Summary
Conversion between a cyst and trophozoite stage is essential to disease transmission and pathogenesis in the parasitic protist Entamoeba histolytica. A transcriptomic analysis of E. histolytica cysts and trophozoites has recently been accomplished, but the molecular basis of the regulation of encystation is not known. We have now identified a developmentally regulated Myb protein (belonging to the SHAQKY family of Myb proteins), which controls expression of a subset of amebic stage-specific genes. Overexpression of the nuclear localized Myb protein resulted in a transcriptome that overlapped significantly with the expression profile of amebic cysts. Analysis of promoters from genes regulated by the Myb protein identified a CCCCCC promoter motif to which amebic nuclear protein(s) bind in a sequence-specific manner. Chromatin immunoprecipitation demonstrated that the E. histolytica Myb protein binds directly to promoters of genes which contain the CCCCCC motif and which are regulated by the Myb protein. This work is the first identification of a transcription factor, which regulates expression of a subset of stage-specific genes in E. histolytica. Identification of transcriptional regulatory networks that control developmental pathways will provide novel insights into the biology of this important human pathogen.
Keywords: transcription factor, microarray, parasite, chromatin immunoprecipitation, promoter motif, gene expression
Introduction
Developmental regulation and stage conversion is necessary for disease propagation in many pathogenic protozoa. Enteric parasites such as Entamoeba histolytica transition between an environmentally resistant cyst form, which is required for transmission and a trophozoite stage, which proliferates and causes disease but cannot survive outside the host (Eichinger, 2001). The two different stages of the parasite’s life cycle are marked by sharply differing morphologies. Genome-wide studies have shed light on the regulation of gene expression during stage conversion in parasites including Plasmodium (Bozdech et al., 2003), Toxoplasma (Cleary et al., 2002), Giardia (Palm et al., 2005) and Entamoeba (Sanchez et al., 1994).
Developmentally regulated genes can be transcriptionally controlled by a number of conserved pathways. The genomes of apicomplexan parasites appear to have a relative paucity of transcription factors (Aravind et al., 2003), leading to the speculation that chromatin structure may play a key role in regulating gene expression (Hakimi and Deitsch, 2007). In Toxoplasma, changes in histone acetylation in the promoters of stage-specific genes correlate with the transition from the tachyzoite to bradyzoite stage and expression of stage-specific genes is modulated by addition of histone deacetylase (HDAC) inhibitors (Boyle et al., 2006; Saksouk et al., 2005). Sequence-specific binding proteins also regulate parasite gene expression. In Plasmodium members of the AP2 transcription factor family bind specific DNA sequences and have developmentally regulated expression, implying that they may regulate expression of stage-specific genes (De Silva et al., 2008).
The Myb domain is a sequence-specific DNA binding domain that was originally identified in vertebrates (Biedenkapp et al., 1988). Myb domain proteins control stage conversion and cell fate in a broad range of systems. Vertebrate Myb proteins directly regulate gene expression and are involved in both cell proliferation and differentiation (Oh and Reddy, 1999). Subsequently, Myb domain containing proteins have been identified in insects (Katzen et al., 1985), fungi (Morrow et al., 1993) and plants (Jin and Martin, 1999). In plants the Myb family is highly expanded and plays crucial roles in such diverse functions as circadian clock regulation, small molecule metabolism, and trichome development. A developmentally regulated Myb protein has been identified in the parasitic protist Giardia lamblia, and is involved in the regulation of cyst wall proteins that are expressed during encystation (Huang et al., 2008; Sun et al., 2002).
Entamoeba histolytica is a deep-branching eukaryote and a leading cause of parasitic death in humans (WHO, 1997). The infectious cycle of E. histolytica begins with the ingestion of the cyst, a non-dividing form that is able to survive in the environment due to a protective, chitin-containing cell wall. After ingestion, the cyst excysts in the small intestine to produce the proliferative and invasive trophozoite form. Due to unknown factors, some trophozoites encyst, allowing them to be excreted in the stool and to go on to infect new hosts (Haque et al., 2003). The encystation program is an attractive target for the development of new diagnostic or treatment approaches. Data on the molecular factors that regulate the developmental pathway in E. histolytica are limited due to the lack of an in vitro encystation method. Recent work has identified the transcriptome of E. histolytica cysts and determined that subsets of developmentally regulated genes are affected by histone acetylation (Ehrenkaufer et al., 2007a; Ehrenkaufer et al., 2007b).
We now present data on an E. histolytica Myb gene that is developmentally regulated (EhMyb-dr) and which regulates expression of a number of stage-specific genes. The EhMyb-dr belongs to the SHAKQY family of Myb genes and overexpression of this gene in E. histolytica trophozoites results in parasites that have a transcriptional profile that overlaps significantly with amebic cysts. Analysis of the promoter regions of genes regulated by EhMyb-dr identified conserved promoter motifs. Using electrophoretic mobility shift assays, we demonstrate that protein(s) from amebic nuclear extract binds specifically to a C-rich motif (CCCCCC), and that the binding is increased in EhMyb-dr overexpressing parasites. Using chromatin immunoprecipitation, promoters of EhMyb-dr regulated genes containing the C-rich motif were identified as interacting directly with the EhMyb-dr protein. This work is the first identification of a developmentally regulated transcription factor in E. histolytica and an important advance in understanding the molecular framework that regulates stage conversion in this important human pathogen.
Results
Identification of a developmentally regulated Myb domain gene in Entamoeba
We previously used a whole genome E. histolytica microarray to compare the transcriptomes of E. histolytica cysts and trophozoites and identified 672 cyst-specific and 767 trophozoite-specific genes (Ehrenkaufer et al., 2007b). In order to identify genes, which may potentially be involved in the regulation of stage conversion, we searched for genes with annotations that might indicate regulatory functions and which were developmentally regulated. The gene annotated as 7.m00410 (7.t00022; EHI_038640) encodes a 145aa protein with a single Myb DNA binding domain (Fig 1A). This protein is conserved across Entamoeba species with homologs in both E. dispar (EDI_259480) (E-value = 9.0e-69) and E. invadens (EIN_051670) (E-value = 2.1e-14) (Fig 1B). The presence of only one Myb domain, as well as the THAKQF motif found in this domain, places this gene as a member of the SHAKQY family of Myb proteins (Fig 1C). This group of Myb proteins is largely found in plants (Rubio-Somoza et al., 2006; Rose et al., 1999), though homologs exist in other eukaryotes, such as Dictyostelium (Fukuzawa et al., 2006). Five other SHAQKY family Myb genes are annotated in the E. histolytica genome (Supp Fig 1). All are expressed in trophozoites; one is developmentally regulated with higher expression in trophozoites than in cysts (Ehrenkaufer et al., 2007b).
Figure 1. Schematic of EhMyb-dr.
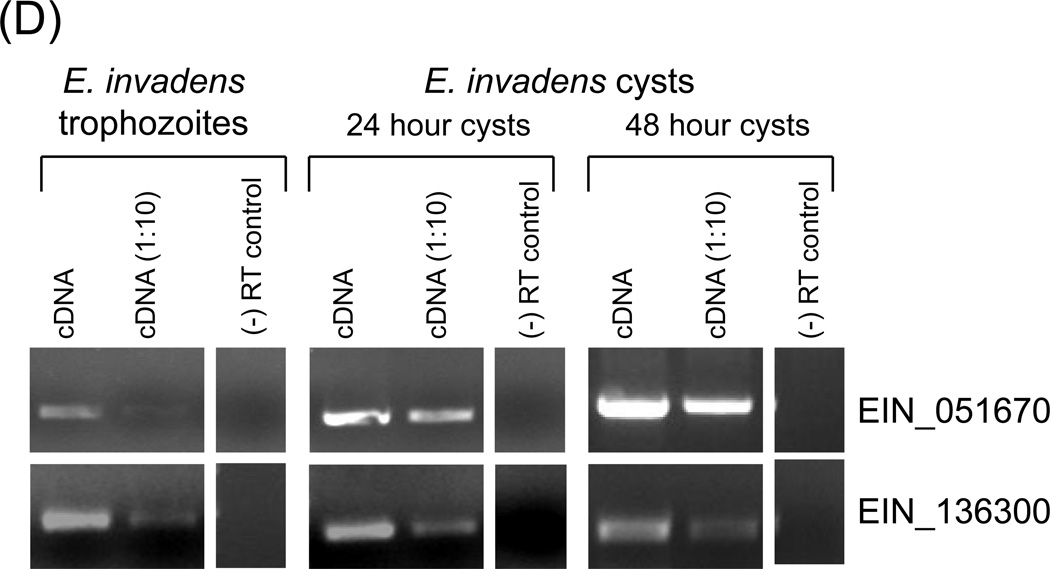
(A) Domain structure of the EhMyb-dr. The Myb domain is approximately 50aa and is located from ~80–130aa and contains the SHAQKY motif. (B) Alignment of EhMyb-dr with its E. dispar (EDI_259480) and E. invadens (EIN_051670) homologs. The SHAQKY domain is indicated by a box. (C) Alignment of the Myb domain of EhMyb-dr with SHAQKY domain proteins from tomato (LeMyb1), Arabidopsis (CCA1), and Dictyostelium (MybE) species. The SHAQKY domain is indicated by a box. All alignments were performed using ClustalW2 (Larkin et al., 2007). (D) RT-PCR showing expression of EIN_051670 (the E. invadens homolog of EhMyb-dr) in trophozoites and cysts after 24 and 48 hours of encystation in E. invadens. A significant increase in expression can be seen at both time points. For each sample a reaction performed with 1µl cDNA, a 1:10 dilution and a minus reverse transcriptase control are shown. EIN_136300 was used as a loading control, as its expression does not change during encystation (Ehrenkaufer et al., 2007b).
Microarray analysis shows low level expression of the EIN_051670 gene in E. histolytica trophozoites with a ~5 fold increase in E. histolytica cysts (Ehrenkaufer et al., 2007b). No other condition including heat shock (Hackney et al., 2007), colonic invasion (Gilchrist et al., 2006), exposure to histone deacetylase inhibitor (Ehrenkaufer et al., 2007a), or oxidative or nitrosative stress (Vicente et al., 2008) resulted in transcriptional regulation of this gene. To confirm the developmental regulation of the homolog of this gene in E. invadens (a reptilian ameba in which in vitro encystation is highly regulated), we monitored expression levels of the E. invadens homolog during encystation. EIN_051670 was expressed in E. invadens trophozoites with sequentially greater upregulation at 24h and 48 hours after transfer to encystation medium (Fig 1D). Some expression of the Myb gene in E. histolytica is also observed in the trophozoite stage, which is consistent with the E. histolytica array data (Ehrenkaufer et al., 2007b). This early induction is consistent with a gene encoding a protein involved in regulation of encystation, but would not likely be true for a protein whose function is required only in mature cysts. We have named this gene EhMyb-dr due to the presence of the Myb domain and its developmental regulation.
EhMyb-dr has nuclear localization in Entamoeba histolytica trophozoites
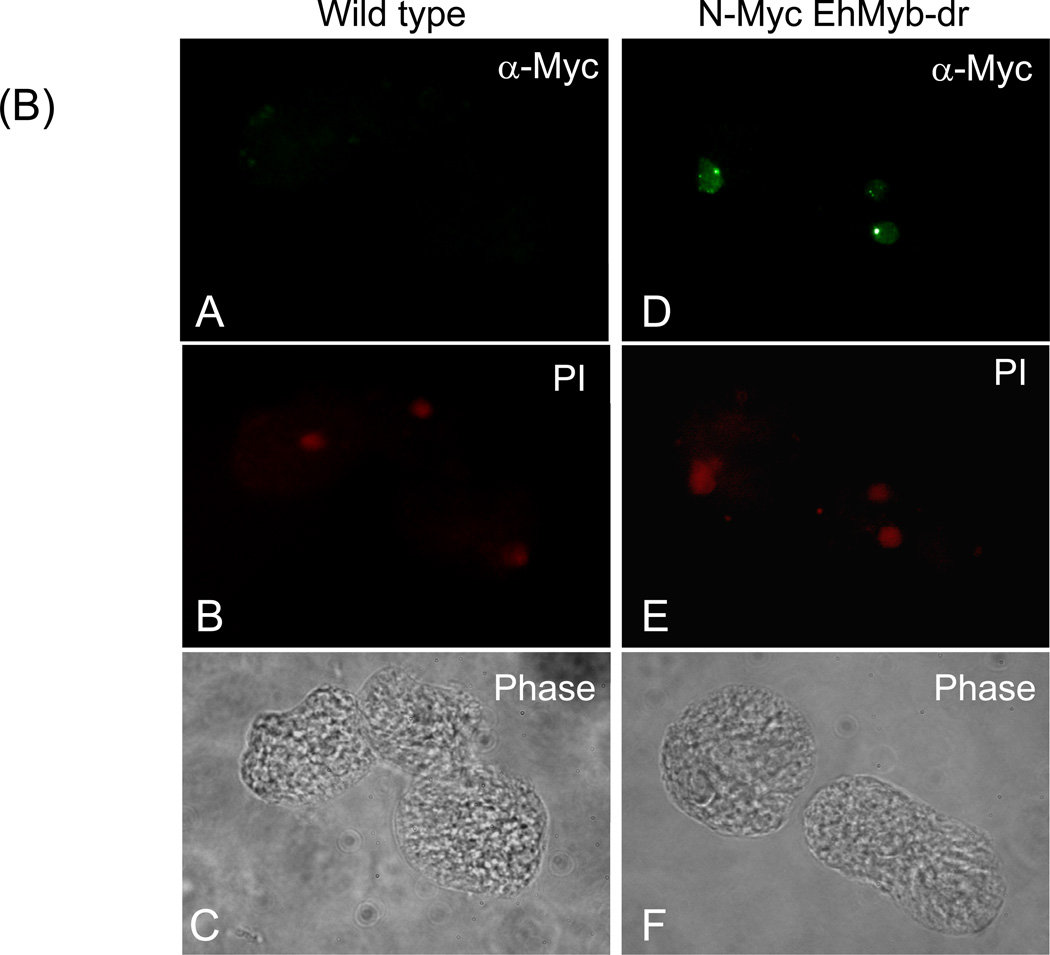
In order to determine the subcellular localization of EhMyb-dr, we created a N-terminal Myc-tagged version of the protein for expression in E. histolytica HM-1:IMSS trophozoites. Stable transfectants were selected and assayed by Western blot using an anti-Myc antibody to ensure that the full-length protein was expressed (Fig 2A). A clear band at 17 kDa was observed specifically in the N-Myc EhMyb-dr parasites, matching the expected size of EhMyb-dr. Stably transfected E. histolytica trophozoites expressing N-Myc EhMyb-dr were stained with anti-Myc antibody and nuclei were labeled with propidium iodide. As can be seen in Fig 2B, the N-Myc EhMyb-dr localized to the nucleus. Control, non-transfected parasites, did not stain with the anti-Myc antibody. From this result we conclude that EhMyb-dr has appropriate localization for its proposed function as a transcription factor.
Figure 2. Localization of EhMyb-dr.
(A) Western blot analysis of N-Myc EhMyb-dr. Using an anti-Myc antibody, single band at ~17 kD is seen. No bands are seen in the extract made from untransfected cells. (B) Immunofluorescence analysis of WT and N-Myc EhMyb-dr parasites. Staining with anti-Myc antibody staining reveals signal in the nuclei of N-Myc EhMyb-dr expressing parasites (B), but not of wild-type (untransfected) parasites (A). Nuclei are marked by propidium iodide staining (E and B). Phase contrast images are shown in F and C.
Overexpression of EhMyb-dr in E. histolytica trophozoites regulates expression of a subset of developmentally regulated genes
In order to identify genes potentially regulated by EhMyb-dr overexpression, we compared gene expression of the EhMyb-dr overexpressing parasites to that of control strains (Supplementary Table 2). Two independent biological samples of N-Myc EhMyb-dr were used for microarray analysis. The expression of EhMyb-dr was increased ~ 40-fold, confirming that we had achieved significant overexpression (Table 1A). Genes were considered regulated by overexpression of N-Myc EhMyb-dr if their expression changed >2-fold, with a p-value of <0.05, compared to control parasites. Using these parameters, we found that 117 genes were upregulated and 88 were downregulated by the overexpression of EhMyb-dr. Semi-quantitative RT-PCR was performed to confirm the array data. All genes tested agreed with the array data (Fig 3).
Table 1. The thirty genes most highly regulated by EhMyb-dr overexpression.
The probe ID, gene annotation, locus ID, expression level, and fold change in cysts are shown. The fold-change is compared to WT parasites. NR is not regulated. (A) Genes up-regulated by EhMyb-dr overexpression. The EhMyb-dr gene (7.m00410) is shown in bold in a grey box and is ~40-fold overexpressed. Genes that have been confirmed to be cyst-specific in E. invadens are shown in grey boxes. (*) the gene was ~6-fold upregulated in E. histolytica cysts but did not meet statistical significance (Ehrenkaufer et al., 2007b). (B) Genes down-regulated by EhMyb-dr overexpression.
| A | |||||
|---|---|---|---|---|---|
|
Probe ID |
Fold-change (EhMyb-dr OX vs. WT) |
p-value |
Fold-change (E. histolytica cysts) |
Gene Annotation |
Locus number |
| 227.m00086_at |
47.02 | 2.26E-02 | NR | conserved hypothetical protein | 227.t00014 |
|
7.m00410_at |
39.50 | 3.20E-02 | 5.09 | hypothetical protein | 7.t00022 |
| 127.m00138_x_at | 16.38 | 1.25E-02 | NR* | hypothetical protein | 127.t00020 |
| 42.m00175_x_at | 15.12 | 4.26E-02 | 4.62 | hypothetical protein | 42.t00010 |
| 168.m00119_s_at | 13.60 | 1.40E-02 | 11.01 | pseudogene, N-acetylmuraminidase | not found |
| 493.m00030_x_at | 11.54 | 2.86E-02 | 14.29 | hypothetical protein | 493.t00001 |
| 432.m00029_at | 9.02 | 1.79E-02 | NR | conserved hypothetical protein | 432.t00004 |
| 395.m00028_s_at | 8.61 | 3.75E-02 | NR | methionine gamma-lyase | 395.t00003 |
| 8.m00393_x_at | 8.46 | 3.93E-02 | 30.94 | late competence protein, putative | 8.t00053 |
| 141.m00082_at | 8.21 | 3.19E-02 | 13.21 | protein kinase, putative | 141.t00005 |
| 114.m00136_at | 7.31 | 4.12E-02 | NR | hypothetical protein | 114.t00010 |
| 418.m00028_at |
7.17 | 2.45E-02 | 102.10 | 70 kDa heat shock protein, putative | 418.t00001 |
| 37.m00199_x_at |
6.97 | 2.76E-02 | 51.08 | protein kinase, putative | 37.t00006 |
| 282.m00066_x_at | 6.56 | 8.52E-03 | NR | hypothetical protein | 282.t00010 |
| 356.m00029_s_at | 6.55 | 2.29E02 | NR | hypothetical protein | 356.t00001 |
| 450.m00030_at | 6.40 | 8.90E-03 | NR | hypothetical protein | 450.t00004 |
| 53.m00179_at | 5.93 | 4.02E-02 | NR | hypothetical protein | 53.t00014 |
| 131.m00151_s_at | 5.86 | 1.62E-02 | 119.70 | Fe-hydrogenase, putative | 131.t00028 |
| 48.m00224_s_at | 5.61 | 1.22E-02 | NR | acetyltransferase, putative | 48.t00042 |
| 373.m00052_s_at | 5.28 | 1.43E-02 | 38.81 | hypothetical protein | 373.t00008 |
| 401.m00029_x_at | 5.22 | 4.07E-02 | NR | hypothetical protein | 401.t00003 |
| 479.m00031_at | 5.15 | 2.79E-02 | 16.27 | hypothetical protein | 479.t00004 |
| 1087.m00005_at | 5.08 | 2.63E-02 | NR | hypothetical protein | 1087.t00001 |
| 21.m00224_at |
4.95 | 3.03E-02 | NR | ubiquitin carboxyl-terminal hydrolase, putative | 21.t00003 |
| 131.m00150_x_at |
4.74 | 6.34E-03 | 40.98 | Fe-hydrogenase, putative | 131.t00027 |
| 71.m00136_x_at | 4.70 | 1.04E-02 | NR | hypothetical protein | 71.t00008 |
| 796.m00013_s_at | 4.68 | 3.53E-03 | NR | conserved hypothetical protein | 796.t00001 |
| 53.m00214_at | 4.67 | 8.02E-03 | 3.79 | glucose ribose porter family protein | 53.t00001 |
| 247.m00077_x_at | 4.53 | 1.58E-03 | NR | hypothetical protein | 247.t00011 |
| 432.m00028_x_at |
4.49 | 3.34E-02 | 3.47 | hypothetical protein | 432.t00003 |
| 34.m00239_at | 3.90 | 1.63E-02 | 40.96 | developmentally regulated protein, putative | 34.t00018 |
| 186.m00117_at | 3.72 | 3.61E-03 | 18.08 | chitin sythase, putative | 186.t00006 |
| 128.m00121_at | 2.86 | 6.09E-03 | 87.37 | chitinase, putative | 128.t00013 |
| 6.m00488_at |
2.73 | 2.41E-03 | 27.09 | hypothetical protein | 6.t00086 |
| B | |||||
|---|---|---|---|---|---|
|
Probe ID |
Fold-change (EhMyb-dr OX vs. WT) |
p-value | Fold-change (E. histolytica cysts) |
Gene Annotation |
Locus number |
| 460.m00024_s_at | −8.70 | 3.37E-02 | NR | hypothetical protein | 460.t00002 |
| 1.m00663_at | −3.95 | 1.74E-02 | −15.70 | myosin calcium-binding light chain, putative | 1.t00066 |
| 34.m00273_at | −3.76 | 3.21E-03 | −2.78 | Rab family GTPase | 34.t00006 |
| 402.m00036_x_at | −3.75 | 4.45E-02 | NR | hypothetical protein | 402.t00006 |
| 174.m00088_at | −3.53 | 4.67E-02 | −8.47 | hypothetical protein | 174.t00015 |
| 19.m00331_at | −3.47 | 8.61E-03 | NR | RNA-binding protein, putative | 19.t00055 |
| 61.m00194_at | −3.42 | 8.33E-03 | −6.13 | hypothetical protein | 61.t00030 |
| 184.m00100_at | −3.33 | 1.02E-02 | −22.88 | Ras family GTPase | 184.t00011 |
| 217.m00081_s_at | −3.33 | 2.78E-02 | −13.89 | hypothetical protein | 217.t00003 |
| 65.m00147_x_at | −3.21 | 1.09E-02 | NR | hypothetical protein | 65.t00009 |
| 2.m00582_at | −3.17 | 3.55E-02 | NR | hypothetical protein | 2.t00092 |
| 131.m00139_at | −3.13 | 4.76E-02 | NR | M-phase inducer phosphatase, putative | 131.t00016 |
| 15.m00350_at | −3.13 | 1.18E-02 | NR | oxidoreductase, aldo keto reductase family | 15.t00059 |
| 83.m00173_x_at | −3.11 | 5.24E-03 | NR | hypothetical protein | 83.t00017 |
| 223.m00069_x_at | −3.09 | 3.88E-02 | −4.42 | hypothetical protein | 223.t00004 |
| 2.m00554_at | −3.08 | 1.29E-02 | NR | DNA polymerase alpha catalytic subunit, putative | 2.t00064 |
| 212.m00092_s_at | −3.07 | 1.77E-02 | −10.13 | thioredoxin, putative | 212.t00014 |
| 10.m00356_x_at | −3.05 | 2.72E-02 | −6.76 | protein kinase, putative | 10.t00040 |
| 6.m00423_x_at | −3.05 | 1.90E-02 | −16.00 | hypothetical protein | 6.t00021 |
| 45.m00160_at | −3.04 | 2.15E-02 | NR | hypothetical protein | 45.t00017 |
| 4.m00703_at | −2.99 | 4.39E-02 | NR | hypothetical protein | 4.t00125 |
| 184.m00098_at | −2.97 | 1.68E-02 | NR | hypothetical protein | 184.t00009 |
| 60.m00169_s_at | −2.95 | 2.78E-02 | NR | hypothetical protein | 60.t00034 |
| 3.m00563_at | −2.94 | 3.11E-02 | NR | hypothetical protein | 3.t00026 |
| 56.m00175_at | −2.93 | 2.44E-02 | NR | elongation factor 1 beta, putative | 56.t00032 |
| 229.m00071_x_at | −2.90 | 3.79E-02 | NR | Ras guanine nucleotide exchange factor, putative | 229.t00009 |
| 30.m00263_s_at | −2.88 | 9.18E-03 | NR | phosphoserine aminotransferase, putative | 30.t00047 |
| 19.m00333_at | −2.87 | 4.48E-02 | NR | cytidine deoxycytidylate deaminase family protein | 19.t00057 |
| 253.m00079_at | −2.86 | 3.62E-02 | NR | hypothetical protein | 253.t00002 |
| 98.m00128_x_at | −2.86 | 3.21E-02 | NR | hypothetical protein | 98.t00001 |
Figure 3. Semi-quantitative RT-PCR confirms microarray data for EhMyb-dr overexpression.
cDNA from WT and EhMyb-dr overexpressing parasites was tested. For all samples, a control reaction without reverse transcriptase was performed and was negative. Small subunit rRNA was used as a control.
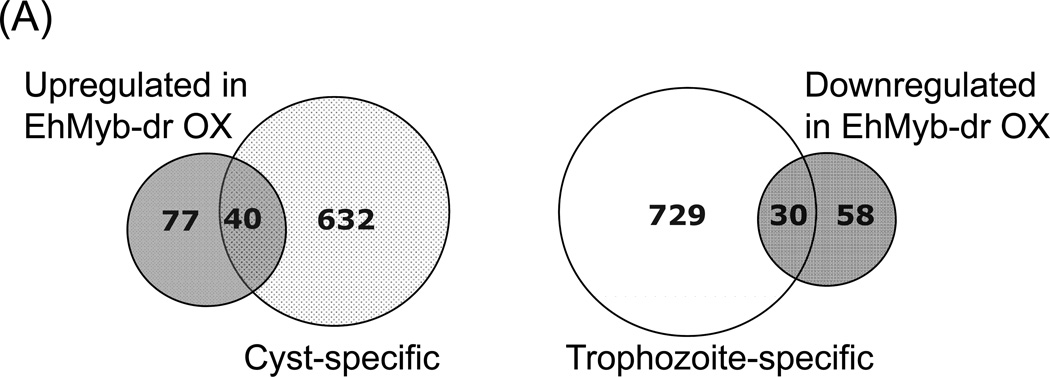
The profile of genes regulated by EhMyb-dr overexpression overlapped significantly with genes regulated in E. histolytica cysts, supporting the idea that EhMyb-dr plays a role in E. histolytica development (Fig 4A). Of the 117 genes upregulated by EhMyb-dr overexpression, 40 overlapped with cyst-specific genes (p-value 6.5e-18); of the 88 genes downregulated by EhMyb-dr overexpression, 30 overlapped with trophozoite-specific genes (p-value 3.0e-12). There was no significant overlap of genes regulated in opposing directions by EhMyb-dr overexpression and in amebic cysts.
Figure 4. Myb domain protein overexpression generated expression pattern similar to Entamoeba cysts.
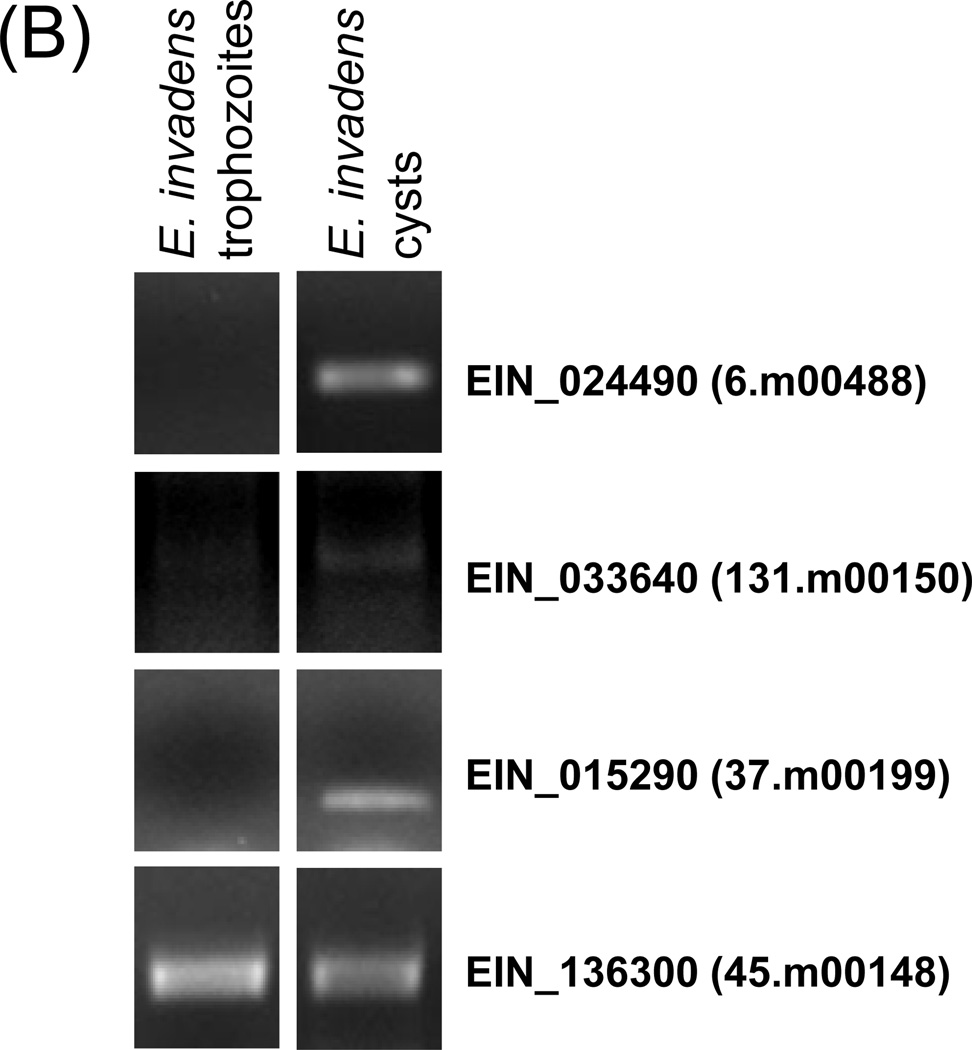
(A) Venn diagram of overlap in transcriptome between N-Myc EhMyb-dr overexpressing trophozoites and E. histolytica cysts. Of the 117 genes with increased expression in EhMyb-dr overexpressing parasites, 40 are also upregulated in E. histolytica cysts (p= 6.5e-18). Of the 88 genes with increased expression in EhMyb-dr overexpressing parasites, 30 are downregulated in E. histolytica cysts (p= 3.0e-12). (B) RT-PCR confirms that genes regulated by N-Myc EhMyb-dr overexpression in E. histolytica trophozoites have encystation-specific homologues in E. invadens. For all samples, a control reaction without reverse transcriptase was performed and was negative. EIN_136300 was a loading control (Ehrenkaufer et al., 2007b).
The 30 most highly up and down-regulated genes are shown (Tables 1A, 1B). Some genes of interest in these lists include the developmentally regulated protein (34.m00239), chitin synthase (186.m00117), and chitinase (128.m00121) all of which have previously been shown to be encystation-specific in Entamoeba (Sanchez et al., 1994; Villagomez-Castro et al., 1992; Das and Gillin, 1991). For the majority of upregulated genes, the level of induction caused by EhMyb-dr was less than the induction in cysts. The level of downregulation caused by EhMyb-dr was also lower compared to that seen in cysts. In order to determine whether genes regulated by EhMyb-dr were developmentally regulated in E. invadens, we identified genes regulated by EhMyb-dr that had single homologs in E. invadens and used RT-PCR to determine their transcript levels in E. invadens trophozoites and cysts. Three genes were tested and all were appropriately upregulated in E. invadens cysts (Fig 4B). For the two most highly regulated genes in EhMyb-dr overexpression (227.m00086 and 127.m00138) we were unable to find a good homologues in E. invadens. The 127.m00138 gene was upregulated ~6-fold in E. histolytica cysts, although the data did not meet statistical significance (Ehrenkaufer et al., 2007b). Overall, these data indicate that the transcriptome effects seen by EhMyb-dr overexpression are at least partially conserved during in vitro encystation of E. invadens. The biological relevance of EhMyb-dr and its role in development is thus conserved in Entamoeba.
Overexpression of EhMyb-dr does not induce cyst wall formation
In order to determine whether parasites that overexpress EhMyb-dr have physical features of amebic cysts, we stained with calcofluor, which stains chitin in the cyst wall. No increase in calcofluor staining was seen in EhMyb-dr overexpressing cells compared to non-transfected controls (data not shown). These findings are consistent with the transcriptome changes, since EhMyb-dr overexpression in trophozoites induced only one out of two chitin synthase genes in the genome and even the upregulated chitin synthase gene was expressed at levels much lower than in cysts (Table 1A). We also noted that other known cell wall components, such as Jacob, were not upregulated by overexpression of EhMyb-dr. In order to determine whether chitin could be induced with further stress, we exposed EhMyb-dr overexpressing parasites to osmotic stress but did not observe significantly more calcofluor staining when compared to controls (data not shown). These results indicate that although overexpression of EhMyb-dr is enough to initiate a portion of the encystation pathway, it is not sufficient to generate mature amebic cysts. Thus, additional regulatory events are likely required for mature cyst formation. Additionally, it should be noted that these experiments were performed in E. histolytica trophozoites of the strain HM-1:IMSS. This is the canonical lab strain and the most amenable to genetic study, but it has been passed for many years in axenic culture, and multiple lines of evidence suggest that it may no longer be competent to undergo encystation (Singh and Ehrenkaufer, 2008; Ehrenkaufer et al., 2007b; Eichinger, 2001). Thus, the data with EhMyb-dr overexpression (lack of mature cysts with chitin-containing cyst walls, despite the transcriptome changes that overlap with cysts) is not surprising. However, the overall features of EhMyb-dr overexpression (a transcriptome that overlaps significantly with E. histolytica and E. invadens cysts) suggest that EhMyb-dr has a role in regulating Entamoeba stage conversion. In the future, development of an in vitro system for studying encystation in E. histolytica, or genetic tools to manipulate gene expression in E. invadens, would allow further genetic dissection and clarification of the role of EhMyb-dr in Entamoeba development.
Conserved promoter motifs can be identified in EhMyb-dr regulated genes
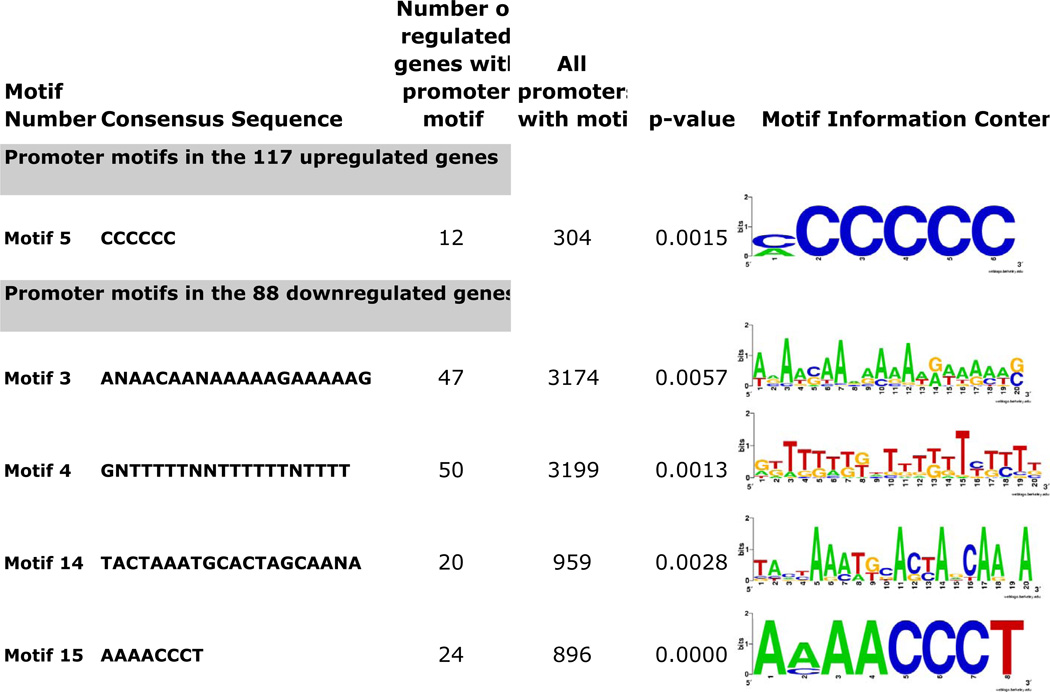
In order to identify the molecular mechanism by which EhMyb-dr regulates gene expression, we used bioinformatic methods to identify DNA motifs present in the promoters of genes regulated by EhMyb-dr. Such motifs could represent binding sites for EhMyb-dr or other transcription factors that regulate EhMyb-dr responsive genes. The promoter regions of genes with altered expression upon EhMyb-dr overexpression were analyzed using the MEME program to identify conserved promoter motifs. The MAST program was used to ascertain which promoter motifs are enriched in the promoters of genes regulated by EhMyb-dr compared to all other genes (9,675 total) in the amebic genome. Of the 30 motifs identified using MEME (15 in promoters of upregulated genes and 15 in promoters of downregulated genes), 5 were significantly enriched (p <0.05) in the promoters of genes with altered expression in EhMyb-dr overexpressing cells (Fig 5).
Figure 5. Promoter motifs significantly enriched in genes regulated by EhMyb-dr overexpression.
The motif number, sequence, number of occurrences in promoters of regulated and unregulated genes, p-value, and information content are shown.
The motif M5 (CCCCCC), was particularly interesting due to its occurrence in the highly A/T rich E. histolytica genome. This motif occurred 304 times in the genome and was highly enriched in the genes upregulated by N-Myc EhMyb-dr (p-value = 0.0015). No genes with this promoter motif were found to be downregulated by N-Myc EhMyb-dr overexpression. In the majority (8/12) of promoters of EhMyb-dr regulated genes in which it was found, at least one occurrence of the CCCCCC motif was present within 200bp of the translation start site, a common feature of the short promoters and transcriptional regulatory motifs that have previously been identified in E. histolytica (Hackney et al., 2007; Purdy et al., 1996). The developmentally regulated genes that were regulated by EhMyb-dr, and which contain the CCCCCC motif, include the developmentally regulated protein (34.m00239) and 432.m00028. In two promoters, multiple copies of the M5 motif were identified: in one promoter (127.m00138) the M5 motifs were at 186bp and 322bp upstream of the start codon; in the other promoter (470.m00032) the motifs were at 163bp, 203bp, and 395bp upstream of the start codon. To ascertain whether the sequences adjacent to the C-rich region might also be involved in EhMyb-dr activity, we aligned the regions surrounding this region in the promoters of EhMyb-dr targets, but found only weak conservation of an A/T rich region (data not shown). The M15 motif (AAAACCCT) has previously been identified in the promoters of genes with high expression in trophozoites and was significantly enriched in the promoters of ribosomal protein and tRNA synthetase genes (Hackney et al., 2007). Since the M15 motif is known to be enriched in the promoters of genes involved in protein synthesis, the downregulation of genes with M15 in their promoters is likely not a direct result of EhMyb-dr expression, but rather indicative of lower translation rates in cysts. Of the 24 genes with the M15 motif that have decreased expression in EhMyb-dr overexpressing cells, 9 genes were also downregulated in cysts (Ehrenkaufer et al., 2007b). The M15 motif was also found in the promoters of 10 genes with increased expression in N-Myc EhMyb-dr expressing cells, but this was not a statistically significant enrichment over its presence in promoters of all genes.
E. histolytica nuclear protein(s) bind preferentially to a CCCCCC promoter motif in EhMyb-dr overexpressing parasites
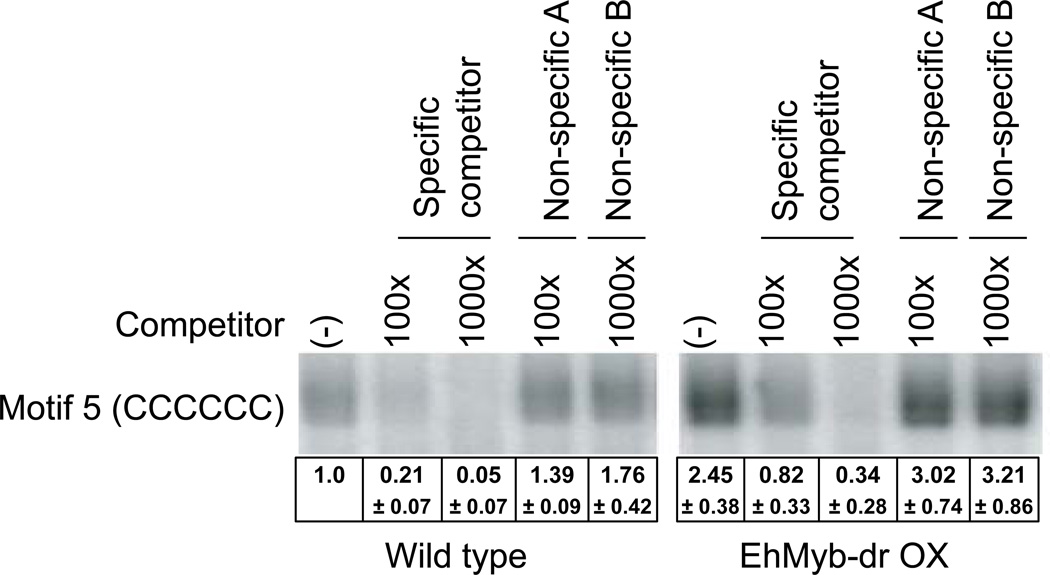
In order to determine if this C-rich motif could act as a binding site for amebic nuclear proteins, we performed electrophoretic mobility shift assay (EMSA) analysis. Labeled oligonucleotide containing the CCCCCC motif was incubated with nuclear extract prepared from either untransfected or EhMyb-dr overexpressing trophozoites. A band was identified which was present using either extract, but which had a significantly higher intensity in extract made from N-Myc EhMyb-dr expressing cells (Fig 6). This band was strongly reduced by the addition of a non-radioactive specific competitor, but not by two non-specific competitors. Densitometric analysis of EMSA bands from multiple experiments indicated that the binding to the C-rich motif was significantly greater in the EhMyb-dr overexpressing parasites. We further confirmed that binding was specific to the C-rich motif of interest and not to the flanking region used in making the oligonucleotides. An EMSA using an oligonucleotide with an altered internal sequence (C content of >50%) and the same flanking region revealed no significant binding to amebic nuclear extract (data not shown). Overall, the data indicate that amebic nuclear protein(s) bind specifically to the CCCCCC motif and that the binding is increased in EhMyb-dr overexpressing parasites. These data are consistent with the hypothesis that the CCCCCC motif can be bound in a sequence-specific manner by EhMyb-dr. However, this interaction could also be indirect with EhMyb-dr forming part of a larger complex that binds this motif.
Figure 6. Electrophoretic mobility shift assays with the M5 motif.
EMSA with a putative EhMyb-dr binding motif, M5 (CCCCCC) in nuclear extract from wild type and N-Myc EhMyb-dr overexpressing parasites. This motif shows a specific band, which can be competed away by specific but not by non-specific competitors and has higher signal from the N-Myc EhMyb-dr parasite nuclear extract. Multiple EMSA experiments were performed and the mean intensity ± standard error are shown. A representative EMSA gel is shown.
ChIP assays demonstrate direct interaction of EhMyb-dr with promoters containing the CCCCCC motif in genes regulated by N-Myc EhMyb-dr
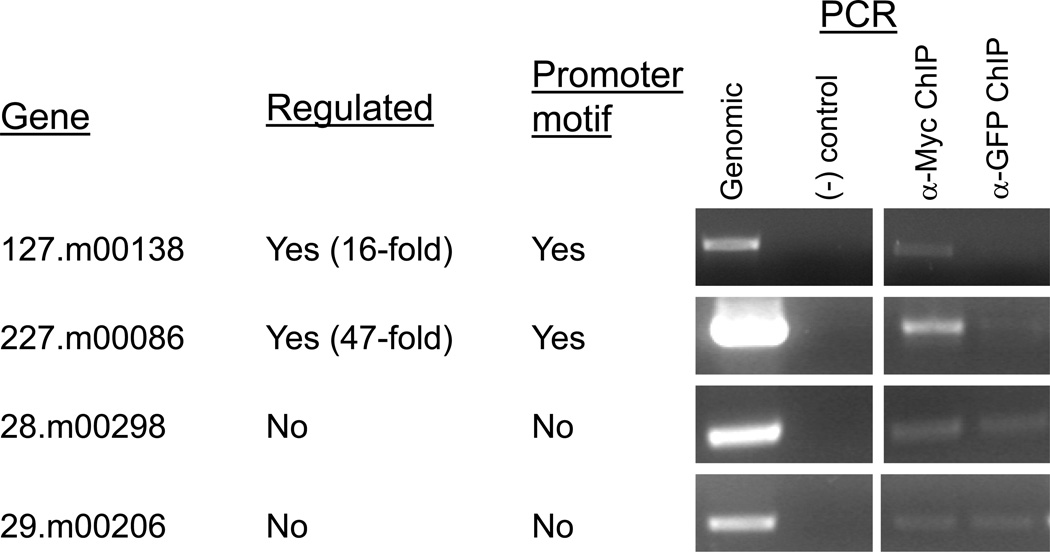
To determine if EhMyb-dr binds the CCCCCC motif in vivo we used ChIP to demonstrate physical interaction of the N-Myc EhMyb-dr to the promoters containing the C-rich motif. Chromatin was prepared from N-Myc EhMyb-dr expressing parasites, spilt into two equal pools, and immunoprecipitated (IP) with either anti-Myc antibody (which will pull down any DNA associated with the N-Myc EhMyb-dr) or with anti-GFP antibody (control). Purified IP DNA was amplified using PCR, in order to determine if promoters containing the CCCCCC motif were enriched in the anti-Myc IP samples as compared to the anti-GFP control. The promoters of two genes (127.m00183 and 227.m00086), which showed increased expression in N-Myc EhMyb-dr transfectants and which contain CCCCCC motifs, were tested. Both showed enrichment in the anti-Myc antibody IP sample when compared to the anti-GFP antibody IP (Fig 7). Two genes not regulated by EhMyb-dr and without CCCCCC motifs, were used as controls; no difference was detected between the samples IP with anti-Myc or anti-GFP antibodies. These data indicate that the N-Myc EhMyb-dr physically interacts in a specific manner with the promoters containing the CCCCCC motifs in genes regulated by EhMyb-dr in vivo. In combination with the EMSA results, the ChIP data provide strong evidence that EhMyb-dr can act as a transcriptional regulator by binding to the CCCCCC motif. Thus, the genes that have the CCCCCC promoter motif and which are regulated by EhMyb-dr overexpression are likely regulated by direct binding of the EhMyb-dr. In contrast the genes that are regulated in EhMyb-dr overexpressing parasites, but which do not have the CCCCCC motif in their promoter, are likely indirectly regulated by EhMyb-dr.
Figure 7. Chromatin immunoprecipitation of N-Myc EhMyb-dr bound to CCCCCC containing promoters.
Chromatin was prepared from N-Myc EhMyb-dr expressing cells and immunoprecipitated with beads bound to either α-Myc antibody or with α-GFP as a control. Samples were assayed using PCR to determine the levels of DNA that had been precipitated from each promoter. Promoter sequences from EhMyb-dr regulated genes containing the motif (127.m00138 and 227.m00086) were enriched in the α-Myc IP samples as compared to the α-GFP samples. Two genes, 28.m00298 and 29.m00206 that are not regulated by N-Myc EhMyb-dr overexpression and do not contain the motif were used as a control, and showed no difference between IP with the α-Myc or the control antibody.
Regulation of developmentally regulated genes by multiple pathways
The transcriptional networks that control encystation in E. histolytica are undoubtedly complex and controlled by a number of factors including promoter occupancy by transcription factors and chromatin modifications. It has previously been demonstrated that exposure to Trichostatin A (an inhibitor of histone deacetylase) induces a transcriptome that overlaps significantly with the developmentally regulated genes (Ehrenkaufer et al., 2007a). In order to determine the extent of overlap of genes upregulated by Trichostatin A exposure by and N-Myc EhMyb-dr overexpression, we compared the transcriptome profiles under those two conditions. Our analysis identified that there was a small, but significant overlap of genes co-regulated under both conditions (Supplementary Table 2B and 2C). Among genes upregulated in the two conditions, 14 genes overlap, 10 of which have increased expression in cysts (p-value= 2e-10). Of the six genes are downregulated by both TSA treatment and by EhMyb-dr overexpression, five are also trophozoite-specific (p-value = 1.8e-6) (Figure 4A). None of the 14 genes upregulated by both TSA and EhMyb-dr has the CCCCCC motif identified earlier. These data are consistent with indirect regulation of these genes by the EhMyb-dr and implicate a common final pathway of gene regulation, which can be initiated independently by EhMyb-dr or histone modifications.
Discussion
Developmental regulation and stage conversion from trophozoites to cysts is key to disease transmission in E. histolytica. However, the molecular framework that regulates this aspect of amebic biology is poorly defined. We have identified the first developmentally regulated transcription factor in E. histolytica, a Myb domain gene. Our findings demonstrate that EhMyb-dr regulates expression of a subset of stage-specific genes in E. histolytica and that it does so partly by interacting with a C-rich motif in the promoters of regulated genes. This work is the first identification of a developmentally regulated transcription factor in E. histolytica.
Myb proteins are important regulators and are found widely in eukaryotes. In animals, a major function of Myb proteins is to promote cell proliferation and progression through the cell cycle, particularly during development (Oh and Reddy, 1999). In contrast, many Myb domain proteins in plants and protists are important for the formation of differentiated cell types, including root hair and pigment producing cells in plants (Ramsay and Glover, 2005). A G. lamblia Myb protein regulates encystation (Huang et al., 2008; Sun et al., 2002), and in Dictyostelium Myb proteins are required at multiple stages in development (Fukuzawa et al., 2006; Guo et al., 1999).
As with their biological roles, the domain structure of Myb proteins is also variable. The founding member of this family, c-Myb, contains three imperfect tandem repeats of the ~50aa Myb domain, called R1, R2 and R3 (Saikumar et al., 1990). However, proteins that contain two repeats or even one Myb repeat have been found and shown to have DNA binding activity (Jin and Martin, 1999). EhMyb-dr is a member of a the SHAQKY subfamily of Myb proteins, which generally have only one or two Myb domains and share a conserved motif (SH[A/L]QKY[R/F]) (Rose et al., 1999). This family is commonly represented in plants, where members help regulate such diverse functions as maintenance of circadian rhythms (Wang et al., 1997) and seed maturation (Rubio-Somoza et al., 2006). A SHAQKY protein from Dictyostelium is developmentally regulated and required for expression of pre-stalk genes, demonstrating that this Myb family can be important for differentiation (Fukuzawa et al., 2006). Hence, the proposed role for EhMyb-dr in regulating expression of stage-specific genes would be consistent with the functions of SHAQKY family of Myb proteins.
While mammalian Myb proteins that contain three Myb domains share a common consensus binding site (C/TAACGG), plant proteins with Myb domains have diverse motifs (Jin and Martin, 1999; Rose et al., 1999; Grotewold et al., 1991), most likely due to divergent sequences in the C-terminal α-helix of the Myb domain (Jin and Martin, 1999). The binding site of a Dictyostelium SHAQKY Myb protein has been identified; a C-rich region (CACCCCAC) was found to be required for both binding and full activity, although additional sequences 5' of the C-rich region were also required (Fukuzawa et al., 2006). We have identified CCCCCC as a putative binding site for EhMyb-dr. The similarity of the DNA motifs bound by Dictyostelium and E. histolytica Myb SHAQKY proteins is intriguing and may indicate that this family of Myb proteins has a conserved binding motif.
The developmentally regulated Myb protein is expressed at low levels in the trophozoite stage and should thus be available, immediately upon receiving the encystation stimuli, to regulate expression of stage-specific genes. This regulation may be direct through binding of the EhMyb-dr to the CCCCCC motif in the genes of promoters that have it. However, there is also likely indirect or secondary induction, as a number of genes induced by EhMyb-dr overexpression do not have this binding motif. Additionally, a number of genes were identified which contain the C-rich motif in their promoter regions but were not regulated by EhMyb-dr overexpression. These genes may be regulated by other transcription modulators or need cofactors not present in the conditions tested.
In summary, we have identified the first developmentally regulated transcription factor, which is likely to be involved in regulating Entamoeba histolytica stage conversion. Our approach identified the transcriptional regulatory network associated with EhMyb-dr as well as the CCCCCC DNA-binding region with which the EhMyb-dr protein interacts. As overexpression of EhMyb-dr was able to induce only a subset of cyst-specific genes, other transcriptional regulators, along with chromatin remolding factors such as histone acetylases, are likely to be required for stage conversion to occur. Further work in characterizing potential regulators of encystation will be necessary to fully understand the molecular and regulatory framework associated with Entamoeba development. Elucidation of these mechanisms will be useful in designing novel means of inhibiting disease transmission.
Experimental Procedures
Entamoeba histolytica culture conditions and plasmid construction
Cultures of E. histolytica HM-1:IMSS were grown under standard axenic culture conditions at 36.5°C (Diamond et al., 1978). E. invadens (IP-1) growth and encystation were previously described (Sanchez et al., 1994). The full-length EhMyb-dr gene was PCR amplified from E. histolytica HM-1:IMSS cDNA, cloned into PCR2.1-TOPO (Invitrogen) and sequence verified. The plasmid was digested with SmaI and XhoI and inserted into the pKT-3M vector (Saito-Nakano et al., 2004), to produce a N-terminal Myc-EhMyb-dr. Trophozoites were transfected using 20µg of plasmid, and 30µl Superfect transfection reagent (Qiagen) in 35mm petri dishes as previously described (MacFarlane and Singh, 2007). Stable transfectants were selected by treatment with 6µg/ml G418, and maintained at 24µg/ml G418.
Immunofluorescence assays and Western blot analysis
Log-phase parasites were chilled for 10min at 4°C, placed in chamber slides (Nunc), and allowed to adhere for 30 min at 37°C. Amebas were rinsed once with room temperature PBS, and fixed in (50%) acetone-(50%) methanol for 5 min. The reaction was blocked with PBT-BSA for 30 min, followed by staining for 1 h with anti-Myc antibody (1:1000) (Cell Signaling). Parasites were rinsed with PBT and incubated with the secondary antibody Alexa 488 anti-mouse 1:1000 (Molecular Probes), and propidium iodide for 1 h. Chitin staining was done with 25 µM calcofluor (Ehrenkaufer et al., 2007b). For Western blot analysis, trophozoites were lysed in NP-40 buffer plus DNase (50 µg/ml), RNase (25 µg/ml) and protease inhibitors (500 µM AEBSF, 1 µM leupeptin, 1 µM E-64). Lysates were blotted onto a PVDF membrane. Mouse anti-Myc antibody (Cell signaling) and anti-mouse HRP (Roche) were used (1:1000) and signal detected with ECL+ (Amersham).
Transcriptional analysis of EhMyb-dr overexpressing parasites
RNA was isolated from N-Myc EhMyb-dr at 24 µg/ml G418 drug selection and prepared for microarray analysis as previously published (Ehrenkaufer et al., 2007b). Samples were hybridized to a custom generated Affymetrix platform full genome microarray, which represents 9,435 open reading frames (Gilchrist et al., 2006). Changes in gene expression caused by the overexpression of N-Myc EhMyb-dr were identified by comparing their expression profile to control parasites. A number of controls were used, including non-transfected HM-1:IMSS trophozoites grown without drug selection (Ehrenkaufer et al., 2007b), as well as HM-1:IMSS trophozoites transfected with plasmids containing a selectable marker and grown with G418 drug selection. Probe sets were considered differentially expressed between the N-Myc EhMyb-dr and control parasites if they had at least a 2-fold change and p-value < 0.05 calculated using a Welch's t-test. cDNA generation and RT-PCR was performed as in (Ehrenkaufer et al., 2007b). PCR primers are in Supp Table 1.
Promoter motif identification in EhMyb-dr coregulated genes
The MEME and MAST programs were downloaded from UCSD (http://meme.sdsc.edu). The MEME motif elucidation program was run with the command line arguments: –dna –mod anr –minw 6 –maxw 20 –minsites 4 –nmotifs 15 (Hackney et al., 2007). Using these parameters, the program identifies 15 non-overlapping motifs found any number of times in each promoter, with a width between 6 and 20 nucleotides. The promoter sequences of all E. histolytica genes were obtained as in (Hackney et al., 2007). We used the MAST program to identify all occurrences of each motif in the promoter sequences of all amebic genes. The e-value cutoff for motif identification was < 500. Information content for promoter motifs logos is generated by weblogo (http://weblogo.berkeley.edu/).
Electrophoretic mobility shift assays (EMSA)
Amebic nuclear proteins were isolated and EMSA performed using the protocol described in (Gilchrist et al., 1998). For each reaction, 5µg of nuclear protein (determined using Bradford reagent) and 50fmol labeled probe were used. Double-stranded oligonucleotide probes (Supp Table 1) were designed for each motif and binding reactions occurred as outlined in (Hackney et al., 2007). For competition, 100 and 1000-fold molar excess unlabeled oligonucleotide were added to the reactions. Quantitation of band intensity from multiple EMSA experiments was performed using Kodak MI software. The mean ± standard error is shown and the results normalized to the signal from the CCCCCC motif binding to the WT extract (set to 100%).
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed as described in (Gilchrist et al., 2003) with modifications. Protein G sepharose beads (Sigma) were bound to 4 µg of either anti-Myc (Cell Signaling) or anti-GFP (Roche) antibodies. Two 75 cm2 flasks of N-Myc EhMyb-dr overexpressing cells were crosslinked in PBS + 1% formaldehyde for 10 min at 37°C and the crosslinking reaction stopped by the addition of glycine to 0.125M. Cells were iced to release from flask, pelleted, washed once in PBS and resuspended in buffer A (10mM HEPES-KOH pH 7.9, 1.5mM MgCl2, 10mM KCl) plus protease inhibitor cocktail (Pierce) and 1µM E-64). After 20 min incubation on ice, cells were pelleted, resuspended in buffer A + 6%NP-40, and passed through a Qiashredder homogenization column (Qiagen). Nuclei were pelleted by spinning 10 min at 1200 rcf, resuspended in sonication buffer (20mM HEPES-KOH pH 7.9, 0.42nM NaCl, 1mM EDTA, 1mM EGTA supplemented with protease inhibitors), and sonicated 4×30s. Whole nuclei and cell membranes were removed by centrifugation at 20,000 rcf, and supernatant was passed 3× through a 30-gauge needle. The resulting extract was diluted 3× in low-pH buffer (10mM HEPES-KOH pH 7.0, 1.5mM MgCl2, 10mM KCl + protease inhibitors). Chromatin was bound to the beads by incubation for 2 hrs at 4°C with gentle agitation. Beads were washed 2× in low salt buffer (10mM HEPES-KOH pH 7.0, 1.5mM MgCl2, 10mM KCl, 140mM NaCl ), 2× in high salt buffer (10mM HEPES-KOH pH 7.0, 1.5mM MgCl2, 10mM KCl, 500mM NaCl), 2× in wash buffer (10mM Tris-HCl pH 8.0, 250mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate, 1mM EDTA) and 1× in TE. Elution occurred by incubation for 10 min at 65°C in 50mM Tris pH 8.0, 10mM EDTA, 1% SDS. Beads were pelleted, and supernatant recovered. Crosslinking was reversed by overnight incubation at 65°C in TE/SDS solution. Finally, samples were subjected to proteinase K digestion and treatment with RNAse A, and cleaned on a Qiagen PCR purification column. From 30 µl of eluate, 5 µl were used for PCR amplification. Primer sequences and conditions are found in Supplementary Table 1.
Supplementary Material
Supplementary Figure 1: Alignment of all E. histolytica SHAQKY domain proteins.
Supplementary Table 1: (A) Primers and PCR conditions utilized. The gene ID, primer name, sequences, and PCR conditions are listed. (B) Primers used in EMSA analysis.
Supplementary Table 2: Microarray data. (A) Microarray data for all genes for all arrays used in the analysis. The probe ID, gene annotation, locus ID, and expression level are shown for E. histolytica HM-1:IMSS, control arrays, and EhMyb-dr overexpressing parasites. (B) Microarray data for all genes up-regulated by EhMyb-dr overexpression. The probe ID, gene annotation, locus ID, fold-change, and p-value are shown. Genes regulated by TSA exposure are indicated. (C) Microarray data for all genes transcriptionally down-regulated by EhMyb-dr overexpression. The probe ID, fold-change, gene annotation, locus ID, fold-change, and p-value are shown. Genes regulated by TSA exposure are indicated.
Acknowledgements
We acknowledge all members of the Singh lab for helpful comments and suggestions. US was supported in part by NIH grants AI-069382 and AI-068899, JAH was supported by a training grant to the Department of Microbiology T32 AI-07328 and GME was supported by NIH grant AI-068899 and a Stanford University McCormick Fellowship.
Abbreviations
- WT
Wild type
- EMSA
Electrophoretic mobility shift assay
- ChIP
Chromatin immunoprecipitation
- IP
Immunoprecipitate
- HDAC
histone deacetylase
Footnotes
All microarray data are available at the GEO database website at NCBI (Accession number GSE13023).
References
- Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- Biedenkapp H, Borgmeyer U, Sippel AE, Klempnauer KH. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988;335:835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- Boyle JP, Saeij JP, Cleary MD, Boothroyd JC. Analysis of gene expression during development: lessons from the Apicomplexa. Microbes Infect. 2006;8:1623–1630. doi: 10.1016/j.micinf.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum . PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary MD, Singh U, Blader IJ, Brewer JL, Boothroyd JC. Toxoplasma gondii asexual development: identification of developmentally regulated genes and distinct patterns of gene expression. Eukaryot Cell. 2002;1:329–340. doi: 10.1128/EC.1.3.329-340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Gillin FD. Chitin synthase in encysting Entamoeba invadens . Biochem J. 1991;280(Pt 3):641–647. doi: 10.1042/bj2800641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva EK, Gehrke AR, Olszewski K, Leon I, Chahal JS, Bulyk ML, Llinas M. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc Natl Acad Sci U S A. 2008;105:8393–8398. doi: 10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba . Trans R Soc Trop Med Hyg. 1978;72:431–432. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- Ehrenkaufer GM, Eichinger DJ, Singh U. Trichostatin A effects on gene expression in the protozoan parasite Entamoeba histolytica . BMC Genomics. 2007a;8:216. doi: 10.1186/1471-2164-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. Identification of developmentally regulated genes in Entamoeba histolytica : insights into mechanisms of stage conversion in a protozoan parasite. Cell Microbiol. 2007b;9:1426–1444. doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- Eichinger D. Encystation in parasitic protozoa. Curr Opin Microbiol. 2001;4:421–426. doi: 10.1016/s1369-5274(00)00229-0. [DOI] [PubMed] [Google Scholar]
- Fukuzawa M, Zhukovskaya NV, Yamada Y, Araki T, Williams JG. Regulation of Dictyostelium prestalk-specific gene expression by a SHAQKY family MYB transcription factor. Development. 2006;133:1715–1724. doi: 10.1242/dev.02327. [DOI] [PubMed] [Google Scholar]
- Gilchrist CA, Mann BJ, Petri WA., Jr Control of ferredoxin and Gal/GalNAc lectin gene expression in Entamoeba histolytica by a cis-acting DNA sequence. Infect Immun. 1998;66:2383–2386. doi: 10.1128/iai.66.5.2383-2386.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist CA, Leo M, Line CG, Mann BJ, Petri WA., Jr Calcium modulates promoter occupancy by the Entamoeba histolytica Ca2+-binding transcription factor URE3-BP. J Biol Chem. 2003;278:4646–4653. doi: 10.1074/jbc.M211271200. [DOI] [PubMed] [Google Scholar]
- Gilchrist CA, Houpt E, Trapaidze N, Fei Z, Crasta O, Asgharpour A, et al. Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol Biochem Parasitol. 2006;147:163–176. doi: 10.1016/j.molbiopara.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Athma P, Peterson T. Alternatively spliced products of the maize P gene encode proteins with homology to the DNA-binding domain of myb-like transcription factors. Proc Natl Acad Sci U S A. 1991;88:4587–4591. doi: 10.1073/pnas.88.11.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo K, Anjard C, Harwood A, Kim HJ, Newell PC, Gross JD. A myb-related protein required for culmination in Dictyostelium . Development. 1999;126:2813–2822. doi: 10.1242/dev.126.12.2813. [DOI] [PubMed] [Google Scholar]
- Hackney JA, Ehrenkaufer GM, Singh U. Identification of putative transcriptional regulatory networks in Entamoeba histolytica using Bayesian inference. Nucleic Acids Res. 2007;35:2141–2152. doi: 10.1093/nar/gkm028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi MA, Deitsch KW. Epigenetics in Apicomplexa: control of gene expression during cell cycle progression, differentiation and antigenic variation. Curr Opin Microbiol. 2007;10:357–362. doi: 10.1016/j.mib.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Haque R, Huston CD, Hughes M, Houpt E, Petri WA., Jr Amebiasis. N Engl J Med. 2003;348:1565–1573. doi: 10.1056/NEJMra022710. [DOI] [PubMed] [Google Scholar]
- Huang YC, Su LH, Lee GA, Chiu PW, Cho CC, Wu JY, Sun CH. Regulation of cyst wall protein promoters by Myb2 in Giardia lamblia . J Biol Chem. 2008 doi: 10.1074/jbc.M805023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Katzen AL, Kornberg TB, Bishop JM. Isolation of the proto-oncogene c-myb from D. melanogaster . Cell. 1985;41:449–456. doi: 10.1016/s0092-8674(85)80018-0. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- MacFarlane RC, Singh U. Identification of an Entamoeba histolytica serine-, threonine-, and isoleucine-rich protein with roles in adhesion and cytotoxicity. Eukaryot Cell. 2007;6:2139–2146. doi: 10.1128/EC.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow BE, Ju Q, Warner JR. A bipartite DNA-binding domain in yeast Reb1p. Mol Cell Biol. 1993;13:1173–1182. doi: 10.1128/mcb.13.2.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- Palm D, Weiland M, McArthur AG, Winiecka-Krusnell J, Cipriano MJ, Birkeland SR, et al. Developmental changes in the adhesive disk during Giardia differentiation. Mol Biochem Parasitol. 2005;141:199–207. doi: 10.1016/j.molbiopara.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Purdy JE, Pho LT, Mann BJ, Petri WA., Jr Upstream regulatory elements controlling expression of the Entamoeba histolytica lectin. Mol Biochem Parasitol. 1996;78:91–103. doi: 10.1016/s0166-6851(96)02614-x. [DOI] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Rose A, Meier I, Wienand U. The tomato I-box binding factor LeMYBI is a member of a novel class of myb-like proteins. Plant J. 1999;20:641–652. doi: 10.1046/j.1365-313x.1999.00638.x. [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Martinez M, Diaz I, Carbonero P. HvMCB1, a R1MYB transcription factor from barley with antagonistic regulatory functions during seed development and germination. Plant J. 2006;45:17–30. doi: 10.1111/j.1365-313X.2005.02596.x. [DOI] [PubMed] [Google Scholar]
- Saikumar P, Murali R, Reddy EP. Role of tryptophan repeats and flanking amino acids in Myb-DNA interactions. Proc Natl Acad Sci U S A. 1990;87:8452–8456. doi: 10.1073/pnas.87.21.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito-Nakano Y, Yasuda T, Nakada-Tsukui K, Leippe M, Nozaki T. Rab5-associated vacuoles play a unique role in phagocytosis of the enteric protozoan parasite Entamoeba histolytica . J Biol Chem. 2004;279:49497–49507. doi: 10.1074/jbc.M403987200. [DOI] [PubMed] [Google Scholar]
- Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, et al. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. . Mol Cell Biol. 2005;25:10301–10314. doi: 10.1128/MCB.25.23.10301-10314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez L, Enea V, Eichinger D. Identification of a developmentally regulated transcript expressed during encystation of Entamoeba invadens . Mol Biochem Parasitol. 1994;67:125–135. doi: 10.1016/0166-6851(94)90102-3. [DOI] [PubMed] [Google Scholar]
- Singh U, Ehrenkaufer GM. Recent insights into Entamoeba development: Identification of transcriptional networks associated with stage conversion. Int J Parasitol. 2008 doi: 10.1016/j.ijpara.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CH, Palm D, McArthur AG, Svard SG, Gillin FD. A novel Myb-related protein involved in transcriptional activation of encystation genes in Giardia lamblia. Mol Microbiol. 2002;46:971–984. doi: 10.1046/j.1365-2958.2002.03233.x. [DOI] [PubMed] [Google Scholar]
- Vicente JB, Ehrenkaufer GM, Saraiva LM, Teixeira M, Singh U. Entamoeba histolytica modulates a complex repertoire of novel genes in response to oxidative and nitrosative stress: implications for amebic pathogenesis. Cell Microbiol. 2008 doi: 10.1111/j.1462-5822.2008.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villagomez-Castro JC, Calvo-Mendez C, Lopez-Romero E. Chitinase activity in encysting Entamoeba invadens and its inhibition by allosamidin. Mol Biochem Parasitol. 1992;52:53–62. doi: 10.1016/0166-6851(92)90035-i. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Amoebiasis. Wkly Epidemiol Rec. 1997;72:97–99. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Alignment of all E. histolytica SHAQKY domain proteins.
Supplementary Table 1: (A) Primers and PCR conditions utilized. The gene ID, primer name, sequences, and PCR conditions are listed. (B) Primers used in EMSA analysis.
Supplementary Table 2: Microarray data. (A) Microarray data for all genes for all arrays used in the analysis. The probe ID, gene annotation, locus ID, and expression level are shown for E. histolytica HM-1:IMSS, control arrays, and EhMyb-dr overexpressing parasites. (B) Microarray data for all genes up-regulated by EhMyb-dr overexpression. The probe ID, gene annotation, locus ID, fold-change, and p-value are shown. Genes regulated by TSA exposure are indicated. (C) Microarray data for all genes transcriptionally down-regulated by EhMyb-dr overexpression. The probe ID, fold-change, gene annotation, locus ID, fold-change, and p-value are shown. Genes regulated by TSA exposure are indicated.