Abstract
Detailed analyses of the chromatin around the BIM promoter has revealed that latent Epstein–Barr virus (EBV) triggers the recruitment of polycomb repressive complex 2 (PRC2) core subunits and the trimethylation of histone H3 lysine 27 (H3K27me3) at this locus. The recruitment is absolutely dependent on nuclear proteins EBNA3A and EBNA3C; what is more, epitope-tagged EBNA3C could be shown bound near the transcription start site (TSS). EBV induces no consistent changes in the steady-state expression of PRC2 components, but lentivirus delivery of shRNAs against PRC2 and PRC1 subunits disrupted EBV repression of BIM. The activation mark H3K4me3 is largely unaltered at this locus irrespective of H3K27me3 status, suggesting the establishment of a ‘bivalent’ chromatin domain. Consistent with the ‘poised’ nature of these domains, RNA polymerase II (Pol II) occupancy was not altered by EBV at the BIM TSS, but analysis of phospho-serine 5 on Pol II indicated that EBNA3A and EBNA3C together inhibit initiation of BIM transcripts. B cell lines carrying EBV encoding a conditional EBNA3C-oestrogen receptor-fusion revealed that this epigenetic repression of BIM was reversible, but took more than 3 weeks from when EBNA3C was inactivated.
INTRODUCTION
Epstein–Barr virus (EBV) is a γ-herpesvirus and as such is characterized by a tropism for lymphocytes and its ability to persist in memory B cells for the lifetime of the infected host. This results in EBV asymptomatically infecting >90% of the human population. In vitro, EBV can very efficiently induce the activation and continuous proliferation of resting human B cells—sometimes called immortalization. The resulting lymphoblastoid cell lines (LCLs) carry the viral genome as extra-chromosomal episomes and express only nine latency-associated EBV proteins. There are six nuclear antigens (EBNAs 1, 2, 3A, 3B, 3C and LP), three membrane proteins (LMP1, LMP2A and LMP2B) and in addition several RNA species—including multiple microRNAs. Together these factors activate the quiescent B cells from G0 into the cell cycle, initiate and sustain proliferation and maintain the viral genome in its extra-chromosomal state [reviewed in (1–3)].
EBV can be the causative agent in the benign self-limiting lymphoproliferation, infectious mononucleosis (IM). Uncontrolled proliferation of similar B cells in the immunocompromised can result in a fatal form of IM, chronic B-lymphoproliferative disease or rarely the development of malignant lymphoma (4). In normal individuals it appears that proliferation of the infected B cells is controlled by potent signals that trigger differentiation and also the action of EBV-specific cytotoxic T lymphocytes that recognize and destroy the proliferating B-blasts (5,6). Individuals co-infected with malaria (mainly children) or HIV (mainly adults) are at increased risk of developing EBV-associated B cell lymphomas, including Burkitt’s lymphoma (BL) and diffuse large B cell lymphoma (DLBCL) (2,4).
EBNA3A, 3B and 3C are considered to comprise a family which probably arose in γ-herpesvirus evolution by a series of gene duplication events since they share limited but significant amino acid sequence homology, have the same gene structure (i.e. a short 5′-coding exon and a long 3′-coding exon) and are arranged in tandem in the EBV genome. All EBNA3 transcripts are alternatively spliced from very long mRNAs generally initiated at the Cp latency promoter; LCLs have only a few copies of these transcripts per cell, suggesting their expression is very tightly regulated and the turnover of the EBNA3s is very slow (2,7). Although they are ancestrally related, have limited homology and share certain features, there is nothing to suggest that these three large (>900 amino acids) proteins have extensively redundant functions [reviewed in (2)]. Genetic studies using recombinant viruses indicated that EBNA3A and EBNA3C are essential for the efficient in vitro transformation of B cells into LCLs, whereas EBNA3B is completely dispensable (2,8).
All three EBNA3 proteins bind to the cellular DNA-binding factor RBP-JK (also known as CBF1). This is the same protein that binds, and targets to DNA, the EBV transactivator EBNA2 and the Notch-IC effector of the Notch signalling pathway. EBNA3A, 3B and 3C bind to the same site on RBP-JK/CBF1 as EBNA2 and can all inhibit EBNA2-mediated activation of the LMP2 promoter and can repress Cp reporter plasmids and plasmids containing multiple RBP-JK/CBF1-binding sites derived from Cp (9–12). Since Cp is generally the promoter for all EBNA mRNA initiation in LCL cells, it is widely assumed that the EBNA3 proteins contribute to a negative auto-regulatory loop. In addition all three EBNA3s exhibit robust repressor activity when targeted directly to DNA by fusion with the DNA-binding domain of Gal4 (9,13–15, and our unpublished data). They also interact with one or more cellular factors involved in transcriptional repression or silencing; these include histone deacetylases (HDACs) and CtBP (15–18, and our unpublished results).
Consistent with EBNA3A and EBNA3C being oncoproteins, both can independently co-operate with oncogenic Ras in the transformation and immortalisation of primary rodent fibroblasts (15,19). Since this type of assay identifies factors that overcome the outcomes of constitutive oncogenic signalling or ‘oncogenic stress’ (20,21), it suggested that EBNA3A and EBNA3C might play similar roles in the transformation of B cells and EBV-associated lymphomagenesis.
Epigenetic [i.e. heritable in the absence of changes to DNA sequence (22)] gene silencing is most commonly associated with methylation of cytosine in CpG dinucleotides of DNA at gene promoters. However, repression mechanisms involving covalent modification of histones that are common in development can sometimes act as primers for DNA methylation (see below). One of the best characterized of these involves repression of transcription by the polycomb repressive complex 2 (PRC2). This multiprotein complex mediates repression through the histone methyltransferase activity of one of its components, EZH2, that catalyzes trimethylation of H3K27 [reviewed in (23–26)]. Other components of PRC2 are SUZ12, EED and RbAp46/48 and recently an ancillary factor, JARID2, has been identified as being essential for recruitment of PRC2 to some polycomb-target genes (27–31). It remains unclear how the polycomb proteins are recruited to specific promoters in mammalian cells, although sequence context is probably important and a preference for regions rich in CpG dinucleotides [CpG-islands (CGIs)] has been reported (32). However, for most target genes, it remains to be determined whether specificity comes from sequence-specific transcription factors, PRC2-interacting RNA species or yet to be identified mechanisms [reviewed in (23,26,33–37)]. H3K27me3 can result in the binding of a second complex, PRC1, which in mammals includes BMI1, MEL18, RING1 and CBX family members [reviewed in (23,26)]. PRC1 mediates chromatin compaction (38) and the local formation of heterochromatin, and together with PRC2, increases the chances of the more stable CpG methylation mark being deposited—particularly in the development of cancer [(39–44), reviewed in (45)]. Although recent evidence suggests H3K27me3 is stable and heritable (46,47), the modification can be rapidly removed by demethylase enzymes such as JMJD3 (48). Moreover, if a promoter carries H3K27me3 and simultaneously has the activation-associated modification H3K4me3 at the same locus, it is repressed but is described as ‘bivalent’ and thought to be poised for rapid reactivation. Genes with such bivalent domains are common in stem cells (49).
We have demonstrated that all three EBNA3 proteins can manipulate the expression of several hundred cellular genes and that they often act co-operatively to induce epigenetic chromatin modifications on target genes (50). In the case of p16INK4A we showed that the repression of transcription involves the covalent K27me3 modification of histone H3 at the p16INK4A promoter and that this requires expression of EBNAs 3A and 3C and involves polycomb complexes (51).
Regulation of the gene encoding BIM (BCL2L11) by EBV involves repression of transcription that in EBV-positive BL can result in CpG methylation of DNA across a large CpG island at the 5′-end of the gene (52). However, the data suggest that DNA methylation was a secondary event following an EBV-induced histone modification that probably involves polycomb proteins. Here, using EBV-negative BL cell lines infected with various BAC-derived recombinant EBVs, we explore the nature of BIM repression and determine the contribution of the EBNA3 proteins and components of the polycomb system.
MATERIALS AND METHODS
Cell culture and treatments
All B cell lines were cultured in RPMI-1640 medium (Invitrogen), supplemented with 10% foetal bovine serum, penicillin, streptomycin, 1 mM sodium pyruvate (Sigma) and 50 µM α-thioglycerol. Hygromycin B (Roche) was added to cultures (100 µg/ml) of BL cell lines containing hygromycin-resistant recombinant EBV [BL31 and BL2 panels, described in (50,53)]. LCLs are described in (53,54).
BL31 WT cell lines with either BMI1, SUZ12 or JARID2 depleted were produced by infection of BL31 WT with MISSION® shRNA Lentiviral Transduction Particles from Sigma (BMI1: #TRCN0000020155; SUZ12: #TRCN0000038728; JARID2: #TRCN0000107367; Non-targeting: #SHC002V). Stable cell lines were grown out with selection in medium containing 1 µg/ml puromycin dihydrochloride (Sigma).
BL31 cells expressing a conditional EBNA3C (BL31 3CHT) were grown in 400 nM 4-hydroxytamoxifen (HT, Sigma). To remove HT, cells were washed twice with medium not containing HT and re-suspended in the same volume of non-HT medium. Production of 3CHT BAC viruses and the protocol for BL31 infections have been described (51,53).
Recombinant tagged EBNA3C viruses and cell lines
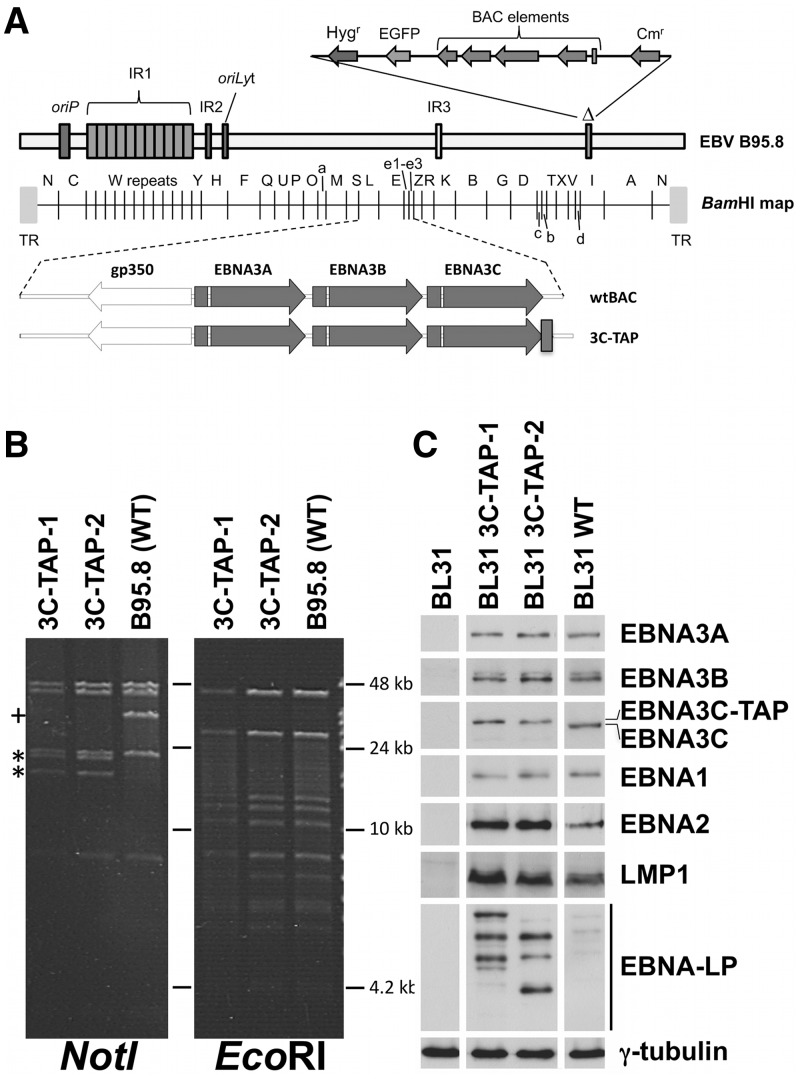
In order to generate cell lines expressing EBNA3C protein tagged with an epitope suitable for ChIP isolation, we have produced recombinant EBVs containing EBNA3C with an extended c-terminus composed of a tandem Strep-tag II followed by a c-terminal FLAG-tag (TAP-tag) (55). The TAP-tag is separated from EBNA3C by the peptide linker PGSQGGDG, with a Not I restriction site after the end of the tag. The tagged gene was introduced into the B95-8 EBV-BAC genome using a markerless RecA-mediated recombination system (56). The two independent but identically produced recombinant EBV-BACs (3C-TAP-1 and 3C-TAP-2) were used to produce recombinant viruses from HEK293 cell lines, and these viruses were used to infect BL31 cells using previously described protocols (53,57,58). For episome rescue, low molecular weight DNA was electroporated into competent DH10B Escherichia coli, and recovered EBV-BAC clones were analysed by pulsed- field gel electrophoresis as previously described (59).
Western immunoblotting
Western immunoblotting was performed essentially as previously (7,53,60). Whole-cell extracts were used when probing for histone H3. Each western blot was performed at least twice and representative results are shown. The antibodies used in this study are given in Supplementary Table S1.
Precipitation of methylated DNA
Genomic DNA was extracted using the Qiagen DNeasy midi kit. DNA re-suspended at 1 mg/ml in 200 µl of dH2O was sonicated in a Diagenode Bioruptor Standard sonicator with 1.5-ml tube holder at high setting for 10 min to produce 200- to 1000-bp fragments. Methylated DNA was precipitated from 1 µg of sonicated DNA using the MethylCollector Ultra kit (Active Motif, 55005), according to the manufacturer’s instructions. Precipitated DNA was analysed by quantitative PCR (Q-PCR) using the Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen). The samples were run in an ABI 7900 HT real time PCR machine. The cycling conditions were 50°C for 2 min, 95°C for 2 min and then 40 cycles of 95°C for 15 s, 60°C for 60 s. The controls and analysis were the same as described previously (52). The primers used are shown in Supplementary Table S2.
Chromatin immnoprecipitations
A chromatin immunoprecipitation (ChIP) assay kit from Millipore (17-295) was used, according to the manufacturer’s protocol. To obtain sheared chromatin with DNA of 200–1000 bp in length, extracted chromatin from 1 × 106 cells per ChIP was sonicated using a Diagenode Bioruptor Standard sonicator with 1.5-ml tube holder at high setting for 12.5 min. The antibodies used for immunoprecipitation are shown in Supplementary Table S1. DNA from precipitated chromatin was quantified as for precipitated DNA above. Controls and analysis were the same as described previously (52). Primers used are given in Supplementary Table S2.
Quantification of mRNA
RNA was extracted from 3 × 106 cells for each line, using the Qiagen RNeasy mini kit, according to the manufacturer’s instructions. Approximately 1 µg of RNA was used to reverse transcribe and produce cDNA with Invitrogen’s SuperScript III First-strand Synthesis Supermix, according to the manufacturer’s protocol. The cDNA was then quantified by Q-PCR with Applied Biosystems’ 7900 HT real time PCR machine under the same conditions as described above. Approximately 20 ng of cDNA was used for each reaction. Standard curves of typically five 10-fold serial dilutions of a mixture of all cDNAs from each experiment were used to determine the efficiency of each primer pair. Values for mRNAs under inspection were normalized for each sample using the values for housekeeping gene GNB2L1 as an internal control. Errors given in graphs are the standard deviations from three replicate reactions for each assay and the corresponding control from each sample. The sequences of the primers used in this study are given in Supplementary Table S3.
RESULTS
Repression of BIM expression by EBV is associated with H3K27me3 at the promoter and requires both EBNA3A and EBNA3C
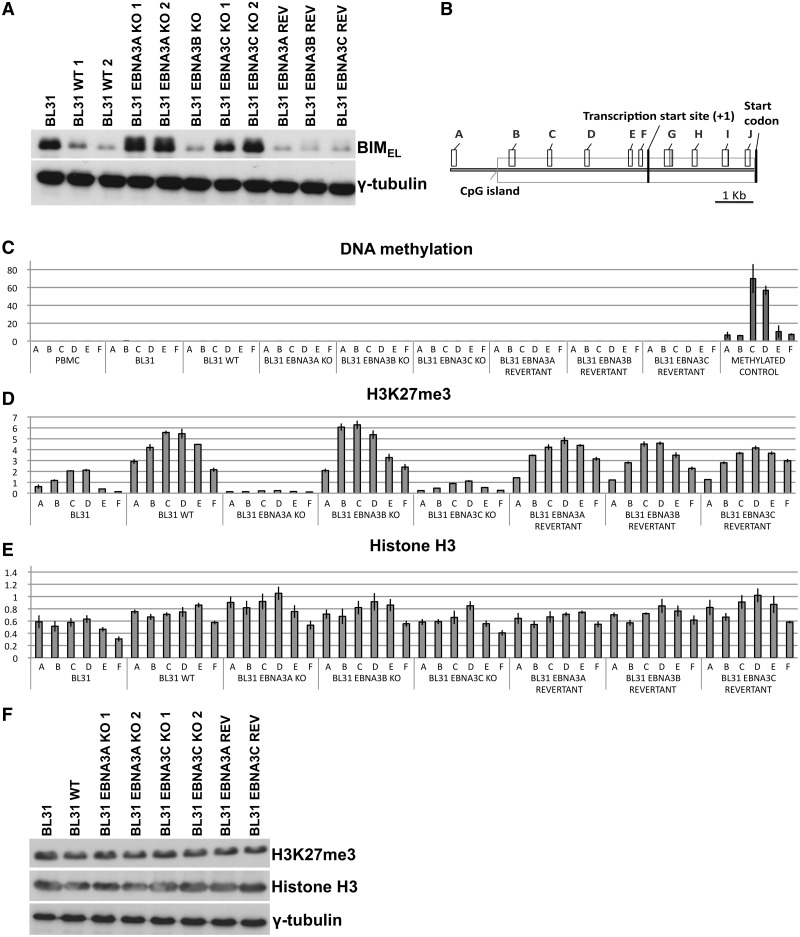
In order to investigate the roles of EBNA3A and EBNA3C in the regulation of cellular gene expression, a panel of cell lines based on the EBV-negative BL-derived line BL31 were established and characterized (50,53). Here a selection of these were subjected to western blot analysis to confirm our previous observation that BL31 expressed high levels of BIM protein and that this was substantially reduced in BL31 latently infected in vitro with wild-type (WT) EBV-BAC-derived virus (Figure 1A). This analysis also confirmed that in BL31 infected with recombinant viruses from which EBNA3A or EBNA3C genes were deleted, BIM expression was not repressed and remained at a similar level to that in BL31 (Figure 1A). Revertant (REV) viruses reconstituted with the genes that had previously been deleted behaved—as expected—like the WT EBV and repressed BIM expression.
Figure 1.
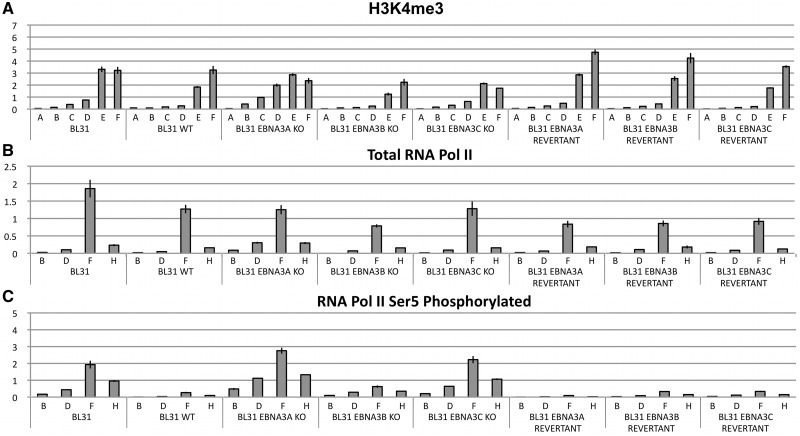
Epigenetic marks that are influenced by EBNA3A and EBNA3C during the repression of BIM by EBV. (A) The levels of BIM protein (whose predominant isoform is the extra-large BIMEL) were assayed by western blotting protein extracts from BL31 cell lines. BIM levels in uninfected BL31 or BL31 infected with the wild-type (B95.8-BAC) EBV (WT), or viruses with each individual EBNA3 knocked out (KO) and the revertant viruses (REV) are shown. In all the EBNA3A and EBNA3C knockouts BIM levels are similar to the levels observed in the uninfected cells. The expression of γ-tubulin is shown as a control for the amount of protein loaded. (B) Schematic of the BIM locus showing the approximate locations of the CpG island, TSS, BIM start codon and the PCR primer pairs used in this study (A–J). (C) Methylated DNA precipitation was used to assess whether DNA methylation at the BIM promoter was responsible for repression of BIM expression. The histogram bars represent fold enrichment for co-precipitated DNA relative to input DNA, as determined by Q-PCR. The height of the bars represents the abundance of the epigenetic mark at the locus assessed by each primer set. Uninfected BL31 cells, BL31 infected with WT EBV, or EBNA3 knockouts (KO) or the revertant viruses were tested. DNA from peripheral blood mononuclear cells (PBMC) and in vitro methylated PBMC DNA were used as negative and positive controls respectively. The error bars represent the standard deviation from triplicate Q-PCR assays. (D) The same panel of BL31 cell lines as in (C) was used to assess the abundance of the H3K27me3 mark across the BIM promoter by ChIP analysis. The histogram bars represent the ratio of chromatin precipitated with an antibody specific for the trimethylated form of H3K27 (H3K27me3), relative to 5% of input, after values from negative control IgG precipitation were subtracted as background. The error bars represent the standard deviation from triplicate PCR assays. (E) ChIP analysis for total histone H3 associated with the BIM promoter, presented as in (D). (F) Western blot with antibody against H3K27me3 showing the total amount of H3 with this modification in BL31 cells, BL31 cells infected with the wild-type virus, EBNA3A KO, EBNA3C KO and revertant viruses. Expression levels of histone H3 and γ-tubulin were used as loading controls.
Flanking the BIM transcription start site (TSS) is a remarkably large CGI that extends for ∼6 kb (http://genome.ucsc.edu/cgi-bin/hgGateway; Figure 1B). We previously showed that in various EBV-positive B cell lines and primary BL samples significant DNA methylation across this region could be detected, but this was absent in EBV-negative equivalents (52). However analysis of several BL cell lines, including BL31, that were newly infected with EBV and also recently established LCLs revealed that although BIM transcription was repressed, there was little or no CpG methylation but rather the epigenetic repressive histone modification H3K27me3 was evident. Because of the apparently progressive nature of DNA methylation across the BIM CGI, a panel of low passage BL31-derived cell lines was subjected to methylated-DNA precipitation (MeDP) coupled with quantitative real-time PCR analysis using primer-pairs (A–F) as indicated (Figure 1B). Primary peripheral blood mononuclear cells and in vitro methylated genomic DNA were used as negative and positive controls respectively. All the lines analysed, irrespective of their EBV status, showed no detectable DNA methylation, as judged by MeDP, across a region extending from ∼8 kb upstream of the BIM TSS to the start codon—indicating that here histone modification was the most likely mode of repression (Figure 1C).
Having shown in these EBV-infected BL31 cells that CpG methylation was not established at BIM, ChIP with an antibody directed against H3K27me3 and quantitative PCR analyses across the CGI was performed using the same primer-pairs as for MeDP. This showed that repression correlated with deposition of the H3K27me3 mark and expression of EBNA3A and EBNA3C (in the WT-, EBNA3BKO- and the revertant-infected cell lines in Figure 1D). In contrast, when either EBNA3A or EBNA3C were deleted from EBV (EBNA3AKO and EBNA3CKO) BIM expression remained high and, significantly, the amount of H3K27me3 detected across the BIM locus was below the background in uninfected BL31 cells. Occupancy of H3K27me3 on promoters of genes encoding Myoglobin and GAPDH were analysed in a similar manner as positive and negative controls respectively (Supplementary Figure S1A). ChIP for total histone H3 at BIM, Myoglobin and GAPDH revealed little difference between the cell lines used (Figure 1E and Supplementary Figure S1B). This indicates that any changes in the distribution of covalent histone modifications detected are not due to alterations in nucleosome density. The effects on H3K27me3 distribution at the BIM promoter induced by EBNA3A and EBNA3C are specific, since the overall levels of H3K27me3 were unaffected by EBV, EBNA3A or EBNA3C (Figure 1F).
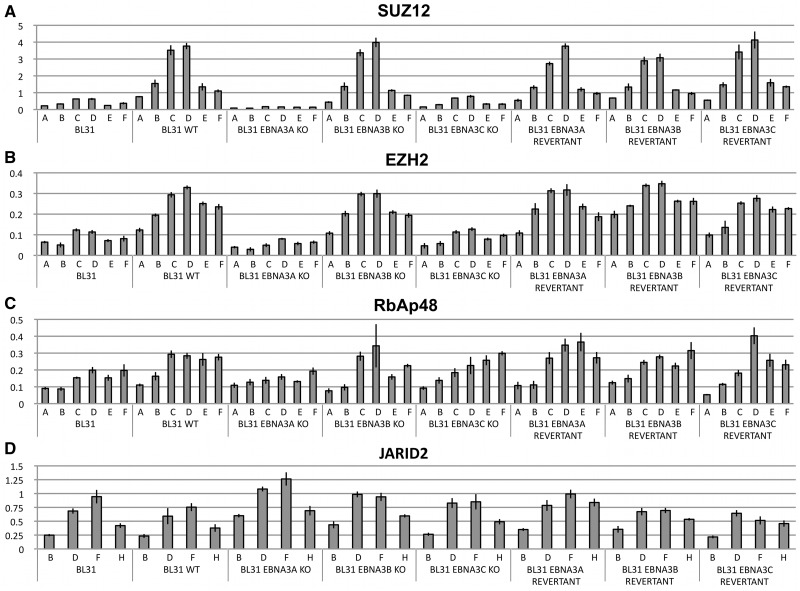
EBNA3A and EBNA3C are necessary for recruitment of the PRC2 subunits SUZ12 and EZH2 to BIM regulatory elements
The protein complex responsible for establishing the H3K27me3 mark is PRC2. In mammals this is generally composed of a catalytic core made up of the histone methyltransferase EZH2 and subunits SUZ12, EED and RbAp46/48 (see ‘Introduction’ section). ChIPs for SUZ12, EZH2 and RbAp48 were performed on the panel of BL31 cell lines and analysed using the same primer-pairs as in Figure 1. SUZ12 was found occupying sites throughout the BIM CGI with a peak corresponding to the H3K27me3 peak (primer-pairs C and D) only when wild-type EBV, the EBNA3B-knockout virus or the revertant viruses were present (Figure 2A). SUZ12 binding at this locus was almost undetectable in the cells infected with EBNA3A- and EBNA3C-knockout viruses (Figure 2A). These viral proteins appear to be necessary for assembly of PRC2 and the resulting deposition of H3K27me3 on the BIM promoter. This was confirmed by the ChIP analysis for EZH2, which had a similar distribution to SUZ12—albeit with a higher level of background binding in the uninfected BL31 cells (Figure 2B). In contrast RbAp48—a chaperone that binds directly to histones H3 and H4 but is involved in targeting PRC2 to nucleosomes (61–63)—could be detected across the locus irrespective of whether EBV (or EBNA3A and EBNA3C) were present. The levels of bound RbAp48 in the uninfected BL31 cells and BL31-EBNA3AKO were perhaps slightly lower in the experiment shown (Figure 2C); however this was not a consistent finding in multiple ChIP experiments. In order to verify that the amount of RbAp48 detected at BIM were above background, we compared it to the levels seen at a known PRC2-target (IRX4) (27) and a non-PRC2-targeted gene (MCM6) (29) (Supplementary Figure S1C). We were unable to identify satisfactory anti-EED antibodies for ChIP analysis of this remaining PRC2 core component.
Figure 2.
PRC2 occupancy across the BIM promoter region. The presence and abundance of PRC2 proteins was assayed by ChIP analysis of a BL31 panel of cells, as indicated. Presentation is the same as for ChIP analysis in Figure 1. The region of amplification for each primer set (A–F) can be seen in Figure 1B. The error bars represent the standard deviation from triplicate PCR assays. ChIP was performed using an antibody specific for (A) PRC2 core subunit SUZ12, (B) PRC2 catalytic subunit EZH2, (C) core subunit RbAp48, (D) ancillary factor JARID2.
JARID2 is a trans-acting factor that has been found co-localized with PRC2 core subunits at many loci in the mammalian genome—particularly in embryonic stem cells (27–30). It is thought that JARID2 can aid in the recruitment of PRC2 and perhaps also PRC1 (64) (see below). Similar ChIP analyses to those described above (but using primers corresponding to sites located more proximal to exon 1) were performed. JARID2 was found distributed around the BIM TSS in a similar pattern to RbAp48 and binding was not dependent on EBV, EBNA3A or EBNA3C (Figure 2D). JARID2 levels at BIM were found to be comparable to those at the PRC2-target IRX4 (27) and significantly higher than at the non-PRC2-targeted gene MCM6, (29) (Supplementary Figure S1D).
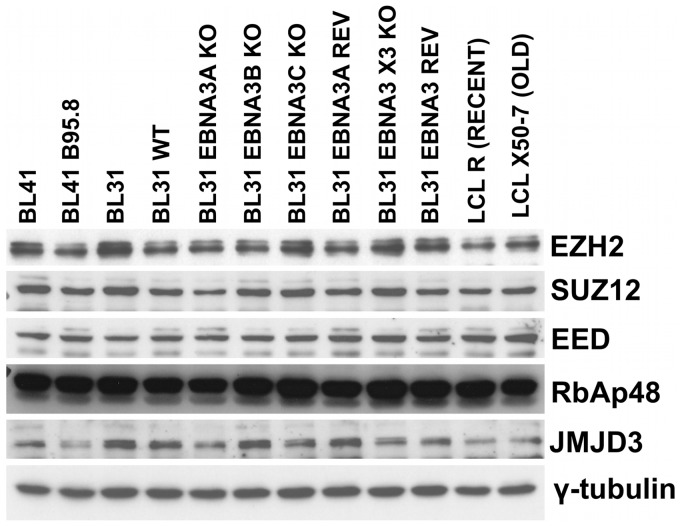
Latent EBV does not consistently alter the expression of PRC2 components or that of the H3K27me3-demethylase JMJD3
Since EBV, and specifically EBNA3A and EBNA3C influence the recruitment of PRC2 and the pattern of H3K27me3 across the BIM regulatory region, it is possible that the virus may alter the expression of PRC2 subunits or of the histone demethylase JMJD3 that specifically removes the H3K27me3 mark (48). The steady-state levels of EZH2, SUZ12, EED, RbAp48 and JMJD3 were therefore assessed by western blotting protein extracts from a variety of EBV-negative and EBV-positive cell lines (Figure 3). Although there were minor differences in the amounts of proteins expressed, none of these were consistently associated with the EBV or EBNA3 status of the cells. These data are consistent with the published transcriptome analyses of the BL31 cell lines [(50), http://www.epstein-barrvirus.org.uk/]. The changes in PRC2 binding across the BIM CGI promoter cannot be explained by the general availability of PRC2 core subunits in the cell or the amount of H3K27me3 demethylase JMJD3 expressed.
Figure 3.
Steady-state levels of PRC2 subunits and the H3K27me2/3 demethylase JMJD3. Protein was extracted from exponentially proliferating cultures of BL41 and BL31 cells, or cells infected with EBV. BL31 cells infected with recombinant EBV with each of the EBNA3s deleted (KO) and a EBNA3 triple knockout (shown as X3) from which the whole EBNA3 locus was deleted and revertants (REV) were also used. Two LCLs, one recently transformed (RECENT) and one that has been grown in culture for several years (OLD) were included. Western blots of protein extracts were used to assess the relative amounts of PRC2 core subunits EZH2, SUZ12, EED and RbAp48 expressed. Expression of the lysine demethylase JMJD3 was also analysed and expression of γ-tubulin was used as loading control.
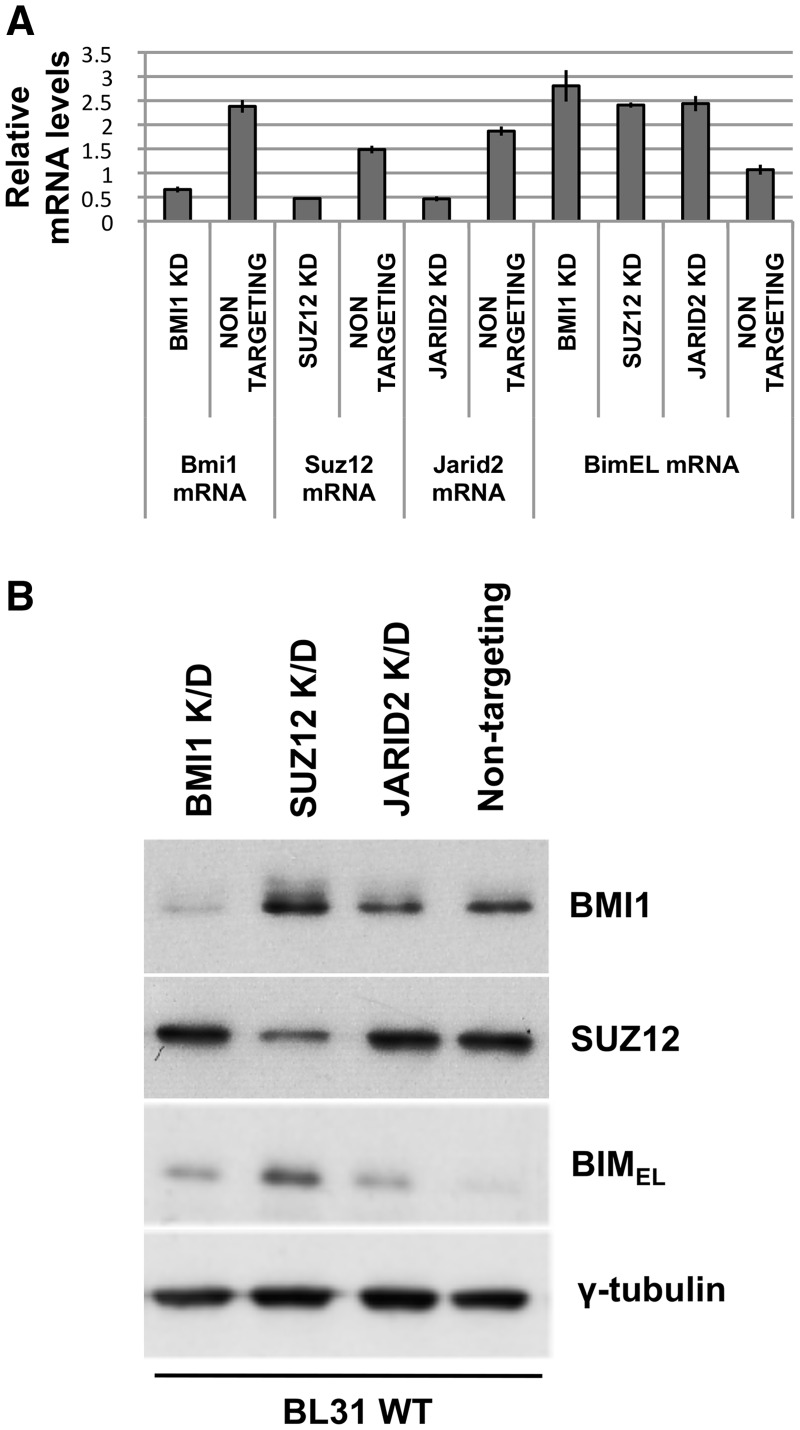
Short-hairpin ShRNA-mediated depletion of PcG subunits shows they are factors required for EBV-mediated repression of BIM
Roles for SUZ12 and JARID2 in the repression of BIM in EBV-positive B cells were suggested by their distribution across the promoter region. In addition it has been reported that, although PRC2 alone is sufficient for H3K27me3 (65), effective repression at many loci probably requires the subsequent recruitment of PRC1, a complex that has been suggested to contribute to repression by preventing transcriptional elongation from some bivalent loci (66). Therefore in order to formally assess the contribution of SUZ12 (an essential component of PRC2), JARID2 and the other polycomb complex PRC1 in the EBV-mediated repression of BIM expression, BL31 cells infected with EBV (BL31 WT) were stably transduced with lentiviruses that produce shRNAs specific for SUZ12, JARID2 or the PRC1 core subunit BMI1. These produced cell lines from which the targeted RNA and protein were depleted, but never completely absent (Figure 4). Cells transduced with viruses producing a non-targeted RNA were included as a control. Independently depleting any of these three factors produced an increase in both BIM mRNA and protein, indicating an essential role for each in the repression achieved by EBV via EBNA3A and EBNA3C (Figure 4). In the case of JARID2, only the level of mRNA was measured, since we were unable to identify a reliable antibody for western blotting the human JARID2 protein (data not shown). However, JARID2 was found to be involved in EBV-mediated recruitment of SUZ12 at BIM, since depleting JARID2 in EBV-infected cells significantly reduced SUZ12 occupancy; in the SUZ12 knock-down cells binding of SUZ12 at BIM was nearly completely ablated (Supplementary Figure S2).
Figure 4.
BMI1, SUZ12 and JARID2 are necessary for robust repression of BIM in EBV-infected cells. BL31 cells infected with wild-type (B95.8-BAC) EBV were superinfected with lentiviruses expressing shRNAs specifically targeting BMI1, SUZ12 or JARID2 mRNAs. Cells superinfected with a lentivirus expressing a ‘scrambled’, non-targeting shRNA were used as negative controls. (A) Quantitative real-time RT–PCR was performed to show the effects on mRNA levels for each factor. The histogram bars represent values relative to housekeeping gene GNB2L1. The error bars represent standard deviation from three replicate assays. (B) Western blots showing the depletion of the relevant proteins and effects on BIM expression. It should be noted that in our hands human JARID2 is not reliably detected with any of the antibodies available. The predominant isoform of BIM protein (BIMEL) was western blotted to show the effect on its abundance resulting from the depletion of BMI1, SUZ12 or JARID2.
H3K4me3 modification and RNA polymerase II (Pol II)-binding to BIM remain unaffected by EBV, but EBNA3A and EBNA3C together inhibit the initiation of transcription
In mammalian development, the promoters of genes regulated by the polycomb system are often found in a ‘bivalent’ state (see ‘Introduction’ section). That is, in addition to carrying repressive H3K27me3 marks that are associated with non-expressing genes, they simultaneously carry the H3K4me3 mark, generally associated with activation, and Pol II phosphorylated on Ser5 bound at the TSS (67,68). Such promoters are considered to be poised for rapid reactivation.
ChIP analysis was performed for the H3K4me3 modification and analysed using the same primer set as for H3K27me3 and PRC2 components (Figure 5A). The apparent distribution of H3K4me3 was closer to the TSS (primers E and F) and, in contrast to H3K27me3, was consistently found in all the cells examined irrespective of EBV or EBNA3 status. These data suggested that the BIM promoter might be maintained in a bivalent, poised state by latent EBV. ChIP analyses were performed using pan-specific anti-Pol II antibodies and primer pairs located upstream and downstream of the TSS. This showed that Pol II was bound at or near the TSS in all the cells, irrespective of the level of BIM expressed, also consistent with the promoter being in a bivalent state (Figure 5B). However, further ChIP analyses using antibodies directed against Pol II phosphorylated at Ser5 consistently showed that EBV suppressed the establishment of this modification to the bound Pol II (Figure 5C), but EBNA3A- or EBNA3C-deleted EBV failed to suppress phosphorylation on Serine 5 of Pol II. These data show that the BIM promoter is not a classical bivalent domain and that EBV via EBNA3A and EBNA3C blocks initiation of BIM transcripts rather than Pol II recruitment or transcript elongation (discussed in more detail below). Genes encoding Myoglobin and GAPDH were used as controls for the efficiency and specificity of H3K4me3, Pol II and Phospho-Ser5 Pol II ChIPs (Supplementary Figure S1E–G).
Figure 5.
H3K4me3 and RNA polymerase (Pol) II occupancy on the BIM promoter. ChIP was performed on extracts from the BL31 panel of cells. Values represent ratio of chromatin precipitated relative to 5% of input. (A) An antibody specific for the tri-methylated form of H3K4 (H3K4me3) was used for ChIP. (B) N20 antibody that recognises an epitope located at the N-terminal of the largest subunit of Pol II, RPB1 was used. This antibody precipitates total Pol II, irrespective of the phosphorylation state of the C-terminal repeat domain (CTD) of RBP1. The regions of BIM investigated included one adjacent to the TSS (F) and one located downstream (H). (C) An antibody that specifically detects phospho-Ser5 Pol II was used.
Data from ChIP analysis of BIM with antibodies against phosphorylated Pol II at Ser2 were consistent with these results and indicate that when EBNA3A or EBNA3C are absent, as one would predict, not only transcription initiation but also productive elongation of the transcripts occurs (Supplementary Figure S3).
In order to establish that these EBV-associated epigenetic modifications of BIM were not restricted to BL31, a second EBV-negative BL-derived line (BL2) and its EBV-carrying ‘converts’ were analysed in a similar manner (Supplementary Figure S4). Although parental BL2 fails to express detectable BIM protein due to homozogous deletion (69), the CGI and 5′-regulatory region are intact and regulation of chromatin and Pol II modification by EBV appears to be essentially the same as in BL31.
PcG-mediated repression of BIM by EBV is only slowly reversed
Bivalent domains are said to be ‘poised’ and able to rapidly switch between repression and productive transcription [reviewed in (67,68)]. It is believed that this fast switching between states can be facilitated in part because the Pol II at these domains is already phosphorylated at Ser5, thus being only one step from productive elongation of transcripts. H3K27me3 appears to be a bona fide stable epigenetic mark that can facilitate its own propagation through cell divisions (46,47), so it may be important that activating a repressed or silenced promoter can be triggered by the enzymatic removal of H3K27me3 by a specific demethylase such as JMJD3 (70,71). EBV-mediated repression of BIM appears to be regulated by PcG complexes and the H3K27me3 modification, but BIM is not classically ‘poised’, so we wanted to investigate whether the repressed state was reversible.
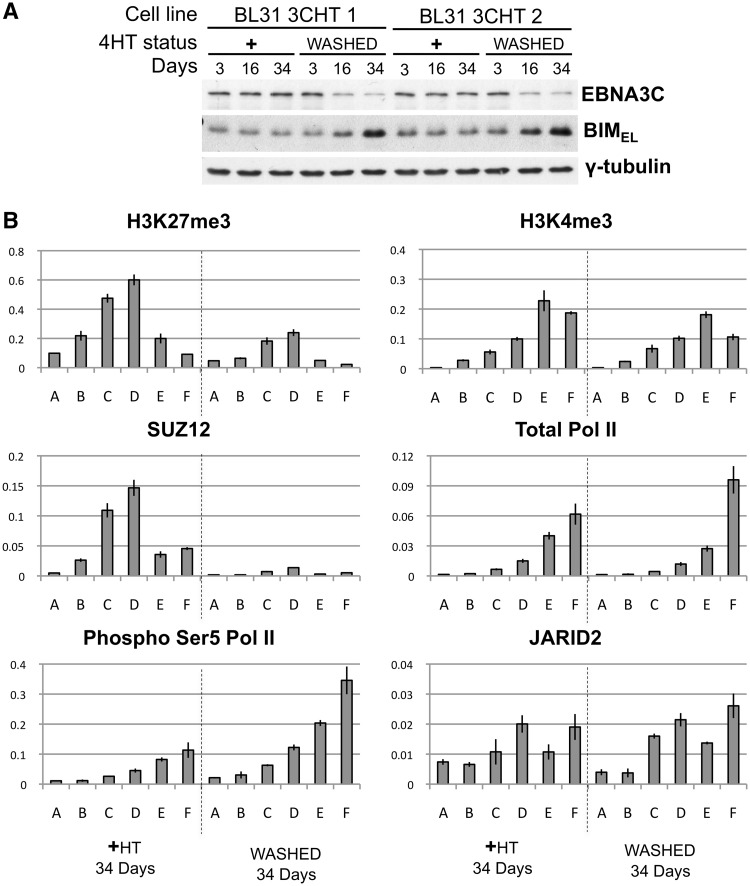
In order to study de-repression of BIM we made use of an EBV-BAC virus carrying a gene encoding EBNA3C fused to a modified oestrogen receptor (51). The activity of the EBNA3C protein produced (EBNA3C-HT) is dependent on the presence of the activating ligand 4-HT in the growth medium. This virus was used to infect BL31 cells and produce stable lines BL31-3CHT 1 and 2. These were established with HT in the medium to maintain EBNA3C function. For the analysis of the chromatin changes during BIM de-repression, BL31-3CHT cells were washed in fresh medium and grown without HT for up to 34 days. In the absence of HT the EBNA3C fusion is inactivated by nuclear exclusion, association with heat-shock proteins and proteolytic degradation (51,72). Western blots of protein extracts from cells without HT and similar cells grown in parallel with HT showed that inactivation of EBNA3C was indeed associated with a very gradual increase in the level of BIM protein (Figure 6A). ChIP assays (quantified with PCR primers A–F), were used to assess the distribution of H3K27me3, SUZ12, H3K4me3, total Pol II, Pol II phospho-Ser5 and JARID2 (Figure 6B). Consistent with the gradual de-repression and accumulation of BIM, after 34 days without HT, SUZ12 was no longer detectable on the BIM promoter and H3K27me3 was considerably reduced. Concomitantly Pol II phospho-Ser5 increased. Consistent with the results shown above the H3K4me3 modification and the amount of bound Pol II and JARID2 remained relatively unchanged. Since these cells complete the cell cycle about once every 24 h, they would probably have undergone more than 30 divisions before the H3K27me3 mark was completely erased. This suggests that the epigenetic repression of BIM is relatively stable through multiple generations and unlikely to involve the rapid active removal of H3K27me3 by JMJD3.
Figure 6.
H3K27me3-associated repression of BIM is reversible in BL31 cells. Two independent lines of BL31 infected with BAC-derived EBV expressing conditional EBNA3C-HT were used. Half of the cells established and cultured in the presence of the activating ligand 4-HT (+HT) were washed and re-suspended in medium without HT (WASHED) and cultured without HT for the number of days indicated. Cells from the original culture were grown in parallel with HT as a control population. (A) EBNA3C-HT and BIMEL expression were followed by western blot analysis, as before γ-tubulin was used as loading control. (B) ChIPs were performed on samples taken after 34 days in the presence or absence of HT to assess the occupancy of the promoter by H3K27me3, SUZ12, Pol II phosphorylated at Ser5, H3K4me3, total Pol II and JARID2. Values represent ratio of chromatin precipitated, after correction for IgG, relative to 50% of input. ChIPs were performed as described above using the primer sets indicated.
Epitope-tagged EBNA3C expressed at physiological levels in the context of latent infection directly targets EBV-repressed promoters and physically interacts with EBNA3A
It has been shown that both EBNA3A and EBNA3C associate with chromatin modifiers, including HDACs (see ‘Introduction’ section) and that HDACs are involved in the repression of BIM (52), but so far we have been unable to show a direct association of EBNA3A or EBNA3C with any PRC2 subunits. Furthermore, the slow process of de-repression following EBNA3C-HT inactivation could mean that the effects of EBNA3A and EBNA3C are indirect.
It was therefore necessary to test whether EBNA3A and EBNA3C directly target the BIM promoter, but we were unable to perform reliable ChIPs using existing antibodies against either EBNA3A or EBNA3C. Consequently the B95.8 EBV-BAC system was used to generate a tandem-affinity-purification (TAP) tag, [consisting of a Strep and a FLAG tag (55)], at the c-terminus of EBNA3C in the virus (Figure 7A). The BACs were analysed by restriction enzyme digestion and pulsed-field gel electrophoresis (Figure 7B) and then used to produce virus to infect BL31 cells. The infected BL31 cells (BL31 3C-TAP-1 and -2), expressing tagged EBNA3C (EBNA3C-TAP) were validated by western blot for the expression of EBV latent proteins (Figure 7C). Viral genome integrity in these cell lines was confirmed by episome rescue (Supplementary Figure S5).
Figure 7.
Creation and validation of cell lines infected with EBV that expresses epitope-tagged EBNA3C. (A) Schematic showing the addition of the tandem-affinity-purification (TAP) tag at the c-terminus of EBNA3C (3C-TAP) in the context of the B95.8-BAC-derived virus used in this study. (B) Validation of BAC DNA. Two independent BACs were created (3C-TAP-1 and -2). BAC DNA for each was analyzed by restriction digestion with NotI and EcoRI, followed by pulsed-field gel electrophoresis. Addition of the tag creates a NotI site in the 36.8-kb fragment containing the EBNA3s in the wild-type BAC, marked with (+). NotI digestion of the tag-containing BACs results in two new bands (20.8 and 16.1 kb), marked with asterisk. A diagnostic EcoRI digestion shows no unintended changes were introduced during addition of the tag. (C) Validation of cell lines after infection with the TAP-tagged viruses. Protein was extracted from uninfected BL31 cells, cells infected with wild-type virus, or cells infected with the two tagged-viruses and assayed by western blot for the presence of the EBV latent proteins as indicated. Expression of γ-tubulin was used as loading control.
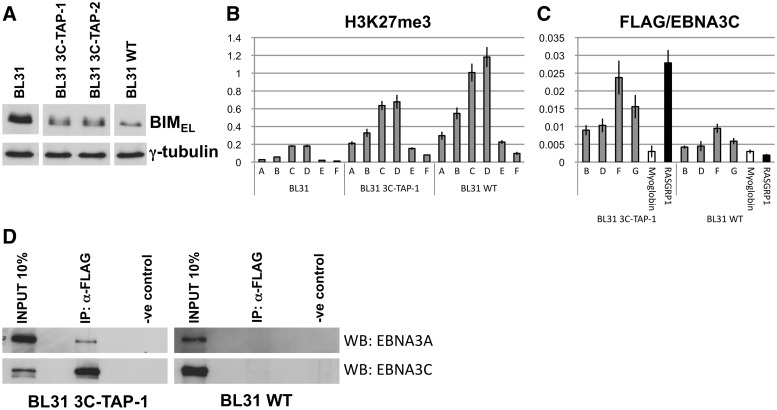
EBNA3C-TAP was able to successfully participate in the repression of BIM and reduce the level of the protein, although not quite as efficiently as unmodified EBNA3C (Figure 8A). However, the mode of repression was the same as for wild-type, with a significant increase in H3K27me3 at the BIM promoter, relative to uninfected cells (Figure 8B).
Figure 8.
TAP-tagged EBNA3C binds promoters of BIM and RASGRP1 and is co-precipitated with EBNA3A. (A) Western blot showing BIM levels in BL31 and after infection with B95.8-BAC (WT) or EBNA3C-TAP tagged viruses. Expression of γ-tubulin used as loading control. (B) ChIP analysis showing H3K27me3 levels at the BIM promoter in BL31 and following infection with WT or an EBNA3C-TAP virus. Values represent ratio of chromatin precipitated, after correction for IgG, relative to 5% of input. (C) ChIP analysis using an anti-FLAG antibody to precipitate EBNA3C-TAP and chromatin associated with it in BL31 3C-TAP-1 cells. Similar ChIP was performed in cells infected with the B95.8-BAC virus (BL31 WT) as a control for antibody specificity. Occupancy of EBNA3C-TAP across the BIM promoter is shown (primer sets B, D, F, G). No binding was detected at a negative control promoter (Myoglobin), but it was at the promoter of RASGRP1, another gene that is repressed by EBNA3C (50). (D) Immunoprecipitation was performed with a mouse anti-FLAG antibody using BL31 3C-TAP-1 cells and BL31 WT cells (as a control for antibody specificity). Mouse IgG was used for negative control immunoprecipitations. Precipitates were compared to 10% of input after western blots were probed for EBNA3A (WB: EBNA3A) and EBNA3C (WB: EBNA3C).
Anti-FLAG antibody was then used to immunoprecipitate EBNA3C-TAP from BL31 3C-TAP-1 cells. Q-PCR analysis of the ChIPed DNA showed that EBNA3C-TAP was enriched at BIM, close to the TSS (primers F and G), relative to background established by analysis of the Myoglobin gene and the signal from ChIP of cells expressing non-tagged EBNA3C (Figure 8C). EBNA3C-TAP also localized at the promoter of RASGRP1, a gene we have shown previously is epigenetically repressed by EBV when EBNA3C is expressed (50) (Figure 8C). Similar results were obtained when a second independently established BL31 line expressing EBNA3C-TAP (BL31 3C-TAP-2) was analysed (Supplementary Figure S6A).
Lacking the appropriate reagents to perform ChIP for EBNA3A directly, we tested whether there was any physical interaction between EBNA3A and EBNA3C-TAP in infected cells. Immunoprecipitation with an anti-FLAG antibody showed that not only was EBNA3C-TAP precipitated, but EBNA3A was co-precipitated (Figure 8D). The anti-FLAG antibody did not cross-react with or precipitate EBNA3A or EBNA3C from BL31 WT cells (Figure 8D). The interaction between EBNA3A and EBNA3C was confirmed by showing an anti-EBNA3A antibody could precipitate both proteins from an LCL (Supplementary Figure S6B).
DISCUSSION
In this study we have provided formal proof that when EBV represses BIM transcription in B cells, expression of both EBNA3A and EBNA3C is necessary for the assembly of PRC2 on the BIM promoter and the deposition of the epigenetic repressive mark H3K27me3. It appears that in uninfected BL31 cells, RbAp48 and JARID2 associate with the chromatin proximal to the BIM TSS and that EBV infection is necessary to recruit SUZ12 and EZH2 to establish functional PRC2. Furthermore we show that when B cells are infected with EBV co-expressing EBNAs 3A and 3C, the BIM CGI adopts characteristics of a bivalent promoter with both H3K27me3 and H3K4me3 present. These data are consistent with genome-wide ChIP screens that have identified the BIM locus as a PRC2 target with a promoter showing the characteristics of a bivalent domain in embryonic stem cells [e.g. (73)]. Importantly, we have shown for the first time that EBNA3C is directly targeted to the BIM locus and another promoter it regulates. Furthermore, EBNA3C physically interacts with EBNA3A in infected cells.
Although PRC2 binding represses gene expression, the promoters of active genes and those with bivalent chromatin are occupied by similar amounts of Pol II, but on bivalent promoters it is generally in a paused state that fails to elongate productive transcripts. This is because Pol II bound at bivalent genes is usually characterized by high levels of serine 5 phosphorylation, but not the serine 2 phosphorylation that is necessary for transcript elongation (31,74). It was therefore surprising to find that when BIM is repressed by EBV, EBNA3A and EBNA3C are not only responsible for the recruitment of PRC2, but also the inhibition of serine 5 phosphorylation on Pol II bound at the TSS of this apparently bivalent promoter. This suggests that EBNA3A and EBNA3C co-operate to influence both polycomb complex assembly on BIM and inhibition of the TFIIH kinase activity normally catalyzed by CDK7 (75–77). Although it is presently unclear whether PRC2 assembly and TFIIH kinase activity are linked—either generally or specifically in latent EBV infection—we have seen that even when EBV is present, inhibition of the kinase activity here requires PRC2. Specifically, when SUZ12 was knocked-down in WT EBV-infected cells BIM expression increased, hence transcript elongation must have taken place. This was accompanied by an increase in the abundance of the initiated Ser5 phosphorylated form of Pol II at BIM (data not shown).
Although the regulation of BIM by EBV has been dissected in two EBV-negative BL cell lines infected in vitro with EBV, an equivalent distribution of H3K27me3 on BIM was found in several LCLs that were immortalized by B95.8 EBV (52, and data not shown). Furthermore an unbiased global ChIP-seq analysis identified the human BIM promoter as bivalent in the LCL GM12878 as well as in embryonic stem cells (UCSC Genome Browser v257/ENCODE/Broad—http://genome.ucsc.edu/cgi-bin/hgTracks).
It remains to be determined precisely how EBNA3A and EBNA3C co-operate to induce the assembly of PRC2 on the BIM promoter and also block phosphorylation of serine 5 on Pol II, but EBNA3C, at least, is physically present at the promoter so the process is probably direct. A recent report showed that EBNA3A and EBNA3C have the capacity to interact in the yeast-two-hybrid system (78) and here we have shown their physical interaction by co-immunoprecipitation from EBV infected cells, so it is reasonable to speculate that they act together as part of a multiprotein complex at genes they repress.
It is well established that EBNA3A and EBNA3C interact with RBP-JK/CBF1, with the co-repressor CtBP and can co-precipitate histone deacetylases (see ‘Introduction’ section), but we have to date been unable to show a similar physical interaction with any PRC2 subunits (our unpublished data). Site-specific mutations in EBNA3A and EBNA3C showed their interaction with CtBP is functionally important for the repression of p16INK4A and efficient transformation of primary B cells (51). Data from CtBP-binding mutant viruses in BL31 suggest these interactions also make a contribution to the repression of BIM (Supplementary Figure S7). The mechanism of CtBP action in EBNA3A/C-mediated epigenetic repression has not yet been identified. We cannot at this stage say whether EBNA3C is targeted to BIM by RBP-JK/CBF1. There is no canonical RBP-JK/CBF1-binding site proximal to the BIM TSS. However, if a more flexible interpretation of the required DNA sequence is taken, then there are potential sites both proximally and distally located. High resolution mapping of EBNA3-binding sites by ChIP-seq and/or the construction of viruses expressing EBNA3C (and perhaps EBNA3A) that no longer bind RBP-JK/CBF1 will be needed to resolve this issue.
Because of its potency as an inducer of apoptosis, BIM is a uniquely important tumour suppressor in both mouse and human B cells [(79) reviewed in (80–82)]. Therefore we have previously suggested that repression of its expression by EBV, via EBNA3A and EBNA3C is likely to be a major contributory factor in the pathogenesis of EBV-associated B lymphomas (52,53,83). Our demonstration that in a number of African EBV-positive BL the CGI around the BIM promoter is heavily methylated prompted us to ask whether EBV initially represses BIM expression by manipulating the host polycomb system. This has now been established beyond reasonable doubt. Polycomb-mediated repression is itself relatively stable, is thought to be heritable and can probably create a platform for methyl cytosine-mediated silencing during the development of cancer (see ‘Introduction’ section). We have demonstrated here that extinguishing EBNA3C function slowly reverses the H3K27me3-mediated repression of BIM. However even after more than 30 cell divisions without functional EBNA3C, BIM expression did not appear to return to the level seen in uninfected cells.
Taken together all these data indicate that polycomb-mediated repression of BIM plays an important role in the transformation of resting B cells into LCLs, in normal EBV persistence and in increasing the likelihood of B cell lymphomagenesis. EBNA3A and EBNA3C are clearly essential in the process and this is consistent with their abilities to also co-operate in the polycomb-mediated repression of p16INK4A and to act as oncoproteins. They subvert, by a yet to be determined mechanism, the polycomb system of gene regulation to repress transcription of two very important tumour suppressors whose expression is triggered by activated oncogenes such as MYC.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–7, and Supplementary References [84–87].
FUNDING
The Wellcome Trust [programme 077489]. Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the members of the Allday lab for their helpful comments on the manuscript.
REFERENCES
- 1.Bornkamm GW, Hammerschmidt W. Molecular virology of Epstein-Barr virus. Philos. Trans. Roy. Soc. Lond. B Biol. Sci. 2001;356:437–459. doi: 10.1098/rstb.2000.0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kieff E, Rickinson AB. In: Fields Virology. David PMH, Knipe M, editors. Philadelphia, PA: Lippincott-Raven Publishers; 2007. [Google Scholar]
- 3.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford DH. Biology and disease associations of Epstein-Barr virus. Philos. Trans. Roy. Soc. Lond. B Biol. Sci. 2001;356:461–473. doi: 10.1098/rstb.2000.0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babcock GJ, Decker LL, Freeman RB, Thorley-Lawson DA. Epstein-barr virus-infected resting memory B cells, not proliferating lymphoblasts, accumulate in the peripheral blood of immunosuppressed patients. J. Exp. Med. 1999;190:567–576. doi: 10.1084/jem.190.4.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roughan JE, Thorley-Lawson DA. The intersection of Epstein-Barr virus with the germinal center. J. Virol. 2009;83:3968–3976. doi: 10.1128/JVI.02609-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Touitou R, O'Nions J, Heaney J, Allday MJ. Epstein-Barr virus EBNA3 proteins bind to the C8/alpha7 subunit of the 20S proteasome and are degraded by 20S proteasomes in vitro, but are very stable in latently infected B cells. J. Gen. Virol. 2005;86:1269–1277. doi: 10.1099/vir.0.80763-0. [DOI] [PubMed] [Google Scholar]
- 8.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cludts I, Farrell PJ. Multiple functions within the Epstein-Barr virus EBNA-3A protein. J. Virol. 1998;72:1862–1869. doi: 10.1128/jvi.72.3.1862-1869.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Roux A, Kerdiles B, Walls D, Dedieu JF, Perricaudet M. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology. 1994;205:596–602. doi: 10.1006/viro.1994.1687. [DOI] [PubMed] [Google Scholar]
- 11.Radkov SA, Bain M, Farrell PJ, West M, Rowe M, Allday MJ. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J. Virol. 1997;71:8552–8562. doi: 10.1128/jvi.71.11.8552-8562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waltzer L, Perricaudet M, Sergeant A, Manet E. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-J kappa-EBNA2-activated transcription by inhibiting the binding of RBP-J kappa to DNA. J. Virol. 1996;70:5909–5915. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bain M, Watson RJ, Farrell PJ, Allday MJ. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol. 1996;70:2481–2489. doi: 10.1128/jvi.70.4.2481-2489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourillot PY, Waltzer L, Sergeant A, Manet E. Transcriptional repression by the Epstein-Barr virus EBNA3A protein tethered to DNA does not require RBP-Jkappa. J. Gen. Virol. 1998;79(Pt 2):363–370. doi: 10.1099/0022-1317-79-2-363. [DOI] [PubMed] [Google Scholar]
- 15.Hickabottom M, Parker GA, Freemont P, Crook T, Allday MJ. Two nonconsensus sites in the Epstein-Barr virus oncoprotein EBNA3A cooperate to bind the co-repressor carboxyl-terminal-binding protein (CtBP) J. Biol. Chem. 2002;277:47197–47204. doi: 10.1074/jbc.M208116200. [DOI] [PubMed] [Google Scholar]
- 16.Knight JS, Lan K, Subramanian C, Robertson ES. Epstein-Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J. Virol. 2003;77:4261–4272. doi: 10.1128/JVI.77.7.4261-4272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radkov SA, Touitou R, Brehm A, Rowe M, West M, Kouzarides T, Allday MJ. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 1999;73:5688–5697. doi: 10.1128/jvi.73.7.5688-5697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Touitou R, Hickabottom M, Parker G, Crook T, Allday MJ. Physical and functional interactions between the corepressor CtBP and the Epstein-Barr virus nuclear antigen EBNA3C. J. Virol. 2001;75:7749–7755. doi: 10.1128/JVI.75.16.7749-7755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker GA, Crook T, Bain M, Sara EA, Farrell PJ, Allday MJ. Epstein-Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene. 1996;13:2541–2549. [PubMed] [Google Scholar]
- 20.Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 21.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 22.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 24.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 2010;35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Simon JA, Kingston RE. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009;10:1–12. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 26.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasini D, Cloos PAC, Walfridsson J, Olsson L, Bukowski J-P, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:1–6. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 29.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji Coordinates Control of PRC2 Enzymatic Activity and Target Gene Occupancy in Pluripotent Cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan G-C, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jørgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat. Cell Biol. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grau DJ, Chapman BA, Garlick JD, Borowsky M, Francis NJ, Kingston RE. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011;25:2210–2221. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 40.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakamoto Y, Watanabe S, Ichimura T, Kawasuji M, Koseki H, Baba H, Nakao M. Overlapping roles of the methylated DNA-binding protein MBD1 and polycomb group proteins in transcriptional repression of HOXA genes and heterochromatin foci formation. J. Biol. Chem. 2007;282:16391–16400. doi: 10.1074/jbc.M700011200. [DOI] [PubMed] [Google Scholar]
- 42.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 43.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 44.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat. Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 45.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 46.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen KH, Helin K. Epigenetic inheritance through self-recruitment of the polycomb repressive complex 2. Epigenetics. 2009;4:133–138. doi: 10.4161/epi.4.3.8483. [DOI] [PubMed] [Google Scholar]
- 48.Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 49.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 50.White RE, Groves IJ, Turro E, Yee J, Kremmer E, Allday MJ. Extensive co-operation between the Epstein-Barr virus EBNA3 proteins in the manipulation of host gene expression and epigenetic chromatin modification. PLoS One. 2010;5:e13979. doi: 10.1371/journal.pone.0013979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skalska L, White RE, Franz M, Ruhmann M, Allday MJ. Epigenetic repression of p16(INK4A) by latent Epstein-Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog. 2010;6:e1000951. doi: 10.1371/journal.ppat.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paschos K, Smith P, Anderton E, Middeldorp JM, White RE, Allday MJ. Epstein-barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene bim. PLoS Pathog. 2009;5:e1000492. doi: 10.1371/journal.ppat.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderton E, Yee J, Smith P, Crook T, White RE, Allday MJ. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt's lymphoma. Oncogene. 2008;27:421–433. doi: 10.1038/sj.onc.1210668. [DOI] [PubMed] [Google Scholar]
- 54.O'Nions J, Allday MJ. Epstein-Barr virus can inhibit genotoxin-induced G1 arrest downstream of p53 by preventing the inactivation of CDK2. Oncogene. 2003;22:7181–7191. doi: 10.1038/sj.onc.1206838. [DOI] [PubMed] [Google Scholar]
- 55.Gloeckner CJ, Boldt K, Schumacher A, Roepman R, Ueffing M. A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics. 2007;7:4228–4234. doi: 10.1002/pmic.200700038. [DOI] [PubMed] [Google Scholar]
- 56.White RE, Calderwood MA, Whitehouse A. Generation and precise modification of a herpesvirus saimiri bacterial artificial chromosome demonstrates that the terminal repeats are required for both virus production and episomal persistence. J. Gen. Virol. 2003;84:3393–3403. doi: 10.1099/vir.0.19387-0. [DOI] [PubMed] [Google Scholar]
- 57.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl Acad. Sci. USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yee J, White RE, Anderton E, Allday MJ. Latent Epstein-Barr virus can inhibit apoptosis in B cells by blocking the induction of NOXA expression. PLoS One. 2011;6:e28506. doi: 10.1371/journal.pone.0028506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wade-Martins R, Frampton J, James MR. Long-term stability of large insert genomic DNA episomal shuttle vectors in human cells. Nucleic Acids Res. 1999;27:1674–1682. doi: 10.1093/nar/27.7.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O'Nions J, Allday MJ. Proliferation and differentiation in isogenic populations of peripheral B cells activated by Epstein-Barr virus or T cell-derived mitogens. J. Gen. Virol. 2004;85:881–895. doi: 10.1099/vir.0.19704-0. [DOI] [PubMed] [Google Scholar]
- 61.Lejon S, Thong SY, Murthy A, AlQarni S, Murzina NV, Blobel GA, Laue ED, Mackay JP. Insights into association of the NuRD complex with FOG-1 from the crystal structure of an RbAp48.FOG-1 complex. J. Biol. Chem. 2011;286:1196–1203. doi: 10.1074/jbc.M110.195842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Justin N, De Marco V, Aasland R, Gamblin SJ. Reading, writing and editing methylated lysines on histone tails: new insights from recent structural studies. Curr. Opin. Struct. Biol. 2010;20:730–738. doi: 10.1016/j.sbi.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 64.Herz H-M, Shilatifard A. The JARID2-PRC2 duality. Genes Dev. 2010;24:857–861. doi: 10.1101/gad.1921610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 67.Brookes E, Pombo A. Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep. 2009;10:1213–1219. doi: 10.1038/embor.2009.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richter-Larrea JA, Robles EF, Fresquet V, Beltran E, Rullan AJ, Agirre X, Calasanz MJ, Panizo C, Richter JA, Hernandez JM, et al. Reversion of epigenetically mediated BIM silencing overcomes chemoresistance in Burkitt lymphoma. Blood. 2010;116:2531–2542. doi: 10.1182/blood-2010-02-268003. [DOI] [PubMed] [Google Scholar]
- 70.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, Maertens G, Banck M, Zhou M-M, Walsh MJ, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maruo S, Wu Y, Ishikawa S, Kanda T, Iwakiri D, Takada K. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc. Natl Acad. Sci. USA. 2006;103:19500–19505. doi: 10.1073/pnas.0604919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 74.Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roy R, Adamczewski JP, Seroz T, Vermeulen W, Tassan JP, Schaeffer L, Nigg EA, Hoeijmakers JH, Egly JM. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 76.Lu H, Zawel L, Fisher L, Egly JM, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 77.Yankulov KY, Bentley DL. Regulation of CDK7 substrate specificity by MAT1 and TFIIH. EMBO J. 1997;16:1638–1646. doi: 10.1093/emboj/16.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, Hill DE, et al. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl Acad. Sci. USA. 2007;104:7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc. Natl Acad. Sci. USA. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dang CV, O'Donnell KA, Juopperi T. The great MYC escape in tumorigenesis. Cancer Cell. 2005;8:177–178. doi: 10.1016/j.ccr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 81.Strasser A. The role of BH3-only proteins in the immune system. Nat. Rev. Immunol. 2005;5:189–200. doi: 10.1038/nri1568. [DOI] [PubMed] [Google Scholar]
- 82.Allday MJ. How does Epstein-Barr virus (EBV) complement the activation of Myc in the pathogenesis of Burkitt's lymphoma? Semin. Cancer Biol. 2009;19:366–376. doi: 10.1016/j.semcancer.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thorley-Lawson DA, Allday MJ. The curious case of the tumour virus: 50 years of Burkitt's lymphoma. Nat. Rev. Microbiol. 2008;6:913–924. doi: 10.1038/nrmicro2015. [DOI] [PubMed] [Google Scholar]
- 84.Delbarre E, Jacobsen BM, Reiner AH, Sorensen AL, Kuntziger T, Collas P. Chromatin environment of histone variant H3.3 revealed by quantitative imaging and genome-scale chromatin and DNA immunoprecipitation. Mol Biol Cell. 2010;21:1872–1884. doi: 10.1091/mbc.E09-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nowak K, Kerl K, Fehr D, Kramps C, Gessner C, Killmer K, Samans B, Berwanger B, Christiansen H, Lutz W. BMI1 is a target gene of E2F-1 and is strongly expressed in primary neuroblastomas. Nucleic Acids Res. 2006;34:1745–1754. doi: 10.1093/nar/gkl119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang X, Ding L, Sandford AJ. Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Mol. Biol. 2005;6:4. doi: 10.1186/1471-2199-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, Kim SK. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23:975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.