Abstract
Werner’s syndrome (WS) and Bloom’s syndrome (BS) are cancer predisposition disorders caused by loss of function of the RecQ helicases WRN or BLM, respectively. BS and WS are characterized by replication defects, hyperrecombination events and chromosomal aberrations, which are hallmarks of cancer. Inefficient replication of the G-rich telomeric strand contributes to chromosome aberrations in WS cells, demonstrating a link between WRN, telomeres and genomic stability. Herein, we provide evidence that BLM also contributes to chromosome-end maintenance. Telomere defects (TDs) are observed in BLM-deficient cells at an elevated frequency, which is similar to cells lacking a functional WRN helicase. Loss of both helicases exacerbates TDs and chromosome aberrations, indicating that BLM and WRN function independently in telomere maintenance. BLM localization, particularly its recruitment to telomeres, changes in response to replication dysfunction, such as in WRN-deficient cells or after aphidicolin treatment. Exposure to replication challenge causes an increase in decatenated deoxyribonucleic acid (DNA) structures and late-replicating intermediates (LRIs), which are visible as BLM-covered ultra-fine bridges (UFBs) in anaphase. A subset of UFBs originates from telomeric DNA and their frequency correlates with telomere replication defects. We propose that the BLM complex contributes to telomere maintenance through its activity in resolving LRIs.
INTRODUCTION
Linear chromosomes are capped by telomeres, highly specialized structures that protect chromosome ends from degradation and damage (1). Mammalian telomeres are composed of several kilobases of TTAGGG repeats and associated proteins: a core complex of six telomere-specific proteins (Shelterin complex) and a growing number of accessory proteins that assist with proper chromosome end protection, telomere length regulation and telomere processing (2–4).
The WRN RecQ helicase is an enzyme with multiple roles in essential pathways of deoxyribonucleic acid (DNA) repair, homologous recombination, replication (5) and a well-described activity in telomere replication (6). WRN readily alleviates G-quadruplex secondary structures, which are predicted to form in the G-rich telomeric regions (5). These structures likely impede progress of the lagging-strand replication machinery, and if unresolved, they prevent complete synthesis of the daughter strand (6). Cells lacking a functional WRN RecQ helicase experience genome-wide replication defects and undergo more rapid telomere shortening that occurs stochastically with each cell cycle (7). Compared with normal fibroblasts, Werner’s syndrome (WS) cells exhibit an increase in sister-telomere loss (STL) or telomere-free ends (TFE) at some of chromosome ends. Although infrequent, telomere defects (TDs) significantly impair cell viability and activate damage signaling and subsequent processing by non-homologous end joining, potentially forming dicentric chromosomes and causing genome instability (6,8,9).
A second caretaker RecQ helicase, BLM, processes a range of DNA substrates throughout the genome and participates in several critical DNA metabolic pathways that ensure proper genome maintenance (10). BLM assists in replication fork stabilization, branch migration of homologous recombination intermediates, resolution of inappropriate crossover events and DNA end resection (11–14). BLMs activity at telomeres has only been clearly described in tumor cells that use the homologous recombination and copy-switching mechanism, alternative lengthening of telomeres (ALT) and in mouse embryonic fibroblsts (MEFs), which require BLM activity to maintain telomeres and to suppress activation of fragile sites, which include telomeric sequences (15–17). Although these studies contribute to understanding the role of RecQ helicase in telomere maintenance, a telomere-specific function for BLM in primary human fibroblasts remains unclear (16–21).
In addition to its well-described activities in homologous recombination and DNA repair, BLM has most recently been identified in a protein complex that ensures faithful chromosome segregation by dissolution of residual secondary structures observed as ultra-fine bridges (UFBs) in anaphase cells (22). UFBs are dissolved by the BTR complex, which consists of BLM-TOPOIIIα-hRMI1/2 (22–24), and may be visualized by immunofluorescence with any of these protein-specific antibodies. However, how these structures are formed and the definitive mechanisms for their resolution are unclear (25). Examining molecular mechanisms of UFB formation led to the predictions that they arise from two classes of DNA structures: catenane structures that mainly form at centromeres and are generally resolved in early anaphase (22,26) and incompletely replicated intermediate structures associated with Fanconi anemia (FA) proteins, which primarily form at fragile sites in response to replication challenge (27–29). Although genomic fragile sites lack a clear distinguishing feature, they are defined by sensitivity to aphidicolin-dependent partial inhibition of replication, which leads to breaks, deletions and recombination events (26,27). Telomeres have demonstrated fragile site behavior in cells exposed to aphidicolin or in cells lacking the telomeric protein TRF1. These conditions induced telomere aberrations and replication fork stalling, which indicates that telomeres pose an inherent challenge to replication machinery (17).
Herein, we show that BLM contributes to telomere maintenance in normal human fibroblast cells, and we propose that this is accomplished through BLMs capacity for dissolution of late-replicating structures that occur throughout the genome. Cells lacking BLM helicase activity display a greater frequency of TDs, which are defined by absence of fluorescent signal at one (STL) or both (TFE) chromatid arms. We find that the presence of BLM at telomeres is more prominent in a genetic background of telomere dysfunction, such as in WS cells. BLM is recruited to difficult-to-replicate regions, where it facilitates resolution of latent DNA structures in anaphase. Compared with normal IMR90 cells, anaphases from WRN-deficient cells exhibit a higher incidence of persistent UFBs and UFBs that extend from telomeric foci. We propose that BLM contributes to efficient telomere maintenance through its genome-wide activity in resolving difficult-to-replicate regions, which include telomeres.
MATERIALS AND METHODS
Cell culture and production of cell lines
Cell culture and retroviral infections were performed as described (6). For all experiments, we compare cell lines with matched accumulated population doublings, therefore, accounting for variables caused by age-related damage or the gradual telomere shortening that is observed in cultured cell lines over time.
BLM complementary DNA (cDNA; Open Biosystems) was cloned into the pBabe-puro retroviral vector (Addgene). BLM and WRN were stably knocked down by cloning shBLM (TGCCAATGACCAGGCGATC) and shWRN (AATTCTCCGAACGTGTCACGT) sequences in pSuper.retro (Oligoengine). Transient BLM knockdown was also achieved by transfecting IMR90-E6E7 and AG05229-E6E7 cell lines with ON-TARGET plus SMARTpool BLM small interfering RNA (siRNA) (Dharmacon, Thermo Scientific) for 72 h.
Antibodies and western blotting
Anti-BLM (Rabbit 7099) (30), Anti-TRF1 (Rabbit 6839) and Anti-TRF2 (Rabbit 6841) were produced at the Salk Institute. Commercial antibodies: anti-γH2AX (Cell Signaling), anti-53BP1 (A300-273A; Bethyl Laboratories) and anti-WRN (ab200, Abcam). Western analysis was performed as described (6).
Metaphase analysis
Metaphase preparation, fluorescence in situ hybridization (FISH), immunofluorescence-FISH (IF-FISH) and chromosome orientation-FISH (CO-FISH) were performed as described (6,31).
Fibroblast synchronizations and chromatin immunoprecipitation
IMR90 and WI-38 fibroblasts were synchronized at the G1/S boundary by a double-thymidine block, and synchronization efficiency was evaluated by FACS. IMR90 cell lysate preparation and ChIP analysis were performed as described (32).
Aphidicolin treatment
Fibroblast cell lines were treated with 0.3 µM aphidicolin or dimethyl sulfoxide for 24 h. Cells were washed with phosphate-buffered saline and grown in fresh, untreated media for 5 h, then fixed with 4% paraformaldehyde (PFA) before IF-FISH.
Anaphase and metaphase analysis following TRF1 knockdown
TRF1 was targeted by transfecting cells with ON-TARGET plus SMARTpool TERF1 siRNA (Dharmacon, Thermo Scientific) for 72 h.
Image capture and analysis
Slides and coverslips were examined with a Zeiss Axio Imager Z1 fluorescence microscope with apotome. Z-stack projections were captured with an external Hamamatsu ORCA-ER digital camera, and images were analyzed in AxioVision software. For experiments that assessed colocalization events in interphase cells, slides were also imaged on a Zeiss fluorescence microscope with MetaSystems slide scanner. This software analyzed captured images and acquired cumulative descriptive data that were combined to generate population statistics for each cell line.
To examine UFBs, each anaphase was captured using the Zeiss apotome as an image composed of at least seven Z-stacks at a minimal distance of 0.24 µm. Rather than generating a merged image, anaphases were analyzed as Z-stack projections for BLM-positive UFBs and colocalizations between UFBs and telomeric foci. Due to spatial constraints, we did not include colocalization data from anaphases that had more than six UFBs, which primarily occurred in early anaphases (≥1 µm distance). These measures ensured optimal resolution and reduced false-positive colocalizations.
RESULTS
BLM localizes to telomeres in a cell cycle-dependent manner
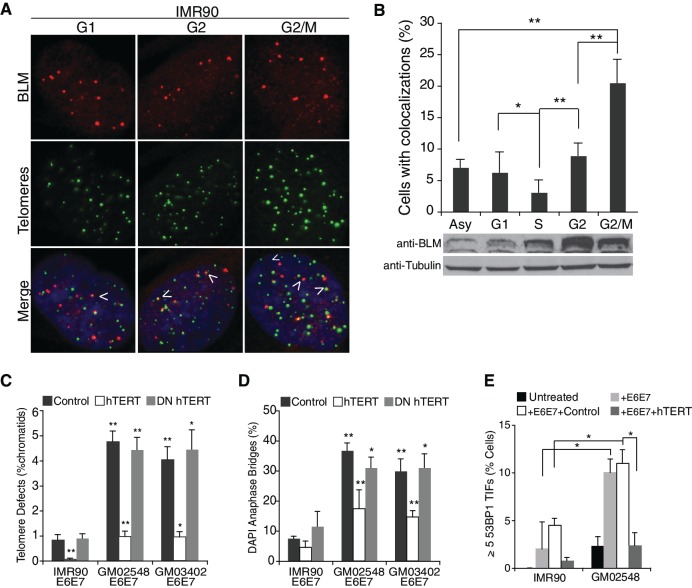
BLM protein expression and nuclear localization patterns have been demonstrated to vary in a cell cycle-dependent manner (33). To accurately determine a possible telomere-specific role for BLM in primary fibroblasts, we synchronized IMR90 cells by a double-thymidine block and analyzed BLM binding to telomeres during the cell cycle by IF-FISH or ChIP. IF-FISH revealed that although BLM was visible at a small subset of telomeres, cells in late G2 displayed a significant increase in multiple colocalization events (Figure 1A and B). WI38 fibroblasts displayed a similar colocalization pattern (Supplementary Figure S1A). ChIP analysis of BLM localization confirmed that BLM occupation at telomeric chromatin was enriched in G2 and G2/M fractions (Supplementary Figure S1B). As previously described (33), and as shown by western blot of synchronized IMR90 cells (Figure 1B), BLM proteins levels increase from late S to late G2/M stage, though this does not correlate with the increase in BLM-telomere interactions observed in G2 and late G2/M, suggesting that the G2-associated increase of BLM at telomeres was not just simply a result of increased expression levels.
Figure 1.
BLM localizes to a subset of telomeres preferentially in G2/M. (A) IF-FISH images of synchronized IMR90 cells, using a BLM specific antibody (red) and a telomeric probe, FITC-[CCCTAA]4 (green). Arrows indicate colocalization events between BLM and telomeric foci. (B) Top panel: quantification of IF-FISH data, showing the percentage of IMR90 cells with over two colocalization events. At least 100 cells were counted per time-point and values were averaged from four independent experiments. Error bars represent the standard deviation and P-values were calculated with a Student’s t-test (*P < 0.05 and **P < 0.005). Bottom panel: western blot of synchronized IMR90 cells probed with BLM antibody or a tubulin control. IMR90 lysates were collected at the indicated stages of the cell cycle. (C) Percentage of chromatids with TDs in IMR90 or BS fibroblast cell lines expressing an empty vector control, hTERT or DN_hTERT. At least 1500 chromatids per cell line, per experiment were counted from three independent experiments. (D) DAPI-positive anaphase bridges in IMR90 and BS fibroblast cells expressing an empty vector control, hTERT or DN_hTERT. At least 55 anaphases per cell line were counted from three independent experiments. (E) Percentage of IMR90 and GM02548 BS cells with over five TIF. Cells were fixed with 4% PFA and prepared for IF-FISH with a 53BP1-specific antibody and the telomeric probe FITC-[CCCTAA]4. At least 100 cells from each cell line were counted in two independent experiments. Error bars represent the standard deviation and P-values were calculated with a Student’s t-test (*P < 0.05 and **P < 0.005).
These data demonstrate that BLM localizes to telomeres in primary cells late in the cell cycle, potentially indicating a role in postreplicative processing of these regions.
Cells lacking BLM exhibit TDs and telomere-dependent chromosome fusions
Since BLM helicase activity is critical for genome integrity, BLM-deficient cells are characterized by chromosomal aberrations, which may arise from internal breaks and fusions (34,35). A straightforward method to assess genome instability is to count 4′,6-diamidino-2-phenylindole (DAPI)-positive anaphase bridges, which are generated by diverse DNA damage and repair pathways. As expected Bloom’s syndrome (BS) cells exhibited a significantly greater frequency of anaphase bridges than normal IMR90 fibroblasts, regardless of E6 or E7 oncoprotein expression (Table 1).
Table 1.
Frequency of TDs and anaphases with DAPI-positive anaphase bridges in IMR90 and Bloom’s syndrome fibroblast cell lines after previously described knockdown or overexpression experiments
| TDs (percent chromatids) | Anaphase bridges (percent anaphases) | |
|---|---|---|
| IMR90 | 1.0 | 1.1 |
| IMR90-E6E7 | 1.2 | 4.6 |
| IMR90-E6E7 NS | 1.5 | 7.3 |
| IMR90-E6E7 shBLM | 3.1* | 25* |
| GM02548 | 2.5** | 23** |
| GM02548-E6E7 | 3.4** | 29* |
| GM02548-E6E7 NS | 3.3 | ND |
| GM02548-E6E7 shWRN | 5.8* | 43.5 |
| GM02548-E6E7 Control | 3.9 | 36 |
| GM02548-E6E7 BLM | 2.9 | 21* |
| GM03402B | 3.1** | 21** |
| GM03402B-E6E7 | 3.7** | 33** |
| GM03402B-E6E7 NS | 5.3 | ND |
| GM03402B-E6E7 shWRN | 7.1* | ND |
| AG05229-E6E7 NS | 4.1 | 36 |
| AG05229-E6E7 shBLM | 7.1* | 45 |
Values shown are the percentage of chromatids with TDs, which were counted by telomeric FISH of metaphase chromosomes. At least 1500 chromatids were counted for each cell line, and average values were obtained from three FISH-independent experiments. Where provided, the percentages of DAPI-positive anaphase bridges were determined by counting at least 75 anaphases from each cell line, from three independent experiments. For all cell lines, the P-values were calculated using a Student’s t-test. *P < 0.05; **P < 0.005. ND, not determined.
To identify whether telomere dysfunction contributes to the instability in BS cells, we first examined individual telomere phenotypes by performing FISH on metaphase chromosomes. The frequency of covarying TDs, such as STL or TFE, was significantly elevated in BS cell lines (2.5% and 3.1% chromatids; P < 0.005) compared with normal IMR90 fibroblasts (1%) (Table 1). Expression of E6 and E7 oncoproteins suppressed p53 and pRb-mediated cell cycle checkpoints, which allowed cells to continue growing even in the presence of critically shortened telomeres or DNA damage. On E6 and E7 expression, we observed an elevation of TDs in BS cell lines (3.4% and 3.7% TDs) but not in IMR90-E6E7 (1.1% TDs) (Table 1).
Subsequent experiments established that TDs, such as telomere-free chromosome ends and DAPI-positive anaphase bridges were a direct consequence of BLM helicase deficiency and not a secondary characteristic of BS cells. Retroviral delivery of a BLM shRNA efficiently suppressed protein expression in IMR90-E6E7 fibroblasts (Supplementary Figure S2A), which exhibited TDs (3.1%; P < 0.05) and anaphase bridges (25%; P < 0.05) at levels observed in BS fibroblasts (Table 1). Next, we reconstituted BLM by expressing full-length cDNA in GM02548-E6E7 BS fibroblasts, which reduced TDs (from 3.9% to 2.9%) and anaphase bridges (from 36% to 21%; P < 0.05). The limited efficiency in correcting these phenotypes is presumably because exogenous BLM protein expression reaches approximately half of normal levels (Supplementary Figure S2B). This incomplete overexpression is typical of BLM-reconstituted BS cell lines and has been previously demonstrated to only partially restore the chromosomal defects that characterize BS cells (36).
To confirm that exposed metaphase chromosome ends are bone fide TDs, we treated fibroblast cell lines with the catalytic subunit of telomerase human telomerase reverse transcriptase (hTERT), an empty vector control or an inactive dominant-negative hTERT (DN_hTERT). hTERT expression will specifically elongate only telomeres but will not repair non-telomeric lesions (37). TDs (Figure 1C) and anaphase bridge formation (Figure 1D) were significantly reduced in BS fibroblasts expressing wild-type hTERT, which indicates that BS cells have a greater frequency of critically shortened telomeres than age-matched normal fibroblast cells. Furthermore, these dysfunctional telomeres are potential substrates for DNA repair pathways that produce fusion events and compromise genome integrity (Figure 1D).
Critically, short or dysfunctional telomeres are recognized by the DNA damage response pathway and form telomere damage-induced foci (TIF) (38), which are cytologically visible as colocalization events between 53BP1 foci and telomeric foci (Supplementary Figure S3A) (38). IF-FISH analysis revealed that 10% of GM02548-E6E7 cells had at least five TIF, which was reduced to <2% on hTERT expression (Figure 1E). These findings confirm that a subset of DNA damage events in BS cells occur at telomeres, even in cells with intact checkpoints. In summary, BLM deficiency leads to a low, but consistent, level of telomere loss that is sufficient to expose chromosome ends and then activates a DNA damage response that ultimately generates chromosomal aberrations.
TDs occur at remarkably similar frequencies in BS cells and WS cells (Table 1) (6). However, CO-FISH in BLM-deficient HeLa cells (Supplementary Figure S3B) exhibited no difference between leading-strand STL and lagging-strand STL (Supplementary Figure S3C). These findings contrast to the lagging-strand STL observed in cells lacking a functional WRN helicase (Supplementary Figure S3B and S3C) and indicate that BLM and WRN have distinct functions in telomere maintenance.
Loss of both BLM and WRN helicases exacerbates TDs
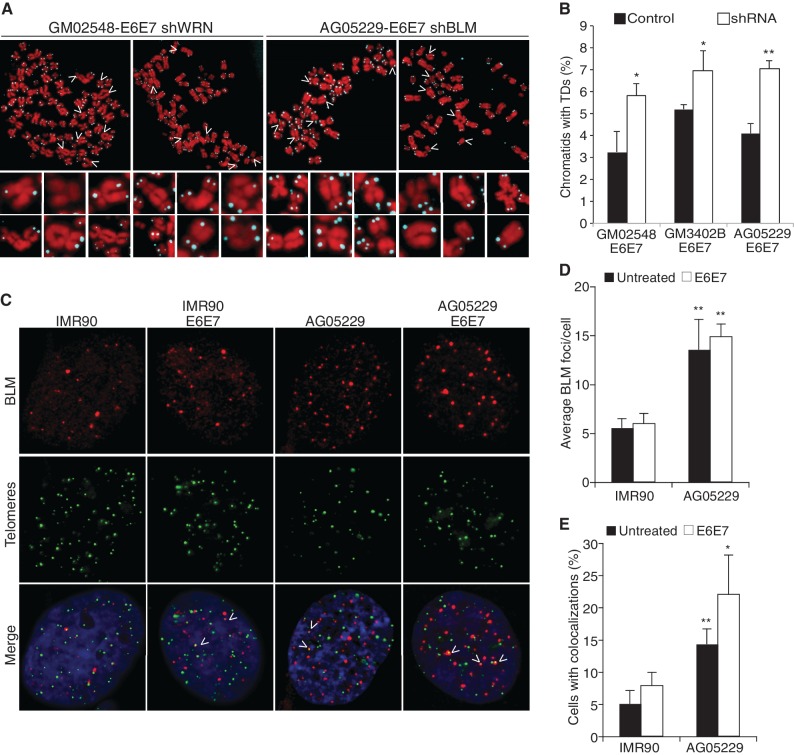
Our data reveal similarities and subtle differences between the telomere phenotypes and cell cycle-regulated telomere-binding patterns of WRN and BLM (Figure 1A and B and S1) (6,7,9). WRN localizes to chromosome ends in late S and facilitates lagging-strand replication, whereas BLM performs a downstream activity in G2 that is necessary to maintain both leading and lagging telomeres. To investigate the possible redundancy in WRN and BLM activity at telomeres, WS and BS fibroblast cells were stably transduced with shRNA targeting BLM and WRN, respectively (Supplementary Figure S2C). Metaphase spreads from WRN- and BLM-deficient cell lines exhibited an increase in critically shortened telomeres and chromosomal aberrations (Figure 2A). Quantification confirmed that the absence of both helicases considerably exacerbates telomere dysfunction (Figure 2B). These data indicate non-redundant functions for BLM and WRN in chromosome end maintenance (6,7), which is further supported by the report that both WRN and BLM mutations enhance the pathology in later-generation mice lacking the telomerase RNA (39). However, they do not exclude a reciprocal or inter-dependent relationship.
Figure 2.
Loss of both BLM and WRN RecQ helicases exacerbates TDs. (A) Images of FISH of metaphase chromosomes from GM02548-E6E7 (BS) fibroblasts after WRN knockdown and AG05229-E6E7 (WS) fibroblasts after BLM knockdown. Arrows indicate chromosomes in the magnified panels below each metaphase spread. (B) Percentage of chromatids with TDs. At least 2000 chromatids from each cell line were counted, and average values were compiled from two independent experiments. (C) IMR90 and WS cells were fixed with 4% PFA and used for IF-FISH with a BLM antibody and the telomeric probe FITC-[CCCTAA]4. IF-FISH images of indicated fibroblast cell lines, showing BLM (red) and telomeres (green); arrows show colocalization events. (D) Average number of BLM foci per cell and (E) percentage of cells with more than two colocalization events between BLM and telomeres. The values for each cell line were averaged from three independent IF-FISH experiments. Arrow bars represent standard deviation, and P-values were calculated by a Student’s t-test (*P < 0.05 and **P < 0.005).
WS cells exhibit an increase in BLM foci and UFB formation
Although it is unlikely that BLM and WRN perform overlapping functions in telomere replication and processing, it is feasible that they cooperatively enhance each other’s recruitment or activity. The presence of WRN could assist or promote the downstream BLM localization to telomeres. Alternatively, BLMs recruitment to telomeres could be a response to the replication defects caused by WRN deficiency.
To test the theory that BLM recruitment to telomeres is greater in the absence of WRN, we assessed BLM localization in IMR90 and WRN-deficient fibroblasts by IF-FISH with BLM antibody and a telomeric probe (Figure 2C). WS cells displayed a remarkable elevation in the average number of BLM foci per cell (Figure 2C and D) and an increase in cells exhibiting multiple colocalization events between BLM and telomeric foci (Figure 2E), irrespective of BLM expression levels. Not only does BLM localize to telomeres independently of WRN but also it is more frequently detected at chromosome ends in WRN-deficient cells.
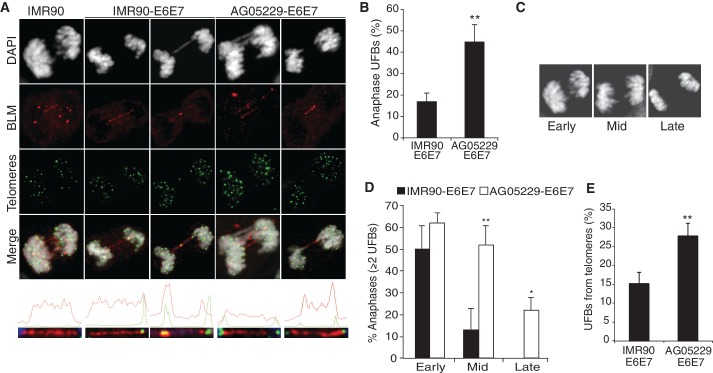
Although examining BLM localization patterns, we noted the presence of anaphase cells with BLM-positive UFBs (Figure 3A). UFBs composed of fully replicated but unwound catenane structures and are commonly shown to extend from centromeric regions and tend to be resolved by mid-anaphase (22,40,41). The second class of UFBs is generally caused by persistent, incompletely replicated structures that arise at particular DNA sequences as a result of some form of replication challenge. This subset of UFBs is distinguished by association with FA proteins and a proclivity to originate from fragile sites or other regions experiencing replication delays (26,28).
Figure 3.
Ultra-fine bridges (UFBs) are more frequent and persist longer in WS fibroblasts. (A) IF-FISH images of UFB-positive anaphases representing UFBs that do not localize with telomeric foci (left panel), UFBs that colocalize with a telomeric foci at one end or at the center (second and third panels) and UFBs that colocalize with telomeres at both ends (final two panels). Graphs illustrating the signal intensities generated by the profile function of the AxioVision software are shown below each anaphase. (B) The percentage of anaphases with at least one UFB and (C) DAPI-stained anaphases representing three stages of anaphase, determined by distance (in µm) between dividing cells: early, mid and late anaphases. (D) Percentage of UFB-positive anaphases with multiple UFBs per anaphase. Data from IMR90-E6E7 and AG05229-E6E7 cells were grouped according to progress through anaphase (panel C). At least 25 anaphases were analyzed for each cell line per experiment, from four independent experiments. Arrow bars show standard deviation, and P-values were calculated by a Student’s t-test (*P < 0.05 and **P < 0.005). (E) The percentage of UFBs extending from telomeres (T-UFBs). The values were calculated from four independent IF-FISH experiments, where at least 30 anaphases were analyzed from each cell line.
UFB frequency was assessed in IMR90 and WS fibroblasts to examine whether their formation correlated with defective telomere replication in WS cells, thus suggesting the presence of late-replicating intermediate (LRI) structures. As previously reported, UFBs were common events in normal IMR90 cells (22,26), but WRN-deficient cells exhibited a significantly greater number of anaphases with at least one UFB (Figure 3B). The events that lead to UFB formation may be in part characterized by how rapidly they are resolved in anaphase. Therefore, UFB-positive anaphases were analyzed according to duration in anaphase by combining data from four independent IF-FISH experiments and grouping UFB-positive anaphases into three categories (early, mid and late stages) according to distance (micrometer) between separating DAPI clusters (Figure 3C). Although IMR90-E6E7 and AG05229-E6E7 cells had similar frequencies of multiple UFBs in early anaphase, most UFBs in IMR90-E6E7 cells are resolved by mid-anaphase. In contrast, a significantly higher portion of AG05229-E6E7 anaphases contained multiple UFBs into mid- and late-stage anaphases (Figure 3C and D).
Interestingly, a number of IMR90 and WS anaphases contained UFBs extending from telomeric foci at one or both ends, in a pattern similar to late-replicating UFBs stretched between fragile site foci (26,28), which signifies inefficient telomere replication even in unchallenged conditions (Figure 3A and E). Nonetheless, telomere-UFBs (T-UFBs) were significantly more common in the telomere replication-defective WS cells (Figure 3E), supporting our hypothesis that UFBs extend from telomeres in response to replication dysfunction. We performed FISH to visualize telomeric sequence along the length of UFBs, but these attempts were largely unsuccessful. This is not surprising given the fragility of UFBs, which lack histones, and the stringency of fixation during the FISH protocol (22).
To confirm that WRN activity is particularly important to prevent UFB formation at defective telomeres, we assessed BLM-positive UFBs in WS cells stably expressing the full-length WRN or an empty vector control. Restoring WRN considerably reduced UFB-positive anaphases and T-UFBs (Supplementary Figure S4A and S4B), suggesting that impaired telomere replication may generate incompletely replicated DNA structures that eventually require processing by BLM and interacting proteins. This specialized complex (BTR) mediates dissolution of late-replicating DNA structures and facilitates faithful chromosome segregation without activating checkpoints or disturbing cell cycle progression (42).
An unrelated protein, PICH (polo-like kinase-1 interacting checkpoint helicase) is also found at these cytological bridge-like structures and colocalizes with BLM at UFBs (40,41). This offers an alternative detection method for UFB, and we examined UFB frequency in BLM-deficient fibroblast cells by staining against PICH. BLM knockdown also increased UFB and T-UFB frequency, providing confirmation that BLM suppresses replication dysfunction and LRIs (43).
Replicative challenge induces changes in BLM localization and increases UFB formation
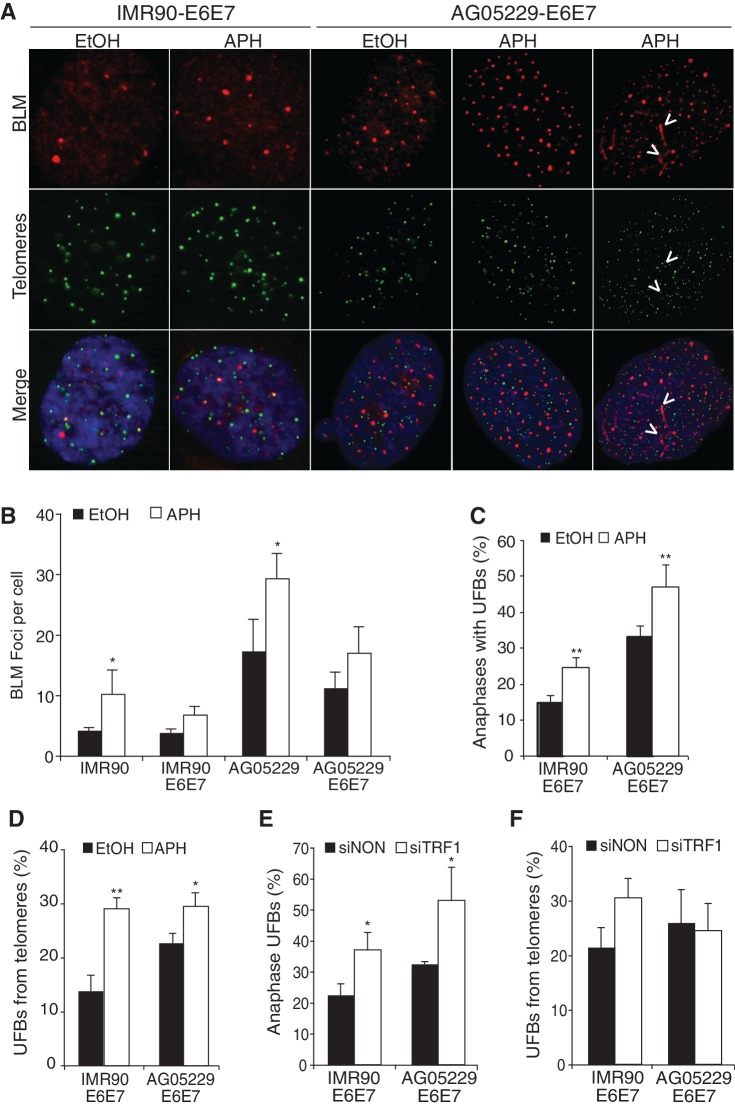
Exposure to the DNA polymerase inhibitor aphidicolin induces genomic fragile site expression, which become origins of chromosome breaks and LRIs (26–28). Telomeres have a demonstrated sensitivity to aphidicolin and are predicted to behave as fragile sites, particularly in the absence of TRF1 (17,44). Telomeres in WRN-deficient cells also suffer from frequent replication defects that appear to cause a greater number of UFBs (Figure 3B and D). Therefore, we asked whether the WS telomere phenotypes or T-UFB events were affected by aphidicolin exposure and whether BLM localization with telomeres responded to replication dysfunction. Aphidicolin treatment significantly increased the number of BLM foci per cell (Figure 4A and B) in IMR90 and WS fibroblasts (not expressing E6 and E7), though this effect was more pronounced in AG05229 WS cells, due to a higher ‘baseline’ level of BLM foci (Figure 2D). UFB-positive anaphases and T-UFBs were also significantly increased in IMR90-E6E7 and AG05229-E6E7 WS cells after aphidicolin treatment (Figure 4C and D). These data are supported by previous reports that exposure to aphidicolin enhances BLM expression and nuclear localization, in addition to inducing frequent replication defects and UFB formation (26,28,33).
Figure 4.
Replication challenge increases UFB formation. (A) IF-FISH images from IMR90-E6E7 and AG05229-E6E7 fibroblast cells. Arrows indicate colocalization events at BLM bridges observed in interphase cells. (B) Average number of BLM foci per cell. At least 100 cells were counted per condition from three independent experiments. (C) Percentage UFB-positive anaphases and (D) percentage of UFBs extending from telomeres (T-UFBs) in IMR90-E6E7 and AG05229-E6E7 cells treated with ethanol or aphidicolin. At least 30 anaphases were counted per condition from three independent experiments. (E) Percentage of UFBs and (F) UFBs that extend from telomeric foci (T-UFBs) on TRF1 suppression. At least 35 anaphases were counted per cell line from two independent experiments. Arrow bars show standard deviation, and P-values were calculated from a Student’s t-test (*P < 0.05 and **P < 0.005).
Induction of telomere replication defects by TRF1 knockdown increases UFB formation
Aphidicolin treatment induces a genome-wide replication challenge, but we wanted to generate a telomere-specific replication dysfunction and assess recruitment of the BLM processing complex to chromosome ends. It has been shown that TRF1 facilitates telomere replication and suppresses unresolved DNA structures (17,44). By downregulating TRF1 expression in IMR90-E6E7 and particularly in AG05229-E6E7 cells, we intended to further obstruct telomere replication and then determine whether greater replicative challenge at telomeres correlated with the frequency of T-UFBs. siRNA-mediated reduction of TRF1 (Supplementary Figure S5A and S5B) caused a remarkable increase of UFB-positive anaphases in IMR90-E6E7 and AG05229-E6E7 cell lines (Figure 4E) and marginally increased T-UFBs in IMR90-E6E7 cells but actually reduced T-UFBs in WS cells (Figure 4F).
We predicted that the decrease of T-UFBs was a consequence of spontaneous telomere loss induced by TRF1 and WRN deficiencies, which would render telomeric signals below detection limits. Examination of metaphase chromosomes from these cell lines confirmed that TRF1 knockdown in AG05229-E6E7 cells increased the frequency of telomere-free chromatids from 7.2% to 13% (Supplementary Table S1). These T-UFB data are difficult to interpret, as a subset of BLM-positive UFBs may extend from critically shortened telomeres below the FISH detection limit. Nonetheless, TRF1 knockdown in IMR90-E6E7 showed compelling evidence that inhibiting telomere replication correlates with BLM colocalization events and the incidence of T-UFB formation.
DISCUSSION
The data presented in this study support a role for the BLM complex in telomere maintenance as a response to the difficult-to-replicate nature of chromosome ends. It has been well established that the telomeric sequence challenges the replication machinery, partially due to the G-rich nature of the telomeric leading strand (6), which is predicted to form stable energy-rich G structures, such as G quadruplexes (45). RecQ helicases are predicted to alleviate ‘roadblocks’ that persistently pose a challenge to replication fork progression through the telomeric strands.
Interestingly, a recently published report on telomere replication efficiency led to the proposal that telomeres in fact behave as fragile sites and are particularly susceptible to replicative stress (17,44). TRF1 deletion, BLM deletion and partial inhibition of DNA polymerases by aphidicolin each caused an elevated frequency of fragile telomeres in MEFs, visible by FISH of metaphase spreads. Further evidence demonstrated an epistatic relationship between TRF1 and BLM in regulating the metaphase fragile telomere phenotype, suggesting that TRF1 is required to recruit BLM and suppress replication delays (17). Although these data add credence to the argument that telomeres behave as fragile sites, they are not transferable to the present analysis of T-UFB formation in untransformed primary fibroblasts. Our data show that TRF1-deficient IMR90 fibroblasts exhibit a markedly greater number of BLM-positive UFBs and elevated frequency of T-UFBs, which may potentially be the anaphase version of fragile telomere phenotypes (Figure 4E and F). The observations that suppressing TRF1 affects UFB and T-UFB frequency may prompt further exploration that will provide valuable information about the fragile behavior of mammalian telomeres.
Inhibition of BLM leads to telomere dysfunction and cosuppression of WRN exacerbates the phenotype. Therefore, we predict distinct roles for BLM and WRN at telomeres, which are corroborated by previous research demonstrating that Terc-deficient mice lacking both BLM and WRN helicases experienced a greater disease pathology than loss of only one helicase. These RecQ-double mutants with Terc-deficiency exhibited greater genome-wide chromosomal abnormalities, which were worsened by telomere-dependent fusions (39).
On the basis of our analysis of UFB formation, we propose that in G2 BLM and its interacting proteins, TOPO IIIα-RMI1-RMI2 (BTR complex) (23,24,46) are recruited to any genomic regions with persistent replication intermediates or catenane structures, including telomeres that have experienced a delay in replication fork progression, which is a common defect of TTAGGG repeats. The presence of UFBs in anaphases of normal, healthy fibroblasts with intact genomes is evidenced that cells are equipped to deal with late-replicating sequence or catenane structures without activating cycle checkpoints and sacrificing efficiency.
Severe delays in replication completion lead to the generation of ultra-fine telomeric DNA bridges that persist into mitosis; these residual indicators of replication delay are more frequent in cells with inherent replication problems, such as impaired telomere replication in WS-E6E7 cells (∼28%). The relatively high number of UFBs extending from telomeric foci in IMR90-E6E7 cells (∼15%) indicates that telomeres regularly, if not frequently, encounter replication defects (Figure 3E).
In summary, these findings predict that persistent intermediate structures in telomeric regions are resolved by BTR-mediated dissolution, which ensures proper chromosome segregation. To determine whether the mechanisms employed at chromosome ends are required for dissolution of late replication intermediates at fragile sites or for simple decatenation, it will be necessary to confirm whether FA protein colocalizes with telomeric foci located at BLM UFB ends.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–5.
FUNDING
The Cellular and Molecular Genetics Training Grant of University of California, San Diego (to C.B.); Aileen Andrews Grant Foundation (to C.B.); Cancer Center Core Grant [P30 CA014195-38]; National Institutes of Health [AG025837 and GM087476 to J.K.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ian D. Hickson, for generous samples of BLM antibody, and Laure Crabbe and Clodagh O’Shea for reagents and advice.
REFERENCES
- 1.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 3.O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 5.Opresko PL, Cheng WH, von Kobbe C, Harrigan JA, Bohr VA. Werner syndrome and the function of the Werner protein; what they can teach us about the molecular aging process. Carcinogenesis. 2003;24:791–802. doi: 10.1093/carcin/bgg034. [DOI] [PubMed] [Google Scholar]
- 6.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 7.Arnoult N, Saintome C, Ourliac-Garnier I, Riou J, Londono-Vallejo A. Human POT1 is required for efficient telomere C-rich strand replication in the absence of WRN. Genes Dev. 2009;23:2915–2924. doi: 10.1101/gad.544009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat. Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 9.Crabbe L, Jauch A, Naeger CM, Holtgreve-Grez H, Karlseder J. Telomere dysfunction as a cause of genomic instability in Werner syndrome. Proc. Natl Acad. Sci. USA. 2007;104:2205–2210. doi: 10.1073/pnas.0609410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu W, Hickson I. RecQ helicases: multifunctional genome caretakers. Nat. Rev. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 11.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom's syndrome gene product promotes branch migration of holliday junctions. Proc. Natl Acad. Sci. USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restrart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- 14.Kikuchi K, Abdel-Aziz HI, Taniguchi Y, Yamazoe M, Takeda S, Hirota K. Bloom DNA helicase facilitates homologous recombination between diverged homologous sequences. J. Biol. Chem. 2009;284:26360–26367. doi: 10.1074/jbc.M109.029348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 16.Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum. Mol. Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 17.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yankiwski V, Marciniak RA, Guarente L, Neff NF. Nuclear structure in normal and Bloom syndrome cells. Proc. Natl Acad. Sci. USA. 2000;97:5214–5219. doi: 10.1073/pnas.090525897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, Bohr VA. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J. Biol. Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- 20.Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- 21.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan K, North P, Hickson I. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26 doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cejka P, Plank J, Bachrati C, Hickson I, Kowalczykowski S. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Bachrati C, Ou J, Hickson I, Brown G. Human Topoisomerase IIIa is a single stranded decatenase that is stimulated by BLM and RMI1. J. Biol. Chem. 2010;285:21426–21436. doi: 10.1074/jbc.M110.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan KL, Hickson ID. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin. Cell Dev. Biol. 2011;22:906–912. doi: 10.1016/j.semcdb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Chan K, Palmai-Pallag T, Ying S, ID H. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 27.Glover T, Berger C, Coyle J, Echo B. DNA polymerase a inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 1984;67:136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- 28.Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Biol. 2009;11:761–769. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- 29.Pichierri P, Franchitto A, Rosselli F. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J. 2004;23:3154–3163. doi: 10.1038/sj.emboj.7600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orazio NI, Naeger CM, Karlseder J, Weitzman MD. The adenovirus E1b55K/E4orf6 complex induces degradation of the Bloom helicase during infection. J. Virol. 2011;85:1887–1892. doi: 10.1128/JVI.02134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett HA, Neumann AA, Reddel RR. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat. Struct. Mol. Biol. 2009;16:1244–1251. doi: 10.1038/nsmb.1725. [DOI] [PubMed] [Google Scholar]
- 32.O'Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutertre S, Ababou M, Onclercq R, Delic J, Chatton B, Jaulin C, Amor-Gueret M, German J. Cell cycle regulation of the endogenous wild type Bloom's syndrome DNA helicase. Oncogene. 2000;19:2731–2738. doi: 10.1038/sj.onc.1203595. [DOI] [PubMed] [Google Scholar]
- 34.Chaganti R, Schonberg S, German J. A manifold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc. Natl Acad. Sci. USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- 36.Rassool F, North P, Mufti G, Hickson I. Constitutive DNA damage is linked to DNA replication abnormalities in Bloom's synrome cells. Oncogene. 2003;22:8749–8757. doi: 10.1038/sj.onc.1206970. [DOI] [PubMed] [Google Scholar]
- 37.Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Beijersbergen RL, Knoll JH, Meyerson M, Weinberg RA. Inhibition of telomerase limits the growth of human cancer cells. Nat. Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 38.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 39.Du X, Shen J, Kugan N, Furth EE, Lombard DB, Cheung C, Pak S, Luo G, Pignolo RJ, DePinho RA, et al. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol. Cell Biol. 2004;24:8437–8446. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baumann C, Korner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 41.Wang LH, Schwarzbraun T, Speicher MR, Nigg EA. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 2008;117:123–135. doi: 10.1007/s00412-007-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres-Rosell J, De Piccoli G, Cordon-Preciado V, Farmer S, Jarmuz A, Machin F, Pasero P, Lisby M, Haber JE, Aragon L. Anaphase onset before complete DNA replication with intact checkpoint responses. Science. 2007;315:1411–1415. doi: 10.1126/science.1134025. [DOI] [PubMed] [Google Scholar]
- 43.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 46.Wu L, Davies S, North P, Goulaouic H, Riou J, Turley H, Gatter K, Hickson I. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 2000;275:9636. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.