Abstract
Tuberculosis, caused by Mycobacterium tuberculosis, remains a leading infectious disease taking one human life every 15 s globally. The two well-characterized strains H37Rv and H37Ra, derived from the same parental strain M. tuberculosis H37, show dramatically different pathogenic phenotypes. PE/PPE gene family, comprising of 176 open reading frames and present exclusively in genus Mycobacterium, accounts for ∼10% of the M. tuberculosis genome. Our comprehensive in silico analyses of PE/PPE family of H37Ra and virulent H37Rv strains revealed genetic differences between these strains in terms of several single nucleotide variations and InDels and these manifested in changes in physico-chemical properties, phosphorylation sites, and protein: protein interacting domains of the corresponding proteomes. Similar comparisons using the 13 sigma factor genes, 36 members of the mammalian cell entry family, 13 mycobacterial membrane protein large family members and 11 two-component signal transduction systems along with 5 orphaned response regulators and 2 orphaned sensor kinases failed to reveal very significant difference between H37Rv and H37Ra, reinforcing the importance of PE/PPE genes. Many of these changes between H37Rv and H37Ra can be correlated to differences in pathogenesis and virulence of the two strains.
INTRODUCTION
According to the 2011 WHO Report (1), there were 8.8 million incidences of tuberculosis (TB) globally, with 1.1 million deaths among HIV-negative people and an additional 0.35 million deaths from HIV-associated TB, mostly in the productive age group. Despite the availability of effective chemotherapy and BCG vaccine, TB continues to be the leading infectious bacterial pathogen till date. The causal organism for TB, Mycobacterium tuberculosis (M.tb), is a highly successful intracellular pathogen that has evolved by successive genomic reduction events (2). H37Rv and H37Ra are well-characterized laboratory strains of M.tb, both of which are derived from the same parental strain, H37 (3), but show dramatically different pathogenic phenotypes.
Complete genome sequencing of M.tb revealed the existence of unique family of protein, PE/PPE, found exclusively in the genus Mycobacterium (4). These genes are so named due to the presence of Pro-Glu (PE) and Pro-Pro-Glu (PPE) signature motifs near the N-terminus of their gene products (5). This gene family has ∼107 PE and ∼69 PPE proteins. The PE family has been further classified into two subgroups: PE and PE_PGRS. PE_PGRS has a conserved PE domain and variable C terminal domain (PGRS domain). PE/PPE multigene families are usually organized in a bicistronic manner with a PE gene, followed by a PPE gene (6). They range in size from small peptide (Rv3018A–28 amino acids) to large proteins (Rv0151–588 amino acids) and are highly polymorphic within the Mycobacterium complex and even between different strains of the same species. Most of these proteins are found to be localized at cell surface and/or secreted and have been reported to be antigenic in nature (7–9). They play a role in regulating pathogenesis and virulence, in modulating the host immune response (9–12) and in generating antigenic variations (4), possibly due to the higher number of single-base substitution in these genes as compared to the rest of the genome (13). We therefore, focused on the PE/PPE gene family and the encoded proteins for our comparative study of H37Rv and H37Ra. For a meaningful interpretation of the comparative methodology, we also investigated some key non-PE/PPE gene families known to be associated with pathogenesis. These were as follows: (i) mce (mammalian cell entry) family, (ii) MmpL (Mycobacterial membrane protein Large) family, (iii) Sigma factors family and (iv) two-component signal transduction systems (TCSSs). The mce proteins play an important role in entry and survival of the bacteria in the host. This family consists of 36 members excluding Rv0590A (a probable continuation of mce2B (14). Mutation analyses of mce operons had been linked to attenuation of M.tb virulence (15). MmpL proteins comprising of 13 members (mmpl13a and mmpl13b are the two putative Open Reading Frame (ORFs) of mmpl13 gene) belong to RND (resistance, nodulation and cell division proteins) family of transporter proteins that are mainly involved in multidrug resistance (16). The association of mmpL7, mmpL4, mmpL11, mmpL8, mmpL5 and mmpL10 genes in virulence of mycobacteria has been described by a mutational approach (17). The 13-membered Sigma factor family (18) allows M. tuberculosis to sustain multiple stages of host–pathogen interactions, including adhesion, invasion, intracellular replication and dissemination to other sites by controlling the temporal expression of specific regulons (19,20). The TCSSs include a membrane-localized histidine sensor kinase (SK) that recognizes external signal and an internal cytoplasmically localized response regulator (RR). There are 11 TCSSs along with 5 orphaned RRs and 2 orphaned SKs, many of which have been found to be associated with virulence (21).
Our results indicate presence of several differences, many of which are novel, between H37Rv and H37Ra. Remarkably, these differences are found only in certain characteristics specific to the PE/PPE family and do not exist in any of the other families. These results provide strong evidence of a key role played by PE/PPE family in virulence and pathogenesis of M.tb. Furthermore, these results provide many putative candidates for functional studies related to virulence and pathogenesis of M.tb.
MATERIALS AND METHODS
Search for corresponding genes of H37Rv in H37Ra
All the PE/PPE genes of M.tb H37Rv analyzed in our study, listed in Supplementary Table S1, were based on search of the genes from gene database of National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/gene). FASTA sequences of the genes were used to search for corresponding genes in H37Ra in the European Nucleotide Archive sequence database (www.ebi.ac.uk/ena). Nucleotide position number (Base Range) was used as a reference point to compare the corresponding loci in H37Ra using the H37Ra genome database of EMBL (www.ebi.ac.uk/ena/data/view/CP000611). Furthermore, BLASTn was used to search for differences between H37Rv and H37Ra in terms of single nucleotide variations, insertions and deletions in the PE/PPE genes and non-PE/PPE gene families namely, members of the sigma factor family, the mce family, the MmpL family and genes belonging to the TCSS family.
Computational analyses of PE/PPE and non-PE/PPE proteomes of H37Rv and H37Ra
A list of Primary (citable) accession numbers in UniProtKB (Universal Protein Resource) of PE/PPE genes and non-PE/PPE genes in H37Rv and the corresponding genes in H37Ra was prepared. T-coffee align program was used to compare the amino acid sequences of these genes in both strains. Genes having substitutions, insertions and deletions in amino acid sequence were selected for further analysis.
Prediction of protein stability
Comparison of various physical and chemical parameters of proteins coded by the PE/PPE and non-PE/PPE genes in both strains was carried out with the help of ProtParam tool from ExPASy portal (http://web.expasy.org/protparam/). The computed parameters include the instability index, aliphatic index and grand average of hydrophobicity (GRAVY).
Prediction of structure in proteins
GlobPlot (http://globplot.embl.de/cgiDict.py) tool was used to predict globularity in the proteins showing difference between H37Rv and H37Ra.
Prediction of phosphorylation sites
NetPhosBac (http://www.cbs.dtu.dk/services/NetPhosBac-1.0/) was used to predict serine and threonine phosphorylation sites in the PE/PPE and non-PE/PPE proteins displaying difference between H37Rv and H37Ra.
RESULTS
Changes in the PE/PPE and non-PE/PPE genes of H37Rv and H37Ra
Those PE/PPE genes, as well as non-PE/PPE genes, which are similar or different between H37Rv and H37Ra, are listed in Supplementary Table S1. A comparison of all the PE, PPE and PE_PGRS protein sequences between H37Rv and H37Ra highlighted remarkable differences in these proteins (Supplementary Table S2). It can be seen that for PE/PPE gene family, of the 176 proteins, 109 were found to have similar amino acid sequence in H37Rv and H37Ra. There were 21 genes whose T-Coffee alignments of their protein sequence exhibited differences due to CDS mismatch of the genes between H37Rv and H37Ra despite having 100% nucleotide sequence similarity based on BLASTn analysis. PE10, PE_PGRS 49, PE_PGRS56, PE_PGRS60 gene sequences of H37Rv were found to have 100% sequence similarity with the H37Ra counterpart. However, corresponding CDS and annotated proteins were not found in H37Ra. Further it was noticed that the NCBI gene records of PE21 (Rv2099c) and PE27a (Rv3018A), PPE (Rvnp03) and PE_PGRS (Rvnp02) have been discontinued; and PPE47 (Rv3021c) and PPE48 (Rv3022c) have been merged into single gene in H37Rv.
The dissimilar ones displayed changes at N-terminal, some at C-terminal and the rest showed substitutions, InDels within the protein sequence. Further analyses of the region of differences in sequence of these proteins in H37Ra did not reveal any conserved domains. MRA_1205A has a single nucleotide deletion when compared to Rv1196, thereby resulting in frame shift mutation which leads to the loss of conserved C-terminal domain of Rv1196 producing a truncated protein. Insertion of a nucleotide in MRA_1102 resulted in a stop codon leading to a shortened C-terminal end. Other members like MRA_3548, MRA_3553 were hot spots for mutations. Multiple nucleotide insertions, deletions and single nucleotide variations were observed in these genes which are in concordance with the polymorphisms earlier reported for these genes (22). Extensive alterations in amino acids sequences were evident upon comparison of Rv3508 and Rv3514.
PE_PGRS59 gene of H37Rv showed 99% identity with H37Ra genome sequence with one deletion at nucleotide position 488 of H37Rv resulting into coding of two PE_PGRS proteins in H37Ra (MRA_3634 and MRA_3635). Among these two proteins only MRA_3635 showed similarity to Rv3595c. Differences in these three proteins were further followed up by computational physico-chemical analyses. The protein sequences of Rv0387c (244aa) and Rv0388c (180aa) matched with the C-terminal and N-terminal end, respectively of MRA_0395 (443aa). The corresponding nucleotide sequence of Rv0388c in H37Ra revealed insertions and single nucleotide variations, while Rv0387c nucleotide sequence in H37Ra showed 100% similarity. Differences in Rv0388c and MRA_0395 were followed up by computational physico-chemical analyses of the proteins.
Similar analyses were carried out for non-PE/PPE family of genes known to be associated with virulence in mycobacteria. All 36 members of the mce family showed 100% nucleotide and amino acid sequence similarity between H37Rv and H37Ra. In case of MmpL family, of the 13 members, one gene mmpl13b (Rv1146) exhibited an N-terminal extension in H37Ra protein (MRA_1156). In the Sigma factor family, only one of the 13 members, SigM (Rv3911), had a single nucleotide insertion which resulted in a frame shift prematurely terminating the translated protein. In the TCSS family, 10 of the 11 TCSSs along with 5 orphaned RRs and 2 orphaned SKs exhibited 100% nucleotide and amino acid sequence similarity. phoP (Rv0757), which is the RR of phoP-phoR TCSS, exhibited a single nucleotide variation, which resulted in amino acid substitution in H37Ra. In conclusion, these comprehensive comparative analyses revealed the presence of several interesting, novel and potentially significant differences between H37Rv and H37Ra genome in the context of PE/PPE genes. These differences were then analyzed in terms of changes in physico-chemical properties of the corresponding protein sequences.
Analyses of aliphatic index and GRAVY of PE/PPE and non-PE/PPE proteins in H37Rv and H37Ra
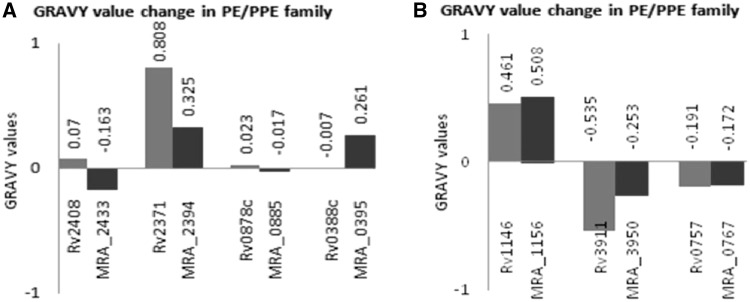
The aliphatic index of a protein signifies the relative volume occupied by aliphatic side chains. Aliphatic hydrophobicity increases with increase in temperature and hence it is a positive factor for increase in thermal stability of globular proteins (23). GRAVY is an indication of protein solubility where a positive value correlates with hydrophobicity and negative with hydrophilicity. More hydrophilic the protein greater will be the extent of hydrogen boding with water molecules and higher will be the solubility. The analysis of GRAVY by ProtParam (Figure 1A) shows that PE24 (Rv2408) is hydrophobic in H37Rv whereas its counterpart in H37Ra (MRA_2433) is hydrophilic, thereby rendering it more soluble. These analyses reveal that N-terminal extension in MRA_2433 might play a role in this change. A similar transition from hydrophilic PPE9 (Rv0388c) to hydrophobic MRA_0395 was observed. On the other hand, the N-terminal extension in PE_PGRS 40 (MRA_2394) renders it less hydrophobic as indicated by the decreased positive value of GRAVY for the corresponding protein in H37Rv (Rv2371) (Figure 1A). In case of PE_PGRS59 (Rv3595c), a single-nucleotide deletion resulted in two different proteins in H37Ra (MRA_3634 and MRA_3635) both are more hydrophobic than Rv3595c (Supplementary Table S3). Along with N-terminal extension, C-terminal changes also played a role in such modifications. This is also evident in MRA_0885 of H37Ra, where a C-terminal extension in PPE13 (Rv0878c) renders the H37Ra protein slightly hydrophilic. Analyses of aliphatic index (Supplementary Table S3) showed marginal changes: ∼10% in PE13 (Rv1195/MRA_1205), PE24 (Rv2408/MRA_2433), PPE7 (Rv0354c/MRA_0363), PPE9 (Rv0388c/MRA_0395) and PPE18 (Rv1196/MRA_1205A), whereas PE_PGRS40 (Rv2371/MRA_2394), wag22 (Rv1759c/MRA_1772) and PE_PGRS59 (Rv3595c/MRA_3634, Rv3595c/MRA_3635) exhibited ∼20% change in aliphatic index (Supplementary Table S3). No major changes in GRAVY values and aliphatic index (Supplementary Table S3) were observed in other proteins. Substitutions alone were able to affect the indices to negligible degree only. No significant changes in GRAVY values (Figure 1B) and aliphatic index (data not shown) were observed in non-PE/PPE protein, mmpl13b (Rv1146) and phoP (Rv0757) proteins (Figure 1B). However, SigM in H37Ra (MRA_3950) showed ΔGRAVY >50% and aliphatic index change >10% between H37Rv and H37Ra (Supplementary Table S3).
Figure 1.
Nucleotide variations between H37Rv and H37Ra manifest in changes in GRAVY values. GRAVY values of PE/PPE proteins in H37Rv and their homologs in H37Ra were plotted where ΔGRAVY was >50%. (A) A major transition from hydrophobic to hydrophilic protein is observed in Rv2408/MRA_2433 and Rv0878c/MRA_0885, whereas Rv2371/MRA_2394 only showed a significant difference in GRAVY value. A hydrophilic to hydrophobic transition was observed in Rv0388c/MRA_0395. (B) Non-PE/PPE proteins. H37Rv proteins MMPL13b (Rv1146), SigM (Rv3911), PhoP (Rv0757) and their homologs in H37Ra (MRA_1156, MRA_3950, MRA_0767), respectively. ΔGRAVY value >50% was observed only in case of Rv3911/MRA_3950.
Thus, the changes in amino acid composition of proteins between H37Rv and H37Ra could alter the solubility of these proteins. These results clearly show that the molecular sequence variations seen between H37Rv and H37Ra PE/PPE protein quite often reflected in physico-chemical changes in the encoded protein.
Analyses of instability index of PE/PPE and non-PE/PPE proteins in H37Rv and H37Ra
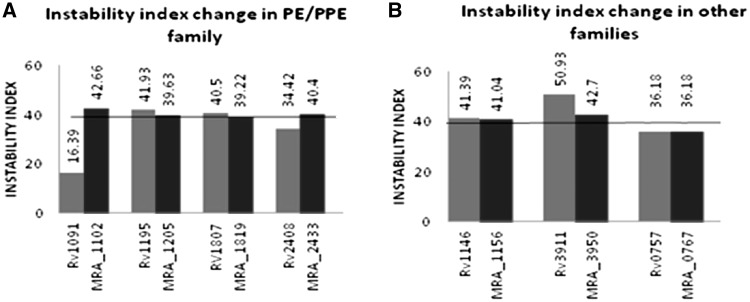
The instability index is indicative of the stability of the protein under in vitro conditions. Instability index >40 is a sign of unstable protein and <40 is an indication of stable protein. The in vivo instability of proteins is possibly determined by the order of certain amino acids in its sequence, some dipeptides occurring differently in unstable and stable proteins. Presence of such dipeptides helps in analysis of protein stability (24). Examples of instability index analyses of PE/PPE proteins in H37Rv and their homologs in H37Ra are shown (Figure 2A). A perusal of the same clearly reveals that a few of the PE/PPE proteins are stable in H37Rv (Rv2408, Rv1091) but the corresponding protein in H37Ra are unstable (MRA_2433 and MRA_1102). Other examples were found where the proteins are unstable in H37Rv (Rv1195 and Rv1807) but stable in H37Ra (MRA_1205 and MRA_1819). In yet another case of PE_PGRS59, Rv3595c and MRA_3635 are stable while MRA_3634 is unstable (Supplementary Table S3). In few other instances, nucleotide changes between H37Rv and H37Ra were observed but without any corresponding influence on instability index (Supplementary Table S3). Proteins like PPE9 (Rv0388c/MRA_0395), PPE13 (Rv0878c/MRA_0885) and PE_PGRS40 (Rv2371/MRA_2394), which were showing significant changes in GRAVY values, failed to show any significant change in instability index.
Figure 2.
Variations in nucleotide sequences between H37Rv and H37Ra affect the stability of PE/PPE proteins but not the non-PE/PPE proteins. Instability index value >40 is indicative of unstable protein and value <40 means the protein is stable. (A) PE/PPE proteins in H37Rv and their homologs in H37Ra show a change from stable to unstable (Rv1091/MRA_1102 and Rv2408/MRA_2433) or the reverse (Rv1195/MRA_1205 and Rv1807/MRA_1819). (B) Non-PE/PPE proteins MMPL13b (Rv1146), SigM (Rv3911), PhoP (Rv0757), and their homologs in H37Ra (MRA_1156, MRA_3950, MRA_0767), respectively, did not show any transition from stable to unstable or reverse.
In the case of non-PE/PPE proteins, no change in instability was observed (Figure 2B). These results therefore suggest that changes in amino acid composition of PE/PPE proteins between H37Rv and H37Ra can be a cause for discrete stability of these proteins.
Analyses of globular domains of PE/PPE and non-PE/PPE proteins in H37Rv and H37Ra
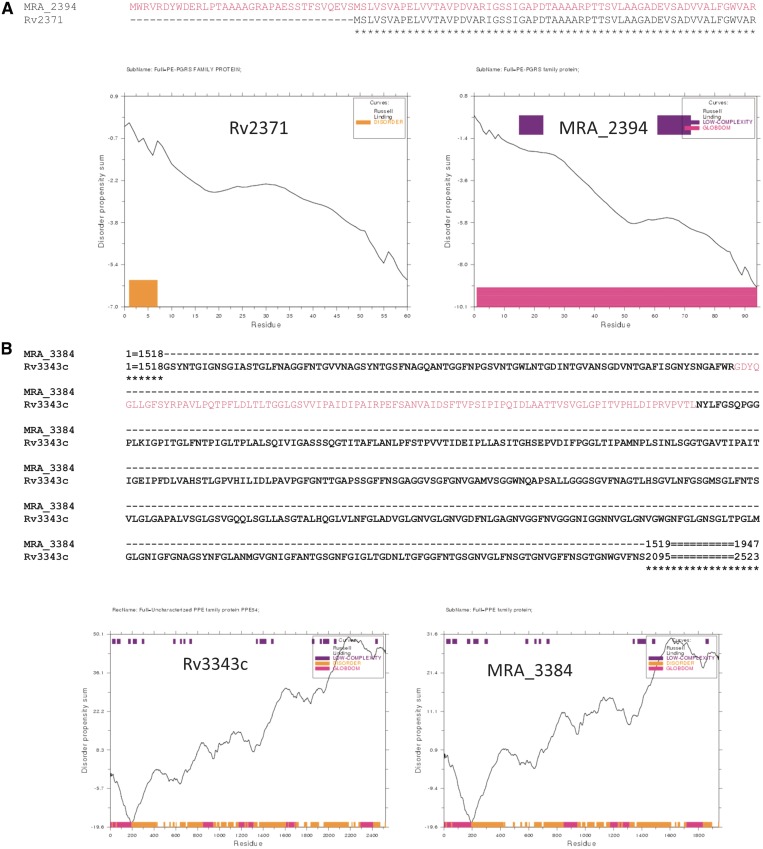
Globular domains in protein molecules confer special functions to a protein. Thus, addition or deletion of globular domains in a protein might lead to a loss or gain of function. In H37Rv, PPE5 (Rv0304c) has four globular domains, an N-terminal extension in its H37Ra homologue (MRA_0313) introduces an additional globular domain (Supplementary Figure S1-A). Similarly, an N-terminal extension in MRA_2394 (Figure 3A), renders the protein globular when compared to PE_PGRS40 (Rv2371). Likewise deletion of a stretch of amino acids in the centre of MRA_3384 compared to PPE54 (Rv3343c) results in deletion of the globular domain, probably leading to a loss of function in H37Ra (Figure 3B). InDels and substitutions in Rv2098c lead to a deletion of the entire globular domain in MRA_2113 of H37Ra (Supplementary Figure S1-B). Along with the N-terminal variation, C-terminal end variation also leads to change in globular domains of the protein. This is illustrated by acquisition of a globular domain in MRA_1102 resulting from deletions in C-terminal end compared to Rv1091 (Supplementary Figure S1-C).
Figure 3.
Gain or loss of globular domain is a function of variation between H37Rv and H37Ra. Disorder propensity of the protein stretch was calculated by GlobPlot analyses to identify the globular domain (shown in pink). Amino acid sequence alignment above the figure illustrates the differences between H37Rv and H37Ra. (A) N-terminal extension in MRA_2394 (shown in right panel) renders the protein globular. (B) Deletion in MRA_3384 leads to a loss of globular domain.
Along with examples of addition and deletion of globular domain in proteins there were several proteins where the globular domain was either shrinking or expanding in the homologous counterpart. For example, the proteins PE13 (Rv1195/MRA_1205), PE24 (Rv2408/MRA_2433), PE_PGRS25 (Rv1396c/MRA_1405), PE_ PGRS47 (Rv2741/MRA_2767) and wag22 (Rv1759c/MRA_1772) have more than 10 amino acids increase in globular domain, while PPE9 (Rv0388c/MRA_0395) have more than 10 amino acids decrease in globular domain (data not shown). In case of PE_PGRS59, MRA_3635 and Rv3595c exhibit similar globular domain, whereas MRA_3634 has a single globular domain (Supplementary Figure S1-D and E). In case of non-PE/PPE proteins, mmpl13b (Supplementary Figure S1-F) and sigM (Supplementary Figure S1-G) extension in already existing globular domain was observed while there was no difference in globular domain in phoP (Supplementary Figure S1-H) protein when compared between H37Rv and H37Ra.
These results once again reiterate that the changes in the sequences of PE/PPE genes of the two strains could consequently impact the likely function of the encoded proteins as evident from gain/loss of globular domain.
Analyses of protein phosphorylation sites in PE/PPE and non-PE/PPE proteins in H37Rv and H37Ra
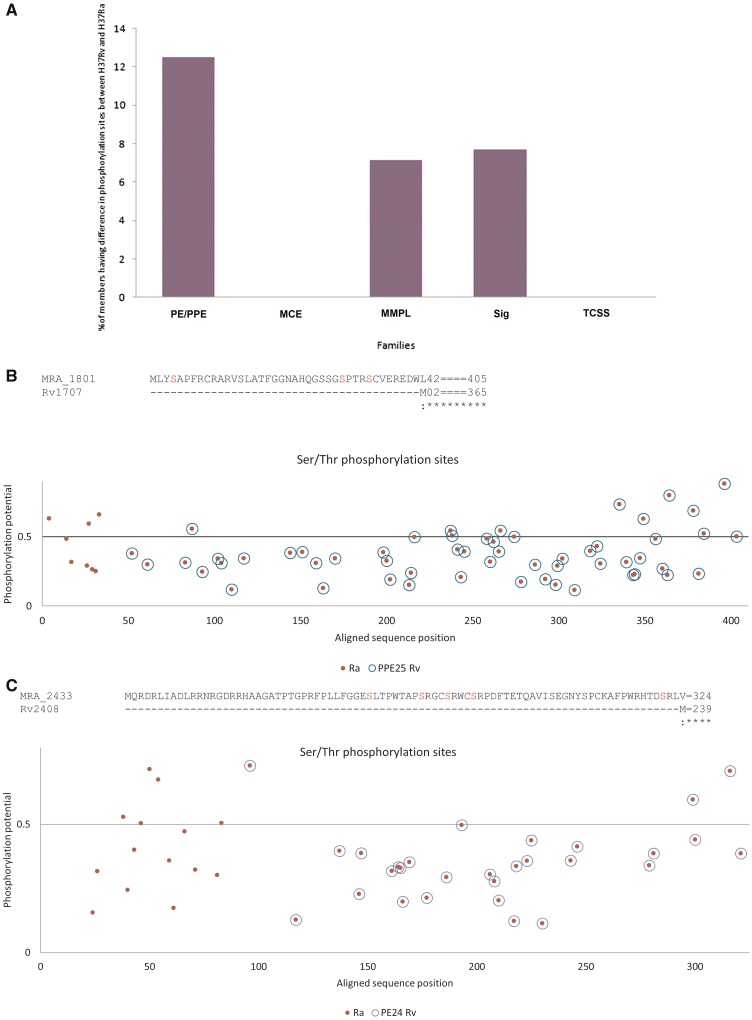
The role of phosphorylation of serine/threonine residues in regulating cell signaling and host-pathogen interaction is well documented. Thus, differences in these sites, as a consequence of variations in nucleotide sequences between H37Rv and H37Ra, will highlight the likely differences in protein–protein interactions and binding of different domains consequently leading to altered downstream effects. Analysis of potential serine/threonine phosphorylation sites in PE/PPE proteins, and non-PE/PPE proteins, revealed a number of differences between H37Rv and H37Ra. These are summarized in Figure 4A. More than 12% of PE/PPE protein family displayed either gain or loss of phosphorylation sites. Loss in serine/threonine phosphorylation sites due to N-terminal shortening was observed in PPE2 (MRA_0265) (Supplementary Figure S2-A), PPE4 (MRA_0295) (Supplementary Figure S2-B) and PE_PGRS11 (MRA_0763) (Supplementary Figure S2-C). Conversely, gain of serine/threonine phosphorylation sites was noticed in PPE5 (MRA_0313) (Supplementary Figure S2-D), PE_PGRS47 (MRA_2767) (Supplementary Figure S2-E), PPE25 (MRA_1801, Figure 4B), PE13 (MRA_1205) (Supplementary Figure S2-F) and PE24 (MRA_2433) (Figure 4C) due to N-terminal extension or due to substitution (Supplementary Figure S2-J). Substitutions and InDels introduced both gain and loss of phosphorylation sites in few proteins namely PE_PGRS22 (MRA_1102) (Supplementary Figure S2-G) and other ORFs (Supplementary Figure S2-K, S2-P, S2-Q and S2-R), and loss of these sites, such as PE_PGRS52 (MRA_3428, (Supplementary Figure S2-H), PE_PGRS36 (MRA_2113) (Supplementary Figure S2-I) and many others (Supplementary Figure S2-L, S2-M, S2-N, S2-O). Gain of phosphorylation sites was observed in MRA_3634 and MRA_3635. Two additional sites were present in MRA_3635 (Supplementary Figure S2-S), whereas MRA_3634 exhibits new seven phosphorylation sites when compared to Rv3595c (Supplementary Figure S2-T).
Figure 4.
Nucleotide variations between H37Rv and H37Ra result in gain or loss of serine/threonine phosphorylation sites. (A) The relative percentage of members of PE/PPE, mce, MmpL, Sigma, TCSS families exhibiting difference in phosphorylation sites between H37Rv and H37Ra is shown. (B, C) Gain of serine/threonine phosphorylation sites. The predictive phosphorylation site(s) in proteins is represented by blue circle (in the case of H37Rv) or red dots (in case of H37Ra). Overlap of blue and red circle indicates no gain or loss of phosphorylation sites in H37Rv and H37Ra proteins. Horizontal line represents the cutoff (threshold) above which it is considered as a putative phosphorylation site. The actual sequence of amino acids and the potential phosphorylation site (highlighted in red) is shown above the respective figures. N-terminal extension in MRA_1801 (B) or MRA_2433 (C) leads to gain of additional serine/threonine phosphorylation sites.
Of the non-PE/PPE proteins, any gain or loss of phosphorylation site was not observed but only change in position of potential phosphorylation sites was noticed (phoP, Supplementary Figure S2-U). However, in sigM, there was both loss and gain of phosphorylation sites in H37Ra (Supplementary Figure S2-V), whereas gain of one phosphorylation site was observed in Mmpl13b in H37Ra (Supplementary Figure S2-W).
The results presented earlier clearly highlight the important consequence of nucleotide sequence changes between H37Rv and H37Ra in PE/PPE genes. Such changes manifest in major structural and physico-chemical alterations in the corresponding proteomes thereby likely impacting their ability to bring about protein–protein interactions that are very important for host pathogen cross talks and consequent virulence and pathogenesis.
DISCUSSION
TB is still a major cause of mortality and morbidity in the world. Co-infection with HIV and emergence of drug resistance is further adding to the problem (25,26). The presence of PE/PPE genes exclusively in genus mycobacterium (4) and their role in pathogenesis, virulence and latency is becoming increasingly evident (9–12,27,28). Previous studies have pointed to the genetic differences between M.tb. H37Rv and H37Ra in the context of virulence (27). We carried out genomics and amino acids sequence comparison of PE/PPE gene families across the two strains of M.tb. From our analyses, it appears that substitutions alone were not very significant but InDels along with substitutions resulted in remarkable differences in the properties of these proteins. We also found absence of five proteins PE10, PE_PGRS49, PE_PGRS56, PE_PGRS60 in H37Ra even though the nucleotide sequences were present in the genome, possibly a reflection of unannotated proteins. We also found difference in annotation of proteins between the two genomes.
Among the PE_PGRS family of proteins, we found complete similarity at amino acid sequence level of PE_PGRS14, PE_PGRS24, PE_PGRS30, PE_PGRS33, PE_PGRS34, PE_PGRS35 and PE_PGRS45 in both the strains. These genes, constitutively expressed under various in vitro conditions (29), possibly have housekeeping functions. Interestingly, differences in amino acid sequence of other genes, many of which have been linked to pathogenic function of the H37Rv strain were evident. One such example was wag22 protein in H37Rv (Rv1759c) which has been shown to have fibronectin-binding properties and also known to be expressed during infection (30). In a previous study, it was shown that inactivation of wag22 homologs in Mycobacterium marinum resulted in decreased survival in granulomas due to defective replication in macrophages (28). C-terminal end of this protein, in H37Rv, has been found to be antigenic (30); however in H37Ra, a truncation at C-terminal end in MRA_1772 caused a change in aliphatic index, loss of phosphorylation sites and expansion in globular domain. The role of PGRS domain, present toward the C-terminal end of PE-PGRS protein, in protection from ubiquitin–proteasome-dependent (UPD) proteolysis has been reported (31). Using green fluorescence protein fusion recombinants with PE_PGRS, the importance of the PGRS domain in the stability of PE_PGRS proteins have been earlier shown (32). Truncation of C-terminal was found in two PE_PGRS proteins namely PE_PGRS22 and wag22, which may have altered the in vivo stability of these proteins in H37Ra. PE_PGRS59 (Rv3595c) exhibited 99% identity with H37Ra but a single-nucleotide deletion in H37Ra corresponding to the nucleotide position 488 of H37Rv, resulted in two PE_PGRS proteins namely MRA_3634 and MRA_3635. Among these two proteins, only one showed similarity to Rv3595c. The truncation of Rv3595c protein and formation of new protein in H37Ra is yet another significant observation. MRA_3635 and Rv3595c share same globular domain position and are both stable. Additionally, both MRA_3635, MRA_3634 are more hydrophobic and exhibit additional phosphorylation sites when compared with Rv3595c.
PPE proteins are found to be associated with secretion of proteins responsible for virulence of M.tb such as ESAT-6 and CFP-10. These proteins together with PE family are potent T-cell and B-cell antigens (7,8) and could help the bacterium evade the host immune response and misdirect the adaptive immune response (10). PPE18 is found to be expressed on the surface and interacts with TLR2 receptor on macrophages modulating the innate immune balance of the host. It represses production of IL-12 and promotes IL-10 production thereby helping persistence of bacteria in the host (11,12). It also interacts differentially with PE13 and PE31 to bring about modulation in host pathogen interactions (33). Expression of PE13 and PPE18 was repressed in H37Ra in macrophages (34). In our study, we observed differences in amino acids sequences of PPE18 protein between H37Rv and its H37Ra homologue. Specific InDels brought about differences in serine/threonine phosphorylation sites in H37Rv and H37Ra, which might lead to, altered protein–protein interactions. Also, as evident from the GlobPlot analysis, the normally conserved N-terminal globular domain in H37Rv PE13 homologue in H37Ra appeared to be extended which might play a role in altering its function. Similarly, N-terminal extension in MRA_1801 (homologue of Rv1787) resulted in introduction of new serine/threonine phosphorylation sites, with possible implications in signaling and host–pathogen interactions. Strains of M. avium lacking this functional PPE protein, MAV_2928 (homologue of Rv1787), have been found to be compromised in virulence by inhibiting acidification of the vacuole and phagosome-lysosome fusion (35). It is also known that PPE31 (Rv1807) is required for the growth of the bacterium in vivo during infection in mice (36). Interestingly, while this protein has been found to be stable in H37Ra, our results with H37Rv are contradictory indicating a decreased stability with obvious physiological implications.
In addition to PPE gene family, PE_PGRS family has also been implicated in bacterial pathogenesis. Previous studies have shown the association of duplication event of PE_PGRS family with the esx gene cluster region 5 (37). ESX-5, the product of esx gene has been found to be involved in virulence of pathogenic mycobacterium (38). PE_PGRS proteins may play a role in pathogenesis of mycobacterium, by modulating interactions of some of the cell-wall–associated virulence factors with the host cell (22). A member of PE_PGRS gene family, Rv3508, has been found to be involved in hypoxia and nitric oxide stress response of M.tb (22). In our analyses, we found insertions, deletions and substitutions in the amino acid sequence of H37Ra homologue of Rv3508. Further, the NetPhosBac analyses of this protein showed loss of phosphorylation sites in H37Ra. As stated earlier, phosphorylation plays a role in signaling process therefore, loss of such sites may render protein insensitive to such stress responses leading to attenuation.
In our analyses, in addition to the functionally known PE/PPE proteins described earlier, there were several other proteins, such as PE24, PPE5, PPE9, PPE13, PE_PGRS22, PE_PGRS25, PE_PGRS36, PE_PGRS40, and PE_PGRS47, which have not yet been assigned any function, that were found to differ significantly between the two strains. As compared to PPE5 of H37Rv, MRA_0313 has N-terminal extension in the amino acid sequence. The consequent acquisition of an additional globular domain in this protein might impart a supplementary role to the protein. Similarly in MRA_2394, N-terminal extension rendered the protein globular when compared to PE_PGRS40 (Rv2371) along with making it less hydrophobic as indicated by the decreased positive value of GRAVY in H37Ra. Likewise deletion of a stretch of amino acids in the central region of MRA_3384, compared to PPE54 (Rv3343c), resulted in loss of a globular domain, probably leading to a loss of function in H37Ra. InDels and substitutions in Rv2098c resulted in deletion of parts of its globular domain or the entire globular domain in MRA_2113. Along with the N-terminal variation, C-terminal variation also resulted in change in globular domains of the protein. This is illustrated by acquisition of a globular domain in MRA_1102 resulting from deletions in C-terminal end compared to Rv1091. These examples of likely functional consequence, however, remain theoretical in the absence of knowledge about their physiological role.
GRAVY values by ProtParam analysis revealed a hydrophobic to hydrophilic transition in case of PPE13 (Rv0878c) and PE24 (Rv2408) in H37Ra. This may alter the solubility of proteins resulting in a change in their functions. Along with N-terminal extension, C-terminal changes also play a role in such modifications. This is illustrated in MRA_0885 of H37Ra, where a C-terminal extension in PPE13 (Rv0878c) rendered the H37Ra protein hydrophilic. Thus, changes in amino acid composition of proteins between H37Rv and H37Ra can alter the solubility of these proteins. In our analysis, ∼10% change in aliphatic index of PE13, PE24, PE_PGRS40, wag22, PPE7 and PPE18 was also observed while comparing proteins of H37Rv and H37Ra. No significant changes in aliphatic index were observed in the other proteins. Substitutions alone were able to affect the indices to negligible degree only.
Non-PE/PPE gene families were also used in our analyses for comparison with PE/PPE gene family. The mce proteins are required for mycobacterium to enter and survive in mammalian cells. While many genes belonging to this large family of 36 members (14) have no known function, mutation of mce operons had been linked to attenuation of M.tb virulence (15). It has been suggested that mce operons in M.tb may not be a direct indicator of pathogenicity due to their wide distribution in non-pathogenic and pathogenic mycobacterium (39). Interestingly in our study, we were unable to observe differences that could be attributed to the attenuation of the H37Ra strain. MmpL proteins belong to RND (resistance, nodulation and cell division proteins) family of transporter proteins that are mainly involved in multidrug resistance (16). Based on mutation analysis, the association of mmpL7, mmpL4, mmpL11, mmpL8, mmpL5 and mmpL10 genes in virulence of mycobacteria has been described (17). We failed to observe any difference in this gene family between H37Rv and H37Ra except for one member. The mmpl13 gene of H37Rv and H37Ra was found to be split into two contiguous putative open reading frames, mmpL13a and mmpL13b. We detected a CDS mismatch in mmpl13b gene that resulted in extension of globular domain and gain of phosphorylation site in H37Ra. Because of the absence of any designated functional role of mmpl13b in pathogenesis or virulence of mycobacteria, we are unable to comment on the likely significance of this observation.
Alternative sigma factors regulate transcription in mycobacteria in response to specific stimuli (19). This fact is highlighted by the ratio of alternative sigma factors to genome size which is highest amongst the obligate pathogens (18). There are 13 members of this transcription family namely, sigma A to sigma M. SigA, SigC, SigE, SigF, SigH and SigL were suggested to be involved in virulence (20). In our analysis, only SigM (Rv3911) was found to have an insertion of one nucleotide which resulted in a frame shift leading to a truncation of the protein. This resulted in extension of globular domain and loss of phosphorylation site in H37Ra. It has been suggested that sigM might be playing a role in long term in vivo adaptation rather than in virulence (40,41). The TCSSs consist of a membrane-localized histidine SK that recognizes external signal and an internal cytoplasmically localized RR. There are 11 TCSSs along with 5 orphaned RRs and 2 orphaned SKs (21). In our study, we found a single-nucleotide polymorphism only in the case of phoP which is the RR of phoP-phoR TCSS. This variation resulted in an amino acid substitution in H37Ra. However, this did not manifest in any significant change in the physico-chemical property of the protein.
In summary, through a comparative genomic analysis, we identified several key changes in the nucleotide sequence between PE/PPE homologs of H37Rv and H37Ra. These changes mainly included insertions and deletions which affected the ORFs of the gene. These alterations in the genes quite often resulted in major physico-chemical changes in the encoded proteins. Search of literature established correlation of many such changes to in vivo function including virulence and pathogenesis. Further biochemical and functional studies are required to establish the role of such changes in amino acid sequence in the attenuation of H37Ra. Such studies along with combination of clinical sample data will not only enhance our understanding of the mechanisms of virulence, pathogenesis and latency of TB caused by M.tb but could also assist in designing new interventions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1 and 2.
FUNDING
‘The Department of Biotechnology, Government of India, Centre of Excellence’ research [BT/01/COE/07/02 to S.E.H./N.Z.E.]; Council of Scientific and Industrial Research (CSIR) (Junior Research Fellowship); Indian Institute of Technology, Delhi (to S.K., K.S. and Y.S.); S.E.H. is a J.C. Bose National Fellow, Government of India and Robert Koch Fellow, Robert Koch Institute, Berlin, Germany. S.E.H. is a Visiting Professor at the King Saud University, Riyadh, Saudi Arabia. Funding for open access charge: Indian Institute of Technology, Delhi.
Conflict of intertest statement. None declared.
Supplementary Material
REFERENCES
- 1.World Health Organization. WHO Report. Geneva, Switzerland: World Health Organization; 2011. Global tuberculosis control: surveillance, planning, financing. [Google Scholar]
- 2.Ahmed N, Dobrindt U, Hacker J, Hasnain SE. Genomic fluidity and pathogenic bacteria: applications in diagnostics, epidemiology and intervention. Nat. Rev. Microbiol. 2008;6:387–394. doi: 10.1038/nrmicro1889. [DOI] [PubMed] [Google Scholar]
- 3.Steenken W, Jr, Gardner LU. History of H37 strain of tubercle bacillus. Am. Rev. Tuberc. 1946;54:62–66. doi: 10.1164/art.1946.54.1.62. [DOI] [PubMed] [Google Scholar]
- 4.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Gordon SV, Eiglmeier K, Garnier T, Brosch R, Parkhill J, Barrell B, Cole ST, Hewinson RG. Genomics of Mycobacterium bovis. Tuberculosis (Edinb) 2001;81:157–163. doi: 10.1054/tube.2000.0269. [DOI] [PubMed] [Google Scholar]
- 6.Tundup S, Akhter Y, Thiagarajan D, Hasnain SE. Clusters of PE and PPE genes of Mycobacterium tuberculosis are organized in operons: evidence that PE Rv2431c is co-transcribed with PPE Rv2430c and their gene products interact with each other. FEBS Lett. 2006;580:1285–1293. doi: 10.1016/j.febslet.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 7.Chakhaiyar P, Nagalakshmi Y, Aruna B, Murthy KJ, Katoch VM, Hasnain SE. Regions of high antigenicity within the hypothetical PPE major polymorphic tandem repeat open-reading frame, Rv2608, show a differential humoral response and a low T cell response in various categories of patients with tuberculosis. J. Infect. Dis. 2004;190:1237–1244. doi: 10.1086/423938. [DOI] [PubMed] [Google Scholar]
- 8.Choudhary RK, Mukhopadhyay S, Chakhaiyar P, Sharma N, Murthy KJ, Katoch VM, Hasnain SE. PPE antigen Rv2430c of Mycobacterium tuberculosis induces a strong B-cell response. Infect. Immun. 2003;71:6338–6343. doi: 10.1128/IAI.71.11.6338-6343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akhter Y, Ehebauer MT, Mukhopadhyay S, Hasnain SE. The PE/PPE multigene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: perhaps more? Biochimie. 2012;94:110–116. doi: 10.1016/j.biochi.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 10.Sampson SL. Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin. Dev. Immunol. 2011;2011:497203. doi: 10.1155/2011/497203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nair S, Ramaswamy PA, Ghosh S, Joshi DC, Pathak N, Siddiqui I, Sharma P, Hasnain SE, Mande SC, Mukhopadhyay S. The PPE18 of Mycobacterium tuberculosis interacts with TLR2 and activates IL-10 induction in macrophage. J. Immunol. 2009;183:6269–6281. doi: 10.4049/jimmunol.0901367. [DOI] [PubMed] [Google Scholar]
- 12.Nair S, Pandey AD, Mukhopadhyay S. The PPE18 protein of Mycobacterium tuberculosis inhibits NF-κB/rel-mediated proinflammatory cytokine production by upregulating and phosphorylating suppressor of cytokine signaling 3 protein. J. Immunol. 2011;186:5413–5424. doi: 10.4049/jimmunol.1000773. [DOI] [PubMed] [Google Scholar]
- 13.Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang F, Xie JP. Mammalian cell entry gene family of Mycobacterium tuberculosis. Mol. Cell Biochem. 2011;352:1–10. doi: 10.1007/s11010-011-0733-5. [DOI] [PubMed] [Google Scholar]
- 15.Gioffré A, Infante E, Aguilar D, Santangelo MP, Klepp L, Amadio A, Meikle V, Etchechoury I, Romano MI, Cataldi A, et al. Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect. 2005;7:325–334. doi: 10.1016/j.micinf.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Domenech P, Reed MB, Barry CE., III Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 2005;73:3492–3501. doi: 10.1128/IAI.73.6.3492-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamichhane G, Tyagi S, Bishai WR. Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect. Immun. 2005;73:2533–2540. doi: 10.1128/IAI.73.4.2533-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodrigue S, Provvedi R, Jacques PE, Gaudreau L, Manganelli R. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2006;30:926–941. doi: 10.1111/j.1574-6976.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- 19.Bashyam MD, Hasnain SE. The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infect. Genet. Evol. 2004;4:301–308. doi: 10.1016/j.meegid.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 2003;16:463–496. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bretl DJ, Demetriadou C, Zahrt TC. Adaptation to environmental stimuli within the host: two-component signal transduction systems of Mycobacterium tuberculosis. Microbiol. Mol. Biol. Rev. 2011;75:566–582. doi: 10.1128/MMBR.05004-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu G, Fu X, Jin K, Zhang L, Wu W, Cui Z, Hu Z, Li Y. Integrative analysis of transcriptome and genome indicates two potential genomic islands are associated with pathogenesis of Mycobacterium tuberculosis. Gene. 2011;489:21–29. doi: 10.1016/j.gene.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 23.Ikai A. Thermostability and aliphatic index of globular proteins. Biochem. 1980;88:1895–1898. [PubMed] [Google Scholar]
- 24.Guruprasad K, Reddy BV, Pandit MW. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990;4:155–161. doi: 10.1093/protein/4.2.155. [DOI] [PubMed] [Google Scholar]
- 25.Ahmed N, Hasnain SE. Genomics of Mycobacterium tuberculosis: old threats and new trends. Indian J. Med. Res. 2004;120:207–212. [PubMed] [Google Scholar]
- 26.Dye C, Williams BG. The population dynamics and control of tuberculosis. Science. 2010;328:856–861. doi: 10.1126/science.1185449. [DOI] [PubMed] [Google Scholar]
- 27.Zheng H, Lu L, Wang B, Pu S, Zhang X, Zhu G, Shi W, Zhang L, Wang H, Wang S, et al. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One. 2008;3:e2375. doi: 10.1371/journal.pone.0002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramakrishnan L, Federspiel NA, Falkow S. Granuloma specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 2000;288:1436–1439. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- 29.Dheenadhayalan V, Delogu G, Sanguinetti M, Fadda G, Brennan MJ. Variable expression patterns of Mycobacterium tuberculosis PE_PGRS genes: evidence that PE_PGRS16 and PE_PGRS26 are inversely regulated in vivo. J. Bacteriol. 2006;188:3721–3725. doi: 10.1128/JB.188.10.3721-3725.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espitia C, Laclette JP, Mondragon-Palomino M, Amador A, Campuzano J, Martens A, Singh M, Cicero R, Zhang Y, Moreno C. The PE-PGRS glycine-rich proteins of Mycobacterium tuberculosis: a new family of fibronectin-binding proteins? Microbiology. 1999;145:3487–3495. doi: 10.1099/00221287-145-12-3487. [DOI] [PubMed] [Google Scholar]
- 31.Koh KW, Lehming N, Seah GT. Degradation-resistant protein domains limit host cell processing and immune detection of mycobacteria. Mol. Immunol. 2009;46:1312–1318. doi: 10.1016/j.molimm.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Brennan MJ, Delogu G. The PE multigene family: a ‘molecular mantra’ for mycobacteria. Trends Microbiol. 2002;10:246–249. doi: 10.1016/s0966-842x(02)02335-1. [DOI] [PubMed] [Google Scholar]
- 33.Mukhopadhyay S, Balaji KN. The PE and PPE proteins of Mycobacterium tuberculosis. Tuberculosis. 2011;91:441–447. doi: 10.1016/j.tube.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Li AH, Waddell SJ, Hinds J, Malloff CA, Bains M, Hancock RE, Lam WL, Butcher PD, Stokes RW. Contrasting transcriptional responses of a virulent and an attenuated strain of Mycobacterium tuberculosis infecting macrophages. PLoS One. 2010;5:e11066. doi: 10.1371/journal.pone.0011066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jha SS, Danelishvili L, Wagner D, Maser J, Li YJ, Moric I, Vogt S, Yamazaki Y, Lai B, Bermudez LE. Virulence-related Mycobacterium avium subsp hominissuis MAV_2928 gene is associated with vacuole remodeling in macrophages. BMC Microbiol. 2010;10:100. doi: 10.1186/1471-2180-10-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 37.Gey VPN, Sampson SL, Lee H, Kim Y, van Helden PD, Warren RM. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 2006;6:95. doi: 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. Type VII secretion–mycobacteria show the way. Nat. Rev. Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 39.Haile Y, Caugant DA, Bjune G, Wiker HG. Mycobacterium tuberculosis mammalian cell entry operon (mce) homologs in Mycobacterium other than tuberculosis (MOTT) FEMS Immunol. Med. Microbiol. 2002;33:125–132. doi: 10.1111/j.1574-695X.2002.tb00581.x. [DOI] [PubMed] [Google Scholar]
- 40.Agarwal N, Woolwine SC, Tyagi S, Bishai WR. Characterization of the Mycobacterium tuberculosis sigma factor SigM by assessment of virulence and identification of SigM-dependent genes. Infect. Immun. 2007;75:452–461. doi: 10.1128/IAI.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raman S, Puyang X, Cheng TY, Young DC, Moody DB, Husson RN. Mycobacterium tuberculosis SigM positively regulates Esx secreted protein and nonribosomal peptide synthetase genes and down regulates virulence-associated surface lipid synthesis. J. Bacteriol. 2006;188:8460–8468. doi: 10.1128/JB.01212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.