Abstract
‘Ribosome scanning’ is the generally accepted mechanism for explaining how a ribosome finds an initiation codon located far removed from the ribosome recruiting site (cap structure). However, the molecular characteristics of ribosome scanning along 5′ untranslated regions (UTRs) remain obscure. Herein, using a rabbit reticulocyte lysate (RRL) system and artificial ribonucleic acid (RNA) constructs composed of a capped leader RNA and an uncapped reporter RNA annealed through a double-stranded RNA (dsRNA) bridge, we show that the ribosome can efficiently bypass a stable, dsRNA region without melting the structure. The insertion of an upstream open reading frame in the capped leader RNA impaired the translation of reporter RNA, indicating that a ribosome associated with the 5′-end explores the regions upstream of the dsRNA bridge in search of the initiation codon. These data indicate that a ribosome may skip part(s) of an messenger RNA 5′UTR without thoroughly scanning it.
INTRODUCTION
The selection of the initiation codon from among numerous AUG codons in an messenger ribonucleic acid (mRNA) molecule is the pivotal step in translation, ensuring that the correct polypeptide encoded by the mRNA is produced. In prokaryotes, the 30 S ribosomal subunit binds directly to mRNA near the initiation codon through base-pairing between the 16 S rRNA and the Shine-Dalgarno sequence located near the initiation codon (1). However, in eukaryotes, the 40 S ribosomal subunit is generally recruited to the cap structure at the 5′-end of an mRNA, which is often far removed from the initiation codon (2). ‘Ribosomal scanning’ is the generally accepted hypothesis for explaining how the 40 S ribosomal subunit, recruited to the 5′-end of an mRNA molecule, finds the initiation codon located far downstream of the cap structure (3). According to this model, the 40 S ribosomal subunit migrates along the mRNA toward the 3′-end, linearly scanning it until it encounters the first AUG codon, which is the most frequently selected initiation codon. If the first AUG contains the preferred surrounding sequence (A/GxxAUGG), called a favourable Kozak context, it is used as the initiation codon. If the first AUG contains an unfavourable context, some of the 40 S ribosomal subunits bypass this codon and continue scanning toward downstream AUGs (4).
Since it was first suggested by Kozak (5), most phenomena observed in translation initiation events, with the exception of some cases such as internal ribosome entry site or ribosome shunting (6–8), have been adequately explained by the ribosome scanning model. However, it remains unclear how ribosomes move along 5′ untranslated regions (5′UTRs) of mRNA. A number of recent elegant studies have greatly increased our understanding of ribosome scanning (9–12). As originally proposed by Kozak (3), the movement of the scanning ribosome was unidirectional. Although the possibility of backward scanning has also been suggested (13,14), Spirin et al. (9,15) argued that unidirectional movement is a molecular characteristic of ribosome scanning. By analyzing LINE-1 mRNA, which contains a 900-nt, highly structured 5′UTR, these authors showed that the translation–initiation rate is directly proportional to the length of RNA but independent of a particular mRNA sequence or secondary structure (15). Interestingly, the insertion of a stable stem loop (−30 kcal/mol) in the middle of the 5′UTR did not alter the translation rate, consistent with a previous report by Kozak (16). This raises an intriguing question: Are all sequences in the 5′UTR inspected base-by-base by the migrating ribosomes, or can some regions be bypassed? Recent articles have provided insights into this question. In the absence of eIF1 and DHX29, a ribosome can bypass stems but forms an aberrant 48 S complex to be dissociated by eIF1 (11). Moreover, when the first AUG is blocked by complementary oligonucleotides or an exon junction complex (EJC), the second AUG is preferentially used as the initiation codon (12), indicating that a portion(s) of the mRNA can be ignored by ribosomes searching for the initiation codon.
On the basis of these observations, we hypothesized that some parts of an mRNA can be bypassed by a ribosome searching for the initiation codon. To test this hypothesis, we designed artificial RNAs composed of leader and reporter RNAs that can be annealed to each other to form a stem structure. Surprisingly, the translation of reporter RNA was augmented when a capped leader RNA was annealed with the reporter RNA, indicating that a ribosome recruited at the 5′-end of the leader RNA can bypass the double-stranded RNA (dsRNA) bridge to reach the initiation codon located in the reporter RNA. These data indicate that, in the presence of a full complement of initiation factors, some parts of the 5′UTR can be ignored by a ribosome searching for the initiation codon.
MATERIALS AND METHODS
RNA constructs
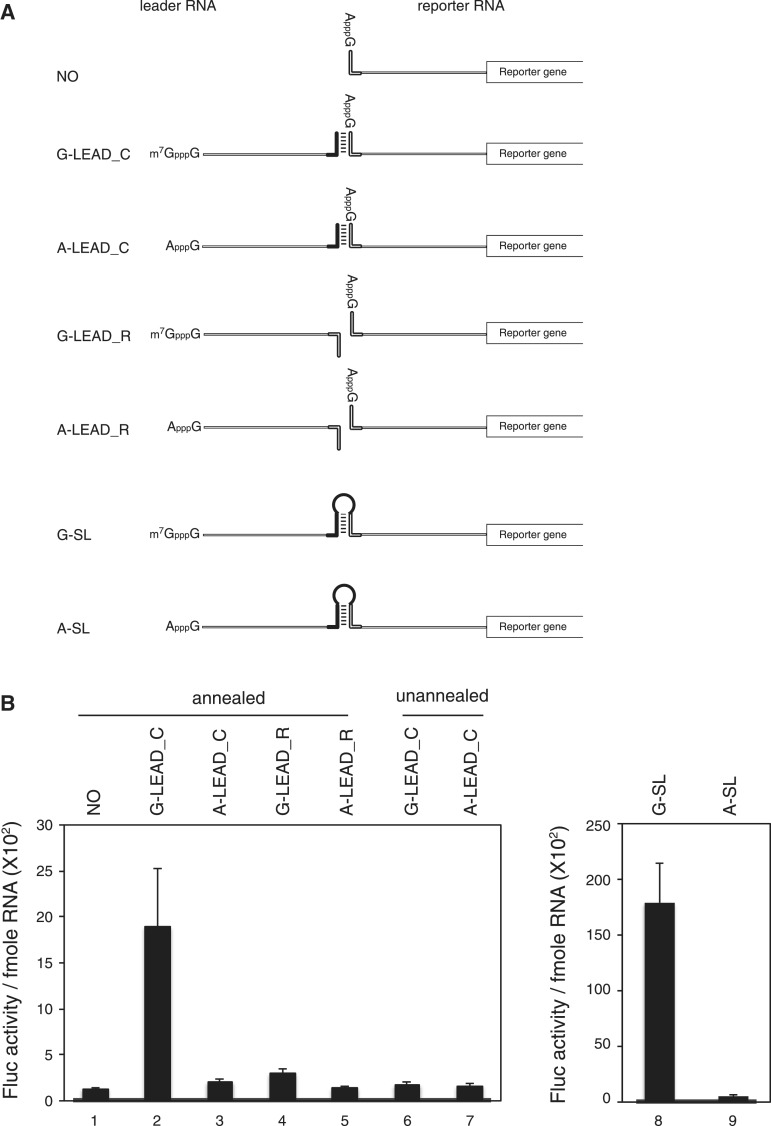
RNA sequences used in the in vitro translation experiments are depicted in Supplementary Table S1. The reporter RNA contains a 19-nt annealing sequence (Figure 1A, NO) or 34-nt annealing sequence (Supplementary Figure S5A, NO+) followed by a 50-nt noncoding sequence originating from plasmid pGL3. The leader RNAs contain four copies of the β-globin 5′UTR followed by a 19-nt sequence (Figure 1A) or 34-nt sequence (Supplementary Figure S5A) complementary to the annealing sequence (LEAD_C or LEAD_C+) or a sequence that is the reverse complement (LEAD_R or LEAD_R+). Therefore, LEAD_C can form a dsRNA bridge with the reporter RNA but LEAD_R cannot (Figure 1F). Four copies of the β-globin 5′UTR were chosen for two reasons: (1) the translation of β-globin is obligatorily end-dependent; and (2) the leader RNA with a UTR of 200 nt (the length of four copies of β-globin 5′UTR) showed high annealing efficiency with the reporter RNA (Figure 1F). The 5′-ends of the reporter RNAs contain the ApppG cap (A cap) structure, which does not interact with eIF4E. The 3′-end of the reporter RNA does not harbour poly(A) to avoid complication of translational enhancement by poly(A) tail. Radiolabelled reporter RNA, which bears the annealing site followed by the 50-nt noncoding sequence and a part of firefly luciferase gene encoding the N-terminal 20 amino acids, used in Figures 1C, 1E and 2 was prepared from PCR-amplified DNA from reporter RNA gene (with primers 5′-TACTTAATACGACTCACTATAGG-3′ and 5′-CCATCTTCCAGCGGATAGAA-3′) to increase resolution of sucrose gradient analysis and gel shift assay.
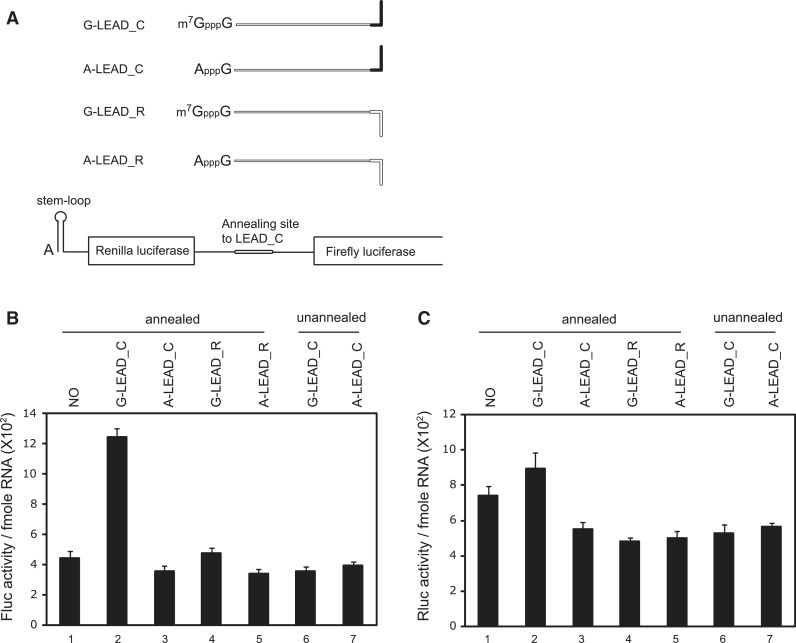
Figure 1.
Enhancement of translation by an m7GpppG-capped (G-capped) leader RNA associated with a reporter RNA via a dsRNA bridge. (A) Schematic diagram of the leader and reporter RNAs. The reporter RNA contains a reporter gene (firefly luciferase). The leader RNAs contain 3′ sequences that are the complements (G-LEAD_C and A-LEAD_C, solid bar) or reverse complements (G-LEAD_R and A-LEAD_R, open bar) of 5′ sequences in the reporter RNAs. The leader RNAs are either m7GpppG-capped (G-LEAD_C and G-LEAD_R) or ApppG-capped (A-LEAD_C and A-LEAD_R). RNA complexes containing both leader and reporter RNAs covalently connected by a stem-loop structure, thus within a single molecule, and with an m7GpppG-cap (G-SL) or an ApppG-cap (A-SL), were constructed to serve as control mRNAs for experiments involving G-LEAD_C and A-LEAD_C, respectively. (B) Reporter translational efficiencies were determined by measuring firefly luciferase activity after pre-annealing of leader and reporter RNAs (lanes 1–5), or with omission of the pre-annealing step (lanes 6 and 7). The luciferase activity in each translation mixture was monitored after 1 h incubation. The translation efficiencies of G-SL and A-SL were also measured and depicted in a separate graph. Similar relative translation efficiencies of the reporter and lead RNA combinations were observed from translation reactions for 30 min instead of 1 h (Supplementary Figure S4). The error bars indicate the standard deviations of nine independent experiments. (C) Leader RNAs did not affect the stability of reporter RNA in translation mixtures. Such mixtures, containing leader and radiolabelled reporter RNAs, were incubated on ice (lanes 1–5; t = 0 h) or subjected to in vitro translation at 30°C for 1 h (lanes 6–10, t = 1 h). (D) The effect of a cap analogue (0.2 mM) on cap-dependent translation mediated by capped leaders associated with reporter RNAs through dsRNA bridges was analyzed relative to the luciferase activity from a reporter RNA lacking a leader RNA (lane 1). (E) Effects of various leader RNAs on 80 S complex formation on reporter RNAs. The amounts of 80 S complex on reporter RNAs labelled with [32P] were monitored by sucrose density gradient analysis after incubation of leader and reporter RNAs in RRL for 15 min in the presence of cycloheximide. (F) Confirmation of hybridization between leader RNAs and reporter RNAs. Annealing reactions with reporter RNA only (NO), or with mixtures of leader RNAs (G-LEAD_C and G-LEAD_R) and reporter RNA, were performed using a thermocycler (lanes 2–4). Reporter RNA was used without an annealing step (lane 1), and the reporter RNA G-SL, which contains covalently linked leader RNA (lane 5), was used as controls. The relevant reporter and leader RNAs are shown in panel A. RNAs were resolved by electrophoresis in a 1% (w/v) agarose gel. Most reporter RNAs became annealed with complementary leader RNAs (G-LEAD_C), as indicated by arrows (lane 3). Reporter RNAs mixed with leader RNAs lacking complementary sequences remained in monomer form (lane 4).
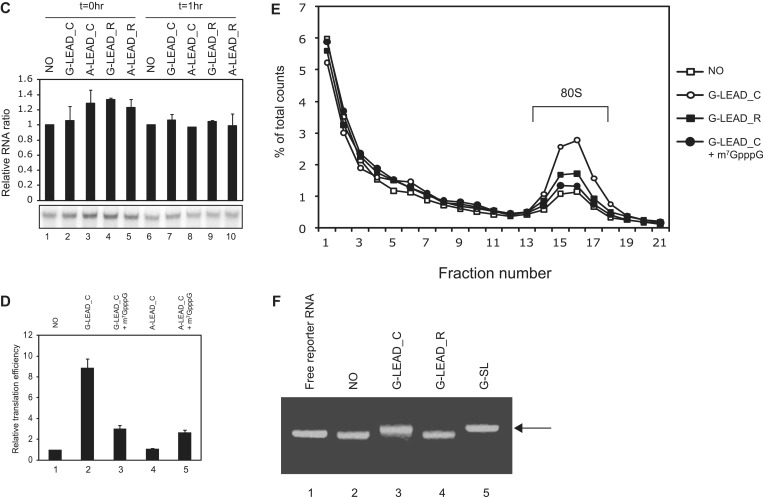
Figure 2.
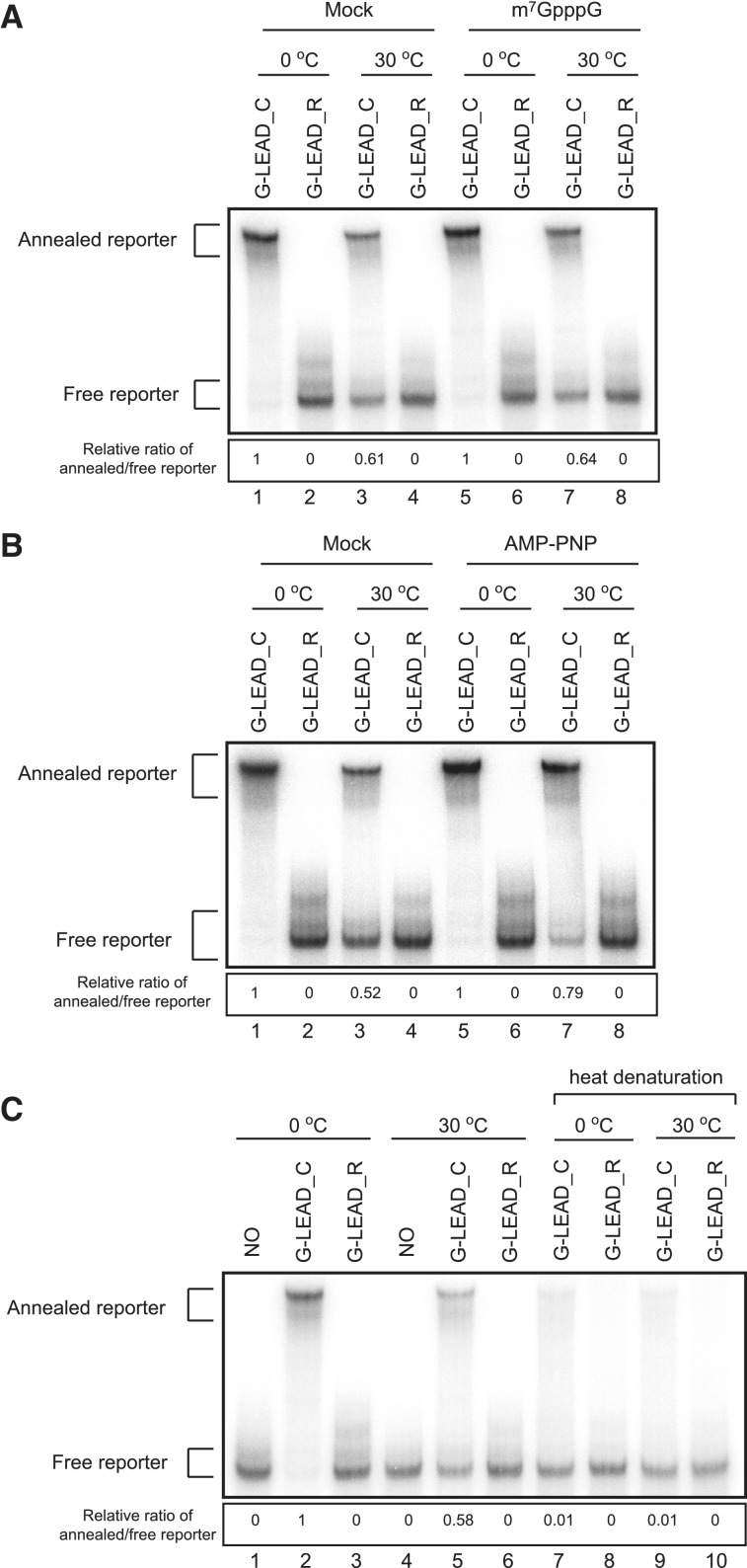
Integrity of the leader–reporter RNA complexes after incubation in translation mixtures. (A) The integrity of leader–reporter RNA complexes after incubation in translation mixtures (RRLs) on ice (lanes 1–2 and 5–6) or at 30°C for 15 min (lanes 3–4 and 7–8) was monitored on an 8% (w/v) non-denaturing acrylamide gel. In some mixtures, 0.2 mM cap analogue was added to translation mixture to block translation (lanes 5–8). The ratio of annealed-to-free reporter G-LEAD_C, after incubation at 0°C, in mock- or cap analogue-treated RRL, was set to unity, to permit comparisons (lanes 1–4 and lanes 5–8, respectively). (B) Inhibition of helicase in RRL stabilizes leader–reporter RNA complex. RNA integrity was monitored in the absence (lanes 1–4) or presence (lanes 5–6) of 2 mM AMP-PNP that inhibits ATP-dependent helicase activities. (C) Putative covalent bond formation between leader and reporter RNAs. To test whether translational enhancement by capped leader RNA was mediated by putative covalent bond formation between leader and reporter RNAs, leader (G-LEAD_C and G-LEAD_R) and reporter RNAs were incubated in translation mixtures at 30°C (lanes 4–6 and 9–10) or on ice (lanes 1–3 and 7–8) for 15 min. The RNAs in RRLs were extracted and then resolved on an 8% (w/v) non-denaturing acrylamide gel with (lanes 7–10) or without (lanes 1–6) heat denaturation at 95°C for 5 min.
RNA annealing and in vitro translation
Leader (1 µM) and reporter RNAs (500 nM) in 20 mM HEPES buffer (pH 7.4) containing 100 mM KCl and 2 mM MgCl2 were annealed by heating at 65°C for 15 min and then gradually cooled at a rate of 1°C/min to 20°C using a thermocycler. The specificity of RNA annealing and the annealed position were evaluated by primer extension inhibition method (Supplementary Figure S1). The annealed RNAs were translated in vitro with RNase-untreated rabbit reticulocyte lysates (RRLs; Promega) to maximize cap-dependency of translation (17). Translation reaction mixtures (10 µl) contained 1 µl of RNA annealing mixture (5 nM as final concentration) and 5 µl of RRL. Translation reactions were performed for indicated times in figure legends at 30°C in the presence of 120 mM KCl, 0.5 mM MgCl2 and 20 mM amino acids. To achieve maximal cap-dependency of translation, 0.5 mM MgCl2 was used in in vitro translation reactions (17). Cap analogue (0.2 mM, New England Biolab) was added prior to reaction to block cap-dependent translation where indicated.
Luciferase assay
Renilla and firefly luciferase activities in RRLs were monitored by a GloMax20/20 luminometer (Promega). Luciferase assay was performed according to the protocol provided by the reagent supplier (Promega).
Sucrose density gradient analysis
80 S ribosomal complexes were assembled onto radiolabelled reporter RNA pre-annealed with the indicated leader RNAs, in 50 µl reaction mixtures. Translation reactions were performed for 15 min at 30°C in the presence of 20 mM cycloheximide, to stall the 80 S complex. The reactions were stopped by the addition of the same volume of ice-cold stop solution (sucrose buffer with 0.5 mg/ml cycloheximide) and next loaded onto a 5–20% (w/v) sucrose density gradient prepared in sucrose buffer [50 mM HEPES (pH 7.4), 100 mM KOAc, 100 mM NH4Cl, 10 mM MgCl2, 2 mM DTT and 0.1 mM EDTA]. After centrifugation at 37 000 rpm (170 000g) for 2 h 30 min at 4°C in a SW41Ti rotor, fractions were collected and analyzed using a scintillation counter.
Quantification of RNA
For real-time RT-PCR, total RNAs were purified from RRL after translation reactions using the TRIzol reagent (Invitrogen). Total RNAs (2 µg) were reverse transcribed employing the Improm-II Reverse Transcription System (Promega) with firefly luciferase- or GAPDH-specific primers (for firefly luciferase 5′-CCGGTATCCAGATCCACAAC-3′, for GAPDH 5′-GATGCAGGGATGATGTTC-3′). The cDNAs were quantified by real-time PCR using SYBR premix Ex Taq (Takara) as previously described (for firefly luciferae 5′-GAGGTTCCATCTGCCAGGTA-3′ and 5′-CCGGTATCCAGATCCACAAC-3′, for GAPDH 5′-TGCACCACCAACTGCTTAG-3′ and 5′-GATGCAGGGATGATGTTC-3′) (18). For measuring RNA stability after translation reaction, translation mixture harbouring annealed leader and radiolabelled reporter were incubated on ice or at 30°C for 1 h. Total RNAs were extracted with phenol and resolved on an 8% acrylamide gel. The intensity of [32P]-labelled reporter RNAs in each band was measured by a phosphoimager.
Observation of the integrity of reporter–leader RNA complex
Leader and [32P]-labelled reporter RNA complexes were subjected to in vitro translation for 15 min at 0°C (on ice) or at 30°C in the presence of 200 mM cycloheximide. Cap analogue (0.2 mM) or AMP-PNP (2 mM) were added to reaction mixtures where indicated. The RNA complexes in translation mixtures were extracted with phenol, and RNAs in aqueous solution were resolved on an 8% acrylamide gel. To calculate the ratios of annealed to free reporter RNAs, the intensity of [32P]-labelled reporter RNAs in each band was measured by a phosphoimager. For monitoring of covalent bond formation between leader and reporter RNAs, RNAs were extracted from translation mixtures and all the samples were divided into two aliquots. Samples incubated on ice or at 30°C were heat-treated at 95°C for 5 min to dissociate leader and reporter RNAs before resolution on an 8% acrylamide gel.
Primer extension inhibition (toe printing)
Primer extension with annealed RNA was performed as described previously with minor modifications (19). A primer corresponding to nucleotides 83–102 (5′-AGGCTATGAAGAGATACGCC-3′) of the firefly luciferase-coding sequence was end-labelled with [γ-32P]ATP using T4 polynucleotide kinase (Fermentas). Reaction mixtures were resolved on a 6% sequencing gel, and radioactive signals were monitored using a phosphorimager.
RESULTS
Cap-dependent translation through a dsRNA bridge
To investigate whether a ribosome, which is associated with the cap structure at the 5′-end of an mRNA, thoroughly scans the 5′UTR of an mRNA base-by-base in search of the initiation codon, we made artificial RNAs composed of leader and reporter RNAs (Figure 1A). Leader RNA can form dsRNA bridge with reporter RNA if proper annealing step is applied. If base-by-base inspection of the 5′UTR by a scanning ribosome was the only mechanism for finding the initiation codon, a ribosome attached at the cap structure would be unable to reach the initiation codon in the reporter RNA because it would pass through and dissociate the dsRNA bridge. In contrast, if a ribosome was able to bypass the dsRNA bridge, the translation of reporter RNA would be enhanced when the capped leader RNA is annealed though not linked by a covalent bond.
In vitro translation reactions with reporter mRNA only exhibited a basal level of translation (Figure 1B and D, lane 1). Annealing the ApppG-capped (A-capped) leader (Figure 1A, A-LEAD_C) had no effect on reporter mRNA translation (Figure 1B, lane 3). However, replacement of the A-capped leader with an m7GpppG-capped (G-capped) leader (Figure 1A, G-LEAD_C) increased the translation of the annealed reporter gene by approximately 9-fold (Figure 1B, lane 2). G-capped (G-LEAD_R) leader RNAs with 3′-ends that are the reverse complement of the annealing sequence and thus cannot anneal to the reporter slightly enhanced the translation of the reporter RNA (Figure 1B, lanes 4), possibly through a weak, non-specific interaction(s) with reporter RNA which was not detected in our experimental confirmation of the interaction (Figures 1F and 2 and Supplementary Figure S1). Moreover, omission of the annealing step eliminated the reporter gene translation-enhancing effect of G-capped leader RNA (Figure 1B, compare lanes 6 and 1 vs. lanes 2 and 1). Enhancement of firefly luciferase activity by annealing of G-LEAD_C was not attributable to an alteration in RNA stability caused by RNA annealing because the RNA level was not changed after the translation reaction had proceeded for 1 h (Figure 1C and Supplementary Figure S2).
The cap dependency of translational enhancement by the leader RNA, G-LEAD_C, was further confirmed by adding a cap analogue to the in vitro translation reaction mixtures. In these experiments, the addition of cap analogue (0.2 mM) dramatically reduced (∼10-fold) the translation of a reporter RNA containing the m7GpppG-cap structure at the 5′-end (Supplementary Figure S3, lanes 1 and 2). In contrast, the addition of cap analogue to a mixture containing reporter RNA with the ApppG-cap structure at the 5′-end enhanced translation by ∼2.5-fold (Supplementary Figure S3, lanes 3 and 4). This phenomenon has been reported elsewhere, although its molecular basis is obscure (20–22). Similar results were obtained from the complex composed of G-LEAD_C and A-LEAD_C; the translational enhancement induced by the G-capped leader RNA was substantially, but not completely, reduced by the addition of cap analogue (Figure 1D, compare lane 3 with lane 2). The residual enhancement of translation by the addition of cap analogue is probably attributed, at least in part, to the translation-enhancing effect of cap analogue (Supplementary Figure S3) on the A-capped reporter RNA which remains unannealed or is dissociated from the leader–reporter RNA complex in the translation reaction mixture (see later). As previously reported (20–22), the addition of the cap analogue increased the translation of the reporter gene annealed with A-capped leader RNA (Figure 1D, compare lane 5 with lane 4).
It is possible that a putative scanning-skip signal was present in the dsRNA bridge we used. Moreover, because the stem we used (−30 kcal/mol) could be separated by a scanning ribosome (15,16), once scanning of leader RNA was completed and the dsRNA bridge was dissociated, the ribosome might associate with the 5′-end of reporter RNA. To exclude this latter possibility, we replaced with a stronger one (−75 kcal/mol) that is sufficient to inhibit translation when introduced into the middle of the mRNA (16) and tested the annealed leader and reporter RNAs in an in vitro translation assay. The results revealed cap-dependent translational enhancement by G-capped leader RNA (Supplementary Figure S5) similarly to the results shown in Figure 1, indicating that cap-dependent translation through the dsRNA bridge is not directed by a ribosome thoroughly scanning through the 5′UTR. To test whether cap-dependent translation was enhanced by G-LEAD_C at the initiation step, we investigated the effect of leader RNA on 80 S complex formation, using sucrose density gradient analysis (Figure 1E). The extent of 80 S complex formation was greatly increased in the presence of G-LEAD_C, but not when G-LEAD_R was added (Figure 1E, compare the G-LEAD_C and G-LEAD_R with the NO). Moreover, 80 S complex formation was reduced to background level when the cap analogue was added to the reaction mixture (Figure 1E, compare the G-LEAD_C with that of G-LEAD_C+m7GpppG). Collectively, these results indicate that (1) the cap on leader RNA functions in a manner similar to that of a cap at the 5′-end of an mRNA, even though the cap is connected to the reporter RNA through a dsRNA bridge; and, (2) translational enhancement by G-LEAD_C is mediated by the eIF4F complex by interactions between the cap structure and eIF4E and occurs at the level of translation initiation.
The relative translational enhancement by a cap structure on a single mRNA containing a stem-and-loop structure (G-SL in Figure 1A), compared to that of a cap on a leader RNA associated with the reporter RNA by a dsRNA bridge (G-LEAD_C), was investigated using the in vitro translation system (Figure 1B and Supplementary Figure S4). When the cap was present on the RNA with a stem-loop structure, translation efficiency was enhanced 40-fold (compare lanes 8 and 9 in Figure 1B). However, the translational enhancement seen when the cap was on a leader RNA was about 9-fold (compare lanes 2 and 3 in Figure 1B). Considering the annealing efficiency of the leader to the reporter RNA was almost 100% (Figure 1F), this result may indicate that only a portion (25% analyzed by fold enhancement before and after capping) of ribosomes recruited to the 5′-end of an mRNA participate in the translation via bypassing of a stem-loop. It is noteworthy that the translational enhancement by the G-capped leader RNA was also observed from a similar set of experiments performed in HeLa lysates instead of RRL (Supplementary Figure S7A). The reporter RNA annealed with G-LEAD_C showed about 30% of translation efficiency of G-SL in HeLa lysates indicating that the translational enhancement by a G-capped leader is not a specific phenomenon of RRL.
Translational efficiency of the reporter RNA annealed with G-LEAD_C was about 12% of that of G-SL mRNA (Supplementary Figure S6). Curiously, the translation efficiency of A-SL is about 2-fold higher than that of the reporter annealed with A-capped leader RNA (Supplementary Figure S6, compare lane 8 with lane 3), for unknown reason. Therefore, simple comparison of translation efficiency of G-capped G-SL mRNA with that of the reporter RNA annealed with G-capped leader (G-LEAD_C) is not appropriate to acquire the degree of translation enhancement by the cap structure. Considering the 2-fold enhancement of the translation of A-SL compared with the translation of the reporter annealed with A-LEAD_C, we can estimate that translation level of the reporter RNA annealed with the G-capped leader is equivalent to ∼24% (12% × 2) of that of G-SL mRNA. Taken these aspects together, about 24% of ribosomes recruited to the 5′-end of an mRNA participate in the translation via bypassing of a stem-loop in RRL.
As a control experiment, we also generated a reporter RNA that is similar to G-SL, but lacks the stem-loop in the 5′UTR (designated as G-w/o SL) to test the effect of the stem-loop on translation. The mRNA G-w/o SL showed the same translational efficiency as G-SL (Supplementary Figure S4, lanes 8 and 10) indicating that the stem-loop (−30 kcal/mol) in the 5′UTR of G-SL does not affect translation, which reconfirms the previous reports suggesting that some stem loop structures do not inhibit translation (15,16). Moreover, the G-capped reporter (G-NO) without annealing of leader RNA, which contains four copies of β-globin 5′UTR sequence, showed about 2.5-fold higher translational efficiency than G-SL and G-w/o SL (compare lanes 8 and 10 with 12 in Supplementary Figure S4). The difference in translation efficiencies of these mRNAs is most likely attributed to the length of mRNAs (G-SL and G-w/o SL are much longer than G-NO). The data also indicate that β-globin sequence per se does not augment the translation of the reporter mRNA annealed to the G-capped leader RNA.
The RNA bridge between leader and reporter RNAs may have been dissociated by the ribosome scanning through it or by a RNA helicase(s) existing in the lysate before the execution of translation causing decreased translation efficiency of G_LEAD_C compared with G-SL. The degree of dissociation of RNA bridge between leader and reporter RNA in translation mixtures was sought by measuring the integrity of RNA association after incubation in such mixtures (Figure 2). About 60% of RNAs remained as RNA complexes in in vitro translation mixtures when leader and reporter RNA complexes were incubated under the translation-competent conditions described in the legend to Figure 1E (Figure 2A, lane 3). Similar amount of RNA remained in complex form in in vitro translation mixtures when leader and reporter RNA complexes were incubated under translation-prohibitive conditions, thus after the addition of the cap analogue described in the legend to Figure 1D (Figure 2A, lane 7). The data indicate that ribosome scanning does not detectably contribute to dissociation of dsRNAs. On the other hand, the addition of AMP-PNP to the translation mixtures, which block the activity of RNA helicase including eIF4A, significantly, but not entirely stabilized the leader–reporter RNA complex (Figure 2B, compare lanes 3 and 7). These data indicate that dsRNAs are dissociated into two distinct RNAs in translation mixtures, either by thermal energy or by the action(s) of RNA helicases present in the reaction mixtures. In addition, the data suggest that the lower translation efficiency seen in the presence of the G-LEAD_C-reporter RNA complex, compared to that noted when G-SL is added, is at least in part attributable to dissociation of annealed RNA in the translation mixture.
Fortuitous covalent bond formation between leader and reporter regions in translation reaction mixtures, which might generate mRNAs containing both the m7G cap and the reporter, was also investigated (Figure 2C). Only very small amounts of annealed RNA (∼1% of total) were observed, after heating, in the region where annealed RNAs band (Figure 2C, compare lanes 5 and 9). The RNAs in this area may be covalently linked or may have re-annealed during cooling. This indicates that either no, or very few, covalent linkages formed between leader and reporter RNAs [mediated by putative pyrophosphatase(s) or RNA ligase(s) in the lysate].
Taken together, these data suggest that cap-dependent translation occurs through a dsRNA bridge that holds the leader and reporter RNAs together, even though the two RNAs are not covalently connected (Figure 2C). These data indicate that some parts of 5′UTR of an mRNA, including stem-loop structures, can be bypassed by a ribosome searching for the initiation codon.
Addition of an upstream open reading frame to the leader RNA impairs translation enhancement by cap
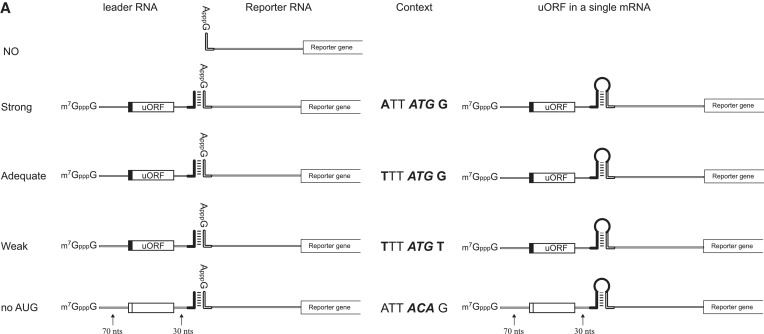
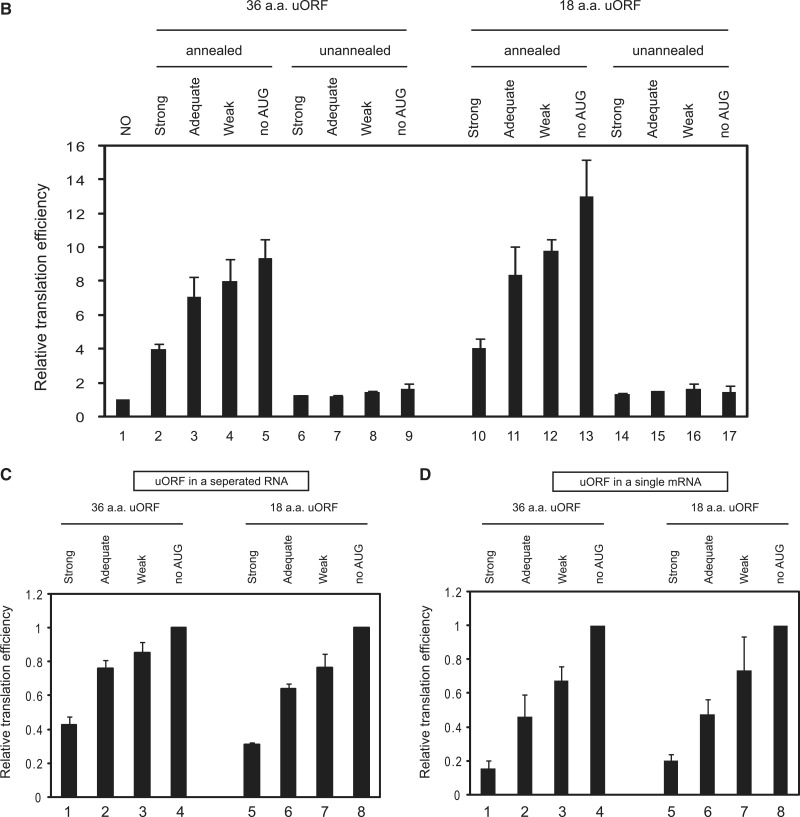
Generally, introduction of an upstream open reading frame (uORF) in the 5′UTR of an mRNA inhibits cap-dependent translation from the authentic initiation codon (23,24). The inhibitory effect of the uORF depends on both the surrounding sequence of the AUG in the uORF and the length of uORF (23,24). An AUG codon with a favourable Kozak context exerts a strong inhibitory effect on translation from the authentic initiation codon (23). Moreover, longer uORFs were reported to exert stronger inhibitory effects on translation, whereas shorter uORFs were more competent to reinitiate translation (24). We thus tested whether the enhancement of translation by capped leader RNA also shows this phenomenon (Figure 3). The uORF with a more favourable Kozak context showed a stronger inhibitory effect on the translation of the reporter gene in a single mRNA, as previously reported (23,24) (Figure 3D). Importantly, similar translational repression by uORF was observed from the reporter gene in a separated reporter RNA when it was annealed with a leader RNA containing a uORF (Figure 3B and C). Moreover, the extent of translational repression by the uORF followed the rule observed from mRNAs containing the uORFs and reporter gene in a single RNA: namely, the uORF with a stronger Kozak context exerted a stronger inhibitory effect on the translation of the reporter gene (Figure 3C, compare lanes 1–3 and 5–7). However, translational repression by uORFs was not affected by the length of uORFs (Figure 3C, compare lanes 1, 2 and 3 with 5, 6 and 7, respectively). These data strengthen the conclusion that a ribosome recruited at the leader RNA is responsible for the translation of the authentic AUG in the reporter RNA. These data also argue against putative ribosome ‘shunting’ from the 5′ proximal region of an mRNA to a point near the initiation codon as a plausible explanation of this phenomenon since such ribosome shunting would ignore the presence of a uORF, and translation from the reporter RNA would be unaffected by the presence of a uORF (8).
Figure 3.
Effects of uORFs in leader RNAs on the translation of reporter RNAs associated with leader RNAs via dsRNA bridges. (A) Schematic diagram of leader and reporter RNAs. The leader RNA contained uORFs encoding oligopeptides [36 or 18 amino acids (a.a.)] preceded by sequences with a strong (ATT ATG G), adequate (TTT ATG G) or weak (TTT ATG T) Kozak context, or no AUG (ATT ACA G) (23). The 5′ terminus of the leader RNA was G-capped, and the 3′-end contained a sequence complementary to the 5′ sequence in the reporter RNA. The reporter RNA is the same as that described in Figure 1A. The uORFs are located 70 nt downstream of the cap structure and 30 nt upstream of the complementary sequence. The spacer regions were originated from the human β-globin gene (left panel). RNA complexes containing both leader and reporter RNAs covalently connected by a stem-loop structure (i.e. within a single molecule) were constructed to serve as control mRNAs (right panel). (B) The RNA annealing process and in vitro translation experiments were performed as described in Materials and Methods, except that the reaction time for in vitro translation was 15 min instead of 60 min. Reporter translational efficiencies were determined by measuring firefly luciferase activity after pre-annealing leader and reporter RNAs (lanes 1–5 and 10–13), or with omission of the pre-annealing step (lanes 6–9 and 14–17). The relative luciferase activity in each translation mixture is shown. The luciferase activity of the reaction without leader RNA was set to unity to permit comparisons. The error bars indicate the standard deviations in six independent experiments. (C) The luciferase activities in the reaction mixtures were further analyzed after normalization to the luciferase activity in the reaction mixture containing leader RNAs with no AUG. Specifically, the luciferase activities in lanes 2–5 were normalized to that in lane 5 in panel (B), and the luciferase activities in lanes 10–13 were normalized to that in lane 13 in panel (B). (D) The translation of reporter RNAs containing both leader and reporter RNAs in a single RNA molecule was monitored by measuring luciferase activity in reaction mixtures. Relative translational efficiency is depicted after normalization to luciferase activity in the reaction mixture containing no uORF (ATT ACA G). Lanes 1–3 and 5–7 depict relative translational efficiencies of mRNAs containing uORFs encoding oligopeptides of 36 and 18 amino acids, respectively.
Cap-dependent translation through a dsRNA bridge in the intercistronic region of a dicistronic mRNA
One possible explanation for the cap-dependent translation through a dsRNA bridge shown in Figures 1 and 3 is the putative ‘scanning-transfer mechanism’. According to this mechanism, a 40 S ribosome reaching the 3′-end of the leader RNA might interact with the reporter RNA through the A-capped 5′-end of the reporter RNA owing to the close proximity of the 3′-end of the leader RNA and the 5′-end of the reporter RNA, even though melting of the dsRNA bridge occurs independent of translation (Figure 2A). To test whether translational enhancement by a capped leader RNA occurs in a 5′-end-independent manner, we used a dicistronic reporter RNA in which a leader RNA was annealed to the intercistronic region (Figure 4A). The dicistronic reporter RNA contains, in order, an A-cap followed by a strong stem-loop structure, the Renilla luciferase gene, an intercistronic region that includes the annealing site and the firefly luciferase gene. The A-cap and stem-loop structure, which inhibits end-dependent translation, was introduced to prevent (or minimize) ribosome effects from reaching the termination codon of the first gene (Figure 4A). In this construct, a ribosome that is dissociated from the leader RNA cannot re-enter the reporter RNA. Attachment of the G-capped leader RNA (G-LEAD_C) to the reporter RNA resulted in more than a 3-fold increase in the translation of a downstream reporter gene (Figure 4B, lane 2). In contrast, A-capped leader RNAs or unassociated G-capped leader RNAs did not affect or just slightly enhanced reporter gene translation (Figure 4B, lanes 3 and 4, respectively). Moreover, translation enhancement was not observed when the annealing step was omitted (Figure 4B, lanes 6 and 7), and the translation of the first gene was not influenced by the association of leader RNA with the reporter RNA (Figure 4C). Translational enhancement of the reporter gene by the capped leader RNA in a dicistronic mRNA was weaker than that in a monocistronic mRNA (compare lane 2 in Figure 4B with lane 2 in Figure 1B). The bulky RNA region upstream of the dsRNA bridge appears to interfere with the skipping of the dsRNA bridge by the ribosome recruited to the cap. Taken together, these data indicate that translational enhancement by capped leader RNA through a dsRNA bridge occurs in a 5′-end-independent manner.
Figure 4.
Enhancement of translation by a G-capped leader RNA associated with a dicistronic reporter RNA via a dsRNA bridge. (A) Schematic diagram of the dicistronic reporter RNA used in this study. The translation of the first gene (Renilla luciferase) was minimized by introducing a stable stem-loop structure at the 5′-end of the reporter RNA. (B and C) Translation reactions were carried out with (lanes 1–5) or without (lanes 6 and 7) a pre-annealing step with A-capped reporter. Translation activity was expressed as light units per fmole reporter RNA in firefly luciferase activity (B) or in Renilla luciferase activity (C). Error bars indicate the standard deviations in three independent experiments.
DISCUSSION
Base-by-base inspection of 5′UTRs by scanning 40 S ribosomal subunits is the generally accepted paradigm for cap-dependent translation of eukaryotic mRNAs. Recently, several reports have suggested that a ribosome may not thoroughly trawl the 5′UTR in search of the initiation codon under certain conditions (11,12). Here, we provide evidence that parts of mRNA are frequently bypassed by ribosomes searching for the initiation codon. We designed an artificial mRNA composed of leader and reporter RNAs that can be associated via a dsRNA bridge. Surprisingly, the translation of the reporter RNA was enhanced up to 9-fold when a G-capped leader RNA was annealed with the reporter RNA (Figure 1B), regardless of the sequence and stability of the annealed region (Supplementary Figure S5). The cap-dependent translation from the uncapped reporter RNA, which is annealed with a capped leader RNA, cannot occur by a ribosome inspecting base-by-base through the dsRNA bridge since the two RNAs are not covalently linked and the annealed RNA would be dissociated by the ribosome passing through it (Figure 1A).
One plausible explanation for the cap-dependent translation by a dsRNA bridge is that the dsRNA bridge was bypassed by a scanning ribosome (11). However, it is not clear how a ribosome could bypass the dsRNA bridge in the presence of eIF1 and RNA helicases such as DHX29, which prevent bypassing of dsRNAs located upstream of the initiation codon (11). In support of this, the rabbit RRL used in these studies contains a substantial amount of DHX29 (11), and the eIF1 in RRL functions adequately, as indicated by the Kozak context-dependent inhibition of reporter gene translation by the uORFs (Figure 3D). Although not tested in these reconstitution experiments (11), it is conceivable that an unidentified protein assists a ribosome in bypassing the dsRNA region. However, the identity of the putative protein remains obscure. It is noteworthy that the translational enhancement of reporter gene by G-capped leader RNA was observed not only in RRL but also HeLa lysates (Figure 1B and Supplementary Figure S7). In fact, translation efficiency of the reporter RNA annealed with G-LEAD_C in HeLa lysates reached up to 30% of that of control mRNA G-SL containing the leader RNA covalented conjugated with the reporter as a sigle mRNA. This indicates that various mammalian cells support bypassing parts of mRNAs by ribosomes searching for the initiation codon. Intriguingly, wheat germ extracts, which were originated from a plant, did not show translational enhancement by G-capped leader annealed with reporter RNA (Supplementary Figure S7B). This indicates that plant cells may lack the putative protein(s) assisting a ribosome in bypassing parts of mRNA. Alternatively, plant cells might contain a protein(s) preventing ribosomal bypassing or a protein(s) strongly dissociating the dsRNA bridge that connects the leader RNA with the reporter RNA.
Skipping of parts of mRNAs by ribosomes was reported recently by Matsuda and Mauro (12), who showed that a specific region of mRNA was bypassed by a ribosome under physiological conditions. Specifically, these authors showed that masking of the first AUG of an mRNA by an antisense, locked nucleic acid oligonucleotide or an EJC resulted in preferential use of a second AUG in COS7 cells (12). These data may indicate that the masked AUG codon was skipped by the scanning ribosome under physiological conditions. In other report, the insertion of a CGG repeat, which forms a highly stable secondary structure potentially capable of blocking ribosomal scanning, showed only mild inhibition of translation in vivo and in vitro (25). Moreover, Dmitriev et al. (26) showed that deletions in the long structured 5′UTR of LINE-1 mRNA have little effect on cap-dependent translation, suggesting neither its large size nor its stem-loop structures appear to be obstacles to the 40 S ribosomal subunit movement.
It is noteworthy that the translation efficiency of reporter RNA non-covalently associated with capped leader RNA reached 20–30% of that of covalently connected RNA (Figure 1B). Considering the rapid dissociation of annealed RNAs by helicases in RRL (Figure 2), this indicates that a substantial level of mRNA translation (20% or more) may occur through ribosomes that skip over a part of the 5′UTR. In conclusion, ribosomes may not always thoroughly scan the 5′UTRs of mRNAs base by base. Instead, scanning ribosomes appear to skip parts of the 5′UTR of an mRNA in search of the initiation codon. The molecular features of the 5′UTR necessary for ribosomal skipping and resumption of ribosomal scanning remain to be elucidated.
SUPPLEMETNARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–7.
FUNDING
Funding for open access charge: NRF BRL [2010-0019706]; Bio R&D program through the NRF [2010-0018167]; WCU program through the NRF [R31-2008-000-10105-0] funded by the Korea government (MEST).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are indebted to Drs. Nahum Sonenberg and Yuri Svitkin (Mcgill University) for critical reading of the manuscript and helpful discussion. The authors also thank Dr. V.P. Mauro (The Scripps Research Institute) for providing us plasmids pGL3c containing various copies of β-globin leader.
REFERENCES
- 1.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl Acad. Sci. USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 3.Kozak M. The scanning model for translation: an update. J. Cell. Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell. Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978;15:1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- 6.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 7.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yueh A, Schneider RJ. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- 9.Spirin AS. How does a scanning ribosomal particle move along the 5′-untranslated region of eukaryotic mRNA? Brownian Ratchet model. Biochemistry. 2009;48:10688–10692. [Google Scholar]
- 10.Spirin AS. The ribosome as a conveying thermal ratchet machine. J. Biol. Chem. 2009;284:21103–21119. doi: 10.1074/jbc.X109.001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abaeva IS, Marintchev A, Pisareva VP, Hellen CU, Pestova TV. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J. 2011;30:115–129. doi: 10.1038/emboj.2010.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda D, Mauro VP. Determinants of initiation codon selection during translation in mammalian cells. PLoS One. 2010;5:e15057. doi: 10.1371/journal.pone.0015057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuda D, Dreher TW. Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA. 2006;12:1338–1349. doi: 10.1261/rna.67906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams MA, Lamb RA. Effect of mutations and deletions in a bicistronic mRNA on the synthesis of influenza B virus NB and NA glycoproteins. J. Virol. 1989;63:28–35. doi: 10.1128/jvi.63.1.28-35.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vassilenko KS, Alekhina OM, Dmitriev SE, Shatsky IN, Spirin AS. Unidirectional constant rate motion of the ribosomal scanning particle during eukaryotic translation initiation. Nucleic Acids Res. 2011;39:5555–5567. doi: 10.1093/nar/gkr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak M. Influence of mRNA secondary structure on binding and migration of 40S ribosomal subunits. Cell. 1980;19:79–90. doi: 10.1016/0092-8674(80)90390-6. [DOI] [PubMed] [Google Scholar]
- 17.Soto Rifo R, Ricci EP, Decimo D, Moncorge O, Ohlmann T. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 2007;35:e121. doi: 10.1093/nar/gkm682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paek KY, Kim CS, Park SM, Kim JH, Jang SK. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J. Virol. 2008;82:12082–12093. doi: 10.1128/JVI.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SM, Paek KY, Hong KY, Jang CJ, Cho S, Park JH, Kim JH, Jan E, Jang SK. Translation-competent 48S complex formation on HCV IRES requires the RNA-binding protein NSAP1. Nucleic Acids Res. 2011;39:7791–7802. doi: 10.1093/nar/gkr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarun SZ, Jr, Sachs AB. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 22.De Gregorio E, Preiss T, Hentze MW. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA. 1998;4:828–836. doi: 10.1017/s1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 24.Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludwig AL, Hershey JW, Hagerman PJ. Initiation of translation of the FMR1 mRNA Occurs predominantly through 5′-end-dependent ribosomal scanning. J. Mol. Biol. 2011;407:21–34. doi: 10.1016/j.jmb.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dmitriev SE, Andreev DE, Terenin IM, Olovnikov IA, Prassolov VS, Merrick WC, Shatsky IN. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Mol. Cell. Biol. 2007;27:4685–4697. doi: 10.1128/MCB.02138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]