Abstract
DNA methytransferases (MTs) in bacteria are best understood in the context of restriction–modification (R–M) systems, which act as bacterial immune systems against incoming DNA including phages, but have also been described as selfish elements. But several orphan MTs, which are not associated with any restriction enzyme, have also been characterized and may protect against parasitism by R–M systems. The occurrence of MTs in these two contexts, namely as part of R–M systems or as orphans, is poorly understood. Here we report the results of a comparative genomic survey of DNA MTs across ∼1000 bacterial genomes. We show that orphan MTs overwhelm R–M systems in their occurrence. In general, R–M MTs are poorly conserved, whereas orphans are nearly as conserved within a genus as any average gene. However, oligonucleotide usage and conservation patterns across genera suggest that both forms of MTs might have been horizontally acquired. We suggest that many orphan MTs might be ‘degradation’ products of R–M systems, based on the properties of orphan MTs encoded adjacent to highly diverged REs. In addition, several fully degraded R–M systems exist in which both the MT and the RE are highly divergent from their corresponding reference R–M pair. Despite their sporadic occurrence, conserved R–M systems are present in strength in two highly transformable genera, in which they may contribute to selection against integration of foreign DNA.
INTRODUCTION
DNA methyltransferases (MTs) in bacteria are best studied as part of restriction–modification (R–M) systems, which may contribute to bacterial immune response against incoming DNA including phages (1,2). However, the evolutionary significance of the above property has been contested (3). It has been suggested that R–M systems may be selfish elements (4–7), which protect themselves by post-segregational killing of cells (in which they have been disturbed) by pre-existing and stable restriction enzymes. Several R–M systems are located on plasmids, EcoRII being an example, and a few others have been demonstrated to be acquired by horizontal gene transfer (HGT) (8–10), followed by integration into the host chromosome.
Also known are orphan MTs, which are not associated with any restriction enzyme. These may have core cellular functions in various DNA transactions (11): examples include the Dam methylase in Escherichia coli (12–14) and CcrM in Caulobacter crescentus (15). Other MTs provide immunity against parasitism by R–M systems, by targeting the same DNA site as the corresponding R–M system (16): an example is the Dcm cytosine methyltransferase in E. coli that recognizes the same target site as the EcoRII R–M system.
R–M and orphan MTs are different from one another: whereas the former can cleave unmethylated incoming DNA, its evolutionary significance notwithstanding, the latter cannot. Furthermore, the selective pressures that lead to the maintenance of each of these two systems could be different: whereas this could be protection against extraneous DNA, or more prominently, post-segregational lethality for R–M systems, it could be core regulatory functions or immunity against selfish R–M systems for the latter. Alternatively, many orphan systems could simply be under neutral selection.
Despite the widespread use of components of R–M systems as molecular biology tools and the deep understanding of the functions of selected orphan MTs, there is little information on their occurrence across the large number of fully sequenced bacterial genomes. As a result, there is a lack of understanding of whether orphan MTs might have evolved from R–Ms thus serving to fight parasitism by selfish R–Ms. Here we present a comparative genomic survey of MTs in >1000 completely sequenced bacterial genomes, and compare orphans and R–M components in terms of their occurrence and conservation.
MATERIALS AND METHODS
Sequences of class II R–M components were obtained from REBASE (17). Target site information and pairing between REs and MTs were also obtained from REBASE. Sequences of all annotated proteins in completely sequenced bacterial genomes were downloaded from the KEGG database (18).
Sequence homologues were identified using phmmer [HMMER 3; http://hmmer.janelia.org; (19)], using an e-value cut-off of 10−100 unless stated otherwise (Supplementary data set). Most (>98%) hits thus obtained had a sequence similarity of 50% or more with the query sequence, as determined using Needleman–Wunsch global alignment algorithm (20) implemented in EMBOSS (21) (Supplementary Figures S1–S2). Target site information was transferred from the query to the hit. A MT thus identified was called an orphan if we were unable to detect a RE with the same target site as the MT in its neighbourhood (less than 10 genes away, based on genomic coordinates). We also identified ‘degraded’ REs using a borderline e-value cut-off of 0.01, and found that these were encoded adjacent to a subset of orphan MTs; the properties of these orphan MTs are also described.
Orthologues were defined using bidirectional best-hit (BBH) phmmer, at an e-value threshold of 10−50. These identifications were performed either within a genus or across genera and used to calculate ‘degree of conservation’ and ‘orthologue consistency’ respectively, as described in the ‘Results’ section. For calculating orthologue consistency, we defined all members of a phylum as a ‘group’; for the Proteobacterial phylum, which is highly represented in the genome database, we used the order instead.
Various calculations were also performed on sets of randomly picked genes. These genes were picked such that the numbers of genes from each genome was equal to that in the actual data set.
Horizontally-acquired genes, based on abnormal oligonucleotide usage, were identified using Alien Hunter (22). Orthologue consistency was used as an additional measure to infer the extent of HGT in a set of genes.
All statistical tests were performed in R (http://r-project.org).
RESULTS
Most MTs are orphans
We assembled a data set of 208 DNA MTs and their cognate restriction endonucleases (RE) from REBASE. This included protein sequence and target DNA site information for each pair. Using phmmer, we searched the annotated protein sequences from 1227 prokaryotes (1134 bacterial), with fully sequenced genomes, for homologues of MTs and REs, using e-value ≤ 10−100 (Supplementary Figure S1). The stringency of the search and the use of known R–M pairs as queries mean that our hits are predominantly relatives of R–M systems.
A standard e-value cut-off of 0.01 will help us reduce false negatives and thus expand on our hit list; in fact, it might help us identify additional ‘bonafide’ MTs and REs, which are not covered by our starting query set. However, such low thresholds are also beset by other issues, which made us shun them. One problem is that it is difficult to make a one-to-one or even a few-to-one mapping between a query and a hit. And such thresholds also return many false positive hits, which are difficult to weed out in the absence of manual inspection, especially for orphan systems (for which we do not have the confidence offered by the identification of a neighbouring RE). For example, at an e-value ≤ 0.01, these sequence searches return RNA methyltransferases and even protein methyltransferases (e.g. RlmL and PrmC in E. coli respectively).
We identified 914 MTs (883 bacterial) but only 215 REs, across 559 genomes (535 bacterial; Table 1). Thus, many MTs are orphans (n = 718, 79%), defined by the absence of a neighbouring RE. Among orphans, we define two types: (i) class 1 orphan MT, present in genomes with no detectable RE (n = 558); (ii) class 2 orphan MT, from genomes encoding other R–M systems (n = 160). There were 196 R–M MTs. These are the list A or ‘bonafide’ genes discussed in the rest of the paper, unless indicated otherwise.
Table 1.
Numerical summary of results
| Total number | Number in genera with more than one sequenced genome | Number conserved in at least one other genome within genus | Number predicted to be horizontally acquired by Alien Hunter | |
|---|---|---|---|---|
| Class 1 orphan MTa | 558 | 367 | 269 | 189 |
| Class 2 orphan MTa | 160 | 132 | 110 | 31 |
| R–M MT | 196 | 169 | 112 | 80 |
| R-E | 215 | N/A | N/A | N/A |
| Orphan MTs associated with degraded REb | 174 | 121 | 68 | 83 |
| Orphan REs associated with degraded MTb | 15 | N/A | N/A | N/A |
| Fully degraded R–M systemsc | 222 | N/A | N/A | N/A |
aOne gene from each of these headings also match against an RE in the reference sequence. However, these two instances do not affect any of our results.
bThese genes are present in the set of MT or RE homologues identified at e-value ≤ 10−100.
cThese genes are NOT present in the set of MT or RE homologues identified at e-value ≤ 10−100.
N/A represents numbers that were not calculated.
The following indicate the reliability of our automated procedures: there are few instances (<2%) where a neighbouring MT–RE pair is not annotated with the same target site. As expected, orphan REs are rare (<10%) and should be considered largely as false negative errors in calling MTs, or non-functional molecules. The large number of orphans is not entirely a consequence of false negatives in identifying REs, as follows:
when MTs and REs are both identified at e-value ≤ 10−20, a total of 1755 MTs could be found, of which only 430 are associated with an adjacent RE and
though ∼25% of our orphan MTs are located adjacent to weaker homologues of REs, identified at e-value ≤ 0.01, we suggest that these belong to a different evolutionary regime than the bonafide R–M MTs described above; this is discussed later. Therefore, these MTs are considered as orphans. Note, however, that even the inclusion of these among R–M systems will not detract from our primary observation that a majority of MTs, identified as relatives of R–M systems, are orphans.
R–M MTs are poorly conserved
We investigated the degree to which MTs are conserved by searching for one-to-one BBH orthologues for each MT within the respective genus. We measured the degree of conservation for each gene in our data set by calculating the proportion of genomes (from the same genus as the query) in which an orthologue is present. The significance of the distribution of degree of conservation for each of R–M and classes 1 and 2 orphan MTs was calculated using sets of randomly picked genes in which each genome was represented to the same level as in the actual data set. In general, R–M MTs are poorly conserved when compared to their respective sets of random genes, whereas this is not true of orphan MTs (Figure 1A and B). A similar distinction between orphan and R–M MTs is seen even when sequence searches are performed at a e-value ≤ 10−20 (data not shown). Thus, in many organisms there is little pressure for the maintenance of R–M systems.
Figure 1.

Conservation of orphan and R–M MTs (A) Probability density of the degree of conservation (defined as the proportion of genomes—within the genus—in which a MT is conserved) of class 1 (red, solid line) and class 2 orphan MTs (magenta, solid line) and R–M MTs (blue solid line). The dashed lines, in colours matching that for the real data set described above, show the distributions for an example simulation in which random sets of genes were chosen as controls such that the number of genes from each genome equals that in the real data set. (B) Distribution of P-values (Wilcoxon test; negative logarithm, base10) comparing the distribution of the degree of conservation of genes for the three types of MTs with that from random simulations (n = 30). A dotted horizontal line is drawn at P = 0.01.
Many R–M and orphan MTs are horizontally acquired
It has been previously shown that selected R–M systems are horizontally acquired (8–10). To test the generality of this across our set of orphan and R–M MTs, we first used Alien Hunter, a program that identifies horizontally acquired genes as those with atypical higher-order-oligonucleotide usage (22). We also performed this search across random sets of genes, assembled as described earlier. We find that many MTs, including both orphan and R–M MTs, have atypical oligonucleotide usage, and are likely to have been acquired by recent HGT (Figure 2A). These numbers are statistically significant, in comparison to random expectation. This is true of MTs that lack BBH orthologues in the same genus, and to a substantially lesser extent of those conserved in at least one other genome from the same genus. Similar enrichments are obtained for both orphan and R–M MTs identified at a lower e-value threshold of 10−20. We do note that the number of class 2 orphans that have atypical oligonucleotide composition is not statistically significant. However, given the evidence below, which does suggest their horizontally acquisition, these orphans might be products of gene transfer between closely related species, which will not differ much from each other in terms of oligonucleotide usage.
Figure 2.
Horizontal acquisition of MTs. (A) Numbers of horizontally acquired MTs, as predicted by Alien Hunter, for the three types of MTs (coloured as in Figure 1), conserved (column 1) or not (column 2) in at least one other member of its genus. The vertical lines are drawn at the actual number, whereas the boxplots show the distribution of numbers obtained for 1000 sets of randomly chosen genes (see legend to Figure 1). The P-values were calculated from the Z-score obtained from the random permutations. (B) Probability densities of orthologue consistency score for the three types of MTs (solid lines), and a corresponding set of random genes (dashed lines); colours are as in Figure 1. For this, we used only MTs with restricted orthologue occurrence, i.e. the number of genomes in which an orthologue is present is less than that representing the respective phylogenetic phylum (Firmicutes for example; for the large Proteobacteria phylum, the order was used instead). Though class 2 orphans are not particularly enriched for Alien Hunter-predicted horizontally acquired genes, their poor orthologue consistency scores suggest that they may also have been so acquired. (C) Boxplots showing the distribution of orthologue consistency score for MTs defined as horizontally acquired (or not) by AlienHunter. The probability density curve shows the distribution expected for a random set of genes.
Abnormal oligonucleotide usage can detect only recent horizontal transfers. It further cannot identify gene transfers between closely related organisms, which will not leave abnormal oligonucleotide usage marks. To identify signatures of more ancient HGTs, we identified BBH ‘orthologues’ of MTs across all genera in our data set, using one representative genome (with the largest size) per genus (n = 450 genomes). We first defined MTs with restricted orthologue occurrence as those for which the number of orthologues thus identified is less than the number of genomes belonging to the same phylogenetic group (phylum or order, as defined in ‘Materials and Methods’ section). For those MTs with restricted orthologue occurrence, we calculated ‘orthologue consistency’, defined by the proportion of orthologues from the same phylogenetic group as the query MT; this value will be meaningless for genes with wide orthologue presence. Figure 2B shows that orthologue consistencies for both orphan and R–M MTs (<0.25) are significantly less than for randomly selected genes (∼0.75). This is true of both classes 1 and 2 orphans. Even for MTs not predicted to be horizontally acquired by Alien Hunter, orthologue consistency is low (Figure 2C). This suggests that MTs with restricted prevalence occur sporadically across phylogenetic groups. This lack of consistency in terms of their occurrence within related organisms might indicate that these were horizontally acquired. Additionally, the relatively poor within-genus conservation of extant R–M MTs, when compared to orphan MTs, might suggest that they are in general more recent acquisitions.
Orphan MTs may be products of RE degradation
Do orphan MTs emerge from R–M systems? We hypothesized that orphan MTs could originate by degradation of R–M MT genes based on the following: (i) Several R–M systems in Helicobacter pylori have either both components inactivated by mutation or have only the MT activity (23). (ii) Furthermore, ∼15% of within-genus orthologues of class 2 orphan MTs are R–M MTs (Supplementary Figure S3). These represent instances (n = 84) in which one organism retains a R–M system, whereas another from the same genus has ‘replaced’ the R–M system with an orphan MT of the same target site. Such orphan MTs might be extreme examples of products of R–M degradation. This is not exclusive of orphan systems being products of recent HGT, as they might have been originally acquired in the form of R–M systems and then evolved into an orphan while retaining signatures of their ‘acquired ancestry’.
In the above paragraph, ‘(i)’ represents only a few examples from one species, whereas the conclusions from ‘(ii)’ are also limited to a few genera with Helicobacter being the most prominent. To search for evidence for RE degradation, we scanned for intermediates in which an orphan MT is encoded adjacent to a RE with relatively weaker sequence similarity to query R–M systems, which might be REs undergoing a process of decay (Figure 3A). To identify such intermediates, we searched for REs at a relaxed e-value cut-off of 0.01. That these are relatively well diverged from the query RE might suggest that they tend to be non-functional, akin to those described in H. pylori by Lin and colleagues (23). Then we tested whether any of our orphan MTs could be paired with these REs, which we called as ‘degraded’.
Figure 3.
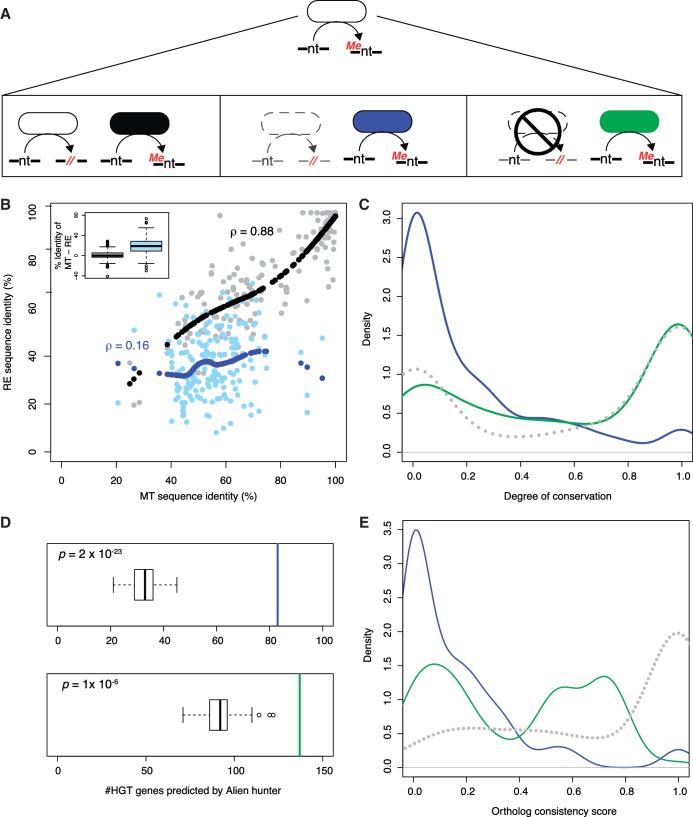
Partially degraded R-M systems (A) Classification of MTs based on the type of RE it is associated with. Black stands for bonafide R–M systems, blue for MTs associated with a degraded RE and green for bonafide orphans. (B) Scatter plot (grey and light blue) showing the association between the sequence identity of the MT and the RE forming a bonafide R–M pair (grey/black), and for those R–M systems with a degraded RE (light blue/blue). The black (and blue) points track the LOESS fit for these correlations for the grey (and light blue) scatters. The inset shows the distribution of the differences between the sequence identities, with the reference query sequences, of MT and RE for bonafide (grey) and degraded (light blue) REs. (C) Degree of conservation for orphans associated with a degraded RE (blue) and those that are not (green). The grey dotted line is for an example random simulation for all orphans taken together. (D) As in Figure 2A, showing the number of horizontally acquired genes among orphans with a degraded RE (blue, top) and those that are not (green, bottom). (E) As in ‘C’ above, except that the distributions are for orthologue consistency.
We find that 174 orphan MTs (24%) in our data set are positioned adjacent to degraded REs. By definition, the REs in these systems are more divergent from the query than the MT, in comparison to bonafide R–M systems (Figure 3B, inset). To test whether these R–M pairs are different from the bonafide R–M systems, we calculated the correlation between the divergences of the MT and RE forming an R–M pair from their respective query sequences (from REBASE). The divergence was measured by Needleman–Wunsch global sequence alignment. If the two proteins were functionally linked, then any measure of their rates of sequence change should be correlated. For the bonafide R–M pairs, we observe a near-linear correlation (ρ = 0.88) between the divergences of the MT and the RE from their respective references (Figure 3A). On the other hand, the correlation, if any, for R–M pairs with a degraded RE is weak (ρ = 0.16). If the assignment of R–M pairs to ‘bonafide’ and ‘degraded’ is randomized, then such a difference vanishes, with both classes characterized by high correlation coefficients (Supplementary Figure S4). Therefore, whereas in bonafide R–M pairs any divergence in the sequence of the RE is largely matched by that in the MT, this is not true of those with a degraded RE. We therefore suggest that orphan MTs associated with degraded REs are intermediates in the formation of fully orphan MTs from previously intact R–M systems.
We then compared the conservation properties of bonafide orphan MTs and those associated with a degraded RE. The latter, when compared to the former, are (i) poorly conserved (Figure 3C); (ii) more likely to be horizontally acquired, though both classes of orphans are significantly enriched for HGT in comparison to randomly selected genes (Figure 3D); and (iii) have lower orthologue consistency (Figure 3E). This minority subset of orphan MTs associated with degraded REs is similar to bonafide R–M systems in these properties.
Finally, we obtained a set of 222 R–M pairs in which both the MT and the RE are ‘degraded’ by performing our sequence searches for both MT and RE at e-value ≤ 0.01. Given that they are both ‘degraded’ and presumably inactive, along the lines of several R–M systems described in H. pylori (23), we would expect little correlation between the divergences of the two components from their respective queries; and this what we observe (Figure 4A). Furthermore, the divergence of REs of these systems is significantly more than that for those systems in which only the RE is degraded, suggesting further degradation (Figure 4B).
Figure 4.

Fully degraded R–M systems (A) Scatter plot (light green) showing the association between the sequence identity of the MT and the RE for those R–M systems where both the MT and the RE are degraded. The dark green points track the LOESS fit for the correlation. (B) Sequence identity (%) of REs in bonafide R–M systems (black), in R–M systems in which only the RE is degraded (blue), and in R–M systems where both the MT and the RE are degraded (green).
Therefore, to summarize, we suggest that many orphans could be products of degradation of R–M systems, which were originally acquired by horizontal transfer. We observe many intermediates in which the MT is likely to be functional, whereas its partner RE is degraded. We suggest that these MTs are more recent acquisitions than bonafide orphan MTs, among which may be ones with core cellular functions. On the other hand, there are very few instances (n = 15) in which the MT is more divergent than the RE; these would make sense if the RE is inactive or if the divergent MT is in fact active. In addition are R–M system relics in which both components are substantially diverged from the reference R–M system.
Thus, MTs are first acquired in the form of R–M systems by horizontal transfer. The pressure of post-segregational killing by the RE, in the event that the MT or the entire R–M system is disturbed, might lead to selective degradation and subsequent loss of the RE [see Figure 5 in (23)]. The resulting orphan MT would confer resistance against further invasion and parasitism by the same R–M system. Given enough time, it might even acquire core cellular functions. In other cases, the MT might undergo further degradation and probable loss where there is no selective pressure for its maintenance.
DISCUSSION
Conserved R–M systems and their role in shaping bacterial genome structure
Our observation that R–M systems are poorly conserved, while suggesting a lack of selective pressure for their maintenance, does not entirely rule out a role for them (i) in shaping the host genome composition or (ii) in controlling HGT.
The fact that many bacterial genomes avoid short palindromic sequences has been interpreted as a consequence of lethal pressure imposed by R–M systems (3). We notice that palindromic sequences targeted by MTs are in general avoided; however, target sites of recently acquired R–M MTs are less constrained (Supplementary Figure S5). This might indicate that selection has not had enough time / generations to act on the host genome’s oligonucleotide usage to respond to pressure from recently acquired R–M systems. In fact, the strongest site avoidance, among all MTs, is seen for R–M MTs not predicted to be products of recent HGTs. This, in addition, might suggest that orphan MTs might begin to relax some pressure for R–M-dependent constraints on palindrome occurrence.
Organisms encoding conserved R–M MTs encode multiple R–M systems with distinct target sites. These organisms belong to a few genera—Helicobacter and Neisseria (Supplementary Figure S6), which are highly transformable (24,25). This, following from the conservation of these R–M MTs, could promote HGT specifically within the genus. Additionally, a phylogenetically distant source organism might evolve mechanisms to methylate sites targeted by the host's R–M system(s), or select against the presence of these target sites in the DNA itself; however, the chances that this would be productive would decrease exponentially with every additional specificity in the host's R–M repertoire.
Orphan versus R–M MT
In E. coli, the orphan MT Dcm offers protection against parasitism by the EcoRII R–M system with which it shares a target site (16). Most genomes with an orphan MT do not have an R–M system. Furthermore, among genomes with both orphan and R–M MTs, there is only a small overlap between the sites targeted by multiple MTs (2%; Supplementary Figure S7). Though the statistical significance of this is weak, it might suggest avoidance of R–M systems in genomes encoding an orphan MT. This might further indicate that the emergence of orphan MTs might relax any selection pressure for the maintenance of R–M systems.
Origins of orphan MTs
Orphan MTs could originate by degradation of R–M MT genes. This is evidenced by our finding that there are intermediates in which the RE component of an R–M system is degraded. In several cases, the entire R–M system has diverged considerably from the query sequence, suggesting full inactivation. There is experimental evidence that several R–M systems in H. pylori have either both components inactivated by mutation or have only the MT activity (23). The fact that we find only a few REs associated with degraded MTs gives credence to our approach. Orphan MTs with R–M MTs as orthologues described above might be examples of products of R–M degradation, wherein the RE is completely lost. These results can only be substantiated with experiments.
Many well-conserved orphan MTs have developed selectable functions involving DNA transactions in several bacteria [e.g. Dam in E. coli, CcrM in C. crescentus and Dcm in E. coli and V. cholerae (26)], besides providing immunity against parasitism by R–M systems with the same target specificity as the orphan MT. These are likely to be bonafide orphans, not even associated with a degraded RE, well conserved, and less likely to be detected as horizontally-acquired.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–7 and Supplementary Dataset 1.
FUNDING
Ramanujan Fellowship, Department of Science and Technology, India (to A.S.N.S.); National Centre for Biological Sciences (NCBS) core funding (to A.S.N.S. and S.K.); Junior Research Fellowship, University Grants Commission, India (to P.S.). Funding for open access charge: National Centre for Biological Sciences (NCBS) core funding.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Arber W. Host specificity of DNA produced by Escherichia coli V. The role of methionine in the production of host specificity. J. Mol. Biol. 1965;11:247–256. doi: 10.1016/s0022-2836(65)80055-9. [DOI] [PubMed] [Google Scholar]
- 2.Rambach A, Tiollais P. Bacteriophage lambda having EcoRI endonuclease sites only in the nonessential region of the genome. Proc. Natl Acad. Sci. USA. 1974;71:3927–3930. doi: 10.1073/pnas.71.10.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rocha EP, Danchin A, Viari A. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res. 2001;11:946–958. doi: 10.1101/gr.gr-1531rr. [DOI] [PubMed] [Google Scholar]
- 4.Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. Shaping the genome–restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev. 1999;9:649–656. doi: 10.1016/s0959-437x(99)00026-x. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handa N, Kobayashi I. Post-segregational killing by restriction modification gene complexes: observations of individual cell deaths. Biochimie. 81:931–938. doi: 10.1016/s0300-9084(99)00201-1. [DOI] [PubMed] [Google Scholar]
- 8.Jeltsch A, Pingoud A. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J. Mol. Evol. 1996;42:91–96. doi: 10.1007/BF02198833. [DOI] [PubMed] [Google Scholar]
- 9.Sekizaki T, Otani Y, Osaki M, Takamatsu D, Shimoji Y. Evidence for horizontal transfer of SsuDAT1I restriction-modification genes to the Streptococcus suis genome. J. Bacteriol. 2001;183:500–511. doi: 10.1128/JB.183.2.500-511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kita K, Tsuda J, Kato T, Okamoto K, Yanase H, Tanaka M. Evidence of horizontal transfer of the EcoO109I restriction-modification gene to Escherichia coli chromosomal DNA. J. Bacteriol. 1999;181:6822–6827. doi: 10.1128/jb.181.21.6822-6827.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wion D, Casadesús J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer BR, Marinus MG. The dam and dcm strains of Escherichia coli–a review. Gene. 1994;143:1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- 13.Boye E, Løbner-Olesen A. The role of dam methyltransferase in the control of DNA replication in E. coli. Cell. 1990;62:981–989. doi: 10.1016/0092-8674(90)90272-g. [DOI] [PubMed] [Google Scholar]
- 14.Peterson SN, Reich NO. Competitive Lrp and Dam assembly at the pap regulatory region: implications for mechanisms of epigenetic regulation. J. Mol. Biol. 2008;383:92–105. doi: 10.1016/j.jmb.2008.07.086. [DOI] [PubMed] [Google Scholar]
- 15.Reisenauer A, Shapiro L. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 2002;21:4969–4977. doi: 10.1093/emboj/cdf490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi N, Naito Y, Handa N, Kobayashi I. A DNA methyltransferase can protect the genome from postdisturbance attack by a restriction-modification gene complex. J. Bacteriol. 2002;184:6100–6108. doi: 10.1128/JB.184.22.6100-6108.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 21.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. TIG. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 22.Vernikos GS, Parkhill J. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics. 2006;22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- 23.Lin LF, Posfai J, Roberts RJ, Kong H. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl Acad. Sci. USA. 2001;98:2740–2745. doi: 10.1073/pnas.051612298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koomey M. Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer. APMIS Suppl. 1998;84:56–61. doi: 10.1111/j.1600-0463.1998.tb05649.x. [DOI] [PubMed] [Google Scholar]
- 25.Hofreuter D, Odenbreit S, Püls J, Schwan D, Haas R. Genetic competence in Helicobacter pylori: mechanisms and biological implications. Res. Microbiol. 2000;151:487–491. doi: 10.1016/s0923-2508(00)00164-9. [DOI] [PubMed] [Google Scholar]
- 26.Banerjee S, Chowdhury R. An orphan DNA (cytosine-5-)-methyltransferase in Vibrio cholerae. Microbiology. 2006;152:1055–1062. doi: 10.1099/mic.0.28624-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.