Abstract
Tuberculosis (TB) has been a disease affecting almost all parts of the world since ages. Lot many efforts came in the past for improving diagnosis and treatment. Also, an effective vaccine has been sought after for long. With the emergence of resistant strains of Mycobacterium tuberculosis, the causal organisms of tuberculosis, and complexities emerging due to other associated infections and disease conditions, there is a desperate need for further research input in the field. Be it the better medication and care or better resistance management, proper diagnostics holds the key to success. It has been observed that a high burden of the disease was accompanied by resource limitations and poor research set-up. The scenario remained like this for several decades. With the refreshed vision of resourceful countries and funding agencies, funding is being provided in many areas of research in tuberculosis diagnosis and treatment. This review has been written with an aim to bring forth the limitations of available methods in the field of diagnostics and making researchers aware about the changing scenario with better funding opportunities and support. The author visualizes an enthusiasm from all over the world for the development of better modalities and urges scientists to join the struggle at this very perfect time to take the challenge and come forward with innovations in this field.
KEY WORDS: Diagnostic techniques and procedures, innovation, tuberculosis
INTRODUCTION
A report, published in September 2009, from the World Health Organization (WHO)-sponsored study includes tuberculosis (TB) and AIDS with a share of 5.5% each in the list of top 10 causes of death in all 10–24 year olds, males and females combined, globally.[1] This has revealed an urgency to act on these issues with all our available resources and an approach to improve upon them. Stop TB partnership announced the theme for World Tuberculosis Day 2010 as “On the move against tuberculosis: Innovate to accelerate action.” This calls for ideas and projects to come up with something other than what we have been practicing for long. We are still not well equipped to fight TB. Improving the research in TB has been a neglected area for long, in part due to the complex characteristics of the causal organism Mycobacterium tuberculosis, the variable host response to infection, and, perhaps, complacency as this disease was nearly eradicated in high-income settings.[2] With increasing HIV–TB co-infections, TB has become a high priority for action and research in international health again. Looking into this, WHO and the World Bank, with International Union against Tuberculosis and Lung Disease (IUATLD), Centre for Disease Control and Prevention (CDC), and other organizations, are reassessing their approaches to the prevention and control of TB.[3] Of late, agencies such as the Foundation for Innovative New Diagnostics (FIND), Stop TB Partnership's New Diagnostics Working Group (NDWG), Global Laboratory Initiative (GLI, another Stop TB Partnership Working Group), WHO, and the Special Program for Research and Training in Tropical Diseases (TDR) have shown increased interest toward the development of better diagnostics for TB.[4] Funding agencies such as the Bill and Melinda Gates Foundation, the Global Fund to Fight AIDS, TB and Malaria (GFATM), and UNITAID have emerged as resource providers for work on this disease that was being neglected in terms of sponsorship and interest of the private sector till now.
With this, lots of efforts and funds are being brought up for the development of newer diagnostic methods in mycobacteriology. But it remains to be a major challenge till date. There are multiple hurdles to cross before we reach the ultimate goal of having an efficacious vaccine, easy and affordable diagnosis, and short-term treatment regimens with minimal side effects.
SMEAR MICROSCOPY
In the field of TB, problem starts at the very first step, i.e., diagnosing the case. Conventional TB diagnosis has been relying on medical history, tuberculin skin test, chest X-rays, and bacteriological examination. M. tuberculosis grows slowly, and clinical specimens submitted to the TB microscopy and culture are contaminated to varying degrees by more rapidly growing unwanted normal flora. Microscopy and culturing both become difficult in such cases. Moreover, the hydrophobic nature of the cell wall due to a high concentration of mycolic acid makes it more difficult to stain mycobaterial cells. The poor sensitivity of conventional smear microscopy has been a major concern. With various samples and sample processing methods, it has been found to be around 36–43% sensitive.[2] In general, direct smear reportedly detects AFB only at concentrations of around 10,000 bacilli/ml of the specimen. Conversely, as few as 100 bacilli/ml may be required for a positive culture.[5] Various modifications to the technique and higher level of sophistications have been introduced in this field to get the highest level of sensitivity. With certain variations in the sample processing methods for smear preparation, a few reports of improvement in detection limits are there. Samples were subjected to different centrifugation forces and centrifugation times after decontamination and liquefaction and the centrifuged deposits were examined by smear and culture. The sensitivity of detection at an RCF of 4000 g for 15 min was 5000 organisms/ml for a smear.[6]
Various workers have demonstrated the improved detection of direct smear-negative cases by universal sample processing (USP) smear microscopy.[7,8] The highest limit of detection in smear microscopy after employing the USP methodology and observing 400–500 fields was experimentally found to be 250–300 bacilli/ml of the sample. Under the best conditions for culture on solid and liquid media with USP-treated spiked sputum, the detection limit was found to be 400 CFU/ml after 8 weeks of incubation. Hence, although it could reduce the bacilli count needed to be detected using smear, it has not helped improving detection by culture. The reason may be the deleterious effect of sample treatment. However, the researchers felt a need for a more sensitive test to detect samples with a low bacterial load. Steingart and co-workers did a systematic review to assess the ability of different processing methods to improve the sensitivity of microscopy.[9] Their search suggested that centrifugation with any of several chemical methods (including bleach) is more sensitive, that overnight sedimentation preceded by chemical processing is more sensitive, and that the specificity is similar. Recently, a pilot study has compared the diagnostic accuracy and incremental yield of two short-duration (<1 h) sputum pretreatment procedures involving pretreatment with bleach and USP centrifugation and concluded that both did not increase yield as compared to direct sputum smears.[10] People tried hard for developing field-based methods to avoid the use of centrifugation during decontamination. Culturing the sputum directly without decontamination or centrifugation has also been tried.[11] But it was found to have a lower sensitivity as compared to the conventional method of centrifuge decontamination. Fluorescence microscopy is credited with increased sensitivity and lower work effort but has a rider of increased cost and technical complexity. Pai and co-workers[4] have reported an average 10% increment in sensitivity of microscopy using fluorescence microscopy with no difference in specificity. Lange and Mori have given an update on the developments being made in the field of fluorescent microscopy.[12] These include light-emitting diode (LED)-based fluorescent microscopy, mobile phone-based microscopy, and automated detection systems using image processing. With many more other studies, it is very apparent that we are still struggling hard to get a sensitive, easy, cost-effective, and faster method for smear microscopy. It seems possible to decrease mortality and morbidity due to TB using smear microscopy diagnosis and DOTS coverage, as being done in most of the high prevalence countries like India.[13] But as many smear negative cases may remain undiagnosed, cutting transmission of infection does not seem to be an achievable target without having better diagnostic amenities.
THE CULTURE OF CULTURING
Just the presence of acid-fast bacilli (AFB) on a sputum smear or other specimens does not confirm a diagnosis of TB as all acid-fast-bacilli are not M. tuberculosis. Culture remains the gold standard for the laboratory confirmation of TB. Also, bacterial isolation for drug-susceptibility testing and genotyping is required using a solid or liquid medium. Therefore, a culture is expected on all initial samples to confirm the diagnosis followed by biochemical tests like catalase test at 68°C, nitrate reduction, and niacin accumulation for speciation. Although culture-based diagnosis of TB is recommended in International Standards of Tuberculosis Care, lack of resources and technical expertise poses as a major limitation in most of the high prevalence countries.[14] Traditionally, primary isolation and culture of mycobacteria is performed on agar or Lowenstein–Jensen (LJ) media. Drug susceptibility testing of isolates is done using the same medium following the proportion method. This method compares the mycobacterial growth levels in clinical isolates to be tested with known standard culture in the presence of different concentrations of anti-TB drugs in a proportional way. This method enables a determination of the concentration that inhibits more than 99% of the inoculum and is reported as the minimal inhibitory concentration (MIC) of the given drug. This is considered as a “gold standard.”[15] But it is cumbersome, technically demanding, lacks reproducibility, and it generally takes at least 21 days for a result after assay set-up. Liquid culturing with radioisotopic detection or with the incorporation of fluorescent dyes was introduced in the past as a confirmatory method (Bactec 460, BACTEC MGIT 960 system, MB/BacT, and Versa Trek system). However, looking at the sophistication and issues like contamination, the struggle again started to search something better. Microscopic observation drug susceptibility (MODS) testing developed recently allows both rapid and low-cost TB diagnosis in liquid culture with the simultaneous determination of drug susceptibilities.[16–18] However, MODS requires higher biosafety level facilities, an expert and experienced microbiologist, and the samples are very prone to contaminations. Thus, it needs addition of various antibiotics in culture media and the samples need to be processed before inoculation involving decontamination and centrifugation steps. Some other unconventional methods like thin-layer agar (TLA) and the direct nitrate reductase assay (NRA) have attempted to address the problem of multiple point processing and hence the generation of aerosols by incorporating visual inspection of results in the form of typical colony morphology or color change to identify TB growth.[19,20] Although they have tried to make things simple, they could not avoid the most cumbersome step of specimen processing.
THE EVOLUTION CONTINUES
Present systems for the detection of TB infection totally rely on microscopy and culture. Microbiological detection is possible only once the microbial load in the sample reaches a substantial number. Some other situations like immune suppression and in the case of children, getting a good quality sample and getting the required microbial load in the samples is tough. Culture-based approaches are never going to be as fast as we need. Hence, indirect approaches to trace the causal organism/byproducts or to look into the host response against the infection are also being considered.
The PCR-based amplification of various target nucleic acids has been tried extensively that allows rapid and sensitive detection of target DNA sequences. Amplified sequences accumulate to concentrations that are easily detected using nonisotopic detection methods. Ideally, PCR can detect a single copy of the gene being targeted for amplification but with all the processing to avoid interference, the detection of as few as 4–10 copies has been made possible by several workers.[21,22]
Various targets have been tried and tested to get better identification or speciation till date. Although the 16S rRNA gene is the most commonly used target, other targets have also provided a high-sensitivity and representative species-specific differentiation. The PCR amplification of the entire 16S–23S rRNA spacer region and use of a secondary technique of randomly amplified polymorphic DNA (RAPD) fingerprinting to differentiate strains belonging to the Mycobacterium species has been reported.[23] The amplification of the entire 16S–23S intergenic region, and diagnostic tests for bacterial organisms using probes targeted for sequences within the 16S–23S intergenic region have also been described.[24,25] Other targets include the 16S rRNA gene, the 16S–23S internal transcribed spacer, the 65-kDa heat shock protein, recA, rpoB, and gyrB.[23,26–29] The 16S rRNA gene-based methods are presently widely used for the identification and differentiation of mycobacteria.[30–34] However, some species cannot be differentiated by their 16S sequences because the number of polymorphic sites in the 16S rRNA gene in the genus Mycobacterium is rather small (e.g., Mycobacterium kansasii and Mycobacterium gastri), while others possess a very high degree of sequence similarity (e.g., Mycobacterium marinum and Mycobacterium ulcerans, Mycobacterium abscessus, and Mycobacterium chelonae).[35] The 16S–23S rRNA gene internal transcribed spacer (ITS) region contains both conserved and highly variable signatures and is rather small. The 16S–23S rRNA gene ITS-based PCR produces a relatively small PCR product (200–350 bp). This sequence of the 16S–23S rRNA ITS can distinguish between M. kansasii and M gastri; however, it fails to distinguish between M. marinum and M. ulcerans and needs secondary methods like RFLP or line blot hybridization to get conclusive results as demonstrated by various workers.[24,36] The first marketed test was INNO-LiPA Mycobacteria (Innogenetics) in which the 16S–23S intergenic region was amplified and then hybridized to a membrane on which were attached probes that recognized mycobacterial species revealed by colorimetry.[37] The same principle was used to develop GenoType MTBC tests (Hain Lifescience GmbH, Nehrin, Germany), which use multiplex amplification of DNA fragments (23S rRNA gene, RD1 region, and gyrB gene) to differentiate M. tuberculosis complex species (Mycobacterium tuberculosis, Mycobacterium bovis ssp bovis, M. bovis BCG, bovis ssp caprae, Mycobacterium africanum, Mycobacterium canettii, and Mycobacterium microti).[38]
The hsp65 gene-based PCR restriction pattern analysis (PRA) is also widely used for the identification of Mycobacterium spp. McNabb and co-workers assessed the use of partial sequences of the hsp65 gene for the routine identification of mycobacteria and reported an overall agreement of 85.2% with other identification methods; discrepancies were most frequently encountered with isolates of M. chelonae, Mycobacterium fortuitum, Mycobacterium gordonae, Mycobacterium scrofulaceum, and Mycobacterium terrae.[39] The amplification of the secA1 gene that codes for the essential protein SecA1, a key component of the major pathway of protein secretion across the cytoplasmic membrane, has also been tried.[40] It was demonstrated that the secA1 gene of the genus Mycobacterium can be used for species-level or complex-level (for the MTB complex) identification. It was also found that secA1 is a suitable target for diagnostics as the degree of interspecies variation for gene sequences among mycobacteria was observed to be moderate. These conclusions came from nonexhaustive studies and a need for further inputs for the improvement of the same was felt.
Apart from costly and cumbersome probe-based methods, combining the detection of various characteristic features or the multiple species identification using multiplex PCR also seems to be promising. For instance, to detect three mycobacterial species, the multiplex PCR is estimated to cost at least half that of a commercial DNA probe which is used to identify only one of the three pathogens that can be identified by a multiplex PCR assay. In this regard, the use of the multiplex PCR could provide large savings in time, costs, and laboratory resources compared with the use of the expensive commercial DNA probes, subcloning procedures, and biochemical tests. The use of multiplex PCR to detect members of the Mycobacterium genus and to detect and differentiate M. tuberculosis, Mycobacterium avium, and Mycobacterium intracellulare has also been investigated.[40] However, there exist a few limitations with respect to DNA isolation, species identification, and obtaining cultures from a sophisticated system like BACTEC as a source to begin with.
The recent development of the real-time PCR-based instrument named GeneXpert system, recently endorsed by the WHO, has led to the rapid detection of TB and drug resistance at the point of care. The MTB/RIF test using this system provided sensitive detection of TB and rifampin resistance directly from the untreated sputum in less than 2 h with minimal hands-on time.[41] WHO has recently issued a policy statement for the Xpert MTB/RIF system.[42] This statement provides a brief description of the results obtained from various analytical studies, controlled clinical validation trials, field demonstration studies, etc. Analysis with this system reports the detection of five genomic copies and 131 cfu/ml MTB spiked in sputum. Based on the results, it has been recommended that
Xpert MTB/RIF should be used as the initial diagnostic test in individuals suspected of MDR-TB or HIV-associated TB (strong recommendation).
Xpert MTB/RIF may be used as a follow-on test to microscopy in settings where MDR and/or HIV is of lesser concern, especially in smear-negative specimens (conditional recommendation, recognizing major resource implications).
However, conventional microscopy, culture, and DST, which are required to monitor treatment progress and to detect resistance to drugs other than rifampicin were found to be indispensible at this time. Also several operational conditions need to be maintained for proper functioning of the instrument. Apart from this, the cost of the instrument and the cartridges used is another concern that is being negotiated and taken care of by agencies like FIND.
Currently, there is an urgent need for a highly sensitive and specific diagnostic method for the identification of active M. tuberculosis disease that can be performed at the point of care, i.e., outside the traditional laboratory setting. As serological methods have never shown to be very consistent in terms of their sensitivity and specificity for the detection of M. tuberculosis infection, research has again focused on finding new biomarkers for better detection. Developing a method that uses alternate clues like biomarkers is desperately needed as all available diagnostic methods require a sputum sample limiting applicability to patients with pulmonary disease who are able to provide sputum for analysis. Various molecules from mycobacterial as well as the host origin are being explored and tested for the same to get the desired speed, accuracy, and consistency in diagnosing the disease and restricting it for the benefit of patient and their environment.
There are several reports of new biomarkers being proposed and have shown some promise. In addition to the biomarkers used in the currently available diagnostic tests, the literature is rich with candidate biomarkers like early secretary proteins such as early secretory antigenic 6 kDa (ESAT-6) and culture filtrate protein 10 (CFP-10), malate synthase, etc., and a few molecules emerging from the host in response to infection with varying degrees of validation with the potential for development into new diagnostic tests.[43–46] The emergence of new knowledge about interactions of M. tuberculosis with its host with the advancing technology has provided new concepts of the clinical phenotypes, pathogenesis, and host immune responses in tuberculosis and the resulting opportunities for biomarker discovery. The use of “omics” approaches may be needed for biomarker discovery in individuals who are exposed but not infected, who get infected but do not get the disease and in comparing them with those who succumb to the infection. Also, it is expected that these new biomarkers would be able to differentiate between active and latent tuberculosis. The “omics” approach involves fetching out the signatures from transcriptomic, proteomic, or metabolomic profiling. Hopefully, some biomarkers would provide us with a reliably consistent indication of infection and would help us detect the same before the infection proceeds to a level wherein we could easily detect AFB in sputum or otherwise but late.
THE INDIAN PERSPECTIVE
India bears the highest burden of TB (1.96 million cases annually).[47] Also, with a significantly higher number of HIV patients (2.3 million prevalent cases),[48] it complicates the management of TB in HIV-infected individuals. With about 50% lifetime risk of developing TB disease in HIV-infected people, it is projected that 50%–60% of the HIV-infected persons in India will develop TB disease during their lifetime.[49,50] Comparable data from various Indian studies conducted between 1994 and 2006 also report the prevalence of HIV–TB co-infection in the range of 3%–55%.[51]
Scientific efforts have been put in by academia and research institutes in India for the development of better diagnostic tools. India has been a big market for in vitro diagnostics but has been dominated by imported and generic products, mostly serological, with virtually no innovations. Examples of the development in the field of hepatitis and others from the Indian industry have given a hope that Indian diagnostic companies could also become world's hub for high-quality generic diagnostics in the field of TB diagnostics. For this to happen, the Indian industry needs to venture into genuine innovation in the field. This requires supportive policies, enhanced and timely funding, and greater collaboration between the workers and the funders from the public and private sector. Such efforts are becoming visible now. RNTCP, being an official caretaker in India for TB control, has been very active in the recent past. In line with the WHO 12-point policy package, RNTCP has also adopted strategies to diagnose and manage TB in HIV-infected patients. The program has immediate priorities of restricting TB infection by providing treatment to all infected individuals. For diagnosis, there exist the guidelines for intensive case finding at the community level, but for early diagnosis of TB in the Indian population, not many efforts could be made. This is very justifiable in light of huge numbers of already existing cases of TB. Indian Council of Medical Research (ICMR) has also been working extensively on disease control programs with a support of the continued exploitation of scientific and technological advances from basic to applied research, from biomedical to health sciences, and from laboratory to field research. ICMR is providing significant information through its laboratories engaged in TB research and also provides funding to various academic and research institutions for research in this area.
Researchers from various parts of India have also shown concerted efforts in this direction. They are not only discussing the problem and its solutions among themselves, but also consulting the international community for disseminating their work and getting updates from all over the world. An international symposium on TB diagnostics held at International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi, India, in December 2010 titled “Innovating to Make an Impact” discussed multiple aspects regarding the challenges in TB diagnostics. A very positive feel for support in the field of diagnostic development came out of this.[52] A consultative meeting held in January 2011 at National AIDS Research Institute, India, on “Galvanizing Evidence for HIV Management” also incorporated a full session on TB supported by WHO. This was given a name “HIV and TB: Partners in crime” (report preparation in progress). Policy makers, subject experts, program implementers, researchers, HIV and TB physicians from different parts of India, researchers from another resource-limited setting like South Africa, and researchers from USA took part in the discussions. Exclusive discussions on diagnosing extra-pulmonary TB, childhood TB, and HIV–TB were conducted as these pose serious challenges to developing universally applicable diagnostic tools for TB. The willingness and determination for better diagnosis and management of TB from laboratory workers to the policy makers further have shown a promising future.
CONCLUSIONS
This systematic review has not tried to incorporate the strategy of meta-analysis as the reports or publications being considered here are not sufficient to cover the entire range of efforts being contributed. Recently, private enterprises are putting in a lot for the development of newer and better diagnostic tools in the field of TB. But the data from this end are not available to statistically assemble the results of studies into a single estimate. Although the review has not taken into view all upcoming and sophisticated technologies which are not evidence based, the aim is to land upon an idea of basic problems being identified in due course of diagnostic development. Table 1 reviews the frequently used techniques, problems associated with them, and an approach to solve these problems. Also, in normal diagnostic settings which use conventional methods of diagnosis, it is always difficult to maintain quality and ensuring biosafety in the absence of recommended equipments. In a very recent policy recommendation by WHO, a warning has been issued against the use of inaccurate blood tests for active TB, and these tests have been defined as “substandard tests with unreliable results.”[53] WHO has advised most of the countries which use these methods to ban the inaccurate and unapproved blood tests and instead rely on accurate microbiological or molecular tests, as recommended by WHO. As there is a big market for these tests in India, the focus is now going to be toward alternatives which can be placed in the market for active TB diagnosis. This is going to generate a huge demand of better diagnostic methods. In the light of a recent report “Totally Drug Resistant Tuberculosis in India,”[54] followed by a big controversy about the origin and proper identification, it becomes indispensible for a clinician to confirm about the status of infecting species along with the exact nature of drug resistance. At the same time, a clinician cannot wait for culture-based reports to initiate the treatment. In such cases, PCR/marker-based techniques would help getting concrete evidences of the etiology and would direct the treatment options on the right track.
Table 1.
Overview of diagnostic methods and their limitations
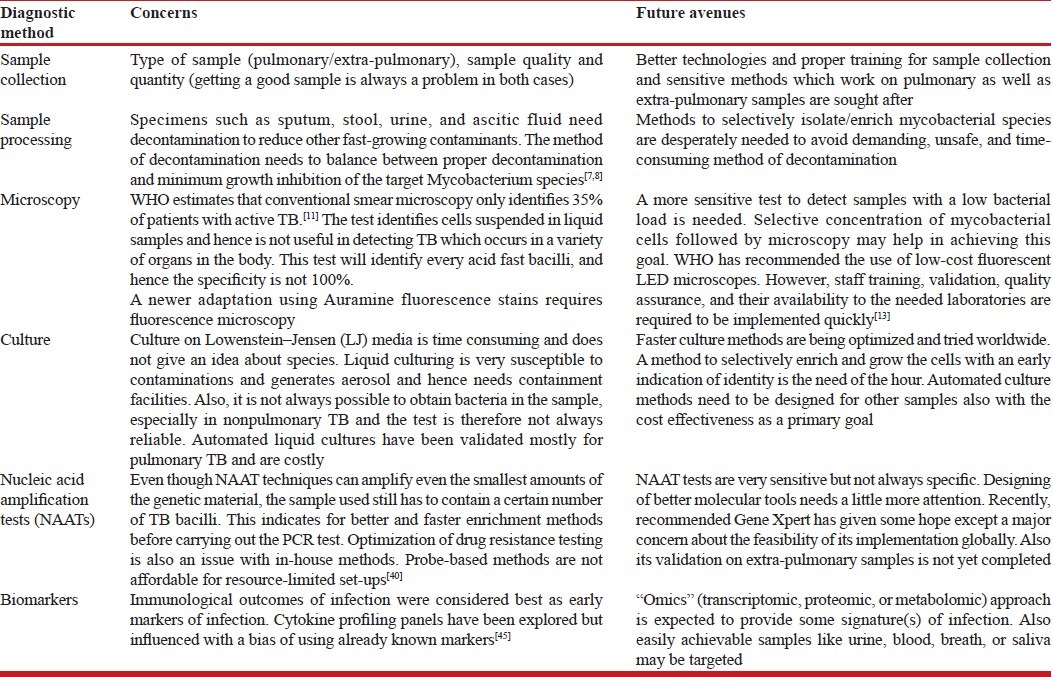
Looking on the other side, wherein commercial automated liquid cultures are recommended for better quality with minimum handling and manipulation of cultures by laboratory technicians, the expense and sophistication required is not a bearable load for all. In such conditions, alternative and novel culture-based approaches with a justifiable balance of cost, biosafety, and sensitivity are highly solicited. Nucleic acid amplification test (NAAT)-based methods seem to be the best and fastest options among all. The big question that arises then is how to facilitate the required resources. We need to upgrade all research and diagnostic units all over the affected regions of the world to equip themselves with basic set-ups of PCR assays and to make confirmative diagnosis a routine and feasible option. In designing assays, sometimes the worker is faced with a significant challenge in selecting the primer and probe combinations that detect all of the organisms to cover a broader range of significant organisms, avoiding cross-reactivity, sequence analysis, etc., to get the best suitable target. Anticipation and removal of inhibitory substances from the samples before the assay set-up is also a major issue to consider. For this, basic knowledge and training about molecular assays can be made a part of the basic training for technicians and scientists working in the field. Also NAA tests are not considered for the evaluation of patients receiving therapy as the technology cannot distinguish between live and dead organisms. This can still be used with the same specificity if followed by short-term culture-based enrichment. A possibility of having some method that incorporates both culture and modern methods of detection has been visualized during this review. A culture enriching the initial viable loads followed by a sensitive and specific NAA test with a minimized need of containment can make identification safer, faster, and more reliable.
With the recent enthusiasm of various policy makers, funding agencies, and grants, the need is now to foster innovations that deliver better tools to diagnose TB with confidence using affordable approaches. After this only, the next step to treat and eradicate the menace of TB and associated risks can be taken with confidence. This review has been written with an intention to make readers aware of the kind of methodological challenges involved in such diagnosis. The author hopes that this will raise inquisitiveness in the brains of true scientific warriors to strive for newer ideas in making things simple and affordable.
ACKNOWLEDGMENT
The author acknowledges the support of Director, NARI, and other colleagues for their valuable suggestions and reviews.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Patton GC, Coffey C, Sawyer SM, Viner RM, Haller DM, Bose K, et al. Global patterns of mortality in young people: a systematic analysis of population health data. Lancet. 2009;374:881–92. doi: 10.1016/S0140-6736(09)60741-8. [DOI] [PubMed] [Google Scholar]
- 2.Reid MJ, Shah NS. Approaches to tuberculosis screening and diagnosis in people with HIV in resource-limited settings. Lancet Infect Dis. 2009;9:173–84. doi: 10.1016/S1473-3099(09)70043-X. [DOI] [PubMed] [Google Scholar]
- 3.Geneva: World Health Organization; 2009. [Last accessed on 2012 Jan 20]. WHO report 2009. Global tuberculosis control: Epidemiology, strategy, financing. Available from: http://whqlibdoc.who.int/publications/2009/9789241563802_eng_doc.pdf . [Google Scholar]
- 4.Pai M, Minion J, Sohn H, Zwerling A, Perkins MD. Novel and improved technologies for tuberculosis diagnosis: progress and challenges. Clin Chest Med. 2009;30:701–16. doi: 10.1016/j.ccm.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Vignesh R, Balakrishnan P, Shankar EM, Murugavel KG, Hanas S, Cecelia AJ, et al. Value of single acid-fast bacilli sputum smears in the diagnosis of tuberculosis in HIV-positive subjects. J Med Microbiol. 2007;56:1709–10. doi: 10.1099/jmm.0.47497-0. [DOI] [PubMed] [Google Scholar]
- 6.Perera J, Arachchi DM. The optimum relative centrifugal force and centrifugation time for improved sensitivity of smear and culture for detection of Mycobacterium tuberculosis from sputum. Trans R Soc Trop Med Hyg. 1999;93:405–9. doi: 10.1016/s0035-9203(99)90135-9. [DOI] [PubMed] [Google Scholar]
- 7.Chakravorty S, Tyagi JS. Novel multipurpose methodology for detection of mycobacteria in pulmonary and extrapulmonary specimens by smear microscopy, culture, and PCR. J Clin Microbiol. 2005;43:2697–702. doi: 10.1128/JCM.43.6.2697-2702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakravorty S, Dudeja M, Hanif M, Tyagi JS. Utility of universal sample processing methodology, combining smear microscopy, culture, and PCR, for diagnosis of pulmonary tuberculosis. J Clin Microbiol. 2005;43:2703–8. doi: 10.1128/JCM.43.6.2703-2708.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steingart KR, Ng V, Henry M, Hopewell PC, Ramsay A, Cunningham J, et al. Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis. 2006;6:664–74. doi: 10.1016/S1473-3099(06)70602-8. [DOI] [PubMed] [Google Scholar]
- 10.Daley P, Michael JS, Kalaiselvan S, Latha A, Mathai D, John KR, et al. A pilot study of short-duration sputum pretreatment procedures for optimizing smear microscopy for tuberculosis. PLoS One. 2009;4:e5626. doi: 10.1371/journal.pone.0005626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grandjean L, Martin L, Gilman RH, Valencia T, Herrera B, Quino W, et al. Tuberculosis diagnosis and multidrug resistance testing by direct sputum culture in selective broth without decontamination or centrifugation. J Clin Microbiol. 2008;46:2339–44. doi: 10.1128/JCM.02476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange C, Mori T. Advances in the diagnosis of tuberculosis. Respirology. 2010;15:220–40. doi: 10.1111/j.1440-1843.2009.01692.x. [DOI] [PubMed] [Google Scholar]
- 13.TB India 2009: RNTCP Status report. Central TB Division, Directorate General of Health Services, Ministry of Health and Family Welfare, Nirman Bhawan, New Delhi. [Last Accessed on 2012 Jan 20]. Available from: http://www.tbcindia.nic.in/pdfs/TB%20India%202009.pdf .
- 14.Hopewell PC, Pai M, Maher D, Uplekar M, Raviglione MC. International standards for tuberculosis care. Lancet Infect Dis. 2006;6:710–25. doi: 10.1016/S1473-3099(06)70628-4. [DOI] [PubMed] [Google Scholar]
- 15.Woods GL. Susceptibility testing for mycobacteria. Clin Infect Dis. 2000;31:1209–15. doi: 10.1086/317441. [DOI] [PubMed] [Google Scholar]
- 16.Caviedes L, Lee TS, Gilman RH, Sheen P, Spellman E, Lee EH, et al. Rapid, efficient detection and drug susceptibility testing of Mycobacterium tuberculosis in sputum by microscopic observation of broth cultures. J Clin Microbiol. 2000;38:1203–8. doi: 10.1128/jcm.38.3.1203-1208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore DA, Mendoza D, Gilman RH, Evans CA, Hollm Delgado MG, Guerra J, et al. Microscopic observation drug susceptibility assay, a rapid, reliable diagnostic test for multidrug-resistant tuberculosis suitable for use in resource-poor settings. J Clin Microbiol. 2004;42:4432–7. doi: 10.1128/JCM.42.10.4432-4437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore DA, Evans CA, Gilman RH, Caviedes L, Coronel J, Vivar A, et al. Microscopic-observation drug-susceptibility assay for the diagnosis of TB. N Engl J Med. 2006;355:1539–50. doi: 10.1056/NEJMoa055524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin A, Munga Waweru P, Babu Okatch F, Amondi Ouma N, Bonte L, Varaine F, et al. Implementation of the thin layer agar for the diagnosis of smear-negative pulmonary tuberculosis in a high HIV prevalence setting in Homa Bay, Kenya. J Clin Microbiol. 2009;47:2632–4. doi: 10.1128/JCM.00264-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shikama ML, Ferro e Silva R, Villela G, Sato DN, Martins MC, Giampaglia CM, et al. Multicentre study of nitrate reductase assay for rapid detection of rifampicin-resistant M.tuberculosis. Int J Tuberc Lung Dis. 2009;13:377–80. [PubMed] [Google Scholar]
- 21.Drosten C, Panning M, Kramme S. Detection of Mycobacterium tuberculosis by real-time PCR using pan-mycobacterial primers and a pair of fluorescence resonance energy transfer probes specific for the M. tuberculosis complex. Clin Chem. 2003;49:1659–61. doi: 10.1373/49.10.1659. [DOI] [PubMed] [Google Scholar]
- 22.Kim JH, Kim YJ, Ki CS, Kim JY, Lee NY. Evaluation of Cobas TaqMan MTB PCR for detection of Mycobacterium tuberculosis. J Clin Microbiol. 2011;49:173–6. doi: 10.1128/JCM.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanduma E, McHugh TD, Gillespie SH. Molecular methods for Mycobacterium tuberculosis strain typing: a users guide. J Appl Microbiol. 2003;94:781–91. doi: 10.1046/j.1365-2672.2003.01918.x. [DOI] [PubMed] [Google Scholar]
- 24.Scheler O, Kaplinski L, Glynn B, Palta P, Parkel S, Toome K, et al. Detection of NASBA amplified bacterial tmRNA molecules on SLICSel designed microarray probes. BMC Biotechnol. 2011;11:17. doi: 10.1186/1472-6750-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim CM, Song ES, Jang HJ, Kim HJ, Lee S, Shin JH, et al. Development and evaluation of oligonucleotide chip based on the 16S-23S rRNA gene spacer region for detection of pathogenic microorganisms associated with sepsis. J Clin Microbiol. 2010;48:1578–83. doi: 10.1128/JCM.01130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ringuet H, Akoua-Koffi C, Honore S, Varnerot A, Vincent V, Berche P, et al. hsp65 sequencing for identification of rapidly growing mycobacteria. J Clin Microbiol. 1999;37:852–7. doi: 10.1128/jcm.37.3.852-857.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blackwood KS, He C, Gunton J, Turenne CY, Wolfe J, Kabani AM. Evaluation of recA sequences for identification of Mycobacterium species. J Clin Microbiol. 2000;38:2846–52. doi: 10.1128/jcm.38.8.2846-2852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim BJ, Lee SH, Lyu MA, Kim SJ, Bai GH, Chae GT, et al. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB) J Clin Microbiol. 1999;37:1714–20. doi: 10.1128/jcm.37.6.1714-1720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasai H, Ezaki T, Harayama S. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J Clin Microbiol. 2000;38:301–8. doi: 10.1128/jcm.38.1.301-308.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tortoli E, Bartoloni A, Böttger EC, Emler S, Garzelli C, Magliano E, et al. Burden of unidentifiable mycobacteria in a reference laboratory. J Clin Microbiol. 2001;39:4058–65. doi: 10.1128/JCM.39.11.4058-4065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turenne CY, Tschetter L, Wolfe J, Kabani A. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J Clin Microbiol. 2001;39:3637–48. doi: 10.1128/JCM.39.10.3637-3648.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cloud JL, Neal H, Rosenberry R, Turenne CY, Jama M, Hillyard DR, et al. Identification of Mycobacterium spp.by using a commercial 16S ribosomal DNA sequencing kit and additional sequencing libraries. J Clin Microbiol. 2002;40:400–6. doi: 10.1128/JCM.40.2.400-406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall L, Doerr KA, Wohlfiel SL, Roberts GD. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J Clin Microbiol. 2003;41:1447–53. doi: 10.1128/JCM.41.4.1447-1453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tortoli E. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin Microbiol Rev. 2003;16:319–54. doi: 10.1128/CMR.16.2.319-354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zelazny AM, Calhoun LB, Li L, Shea YR, Fischer SH. Identification of Mycobacterium species by secA1 sequences. J Clin Microbiol. 2005;43:1051–8. doi: 10.1128/JCM.43.3.1051-1058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong L, Kong F, Yang Y, Cheng J, Gilbert GL. Use of PCR and reverse line blot hybridization macroarray based on 16S-23S rRNA gene internal transcribed spacer sequences for rapid identification of 34 Mycobacterium species. J Clin Microbiol. 2006;44:3544–50. doi: 10.1128/JCM.00633-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavusoglu C, Turhan A, Akinci P, Soyler I. Evaluation of the Genotype MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis isolates. J Clin Microbiol. 2006;44:2338–42. doi: 10.1128/JCM.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter E, Weizenegger M, Rüsch-Gerdes S, Niemann S. Evaluation of genotype MTBC assay for differentiation of clinical Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2003;41:2672–5. doi: 10.1128/JCM.41.6.2672-2675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNabb A, Eisler D, Adie K, Amos M, Rodrigues M, Stephens G, et al. Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources. J Clin Microbiol. 2004;42:3000–11. doi: 10.1128/JCM.42.7.3000-3011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin SJ, Lee BS, Koh WJ, Manning EJ, Anklam K, Sreevatsan S, et al. Efficient differentiation of Mycobacterium avium complex species and subspecies by use of five-target multiplex PCR. J Clin Microbiol. 2010;48:4057–62. doi: 10.1128/JCM.00904-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–15. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Automated real-time nucleic acid amplification technology for rapid and simultaneous detection of tuberculosis and rifampicin resistance: xpert MTB/RIF system. 2011. [Last accessed on 2012 Jan 20]. Available from: http://www.tbevidence.org/documents/policies/WHO_Policy_Xpert_2011.pdf . [PubMed]
- 43.Ruhwald M, Bjerregaard-Andersen M, Rabna P, Eugen-Olsen J, Ravn P. IP-10, MCP-1, MCP-2, MCP-3, and IL-1RA hold promise as biomarkers for infection with M.tuberculosis in a whole blood based T-cell assay. BMC Res Notes. 2009;2:19. doi: 10.1186/1756-0500-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith SG, Lalor MK, Gorak-Stolinska P, Blitz R, Beveridge NE, Worth A, et al. Mycobacterium tuberculosis PPD-induced immune biomarkers measurable in vitro following BCG vaccination of UK adolescents by multiplex bead array and intracellular cytokine staining. BMC Immunol. 2010;11:35. doi: 10.1186/1471-2172-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallis RS, Pai M, Menzies D, Doherty TM, Walzl G, Perkins MD, et al. Biomarkers and diagnostics for tuberculosis: Progress, needs, and translation into practice. Lancet. 2010;375:1920–37. doi: 10.1016/S0140-6736(10)60359-5. [DOI] [PubMed] [Google Scholar]
- 46.Walzl G, Ronacher K, Hanekom W, Scriba TJ, Zumla A. Immunological biomarkers of tuberculosis. Nat Rev Immunol. 2011;11:343–54. doi: 10.1038/nri2960. [DOI] [PubMed] [Google Scholar]
- 47.Geneva: World Health Organization; 2008. [Last accessed on 2012 Jan 20]. WHO report 2008. Global tuberculosis control: surveillance, planning, financing. Available from: http://www.who.int/tb/publications/global_report/2008/pdf/fullreport.pdf . [Google Scholar]
- 48.HIV Sentinel Surveillance and HIV Estimation in India 2007: A Technical Brief. National AIDS Control Organisation 2008. [Last accessed on 2012 Jan 20]. Available from: http://www.nacoonline.org/upload/Publication/MandE%20Surveillance,%20Research/HIV%20Sentinel%20Surveillance%20and%20HIV%20Estimation%202007_A%20Technical%20Brief.pdf .
- 49.Lawn SD, Myer L, Edwards D, Bekker LG, Wood R. Short-term and long-term risk of tuberculosis associated with CD4 cell recovery during antiretroviral therapy in South Africa. AIDS. 2009;23:1717–25. doi: 10.1097/QAD.0b013e32832d3b6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharma SK, Mohan A, Kadhiravan T. HIV-TB co-infection: Epidemiology, diagnosis and management. Indian J Med Res. 2005;121:550–67. [PubMed] [Google Scholar]
- 51.Mahajan A, Tandon VR. HIV/AIDS-TB Co-Infection: What prevalence indicates? JK Science. 2007;9:56–7. [Google Scholar]
- 52.Ghanashyam B. Tuberculosis diagnostics: Innovating to make an impact. Expert Rev Anti Infect Ther. 2011;9:381–4. doi: 10.1586/eri.11.18. [DOI] [PubMed] [Google Scholar]
- 53.Commercial serodiagnostic tests for diagnosis of tuberculosis: Policy statement. WHO. 2011. [Last accessed on 2012 Jan 20]. Available from: http://www.whqlibdoc.who.int/publications/2011/9789241502054_eng.pdf . [PubMed]
- 54.Udwadia ZF, Amale RA, Ajbani KK, Rodrigues C. Totally drug-resistant tuberculosis in India. Clin Infect Dis. 2012;54:579–81. doi: 10.1093/cid/cir889. [DOI] [PubMed] [Google Scholar]


