Abstract
Background and Aims:
Recent microbiological researches have revealed the possible role of human cytomegalovirus (HCMV), Epstein barr virus (EBV), and herpes simplex virus (HSV-1 and HSV-2) in the etiopathogenesis of periodontal diseases. The present pilot study has been undertaken to detect the presence of these viruses in chronic periodontitis, aggressive periodontitis, and healthy individuals and to determine the relationship between these viruses and the clinical parameters.
Materials and Methods:
A total of 10 patients belonging to the age group of 18 to 55 years were included. The patients were randomly assigned into periodontally healthy (sulcus depth ≤ 3 mm), chronic periodontitis, and aggressive periodontitis with pockets measuring ≥6 mm. Seventy-five subgingival plaque samples (25 samples from each group) were collected and subjected to multiplex polymerase chain reaction for the detection of presence of HCMV, EBV, HSV-1, and HSV-2. The results were analyzed using one-way ANOVA for multiple group comparisons followed by Student's t-test for pair-wise comparisons. Categorical data was analyzed by Fisher's exact test.
Results:
HSV-1 was detected in 76% (P<0.001) of sites with chronic periodontitis and 80% (P<0.001) sites with aggressive periodontitis. EBV was detected in 32% (P<0.05) of sites with chronic periodontitis and aggressive periodontitis. The probing pocket depth and clinical attachment level was statistically significant in HSV-1 detected sites compared with undetected sites in aggressive periodontitis patients
Conclusion:
Among these viruses HSV-1 and EBV were found to be significantly associated with destructive periodontal disease, including chronic and aggressive periodontitis. Further, HSV-1 was found to be associated with severity and progression of destructive periodontal disease.
Keywords: Aggressive periodontitis, chronic periodontitis, herpes virus, polymerase chain reaction
INTRODUCTION
The pathogenesis of periodontal disease is a multifactorial process involving a complex interaction between the microbial and host factors and a variety of disease-modulatory environmental factors. However, the bacterial etiology has not yet been able to substantiate various aspects of periodontal disease. Recent microbiological researches on periodontal disease have focused on the possible involvement of human viruses in the etiology and pathogenesis of destructive periodontal diseases.[1–4]
Recent studies have implicated human cytomegalovirus (HCMV) and Epstien barr virus 1 (EBV-1) in the pathogenesis of human periodontal disease. These viruses are capable of infecting and impairing polymorphonuclear leukocytes (PMNs), macrophages, and lymphocytes. Herpes virus-infected cells can reduce the host defense and give rise to overgrowth of pathogenic bacteria and invade the cells more efficiently.[1] Contreras and Slots[2] also demonstrated the presence of EBV-1, HCMV, and other herpes viruses in juvenile and chronic periodontitis lesions. Association of herpes viruses with bacteria was also assessed to improve our understanding of etiopathogenesis of periodontal disease.[1] Furthermore, recent studies have quantified the herpes viruses in periodontal pockets to show their positive association with severity of periodontal disease. The prevalence and number of herpes viruses in periodontal pockets may vary according to type of periodontal disease.[3]
Studies have confirmed the frequent presence of HCMV, EBV, and HSV in crevicular samples of chronic periodontitis lesions and suggested a strong relationship between the presence of these viruses and measurement of probing depth, clinical attachment loss, and the severity of the disease.[4] Evidence from studies also indicate the subgingival presence of EBV-1 and HCMV is strongly associated with aggressive periodontitis,[3,5] and co-infection with HCMV and EBV appears to be particularly deleterious to periodontal health.[1]
The present study aims at detecting and comparing the presence of HCMV, EBV, HSV-1, and HSV-2 in subgingival plaque samples from chronic periodontitis, aggressive periodontitis, and periodontally healthy individuals and its relation with the clinical parameters.
MATERIALS AND METHODS
The patients participating in the study were selected from the Outpatient Department of Periodontics, College of Dental Sciences, Davangere, Karnataka, India. The study was carried out after obtaining the ethical clearance from the institutional review board. This cross-sectional study included 10 patients (6 women and 4 men within age group 18–55 years). All the patients were systemically healthy and had not received any antibiotics or periodontal therapy in the past 6 months prior to the sampling and recording. Smokers, pregnant women, and lactating mothers were excluded from the study. Written informed consent was obtained from each study subject after all the procedures had been fully explained.
Patients were grouped as follows:
Group 1: Periodontally healthy individuals.[6] (Included volunteered dental students and paradental staff with age ≥ 21 years – 45 years, presence of sulcus depth ≤ 3 mm, radiologically without crestal bone loss [CEJ as landmark]).
Group 2: Chronic periodontitis.[7] (Patients with age ≥ 35 years, 30% of sites involved, periodontal pocket depth ≥ 6 mm, and attachment loss ≥ 3 mm, with severe bone loss [≥50% of root length]).
Group 3: Aggressive periodontitis.[7] (Patients with age ≥ 18 years, 30% of sites involved, periodontal pocket depth ≥ 6 mm, and attachment loss ≥ 3 mm, with severe bone loss [≥50% of root length]).
Clinical procedures
A total of 75 sites were selected, 25 sites belonged to each of the three groups. The selected sites were subjected to clinical evaluation of plaque index (Silness and Loe, 1964),[8] gingival index (Loe and Silness, 1963),[9] probing pocket depth, and clinical attachment level. All clinical parameters were measured with a UNC-15 probe calibrated in millimeters and were performed on four sites per tooth (mesiobuccal, midbuccal, distobuccal, and lingual). Prior to sampling, supragingival plaque was gently removed with sterile cotton pellets and the sample sites were isolated with cotton rolls and dried. In the periodontitis patients (Groups 2 and 3), subgingival samples were collected from the deepest periodontal pocket sites of the dentition, and for the periodontally healthy individuals (Group 1) samples were collected from sites with probing depth measuring ≤3 mm. Subgingival samples were collected by a sterile curette. After gently inserting the curette to the bottom of the sample site, subgingival plaque was removed with a single stroke of the curette. The collected plaque samples were transferred into separate sterile plastic vials containing TE (Tris EDTA) buffer, which contains Trishydrochloride (pH – 8.0) 10 mM and Ethylene diamine tetraacetic acid (EDTA) 1 mM. All plaque samples were collected by a single examiner and the collected samples were coded and sent for microbiological analysis. Thus, the analyzer was blinded regarding the source of the sample.
Deoxyribonucleic acid extraction
On reaching the laboratory, the sample is immediately processed for Deoxyribonucleic acid (DNA) extraction. The sample is washed twice in TE buffer at 6000 rpm for 5 minutes each, followed by one wash in Lysis buffer-1 [Tritonx 100 (1%), EDTA – 1 mM, Trihydrochloride 10 mM]. 100 ml of Lysis buffer – 2 is added, which contains (Tri hydrochloride 50 mM, Potassium chloride (50 mM), Magnesium chloride (2.5 mM), Tween–20 (0.45%), Nonidet P-40 (0.45%) with Proteinalase-K (100 mic gms / ml). This is kept in an incubator at 65°C for 2 h followed by 10 min at 95°C. Later, this is kept at -20°C until processing. Multiplex polymerase chain reaction (PCR) is performed for all the samples.[10]
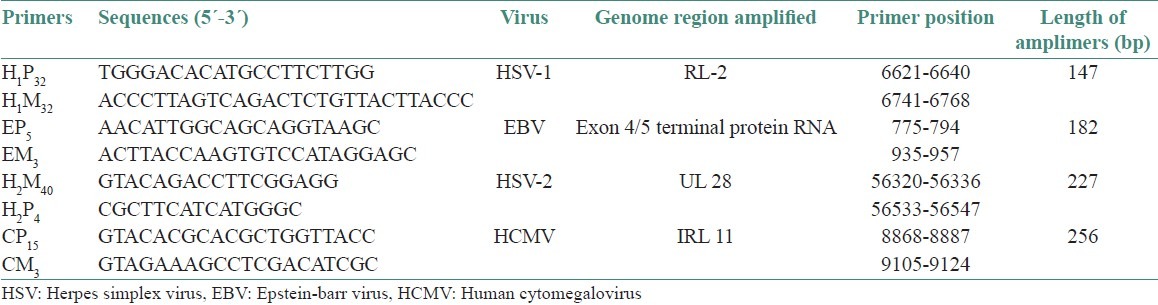
Primers[11] - The sequence of PCR primers were synthesized with an Oligonucleotide synthesizer (Applied biosystems, Fostercity, California, USA). Primer pair H1P32 / H1M32, H2M40 / H2P4, CP15 / CM3, and EP5 / EM 3 were chosen from DNA sequences of HSV-1, HSV-2, HCMV, and EBV complete genomes, respectively. Sequences were accessible with the following Gen Bank accession numbers- X 14112 for HSV-1, Z 86099 for HSV-2, X 17103 for HCMV, and V 01555 for EBV. Primer for a-tubulin sequences were accessible from Gen Bank, accession number X 01703. The amplified PCR fragment was 527 bp. Primer sequences were designed by Primer 3, Whitehead Institute [Table 1].
Table 1.
The sequences of the primers
Viruses and controls
HSV-1 strain F, HSV-2 stain G, HCMV strain AD169, and EBV stain P-3 were used as positive controls. Genetically related viruses such as HHV-8 DNA isolated from infected lymphoma cells and genetically related viruses such as enteroviruses were used in control experiments as negative control.
Polymerase chain reaction amplification
Polymerase chain reaction (PCR) refers to a highly sensitive technique for amplifying a short stretch of DNA. The method depends on the use of two flanking oligonucleotide DNA primers and repeated cycles of primer extension using DNA polymerase. Multiplex PCR is a type of PCR technique in which multiple primer pairs for different target molecules are included in the same amplification mixture. This mixture is used to simultaneously amplify target sequences for several pathogenic microorganisms in a single reaction vial. To optimize the multiplex PCR, a series of titrations of primer concentrations and deoxynucleotide triphosphate (dNTP) levels were performed. Primer concentrations of 10, 25, 50, and 100 pmol from each primer pair were titrated simultaneously with dNTP (0.1, 0.2 and 0.3 mM concentrations of each dNTPs).[10]
The amplifications were performed with DNA thermalcycler (Corbett Research, Mortlake, NSW, Australia). After placing the PCR vials inside the thermalcycler, 40 amplification cycles of 30 s at 94°C, 40 s at 60°C, and 50 s at 72°C were carried out. In a PCR vial (Axygen Inc., Union city, California, USA) with a 50 mL final volume containing 10 p mol of each of the 8 primers (Sigma-Aldrich Corp, USA), 5 mL of 10X reaction buffer, 0.2 mM concentrations of each dNTP, and 2.5 U of cloned pfu DNA polymerase enzyme (Genei Bangalore, India). Five mL of appropriate DNA sample (positive controls, HSV-1 strain F, HSV-2 strain G, HCMV strain AD169, and EBV strain P-3, negative controls, plaque sample) was added to the reaction mixture. After the last cycle, the samples were incubated for 15 min at 78°C to complete the extension of primers. Ten mL of each amplified product was analyzed by agarose gel electrophoresis on 3.5% agarose containing 1 mg of ethidium bromide / mL in 1x TBE buffer and was visualized in a UV transilluminator (Bio- Imaging Systems, NY, USA). The DNA bands were photographed under 200 nm ultraviolet light [Figures 1–3].
Figure 1.
DNA Transcription of viruses in periodontally healthy sample
Figure 3.
DNA transcription of viruses in aggressive periodontitis sample
Figure 2.
DNA transcription of viruses in chronic periodontitis sample
Statistical analysis
Descriptive data that included Mean and Standard Deviations were determined for each clinical parameter in each group and were used for analysis. One-way ANOVA was used for multiple group comparisons followed by Student's t-test for pair-wise comparisons. Whenever assumption of normality was not observed, Mann-Whitney test was also used. Categorical data was analyzed by Fisher's exact test. For all the tests, a P value of 0.05 or less was considered for statistical significance. The statistical analysis was done using SPSS software 15.0.1 version.
RESULTS
Table 1 shows the sequence of primers used in PCR to detect human viruses. Table 2 shows frequency of detection viruses in various study groups. Chronic periodontitis (Group 2) sites revealed HSV-1 in 19 (76%) samples, HSV-2 in three samples (12%) samples, and EBV in eight (32%) samples and HCMV in three (12%) samples. Aggressive periodontitis (Group3) site revealed HSV-1 in 20 (80%) samples. HSV-2 in two (12%) samples, and EBV in eight (32%) samples and HCMV in three (12%) samples. On comparison between groups, HSV-1 and EBV detected viruses in both chronic and aggressive periodontitis was statistically significant compared with healthy group (Group 1).
Table 2.
Detection frequency of virus in periodontal pockets of various study groups
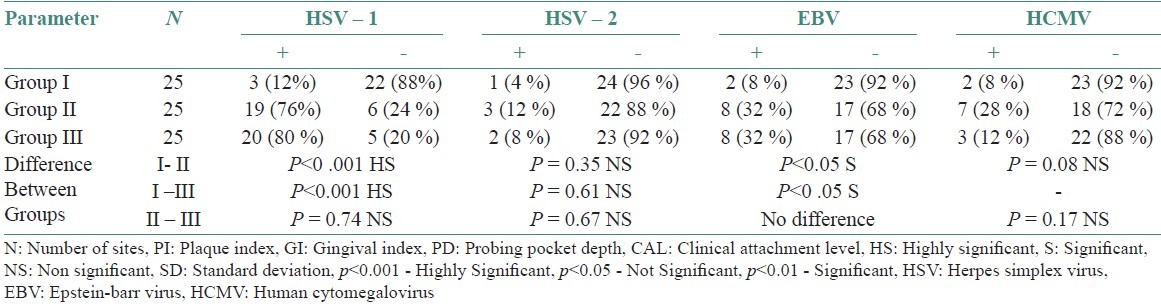
Table 3 shows comparison of clinical parameters between HSV-1 sites, detected and undetected sites in various study groups. Tables 4–6 shows comparison of clinical parameters between HSV-2, EBV, and HCMV detected (+) and undetected (-) sites in various study groups.
Table 3.
Comparison of clinical parameters between HSV-1 detected (+) and undetected (-) sites in various study groups
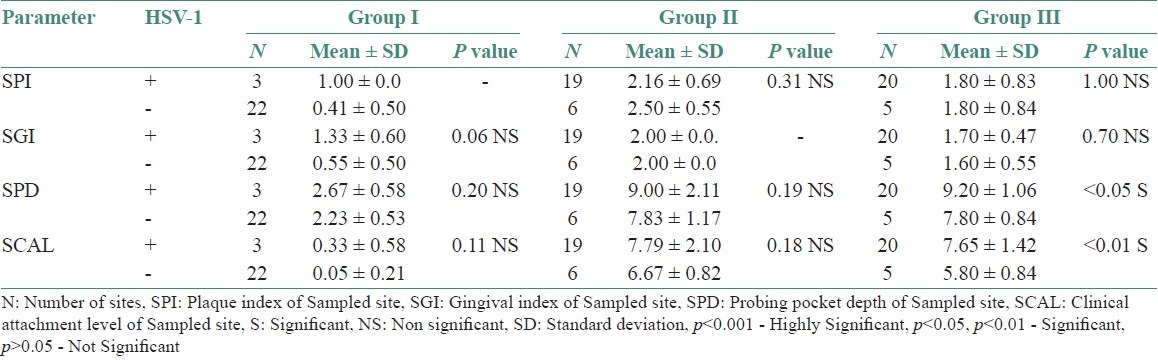
Table 4.
Comparison of clinical parameters between HSV-2 detected (+) and undetected (-) sites in various study groups
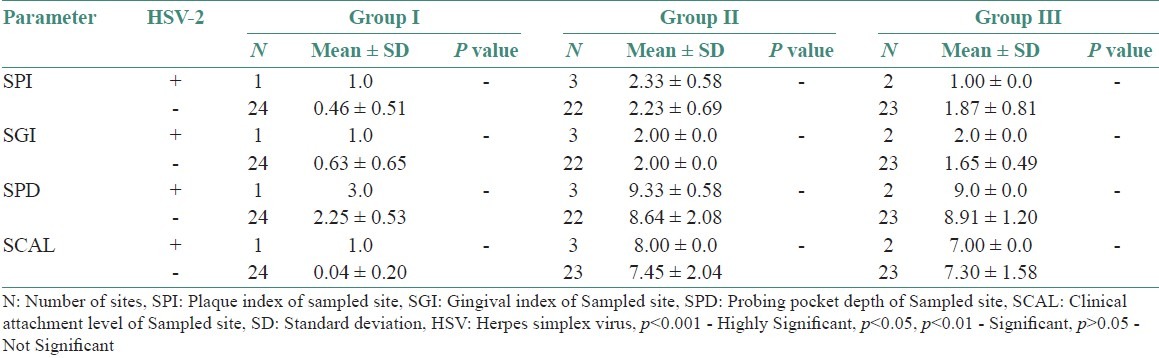
Table 6.
Comparison of Clinical parameters between HCMV detected (+) and undetected (-) sites in various study groups
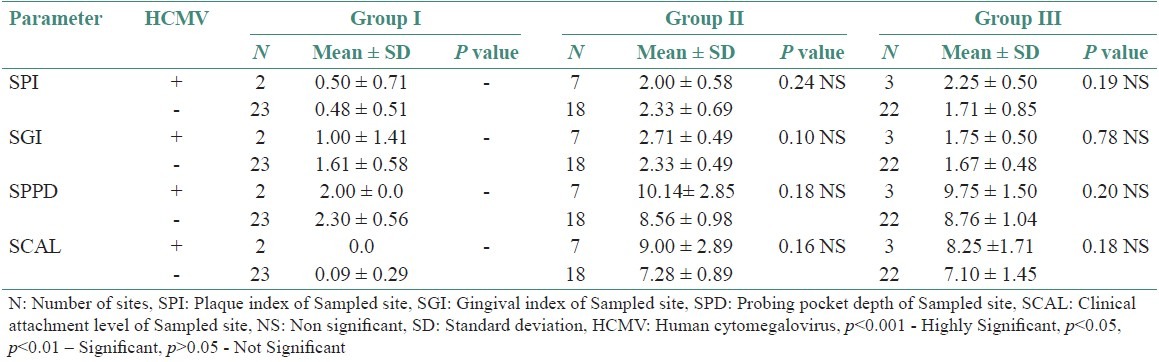
Table 5.
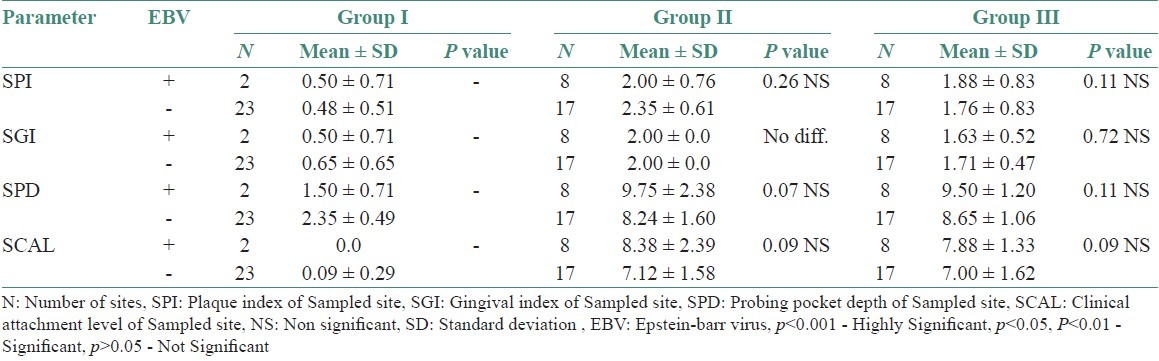
Comparison of clinical parameters between EBV detected (+) and undetected (-) sites in various study groups
DISCUSSION
Herpes simplex viruses are transmitted by close person-to-person contact by mucosal secretions or lesions. HSV-1 usually causes orolabial disease and HSV-2 is associated more frequently with genital and newborn infections. At the site of epithelial infection, viral antigens induce a cell-mediated immune response. Following replication in epithelial cells, some viral nucleocapsids ascend the local sensory neurons by retrograde axonal transport and establish a lifelong latency in the corresponding spinal and cerebral ganglions. Reactivation of herpes simplex virus can occur subclinically, but about one-third of the individuals become prone to clinical recurrences.[11] Cultured epithelial cells and fibroblasts from clinically healthy human gingiva are susceptible to HSV infection,[12] suggesting that those cells could be a reservoir for the latent virus. Using an indirect immunofluorescence assay, HSV-1 antigens could be detected in 4 out of 14 gingival biopsies from periodontally diseased patients.[13] The presence of HSV antigens could also be shown in 26 out of 66 specimens of clinically healthy gingiva using a similar method.[14]
In this study, which was studied on Indian population, a highly statistically significant prevalence of HSV-1 in subgingival samples from chronic periodontitis patients (76%) and aggressive periodontitis patients (80%) compared to healthy controls (12%) was seen. These results were correlated with the study by Saygun et al.,[15] where HSV-1 was found to be statistically significant in aggressive periodontitis patients compared with healthy individuals. However, there was no statistically significant detection of HSV-1 between chronic periodontitis patients (76%) and aggressive periodontitis patients (80%). In our study, HSV-2 was not found to be statistically significant. Similar results were observed in the study by Contreras and Slots,[16] wherein presence of HSV-1 was seen in all the samples but HSV-2 was not detected in any of the samples. HSV-2 is usually transmitted through genital infection and is rarely found in the oral cavity.
EBV affects over 90% of humans and is usually transmitted by oral secretions or blood. The virus replicates in epithelial cells or B-cells of the oropharynx. Resting memory B cells are the main site of persistence of EBV in the body. The number of latently infected cells in a person remains stable for over years.[17] Latent EBV infection can be reactivated, leading to viral shedding into oral mucosa.[11] In our study, EBV was found to be significant in chronic periodontitis patients (32%) compared with healthy controls (8%). Saygun et al.[4] found EBV to be present in 44.3% of sites from patients with chronic periodontitis and only 14.3% of the sites obtained from healthy controls. Konstantinidis et al.[18] showed EBV to be present in 11 out of 12 chronic periodontitis patients and only one in healthy controls. Similarly, in aggressive periodontitis patients, EBV was found to be significant (32%) compared with healthy controls (8%). Studies conducted by Yapar et al.[5] and Saygun et al.[15] showed EBV to be significant in the sites of aggressive periodontitis patients compared with healthy control sites. However, no difference was found between chronic periodontitis and aggressive periodontitis patients in this study. This is contradictory to the study results obtained by Kubar et al.[3] where EBV was seen to be present in 89% sites in aggressive periodontitis patients compared with 46% in chronic periodontitis patients, which was statistically significant. So with these findings from our study, we can infer that there is a close association of HSV-1 and EBV with chronic and aggressive periodontitis.
HCMV, the largest member of herpes family of viruses, is responsible for a significant percentage of asymptomatic viral infections worldwide. HCMV exhibits marked tropism for cells of the immune system. HCMV interferes with innate and adaptive cellular and humoral immune effector mechanisms by activation and silencing of natural killer cells, by down-modulating antigen presentation in the MHC class I and II pathways and by impairing apoptosis. HCMV infection induces abnormalities in adherence, chemotaxis, phagocytic, oxidative, secretory, and bactericidal activities of polymorphonuclear neutrophils. It also diverts potent antiviral cytokine response or even interferes with cytokine production.[19]
In this study, HCMV detection was not found to be statistically significant in all the three groups. This is contradictory to the previous studies conducted by Kamma et al.,[20] Yapar et al.,[5] Contreras et al.,[21] and Kubar et al.,[22] which has found HCMV in periodontitis lesions. This variation of results among various studies could be due to geographical, economic background, and ethnicity.
In chronic and aggressive periodontitis, HSV-1 detected sites are more compared with undetected sites indicating that HSV-1 is associated with the progression and severity of the periodontal disease. In this study, EBV-1, HCMV, and HSV-2 have not shown any correlation with the clinical parameters of periodontal disease. However, studies conducted by Contreras and Slots et al.,[16] Ling et al.,[23] and Kamma et al.,[20] have found contradictory results.
This observation that herpes virus tends to be associated with initiation and progression of periodontitis offers testable hypothesis for explaining the periodontopathic effects of these viruses. It is hoped that such information will help bridge the gap between our clinical understanding of destructive periodontal disease and our lack of knowledge at the molecular levels of mechanisms involved in periodontal tissue breakdown. Further studies should include longitudinal, clinical, and microbiological studies and further research on the role of herpes viruses in different forms of periodontitis.
CONCLUSIONS
The following conclusions could be made from the findings of this study.
HSV-1 and EBV are significantly associated with destructive periodontal disease including chronic and aggressive periodontitis.
HSV-1 detected sites were more in relation to sites of deep pocket depth and increased clinical attachment loss, and this signifies that these viruses might be associated with severity and progression of destructive periodontal disease.
Although a cause and effect relationship has yet to be proven, which helps to improve in diagnosis, determining a more specific treatment plan, and preventing disease. Further studies needs to include larger sample size and bacterial viral relationship.
ACKNOWLEDGMENTS
Dr. Kishore Bhat, Department of Microbiology, Maratha Mandal Dental College, Belgaum, Karnataka, India. Mr. Sangam, Department of Community Medicine, J.J.M. Medical College, Davangere, Karnataka, India.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Contreras A, Umeda M, Chen C, Bakker I, Morrison JC, Slots J. Relationship between herpesviruses and adult periodontitis and periodontopathic bacteria. J Periodontol. 1999;70:478–84. doi: 10.1902/jop.1999.70.5.478. [DOI] [PubMed] [Google Scholar]
- 2.Contreras A, Slots J. Mammalian viruses in human periodontitis. Oral Microbiol Immunol. 1996;11:381–6. doi: 10.1111/j.1399-302x.1996.tb00199.x. [DOI] [PubMed] [Google Scholar]
- 3.Kubar A, Saygun I, Ozdemir A, Yapar M, Slots J. Real-time polymerase chain reaction quantification of human cytomegalovirus and Epstein Barr virus in periodontal pockets and the adjacent gingiva of periodontitis lesions. J Periodontal Res. 2005;40:97–104. doi: 10.1111/j.1600-0765.2005.00770.x. [DOI] [PubMed] [Google Scholar]
- 4.Saygun I, Sahin S, Ozdemir A, Kurtiş B, Yapar M, Kubar A, et al. Detection of human viruses in patients with chronic periodontitis and the relationship between viruses and clinical parameters. J Periodontol. 2002;73:1437–43. doi: 10.1902/jop.2002.73.12.1437. [DOI] [PubMed] [Google Scholar]
- 5.Yapar M, Saygun I, Ozdemir A, Kubar A, Sahin S. Prevalence of human herpesviruses in patients with aggressive periodontitis. J Periodontol. 2003;74:1634–40. doi: 10.1902/jop.2003.74.11.1634. [DOI] [PubMed] [Google Scholar]
- 6.Newman MG, Takei HH, Carranza FA. 9th ed. California: Saunders; 2003. Carranza's clinical peirodontology; pp. 398–415. [Google Scholar]
- 7.Armitage GC. Periodontal diagnoses and classification of periodontal diseases. Periodontol 2000. 2004;34:9–21. doi: 10.1046/j.0906-6713.2002.003421.x. [DOI] [PubMed] [Google Scholar]
- 8.Silness J, Loe H. Periodontal disease in pregnancy. II. correlation between oral hygiene and periodontal conditions. Acta Odontol Scand. 1964;22:121–35. doi: 10.3109/00016356408993968. [DOI] [PubMed] [Google Scholar]
- 9.Loe H, Silness J. Periodontal disease in pregnancy. I. Prevalence and severity. Acta Odontol Scand. 1963;21:533–51. doi: 10.3109/00016356309011240. [DOI] [PubMed] [Google Scholar]
- 10.Markoulato P, Georgopoulou A, Siafakas N, Plakokefalos E, Tzanakaki G, Kourea-Kremastinou J. Laboratory diagnosis of common herpesvirus infections of the central nervous system by multiplex PCR assay. J Clin Microbiol. 2001;39:4426–32. doi: 10.1128/JCM.39.12.4426-4432.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitley RJ. Herpes simplex viruses. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. 3rd ed. Philadelphia: Lippincott – Raven Publishers; 1996. pp. 2297–342. [Google Scholar]
- 12.Zackay-Rones Z, Hochman N, Rones Y. Immunological response to herpes simplex virus in human gingival fluid. J Peridontol. 1982;53:42–5. doi: 10.1902/jop.1982.53.1.42. [DOI] [PubMed] [Google Scholar]
- 13.Ehrlich J, Cohen GH, Hochman N. Specific herpes simplex virus antigen in human gingiva. J Periodontol. 1983;54:357–60. doi: 10.1902/jop.1983.54.6.357. [DOI] [PubMed] [Google Scholar]
- 14.Amit R, Morag A, Ravid Z, Hochman N, Ehrlich J, Zakay-Rones Z. Detection of herpes simplex virus in gingival tissue. J Periodontol. 1992;63:502–6. doi: 10.1902/jop.1992.63.6.502. [DOI] [PubMed] [Google Scholar]
- 15.Saygun I, Kubar A, Ozdemir A, Yapar M, Slots J. Herpesviral-bacterial interrelationships in aggressive periodontitis. J Periodontal Res. 2004;39:207–12. doi: 10.1111/j.1600-0765.2004.00728.x. [DOI] [PubMed] [Google Scholar]
- 16.Contreras A, Slots J. Typing of herpes simplex virus from human periodontium. Oral Microbiol Immunol. 2001;16:63–4. doi: 10.1034/j.1399-302x.2001.160111.x. [DOI] [PubMed] [Google Scholar]
- 17.Rickinson E, Kieff E. Epstein Barr virus. In: Knipe DM, Howley PM, editors. Fields Viriology. 4th ed. Philadelphia: Lippincott Williams and Wilkins; 2001. pp. 2575–627. [Google Scholar]
- 18.Konstantinidis A, Sakellari D, Papa A, Antoniadis A. Real-time polymerase chain reaction quantification of epstein-barr virus in chronic periodontitis patients. J Periodontal Res. 2005;40:294–8. doi: 10.1111/j.1600-0765.2005.00796.x. [DOI] [PubMed] [Google Scholar]
- 19.Pass RF. Cytomegalovirus. In: Knipe DM, Howley PM, editors. Fields Virology. 4th ed. Philadelphia: Lippincott, Williams & Wilkins; 2001. pp. 2675–05. [Google Scholar]
- 20.Kamma JJ, Slots J. Herpes viral-bacterial interactions in aggressive periodontitis. J Clin Periodontol. 2003;30:420–6. doi: 10.1034/j.1600-051x.2003.20002.x. [DOI] [PubMed] [Google Scholar]
- 21.Contreras A, Nowzari H, Slots J. Herpesviruses in periodontal pocket and gingival tissue specimens. Oral Microbiol Immunol. 2000;15:15–8. doi: 10.1034/j.1399-302x.2000.150103.x. [DOI] [PubMed] [Google Scholar]
- 22.Kubar A, Saygun I, Yapar M, Ozdemir A, Slots J. Real-time PCR quantification of cytomegalovirus in aggressive periodontitis lesions using TaqMan technology. J Periodontal Res. 2004;39:81–6. doi: 10.1111/j.1600-0765.2004.00707.x. [DOI] [PubMed] [Google Scholar]
- 23.Ling LJ, Ho CC, Wu CY, Chen YT, Hung SL. Association between human herpesviruses and severity of periodontitis. J Periodontol. 2004;75:1479–85. doi: 10.1902/jop.2004.75.11.1479. [DOI] [PubMed] [Google Scholar]


