Abstract
We present a simple yet efficient technique to monitor the dynamics of DNA-based reaction circuits. This technique relies on the labeling of DNA oligonucleotides with a single fluorescent modification. In this quencher-free setup, the signal is modulated by the interaction of the 3′-terminus fluorophore with the nucleobases themselves. Depending on the nature of the fluorophore's nearest base pair, fluorescence intensity is decreased or increased upon hybridization. By tuning the 3′-terminal nucleotides, it is possible to obtain opposite changes in fluorescence intensity for oligonucleotides whose hybridization site is shifted by a single base. Quenching by nucleobases provides a highly sequence-specific monitoring technique, which presents a high sensitivity even for small oligonucleotides. Compared with other sequence-specific detection methods, it is relatively non-invasive and compatible with the complex dynamics of DNA reaction circuits. As an application, we show the implementation of nucleobase quenching to monitor a DNA-based chemical oscillator, allowing us to follow in real time and quantitatively the dephased oscillations of the components of the network. This cost-effective monitoring technique should be widely implementable to other DNA-based reaction systems.
INTRODUCTION
Various implementations of nucleic acid-based reaction circuits have been demonstrated since DNA was first used as a substrate for in vitro computation of a Hamiltonian path in 1994 (1). DNA was used to encode complex systems such as interactive molecular automata (2,3), as well as computation mimicking neural networks (4), a square-root calculator (5) and robust chemical oscillators (6,7). These information processing systems are composed of many interacting DNA species and yield one or more outputs, typically encoded in the dynamic (6–8) or end-point concentrations (2,4,5) of some oligonucleotides. In order to read out the results of such molecular systems, as well as for the purpose of rationally designing and troubleshooting these DNA reaction circuits, it is desirable to distinguish their different components and monitor the evolution of their concentrations as the reactions proceed.
Methods to observe nucleic acid-based reactions have evolved from post-experiment gel analysis to real-time sequence-specific monitoring. Real-time monitoring of DNA-based reactions is possible thanks to the development of fluorescence techniques that allow detection and quantification of nucleic acids. In the case of isothermal conditions—as generally used for DNA reaction circuits—a further constraint is that the monitoring technique does not interfere too much with the reaction that is monitored. Ideally, the presence or absence of the fluorescent probe has no influence on the kinetics and thermodynamics of the DNA-based reaction circuit under scrutiny.
DNA-binding fluorophores, such as the SYBR family, become highly fluorescent when bound to single- or double-stranded DNA. They can be used to monitor DNA amplification reactions such as polymerase chain reaction (PCR). Some of them, such as SYBRGreen II or Evagreen (9), can also be used to observe isothermal amplification [EXPAR (10,11)]. However, they only provide sequence-unspecific monitoring; in many cases, it is necessary to obtain more detailed information than the total amount of double-stranded DNA in solution. Probes that are specific to a given, arbitrarily selected sequence are then required.
Sequence-specific monitoring can be obtained with fluorescent probes that hybridize to target sequences, leading to a modification of the intensity of their fluorescence. Such fluorescent probes usually consist in oligonucleotides that are dual-labeled with a donor and an acceptor fluorophore. Through fluorescence resonance energy transfer (12), the acceptor acts as a quencher of the donor and the quenching efficiency strongly depends on the distance between the two fluorophores. Probes bear the complementary sequence of their target, which allow them to hybridize to it. Hybridization and following reactions lead to the separation of donor and acceptor, subsequently dequenching the fluorescence of the donor. For instance, in the case of PCR TaqMan probes (13), depolymerization of the hybridized probe separates donor and acceptor. For Molecular Beacon (14), donor and acceptor are initially brought close to each other by the probe's hairpin structure. The probe opens as it hybridizes to its target, which increases the distance between donor and acceptor.
Besides classic DNA amplification techniques [such as real-time PCR (13) or EXPAR (10,11)], other types of DNA systems also require sequence-specific real-time monitoring. This work focuses on DNA reaction circuits, which are complex reactive assemblies of many DNA strands able to perform some form of pre-encoded program (5–7). Such systems generally require the design of custom monitoring solutions. In some cases, it is still possible to readapt the conventional donor/acceptor pair of fluorophores: DNA-based molecular automaton MAYA (2) uses a fluorogenic substrate with a donor at one end and an acceptor at the other. Cleavage of this substrate separates donor and acceptor, which produces an irreversible dequenching of the donor fluorescence. Also, most DNA-based molecular machines (15,16) use various donor/acceptor pairs of fluorophores to monitor the molecular motions associated with the machine functioning (17).
Donor and acceptor can also be placed on two separate and complementary DNA molecules. In this case, hybridization of the two strands brings donor and acceptor close to each other, which quenches the fluorescence of the donor (4,5,18,19). However, this technique significantly impacts the thermodynamics of the labeled complementary strands (20).
The most complex DNA reaction circuits are out-of-equilibrium systems that are able to display emergent behaviors such as oscillations (6,7), multi-stability (21) or—theoretically—chaotic trajectories (8). Such circuits display non-monotonous concentration evolutions and generally require reversible fluorescence reporting. Moreover, labeled probes can be difficult to use in these systems because some DNA strands are continuously produced and destroyed (7); therefore, a simple, general and non-disruptive monitoring technique would be a welcome addition to the field of molecular programming.
Direct quenching by adjacent nucleobases is a somehow neglected effect where the fluorescence of a single DNA-bound fluorophore is modulated by interactions with the neighboring DNA sequence (17,20,22). Each nucleoside has a different quenching effect on nearby fluorophores, with guanosine exhibiting the highest quenching efficiency (22). Moreover, the quenching ability of each base strongly depends on its paired or unpaired status, leading to fluorescence changes upon duplex formation. Using this property, DNA hybridization (23) and PCR (24,25) have been monitored.
In this work, we show that nucleobase quenching (referred to as ‘N-quenching’ hereafter) provides an efficient method for real-time multiplexed monitoring of DNA reaction circuits with complex dynamics. By labeling the 3′ end of a ‘template’ oligonucleotide with a single fluorophore, hybridization and separation of the complementary ‘signal’ oligonucleotide can be monitored. N-quenching is highly sequence-specific: a non-complementary sequence or a sequence hybridizing a few bases away from the fluorophore is readily distinguished from the target sequence. Regarding short oligonucleotides, N-quenching sensitivity is relatively high compared with DNA-intercalating fluorophores. It is a cost-effective technique that only requires one fluorophore per target oligonucleotide, with no need for additional probes. N-quenching is thus non-invasive and compatible with dynamic DNA reaction circuits. As an implementation example, we monitored the signal oligonucleotides of an autonomous DNA-based chemical oscillator by directly labeling the sequences of interest. N-quenching is a versatile monitoring technique that should be easily implemented to various DNA-based reaction systems.
MATERIALS AND METHODS
Oligonucleotides
All DNA oligonucleotides were purchased from either Integrated DNA Technologies (IDT, Coralville, IA, USA) or biomers.net (Ulm, Germany), with high performance liquid chromatography purification. Concentrations were determined by measuring the absorbance at 260 nm using a GeneQuant Pro RNA/DNA Calculator (GE Healthcare). Using DinaMelt (26), we checked that all sequences used in this study did not display secondary structures at the working temperatures.
Fluorescence shift measurement
For fluorescence intensity shift curves upon temperature-induced hybridization, oligonucleotides were diluted in a buffer containing 100 mM NaCl and 0.1% Synperonic F108 (Sigma-Aldrich) in TE buffer (pH 8.0). Oligonucleotides were used at a concentration of 100 nM for labeled 22-bases long ‘templates’ and 300 nM for 11-bases long ‘signals’. Hybridization and separation were induced by alternating between temperatures lower and higher than the duplexes' melting temperatures. Temperatures were determined so that NUPACK (27) predicts <5% of template strands hybridized at ‘high’ temperature and >95% of template strands hybridized at ‘low’ temperature. Fluorescence of 20 μl samples covered with 15 μl of mineral oil was recorded using an IQ5 real-time thermocycler (Bio-Rad).
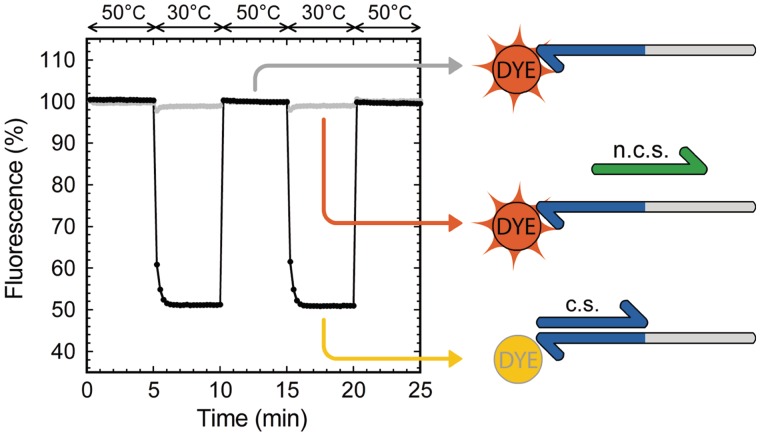
For the experiment shown in Figure 1, a ‘template’ oligonucleotide (5′-AGATGACTCTC-CTTAGACTCAG-3′) bearing a 3′-terminal TAMRA NHS ester modification was used with either a ‘signal’ complementary sequence (5′-CTGAGTCTAAG-3′) or a non-complementary sequence (5′-AACAGACTCGA-3′).
Figure 1.
TAMRA fluorescence quenching upon temperature induced hybridization/separation. Fluorescence intensity is expressed as a percentage of the fluorescence of the TAMRA-labeled template put alone in solution. Presence of the complementary sequence (c.s.) induces a fluorescence shift (black curve) when the temperature is lower than the duplex melting temperature, whereas presence of a non-complementary sequence (n.c.s) does not have influence on the fluorescence of TAMRA (gray curve).
Monitoring of DNA reaction circuits
Reactions were assembled in a buffer containing 10 mM KCl, 10 mM (NH4)2SO4, 50 mM NaCl, 2 mM MgSO4, 45 mM Tris–HCl, 5 mM MgCl2, 6 mM DTT, 100 μg/ml bovine serum albumin (New England Biolabs), 410 mM trehalose, 1× EvaGreen and dNTPs (100 μM each). Bst DNA polymerase, large fragment, Nt.BstNBI nickase and RecJf exonuclease were purchased from New England Biolabs and used at 8, 40 and 12 U/ml, respectively. Samples of 40 μl were observed using an IQ5 real-time thermocycler (Bio-Rad) set at a constant temperature of 38.5°C.
For monitoring the single steady-state network, 50 nM of template A (5′-CTTAGACTCAG-CTTAGACTCAG-3′) with 3′-terminal TAMRA NHS ester modification was put in presence of an initial concentration of 0.1 nM of signal α. In the case of the oscillator, templates A, αtoβ (5′-AGATGACTCTC-CTTAGACTCAG-3′) with 3′-terminal TAMRA NHS ester modification and βtoiα (5′-TTACTCAGCTTAGAC-AGATGACTCTC-3′) with Alexa Fluor 594 NHS ester modification were used at concentrations of 40, 5 and 20 nM. All templates bore two phosphorothioate backbone modifications at their 5′-end to protect them from hydrolysis by RecJf exonuclease (7).
RESULTS
Characterization of nucleobase quenching
We initially observed that adding the complementary strand to a 3′-terminal fluorophore labeled oligonucleotide in solution produced a shift of fluorescence intensity. To further characterize the phenomenon and the possibility to use it in our assay, we selected a 22-bases long oligonucleotide ‘template’ labeled with TAMRA at its 3′-end (Figure 1). This template was put either in the presence of a 11-bases long ‘signal’ complementary sequence or a 11-bases long non-complementary sequence. Signal oligonucleotide hybridized adjacently to the template 3′-terminal dye. We induced hybridization and separation of the strands by applying temperature cycles.
This allowed us to observe the effect of hybridization on the intensity of fluorescence emission of the 3′-terminal dye. Figure 1 shows the fluorescence intensity shift obtained by cycling between temperatures higher and lower than the duplex melting temperature. At ‘low’ temperature, TAMRA showed a 50% drop of fluorescence intensity in the presence of the complementary strand, whereas a non-complementary sequence did not produce any significant shift of fluorescence intensity. Cycling the temperature several times confirmed the reversibility of the phenomenon.
Environmental dependence
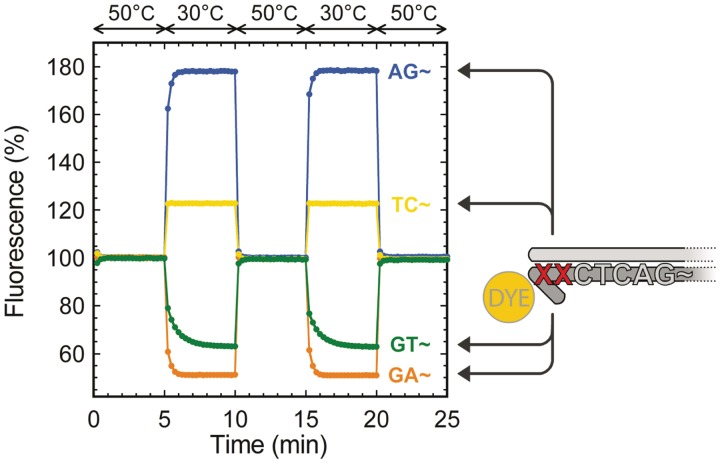
The fluorescence intensity shift upon hybridization depends on the direct environment of the fluorophore. We investigated this property by tuning the fluorophore's nearest bases. In the case of TAMRA, Figure 2 shows the fluorescence intensity shifts for four combinations of the two bases before the 3′-terminal fluorophore. Depending on these last two bases, we observed either a decrease or an increase of the fluorescence of TAMRA upon hybridization of the two complementary strands: the fluorescence increased for terminal 3′-AG and 3′-TC, and decreased for terminal 3′-GT and 3′-GA. These results globally agree with the trends reported by Nazarenko et al. (28) for internally labeled oligonucleotides: the formation of a terminal C–G pair strongly quenches the fluorophore, whereas the hybridization of a complementary strand globally dequenches the fluorophore. We tested other combinations of the last two bases (data not shown), which all produced results consistent with this generalization of the rules reported by Nazarenko et al. (28). Although the position of the dye is not really important in our case, the nature of the 3′-terminal bases determines the direction—positive or negative—of the fluorescence intensity shift.
Figure 2.
TAMRA fluorescence quenching upon temperature induced hybridization/separation for different pairs of 3′-terminal bases. Fluorescence intensity is expressed as a percentage of the fluorescence of the TAMRA-labeled template put alone in solution. Four combinations of the template's two 3′-terminal bases XX (AG, TC, GT, GA) show negative or positive fluorescence intensity shifts upon hybridization of the complementary sequence.
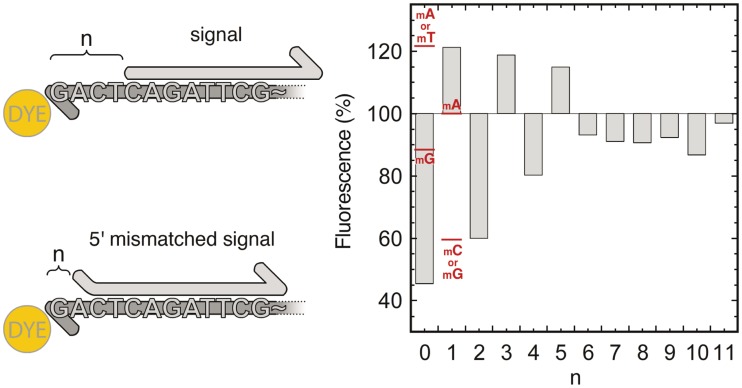
To assess the specificity of N-quenching for the target 11-bases long signal, we compared the shift of fluorescence intensity induced by the target signal (blunt end) with that of a signal strand moved from 1 to 11 bases away (dangling end) from the fluorophore (Figure 3). The amplitude of the intensity shift effectively decreased as the distance between the dye and the first base pair increased. As this distance n increased, we observed positive or negative fluorescence intensity shifts depending on the nature of the dye's nearest base pair (i.e. the terminal base pair). The intensity shift was positive for a terminal A–T and negative for a terminal C–G base pair. Using the same assay, this trend was confirmed on another sequence (Supplementary Figure S1). Following this observation, we could very clearly discern a signal oligonucleotide hybridized at position n = 0 (negative shift) from one located a single base away, at position n = 1 (positive shift).
Figure 3.
Fluorescence intensity shift upon hybridization of a signal oligonucleotide moved from n = 0 to 11 bases away from the template 3′-terminal dye. Red marks show the fluorescence intensities for a signal oligonucleotide hybridizing with a single 5′-mismatch (mA, mT, mC or mG). Fluorescence intensity is expressed as a percentage of the fluorescence of the TAMRA-labeled template put alone in solution.
We also explored the case of an imperfect match between the template and the 5′-end of the signal molecule. As can be seen in Figure 3 (red marks), the fluorescence change still primarily depends on the base pair nearest to the fluorophore. Only A–A and G–G mismatches appeared to depart from this rule, with no obvious rationale.
From n = 6 to 11, we observed weak and position-independent fluorescence intensity shifts. We tentatively attribute this to the rigidification of the DNA coil. Our target implementation (described below) only requires distinction between signal molecules that hybridize at the 3′-end of the template from those that hybridize 11 bases away. In this configuration, N-quenching provides a reliable sequence-specific monitoring technique.
Monitoring an elementary DNA reaction circuit
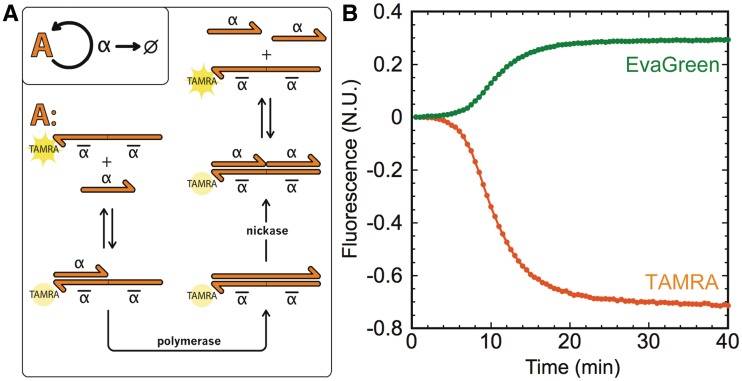
We used N-quenching to monitor the evolution of an elementary DNA-based dynamic system encoding homeostasis. As shown on Figure 4, this network consists of one template ‘A’ that, in presence of a polymerase and a nicking enzyme, encodes for an autocatalytic amplification of its signal α. In the additional presence of an exonuclease that specifically degrades signal molecules α (7), but not template A, this circuit becomes a dynamic, out-of-equilibrium system that possesses a single steady state: as long as dNTPs are available, the concentration of α will always evolve toward a given constant value.
Figure 4.
Monitoring an elementary DNA reaction circuit. (A) Template A encodes the autocatalytic amplification of signal α and bears a TAMRA dye at its 3′-end. As α hybridizes to the 3′-end of template A, it gets elongated by a polymerase. The upper strand of the duplex is then cut in its middle by a nicking enzyme, and signal α and output α are released. The exonuclease specifically degrades single-stranded α. (B) The reaction is triggered with 0.1 nM of α and is monitored with both EvaGreen intercalating dye and the 3′-terminal TAMRA of template A. EvaGreen fluorescence intensity increases as α-A duplexes are formed, and TAMRA fluorescence is quenched as α hybridizes to the 3′-end of A. Both fluorescence intensities are normalized with respect to the maximal shift of fluorescence intensity of EvaGreen for the reaction where the TAMRA modification of template A is replaced by a phosphate.
To test N-quenching, template A was labeled at its 3′-end with TAMRA, allowing us to monitor the concentration of α as it binds to the template. Hybridization of signal α (5′-CT∼) on template A induces a quenching of TAMRA fluorescence. Therefore, the template itself becomes a probe for measuring the concentration of α. As a control, we simultaneously monitored the reaction in the presence of an intercalating dye (EvaGreen) whose fluorescence increases when binding to double-stranded DNA. In Figure 4, we observe that, as expected, the concentration of α evolves toward a steady state: EvaGreen-induced fluorescence increases, TAMRA fluorescence decreases and both eventually reach a plateau that corresponds to the steady state. In this assay, the fluorescence intensity shift observed with N-quenching has twice the amplitude observed for EvaGreen. Also, EvaGreen and N-quenching yielded fluorescent recordings with similar shapes, suggesting that dynamic DNA reaction circuits can be precisely monitored by using N-quenching only.
Monitoring a DNA-based oscillator
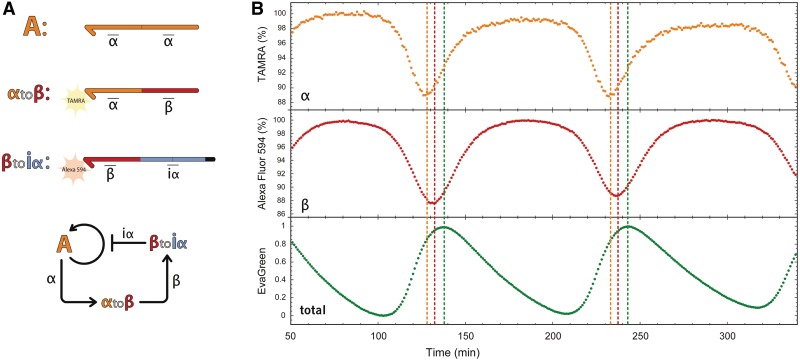
N-quenching was then used to study a network containing more than one dynamic species. We previously reported a DNA-based oscillator (7) that uses the same enzymes cocktail but is encoded in the sequences of three templates (Figure 5). Template A activates the autocatalytic production of signal molecule α. Template αtoβ receives α as input, activating the production of β. Finally, template βtoiα receives β as input, activating the production of iα. iα closes the negative feedback loop: it inhibits α production by blocking the activity of template A. Overall, this chain of reactions produces oscillations of the three signal molecules α, β and iα.
Figure 5.
Multiplexed monitoring of a DNA-based oscillator. (A) 3-Nodes oscillator network and sequence encoding. A is the autocatalytic module of Figure 3. The second template, αtoβ, receives α as input, produces β as output and is labeled in 3′ with TAMRA. The third template, βtoiα, receives β as input, produces iα as output and is labeled in 3′ with Alexa Fluor 594. (B) Time evolution of the oscillator in three colors. α is seen with TAMRA, β with Alexa Fluor 594 and EvaGreen shows the total duplex concentration, roughly corresponding to iα concentration (7). Fluorescence of TAMRA and Alexa Fluor 594 is expressed as a percentage of their respective unquenched fluorescence. Fluorescence of EvaGreen is normalized at 1 for the highest and 0 for the lowest fluorescence intensity. Vertical lines show the peak of each species concentration.
When this reaction circuit is monitored with EvaGreen as previously reported (7), one obtains real time, but non-specific information about the total amount of duplex DNA in the system. In fact, in this case, EvaGreen fluorescence is mainly induced by iα (7), which mostly prevents the observation of α and β. However, because oscillations are produced by the interplay of the three dynamic species, a complete characterization requires individual tracking of α and β concentrations as well. This information is readily obtained using N-quenching: we respectively labeled αtoβ and βtoiα 3′-ends with TAMRA and Alexa Fluor 594, which enabled sequence-specific observation of both α and β. Sequences of α (5′-CTGA∼) and β (5′-GAGA∼) produced a negative shift of fluorescence intensity upon hybridization (with formation of a terminal C–G base pair). With these two labeled oligonucleotides, we could directly observe the phase shifts between α, β and iα concentration peaks: as expected from the structure of the network, the peak of α came first, followed by β and then iα, before the cycle started again.
Using N-quenching, we could also extract quantitative information about the concentrations of α and β throughout the reaction. To do so, we built calibration curves for αtoβ and βtoiα, showing their fluorescence intensity shift as a function of known concentrations of, respectively, α and β. By comparison with these calibration curves, we found that α and β concentrations do not exceed 30 and 55 nM, respectively, at their peak concentration. Also, assuming a linear relationship between the quenching effect and the ratio of hybridized templates, we could deduce that <20% of αtoβ and <25% of βtoiα are in double-strand form at the oscillation peaks.
DISCUSSION
N-quenching sensitivity and quantitative measurement
Using N-quenching, dynamic DNA reaction circuits can be monitored by simply labeling sequences of interest with a single fluorophore. Even though the shifts in fluorescence intensity (up to −50%/ + 80%) are not as high as those obtained by using donor/acceptor pairs of fluorophores (almost 100% quenching for some donor/acceptor pairs), they were sufficient to observe reactions on 20 or 40 μl volumes using a conventional real-time thermocycler (the signal-to-noise ratio in the experiment of Figure 1 is ∼250). In the case of the simple DNA reaction circuit of Figure 4, the relative fluorescence intensity shift produced by a 11-bases long oligonucleotide with TAMRA was higher than the one obtained with EvaGreen. One may remark that in these conditions, the fluorescence of EvaGreen is partially quenched by TAMRA (29). Still, the shift of TAMRA fluorescence intensity is comparable to the one of EvaGreen, using a non-labeled template. This result might be explained by the weak affinity of EvaGreen for short double-stranded DNA (9), and the fact that EvaGreen fluorescence intensity depends on the number of paired bases. Thus, in the case of short DNA strands—here, 11 bases—the sensitivity of N-quenching is comparable to that of EvaGreen intercalating dye. When it comes to monitoring hybridization of strands even shorter than 11 bases, the relative sensitivity should be even greater.
N-quenching allows quantitative measurement of dynamically changing concentrations of target signal molecules. Under the current conditions, we could quantify target signal molecules in concentrations ranging from a few nanomolar up to several hundreds of nanomolar. Because the working temperature is higher than the melting temperature of the target signal molecules, it is possible to quantify concentrations of signal molecule higher than the concentration of labeled template. Moreover, even without calibration, it is possible to quantify the concentration of hybridized template by comparison with the fluorescence of the unoccupied (or saturated) template.
Non-invasive monitoring
In our specific application, single fluorophores are directly attached to the template oligonucleotides that encode the DNA reaction circuit. This way, N-quenching is implemented directly on the circuit rather than being a probe added to the system. Therefore, our expectation was that N-quenching would not significantly interfere with the thermodynamics and kinetics of the system itself.
On the contrary, several studies have reported strong duplex stabilizing effects for donor/acceptor pairs of modifications (18,20,30). For example, Moreira et al. (30) reported that, for dual-labeled oligonucleotide probes, the presence of the two fluorescent modifications increased the melting temperature (Tm) of the probe by up to 4.3°C. In the case of two complementary strands bearing a 5′-terminal donor for one and a 3′-terminal acceptor for the other, an increase of the Tm of the duplex of up to 10°C was reported (20). Such thermodynamic alterations are enough to disrupt the functioning of DNA reaction circuits (19). Moreover, these effects are difficult to predict computationally and may depend on a variety of factors (20). Therefore, specific strategies need to be devised to circumvent this issue: for example, DNA strand displacement reactions usually use a separate ‘probe’ complex rather than directly labeling the sequences involved in the functions of the circuit (4,5,19).
By comparison, the maximal Tm increase found for a single 5′-terminal fluorophore was of only 1.6°C for Cy3 and Cy5 dyes (30). Compared with monitoring techniques that use pairs of donor/acceptor, the stabilizing effect of a single fluorophore is much lower and less disruptive; however, it should be considered when using N-quenching. In our case, whereas the single fluorophore labeling had quantitative effects on the system, it did not modify its global kinetics: oscillations were obtained both with or without the fluorescent modifications.
N-quenching as a general method to monitor position-specific hybridization
Some fluorophores exhibited greater fluorescence intensity shifts than others, and some did not show any change in fluorescence upon hybridization, following the trend previously reported (23,28). Among the fluorophores we tested, N-quenching worked well for FAM, JOE, TAMRA Alexa Fluor 594, DY-530, DY-636 and DY-681. On the other hand, TEX 615, Atto 633 and Cy5 did not exhibit fluorescence intensity shifts upon hybridization and were consequently not used for N-quenching (data not shown). The attachment chemistry of the fluorophore also affects the efficiency of N-quenching: for a given sequence, TAMRA exhibited a larger fluorescence intensity shift when conjugated through NHS ester than when attached with a C6 spacer.
In contrast to other quenching methods, the fluorescence intensity shift can be either positive (terminal base pair C–G) or negative (terminal base pair A–T). By tuning the terminal nucleotides, it is possible to distinguish a signal molecule binding adjacently to the quencher from signal molecules binding one or more bases away. This unambiguous detection could be used to cheaply distinguish single nucleotide polymorphisms (31). It may also be used to distinguish invading strands (16) whose toeholds differ by as little as one nucleotide.
In this work, we devised a monitoring technique that relies on a single fluorophore labeling and quenching by nucleobases. We demonstrated the efficiency of N-quenching by monitoring the hybridization and melting of 11-bases long oligonucleotides in a sequence-specific manner. The sensitivity of N-quenching is lower than that of fluorescent monitoring techniques based on donor/acceptor pairs of fluorophores. However, we showed that it is sufficient to detect nanomolar concentrations of short oligonucleotides in microliter-scale volumes. N-quenching can be easily implemented to dynamic DNA reaction circuits and used to deduce rich quantitative information about the dynamics of the system. Also, by tuning the fluorophore's nearest nucleotides, it is possible to obtain unambiguous position information about the incoming signal oligonucleotide. Moreover, using a single fluorophore is cheaper than using a pair of fluorophore and quencher and also has a lower impact on DNA kinetics and thermodynamics. Therefore N-quenching should be widely implementable to other DNA-based systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
CNRS and a Grant-in-Aid for Scientific Research from MEXT, Japan [No. 23119006]. Funding for open access charge: MEXT grant.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Anthony Genot, Hervé Guillou and Kevin Montagne for discussion and advice.
REFERENCES
- 1.Adleman LM. Molecular computation of solutions to combinatorial problems. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 2.Stojanovic MN, Stefanovic D. A deoxyribozyme-based molecular automaton. Nat. Biotechnol. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- 3.Macdonald J, Li Y, Sutovic M, Lederman H, Pendri K, Lu W, Andrews BL, Stefanovic D, Stojanovic MN. Medium scale integration of molecular logic gates in an automaton. Nano Lett. 2006;6:2598–2603. doi: 10.1021/nl0620684. [DOI] [PubMed] [Google Scholar]
- 4.Qian L, Winfree E, Bruck J. Neural network computation with DNA strand displacement cascades. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- 5.Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Winfree E. Synthetic in vitro transcriptional oscillators. Mol. Syst. Biol. 2011;7:465. doi: 10.1038/msb.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montagne K, Plasson R, Sakai Y, Fujii T, Rondelez Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 2011;7:466. doi: 10.1038/msb.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soloveichik D, Seelig G, Winfree E. DNA as a universal substrate for chemical kinetics. Proc. Natl. Acad. Sci. USA. 2010;107:5393–5398. doi: 10.1073/pnas.0909380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao F, Leung W-Y, Xin X. Characterization of EvaGreen and the implication of its physicochemical properties for qPCR applications. BMC Biotechnol. 2007;7:76. doi: 10.1186/1472-6750-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ness JV, Ness LKV, Galas DJ. Isothermal reactions for the amplification of oligonucleotides. Proc. Natl Acad. Sci. USA. 2003;100:4504–4509. doi: 10.1073/pnas.0730811100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan E, Erwin B, Dames S, Ferguson T, Buechel M, Irvine B, Voelkerding K, Niemz A. Specific versus nonspecific isothermal DNA amplification through thermophilic polymerase and nicking enzyme activities. Biochemistry. 2008;47:9987–9999. doi: 10.1021/bi800746p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stryer L, Haugland RP. Energy transfer: a spectroscopic ruler. Proc. Natl Acad. Sci. USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holland P, Abramson R, Watson R, Gelfand D. Detection of specific polymerase chain reaction product by utilizing the 5′→3′exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl Acad. Sci. USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 15.Mao C, Sun W, Shen Z, Seeman NC. A nanomechanical device based on the B–Z transition of DNA. Nature. 1999;397:144–146. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- 16.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 17.Cooper JP, Hagerman PJ. Analysis of fluorescence energy transfer in duplex and branched DNA molecules. Biochemistry. 1990;29:9261–9268. doi: 10.1021/bi00491a022. [DOI] [PubMed] [Google Scholar]
- 18.Morrison LE, Stols LM. Sensitive fluorescence-based thermodynamic and kinetic measurements of DNA hybridization in solution. Biochemistry. 1993;32:3095–3104. doi: 10.1021/bi00063a022. [DOI] [PubMed] [Google Scholar]
- 19.Zhang DY, Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 2009;131:17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- 20.Marras SAE, Kramer FR, Tyagi S. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res. 2002;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, White KS, Winfree E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2006;2:68. doi: 10.1038/msb4100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seidel C, Schulz A, Sauer M. Nucleobase-specific quenching of fluorescent dyes. 1. Nucleobase one-electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem. 1996;100:5541–5553. [Google Scholar]
- 23.Torimura M, Kurata S, Yamada K, Yokomaku T, Kamagata Y, Kanagawa T, Kurane R. Fluorescence-quenching phenomenon by photoinduced electron transfer between a fluorescent dye and a nucleotide base. Anal. Sci. 2001;17:155–160. doi: 10.2116/analsci.17.155. [DOI] [PubMed] [Google Scholar]
- 24.Crockett AO, Wittwer CT. Fluorescein-labeled oligonucleotides for real-time PCR: using the inherent quenching of deoxyguanosine nucleotides. Anal. Biochem. 2001;290:89–97. doi: 10.1006/abio.2000.4957. [DOI] [PubMed] [Google Scholar]
- 25.Kurata S, Kanagawa T, Yamada K, Torimura M, Yokomaku T, Kamagata Y, Kurane R. Fluorescent quenching-based quantitative detection of specific DNA/RNA using a BODIPY FL-labeled probe or primer. Nucleic Acids Res. 2001;29:e34. doi: 10.1093/nar/29.6.e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 28.Nazarenko I, Pires R, Lowe B, Obaidy M, Rashtchian A. Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res. 2002;30:2089–2195. doi: 10.1093/nar/30.9.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howell WM, Jobs M, and Brookes AJ. iFRET: an improved fluorescence system for DNA-melting analysis. Genome Res. 2002;12:1401–1407. doi: 10.1101/gr.297202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreira BG, You Y, Behlke MA, Owczarzy R. Effects of fluorescent dyes, quenchers, and dangling ends on DNA duplex stability. Biochem. Biophys. Res. Commun. 2005;327:473–484. doi: 10.1016/j.bbrc.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian HKK, Chakraborty B, Sha R, Seeman NC. The label-free unambiguous detection and symbolic display of single nucleotide polymorphisms on DNA origami. Nano Lett. 2011;11:910–913. doi: 10.1021/nl104555t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.