Abstract
Stress response requires the precise modulation of gene expression in response to changes in growth conditions. This report demonstrates that selective nuclear mRNA degradation is required for both the cell wall stress response and the regulation of the cell wall integrity checkpoint. More specifically, the deletion of the yeast nuclear dsRNA-specific ribonuclease III (Rnt1p) increased the expression of the mRNAs associated with both the morphogenesis checkpoint and the cell wall integrity pathway, leading to an attenuation of the stress response. The over-expression of selected Rnt1p substrates, including the stress associated morphogenesis protein kinase Hsl1p, in wild-type cells mimicked the effect of RNT1 deletion on cell wall integrity, and their mRNAs were directly cleaved by the recombinant enzyme in vitro. The data supports a model for gene regulation in which nuclear mRNA degradation optimizes the cell response to stress and links it to the cell cycle.
INTRODUCTION
The cell cycle is a four-step process that leads to the replication of the genetic content and cell division (1). The production of viable progeny requires tight control of each stage, starting with the first gap (G1) in which the cell reaches maturity and acquires the nutrition required for the division (2). Once a critical size is reached, DNA synthesis begins in the start (S) phase. This is followed by a second gap (G2) that ensures the readiness for the final stage of mitosis (M) and division (3). The transitions through the different phases of the cell cycle are driven by factors generated from cyclically transcribed genes (4). The periodicity of the transcriptional activation is achieved largely through post-translationally regulated transcription factors (5), while the impact of transcriptional inhibition is ensured by the generally short half life of the cell cycle regulated mRNAs and proteins (6). In addition, there are a few cases in which the post-transcriptional regulation of RNA stability is associated with the cell cycle. For example, it has been reported that the degradation of the mRNA coding for the G2 cyclin Clb2p is required for the exit from mitosis (7), while the stabilization of the Igo1 and Igo2 mRNAs is required for quiescence upon starvation (8). However, the contribution of RNA stability to the cell cycle remains limited to few examples, and the mechanism by which it contributes to the cell cycle checkpoints remains unclear.
Checkpoints represent quality control mechanisms that ensure that the appropriate conditions are met in order for the cell to divide (9). Under normal conditions, the checkpoints ensure that each phase of the cell cycle is complete before the next one starts. However, when cells are exposed to unfavourable growth conditions, activated checkpoints delay the cell cycle in order to allow the cells to recover from the insult (10). For example, while the inhibitor of B-type cyclins Sic1p normally acts as a part of the checkpoint for the completion of the G1 phase by inactivating the cyclin-dependent kinase Cdc28p (11), it also delays the cell cycle when the cells are exposed to osmotic stress. Hypertonic shock activates the high osmotic glycerol (HOG) pathway that delays cells in the G1 phase by stabilizing Sic1p (12). The HOG pathway also arrests cells in the G2 phase by phosphorylating the morphogenesis checkpoint kinase Hsl1p (13). Similarly, the activation of the cell wall integrity (CWI) pathway delays the G2/M transition through the activity of the mitogen-activated protein kinase (MAPK) Slt2p (14). Indeed, both pathways use similar factors and strategies to regulate the cell cycle, and several links connecting the different stress response networks have been described (15). For example, Hsl1p phosphorylation by Hog1p upon activation of the HOG pathway has recently been shown to contribute to the regulation of Slt2p (16). In contrast, transcription factors, such as Rlm1p, may be activated by both the Slt2p- and Hog1p-dependent pathways (17). Besides protein phosphorylation and transcriptional activation, the cell wall stress results in a global modulation of mRNA stability (18). However, the contribution of the RNA-dependent regulation of gene expression to the stress-dependent cell cycle checkpoints remains largely unexplored.
This study examines the contribution of RNA stability to stress-dependent checkpoints by monitoring the impact of yeast nuclear dsRNA-specific ribonucleases (Rnt1p) on mRNAs associated with the cell cycle. The deletion of RNT1, a kin of the human ribonuclease Dicer (19), delays the G1 phase of the cell cycle (20) and increases the resistance to hypertonic stress (21), suggesting possible links between RNA degradation and the stress-dependent cell cycle checkpoints. A survey of potential Rnt1p substrates associated with the cell cycle identified cleavage signals within the mRNAs coding for the stress-dependent checkpoint-associated proteins Swi4p and Hsl1p. Cells lacking Rnt1p were found to be sensitive to stresses related to the CWI pathway, and many of the genes in this pathway were upregulated by the deletion of RNT1 in vivo and were cleaved by recombinant Rnt1p in vitro. In contrast, the genes implicated in the HOG pathway involved in the hypertonic response were not cleaved by Rnt1p in vitro, and rnt1Δ cells were not found to be sensitive to high osmotic conditions (21). Together, these results add a new layer to the mechanism of the stress-dependent regulation of the cell cycle whereby the selective RNA degradation conditionally alters gene expression.
MATERIALS AND METHODS
Strains and plasmids
All yeast strains used in this study are listed in Supplementary Table S1 and were grown and manipulated using standard procedures (22). The RNT1 and rnt1Δ strains were generated by the replacement of one of the RNT1 alleles by HIS3 in the LLY36 diploid strain, followed by spore dissection (23). The primers used for the PCR product integration are described in Supplementary Table S2. Green fluorescent protein (GFP)-tagged strains were purchased from Invitrogen (Burlington, ON, Canada) and were described in (24). Plasmid pEGH-HSL1 was purchased from Open Biosystems (Huntsville, AL, USA) and was described in (25). The double and single deletion mutants were obtained by crossing single mutants from the Yeast knock out collection obtained from Open Biosystems (26) with the rnt1Δ strain, both of which are derivatives of S288C. Clones isogenic to the rnt1Δ strain were selected after spore dissection.
RNA analysis
RNA extractions and Northern hybridizations were performed as previously described (27). Cleavage of total RNA was performed by mixing 50 µg of total RNA and 4 pmol of purified Rnt1p and incubating for 20 min at 30°C (28). Standard 1% denaturing agarose gels were used to separate 20 µg of the RNAs before their transfer to hybond N+ membrane (GE Healthcare, Baie d’Urfe, QC, Canada). Hybridization signals from probes generated by the random labelling (29) of gene-specific PCR fragments were quantified using a PhosphorImager (GE Healthcare). Mapping of the cleavage sites was performed by primer extension as described previously (30). Rnt1 mRNA levels were assayed by quantitative RT-PCR as described in (31) using 1 ng cDNA in 10 µl reactions with the FastStart Universal SYBR Green Master mix (Roche Diagnostics, Laval, QC, Canada). All oligonucleotides used in this study are listed in Supplementary Table S2.
Microarray analysis
RNA extracted from either W303-1A (32) or Δrnt1 (33) cells grown to either an OD600 of 0.6 in synthetic complete media at 26°C, or following a 4 h shift to 37°C, was hybridized to yeast S98 Affymetrix Oligoarrays (Santa Clara, CA, USA). The microarray experiment was realized and analyzed by the Génome Québec Innovation Center (Montréal, QC, Canada) using the Affymetrix standard software as described in (27).
Search for Rnt1p substrates
Microarray data from (27) and (21) and this study were used to identify mRNAs over-expressed in absence of RNT1. Rnt1p cleavage signals were predicted in these mRNAs as described in (34). Genes with mRNAs over-expressed >20.5 in the three data sets with cleavage signal scores >0.85 were tested for in vitro cleavage of total RNA.
Microscopy
Swi4p- and Hsl1p-GFP fusion proteins were detected by immunofluorescence of yeast cells prepared as described in (20) using a rabbit anti-GFP (Invitrogen Canada, Burlington, ON, Canada) at a dilution of 1:3000 and a Texas-Red-X conjugated goat anti-rabbit antibody (Invitrogen Canada) at a dilution of 1:1000. Nuclei were stained with the DNA dye DAPI. Pictures were acquired on a Zeiss Axio Observer microscope (Carl Zeiss Canada, Toronto, ON, Canada) with a 100×/1.46 oil objective and analyzed using the Columbus software (Perkin Elmer, Woodbridge, ON, Canada). For live cell imaging, cells were spotted on slabs of agarose containing growth media, GFP was excited with a 470 nm LED using the Colibri system (Carl Zeiss Canada) and pictures taken every 10 min. Movie files were assembled from chosen fields and annotated with the relative time in minutes.
DNA content analysis
Cells grown to log phase and fixed overnight at 4°C in 75% ethanol were prepared for flow cytometry analysis by propidium iodide staining as described (20). In general, 10 000 cells were scored in a FACScan (Becton Dickinson, Mississauga, ON, Canada).
Growth assays
Cells grown to log phase in rich media at 26°C were used to inoculate 100 µl cultures in 96-well plates as described previously (35). The cultures were incubated at the appropriate temperature with shaking in a PowerWave microplate scanning spectrophotometer (Bio-Tek, Winooski, VT, USA), and the absorbance at 600 nm was read every 10 min for 80 h. Doubling times for each growth condition were calculated as explained in (35), and were compared to growth in rich media at 26°C. For the genetic interaction analysis, the same procedure was followed for all single and double mutants. The growth of the different strains was evaluated relative to the RNT1 strain grown under the same condition. The expected additive growth defects were estimated for each double mutant by multiplying the relative growth rates of the single mutants (36). A synthetic sickness phenotype was scored when the actual relative growth rate of a double mutant was lower than the expected value by more than 1 standard deviation. All growth assays were performed in duplicate for two independent clones.
RESULTS
Identification of the RNA degradation events required for cell cycle progression
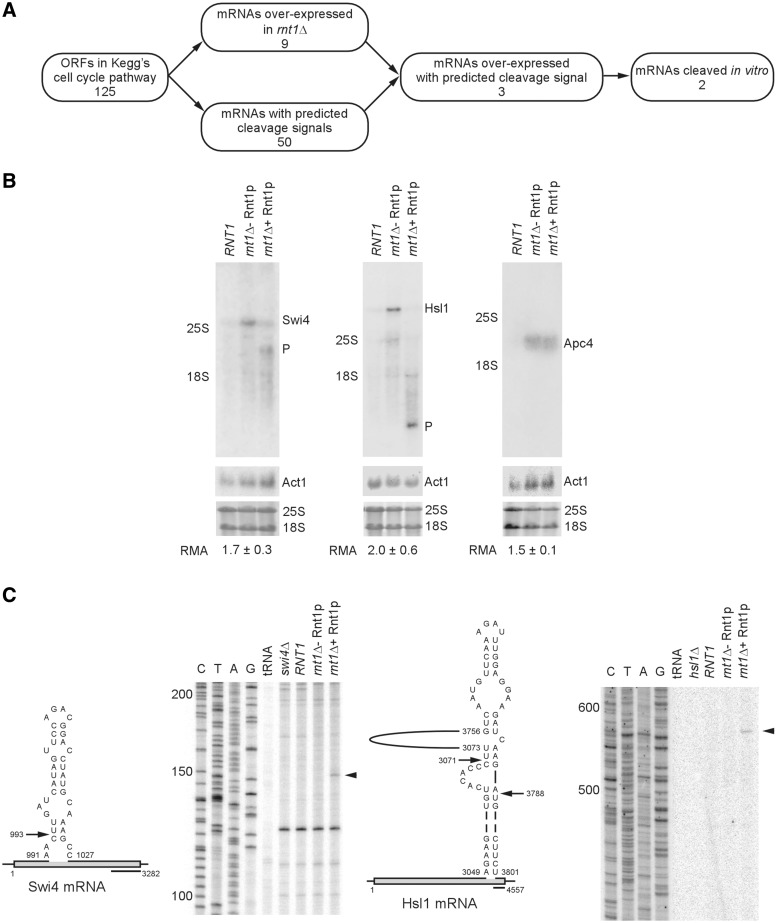
The deletion of RNT1 causes a delay in the G1 phase of the yeast cell cycle, and results in budding defects (20). The delay in the cell cycle and the budding anomalies are largely restored to normal by a RNT1 allele carrying a mutation in the catalytic domain which blocks RNA cleavage without affecting RNA binding, suggesting that RNA cleavage is not essential for cell cycle progression (20). However, it is unclear how the dsRNA-specific ribonuclease may affect the cell cycle in a cleavage independent manner. One possibility is that Rnt1p may reduce gene expression by stably binding its target RNA and preventing its export to the cytoplasm for translation. In this case, the catalytically impaired enzyme may restore the cell cycle phenotype by binding to RNA that would normally be cleaved by the wild-type version. In order to examine this possibility, the impact of Rnt1p on the expression of the mRNAs associated with the yeast cell cycle in the KEGG (Kyoto Encyclopaedia of Genes and Genomes) pathway was monitored in vivo, and their cleavage potentials in vitro were tested using a recombinant enzyme. Out of the 125 cell cycle associated genes in the KEGG pathway, 50 harboured predicted Rnt1p cleavage sites, nine were consistently over-expressed in rnt1Δ cells and three were both over-expressed in vivo and contained a potential cleavage site for Rnt1p (Figure 1A and Supplementary Table S3). Examination of the known functions of these three potential Rnt1p substrates, Apc4, a component of the anaphase promoting complex (37), Hsl1, a kinase regulating the morphogenesis checkpoint (38) and Swi4, the DNA binding component of the SBF transcription factor (39), suggests a common function in regulating stress response and cell cycle progression (16,40,41). Therefore, we propose that Rnt1p represses the expression of a specific and functionally related group of genes that are implicated in stress-dependent cell cycle regulation.
Figure 1.
Identification of post-transcriptionally regulated cell cycle genes. (A) All mRNAs associated with cell cycle genes in the Kegg’s cell cycle pathway were analyzed for the presence of Rnt1p cleavage signals. Genes that are upregulated upon the deletion of RNT1, as indicated by expression microarrays, were validated for cleavage by recombinant Rnt1p in vitro. Genes coding for RNA that are directly cleaved by Rnt1p were deemed candidates for cell cycle-dependent post-transcriptional regulation. (B) Northern blot analysis of total RNA extracted from wild-type cells (RNT1) or cells lacking the RNT1 gene (rnt1Δ) and incubated with or without recombinant Rnt1p in vitro. The different bands were visualized using probes specific to the sequence of Swi4, Hsl1 or Apc4 mRNAs. A probe against the actin mRNA (Act1) was used as negative control of the cleavage reaction. Note the incubation of total RNA with Rnt1p did not change the levels of Act1 mRNA (middle panel). The stained image of the ribosomal RNAs on the membrane prior to hybridization is also shown as loading control. The positions of the mature RNA and the cleavage product (P) are indicated beside the gels. The relative mRNA amount (RMA) in rnt1Δ with respect to RNT1 averaged from at least two experiments ± SD are shown below the gel. The P-values of the increase in the levels of Swi4, Hsl1 and Apc4 are 0.009, 0.007 and 0.02, respectively. (C) Mapping Rnt1p cleavages within the coding sequence of Swi4 and Hsl1 mRNA. RNA extracted from either wild-type or rnt1Δ cells was incubated with purified Rnt1p enzyme. The cleaved RNA was subjected to primer extension using primers downstream of the predicted Rnt1p cleavage sites (illustrated on the left of each panel). Sequence reactions using the same primers were included as markers to identify the cleavage position. The positions of the cleavage sites are identified by arrowheads on the right. The position of the different size markers are indicated on the left of the gel. The contrast of the sequencing and primer extensions parts of the gels has been adjusted independently to permit the reading of the sequence ladders.
Rnt1p directly cleaves the mRNAs of the genes associated with cell cycle regulation
The deletion of RNT1 affects the expression of many genes and, as such, the changes in the expression levels of APC4, SWI4 and HSL1 may simply be an indirect event independent of Rnt1p’s catalytic activities. In order to rule out this possibility, the ability of recombinant Rnt1p to directly and selectively cleave the natural Apc4, Swi4 and Hsl1 mRNAs in vitro was tested. Total RNA was extracted from either wild-type (RNT1), or rnt1Δ cells, and was incubated in either the presence or absence of recombinant Rnt1p. As expected, the deletion of RNT1 increased the mRNA levels of all three genes with P-values ranging from 0.01 to 0.02, confirming the predicted changes in gene expression (Figure 1B). However, only the Swi4 and Hsl1 mRNAs were cleaved when incubated with recombinant Rnt1p in vitro (Figure 1B). The rRNA and Act1 mRNA, which were used as negative controls (Figure 1B, bottom panels), as well as the Apc4 mRNA were not cleaved (Figure 1B, right panels), indicating that Rnt1p selectively cleaves the Swi4 and Hsl1 mRNAs. The cleavage site of Swi4 was mapped by primer extension to the proximity of a canonical Rnt1p cleavage site (Figure 1C, left panel), while that of the Hsl1 mRNA was mapped close to a stem-loop structure formed through long-range interactions within the 3′ end of the gene (Figure 1C, right panel). Consistently, impairing the catalytic activity without affecting either the RNA or protein binding activity of the enzyme (20) was sufficient to increase the expression levels of both Hsl1 and Swi4, confirming that Rnt1p catalytic activity is required for the inhibition of the expression of these genes (Supplementary Figure S1). Similarly, inactivating a thermosensitive mutant of Rnt1p lead to the accumulation of Hsl1 and Swi4 mRNAs (Supplementary Figure S2A) and cleavage products of Rnt1p can be stabilized in exoribonuclease deficient strains (Supplementary Figure S2B and S2C). Clearly, Rnt1p directly triggers the RNA degradation of at least two cell cycle genes implicated in the stress response.
RNT1 deletion alters the levels and cellular localization of Hsl1p
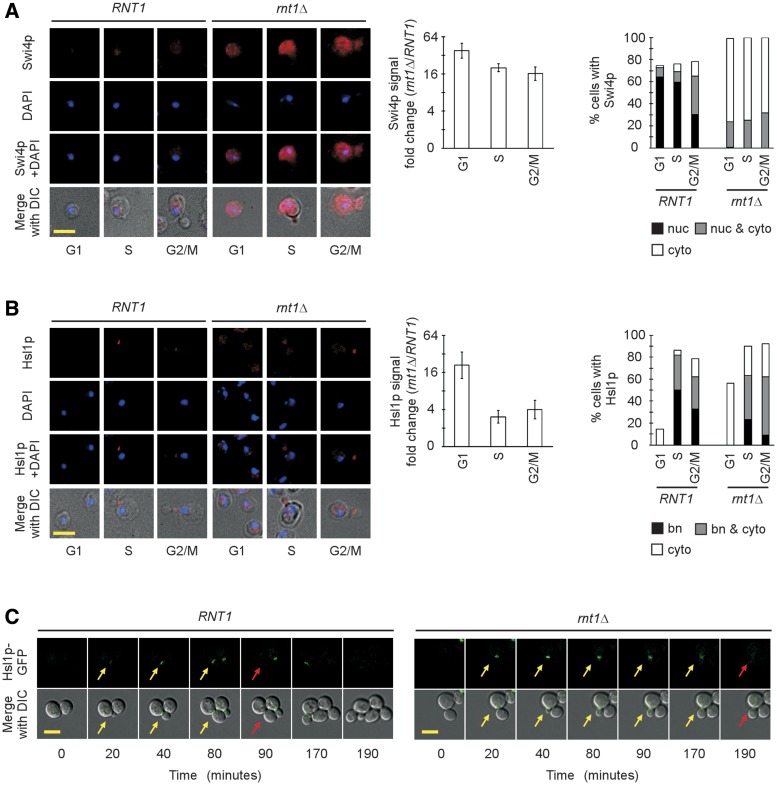
Swi4p is expressed throughout the cell cycle (42), while the expression of Hsl1p is tightly linked to the cell cycle with a maximal protein amount being detected in S phase (38,43). In order to evaluate the impact of RNA degradation on these two different modes of genes expression, the protein levels of both SWI4 and HSL1 were monitored during the cell cycle both before and after the deletion of RNT1. The chromosomal copies of SWI4 and HSL1 were tagged with GFP, and the intracellular signal was quantified using epifluorescence microscopy. The proteins were visualized by immunostaining of cells in each phase of the cell cycle (as judged by the bud size) using antibodies against the GFP tag. As expected, in wild-type cells, GFP-tagged Swi4p expression was detected in all phases of the cell cycle (Figure 2A, left panel). In the G1 and S phases of the cell cycle, Swi4p was mostly nuclear while some cytoplasmic staining was observed in the G2/M phases (Figure 2A). The deletion of RNT1 caused an overall increase in the expression of Swi4p by an average of 22-fold regardless of the cell cycle phase (Figure 2A, center panel). Together the results indicate that Rnt1p-dependent mRNA degradation plays an important role in repressing the expression of Swi4p in all phases of the cell cycle.
Figure 2.
Detecting the impact of RNT1 deletion on the cell cycle-dependent expression of Swi4 and Hsl1 proteins. (A) Expression of GFP-tagged Swi4p as detected by immunofluorescence using tag-specific antibodies. Representative pictures for each phase of the cell cycle as determined by the bud size are shown on the left panel. The tagged protein signals are shown in red, the DAPI staining of the nucleus is shown in blue. The composite images showing the staining signals merged with images of the cell morphology obtained using differential interference contrast (DIC) are included at bottom. Quantitative analysis of the cell cycle-dependent expression of Swi4p is shown in form of bar graph (middle panel). The fold change in the signal after RNT1 deletion was calculated using >200 cells from three independent experiments. The differences in Rnt1p expression had P-values ranging from 0.02 to 0.05. The right panel indicates Swi4p signals detected in the nucleus (nuc), nucleus and cytoplasm (nuc & cyto) or the cytoplasm (cyto) in each phase of the cell cycle. (B) Expression of GFP-tagged Hsl1p as detected by immunofluorescence using tag-specific antibodies. The image acquisition and quantification were conducted as described in A. The distribution of the Hsl1p signals in the cells at the bud neck (bn), the bud neck and cytoplasm (bn & cyto) and the cytoplasm (cyto) is shown on the right. (C) Real-time imaging of Hsl1p accumulation in living cells. The expression of GFP tagged Hsl1p was monitored in normally growing cells from the first appearance of the proteins (yellow arrow) until the signal is no longer detected (red arrow) at the bud neck of dividing cells. Shown is the strongest signal detected for Hsl1p at the bud neck. The scale bars correspond to 5 µm.
In the case of HSL1, the maximum level of the protein in wild-type cells was detected in the S phase as previously reported (38,43), and decreased in G2/M, reaching its minimum level in G1 (Figure 2B, left panel). Almost all of the Hsl1p detected in wild-type cells was localized in the bud neck, except during the G1 phase of the cell cycle where very low levels of the protein were scattered throughout the cytoplasm in about 15% of the cells (Figure 2B, right panel). The deletion of RNT1 increased the expression of Hsl1p in all phases of cell cycle by an average of 4.4-fold. However, the most important increase (21-fold) was observed in G1 when Hsl1p is normally suppressed, indicating that Rnt1p plays an important role in the cell cycle-dependent repression of Hsl1p (Figure 2B, center panel). Like Swi4p, the deletion of RNT1 also affected the localization pattern of Hsl1p rendering a significant proportion of the protein cytoplasmic (Figure 2B, right panel). This indicates that the Rnt1p cleavage of the Hsl1 mRNA plays an important role in controlling the production of Hsl1p during the cell cycle by allowing the protein to properly localize at the bud neck where it conducts its main function in the morphogenesis checkpoint (38). Based on these results, it was hypothesized that Rnt1p increases both the efficiency and the rapidity of the cell cycle-dependent repression of this protein by ensuring that all transcripts that are not needed are degraded in the nucleus. In order to evaluate this possibility, the kinetics of Hsl1p accumulation were followed during the cell cycle in live cells by tracking the Hsl1p-GFP signal in both RNT1 and rnt1Δ cells using epifluorescence. In Figure 2C, Movie 1 (RNT1) and Movie 2 (rnt1Δ), the deletion of RNT1 increases the duration in which Hsl1p-GFP is detected in the bud neck by 90 min. This suggests that the failure to degrade the Hsl1 mRNA in the nucleus delays the shutdown of Hsl1p expression. The conclusion drawn is that Rnt1p inhibits the cytoplasmic localization and promotes the cell cycle repression of Hsl1p by degrading unwanted transcripts in the nucleus, and consequently reduces the amount of protein produced.
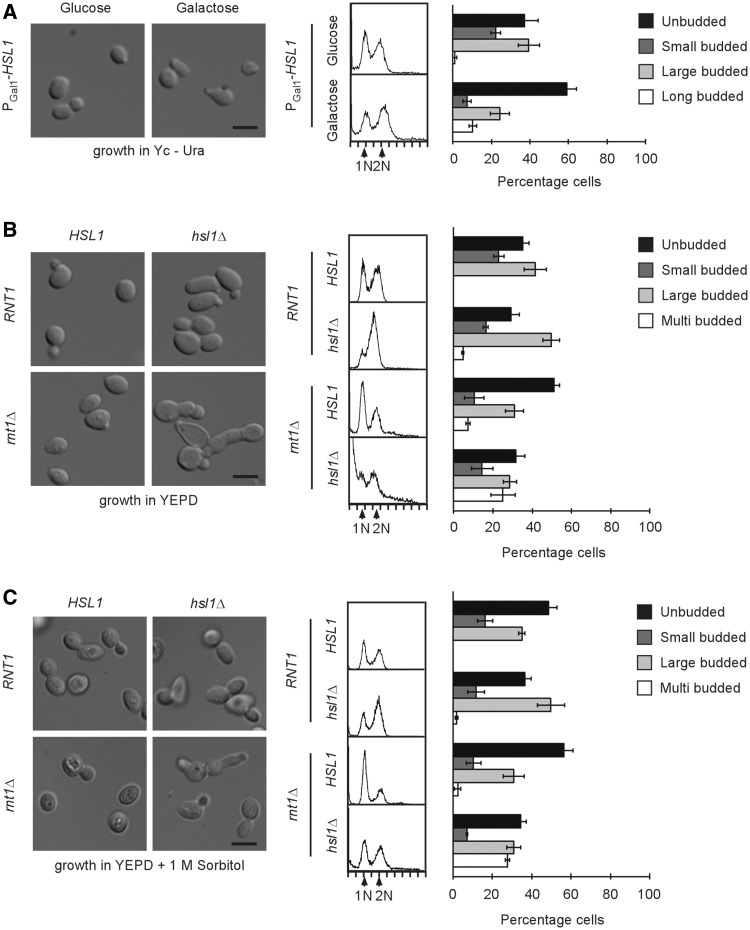
Over-expression of HSL1 partially reproduces the phenotype of the RNT1 deletion
Since both Hsl1p and Swi4p are upregulated in absence of RNT1 (Figure 2A and B), it was hypothesized that the Rnt1p-dependent delay in the G1 phase of the cell cycle might be induced, at least in part, by their over-expression. Accordingly, the impact of HSL1 over-expression on the cell cycle was examined by over-expressing this gene from a plasmid (PGal1-HSL1) carrying the GAL1 promoter (25) in wild-type cells. Cells were grown in the presence of glucose then shifted to galactose containing media for 6 h and the effect on cell cycle was monitored by both microscopy and cytometry (Figure 3A and Supplementary Figure S3A). The expression from the inducible promoter lead to bud neck and cytoplasmic distribution of Hsl1p as observed in the absence of Rnt1p (Supplementary Figure S3A and Figure 2B) and resulted in a lower level of accumulation of Hsl1p than did the deletion of RNT1 (Supplementary Figure S3B). As expected, the cell shape, budding index and the cell cycle-dependent distribution of DNA was not altered in cells transformed with either the PGal1-HSL1 plasmid or an empty vector when the cells were grown in glucose containing media (Figure 3A and Supplementary Figure S3C). In contrast, PGal1-HSL1 cells grown in galactose containing media exhibited a marked increase in the number of unbudded cells, suggesting an increased number of cells in G1. In addition, HSL1 over-expression also induced the formation of cells with elongated buds (Figure 3A). This phenotype is very similar to that previously reported upon the deletion of RNT1 (20) where most of the cells were found to be either unbudded, or to display an aberrant budding pattern. However, unlike rnt1Δ cells, flow cytometry analysis did not reveal an increase in cells with 1N DNA content after the induction of HSL1, as would be expected from the increase in the number of unbudded cells (Figure 3A). Instead, a small increase in the number of cells with 2N DNA was detected as previously reported (44). Therefore, the over-expression of HSL1 appears to reproduce the budding but not the DNA duplication delay associated with the deletion of RNT1 (Figure 3B). The accumulation of unbudded cells that have completed DNA synthesis has been previously observed with other mutants affecting the CWI pathway (45). Accordingly, it appears that a part of rnt1Δ phenotype stems from an increase in the level of Hsl1p that in turn impairs the CWI pathway and inhibits bud formation. The over-expression of SWI4 did not delay cells in G1, as judged by both DNA content and budding index (Supplementary Figure S3D), suggesting that the increase in Swi4p is not the cause of the rnt1Δ cell cycle delay. Interestingly, the over-expression of SWI4 slightly increased the number of cells with long buds (Supplementary Figure S3D) as is the case in rnt1Δ cells (20), suggesting that the Rnt1p-dependent repression of SWI4 is required for normal bud formation and not cell cycle progression. Indeed, it has previously been reported that Swi4p is implicated in the cell wall synthesis pathway that affects bud formation (46).
Figure 3.
HSL1 and RNT1 interact genetically. (A) Over-expression of HSL1 causes a delay in bud formation and the accumulation of long budded cells. RNT1 cells expressing HSL1 under the control of an inducible galactose promoter were grown under repressive (glucose) or inducible conditions (2% galactose for 6 h). Both induced and non-induced cells were collected and analyzed using light microscopy or flow cytometry. The left panel shows image of the cells obtained using light microscopy with the scale bar representing 5 µm. The center panel shows DNA content analysis by flow cytometry with 1N and 2N peaks indicated. The bar graph on the right shows the distribution of cells according to the bud size and morphology. The data presented is an average of three independent experiments with error bars corresponding to SD. (B) Deletion of HSL1 reduces the RNT1-dependent delay in G1 and reduces the CWI. Cells carrying single or double deletions in RNT1 and HSL1 were grown to log phase in YEPD and analyzed as described in A. (C) Sorbitol addition in the growth medium rescues the cell lysis phenotype of rnt1Δ hsl1Δ. Cells carrying single or double deletions in RNT1 and HSL1 were grown to log phase in YEPD + 1 M sorbitol and analyzed as described in A.
The deletion of HSL1 partially suppresses the phenotypic defects of rnt1Δ cells
In order to more directly determine the contribution of Hsl1p to the Rnt1p-dependent phenotype, we tested the impact of deleting HSL1 on budding and the cell cycle in both wild-type and rnt1Δ cells. In Figure 3B, the deletion of HSL1 by itself increased the number of cells with large buds and 2N DNA content, while the deletion of RNT1 resulted in the accumulation of unbudded cells carrying 1N DNA. This confirms a previous report suggesting that while Rnt1p is required for the progression through G1 (20), the expression of Hsl1p is needed for the M/G1 transition (47). Strikingly, the combination of both deletions (rnt1Δ hsl1Δ) suppressed the Rnt1p-dependent accumulation of cells in G1 and restored the cell cycle phase ratio to that observed with wild-type cells (Figure 3B). A few cells exhibited multiple buds and greater than 2N DNA content, suggesting a defect in budding. However, the most pronounced phenotype of the rnt1Δ hsl1Δ cells was the high rate of cell bursting and mortality, as 44% of the cells observed by light microscopy were ghost cells, and that a substantial fraction had less than 1N DNA content (Figure 3B). This phenotype could be rescued by the addition of sorbitol to the growth media (Figure 3C), indicating that Hsl1p and Rnt1p not only contribute to cell cycle progression and bud formation but also influence CWI. The deletion of SWI4 did not affect the phenotype of rnt1Δ cells, once again confirming that the increase in Swi4p is not the cause of the Rnt1p cell cycle defect (Supplementary Figure S3E). This indicates that Rnt1p contributes to both the Hsl1p-dependent cell integrity and bud formation.
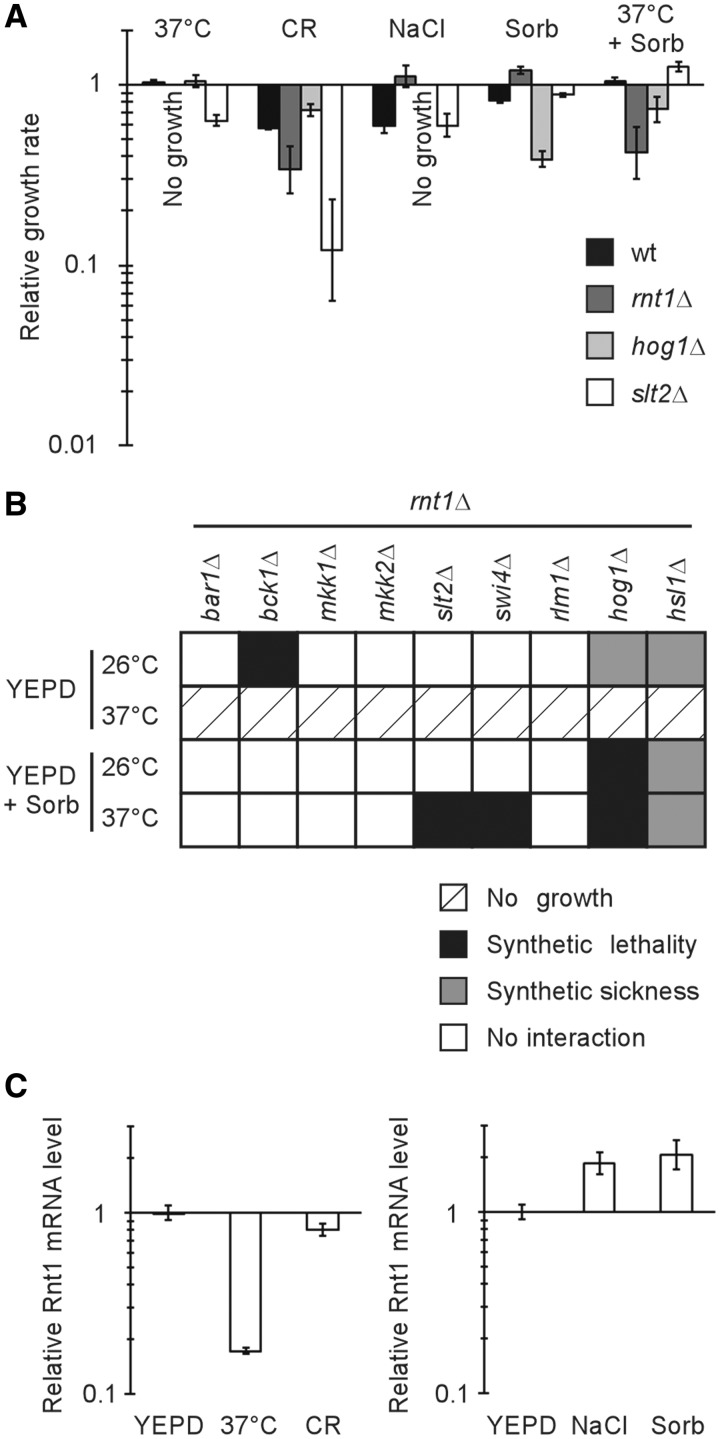
Rnt1p is a key regulator of the cell wall stress pathway
Since the double deletions of RNT1 and HSL1 affected CWI and Hsl1p was previously implicated in the regulation of the Slt2p kinase from the CWI pathway (16), it was hypothesized that Rnt1p may play a role in regulating the CWI pathway. Accordingly, the phenotypic impact of deleting RNT1 was compared to that observed upon the deletion of established cell wall stress response genes like the MAPKs HOG1 and SLT2. Hog1p and Slt2p are part of two separate, but interrelated, protein networks that respond differently to high and low osmotic stresses by remodelling the cell wall and influencing its assembly (48). As expected, the inclusion of Congo red, a classical agent that interferes with cell wall assembly (49), dramatically reduced the growth of the slt2Δ strain when compared with that of wild-type (RNT1) or hog1Δ strains (Figure 4A). Interestingly, the rnt1Δ strain was also more sensitive to Congo red than both RNT1 and hog1Δ strain, suggesting that Rnt1p influences cell wall assembly, as does Slt2p. Indeed, the rnt1Δ strain displayed growth tendencies similar to that of the slt2Δ strain and not like that of the hog1Δ strain, in all growth conditions tested. For example, the inclusion of either NaCl or sorbitol in the media dramatically inhibited the growth of the hog1Δ strain, while not negatively affecting the growth of both the slt2Δ and rnt1Δ strains (Figure 4A). Strikingly, growth in the presence of sorbitol rescued the temperature sensitivity phenotype previously reported for the rnt1Δ strain (20). This means that Rnt1p is required for the cell wall response to high temperature and explains why RNT1 deletion inhibits growth at 37°C. The conclusion drawn is that Rnt1p is required for the optimal response to cell wall stress.
Figure 4.
Rnt1p contributes to the cell wall stress response pathway. (A) Deletion of RNT1 impairs cell wall stress response. Wild-type cells or cells lacking RNT1 (rnt1Δ), the hyperosmotic stress response gene HOG1 (hog1Δ), or the hypo-osmotic stress response gene SLT2 (slt2Δ) were grown in the presence of either 30 µg/ml of cell wall synthesis inhibitor Congo red (CR), 0.5 M sodium chloride (NaCl), 1 M sorbitol (Sorb), or at high temperature (37°C) with or without sorbitol. Growth rates were calculated for each strain relative to wild-type cells grown in YEPD at 26°C. No growth could be detected for rnt1Δ cells grown at 37°C and hog1Δ cells grown in NaCl-containing media. (B) RNT1 genetically interacts with cell wall stress response factors. rnt1Δ cells carrying deletion in eight genes implicated in hyper- and hypo-osmotic stress response or an unrelated gene (BAR1) were generated and assayed for growth in liquid media containing or not 1 M sorbitol at 26 or 37°C. The relative growth rates were calculated and synthetic interactions were scored when the effect was greater than the cumulative effect expected from the product of separate single mutant effects. (C) CWI stress activation causes changes in Rnt1 mRNA accumulation. Rnt1 mRNA level was measured by quantitative RT-PCR from total RNA extracted from RNT1 cells after treatment at 37°C for 20 min, 30 µg/ml Congo Red for 20 min, 0.5 M sodium chloride for 5 min or 1 M sorbitol for 5 min and compared to cells suspended in YEPD at 26°C for the corresponding time. The data is an average of three experiments represented as ratios of expression normalized to the level detected in cells grown in YEPD and to the housekeeping gene ACT1. The difference between normal and stress associated mRNA expression had P-values of 0.001 [in the case of temperature (37°C) shift], 0.05 [in the case of Congo red (CR) induced stress], 0.01 (in the case of NaCl addition) and 0.05 (in the case of sorbitol addition).
In order to place Rnt1p within the protein network regulating the response to cell wall stress, its genetic interaction with the different genes associated with both the CWI and HOG pathways was examined. Different strains carrying either cell wall stress gene deletions or an unrelated gene deletion (BAR1) were crossed to the rnt1Δ strain and the impact on growth monitored under either different temperatures or different osmotic concentrations. As expected, crossing rnt1Δ to the control bar1Δ strain did not modify the growth phenotype, and all of the strains were unable to grow at 37°C (the restrictive temperature for rnt1Δ cells; Figure 4B). In contrast, either strong synthetic sickness or lethality was observed for the rnt1Δ hog1Δ and rnt1Δ hsl1Δ strains in all conditions tested. This indicates that Rnt1p functions in parallel to Hog1p in the cell wall stress response, and suggests that Hsl1p is not the only link between Rnt1p and the cell wall stress response network. The slt2Δ rnt1Δ strain did not show enhanced growth defects at 26°C, despite the fact that rnt1Δ and slt2Δ single mutant phenotypes could be rescued by sorbitol (Figure 4B and Supplementary Figure S4). Consistently, RNT1 did not genetically interact with any of the genes strictly placed in the Slt2p osmotic response-dependent pathways (i.e. like the genes encoding the redundant kinases Mkk1p and Mkk2p). This suggests that Rnt1p is not strictly redundant for the SLT2-dependent low osmotic stress response, at least under certain conditions. On the other hand, deleting either SLT2 or its transcriptional activation partner SWI4 prevented the sorbitol-dependent rescue of rnt1Δ cells at 37°C, suggesting that these two genes may have parallel or redundant functions with RNT1 for growth at high temperature. Indeed, Swi4p activity was shown to be stimulated in a Slt2p-dependent manner in response to heat (40). In contrast, RNT1 interacted genetically with BCK1, a gene encoding the kinase located upstream of Slt2p and implicated in the co-activation of both the HOG and CWI pathways (50), only at 26°C and not at 37°C. Since RNT1 synthetically interacts with BCK1 and not with other genes in the CWI pathway at 26°C, we suggest that Rnt1p and Bck1p are redundant due to the implication of Bck1p in the HOG pathway. Together, the genetic interactions suggest that the Rnt1p contribution to CWI is not restricted to its regulation of Hsl1 mRNA, but extends to a larger network of genetic interactions connecting the Slt2p and Hog1p branches of the pathway (Supplementary Table S4). Indeed, the Slt2p pathway related genes, RHO1, BEM2, FKS1, GSC2, ROM2 and RLM1 were over-expressed upon the deletion of Rnt1p in vivo and their mRNAs were cleaved by the recombinant enzyme in vitro (Supplementary Table S4). Consistently, ectopic over-expression of CWI genes including HSL1 or SWI4 reduced cell growth and rendered cells sensitive to cell wall stress (Supplementary Figure S5). These data suggest that Rnt1p plays an important role in regulating the CWI pathway.
DISCUSSION
The regulation of gene expression can occur at any step along the chain of events leading to protein synthesis. However, most of the well-studied regulatory networks involve transcriptional control coupled with rapid mRNA decay in the cytoplasm. Some of the clearest examples of these regulatory pathways may be found in the cell cycle. In this case, mRNAs coding for phase-specific cyclins (e.g. Cln1p and Cln2p) exhibit very short half lives (6), which makes their cell cycle-dependent transcription an effective measure of gene expression regulation. In this model, the expression cycle is a direct outcome of reciprocal transcriptional cycles, while cytoplasmic mRNA degradation prevents mRNA from entering unwanted rounds of translation. In contrast, the experiments described here highlight a new mechanism of gene regulation in which the nuclear mRNA degradation of the nascent RNA can trigger conditional RNA decay and hence the inhibition of gene expression. Traditionally, nuclear RNA degradation was normally seen as a quality control mechanism preventing the expression of either mis-folded or mutated mRNAs (51) or of mRNAs that are retained in the nucleus (52). The results presented here show that a nuclear ribonuclease can also affect gene expression by degrading normal mRNAs. More specifically, Rnt1p was shown to cleave stem-loop structures within open reading frames resulting in the cell cycle specific decrease in expression, as observed for HSL1. It is not clear at the moment how the cell cycle triggers Hsl1 mRNA degradation. One possibility is that this degradation is regulated by a cell cycle-dependent rhythmic intra-nuclear shuttling of Rnt1p. It has previously been shown that Rnt1p shuttles between the nucleolus and the nucleoplasm in a cell cycle-dependent manner (20). Indeed, Rnt1p exits the nucleolus to the nucleoplasm at the G2/M phase of the cell cycle, exactly when Hsl1 mRNA is expected to be degraded. It is equally possible that Hsl1p may auto-regulate its own expression by regulating either Rnt1p’s activity or its localization. Rnt1p has been identified as a phosphorylation target of Hsl1p in a proteomic assay, but the impact of this phosphorylation on Rnt1p activity has yet to be determined (53). However, other mechanisms of Rnt1p activity regulation could exist since some targets of Rnt1p (e.g. Rom2 and Mkk2) do not seem to be cell cycle regulated (54) and not all of the targets of Rnt1p are kinases. In fact, a cell wall stress, such as Congo red or heat shock, appears to repress Rnt1p expression, thus providing a way of controlling the global activity of Rnt1p in a stress-dependent manner (Figure 4C). Regardless of the specific mechanism triggering conditional mRNA degradation it is now clear that nuclear RNA degradation is not only a means of surveilling RNA quality but also an integral part of the gene regulatory network.
Like bacterial RNase III, Rnt1p was first discovered as ribosomal RNA processing factor (55). Consequently, the effects of deleting these enzymes on cell growth rates were primarily attributed to defects in ribosome biogenesis (56). By monitoring the growth pattern of rnt1Δ cell under different conditions, it was found that the slow growth rate defect could be partially rescued by increasing the osmotic strength of the growth medium using either sorbitol or NaCl (Figure 4A). A resistance to high sodium concentrations was previously observed, however the mechanism underlying this phenotype was not explained (21). Interestingly, increasing the osmotic strength of the growth medium also rescued the inability of rnt1Δ cells to grow at 37°C, suggesting that RNT1 is essential at high temperature because of its contribution to CWI, and not to a role in rRNA processing. Indeed, the osmotic strength-dependent rescue of the rnt1Δ growth defect was blocked by the additional deletions of factors in the CWI pathway such as SLT2 and SWI4. Sensitivity to chitin binding agents like Congo red (Figure 4A) also supports the hypothesis that Rnt1p plays a major role in the cell wall stress response. This notion is further supported by the relatively large number of Rnt1p-dependent CWI-associated genes (Supplementary Table S4). These CWI genes also link Rnt1p to the regulation of stress-dependent cell cycle progression. Rnt1p regulates the expression of the protein kinase Hsl1p, which arrests cells in G2 to promote cell survival under high osmotic conditions (13). However, any possible Rnt1p roles in preventing stress-dependent cell cycle effects cannot fully explain the impact of the enzyme on the cell cycle. The deletion of HSL1 only partially rescues the accumulation of rnt1Δ cells in the G1 phase of the cell cycle and the over-expression of Hsl1p leads to an accumulation of unbudded cells. This indicates that Rnt1p-dependent delay in the G1 phase of the cell cycle may also be caused by other factors like inefficient ribosome synthesis or other activity mediated by the enzyme nucleolar/nuclear shuttling as previously suggested (20). Therefore, we propose that Rnt1p may affect the cell cycle in different ways and that its essential function required for cell growth at high temperature is due to its role in regulating the expression of CWI genes.
Overall, this study provides an example of how post-transcriptional regulation may be integrated into a well-structured regulatory network of gene expression. In the example presented here, nuclear degradation provides a mean complementary to transcriptional and post-translational controls, thereby allowing a rapid and accurate response to stress signals. Unlike transcriptional controls, this alternative regulatory mechanism immediately cuts the supply of de novo message, and rapidly inhibits gene expression regardless of the cytoplasmic stability (i.e. half life) of the target mRNA. These features are not only valuable to the stress response pathway but are also relevant to other pathways responding to changing growth conditions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–5, Supplementary Methods, Supplementary Movies 1 and 2 and Supplementary References [57,58].
FUNDING
The Canadian Institute of Health Research. S.A. is a Chercheur Boursier National of the Fonds de la Recherche en Santé du Québec. Funding for open access charge: Canadian Institute of Health Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We thank Pascal Chartrand, Bernard Turcotte and members of the Abou Elela laboratory for critical reading of the manuscript. We thank Raymund Wellinger for the LLY36 strain and Arlen Johnson for the rat1-1 xrn1Δ strain (AJY856).
REFERENCES
- 1.Hartwell LH. Saccharomyces cerevisiae cell cycle. Bacteriol. Rev. 1974;38:164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberghina L, Rossi RL, Querin L, Wanke V, Vanoni M. A cell sizer network involving Cln3 and Far1 controls entrance into S phase in the mitotic cycle of budding yeast. J. Cell Biol. 2004;167:433–443. doi: 10.1083/jcb.200405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- 4.Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene. 2005;24:2746–2755. doi: 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
- 5.Bähler J. Cell-cycle control of gene expression in budding and fission yeast. Annu. Rev. Genet. 2005;39:69–94. doi: 10.1146/annurev.genet.39.110304.095808. [DOI] [PubMed] [Google Scholar]
- 6.Schneider BL, Zhang J, Markwardt J, Tokiwa G, Volpe T, Honey S, Futcher B. Growth rate and cell size modulate the synthesis of, and requirement for, G1-phase cyclins at start. Mol. Cell. Biol. 2004;24:10802–10813. doi: 10.1128/MCB.24.24.10802-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gill T, Aulds J, Schmitt ME. A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae. J. Cell Biol. 2006;173:35–45. doi: 10.1083/jcb.200512025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talarek N, Cameroni E, Jaquenoud M, Luo X, Bontron S, Lippman S, Devgan G, Snyder M, Broach JR, De Virgilio C. Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5'-3' mRNA decay pathway. Mol. Cell. 2010;38:345–355. doi: 10.1016/j.molcel.2010.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 10.Clotet J, Posas F. Control of cell cycle in response to osmostress: lessons from yeast. Methods Enzymol. 2007;428:63–76. doi: 10.1016/S0076-6879(07)28004-8. [DOI] [PubMed] [Google Scholar]
- 11.Schwob E, Böhm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 12.Escoté X, Zapater M, Clotet J, Posas F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 2004;6:997–1002. doi: 10.1038/ncb1174. [DOI] [PubMed] [Google Scholar]
- 13.Clotet J, Escoté X, Adrover MA, Yaakov G, Garí E, Aldea M, de Nadal E, Posas F. Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J. 2006;25:2338–2346. doi: 10.1038/sj.emboj.7601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison JC, Bardes ES, Ohya Y, Lew DJ. A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat. Cell Biol. 2001;3:417–420. doi: 10.1038/35070104. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs BB, Mylonakis E. Our paths might cross: the role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot. Cell. 2009;8:1616–1625. doi: 10.1128/EC.00193-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simpson-Lavy KJ, Sajman J, Zenvirth D, Brandeis M. APC/CCdh1 specific degradation of Hsl1 and Clb2 is required for proper stress responses of S. cerevisiae. Cell Cycle. 2009;8:3003–3009. [PubMed] [Google Scholar]
- 17.Hahn JS, Thiele DJ. Regulation of the Saccharomyces cerevisiae Slt2 kinase pathway by the stress-inducible Sdp1 dual specificity phosphatase. J. Biol. Chem. 2002;277:21278–21284. doi: 10.1074/jbc.M202557200. [DOI] [PubMed] [Google Scholar]
- 18.Romero-Santacreu L, Moreno J, Pérez-Ortín JE, Alepuz P. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA. 2009;15:1110–1120. doi: 10.1261/rna.1435709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamontagne B, Larose S, Boulanger J, Abou Elela S. The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Curr. Iss. Mol. Biol. 2001;3:71–78. [PubMed] [Google Scholar]
- 20.Catala M, Lamontagne B, Larose S, Ghazal G, Abou Elela S. Cell cycle-dependent nuclear localization of yeast RNase III is required for efficient cell division. Mol. Biol. Cell. 2004;15:3015–3030. doi: 10.1091/mbc.E04-03-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee A, Henras AK, Chanfreau G. Multiple RNA surveillance pathways limit aberrant expression of iron uptake mRNAs and prevent iron toxicity in S. cerevisiae. Mol. Cell. 2005;19:39–51. doi: 10.1016/j.molcel.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 22.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- 23.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 25.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 26.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 27.Ge D, Lamontagne B, Abou Elela S. RNase III-mediated silencing of a glucose-dependent repressor in yeast. Curr. Biol. 2005;15:140–145. doi: 10.1016/j.cub.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Lamontagne B, Abou Elela S. Purification and characterization of Saccharomyces cerevisiae Rnt1p nuclease. Methods Enzymol. 2001;342:159–167. doi: 10.1016/s0076-6879(01)42543-2. [DOI] [PubMed] [Google Scholar]
- 29.Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 30.Ares M, Jr, Igel AH. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990;4:2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- 31.Brosseau JP, Lucier JF, Lapointe E, Durand M, Gendron D, Gervais-Bird J, Tremblay K, Perreault JP, Elela SA. High-throughput quantification of splicing isoforms. RNA. 2010;16:442–449. doi: 10.1261/rna.1877010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 33.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghazal G, Ge D, Gervais-Bird J, Gagnon J, Abou Elela S. Genome-wide prediction and analysis of yeast RNase III-dependent snoRNA processing signals. Mol. Cell. Biol. 2005;25:2981–2994. doi: 10.1128/MCB.25.8.2981-2994.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toussaint M, Levasseur G, Gervais-Bird J, Wellinger RJ, Abou Elela S, Conconi A. A high-throughput method to measure the sensitivity of yeast cells to genotoxic agents in liquid cultures. Mutat. Res. 2006;606:92–105. doi: 10.1016/j.mrgentox.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. Systematic mapping of genetic interaction networks. Annu. Rev. Genet. 2009;43:601–625. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- 37.Thornton BR, Ng TM, Matyskiela ME, Carroll CW, Morgan DO, Toczyski DP. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theesfeld CL, Zyla TR, Bardes EG, Lew DJ. A monitor for bud emergence in the yeast morphogenesis checkpoint. Mol. Biol. Cell. 2003;14:3280–3291. doi: 10.1091/mbc.E03-03-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- 40.Baetz K, Moffat J, Haynes J, Chang M, Andrews B. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell. Biol. 2001;21:6515–6528. doi: 10.1128/MCB.21.19.6515-6528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson-Lavy KJ, Brandeis M. Clb2 and the APC/C(Cdh1) regulate Swe1 stability. Cell Cycle. 2010;9:3046–3053. doi: 10.4161/cc..9.115.12457. [DOI] [PubMed] [Google Scholar]
- 42.Taba MR, Muroff I, Lydall D, Tebb G, Nasmyth K. Changes in a SWI4,6-DNA-binding complex occur at the time of HO gene activation in yeast. Genes Dev. 1991;5:2000–2013. doi: 10.1101/gad.5.11.2000. [DOI] [PubMed] [Google Scholar]
- 43.Burton JL, Solomon MJ. Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol. Cell. Biol. 2000;20:4614–4625. doi: 10.1128/mcb.20.13.4614-4625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Negishi T, Ohya Y. The cell wall integrity checkpoint: coordination between cell wall synthesis and the cell cycle. Yeast. 2010;27:513–519. doi: 10.1002/yea.1795. [DOI] [PubMed] [Google Scholar]
- 46.Igual JC, Johnson AL, Johnston LH. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 1996;15:5001–5013. [PMC free article] [PubMed] [Google Scholar]
- 47.McMillan JN, Longtine MS, Sia RA, Theesfeld CL, Bardes ES, Pringle JR, Lew DJ. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol. 1999;19:6929–6939. doi: 10.1128/mcb.19.10.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roncero C, Durán A. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 1985;163:1180–1185. doi: 10.1128/jb.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.García R, Rodríguez-Peña JM, Bermejo C, Nombela C, Arroyo J. The high osmotic response and cell wall integrity pathways cooperate to regulate transcriptional responses to zymolyase-induced cell wall stress in Saccharomyces cerevisiae. J. Biol. Chem. 2009;284:10901–10911. doi: 10.1074/jbc.M808693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villa T, Rougemaille M, Libri D. Nuclear quality control of RNA polymerase II ribonucleoproteins in yeast: tilting the balance to shape the transcriptome. Biochim. Biophys. Acta. 2008;1779:524–531. doi: 10.1016/j.bbagrm.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Kuai L, Das B, Sherman F. A nuclear degradation pathway controls the abundance of normal mRNAs in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2005;102:13962–13967. doi: 10.1073/pnas.0506518102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 54.de Lichtenberg U, Jensen LJ, Fausbøll A, Jensen TS, Bork P, Brunak S. Comparison of computational methods for the identification of cell cycle-regulated genes. Bioinformatics. 2005;21:1164–1171. doi: 10.1093/bioinformatics/bti093. [DOI] [PubMed] [Google Scholar]
- 55.Abou Elela S, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 56.Takiff HE, Chen SM, Court DL. Genetic analysis of the rnc operon of Escherichia coli. J. Bacteriol. 1989;171:2581–2590. doi: 10.1128/jb.171.5.2581-2590.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Catala M, Tremblay M, Samson E, Conconi A, Abou Elela S. Deletion of Rnt1p alters the proportion of open versus closed rRNA gene repeats in yeast. Mol. Cell. Biol. 2008;28:619–629. doi: 10.1128/MCB.01805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson A. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.