Abstract
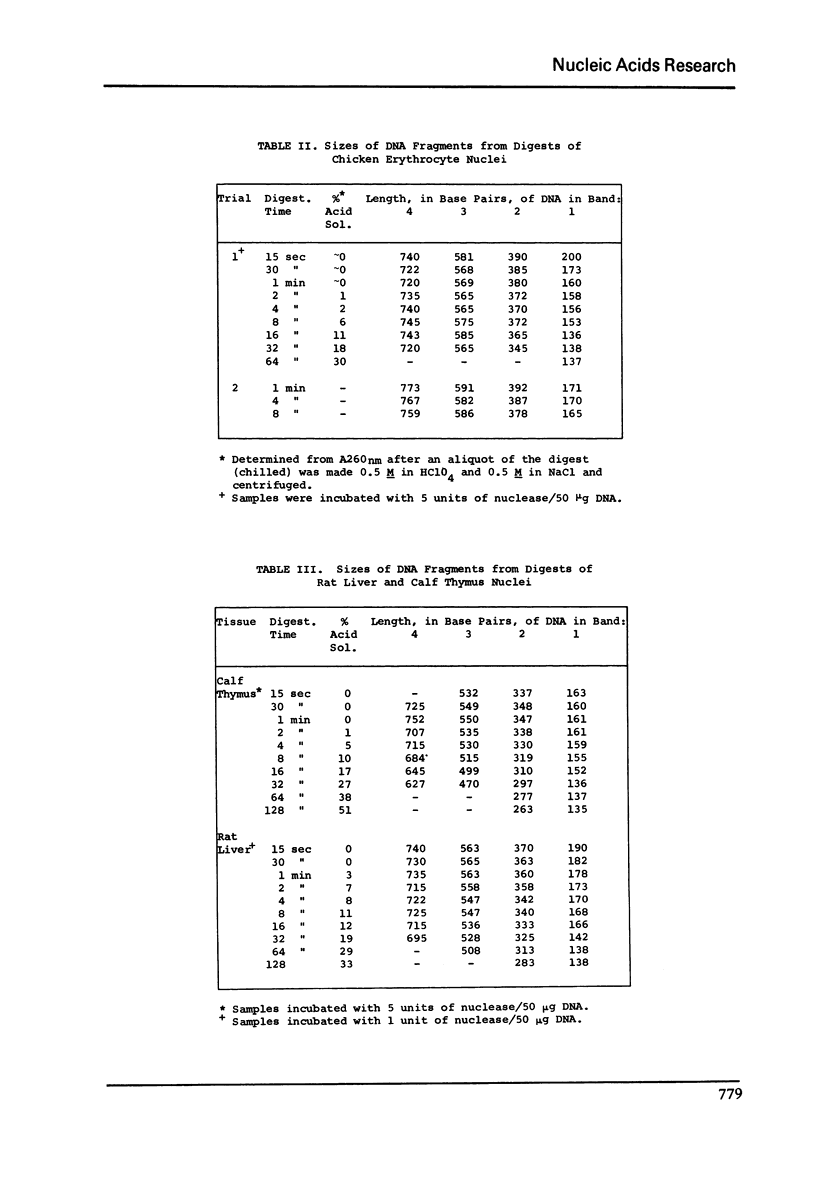
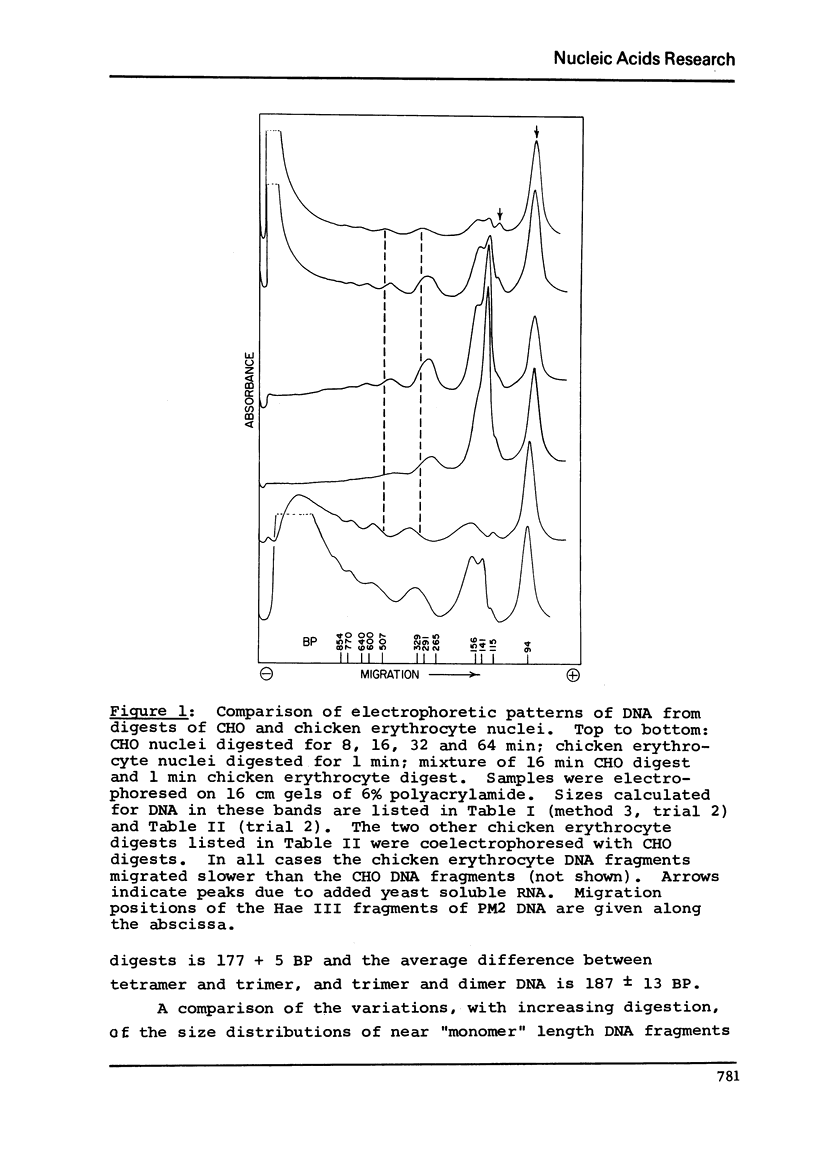
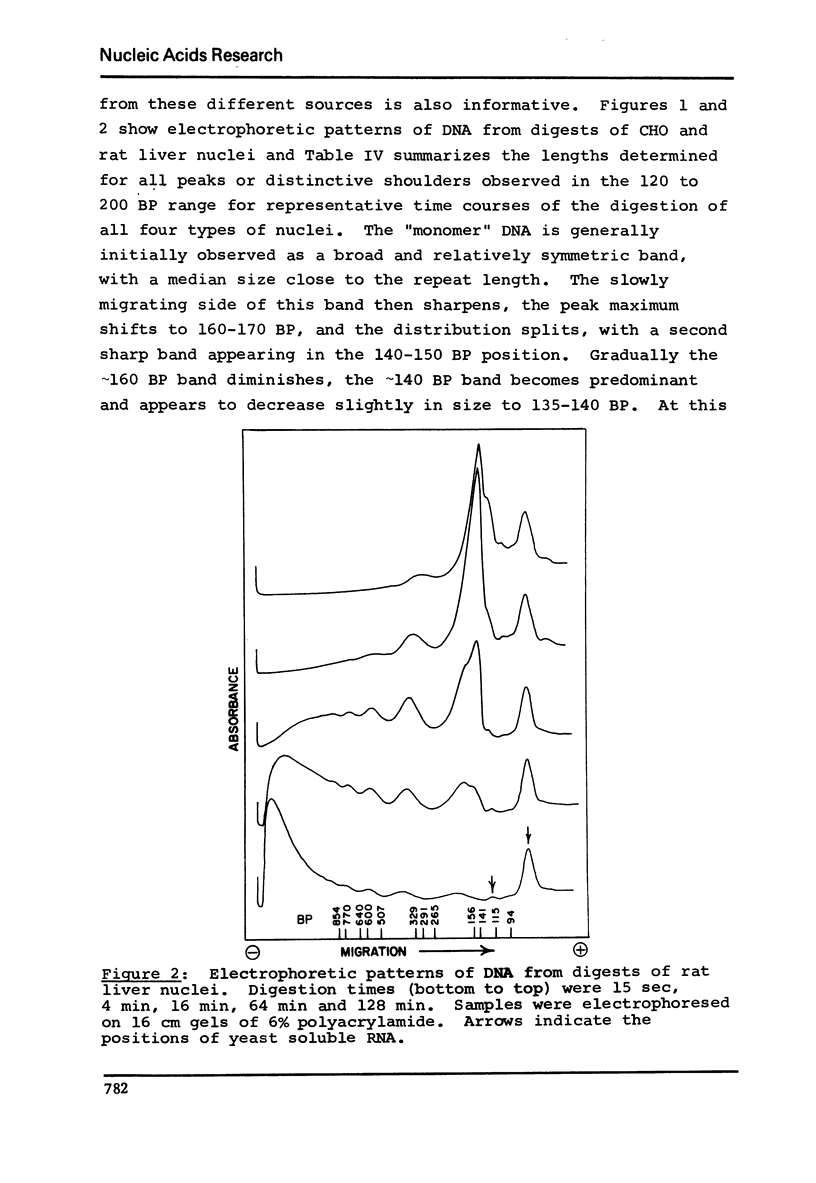
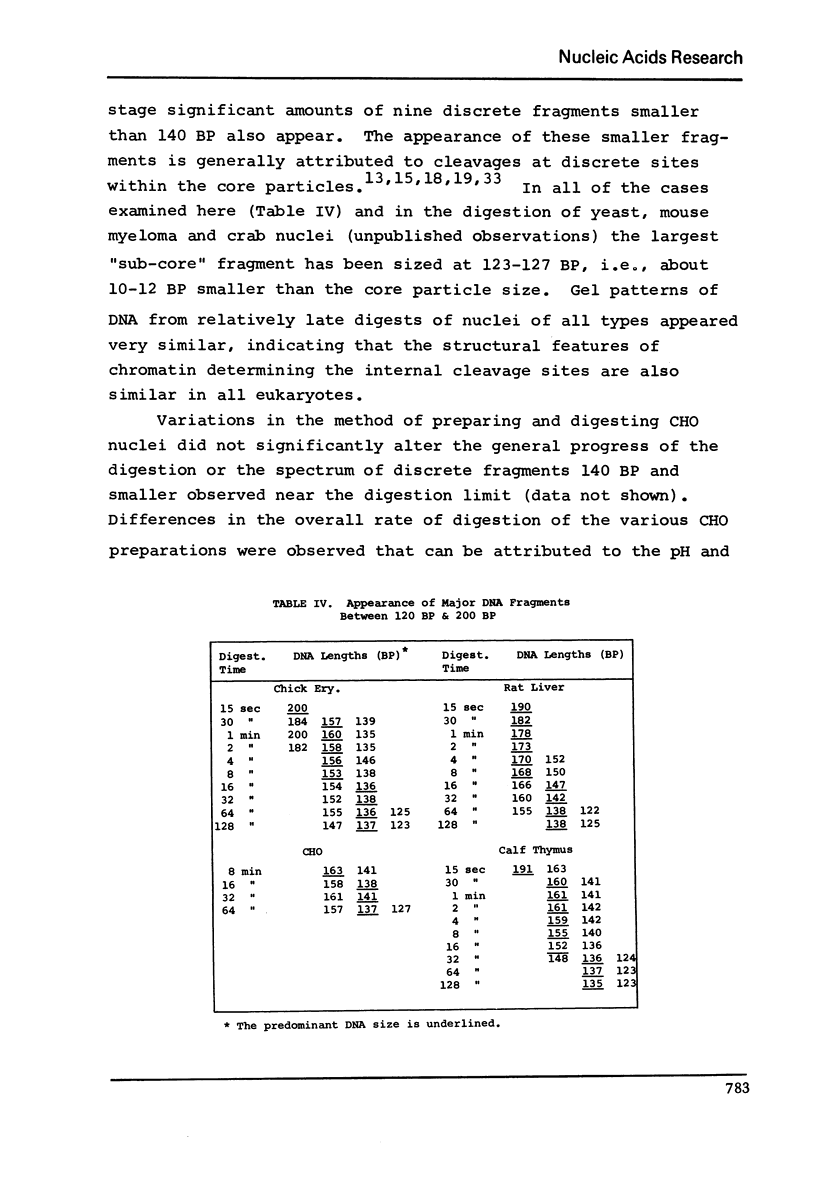
DNA lengths in the structural repeat units of Chinese hamster ovary (CHO) and chicken erythrocyte chromatin were compared by analyzing the sizes of DNA fragments produced after treatment of nuclei with staphylococcal nuclease. The repeat length of CHO chromatin (173 +- 4 BP) is about 20 base pairs (BP) smaller than that of chicken erythrocyte chromatin (194 +- 8 BP). Repeat lengths of rat liver and calf thymus chromatin were found to be about 10 BP shorter than that of chicken erythrocyte chromatin. Thus significant variations occur in repeat units of chromatin of higher eukaryotes. These variations occur in the lengths of "spacer" (or "internucleosomal") DNA segments, not in "core particle" (or "nucleosomal") DNA lengths. The concept of spacer regions and the possible influence of H1 histones is discussed.
Full text
PDF














Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R. Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2921–2925. doi: 10.1021/bi00684a020. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Hewish D. R., Mobbs J. Mammalian chromatin substructure studies with the calcium-magnesium endonuclease and two-dimensional polyacrylamide-gel electrophoresis. Biochem J. 1974 Oct;143(1):67–72. doi: 10.1042/bj1430067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Chemical probes of chromatin structure. Biochemistry. 1974 Aug 13;13(17):3622–3628. doi: 10.1021/bi00714a034. [DOI] [PubMed] [Google Scholar]
- Compton J. L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky M. A., Keevert J. B. Subunit structure of a naturally occurring chromatin lacking histones F1 and F3. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3536–3540. doi: 10.1073/pnas.72.9.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Murphy R. F., Bonner J. Structure of transcriptionally active chromatin. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4404–4408. doi: 10.1073/pnas.72.11.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Honda B. M., Baillie D. L., Candido E. P. Properties of chromatin subunits from developing trout testis. J Biol Chem. 1975 Jun 25;250(12):4643–4647. [PubMed] [Google Scholar]
- Keichline L. D., Villee C. A., Wassarman P. M. Structure of eukaryotic chromatin. Evaluation of periodicity using endogenous and exogenous nucleases. Biochim Biophys Acta. 1976 Feb 18;425(1):84–94. doi: 10.1016/0005-2787(76)90218-5. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Yeast chromatin subunit structure. Science. 1975 Apr 11;188(4184):165–166. doi: 10.1126/science.1090006. [DOI] [PubMed] [Google Scholar]
- Morris N. R. Nucleosome structure in Aspergillus nidulans. Cell. 1976 Jul;8(3):357–363. doi: 10.1016/0092-8674(76)90147-1. [DOI] [PubMed] [Google Scholar]
- Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976 Jul;8(3):349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oosterhof D. K., Hozier J. C., Rill R. L. Nucleas action on chromatin: evidence for discrete, repeated nucleoprotein units along chromatin fibrils. Proc Natl Acad Sci U S A. 1975 Feb;72(2):633–637. doi: 10.1073/pnas.72.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Rill R. L., Oosterhof D. K., Hozier J. C., Nelson D. A. Heterogeneity of chromatin fragments produced by micrococcal nuclease action. Nucleic Acids Res. 1975 Sep;2(9):1525–1538. doi: 10.1093/nar/2.9.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasrabuddhe C. G., Van Holde K. E. The effect of trypsin on nuclease-resistant chromatin fragments. J Biol Chem. 1974 Jan 10;249(1):152–156. [PubMed] [Google Scholar]
- Shaw B. R., Corden J. L., Sahasrabuddhe C. G., Van Holde K. E. Chromatographic separation of chromatin subunits. Biochem Biophys Res Commun. 1974 Dec 23;61(4):1193–1198. doi: 10.1016/s0006-291x(74)80410-9. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P., Jr Chemical evidence that chromatin DNA exists as 160 base pair beads interspersed with 40 base pair bridges. Nucleic Acids Res. 1976 Jan;3(1):117–127. doi: 10.1093/nar/3.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Furber V. Yeast chromatin structure. FEBS Lett. 1976 Jul 15;66(2):274–280. doi: 10.1016/0014-5793(76)80521-2. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Removal of histone H1 exposes a fifty base pair DNA segment between nucleosomes. Biochemistry. 1976 Jul 27;15(15):3307–3314. doi: 10.1021/bi00660a022. [DOI] [PubMed] [Google Scholar]
- van Bruggen E. F., Arnberg A. C., van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. Electron microscopy of chromatin subunit particles. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1365–1370. doi: 10.1016/0006-291x(74)90348-9. [DOI] [PubMed] [Google Scholar]



