Abstract
Telomerase, the enzyme that extends single-stranded telomeric DNA, consists of an RNA subunit (TER) including a short template sequence, a catalytic protein (TERT) and accessory proteins. We used site-specific UV cross-linking to map the binding sites for DNA primers in TER within active Tetrahymena telomerase holoenzyme complexes. The mapping was performed at single-nucleotide resolution by a novel technique based on RNase H digestion of RNA–DNA hybrids made with overlapping complementary oligodeoxynucleotides. These data allowed tracing of the DNA path through the telomerase complexes from the template to the TERT binding element (TBE) region of TER. TBE is known to bind TERT and to be involved in the template 5′-boundary definition. Based on these findings, we propose that upstream sequences of each growing telomeric DNA chain are involved in regulation of its growth arrest at the 5′-end of the RNA template. The upstream DNA–TBE interaction may also function as an anchor for the subsequent realignment of the 3′-end of the DNA with the 3′-end of the template to enable initiation of synthesis of a new telomeric repeat.
INTRODUCTION
Telomeres are complexes of specific DNA, proteins and RNA, which are found at the ends of eukaryotic chromosomes. The telomeres protect the chromosomes from degradation and fusion with other chromosomes (1–4). In most eukaryotes, the telomeric DNA consists of short G-rich repeats (6–8 nucleotides) and ends as a single-stranded 3′-overhang (5,6). Telomerases are cellular reverse transcriptases that catalyze extension of the single-stranded telomeric DNA overhangs, thereby preventing telomere shortening and consequent damage to the cells. They are also capable of catalyzing de novo addition of single-stranded telomeric repeats to chromosome breakage sites (7–10).
The two major components of telomerases are a catalytic protein subunit designated telomerase reverse transcriptase (TERT) (11) and an RNA subunit designated telomerase RNA (TER). TER includes a short template sequence encoding the telomeric DNA repeats (12). A core telomerase consisting of TERT and TER has been reconstituted in some experimental systems and shown to be capable of in vitro extension of short oligodeoxyribonucleotide (oligo) primers by copying the integral RNA template sequence (13–15). Telomerase holoenzymes include, besides the core components, accessory proteins which are essential for their assembly and enzymatic activities in vivo (16).
During the synthesis of each telomeric repeat by telomerase, the RNA template is copied until its 5′-end is reached. At this stage, DNA synthesis is arrested. Then, the 3′-end of the newly synthesized DNA and the active site of the enzyme realign with the 3′-end of the template, and synthesis of the next repeat is initiated. Many telomerases, including those of ciliates and man, generally remain associated with the elongating DNA chains throughout all stages of this reiterative process and are, therefore, highly processive (7,17,18).
The molecular basis for the processivity of ciliate telomerases was initially explored by kinetic techniques (18). These studies led to the hypothesis that processive telomerases possess a site designated anchor site that tethers upstream sequences of growing telomeric DNA chains when their 3′-ends dissociate from the template. To examine this hypothesis, more direct analyses of the interactions between the telomerases and their DNA substrates were carried out. UV cross-linking assays performed with the telomerase of the ciliate Euplotes aediculatus have indicated that upstream sequences of telomeric DNA primers interact with both the protein and the RNA subunits of the telomerase (19). Interference footprinting of the ciliate Tetrahymena thermophila telomerase complexes revealed that this telomerase interacts primarily with the 6-7 3′-terminal nucleotide residues of DNA primers (20–22). This finding is compatible with a recent crystallographic analysis of the interactions of an RNA–DNA duplex with the TERT subunit of the Tribolium castaneum telomerase. The crystallographic data revealed that TERT has a ring configuration with a cavity that can accommodate seven residues of the RNA–DNA duplex, such that the 3′-end of the DNA chain is positioned at the active site of the enzyme (23).
More recently, interaction sites for single-stranded DNA primers in telomerase proteins were mapped by molecular genetics and UV cross-linking methods. Studies conducted with the in vitro reconstituted Tetrahymena core telomerase led to identification of interactions between primer nucleotides aligned at the beginning of the RNA template and the amino acid residues W187 and Q168 in the TERT essential N-terminal (TEN) domain of TERT (Figure 1A) (24). Interestingly, the TEN domain is lacking in the T. castaneum TERT, but is essential for processive synthesis of telomeric DNA by the Tetrahymena telomerase (25,26). Interactions of the DNA with the telomerase RNA-binding domain (TRBD) domain of the Tetrahymena TERT have also been observed (24,27). Other studies carried out with the Tetrahymena telomerase holoenzyme revealed that the accessory proteins Teb1, p75, p45 and p19 are required for processive DNA synthesis by the enzyme (28). Furthermore, it has been shown that DNA primers interact with the phenylalanine residue F351 in the accessory protein p45, as well as the N-terminal region of TERT, including the TEN and the TRBD domains (29). Also, studies of the human and the yeast telomerases revealed interactions between DNA oligo substrates and the N-terminal domains of the human and the yeast TERTs (30–32). These data indicated that the TEN and the TRBD domains of TERT, as well as some accessory proteins, may be components of the anchor site of telomerases.
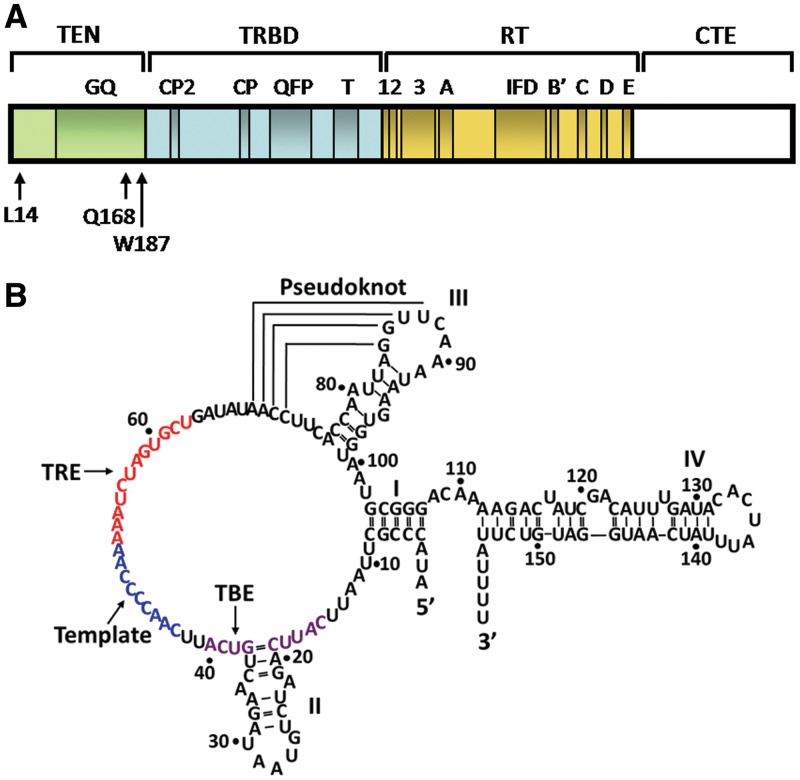
Figure 1.
Schematic illustrations of the Tetrahymena telomerase TERT and TER subunits. (A) A linear map of the Tetrahymena telomerase major catalytic protein TERT, which consists of 1117 amino acids. The four domains: TEN, TRBD, RT (reverse transciptase domain) and CTE (carboxy-terminal element) are indicated. Also indicated are motifs that are conserved in all DNA polymerases and reverse transcriptases (1,2,3,A,B′,C,D,E), and other motifs that are only conserved in telomerases (IFD,T,QFP,CP,CP2,GQ). Of the three amino acids in the TEN domain, which are indicated in the drawing, W187 and Q168 were found by site-directed UV cross-linking and kinetic measurements to contact growing DNA chains in active core telomerase complexes (24). L14 was found to be specifically required for processive extension of telomeric DNA chains by the core telomerase (26). (B) Sequence and secondary structure of the Tetrahymena TER, which consists of 159 nucleotide residues. The template sequence and the regions TBE and TRE (TERT recognition element) are colored blue, violet and red, respectively.
Here, we present a series of experiments designed to further examine the possibility that TER, the telomerase RNA subunit, also possesses an anchor site function. In these experiments we employed a site-specific UV cross-linking procedure to study the interactions of DNA primers with TER in active complexes of the Tetrahymena telomerase holoenzyme. Our data indicate that in the Tetrahymena telomerase complexes upstream nucleotide residues in the elongating DNA interact with nucleotides in the TERT binding element (TBE) region of TER (Figure 1B). The implications of this finding for the mechanism of reiterated telomeric DNA extension by telomerase are discussed below.
MATERIALS AND METHODS
Preparation of the Tetrahymena telomerase holoenzyme
The Tetrahymena telomerase holoenzyme was affinity purified from cells of the T. thermophila strain CU 522 encoding the TERT protein linked to the FZZ tag at the C-terminus (33). Affinity purification of the enzyme was carried out as previously described (29,33).
DNA oligodeoxynucleotides
Oligodeoxynucleotides substituted with 4-thio deoxythymidine (4-thio dT) were purchased from TAG Copenhagen A/S. The nomenclature that we use for such oligos is the position of the 4-thio dT substitution (denoted as S) within the oligo, followed by its length. In this article and in the Supplementary Data, we present the results of experiments conducted with the primer S-5 6-mer, S-20 21-mer, S-15 21-mer, S-13 14-mer and S-16 17-mer having the sequences: GSTGGG, TSGGGGTGCTTGTAGGTTGGG, TTGGGGSGCTTGTAGGTTGGG, TSGGGGTTGGGGTT and TSGGGGTTGGGGTTGGG, respectively. Unsubstituted oligos were purchased from Sigma.
UV cross-linking assays
Binary telomerase–DNA complexes were prepared by incubating purified telomerase holoenzyme with an oligo primer in a buffer containing 50 mM Tris–HCl (pH 8.0), 100 mM sodium acetate, 2 mM MgCl2, 1 mM spermidine, 5 mM β-mercaptoethanol and 20 units of RNasin. The DNA concentrations were 120 µM for the primer S-5 6-mer and 5 µM for the other primers. The concentration of telomerase was 0.02 µg/µl. The volumes of the samples ranged between 5 and 70 µl. UV cross-linking was performed by irradiation of the samples with miniature incoherent light-emitting diodes (LEDs) at 307 nm in a novel apparatus, as previously described (29). The time of irradiation was 2 h. The temperature was maintained at 0–4°C. Subsequently, 0.5 µM of [α-32P]dGTP (3000 Ci/mmol) was added and the samples were incubated for 1 h at 30°C. During this period, the primer molecules were extended with a single radioactively labeled G residue by the cross-linked telomerase enzyme that remained active.
Analysis of the complexes by SDS–PAGE
The complexes were disrupted by addition of Tris–HCl (pH 6.8), sodium dodecyl sulfate (SDS), β-mercaptoethanol, glycerol and bromophenol blue to final concentrations of 62 mM, 1%, 650 mM, 10% and 0.0005%, respectively. The mixtures were frozen at −70°C. Subsequently, the mixtures were thawed at 30°C and further incubated at 30°C for 30 min. SDS–PAGE and phosphorimaging were then performed as previously described (24).
Purification of the cross-linked RNA–DNA molecules
Subsequent to UV irradiation and extension of the primers, the mixtures were incubated 1.5 h at 37°C with 0.2 mg/ml proteinase K in the presence of 0.25% SDS. Nucleic acids were then purified by extraction with a mixture of phenol:chloroform:isoamyl alcohol (25:24:1), followed by ethanol precipitation. The precipitates were resuspended in 10 µl of a solution containing 50 mM Tris–Cl (pH 8.0), 100 mM sodium acetate, 2 mM MgCl2, 1 mM spermidine and 5 mM β-mercaptoethanol. Next, 10 µl of 95% formamide, 5 mM EDTA and 0.025% xylene cyanol were added and the samples were heated at 80°C for 3 min. Then, the nucleic acids were run in a 5%, or 8%, polyacrylamide gel containing 7 M urea and 38.5% formamide. The band containing the cross-linked DNA–RNA molecules was detected by phosphorimaging and was excised from the gel. Nucleic acids were extracted from the gel band by using the Midi GeBAflex-tube Gel Extraction and Dialysis Kit (Gene Bio-Application).
RNase H digestion assays
The cross-linked DNA primer–RNA molecules were dissolved in a solution containing 50 mM Tris–Cl (pH 8.0), 100 mM sodium acetate, 2 mM MgCl2, 1 mM spermidine and 5 mM β-mercaptoethanol. Aliquotes of 10 µl were withdrawn and brought to 16 µl of a solution containing the above components as well as 25 mM KCl, and 40–200 µM complementary oligodeoxynucleotide. The mixtures were heated at 95°C for 5 min and were slowly cooled at 25°C for 1.5 h. Then, 0.05–5 units of RNase H were added and the mixtures were incubated for 15–60 min at 37°C. The reactions were stopped by heating the mixtures for 20 min at 65°C. Next, 17 µl of a solution containing 95% formamide, 5 mM EDTA and 0.025% xylene cyanol were added, and the mixtures were heated at 80°C for 5 min. The RNase H digestion products were analyzed by electrophoresis in 8% polyacrylamide gels containing 7 M urea and 38.5% formamide. Finally, the gels were dried and the radioactively labeled bands were detected by phosphorimaging. The signal strength of a band in each lane was normalized by dividing this signal by the sum of the signals of all bands in the lane. In assays of each primer, the RNA-complementary oligo hybrids were digested with several concentrations of RNase H. The optimal RNase H concentration chosen for mapping the cross-link site(s) was a concentration that generated signals of sufficient strengths for quantitative analysis, but did not generate detectable cuts in the RNA outside RNA–DNA hybrid regions.
Synthesis of uniformly labeled TER
RNA transcription from the TER encoding plasmid pT7159 was carried out with T7 RNA polymerase by using the AmpliScribe™ T7 High Yield Transcription Kit (Epicentre) (24). To radioactively label the RNA, the reaction mixture included, instead of 7.5 mM UTP, 4 mM unlabeled UTP and 0.7 µM [α-32P]UTP (3000 Ci/mmol).
RESULTS
UV cross-linking strategy
Figure 2A illustrates the strategy that we have used for the cross-linking assays. A DNA primer (in this scheme, a 6-mer), in which a single nucleotide has been substituted with 4-thio dT (denoted as S), was incubated with the Tetrahymena telomerase holoenzyme to generate the indicated binary complex. The complex was irradiated with UV light at 307 nm. At this wavelength the substituted DNA nucleotide cross-linked with an RNA nucleotide with which it specifically interacted within the enzyme. We used for the irradiation a novel apparatus that employs miniature LEDs as UV sources. This apparatus allows UV irradiation of biochemical samples in very small volumes (5–10 µl) for up to 3 h, without causing substantial damage to the samples (29). The primer was subsequently elongated with the cross-linked enzyme by a single 32P-dGMP. This reaction could only take place in complexes containing enzyme molecules that remained active after UV irradiation. Moreover, to be extended the primer molecules had to be properly aligned along the RNA template region. Clearly, TER became radioactively labeled by virtue of being cross-linked to the extended primer.
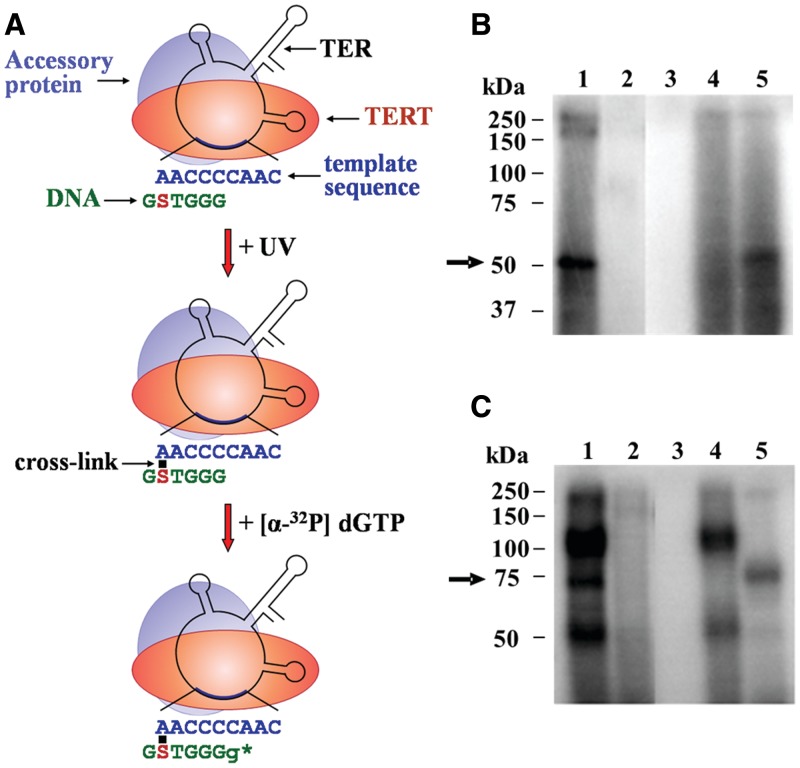
Figure 2.
Assay for site-specific UV cross-linking of DNA primers to the TER subunit in active Tetrahymena telomerase holoenzyme complexes. (A) Schematic drawing of the cross-linking assay (see the text for a detailed description). Only one of several telomerase accessory proteins is shown here for clarity. S denotes a 4-thio dT substitution. Small g denotes a radioactively labeled G residue added to the DNA primer by the cross-linked enzyme. The star indicates the active site. (B) Control assays performed with the primer S-5 6-mer shown in A and with an unsubstituted derivative (see ‘Materials and Methods’ section for primer nomenclature). Cross-linked complexes were prepared by the procedure illustrated in A. The complexes were analyzed by SDS–PAGE, followed by phosphorimaging. Lane 1: complete assay. Lane 2: same primer with no substitution. Lane 3: RNase A treatment before irradiation (0.02 mg/ml RNase A, 45 min incubation at 37°C). Lane 4: RNase A treatment after primer extension (0.02 mg/ml RNase A, 45 min incubation at 37°C). Lane 5: proteinase K treatment after primer extension (0.2 mg/ml proteinase K, 1.5 hr incubation at 37°C in the presence of 0.25% SDS). The arrow indicates the cross-linked TER–primer. (C) Control assays performed with the primer S-20 21-mer and with an unsubstituted derivative. Procedures and lane assignments are the same as in B.
Identification of cross-linked DNA primer–TER molecules
We have first carried out a cross-linking experiment with the primer shown in Figure 2A, which consists of a permutation of a single telomeric repeat and is designated S-5 6-mer (see ‘Materials and Methods’ section for a list and nomenclature of the substituted oligos used for our cross-linking assays). Figure 2B shows a phosphorimage of an SDS-polyacrylamide gel, in which the products obtained in this cross-linking experiment and in control assays were run. Lane 1 shows the products of the experiment. The most prominent 50 kDa band is indicated with an arrow. Two other bands can be seen at the top of the gel. Lane 2 shows a control assay, in which the substituted primer S-5 6-mer has been replaced with a 6-mer primer having the same sequence, but containing no UV-sensitive substitution. It can be seen that no radioactively labeled bands were found in this lane. This result indicated that the irradiation at 307 nm did not produce detectable cross-linking of the RNA, or proteins, with the unsubstituted DNA nucleotides. Lane 3 shows another control assay in which the mixture was digested with RNase A before irradiation. This treatment also resulted in elimination of all the bands, confirming that telomerase, and not traces of other polymerases, catalyzed the primer extension. Lane 4 shows a control assay in which the mixture was digested with RNase A after the primer extension step. Clearly, this treatment removed the 50 kDa band and reduced the amount of radioactivity in the bands found at the top of the gel. In the assay shown in lane 5, the mixture was digested with proteinase K after the primer extension step. As indicated, this treatment caused substantial digestion of the higher molecular weight products, but did not eliminate the 50 kDa band. Taken together, these data indicated that the 50 kDa band contained cross-linked DNA primer–TER molecules, while other radioactively labeled bands contained primarily cross-linked DNA–protein products. It should be noted that in similar assays performed with primers substituted with 5-iododeoxyuridine (5-IdU), instead of 4-thio dT, cross-linked DNA–protein molecules were found to be more abundant than cross-linked DNA–TER molecules (29).
Figure 2C shows a phosphorimage of another SDS-polyacrylamide gel, which displays the results obtained in similar assays performed with the S-20 21-mer primer that contains telomeric repeats at its 3′- and 5′-ends and a non-telomeric sequence in the middle part. The S-20 21-mer primer has been substituted with 4-thio dT at the 20th nucleotide from its 3′-end. This type of primer was employed, rather than primers consisting of telomeric repeats exclusively, because the latter could not be radioactively labeled at sufficiently high efficiency to allow mapping of the cross-link sites (see the last section of ‘Results’). As indicated in the gel presented in Figure 2C, a band of ∼75 kDa was found to contain the radioactively labeled cross-linked DNA–TER molecules. The slower mobility of these molecules compared to that of the corresponding molecules in Figure 2B is likely to be due to retardation in their movement caused by the much longer cross-linked DNA primer (see ‘Discussion’ section).
Low resolution mapping of the cross-links between TER and the DNA primers S-5 6-mer and S-20 21-mer
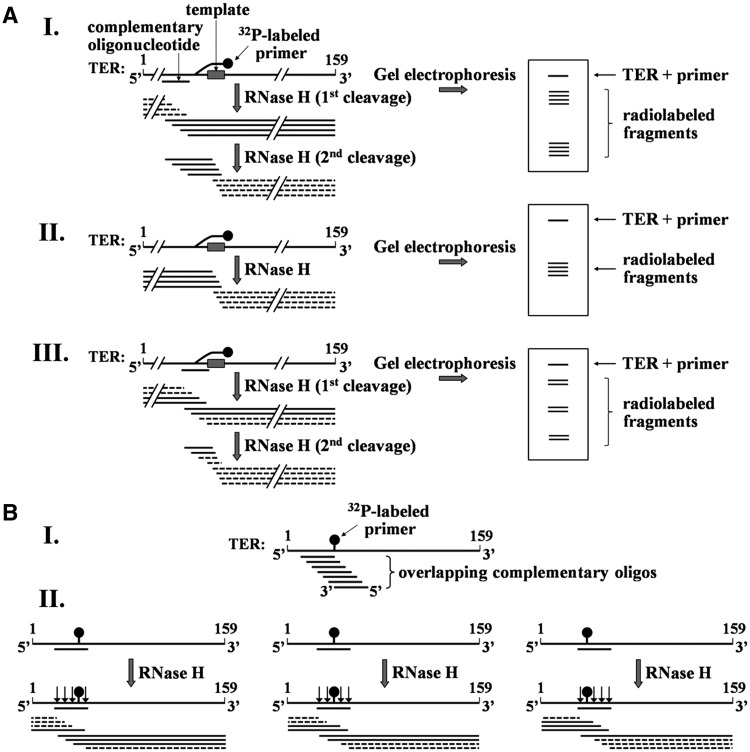
Figure 3A illustrates the strategy that we employed for low resolution mapping of the DNA primer–TER cross-link sites. As panel 3 A–I shows, in each step of the mapping cross-linked TER molecules were annealed with a single complementary oligodeoxynucleotide (in this example the complementary region in TER did not include the cross-link site). The RNA–DNA hybrids were subsequently digested with RNase H, which catalyzed cleavage of the RNA within the hybrid region (RNase H first cleavage). As indicated, this digestion was expected to generate a series of radioactively labeled fragments and another series of unlabeled fragments. In addition, RNase H could also digest RNA–DNA hybrids formed between the template region and the cross-linked primer, thereby producing additional series of smaller fragments (RNase H second cleavage). As shown, gel electrophoresis of the mixture and subsequent phosphorimaging were expected to reveal bands containing the residual undigested RNA and the two series of radioactive digestion products. Panel 3A-II shows that in the absence of a complementary oligo, only fragments generated by RNase H digestion of hybrids formed with the primer would be observed. Panel 3A-III shows that for the case of a complementary oligo that overlapped with the cross-link site, three series of radioactively labeled fragments were expected to be generated.
Figure 3.
Strategies for mapping the cross-link sites in TER. (A) Schematic illustration of the initial mapping procedure. Purified cross-linked DNA primer–TER molecules are hybridized with an oligo complementary to a region of the RNA. The hybrids are digested with RNase H and the products are resolved in 8% polyacrylamide gel containing 7 M urea and 38.5% formamide, followed by phosphorimaging of the gel. Note that only the fragments including solid lines are radioactively labeled. I. RNase H digestion (first cleavage) of hybrid generated by oligo complementary to a region in the RNA that does not include the cross-link site. RNase H (second cleavage) also cleaves the fragments generated by the first digestion within the hybrid formed in these fragments between the primer and the RNA template region. II. An RNase H digestion assay performed in the absence of complementary oligo. Cleavage occurs in the hybrid formed with the primer. III. RNase H digestion performed in the presence of complementary oligo that overlaps with the cross-link site. (B) Strategy for mapping the cross-link site at a high resolution. I. A series of overlapping complementary oligos are employed. These oligos are chosen to cover the region found by the initial mapping to contain the cross-link site. II. Illustration of cleavage patterns obtained with oligos complementary to the left, middle and right sides of the cross-link. As shown in Figure 3A-III, a second cleavage of fragments including the cross-linked site may also occur in these assays within RNA–DNA hybrids formed between the primer and the template region in TER. To save space, this second cleavage is not shown here. A detailed explanation of this procedure is presented in the text.
Figure 4A shows RNase H mapping data obtained in a cross-linking assay performed with the S-5 6-mer primer. Lane 1 shows the gel profile obtained in a control assay in which neither RNase H nor complementary oligo were added to the mixture. The band containing the undigested primer–RNA molecules is indicated with a thick arrow. The smear found at the top of the gel in this lane and in all the other lanes contains aggregation products that appear in some of these gels. Another control assay is shown in lane 5. In this assay, the cross-linked primer–TER molecules were digested with RNase H in the absence of a complementary oligo. A faint band indicated with the thin arrow b contains some fragments generated by cleavage of the short hybrid formed between the RNA template region and the DNA primer (Figure 3A-II). Lane 2 shows the gel profile obtained after RNase H digestion of hybrids made between the cross-linked TER and an oligo spanning the TER nucleotides 57–72. It can be seen that the RNA–DNA hybrid cleavage products are found in a single diffuse band indicated by the thin arrow c. Apparently, no detectable cleavage occurred at the primer-template hybrid region in the presence of the complementary oligo, whose concentration was much larger that of the cross-linked primer (absence of band b). Lane 3 shows the gel profile obtained after RNase H cleavage of hybrids made with an oligo spanning the TER nucleotides 41–60. In this lane two bands of cleavage products (b and d) were obtained. Band b in this lane is considerably stronger than band b in lane 5, since like band d, it contains cleavage products of the hybrid formed with the complementary oligo. Lane 4 shows the gel profile obtained after RNase H cleavage of hybrids made with an oligo spanning the TER nucleotides 24–40. This lane displays a prominent band designated a.
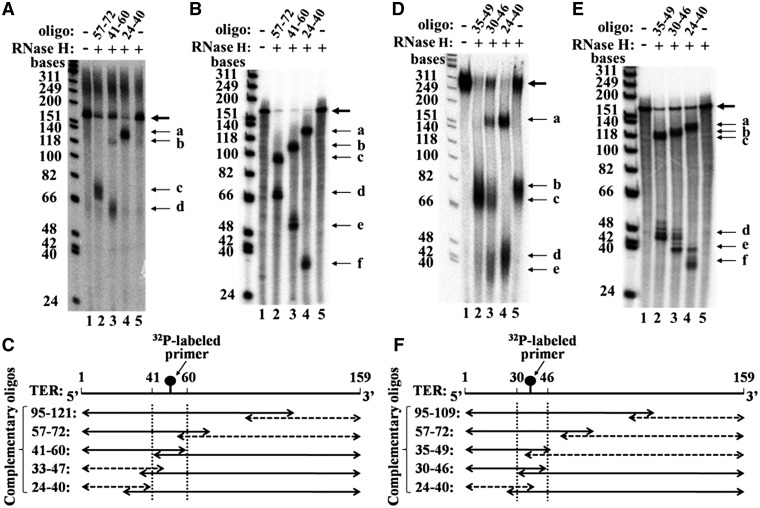
Figure 4.
Initial mapping of cross-link sites by RNase H cleavage. (A) Mapping of cross-links generated with the primer S-5 6-mer. The RNase H cleavage products of RNA–DNA hybrids (thin arrows) were generated by the procedure described in Figure 3A. The bold arrow denotes uncleaved primer DNA–RNA molecules. Lane 1: no RNase H, no complementary oligo. Lane 2: RNase H cleavage products of a hybrid prepared with an oligo complementary to a region of TER spanned by the nucleotides 57–72. Lane 3: RNase H cleavage products of a hybrid formed with an oligo complementary to a region of TER spanned by the nucleotides 41–60. Lane 4: RNase H cleavage products of a hybrid formed with an oligo complementary to a region of TER spanned by the nucleotides 24–40. Lane 5: no complementary oligo. The size markers on the left are φX174 DNA/HinfI fragments (Promega) that were radioactively labeled with [γ-32P] ATP, using T4 polynucleotide kinase. (B) Same RNase H cleavage assays as in A, except that RNA–DNA hybrids were formed with uniformly labeled TER synthesized in vitro, as described in ‘Materials and Methods’ section. These mixtures were not UV irradiated. (C) Schematic drawing of the RNase H cleavage products shown in A and of cleavage products obtained in similar assays performed with other indicated complementary oligos. The solid and the dashed two headed arrows denote the radioactively labeled and the unlabeled fragments, respectively. The dashed vertical lines denote the region including the cross-link. (D) Mapping of cross-link generated with the primer S-20 21-mer. Lane 1: no RNase H, no complementary oligo. Lane 2: RNase H cleavage products of a hybrid prepared with an oligo complementary to a region of TER spanned by the nucleotides 35–49. Lane 3: RNase H cleavage products of a hybrid formed with an oligo complementary to a region of TER spanned by the nucleotides 30–46. Lane 4: RNase H cleavage products of a hybrid formed with an oligo complementary to a region of TER spanned by the nucleotides 24–40. Lane 5: no complementary oligo. (E) Same RNase H cleavage assays as in D, except that RNA–DNA hybrids were formed with the uniformly labeled TER synthesized in vitro. (F) Schematic drawing of the RNase H cleavage products shown in D and of the cleavage products obtained in similar assays performed with other indicated complementary oligos. Notations are as in C.
Figure 4B shows a similar series of RNase H digestion assays performed with uniformly labeled TER molecules. These RNA molecules were synthesized in vitro by transcribing a TER gene cloned into an appropriate plasmid, and were not cross-linked to DNA primers by UV irradiation. It can be seen that two bands were generated by RNase H digestion of hybrids made with each of the complementary oligos (lanes 2-4). As expected, in each of the lanes 2 and 4 one of the two bands corresponded to the single band generated in the assays shown in Figure 4A, while the two bands shown in lane 3 corresponded to the two bands found in lane 3 of Figure 4A. However, the bands in Figure 4B had a slightly faster mobility than the corresponding bands in Figure 4A, since they contained RNA molecules lacking a cross-linked DNA primer.
Figure 4C presents a schematic illustration of the cleavage products obtained in the assays shown in Figure 4A and in similar assays carried out with two additional complementary oligos. Each pair of solid and dashed two-headed arrows represents the radioactively labeled and unlabeled RNase H cleavage products, respectively, which were generated in assays performed with the indicated complementary oligo. The overlapping regions of the arrows in each pair represent the hybrid regions. As indicated, in the experiment performed with the oligo spanning the nucleotides 41–60, both series of fragments were radioactively labeled and were observed in the gel as two bands. This result showed that a cross-link site maps within this overlapping region, which was marked by the two dashed vertical lines. This conclusion is further corroborated below in a detailed high resolution mapping of the cross-link site.
Figure 4D shows similar mapping data that were obtained in a cross-linking experiment performed with the longer primer S-20 21-mer. As in Figure 4A, two control assays were performed, namely incubation in the absence of RNAse H and complementary oligo (lane 1) and RNAse H digestion in the absence of complementary oligo (lane 5). The band b in lane 5 contains fragments generated by RNase H cleavages within the primer–TER hybrid. Clearly, these cleavages occurred at a higher efficiency than the cleavages made in the hybrid between the S-5 6-mer primer and TER (Figure 4A, lane 5). It should also be noted that in this lane and in the other lanes of the gel presented in Figure 4D, the mobilities of the cross-linked TER fragments were affected by the attached (longer) primer to a greater extent than the mobilities observed in Figure 4A (see ‘Discussion’ section). Lanes 2–4 in Figure 4D present the gel profiles obtained after RNase H digestion of hybrids formed with oligos complementary to the TER regions spanned by the nucleotides 35–49, 30–46 and 24–40, respectively. A single major band designated c can be seen in lane 2. Lane 3 displays the three bands a, c and e, besides the band containing the undigested primer–TER molecules. These bands contain digestion products which are schematically drawn in Figure 3A-III. Lane 4 displays just the two bands a and d. These bands correspond to the digestion pattern shown in Figure 3A-I. Figure 4E, like Figure 4B, shows a series of control assays performed with hybrid formed between the complementary oligos and uniformly labeled TER molecules.
The results shown in Figure 4D and data obtained in similar assays performed with two additional complementary oligos, are summarized in the plot shown in Figure 4F. This plot indicates that the major cross-link site for the longer primer S-20 21-mer maps within the TER region 30–46. This conclusion is further corroborated by the fine mapping of this cross-link site shown in Figure 6 below.
Figure 6.
High resolution mapping of the cross-link between the primer S-20 21-mer and TER. (A) Mapping of the cross-link site of the primer S-20 21-mer was performed as illustrated in Figure 3B and described in the legend to Figure 5. The band denoted as c contains fragments generated by secondary cleavage of the fragments denoted as ‘a’ by RNase H, as indicated in the drawing shown in Figure 3A-III. Lane 1: no RNase H, no complementary oligo. Lane 2: oligo complementary to the region of TER spanned by the nucleotides 24–40. Lane 3: oligo complementary to the region of TER spanned by the nucleotides 25–41. Lanes 4–11: overlapping oligos of the same series: 26–42, … ,33–49. Lane 12: no oligo. (B) A plot of the data shown in A. (C) RNase H cleavage assay of hybrids formed between the overlapping oligos used for the assays shown in A and uniformly labeled TER molecules synthesized in vitro. (D) A plot of the data shown in C.
High resolution mapping of the cross-links between TER and the DNA primers S-5 6-mer and S-20 21-mer
To map the primer–TER cross-link sites at a higher resolution, we used the strategy illustrated in Figure 3B. As indicated in panel 3B-I, RNase H cleavage assays were carried out with a series of overlapping complementary oligos which spanned the region found by the lower resolution mapping to include the cross-link site. Panel 3B-II shows the cleavage patterns expected for the assays of three of these oligos which were complementary to the left, middle and right parts, respectively, of this region. As illustrated, both series of large and small fragments produced by RNase H cleavage of the hybrids generated with these oligos were expected to be radioactively labeled. However, the amounts of radioactivity found in each series were expected to decrease or increase as the oligos were shifted from left to right. An assay performed with the oligo located in the middle region was expected to yield two series of fragments (i.e. two broad bands) with equal amounts of radioactivity. The cross-link site should be located in the RNA nucleotide complementary to the middle of this oligo. However, as illustrated in Figure 3A-III, a second cleavage may occur in DNA–RNA hybrids generated by the cross-linked primer itself with the RNA template sequence, thereby generating three broad bands (see below).
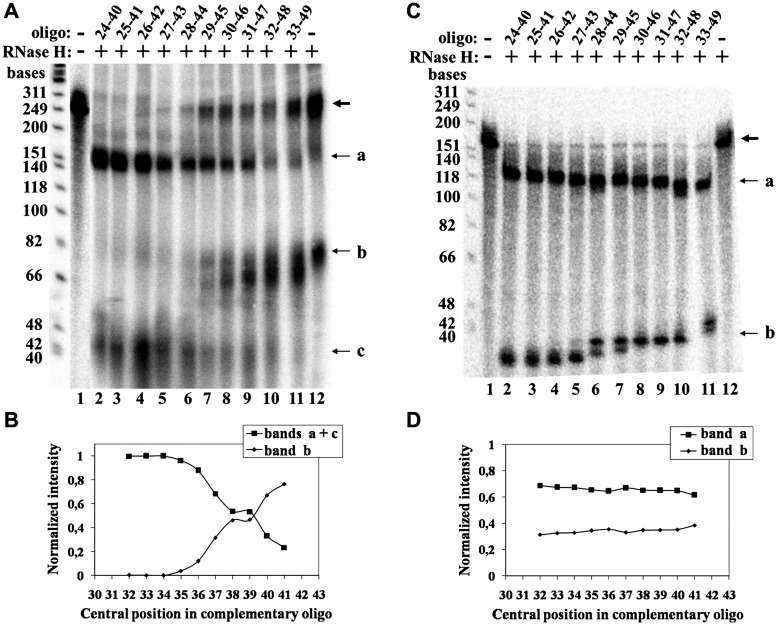
Figure 5A presents a phosphorimage of a gel displaying high resolution mapping data that were obtained in a cross-linking experiment performed with the short S-5 6-mer primer. Lane 1 shows a control assay in which neither RNase H nor complementary oligo were added. A faint band observed in this lane contains non-specific degradation products of about 60 nucleotides. Lane 14 presents a second control assay in which no complementary oligo was added to the mixture. The two faint bands observed in this lane could result from RNase H cleavage of the hybrid formed with the primer. Lanes 2–13 display digestion products denoted a and b that were obtained with the overlapping series of complementary oligos spanning the TER regions 37–53, 38–54, … ,48–64. Note that in this experiment, most of the hybrid molecules were not cleaved at all and the bold arrow designates these undigested molecules. The band containing these molecules appears to be rather broad due to overexposure of the gel. The overexposure was required in order to detect the digestion products that would otherwise be barely visible. Figure 5B shows a plot of the normalized intensities of bands a and b versus the central position of each complementary oligo. The two curves should cross near the site of the cross-link. According to these data, either of the two RNA nucleotide residues, No. 51, or No. 52, might be the site of cross-linking. However, the proper alignment of the primer along the RNA template region, which was required for its extension by the telomerase, supports the notion that the cross-linking occurred with the nucleotide No. 51 (Figure 2A). Figure 5C shows the phosphorimage of a gel in which the RNase H cleavage patterns of hybrids generated between the same complementary oligos and the uniformly labeled synthetic TER were similarly analyzed. In these assays, the populations of smaller fragments were rather heterogeneous. Nevertheless, plots of the sum of radioactivities in both the a and the b series of fragments versus the central position of the complementary oligos gave flat curves, as expected (Figure 5D).
Figure 5.
High resolution mapping of the cross-link between the primer S-5 6-mer and TER. (A) Mapping of the cross-link of the primer S-5 6-mer was performed as illustrated in Figure 3B. The bold arrow indicates the uncleaved primer DNA–TER molecules. a and b denote the bands containing the series of fragments produced by cleavage of the hybrids formed with each complementary oligo. Lane 1: no RNase H, no complementary oligo. Lane 2: oligo complementary to the region of TER spanned by the nucleotides 37–53. Lane 3: oligo complementary to the region of TER spanned by the nucleotides 38–54. Lanes 4–13: overlapping complementary oligos of the same series: 39–55, … ,48–64. Lane 14: no oligo. (B) A plot of the data shown in A. (C) RNase H cleavage assays of hybrids formed between the same overlapping oligos and uniformly labeled TER molecules synthesized in vitro. (D) A plot of the data shown in C. The plotted intensity of band b is the sum of the radioactivities found in all the bands included in the b region of the gel.
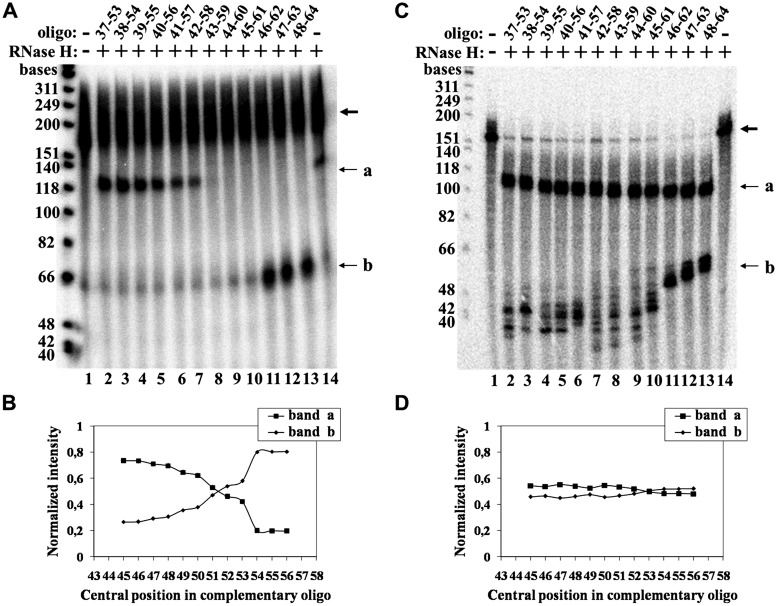
Figure 6A presents a phosphorimage of a gel including data obtained in a similar high resolution mapping experiment performed with the S-20 21-mer primer. Lanes 1 and 12 show the two control assays in which neither RNase H nor complementary oligo, or no complementary oligo, respectively, were added to the mixtures. The two bands in lane 12 resulted from RNase H cleavage of the RNA at hybrids formed with the primer. Lanes 2–11 present the cleavage patterns obtained with the series of complementary oligos. As can be seen, in these lanes there are three bands designated a, b and c that were generated according to the scheme shown in Figure 3A-III. Also note the gradual increase in the intensity of the band containing the undigested molecules, which is denoted by the bold arrow. This could result from steric hindrance of the enzyme at sites that map next to the cross-link. Since the fragments observed in band c were generated by second cleavage of some of the fragments of type a, the data shown in Figure 6A were plotted as the combined radioactivities in bands a+c, or the radioactivity in band b, versus the central position of each complementary oligo (Figure 6B). As in the assays of the shorter primer, RNase H cleavage of uniformly labeled RNA hybridized with the same complementary oligos gave flat curves (Figure 6C and D). Based on the curves shown in Figure 6B, the cross-link site could map at the TER residue C39 and/or the TER residue U38.
Cross-linking experiments performed with other DNA primers
In an attempt to search for additional DNA-binding sites in TER, we have carried out similar cross-linking assays with a DNA primer designated S-15 21-mer. This primer has the same sequence as that of the primer S-20 21-mer, but contains a 4-thio dT substitution at the position No. 15, instead of No. 20. As indicated in Supplementary Figure S1, this primer was radioactively labeled with 32P-dGTP at a considerably lower efficiency than the primer S-20 21-mer. Nevertheless, after finding that the cross-link site of the S-15 21-mer maps within the same region of TER as that of the S-20 21-mer, we were able to carry out a high resolution mapping of this site, as shown in Supplementary Figure S2. It can be seen that the substituted residue No. 15 in the primer S-15 21-mer cross-linked to the TER nucleotides C39 and/or A40.
We also attempted to study the cross-linking between TER and two telomeric primers designated S-13 14-mer and S-16 17-mer (see ‘Materials and Methods’ section for the sequences of these oligos). However, as shown in Supplementary Figure S1, the radioactive labeling of these primers after being cross-linked to TER, was too low to allow mapping of their cross-link sites. This could be due to formation of unusual DNA structures that inhibit the extension of these primers by telomerase. It should be noted that the extension of longer telomeric primers, which may form DNA quadruplexes, was expected to be more strongly inhibited (34).
DISCUSSION
To better understand the mechanism of the reiterative synthesis of telomeric DNA by telomerase, it is essential to obtain a detailed map of the interactions of growing DNA chains with the various components of the enzyme. The data obtained in the present study on the TER–DNA interactions represent a significant contribution towards achievement of this goal. The UV cross-linking procedure illustrated in Figure 2A ensured that the interactions that we detected as specific RNA–DNA cross-links occurred in active telomerase holoenzyme complexes; for primers that cross-linked to the enzyme in inactive complexes were not radioactively labeled and consequently their cross-links were not detected in our assays. However, this procedure did not ensure that all the interactions that actually occur between the DNA and TER were detected in our assays as cross-links. Moreover, our results did not establish that all elongated substrates necessarily travel the path that includes the cross-link sites which we have mapped.
Mapping of the cross-link sites that were detected in our assays was performed by hybridizing the cross-linked DNA–RNA molecules with various complementary oligodeoxyribonucleotides, followed by RNase H cleavage of the hybrids and analysis of the fragments by gel electrophoresis. In particular, we developed a new high resolution mapping technique based on the use of series of overlapping complementary oligos for the RNase H cleavage assays. This mapping technique does not require site-specific cleavage of the RNA by RNase H, for which very expensive oligos containing modified nucleotides are needed (35). Also, the mapping does not require accurate measurements of the lengths of the cross-linked DNA–RNA fragments. This is advantageous because these fragments are branched, rather than linear, and their lengths cannot be accurately deduced by comparison of their mobilities to the mobilities of linear DNA markers of known sizes. Hence, we anticipate that our new methodology will be useful for similar projects in other systems.
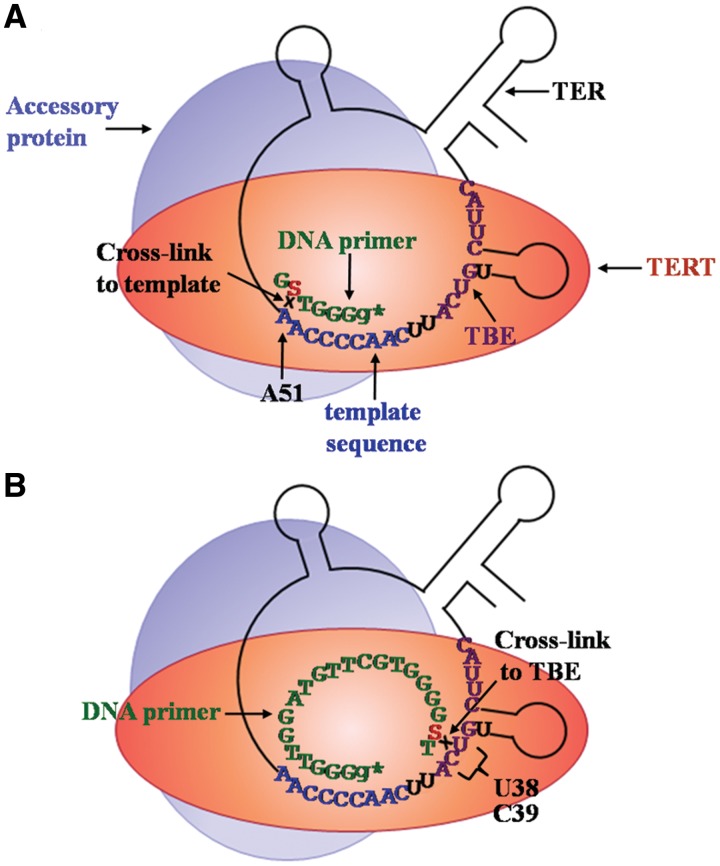
Figure 7 presents a schematic illustration of our results. As Figure 7A indicates, the primer S-5 6-mer had to be properly aligned with the template region in order to be extended by the radioactively labeled G residue. This alignment is also compatible with the direct experimental finding that the 4-thio dT residue in this primer cross-linked with the RNA residue A51. This cross-linking was clearly favored by formation of canonical Watson–Crick hydrogen bonds between these RNA and DNA residues. It is interesting to note, in this connection, that A51 has been previously found to play a role in primer alignment (36).
Figure 7.
Schematic drawing of the alignment and cross-linking of the DNA primers S-5 6-mer and S-20 21-mer in active telomerase holoenzyme complexes. (A) Alignment and cross-linking of the DNA primer S-5 6-mer. Note that the drawing of TER and the protein components of the telomerase is schematic and that only one out of at least five accessory proteins is shown here for simplicity. The template region, the DNA primer and TBE are colored blue, green and violet, respectively. (B) Alignment and cross-linking of the DNA primer S-20 21-mer. Further explanations are presented in the text.
As Figure 7B illustrates, a similar alignment of the 3′-terminal segment of the primer S-20 21-mer could be inferred from its extension by a radioactively labeled G residue. Figure 7B also records the direct experimental observation of cross-linking of the 4-thio dT residue in the primer S-20 21-mer with the TER residues U38 and/or C39. Clearly, neither of these RNA residues could form canonical Watson–Crick hydrogen bonds with the 4-thio dT residue in this primer. However, the cross-linking implied a very close specific contact of the substituted pyrimidine base in the DNA with either of these RNA residues. Interestingly, the 15th nucleotide in the primer S-15 21-mer was found to contact the TER residues C39 and/or A40 (Supplementary Figure S2). Taken together these results indicate that the TER nucleotides U38-A40 may interact with either the 20th, or the 15th DNA residues in the 21-mer primer.
It is noteworthy that the TER nucleotides U38–A40 are included in the region 5′-(U)GUCA-3′ that is highly conserved in ciliate TERs (37). The biological significance of the nucleotides U38–A40 was further highlighted by the discovery that they are essential for definition of the RNA template 5′-boundary and for repeat addition processivity of the telomerase (38,39). Moreover, they were also found to be essential for binding the TRBD of the TERT subunit and were, therefore, included in the TBE of TER (40) (Figures 1B and 7). The observation reported here that these nucleotides also interact with upstream residues of elongating DNA molecules provides a new perspective to the concept of the telomerase anchor site. We propose that these RNA–DNA interactions are not only required for tethering the DNA to the enzyme, but also play an active role in regulation of the entire process of realignment of the 3′-terminal region of the DNA primer with the 3′-terminal region of the RNA template. This process is initiated by termination of DNA elongation at the 5′-end of the template region. Presumably, termination is coupled to the subsequent realignment step. The upstream DNA sequences may be involved in regulation of both steps of this complex process and thereby affect repeat addition processivity by the enzyme.
It has been proposed that the human and the ciliate telomerases share a mechanism for defining the template boundary (41). In particular, human RNA sequences upstream of the template region were found to be involved in the template boundary definition (41). Therefore, it would be interesting to find out whether elongating human telomeric DNA also interacts with these sequences in the human TER.
In conclusion, our present study helps trace the path that newly synthesized telomeric DNA follows within active Tetrahymena telomerase holoenzyme complexes. Besides the interactions with the RNA template region and TBE, the DNA also contacts telomerase proteins. Recent evidence indicates that contact sites for growing DNA chains exist in the N-terminal region of TERT including the TEN and the TRBD domains, and in the accessory protein p45 (29). Other genetic and biochemical data indicate that these TERT domains, as well as several accessory proteins, including p45, play a role in the processive elongation of the telomeric DNA by telomerase (25,26,33). It appears likely that there are additional DNA contact sites with the RNA and proteins of the telomerase holoenzyme, which were not detected so far (29). Clearly, identification and mapping of these sites should lead to a better understanding of the processive mode of synthesis of telomeric DNA by telomerase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
The Israel Science Foundation [378/03 and 967/08]; USA–Israel Binational Science Foundation [2005256]. Funding for open access charge: The Israel Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Prof. Kathleen Collins and Dr Bosun Min of The University of California, Berkeley, for sending us the Tetrahymena telomerase cells encoding the tagged TERT protein. They also thank Prof. Joseph Salzman and Dr Boris Mayler of the Microelectronics Research Center, Department of Electrical Engineering, Technion-Israel Institute of Technology, for their help in the design of our novel apparatus for UV irradiation.
REFERENCES
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Azzalin CM, Lingner J. Telomeres: the silence is broken. Cell Cycle. 2008;7:1161–1165. doi: 10.4161/cc.7.9.5836. [DOI] [PubMed] [Google Scholar]
- 3.Palm W, de Lange LT. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 4.Jain D, Cooper JP. Telomeric strategies: means to an end. Annu. Rev. Genet. 2010;44:243–269. doi: 10.1146/annurev-genet-102108-134841. [DOI] [PubMed] [Google Scholar]
- 5.Jacob NK, Kirk KE, Price CM. Generation of telomeric G strand overhangs involves both G and C strand cleavage. Mol. Cell. 2003;11:1021–1032. doi: 10.1016/s1097-2765(03)00131-x. [DOI] [PubMed] [Google Scholar]
- 6.Sfeir AJ, Chai W, Shay JW, Wright WE. Telomere-end processing the terminal nucleotides of human chromosomes. Mol. Cell. 2005;18:131–138. doi: 10.1016/j.molcel.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 7.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 8.Blackburn EH, Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. 2011;3:a003558. doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekaran VG, Soares J, Jarstfer MB. Structures of telomerase subunits provide functional insights. Biochim. Biophys. Acta. 2010;1804:1190–1201. doi: 10.1016/j.bbapap.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Wyatt HD, West SC, Beattie TL. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010;38:5609–5622. doi: 10.1093/nar/gkq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 12.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 13.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl Acad. Sci. USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryan TM, Goodrich KJ, Cech TR. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem. 2000;275:24199–24207. doi: 10.1074/jbc.M003246200. [DOI] [PubMed] [Google Scholar]
- 15.Harrington L, Zhou W, McPhail T, Oulton R, Yeung DK, Mar V, Bass MB, Robinson MO. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Gene Develop. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallardo F, Chartrand P. Telomerase biogenesis: the long road before getting to the end. RNA Biol. 2008;5:212–215. doi: 10.4161/rna.7115. [DOI] [PubMed] [Google Scholar]
- 17.Greider CW. Telomerase is processive. Mol. Cell. Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins K. Ciliate telomerase biochemistry. Annu. Rev. Biochem. 1999;68:187–218. doi: 10.1146/annurev.biochem.68.1.187. [DOI] [PubMed] [Google Scholar]
- 19.Hammond PW, Lively TN, Cech TR. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell. Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamin S, Baran N, Manor H. Interference footprinting analysis of telomerase elongation complexes. Mol. Cell. Biol. 2000;20:4224–4237. doi: 10.1128/mcb.20.12.4224-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baran N. PhD Thesis. Technion-Israel Institute of Technology; 2000. Novel features of the mechanism of telomere extension by telomerase. [Google Scholar]
- 22.Baran N, Haviv Y, Paul B, Manor H. Studies on the minimal lengths required for DNA primers to be extended by the Tetrahymena telomerase: implications for primer positioning by the enzyme. Nucleic Acids Res. 2002;30:5570–5578. doi: 10.1093/nar/gkf676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat. Struct. Mol. Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 24.Romi E, Baran N, Gantman M, Shmoish M, Min B, Collins K, Manor H. High-resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. Proc. Natl Acad. Sci. USA. 2007;104:8791–8796. doi: 10.1073/pnas.0703157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 26.Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat. Struct. Mol. Biol. 2008;15:870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finger SN, Bryan TM. Multiple DNA-binding sites in Tetrahymena telomerase. Nucleic Acids Res. 2008;36:1260–1272. doi: 10.1093/nar/gkm866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min B, Collins K. Multiple mechanisms for elongation processivity within the reconstituted tetrahymena telomerase holoenzyme. J. Biol. Chem. 2010;285:16434–16443. doi: 10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenfeld KK, Ziv T, Goldin S, Glaser F, Manor H. Mapping of DNA binding sites in the Tetrahymena telomerase holoenzyme proteins by UV cross-linking and mass spectrometry. J. Mol. Biol. 2011;410:77–92. doi: 10.1016/j.jmb.2011.04.040. [DOI] [PubMed] [Google Scholar]
- 30.Moriarty TJ, Ward RJ, Taboski MA, Autexier C. An anchor site-type defect in human telomerase that disrupts telomere length maintenance and cellular immortalization. Mol. Biol. Cell. 2005;16:3152–3161. doi: 10.1091/mbc.E05-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lue NF. A physical and functional constituent of telomerase anchor site. J. Biol. Chem. 2005;280:26586–26591. doi: 10.1074/jbc.M503028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drosopoulos WC, Prasad VR. The telomerase-specific T motif is a restrictive determinant of repetitive reverse transcription by human telomerase. Mol. Cell. Biol. 2010;30:447–459. doi: 10.1128/MCB.00853-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol. Cell. 2009;36:609–619. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 35.Lapham J, Crothers DM. Site-specific cleavage of transcript RNA. Methods Enzymol. 2000;317:132–139. doi: 10.1016/s0076-6879(00)17011-9. [DOI] [PubMed] [Google Scholar]
- 36.Gilley D, Blackburn EH. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol. Cell. Biol. 1996;16:66–75. doi: 10.1128/mcb.16.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCormick-Graham M, Romero DP. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Autexier C, Greider CW. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 1995;9:2227–2239. doi: 10.1101/gad.9.18.2227. [DOI] [PubMed] [Google Scholar]
- 39.Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai CK, Miller MC, Collins K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell. 2003;11:1673–1683. doi: 10.1016/s1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17:2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.