial TEC schematic. Left: Time-resolved measurements of RNA extension by a single CMP nucleotide (olive), release of the reaction by-product PPi (dark teal; simulated as described in Supplementary Figure S3), and translocation of the RNA–DNA hybrid within the RNAP main channel (red). Inset: Translocation curves measured over a 10-min interval in the absence (red) and presence (cyan) of 5 µM TGT. Center: Denaturing polyacrylamide gel of TEC RNAs and fluorescence levels of initial (gray), and CMP (red)-, 2′-dCMP (blue)- and 3′-dCMP (black)-extended TECs. Here and in all subsequent figures, fluorescence levels were normalized as described in Supplementary Methods. Note the faster electrophoretic mobility of 2′- and 3′-dCMP-extended RNAs. Right: Titration of extended TECs with TGT.
Figure 4.
Contributions of the individual steps to the overall transcription rate. The half-lives of nucleotide addition, PPi release and translocation reactions are depicted as olive, turquoise and red bars, respectively. Purple bars depict the combined half-life of PPi hydrolysis into Pi and Pi binding to MDCC-PBP (Supplementary Figure S3), which accounted for the 5.8-ms delay in the fluorescence signal in PPi release measurements. Error bars are SDs of two to three independent experiments. AMP, CMP, GMP and UMP data were obtained with i − 7 6-MI TECs; AMP* data were obtained with i + 2 2-AP TEC (Supplementary Figure S2).
Interestingly, Johnson et al. (28) previously reported that the rate of PPi release from the TEC determined by the PPase-purine ribonucleoside phosphorylase assay was very slow (2.9 s−1) in the absence of the next incoming substrate NTP. In our experiments, the same TEC rapidly released PPi (120 s−1) when assayed using the PPase-MDCC-PBP method. We cannot unequivocally explain this discrepancy, but emphasize that our results were obtained using a method extensively validated for use in a stopped-flow setup (29) and are consistent with the data obtained for several other TECs with different NTPs as cognate substrates. On the other hand, it is not clear how an assay based on purine ribonucleoside phosphorylase, an enzyme with a turnover number of 40 s−1 and Km of 26 µM (30), can be used to monitor the millisecond timescale release of nanomolar concentrations of Pi and output rates as high as 500–600 s−1 (28). Similarly, we argue that the 1–2 nM PPase (0.03 U) in the assay used by Johnson et al. was insufficient for rapid PPi hydrolysis, given that the 10 µM PPase used in our setup still partially limited the PPi to MDCC-PBP·Pi conversion rate of 120 s−1 (Supplementary Figure S3).
TGT induces backward translocation in a TL-dependent fashion
TGT, a bacterial RNAP inhibitor that acts at the elongation stage, has been suggested to influence translocation based on its effects on TEC′s exonuclease footprints (31). Consistent with this, we showed that TGT quantitatively moves extended TECs into the pre-translocated state when added post-catalytically (Figures 3, 5 and 6 and Supplementary Figure S2). TGT acted competitively with the non-hydrolyzable analog of the next substrate NTP: the dependence of GMP extended TEC fluorescence on TGT and CMPcPP concentrations was adequately described by the mutually exclusive binding of TGT and CMPcPP to the TEC (Figure 6). The thermodynamic stabilities of the post-translocated state (presumed to be related to KTGT; Table 1) was dependent on the 3′ RNA nucleotide and decreased in the order G > A > C > U, in full agreement with the results inferred from the sensitivity of TECs to pyrophosphorolysis (32). Given that the forward translocation rate was largely insensitive to the nature of incorporated nucleotide (Figure 4), this result suggests that the nature of the 3′ nucleobase predominantly influences the rate of backward translocation.
Figure 5.
The effects of amino acid substitutions in RNAP on translocation equilibrium. Schematics of the initial scaffolds are indicated above the graphs. (A) TGT titration of AMP-extended TECs assembled using wild-type (red), ΔTL (blue), β′N458D (green), β'R933A (orange), β′H936A (cyan) and β′M932A (black) RNAPs. (B) TGT titration of 2-AP ribonucleoside monophosphate-extended TECs assembled using wild-type and β′M932A RNAPs. Inset: Upon TL folding (dark teal helix) and backward translocation, the sulfur atom of β′Met932 (yellow sphere) closely (∼5 Å) approaches the 2-AP nucleobase (violet) at the RNA 3′ end, thereby quenching its fluorescence. Active-site Mg2+ is depicted as a magenta sphere. Catalytic aspartates and β′Asn458 are shown as sticks. The representation is drawn using the model of the pre-translocated TEC of T. thermophilus RNAP (see Supplementary Figure S4A); residue numbers correspond to E. coli RNAP.
Figure 6.
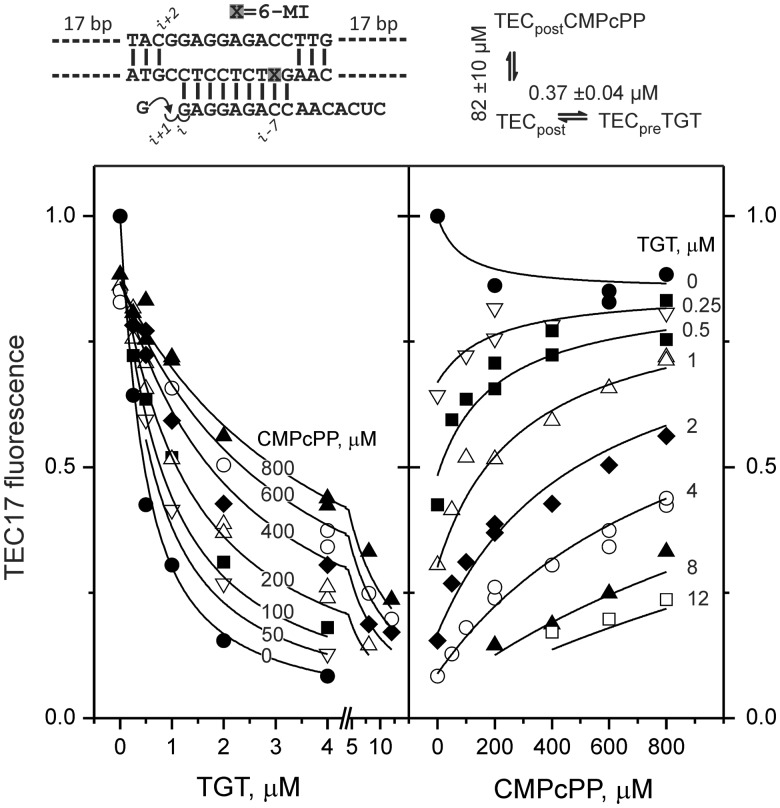
Normalized fluorescence of GMP-extended TEC as a function of TGT and CMPcPP concentrations. A schematic of the initial scaffold and kinetic model used for global analysis of titration data is indicated above the graph. Reaction species in the model are abbreviated as follows: TECpost, post-translocated TEC17; TECpreTGT, pre-translocated TEC17 with bound TGT; TECpostCMPcPP, post-translocated TEC17 with bound CMPcPP. CMPcPP and TGT dissociation constants are indicated. The best-fit value of the TGT dissociation constant is 2-fold higher than the value for GMP-extended TEC reported in Table 1 due to the use of a different batch of TGT.
Table 1.
Apparent TGT dissociation constants
| RNA 3′end | RNAP | Fluorophore |
KTGT (μM) |
||
|---|---|---|---|---|---|
| base\sugar | rRiboside | 2′-Ddeoxy | 3′-Deoxy | ||
| Adenine | Wild-type | 6-MI i − 8 → i − 7 | 0.13 ± 0.04 | 2.1 ± 0.9 | 0.84 ± 0.38 |
| Cytosine | Wild-type | 6-MI i − 8 → i − 7 | 0.11 ± 0.04 | 3.8 ± 1.1 | 1.1 ± 0.7 |
| Guanine | Wild-type | 6-MI i − 8 → i − 7 | 0.18 ± 0.05 | 3.6 ± 1.0 | 1.3 ± 0.3 |
| Uracil | Wild-type | 6-MI i − 8 → i − 7 | 0.050 ± 0.001 | 2.8 ± 0.6 | 2.5 ± 0.2 |
| Adenine | Wild-type | 2-AP i + 1 → i + 2 | 0.15 ± 0.05 | 2.3 ± 0.6 | 1.7 ± 0.5 |
| Adenine | Wild-type | 2-AP i → i + 1 (RNA) | 0.12 ± 0.10 | ||
| Adenine | β′M932A | 6-MI i − 8 → i − 7 | 0.65 ± 0.16 | ||
| Adenine | β′R933A | 6-MI i − 8 → i − 7 | 1.64 ± 0.45 | ||
| Adenine | β′H936A | 6-MI i − 8 → i − 7 | 0.94 ± 0.15 | ||
Errors are standard deviations of the best-fit estimates and were calculated by nonlinear regression of TEC fluorescence versus TGT concentration data using the model described in Supplementary Methods.
In time-resolved measurements, translocation traces with and without saturating amounts of TGT were superimposed up to 200 ms, by which forward translocation and PPi release were essentially completed. Fluorescence returned to near initial levels on a second′s timescale in the presence of TGT, but remained unchanged for at least 10 min in the absence of TGT (Figure 3, left panel, inset). Apparently, TGT does not affect the rate of forward translocation and specifically induces backward translocation of post-translocated, PPi-less TECs.
The effect of TGT was dependent on the presence of the TL domain, as evidenced by the fact that ΔTL RNAP (RNAP with the TL domain replaced by a two-alanine linker) TEC was completely resistant to TGT action (Figure 5A). Recent site-directed mutagenesis studies suggest that TGT acts by stabilizing the TL in a folded state (31). To determine whether the active site of a TGT-saturated TEC is indeed closed by a folded TL, we incorporated a base analog fluorophore at the RNA 3′ end by extending the non-fluorescent TEC with 2-AP ribonucleoside triphosphate. Our prediction was that, upon TGT-induced folding of the TL, 2-AP fluorescence would be quenched by the sulfur atom of TL β′Met932, which stacks against the 3′ RNA base (Figure 5B, inset). Indeed, TGT quenched the fluorescence of 3′ RNA 2-AP by 2-fold, with a KTGT in the submicromolar range (Figure 5B; Table 1). In contrast, addition of TGT did not quench the fluorescence of 3′ RNA 2-AP in the β′M932A TEC. This lack of quenching was attributed entirely to the absence of sulfur proximal to the 2-AP base because a β′M932A TEC identical in sequence to that used in 2-AP experiments but possessing 6-MI in the template DNA (to monitor translocation) did move into the pre-translocated state upon TGT addition (Figure 5A). However, the apparent KTGT for the β′M932A TEC was 5-fold higher than that of the wild-type TEC (Table 1), suggesting that contacts made by β′Met932 with the 3′ RNA base are important for stabilization of the pre-translocated state.
The affinity of 3′ RNA NMPs for nucleophilic and substrate sites determines the translocation state of the TEC
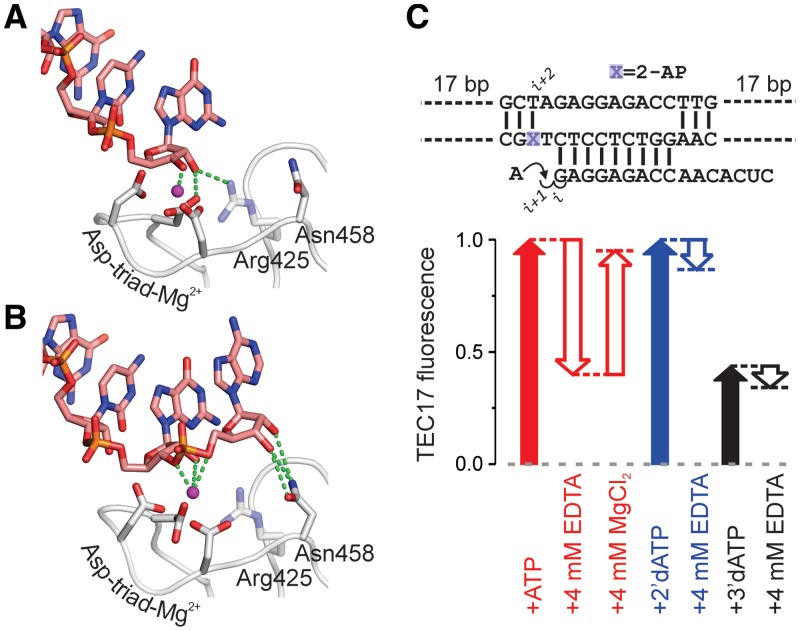
From a structural perspective, interactions of the RNA 3′ NMP with nucleophilic and substrate sites encompass all discriminating contacts between the post- and pre-translocated states. Specifically, in the post-translocated state, 2′ and 3′ OH groups interact with β′Arg425 and the Mg2+-β′Asp-triad, respectively, whereas in the pre-translocated state, both OH groups interact with β′Asn458 (Figure 7A and B). To evaluate the importance of RNA 3′ NMP contacts with nucleophilic and substrate sites, we analyzed the translocation equilibrium in TECs lacking either 2′ or 3′ OH groups at the RNA 3′ end. The TECs were extended with 2′ or 3′ dNTPs and titrated with TGT.
Figure 7.
RNA–protein contacts (green dashed lines) specific for the post- and pre-translocated states. The active site Mg2+ ion is depicted as a magenta sphere. Residue numbers correspond to E. coli RNAP. (A) The post-translocated state (PDB 2O5I). (B) The pre-translocated state model, generated as described in Supplementary Figure S4A. (C) The effects of Mg2+ withdrawal on translocation equilibrium in TECs extended with AMP, 2′ dAMP and 3′ dAMP. A schematic of the initial scaffold is indicated above the graph. The initial concentration of Mg2+ in the assay medium was 2 mM.
TECs lacking a 2′-OH group displayed the same levels of fluorescence as rNMP-extended TECs, but had markedly reduced propensities to occupy the pre-translocated state upon addition of TGT (Figure 3 and Supplementary Figure S2). The apparent TGT binding constant increased ∼10-fold, and TECs were only 30–60% pre-translocated at saturating TGT concentrations (Table 1 and Supplementary Table S2). Apparently, the 2′ OH is dispensable for the stability of the post-translocated TEC, but is critical for the stability of the pre-translocated state. This result is in full agreement with the documented ability of the substrate site to discriminate against 2′ dNTP substrates (33–35).
TECs lacking a 3′ OH group were not only 40% post-translocated (except for 3′ GMP, which was 90% post-translocated; Supplementary Figure S2), but also exhibited reduced propensities to occupy the pre-translocated state upon addition of TGT (approximately 5-fold increase in KTGT; Table 1, Figure 3 and Supplementary Figure S2). An RNA 3′ nucleotide lacking a 3′-OH group thus binds weakly to both substrate and nucleophilic sites and therefore has diminished potential to determine the translocation state and mediate the TEC′s response to TGT. The destabilization of the post-translocated state was anticipated, as the 3′OH group is activated by Mg2+ for nucleophilic attack on substrate NTPs and is therefore expected to interact intimately with the metal ion. Indeed, withdrawal of Mg2+ (with EDTA) from rNMP-extended TECs destabilized the post-translocated state to a similar degree as did the absence of 3′ OH; moreover, the two effects were not additive, consistent with the same Mg2+–3′OH interactions being disrupted (Figure 7C). Mg2+ withdrawal resulted in only a small decrease in fluorescence of 2′ dAMP-extended TEC, suggesting that Mg2+ effects are largely attributable to changes in the translocation state rather than to a direct influence on 2-AP fluorescence. As predicted, β′Asn458 appeared to be an essential constituent of the substrate site: β′N458D TEC did not move into the pre-translocated state by more than 20% at saturating TGT concentrations (Figure 5A).
Assessment of the completeness of translocation following NMP incorporation
The completeness of translocation following NMP incorporation can be evaluated by forward-biasing RNAP with a non-hydrolyzable analog of the next substrate NTP. Indeed, templated CMPcPP was able to counter the effect of TGT on GMP-extended TEC (Figure 6). However, CMPcPP failed to increase fluorescence in the absence of TGT, suggesting that the TGT-free TEC is fully post-translocated. The reason for the ∼15% decrease in fluorescence upon CMPcPP binding to the TEC is not entirely clear, but artifacts of such scale are anticipated in a highly sensitive fluorescent system. For example, binding of CMPcPP in the active site may allosterically induce subtle changes in the geometry of the upstream edge of the transcription bubble or cause the RNA–DNA hybrid to move backwards by a fraction of a register.
Another means of assessing completeness of translocation is to bias RNAP forward with a 2′-dNMP at the RNA 3′ end. Because the pre-translocated state is strongly disfavored in 2′ dNMP-extended TECs, the occupancies of the post-translocated state should be higher than that in rNMP-extended TECs for a system with measurable equilibrium fractions of both states. All TECs used in this study produced the same levels of fluorescence when extended with 2′ dNMP and rNMP, suggesting that they are nearly 100% post-translocated (the pre-translocated-state fractions are below the detection limit of the assay). However, it is entirely possible, if not likely, that the pre-translocated state is populated in a subset of TECs formed during the transcription of diverse DNA sequences (7,9,36).
TGT is a high-affinity PPi analog
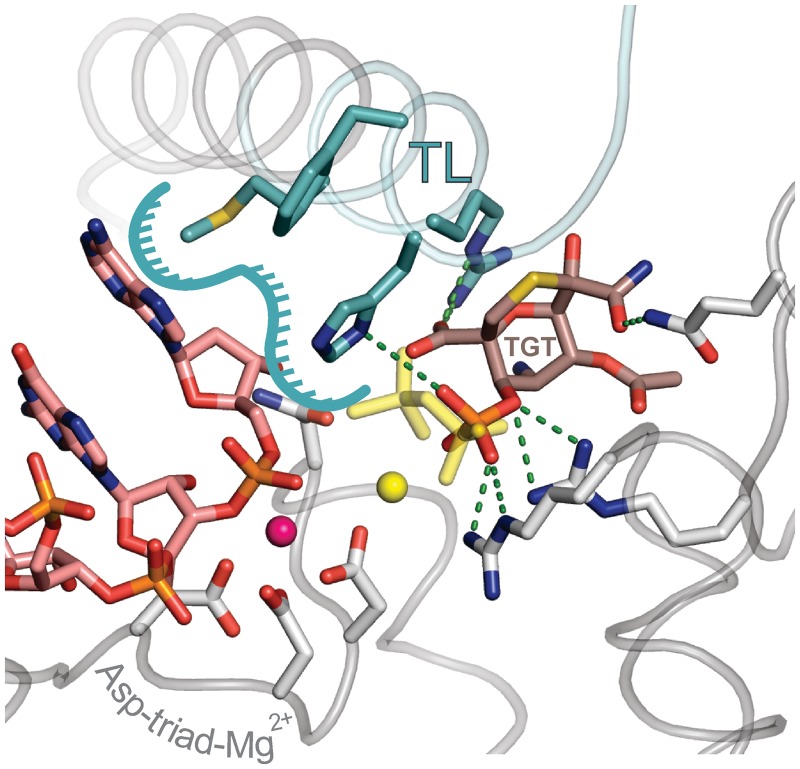
The observation that alterations that compromise the affinity of 3′ NMPs for the substrate site (removal of 2′- and 3′-OH group and N458D substitution) disfavor the pre-translocated state suggests that TEC translocation is fully reversible: RNA 3′ NMPs revisit the substrate site upon backward translocation. To determine whether NMPs can maintain contact with the substrate site after being incorporated into RNA, we performed in silico catalysis using the structure of the Thermus thermophilus TEC with bound NTP analog (2) as a starting point. Indeed, it was possible to join the α-phosphate of NTP to the RNA 3′ OH group while leaving the substrate base and sugar moiety essentially untouched (Supplementary Figure S4A). The resulting model of the closed pre-translocated TEC-PPi complex was clash-free, with bond angles and Mg2+-phosphate oxygen distances commonly found in nucleic acids. To determine whether TGT can be accommodated within the closed active site, we aligned the open TGT–RNAP holoenzyme structure (37) with the closed pre-translocated TEC model. A subtle rotation of TGT and retracing of β′Arg1239 (Arg933 in E. coli RNAP) side chain were necessary to remove clashes and lodge the inhibitor in the protein pocket (Supplementary Figure S4B and S4C). In the resulting model, phosphate and carboxylate groups of TGT made contacts with β′His1242 and β′Arg1239 residues of the TL (His936 and Arg933 in E. coli RNAP), accounting for the toxin’s ability to stabilize the TL in a folded state (Figure 8). Consistent with this, substitution of these residues with alanines resulted in more than a 7-fold increase in KTGT (Table 1). TGT did not interact directly with the RNA, suggesting that its effects on the translocation state of the TEC are mediated exclusively by the TL.
Figure 8.
Structural model of the closed TEC–TGT complex. Phosphate and carboxylate groups of TGT (brown) overlap with the PPi molecule (yellow). TL is colored turquoise and the rest of RNAP is colored gray. RNA is colored rose. The surface contributed by TL residues for interaction with RNA 3′ NMP is contoured by a turquoise line. Asp triad residues and selected side chains that interact with TGT and the 3′ RNA nucleobase are shown as sticks. Active site Mg2+ ions coordinated by the Asp triad and PPi are depicted as magenta and yellow spheres, respectively. TGT–protein contacts are indicated as green dashed lines.
Interestingly, phosphate and carboxylate groups of TGT occupied the volume of PPi molecule (or γ and β phosphates of NTP) (Figure 8), suggesting that binding of the two compounds to the pre-translocated TEC is mutually exclusive. Such a notion is fully consistent with the documented ability of TGT to strongly inhibit pyrophosphorolysis (37), the observation that TGT competes with non-hydrolysable NTP analog under equilibrium conditions (Figure 6), and our kinetic data demonstrating that TGT acts only on post-translocated, PPi-less TEC (Figure 3, left panel, inset). We also argue that the non-competitive effect of TGT on the extension of dinucleotide primer by RNAP holoenzyme–promoter complexes (37) can be explained by an active site closure mechanism that is fully analogous to the action of TGT during elongation. Specifically, we suggest that in the multi-round initiation assay, TGT inhibits the release of tri-nucleotide product from the active site rather than inhibiting dinucleotide extension.
DISCUSSION
This study answers a long-standing question about the nature of the rate-limiting step in transcription by cellular RNAPs, demonstrating that both nucleotide addition and post-catalytic steps involving TL unfolding, PPi release and translocation measurably limit the rate of transcription. Our kinetic analyses also revealed that translocation occurs shortly after or concurrently with PPi release (Figure 4). From a structural perspective, the release of PPi likely reflects active site opening/TL unfolding, as the two processes are mutually dependent. Accordingly, our kinetic data suggest that forward translocation occurs shortly after or concurrently with active site opening.
We then demonstrated that backward translocation is coupled to active site closure in a series of equilibrium experiments using the RNAP inhibitor, TGT, as a tool to stabilize the TL in a folded state. Our data argue that subtle alterations in the sugar of RNA 3′ nucleotides or modifications of nucleophilic and substrate sites have pronounced effects on the propensity of the TEC to occupy either translocation register. We therefore propose that the strength of interactions between the RNA 3′ NMP and nucleophilic and substrate sites is the major factor that determines the translocation state of the TEC. Similarly, Hein et al. (32) recently proposed that the identity of the RNA 3′ nucleotide strongly influences the translocation bias of TECs. The observation that the folded TL is expected to strengthen 3′ NMP binding to the substrate site by contributing additional interactions (Figure 8) provides a trivial thermodynamic explanation for its effect on translocation. In the closed active site, the affinity of the substrate site for the 3′ RNA NMP is invariably higher than that of the nucleophilic site, so the pre-translocated state is strongly favored. In the open active site, the affinity of the substrate site is decreased to less than that of the nucleophilic site, thereby favoring the post-translocated state. Notably, in this context, molecular dynamic simulations and normal mode analysis of eukaryotic RNAP also suggest that forward translocation requires an unfolded TL (38,39). Coupling of translocation to active site opening has also been proposed for structurally unrelated, non-homologous A- and B-family nucleic acid polymerases (40–42). Our finding thus confirms the early prediction of Yin and Steitz (40) that thermodynamic and/or mechanical coupling between these two events is a general feature of nucleic acid polymerases that undergo a conformational change in response to nucleotide binding.
Whereas a folded TL stabilizes a 3′ NMP in the substrate site, the converse is also true. Thus, the RNA 3′ NMP in the substrate site will mediate a subset of interactions contributed by the NTP (natural TL stabilizer), and therefore is expected to stabilize the folded TL, albeit to a lesser extent. Indeed, we found that 3′ NMPs can lock the TL in a folded state when acting in concert with TGT, whereas crystallographic evidence (37) indicates that TGT alone cannot fully stabilize the folded TL, at least when added to preformed RNAP holoenzyme crystals.
Crystallographic and biochemical studies suggest that folding of the TL occurs spontaneously, even in the absence of bound NTP substrate, and is accompanied by relocation of the β-pincer domain, which increases the mobility of downstream DNA (2). The spontaneous folding of the TL thus promotes catalysis if the cognate substrate is present, and induces backward translocation if the substrate is absent or mismatched. Importantly, the latter effect likely contributes to the well-documented role of TL in the fidelity of transcription (3,34,35). Indeed, all proofreading functions of RNAP occur in the pre-translocated or backtracked states.
The TL effect on translocation is exploited by the inhibitor TGT. This compound was initially suggested to inhibit transcription by coordinating an inhibitory Mg2+ ion in the active site (37). Recently, Artsimovitch et al. (31) demonstrated the involvement of TL residues β′Arg933 and β′His936 (E. coli numbers) in the inhibitory mechanism, and proposed that TGT stabilizes the TL in the inactive conformation in TGT–NTP–TEC complex. Our studies involving direct monitoring of TGT effects on the translocation equilibrium complement the findings of Artsimovitch et al. (31), identifying the TL as the key element responsible for inhibition by TGT. However, our results collectively suggest that TGT action is best explained by formation of a pre-translocated TGT–TEC complex with natively folded TL. Specifically, we propose that TGT is a high-affinity pyrophosphate analog that acts in a mechanistically similar manner to phosphonoformic acid, a clinically used antiviral drug that targets B-family nucleic acid polymerases (43,44). Both drugs likely employ adjacent phosphate and carboxylate groups to mimic PPi and function by inducing backward translocation via active site closure, but are specific for non-homologous classes of enzymes. Our results thus provide a universal structural framework for the design of inhibitors that target the translocation step in nucleic acid polymerases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–4, Supplementary Methods and Supplementary Datasets 1 and 2.
FUNDING
Academy of Finland [130581 to G.A.B., 140401 to M.S.J.]. Funding for open access charge: Academy of Finland [130581 to G.A.B.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Irina Artsimovitch (Ohio State University) for providing expression plasmids for wild-type and variant RNAPs and critically reading the manuscript. Armando Alvarez Losada is acknowledged for help with constructing and purifying variant RNAPs. Essential equipment was contributed by the Walter and Lisi Wahl Foundation. Computational infrastructure was provided by Biocenter Finland (Bioinformatics) and by grants from the Sigrid Juselius Foundation; the Joe, Pentti and Tor Borg Memorial Fund; and the Åbo Akademi Center of Excellence in Cell Stress and Aging.
REFERENCES
- 1.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 2.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 3.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamberlin MJ. New models for the mechanism of transcription elongation and its regulation. Harvey Lect. 1992;88:1–21. [PubMed] [Google Scholar]
- 5.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 6.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai L, Fulbright RM, Wang MD. Mechanochemical kinetics of transcription elongation. Phys. Rev. Lett. 2007;98:068103. doi: 10.1103/PhysRevLett.98.068103. [DOI] [PubMed] [Google Scholar]
- 8.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 9.von Hippel PH. An integrated model of the transcription complex in elongation, termination, and editing. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 10.Nudler E. RNA polymerase active center: the molecular engine of transcription. Annu. Rev. Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kireeva M, Kashlev M, Burton ZF. Translocation by multi-subunit RNA polymerases. Biochim. Biophys. Acta. 2010;1799:389–401. doi: 10.1016/j.bbagrm.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Steitz TA. Visualizing polynucleotide polymerase machines at work. EMBO J. 2006;25:3458–3468. doi: 10.1038/sj.emboj.7601211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 14.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat. Struct. Mol. Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 15.Opalka N, Brown J, Lane WJ, Twist KA, Landick R, Asturias FJ, Darst SA. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol. 2010;8:e1000483. doi: 10.1371/journal.pbio.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heikinheimo P, Pohjanjoki P, Helminen A, Tasanen M, Cooperman BS, Goldman A, Baykov A, Lahti R. A site-directed mutagenesis study of Saccharomyces cerevisiae pyrophosphatase. Functional conservation of the active site of soluble inorganic pyrophosphatases. Eur. J. Biochem. 1996;239:138–143. doi: 10.1111/j.1432-1033.1996.0138u.x. [DOI] [PubMed] [Google Scholar]
- 18.Brune M, Hunter JL, Howell SA, Martin SR, Hazlett TL, Corrie JE, Webb MR. Mechanism of inorganic phosphate interaction with phosphate binding protein from Escherichia coli. Biochemistry. 1998;37:10370–10380. doi: 10.1021/bi9804277. [DOI] [PubMed] [Google Scholar]
- 19.Komissarova N, Kireeva ML, Becker J, Sidorenkov I, Kashlev M. Engineering of elongation complexes of bacterial and yeast RNA polymerases. Methods Enzymol. 2003;371:233–251. doi: 10.1016/S0076-6879(03)71017-9. [DOI] [PubMed] [Google Scholar]
- 20.Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 21.Brune M, Hunter JL, Corrie JE, Webb MR. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 22.Pais JE, Bowers KE, Stoddard AK, Fierke CA. A continuous fluorescent assay for protein prenyltransferases measuring diphosphate release. Anal. Biochem. 2005;345:302–311. doi: 10.1016/j.ab.2005.07.040. [DOI] [PubMed] [Google Scholar]
- 23.Hawkins ME. Synthesis, purification and sample experiment for fluorescent pteridine-containing DNA: tools for studying DNA interactive systems. Nat. Protoc. 2007;2:1013–1021. doi: 10.1038/nprot.2007.150. [DOI] [PubMed] [Google Scholar]
- 24.Kashkina E, Anikin M, Brueckner F, Lehmann E, Kochetkov SN, McAllister WT, Cramer P, Temiakov D. Multisubunit RNA polymerases melt only a single DNA base pair downstream of the active site. J. Biol. Chem. 2007;282:21578–21582. doi: 10.1074/jbc.C700098200. [DOI] [PubMed] [Google Scholar]
- 25.Datta K, Johnson NP, von Hippel PH. DNA conformational changes at the primer-template junction regulate the fidelity of replication by DNA polymerase. Proc. Natl Acad. Sci. USA. 2010;107:17980–17985. doi: 10.1073/pnas.1012277107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang GQ, Anand VS, Patel SS. Fluorescence-based assay to measure the real-time kinetics of nucleotide incorporation during transcription elongation. J. Mol. Biol. 2011;405:666–678. doi: 10.1016/j.jmb.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nedialkov YA, Gong XQ, Yamaguchi Y, Handa H, Burton ZF. Assay of transient state kinetics of RNA polymerase II elongation. Methods Enzymol. 2003;371:252–264. doi: 10.1016/S0076-6879(03)71018-0. [DOI] [PubMed] [Google Scholar]
- 28.Johnson RS, Strausbauch M, Cooper R, Register JK. Rapid kinetic analysis of transcription elongation by Escherichia coli RNA polymerase. J. Mol. Biol. 2008;381:1106–1113. doi: 10.1016/j.jmb.2008.06.089. [DOI] [PubMed] [Google Scholar]
- 29.Purich DL. Amsterdam, Holland: Elsevier; 2010. Enzyme kinetics: catalysis & control; pp. 253–254. [Google Scholar]
- 30.Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl Acad. Sci. USA. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Artsimovitch I, Svetlov V, Nemetski SM, Epshtein V, Cardozo T, Nudler E. Tagetitoxin inhibits RNA polymerase through trapping of the trigger loop. J. Biol. Chem. 2011;286:40395–40400. doi: 10.1074/jbc.M111.300889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hein PP, Palangat M, Landick R. RNA transcript 3′-proximal sequence affects translocation bias of RNA polymerase. Biochemistry. 2011;50:7002–7014. doi: 10.1021/bi200437q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svetlov V, Vassylyev DG, Artsimovitch I. Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. J. Biol. Chem. 2004;279:38087–38090. doi: 10.1074/jbc.C400316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat. Struct. Mol. Biol. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuzenkova Y, Bochkareva A, Tadigotla VR, Roghanian M, Zorov S, Severinov K, Zenkin N. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8:54. doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tadigotla VR, O Maoiléidigh D, Sengupta AM, Epshtein V, Ebright RH, Nudler E, Ruckenstein AE. Thermodynamic and kinetic modeling of transcriptional pausing. Proc. Natl Acad. Sci. USA. 2006;103:4439–4444. doi: 10.1073/pnas.0600508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vassylyev DG, Svetlov V, Vassylyeva MN, Perederina A, Igarashi N, Matsugaki N, Wakatsuki S, Artsimovitch I. Structural basis for transcription inhibition by tagetitoxin. Nat. Struct. Mol. Biol. 2005;12:1086–1093. doi: 10.1038/nsmb1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feig M, Burton ZF. RNA polymerase II flexibility during translocation from normal mode analysis. Proteins. 2010;78:434–446. doi: 10.1002/prot.22560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feig M, Burton ZF. RNA polymerase II with open and closed trigger loops: active site dynamics and nucleic acid translocation. Biophys. J. 2010;99:2577–2586. doi: 10.1016/j.bpj.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 41.Berman AJ, Kamtekar S, Goodman JL, Lazaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Golosov AA, Warren JJ, Beese LS, Karplus M. The mechanism of the translocation step in DNA replication by DNA polymerase I: a computer simulation analysis. Structure. 2010;18:83–93. doi: 10.1016/j.str.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchand B, Tchesnokov EP, Gotte M. The pyrophosphate analogue foscarnet traps the pre-translocational state of HIV-1 reverse transcriptase in a Brownian ratchet model of polymerase translocation. J. Biol. Chem. 2007;282:3337–3346. doi: 10.1074/jbc.M607710200. [DOI] [PubMed] [Google Scholar]
- 44.Zahn KE, Tchesnokov EP, Gotte M, Doublie S. Phosphonoformic acid inhibits viral replication by trapping the closed form of the DNA polymerase. J. Biol. Chem. 2011;286:25246–25255. doi: 10.1074/jbc.M111.248864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.