Abstract
DNA built from modular repeats presents a challenge for gene synthesis. We present a solid surface-based sequential ligation approach, which we refer to as iterative capped assembly (ICA), that adds DNA repeat monomers individually to a growing chain while using hairpin ‘capping’ oligonucleotides to block incompletely extended chains, greatly increasing the frequency of full-length final products. Applying ICA to a model problem, construction of custom transcription activator-like effector nucleases (TALENs) for genome engineering, we demonstrate efficient synthesis of TALE DNA-binding domains up to 21 monomers long and their ligation into a nuclease-carrying backbone vector all within 3 h. We used ICA to synthesize 20 TALENs of varying DNA target site length and tested their ability to stimulate gene editing by a donor oligonucleotide in human cells. All the TALENS show activity, with the ones >15 monomers long tending to work best. Since ICA builds full-length constructs from individual monomers rather than large exhaustive libraries of pre-fabricated oligomers, it will be trivial to incorporate future modified TALE monomers with improved or expanded function or to synthesize other types of repeat-modular DNA where the diversity of possible monomers makes exhaustive oligomer libraries impractical.
INTRODUCTION
Custom gene synthesis continues to grow in its importance for molecular and synthetic biology (1). Currently, DNA constructs longer than ∼200 bp are usually produced by assembly polymerase chain reaction (PCR) or ligation (1–3) of multiple oligonucleotides that are themselves synthesized by sequential chemical coupling of nucleotides on a glass column (4) or microarray (5). Recently developed assembly-from-microarray approaches (6,7) in particular promise drastic reductions in time and costs of gene synthesis. However, DNA constructs built from repetitive modules have not been amenable to these methods because of the high likelihood of oligonucleotides incorporating into erroneous parts of constructs (8,9). Therefore, new methods of assembling repeat-module DNA would be highly desirable, especially ones that are automatable or highly scalable. We designed a method, which we refer to as iterative capped assembly (ICA; Figure 1), that involves rapid assembly of repeat-module DNA by sequential ligation of monomers on a solid support together with capping oligonucleotides to increase the frequency of full-length products. The capping oligonucleotides act by ligating to and inactivating incomplete chains generated during the process by imperfect monomer ligation efficiency. We reasoned that one ideal application for ICA would be rapid parallelized construction of transcription activator-like (TAL) effector proteins (10) for genome engineering. These proteins originally from Xanthomonas bacteria (11) carry a DNA-binding domain (referred to here as the TALE) consisting of a repeating chain of nearly identical 34-amino acid monomers. Each monomer addresses a single DNA target base when the TALE anneals to the major groove of DNA (12,13), with four common monomer variants (differing only at the repeat-variable diresidue (RVD) at positions 12 and 13 (10)) each optimally binding one of the four DNA base pairs. TALEs are therefore of great interest to genome engineering since their RVD monomers can be programmably rearranged to form custom DNA-binding domains that, when fused to other user-specified domains, may allow targeting of a vast range of proteins and other molecules to particular genomic locations both in vitro and in vivo. One common application is the design of pairs of TALEs each fused to monomers of the obligate dimer-acting FokI nuclease, which together create a target locus-specific endonuclease (TALE-nuclease or TAL effector nucleases (TALEN) (14)). When two TALENs bind to closely spaced target DNA sites, FokI will dimerize and create a local double-strand DNA break, allowing targeted gene knockout if the cell repairs the break by non-homologous end-joining (NHEJ; (15,16)) or incorporation of donor DNA sequences (17) by homologous recombination or other repair mechanisms. Here, we apply ICA to custom TALEN production and demonstrate efficient assembly of TALEs up to 21 monomers long followed by ligation into a nuclease backbone vector all within 3 h. We show that the capping oligonucleotides are essential for generation of pure full-length products. To investigate the activity of TALENs synthesized by our method, we produced 20 TALEN variants with a range of lengths and measured the ability of 19 of them to stimulate gene editing by a single-stranded donor oligonucleotide of a reporter gene in human cells. All of the TALENs we tested facilitated donor incorporation, providing to our knowledge the first demonstration of TALEN-mediated gene editing by an oligonucleotide. Although several methods for TALE assembly have been reported (15,18–20), ICA is, to our knowledge, unique in its ability to build TALEs in an automatable fashion from individual monomers; this property should allow facile introduction of future improved TALE monomers and application of ICA to other modular constructs built from larger monomer repertoires than that of TALEs. ICA may also allow ultra-high-throughput synthesis of repeat-module DNA using microarray printing technology.
Figure 1.
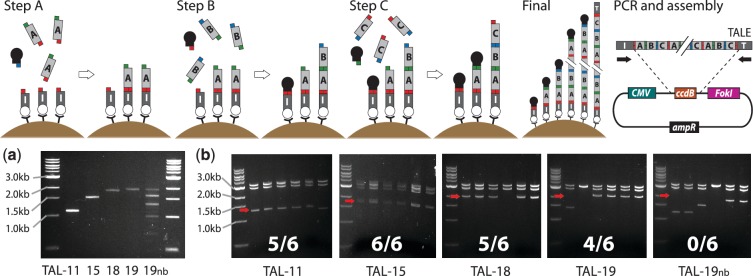
ICA of repeat-module DNA. ICA assembles repeat monomers into chains in a cycling A-B-C ligation-wash pattern with capping oligonucleotides to block incompletely extended chains. In each A step, monomer A is ligated to monomer C (except the first ABC cycle (shown) where A is ligated to a biotinylated initiator oligonucleotide immobilized on streptavidin beads). A hairpin capping oligonucleotide (black) is included to cap unligated monomer B (not relevant in the first cycle). In B steps, monomer B is ligated to monomer A and a capping oligo is included to cap unligated monomer C from the previous step (or unligated initiator in the first cycle). In C steps, monomer C is ligated to monomer B and a capping oligo is included to cap unligated monomer A from the previous step. This ABC cycling pattern continues until a final ligation with a terminator oligonucleotide (T, Final). A single exception occurs in the sixth step where a modified C monomer, Cseq, is used that allows later annealing of a sequencing primer. After the final ligation, products are eluted from the beads and briefly amplified by PCR before Golden-Gate assembly into a backbone vector, e.g. assembly of TALEs into a FokI nuclease-carrying plasmid, using BsaI. (a) Post-elution PCR products for TALES of length 11, 15, 18 and 19 monomers, plus the same 19-mer synthesized without capping oligos (nb). (b) AfeI digestions of TALE-nuclease (TALEN) plasmids from six randomly picked colonies per construct. Expected sizes of full-length TALE insert bands are labeled with red arrows, and the full-length clone frequencies are shown.
MATERIALS AND METHODS
Sequences of all oligonucleotides used in this study are in Supplementary Table S1; double-stranded oligonucleotide pairing schemes are in Supplementary Table S2. All TALEN designs are in Supplementary Table S3. Maps of the TALEN backbone and defective EGFP reporter lentiviral plasmids are in Supplementary Files 2.
Monomer construction
TALE monomers were prepared by PCR and purification followed by BsmBI digestion and purification. Each 100 µl PCR contained 50 µl 2× Phusion HF master mix (NEB), 400 nM each primer and 4 ng template RVD plasmid, taken from (18). PCR was performed with 98°C for 2 min followed by 25 cycles at 98°C for 20 s; 60°C for 30 s and 72°C for 20 s. Products were purified on QIAquick columns (Qiagen) with two buffer PE washes and eluted in 50 µl elution buffer (EB). Typically, three 100 µl PCRs were pooled onto each QIAquick column. Forty-five microliters of each purified product was included in a 100 -µl reaction containing 1× NEBuffer 4, 0.1 mg/ml bovine serum albumin (BSA) and 3 µl BsmBI. Reactions were incubated at 55°C for 3 h and then purified with QIAquick columns, eluting in 50 µl EB.
ICA ligation procedure
Before the ligation steps, all double-stranded oligonucleotides were prepared by mixing equal volumes of the two relevant single-stranded oligonucleotides at 100 µM each, heating at 95°C and then ramping down to 25°C at 0.1°C/s. Capping oligonucleotides were diluted to 5 µM and then subjected to the same heat conditions as above. For each TALEN to be constructed, 5 µl of streptavidin-coated M-270 beads (Life Technologies) were washed twice in 2× Binding and Wash (BW) buffer (2 M NaCl, 10 mM Tris–HCl pH 8.0, 1 mM ethylenediaminetetraacetic acid pH 8.0, 0.2% w/w Tween-20), resuspended in 5 µl 2× BW buffer, mixed with 5 µl of 10 nM initiator oligonucleotide and rotated at room temperature for 30 min. Beads were washed twice in 0.5× BW buffer and resuspended in a 10 -µl reaction mix representing the first ligation step. Each ligation reaction contained 5 µl 2× rapid ligation buffer and 0.5 µl T7 ligase (Enzymatics), 1 µl 5 µM capping oligo and 50 ng of the relevant TALE monomer. The beads were rotated for 2 min at room temperature before being washed twice in 0.5× BW buffer and resuspended in the next ligation reaction. Ligations and washes were repeated for the requisite number of steps (up to 20 total ligations) before a final ligation containing the appropriate terminator oligonucleotide instead of a TALE monomer. After the terminator ligation, beads were washed once in water and either resuspended in 15 µl water with 0.01% Tween-20 and heated to 95°C for 3 min to elute the DNA or resuspended directly in Golden-Gate reaction mix for assembly into the nuclease backbone.
TALE elution and amplification
Eluted TALE ligation products were amplified in 50 -µl PCRs containing 25 µl Phusion 2× HF master mix, 400 nM each post-elution primer Fwd and Rvs and SYBR green I (Invitrogen) at 0.4× concentration. PCR was performed on a BioRad Opticon Monitor real-time PCR machine with 98°C for 2 min followed by cycles of: 98°C, 20 s; 60°C, 30 s and 72°C, 45 s. PCRs were stopped as soon as a SYBR-green amplification signal could be detected, usually after 10–15 cycles. PCR products were visualized by gel electrophoresis and purified by polyethylene glycol precipitation on carboxylated magnetic beads, using a cheaper self-made version (21) of Agencourt Ampure beads.
Assembly of TALEs into the nuclease backbone
Ligated TALE domains were assembled into a nuclease-carrying backbone vector using the Golden-Gate method (22) as applied by (18). 3–5 ng of purified post-elution PCR product (or half of the washed beads using the direct assembly method (see Results)) was added to a 10 -µl reaction containing 1× NEBuffer 4, 1 mM ATP, 0.1 mg/ml BSA, 0.5 µl T7 ligase (Enzymatics), 0.5 µl BsaI (NEB) and 100 ng TALEN backbone plasmid. This plasmid is based on the nuclease plasmid of (18) but modified using PCR and isothermal assembly (23) to remove the C-terminal 0.5 TALE repeat. The Golden-Gate reaction was first subjected to 10 cycles at 37°C, 2 min and 20°C, 3 min; then 10 min at 50°C and 10 min at 80°C. 1.5 µl of each product was transformed into half a vial of TOP10 cells (Life Technologies) and plated onto Luria–Bertani (LB) agar ampicillin.
TALEN verification
After overnight incubation of the TALEN transformation plates, colonies were picked and swirled first in 100 µl LB containing 100 µg/ml ampicillin and then in 100 µl water. Ten microliters of this water was then included in a 50 µl PCR containing 1× Phusion HF master mix, 250 nM each dNTP, 0.4× SYBR green I and 400 nM each primer (TALE-seq-F1 and TALE-seq-F2 for short colony PCR, Whole-TALEN-PCR-Fwd and Whole-TALEN-PCR-Rvs for long colony PCR (See Results)). PCRs were performed at 98°C for 3 min followed by 30 cycles of: 98°C, 20 s; 60°C, 30 s and 72°C, 45 s (2 min for long colony PCR). Products were visualized by gel electrophoresis. Saved colonies in LB corresponding to full-length products were grown overnight in 5 ml LB-amp, and the TALEN plasmids were purified by Qiagen Miniprep. Plasmids or purified long colony PCR products were Sanger-sequenced in three reads using the sequencing primers TALE-seq-F1, TALE-seq-F2 and TALE-seq-R1. Sequences were analyzed with custom scripts to check for perfect TALEN coding sequence.
TALEN activity reporter cell construction
To generate a reporter system for assaying TALEN activity, we constructed a lentiviral vector carrying an EGFP gene driven by the EF1α promoter that is defective due to a non-functional ACG start codon (Figure 2a). EGFP is linked by an internal ribosome entry site to a functional mCherry sequence, to allow fluorescent selection of cells carrying genomically integrated reporter after transduction. The construct also contains an eight-nucleotide degenerate A/C/T (H) barcode at the 5′-untranslated region to allow later identification of each unique genomic insertion into the reporter cell line. The lentiviral vector was created by standard PCR and cloning techniques including isothermal assembly (23). In the final cloning step, a PCR product carrying degenerate bases at the barcode was assembled into the backbone, transformed into Stbl3 cells (Life Technologies) and 90% of the transformation mixture was grown up directly in LB ampicillin and purified by QIAgen Maxiprep. The remaining 10% was plated out and ∼300 colonies were generated, with 10/10 colony PCRs showing the correct insert size. From these results we inferred that the purified library had a diversity of several thousand, making any handful of lentiviral integration events extremely likely to carry unique barcode sequences. The lentiviral plasmid was transfected by Lipofectamine 2000 with psPAX2 (Addgene plasmid 12260) and pMD2.G (Addgene plasmid 12259) packaging plasmids (contributed by the Didier Trono Lab) into cultured Lenti-X 293 T cells (Clontech) to produce lentivirus. Supernatant was collected 48 and 72 h post-transfection, sterile filtered, concentrated with the Lenti-X Concentrator (Clontech) and added to fresh 293 T cells with polybrene. Fluorescence-activated cell sorting (FACS) was used to isolate single mCherry-positive cells after 1 week. A single clonal line was then chosen for TALEN transfection experiments.
Figure 2.
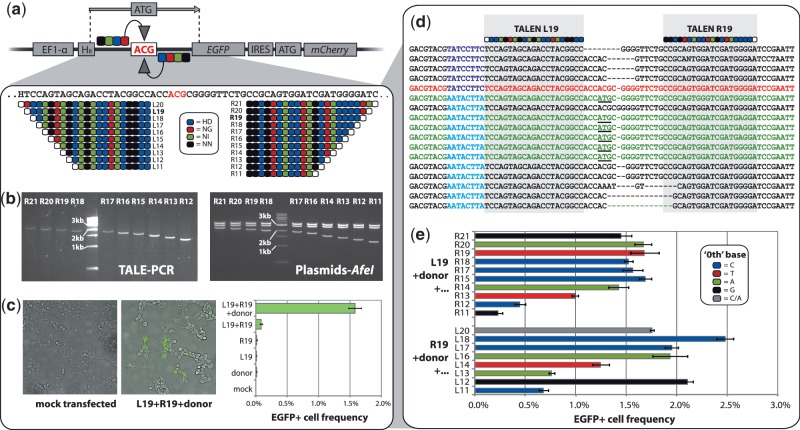
Testing activity of TALEN variants on a human cell reporter system. (a) To generate a TALEN activity reporter system we integrated a barcoded (H8, H = A/C/T) defective EGFP gene carrying a non-functional ACG start codon into the genome of 293 T cells. Gene editing by a 75-nt donor oligonucleotide stimulated by a TALEN-mediated double-strand break near the target site should correct the start codon and rescue EGFP expression. We designed 21 length variants of TALENs designed to flank the defective site. TALEN RVD sequences are shown. White monomer circles represent ‘0th’ positions of the DNA target sequence bound by the TALE N-terminus. (b) Post-elution PCR products of the ligated right TALE domains (for R11 see TAL-11, Figure 1b) and AfeI digestions of corresponding assembled and sequence-verified TALEN plasmids. (c) Epifluorescent microscopy and flow cytometry results after transfection of the reporter cells with the L19/R19 TALEN pair and the donor oligonucleotide. (d) Gene editing events (green sequences, functional start codon underlined) were confirmed by sequencing of the target site in EGFP+ cells, with widespread NHEJ (black sequences) also observed (red = unmodified sequence). Reporter barcodes (blue) identify from which of two genomic reporter copies each sequence is derived. (e) EGFP rescue activity of all successfully synthesized TALEN length variants. Each TALEN was transfected together with the donor and either L19 or R19, whichever was relevant. Bar colors indicate the DNA base located at the ‘0th’ position for each TALEN tested. Error bars show standard error (n = 3 for all).
TALEN transfections
The day before all transfections, reporter cells were trypsinized, centrifuged, resuspended in DMEM + GlutaMax-1 medium with 10% fetal bovine serum and transferred to 24-well poly-l-lysine-coated plates with 250 000 cells and 500 µl medium per well. After 24 h of incubation at 37°C in 5% CO2, TALENs and the donor oligonucleotide EGFP-rescue-75 were transfected into the cells using Lipofectamine 2000 (Life Technologies). DNA mixes containing 300 ng of each plasmid and 200 ng of the donor were made up in 50 µl Opti-MEM reduced serum medium (Life Technologies). 2.5 µl Lipofectamine 2000 was mixed with 50 µl Opti-MEM and left to stand for 5 min. The DNA mix and lipofectamine mix were then combined and incubated at room temperature for 30 min before the whole 100–130 µl transfection mix was added to the cells. After 24 h of incubation, the wells were replaced with fresh medium containing penicillin/streptomycin antibiotic. After a further 3 days of incubation, cells were imaged, dissociated by TrypLE Express (Life Technologies), neutralized in media and then quantified by flow cytometry on a BD Aria Fortessa platform or sorted by FACS.
Sequencing the reporter cell target region
To count the number of reporter construct genomic integrations in the initial reporter cell line, and to sequence the TALEN target region after transfections, a few thousand cells were scraped off the relevant culture dish with a pipette and 8.9 µl of the cells in medium were added to 0.1 µl prepGEM tissue protease enzyme and 1 µl 10× prepGEM gold buffer (ZyGEM). Reactions were incubated for 5 min at 75°C and then 5 min at 95°C. The reactions were then added to 40 µl of PCR mix containing 35.5 µl platinum 1.1× Supermix (Invitrogen), 250 nM each dNTP and 400 nM primers BarcodeF and BarcodeR. Reactions were subjected at 95°C for 3 min followed by 40 cycles of 95°C, 20 s; 60°C, 30 s and 72°C, 20 s. Products were visualized by gel electrophoresis, cloned by the TOPO-TA method (Life Technologies) and sequenced with the M13R primer. Each of 96 random sequences showed one of only two barcodes, indicating that this line carries two genomic copies of the reporter gene.
RESULTS
ICA: rapid, individual monomer-based DNA assembly
For rapid, solid surface-based synthesis of repetitive DNA such as TALEs, we desired an efficient, scalable method of ligating individual DNA monomers into chains at least 20 monomers long. An ideal such method would overcome incomplete ligation efficiency and avoid monomers self-ligating in individual steps without requiring cumbersome preparation of many versions of each monomer carrying different ligation overhangs. We, therefore, designed a method, ICA (Figure 1), that involves successive monomer ligations on a solid support and capping oligonucleotides to inactivate incomplete chains. The method consists of iterative cycles of three ligation steps, each adding one of only three versions (A–C) of the basic monomer, with monomer ‘type’ (such as the required RVD in TALE domain synthesis) selectable in each step. For each monomer type, its A–C versions are identical except they carry distinct four-nucleotide overhangs created by BsmBI digestion of their monomer precursors (PCR products). As in (24) the overhangs differ only in redundant codon positions so ultimately produce identical amino acid sequence. The ‘right’ overhang of the A monomer will ligate only to the ‘left’ overhang of the B monomer. The B monomer right overhang will only ligate to the C monomer left overhang, and the C right overhang only to the A left overhang. Hence, by sequentially introducing monomers and ligase to an immobilized chain in a repeating A, B, C, A, B, C, etc. cycling pattern, with free choice of the monomer type used in each step, a chain of arbitrary length and monomer sequence can be built. The chain is anchored with a biotinylated initiator oligo coupled to streptavidin-coated magnetic beads. The initiator carries a right overhang identical to that of the C monomer and will therefore ligate to the left overhang of the A monomer during the first step. Importantly, the A-B-C pattern of sequential addition allows the use of short hairpin oligonucleotides (capping oligos) to inactivate incomplete chains. For example, after the first ligation step not all initiators will be coupled to A monomers because of imperfect ligation efficiency. By simultaneously introducing in the second step the B monomer and a capping oligo that will ligate to the right overhang of the initiator (in later cycles to the C monomer), residual unligated initiators will be blocked by the capping oligo while the successfully coupled chains from the first step can ligate to the B monomer to form an ‘initiator-A-B’ chain. Similarly, in the third step the C monomer, which will ligate to form ‘initiator-A-B-C’ chains, can be co-introduced with a capping oligo that will ligate to the A monomer and thus block ‘initiator-A’ chains left by incomplete ligation in the second step. This pattern is repeated in every step, where a monomer to extend full-length chains is co-introduced with a capping oligo to block chains unsuccessfully extended in the previous step. For TALE synthesis, a single exception to the A-B-C rule occurs in step 6, where a modified version of the C monomer, Cseq, is used that carries multiple synonymous substitutions allowing later annealing of a specific sequencing primer (18). Once a capping oligo ligates to a chain, further ligations onto that chain are prevented and since the hairpin covalently links the two strands of the DNA duplex, rapid self-annealing will reduce the chance of the chain interfering with downstream PCR. In the final ligation step, a double-stranded terminator oligonucleotide is added to the chain. Both the terminator and initiator oligos carry primer sites that allow PCR amplification of the full-length assemblies after heat elution from the beads, as well as restriction sites that allow Golden-Gate cloning into a backbone vector. The terminator used here carries the coding sequence for the ‘0.5 repeat’ encoding the final base of the DNA recognition site that is a feature of TALE proteins (10,25). In ICA, there are three possible versions (A–C) of the final full monomer depending on the chain length, and for TALE synthesis, four possible RVD types, requiring 12 different terminator oligos to allow termination of TALEs of any length and final DNA target base. Since all DNA target site specificity is encoded in the final ligated TALE chain, only a single backbone vector is needed for all TALENs.
Efficient synthesis of different length TALE domains
To test the ICA method, we performed serial ligations of monomers to construct TALEs of lengths 11, 15, 18 and 19 (not including the ‘0th’ position bound by the TALE N-terminus). We first prepared the TALE monomers by PCR from plasmid monomer templates, using four different primer pairs to produce the A, B, C and Cseq versions for each TALE monomer. PCR products were purified, digested with BsmBI to generate ligatable overhangs and purified again. After coupling a biotinylated initiator oligonucleotide to streptavidin-coated magnetic beads, each ligation cycle consisted of adding ligation mixture to the beads containing monomer and the relevant capping oligo and incubating at room temperature for 2 min, followed by two washes before the next ligation. In this way, 20 consecutive ligations can be performed in <90 min. In the final ligation step for each TALE we added the relevant terminator oligonucleotide, followed by a wash and then heat elution of all DNAs from the beads. We performed PCR of the eluted DNA, using primers complementary to the initiator and terminator oligos, with SYBR green I in a real-time PCR machine in order to monitor amplification and stop it before the plateau phase. This monitoring is necessary because of the repetitive nature of TALE genes; we have observed that over-amplification of TALEs leads to ladder-like PCR products indicating inter-amplicon recombination (also seen in TALEN colony PCRs, see Figure 3). Gel electrophoresis revealed the presence of clean bands of the expected size for every TALE sample except the one (TAL19nb) in which capping oligonucleotides were omitted in synthesis. TAL19nb showed a full-length band and also stronger bands of shorter sizes spaced at ∼300 bp intervals. These bands presumably correspond to TALE chains that were incompletely ligated in one or more steps and became re-extended in a later ABC cycle which would be multiples of three ligation steps later (each monomer is 102 bp). The absence of truncated bands in the samples where capping oligonucleotides were used indicates effective capping of incomplete chains by these oligonucleotides. We then purified all products and assembled them into a FokI-containing backbone plasmid by Golden-Gate ligation (18). After transformation into TOP10 cells, which generated dozens of colonies per TALEN construct, we grew and purified the plasmids of six colonies per TALEN and digested with AfeI to reveal the length of the cloned TALE inserts. The four TALENs where capping oligos were used generated between 4/6 and 6/6 full-length clones whereas the no-capping sample generated 0/6, again showing the benefit of the capping approach. We sequenced four full-length clones per successful construct and found perfect TALEN sequences with the following success rate: TAL11, 1/4 clones; TAL15, 4/4; TAL18, 4/4 and TAL19, 2/4. Observed errors were one stop codon, one coding substitution and three one-base deletions, probably stemming from monomer PCR due to polymerase or primer synthesis errors.
A reporter system to test TALEN pair activity in human cells
To test the scalability of ICA and to investigate how effectively TALENs synthesized with this approach can be used in genome engineering, we constructed a human cell reporter system with which we could measure the activity of multiple TALEN variants. We generated a monoclonal 293 T cell line carrying two lentiviral insertions of a defective EGFP gene driven by the EF1α promoter (Figure 2a). In these cells, EGFP is transcribed but not translated since it carries ACG at the expected ATG start codon site. Gene editing by knock-in of a 75-nt single-stranded donor oligonucleotide carrying a functional ATG start codon should in theory rescue EGFP expression. Since genomic incorporation of homologous single-stranded donor DNA is greatly increased by a DNA cut close to the desired incorporation site (26), we expected that activity of our TALENs could be quickly assayed by measuring EGFP expression frequency of cells transfected with the donor and TALEN pairs designed to flank the target site. We designed 10 left variants and 11 right variants of TALENs of different length (11–21 monomers) that flank the defective ACG start codon on the EGFP gene. We left a constant 17 bp gap between the TALENs. In this design, the TALENs cover a range of DNA bases at their C-terminus encoded ‘0th’ position. Bioinformatic and structural studies have suggested that a thymine is necessary at this base for optimal activity (10,12,15,25), but some contrary evidence exists (13,17,27,28) and empirical testing of this question with multiple custom TALENs has not been reported.
ICA of TALEN variants targeting the reporter gene
We first performed ligations for the 10 right TALE domains. PCR amplification of the eluted ligation products generated clean, full-length products for all TALEN constructs (Figure 2b). We cloned the products using the Golden-Gate method into the nuclease vector and six colony PCRs were performed per TALEN (Supplementary Figure S1). Between 4/6 and 6/6, colonies showed full-length inserts for each TALEN. Two such colonies were grown up per TALEN and sequenced, revealing at least one perfect clone for all except one TALEN. For R15, frameshift mutations or premature stop codons were detected in 26/26 sequenced colonies, suggesting that this TALEN sequence was toxic to Escherichia coli. Ligation and synthesis were then performed for the 10 left TALENs (Supplementary Figure S2). We synthesized L14-L11 with exactly the same protocol as the right TALENs, with similar results and cloning success rate. In contrast, in the production of L20-L15 we halved the amount of monomer (to 25 ng) and ligase (to 0.25 µl) in each step to reduce reagent costs. Although perfect clones were found for L20-L15, aberrant PCR products in gels (Supplementary Figure S2) and the need to screen many colonies (up to 24) suggest that these reagent levels are too low for efficient assembly. L15 was later dropped as the selected sequence-verified clone failed to grow when we prepared all TALENs in parallel for the gene editing experiment.
Confirming activity of TALENs and HR in the reporter cells
Having designed and synthesized the TALEN variants, we first transfected the reporter cells carrying defective EGFP with combinations of the L19/R19 TALEN pair and the donor oligonucleotide (Figure 2c) and incubated for 96 hours. Encouragingly, transfection of the L19/R19 pair plus the donor oligo led to 1.8% of cells expressing EGFP, suggesting the occurrence of TALEN-mediated gene editing. Transfection of the donor, L19 or R19 alone gave negligible EGFP expression. Interestingly, transfection of the L19/R19 pair without donor resulted in a small but measurable number of EGFP-positive cells (0.1%). To confirm occurrence of gene editing and/or NHEJ, we PCR amplified, cloned and sequenced a 189-bp region around the TALEN target site in unsorted and EGFP+ sorted populations of TALEN-pair only and TALEN pair-plus-oligo-treated cells (Figure 2d, Supplementary Figure S3). In cells treated with the TALEN pair only, 18/23 (78%) of clone sequences from unsorted cells showed small deletions indicative of NHEJ, indicating a remarkably high cutting activity of this TALEN pair. Similarly, from the EGFP+ sorted cells from the same treatment, 23/24 clones showed evidence of NHEJ. We could not infer from the sequences why these cells had become EGFP positive. However, rearrangements in one reporter copy that put the EGFP coding sequence into an open reading frame may have removed the PCR priming sites, with the result that all observed sequences stem from the second reporter copy. In unsorted cells treated with the TALEN pair and donor, 19/21 sequences showed evidence of NHEJ with no evidence of gene editing. However, in the EGFP+ sorted cells from the same treatment, 6/17 clone sequences (Figure 2d) matched the donor sequence, indicating successful editing, while 10/17 sequences showed NHEJ. Since there are two genomic insertions in the reporter cells, it is expected that PCR products from the sorted cells will contain a mixture of products, with half showing the gene editing events leading to EGFP expression and the rest showing the second genomic reporter copy, which in this assay mostly shows NHEJ events. We conclude that TALENs can stimulate gene editing by an oligonucleotide donor and therefore that measuring EGFP expression frequency after transfection allows rapid assessment of TALEN activity on our reporter system.
Testing the TALEN length variants and different 0th position bases
Having demonstrated activity of TALENs and gene editing on our reporter system, we then transfected the reporter cells with combinations of our left and right TALEN variants, each time including the EGFP-correcting donor to allow rapid flow cytometry measurement of TALEN activity. Since we had shown that L19 and R19 are functional, we tested all of the right TALENs in combination with L19 and all the left TALENs with R19. All conditions were tested in triplicate. Interestingly, flow cytometry (Figure 2e) revealed that all TALEN pairs tested generated EGFP-positive cells after 96 h and thus possess cutting activity. For the right TALENs tested together with L19, performance was similar among all except the three shortest TALENs. These three TALENs show a pattern of decreasing activity with decreasing length such that R12 generated ∼6-fold fewer EGFP+ cells than R15 and those longer. For the left TALENs tested with R19, a similar pattern was observed except for one exception: L12 showed high activity, whereas L14, L13 and L11 showed reduced activity. Interestingly, most TALENS showed similar results despite overlapping different DNA bases at their C-terminus-encoded 0th position. Although the 0th position of at least one TALEN (L19 or R19) overlapped a T in all pairs tested, average EGFP+ frequencies were not obviously affected by the 0th position base of the second TALEN: T: 1.4% (n = 4); C: 1.7% (n = 7); A: 1.5% (n = 4) and G: 0.8% (n = 3). We conclude that a 0th position thymine is not strictly necessary for TALEN activity although there are several possible explanations for this finding (see ‘Discussion’ section).
Further accelerating ICA
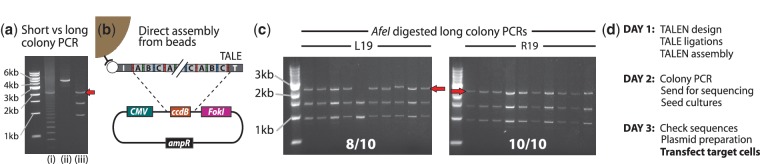
We next sought to further increase the speed and efficiency of ICA. First, we noted that while assembly of a TALEN by ICA takes a single day, 3 days are needed subsequently for colony PCR, overnight clone culture, plasmid purification and sequencing before a sequence-verified construct can be transfected into target cells. Sequencing the TALEN colony PCR product rather than a purified plasmid would save a day and considerable work; however, colony PCR of repetitive DNA such as TALENS tends to generate ladder-like bands, presumably from inter-template recombination (Figure 3a(i)), that prohibit effective sequencing. We reasoned that a much longer colony PCR amplicon would suffer less from this problem, since the TALE sequence will form a much smaller fraction of the amplicon and be situated further from the amplicon ends. We indeed found that a colony PCR amplifying the entire TALEN gene produces much cleaner products (Figure 3a(ii)), with brief AfeI digestion of the raw PCR product allowing easy gel visualization of the TALEN insert size (Figure 3a(iii). We amplified and sequenced long amplicons from two previously verified L19 colonies and found that the products indeed generate high-quality correct sequences. We next noted that the post-elution PCR step in ICA takes some time and introduces some risk of PCR artifacts. Therefore, we investigated whether this step could be eliminated by direct assembly of the ligation products from the beads into the backbone vector (Figure 3b). We resynthesized TALENs L19 and R19 from monomers under the same conditions as before, except this time after the final ligation step, whereas the TALE chains were still attached to the beads, the beads were washed once in TE and resuspended in the Golden-Gate assembly mix with the backbone plasmid. The beads–assembly mixture was subjected to the same Golden-Gate thermal cycling conditions as before and transformed directly into TOP10 cells. Hundreds of colonies were generated from each construct, with 8/10 and 10/10 colony PCRs showing full-length inserts for L19 and R19, respectively. Five full-length clones were sequenced per construct, with 4/5 and 3/5 showing perfect sequence. These results compare very favorably with the results generated from the PCR-then-assembly protocol (Table 1). Therefore, the PCR step after TALE ligation can indeed be bypassed, substantially simplifying TALEN construction by ICA. Taken together, our elimination of the PCR step and the expedited colony PCR and sequencing approach reduces the time between TALE design and transfection into target cells to 3 days, the shortest such time so far reported.
Figure 3.
Accelerating TALEN production by ICA. (a) PCR amplification of repetitive DNA such as TALEs frequently generates artifacts, for example in colony PCR of an assembled TALEN (lane (i)). We found that using PCR primers in the vector far away (∼1400 bp) from the repetitive TALE region dramatically reduces artifacts resulting in clean PCR products (ii) that can be directly sequenced saving a day’s wait for plasmid preparation, or digested with AfeI (iii) to reveal the TALE insert length (red arrow). (b) We tested whether the PCR step after ICA ligation on beads can be omitted by performing Golden-Gate assembly of two 19-monomer long (excluding the 0th position) TALEs into the nuclease vector directly on the beads. (c) After transformation hundreds of colonies were generated, with the L19 and R19 constructs respectively generating 8/10 and 10/10 AfeI-digested colony PCRs with full-length inserts (red arrows). (d) A realistic three-day work schedule using the methods presented here from TALEN design to transfection of target cells.
Table 1.
Summary of cloning and sequencing success using two variants of the ICA protocol
| Full-length clonesa | Perfect sequence clones | ||||
|---|---|---|---|---|---|
| ICA protocol variant | No. constructs made | Worst construct | Best construct | Worst construct | Best construct |
| PCR then assembly | 18b | 4/6 | 6/6 | 1/4c | 4/4 |
| Direct assembly off beads | 2 | 8/10 | 10/10 | 3/5 | 4/5 |
The table shows the range of success rates (full-length clone frequency after TALEN assembly and perfect sequence frequency among selected full-length clones) observed among all constructs built with two variants of the ICA method.
aAlthough dozens to hundreds of colonies were generated per construct, only 6–10 colonies were analyzed.
bThese 18 constructs include the four constructs made with capping oligos in Figure 1 and L14-L11 and R21-R11 from Figure 2. L20-L15 are not included as they were synthesized with reduced amounts of monomer and ligase, which turned out to be detrimental.
cR15 is not included as it produced zero perfect sequences from 26 full-length clones and is therefore likely to be toxic to E. coli.
DISCUSSION
A desirable way to synthesize long repetitive DNA, which is refractory to current high-throughput DNA synthesis methods (6,7), would be to sequentially couple DNA repeat modules by ligation on a solid surface, in a process analogous to chemical oligonucleotide synthesis from single nucleotides (5). However, in contrast to oligonucleotide synthesis, which uses chemical reactions with coupling efficiencies >99.5% (5), enzymatic ligation of relatively large DNA monomers may not be similarly efficient. This is critical since a drop in coupling efficiency just to 90% would result in only ∼12% full-length chains after 20 consecutive ligations. In addition, although nucleotides can be chemically protected and deprotected to prevent self-coupling during single attachment steps (4), analogous processes are not trivial for oligonucleotide ligation, likely requiring further enzymatic steps with sub-optimal efficiencies. Our ICA method overcomes these problems by alternating between three versions (A–C) of each monomer in the ligation steps, which as well as preventing monomer self-ligation permits inclusion in each step of a ‘capping’ oligonucleotide that can ligate to incompletely extended chains from the previous step. The capping oligonucleotides are hairpin-based short single strands, which makes them stoichiometrically perfect, rapidly diffusible and cheap to use in high molar concentrations, leading to high capping efficiency.
Here we have tested ICA on custom TALE genes. Due to their highly repetitive nature, custom TALEs have mostly been synthesized by multi-step hierarchical ligation and cloning. Most recent methods (15,18,20,24) use variations of Golden-Gate cloning (22), in which a Type IIS restriction enzyme allows the creation of position-specific overhangs on monomers (prepared from PCR or plasmids) for multi-piece, defined-sequence ligation in a single reaction. While effective, these methods required extensive initial formatting of the monomers or pre-fabricated dimers into many different position-specific forms whether through PCR (18,24) or construction of large (>70) plasmid libraries (15,20). Due to limits in the number of pieces Golden-Gate ligation can assemble in one reaction, the studies cited above required multiple rounds of assembly and hence between 2 and 5 days to produce new TALEs with target sites >14 bp. Recently, Reyon et al. presented FLASH (19), the first automated method of TALE synthesis which like ICA builds TALEs by sequential ligation on magnetic beads. FLASH is a powerful method that can produce dozens of TALENs per day in parallel using an automated liquid handing station. However, FLASH builds TALEs from oligomers (mostly 4-mers) rather than monomers, picked from an exhaustive pre-fabricated library of 376 plasmids. This reduces the number of ligation reactions needed to make full-length TALEs, which is likely necessary given that capping oligonucleotides are not used in FLASH (although in theory they could be introduced to this method to improve efficiency). However, reliance on the oligomer library will make it cumbersome to replace individual monomers with new ones of improved design and combinatorially virtually impossible to expand the routine repertoire of monomers much beyond 4. We envisage multiple reasons to add new TALE monomers to the standard repertoire, such as incorporation of monomers tagged with fluorescent or activatable moieties or ones that target epigenetically modified bases, or even the creation of hybrid DNA-binding domains that combine TALEs with other motifs such as zinc fingers. ICA is, to our knowledge, unique in providing an automatable platform that can produce full-length TALEs from individual monomers, with only three or four PCRs needed to introduce any new module to the repertoire. In addition, since each ligation step picks from only four standard monomers rather than hundreds of oligomers and takes place rapidly (2 min) at room temperature, ICA is likely to be applicable to current to microarray printing technology to allow production of libraries of thousands of TALE constructs. We note that for effective production of large TALE libraries, the error rate of individual constructs (Table 1) must be reduced. However, since most errors we observed likely stem from primer or polymerase errors introduced during monomer PCR, the majority of these might be removed by preparing monomers in future from digested plasmids (15) rather than PCR.
In testing the TALENs we produced by ICA, we used an oligonucleotide donor to genomically modify a reporter gene after a TALEN-stimulated chromosomal break, representing to our knowledge the first report of TALEN-oligonucleotide gene editing. Sequence analysis showed that the L19/R19 TALEN pair very efficiently cut the target site, with up to 80% of alleles showing evidence of NHEJ 4 days after TALEN transfection. However, the frequency of donor oligonucleotide incorporation was considerably lower, with only 1–2% of cells becoming EGFP positive. Since oligonucleotides carry advantages over longer donor DNA in terms of cost and design simplicity, future work into increasing the ratio of gene editing to NHEJ after TALEN cutting would be extremely useful. By testing our TALEN length variants in human cells, we found that all TALENs tested showed activity, although TALENs shorter than ∼15 monomers long tended to be less effective than longer ones, supporting the need for a synthesis method that can produce TALENs with sufficiently long DNA target sites. In our assay, we observed similar target repair activity among several TALENs with different bases at the 0th position, indicating that a thymine at this position is not strictly required for TALEN activity. This result, which contrasts with previous bioinformatic, crystal structure and some empirical data (10,12,15,25) but agrees with others (13,17,27,28), may have several explanations. TALENs as long as the ones we used may be tolerant of the decrease in binding affinity at any single site including the 0th position. We also note that using EGFP rescue of our reporter system as a measurement of TALEN activity may become less accurate once TALEN cutting efficiency approaches 100%, since donor DNA incorporation will become the limiting factor in the rescue rather than target site cutting. In addition, since at least one of each TALEN pair tested did overlap a 0th position thymine, FOKI recruitment effects might be acting to increase activity of the other TALEN. Nevertheless, our results generally support and extend recent findings (19) that TALENs can be confidently adapted for new targets with minimal design constraints, supporting the usefulness of high-throughput synthesis of TALENs and other TALE-fusion genes. In summary, ICA is a fast, efficient and scalable method of producing repetitive modular DNA of defined sequence. Although here we have applied the method to TALE-gene synthesis, we believe the approach should be useful for production of other modular constructs in synthetic biology (29) where there is increasing demand for rapid DNA assembly-from-parts.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–3 and Supplementary plasmid maps.
FUNDING
National Institutes of Health—National Human Genome Research Institute (NHGRI) [1P50 HG005550]; EMBO long-term fellowship [ALTF 91-2010 to A.W.B.]. Banting Postdoctoral fellowship (to R.C.). Funding for open access charge: NHGRI [1P50 HG005550].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Marc Lajoie, Neville Sanjana, Sriram Kosuri, Nikolai Eroshenko, Keith Joung and John Aach for discussions and advice.
REFERENCES
- 1.Tian J, Ma K, Saaem I. Advancing high-throughput gene synthesis technology. Mol. Biosyst. 2009;5:714–722. doi: 10.1039/b822268c. [DOI] [PubMed] [Google Scholar]
- 2.Smith HO, Hutchison CA, 3rd, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc. Natl. Acad. Sci. USA. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 4.Caruthers MH. Gene synthesis machines: DNA chemistry and its uses. Science. 1985;230:281–285. doi: 10.1126/science.3863253. [DOI] [PubMed] [Google Scholar]
- 5.LeProust EM, Peck BJ, Spirin K, McCuen HB, Moore B, Namsaraev E, Caruthers MH. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 2010;38:2522–2540. doi: 10.1093/nar/gkq163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matzas M, Stahler PF, Kefer N, Siebelt N, Boisguerin V, Leonard JT, Keller A, Stahler CF, Haberle P, Gharizadeh B, et al. High-fidelity gene synthesis by retrieval of sequence-verified DNA identified using high-throughput pyrosequencing. Nat. Biotechnol. 2010;28:1291–1294. doi: 10.1038/nbt.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosuri S, Eroshenko N, Leproust EM, Super M, Way J, Li JB, Church GM. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat. Biotechnol. 2010;28:1295–1299. doi: 10.1038/nbt.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bang D, Church GM. Gene synthesis by circular assembly amplification. Nat. Methods. 2008;5:37–39. doi: 10.1038/nmeth1136. [DOI] [PubMed] [Google Scholar]
- 9.Xiong AS, Peng RH, Zhuang J, Liu JG, Gao F, Chen JM, Cheng ZM, Yao QH. Non-polymerase-cycling-assembly-based chemical gene synthesis: strategies, methods, and progress. Biotechnol. Adv. 2008;26:121–134. doi: 10.1016/j.biotechadv.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 11.Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- 12.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu JK, Shi Y, Yan N. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335:720–723. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun N, Liang J, Abil Z, Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol. Biosyst. 2012;8:1255–1263. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- 18.Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Piatek MJ, Atef A, Piatek A, Wibowo A, Fang X, Sabir JS, Zhu JK, Mahfouz MM. Rapid and highly efficient construction of TALE-based transcriptional regulators and nucleases for genome modification. Plant Mol. Biol. 2012;78:407–416. doi: 10.1007/s11103-012-9875-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundin S, Stranneheim H, Pettersson E, Klevebring D, Lundeberg J. Increased throughput by parallelization of library preparation for massive sequencing. PLoS One. 2010;5:e10029. doi: 10.1371/journal.pone.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 24.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 26.Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 28.Yu Y, Streubel J, Balzergue S, Champion A, Boch J, Koebnik R, Feng J, Verdier V, Szurek B. Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin-3 Os11N3 gene. Mol. Plant Microbe. Interact. 2011;24:1102–1113. doi: 10.1094/MPMI-11-10-0254. [DOI] [PubMed] [Google Scholar]
- 29.Schmid-Burgk JL, Xie Z, Frank S, Virreira Winter S, Mitschka S, Kolanus W, Murray A, Benenson Y. Rapid hierarchical assembly of medium-size DNA cassettes. Nucleic Acids Res. 2012;40:e92. doi: 10.1093/nar/gks236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.