Abstract
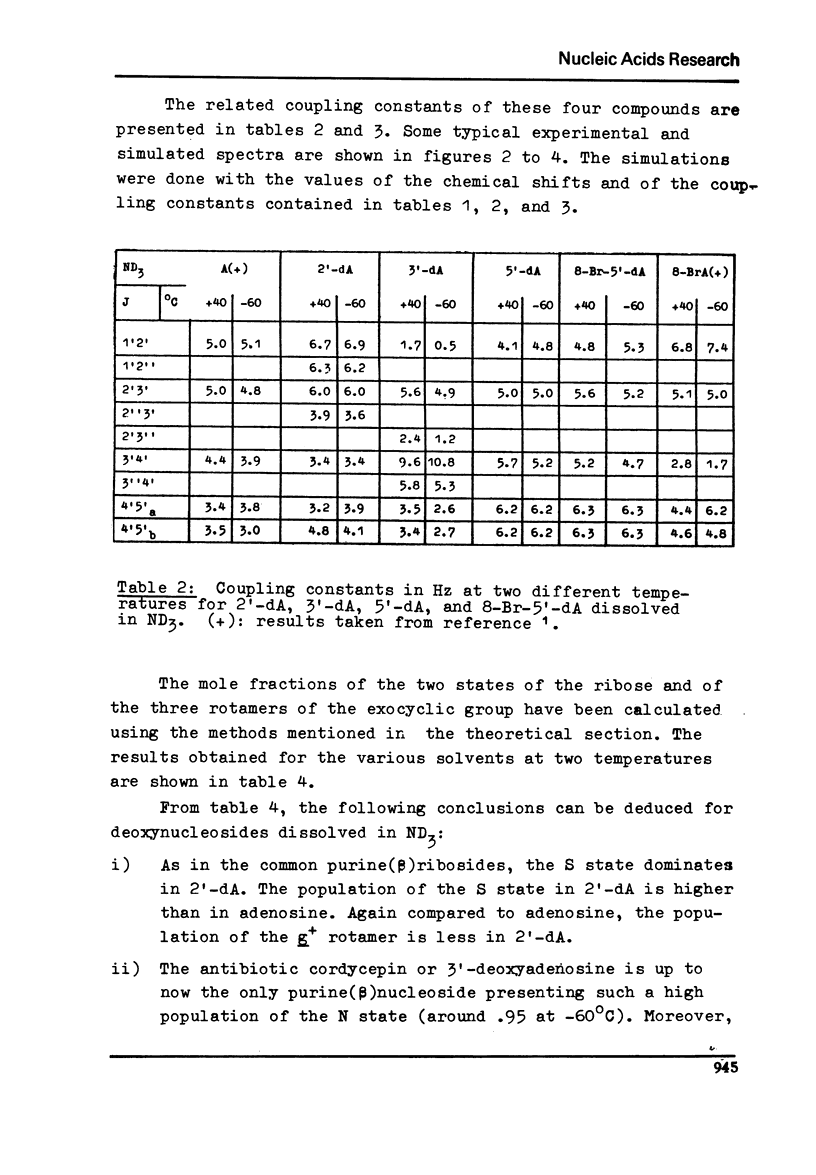
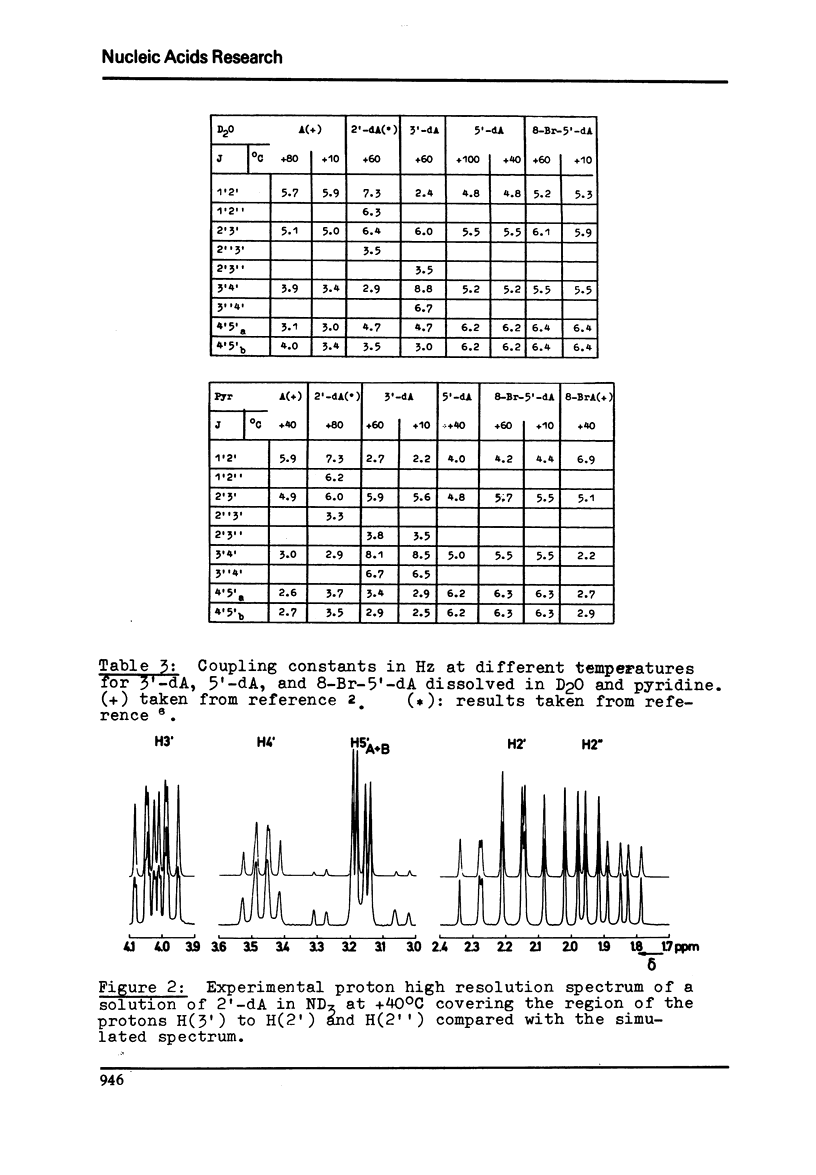
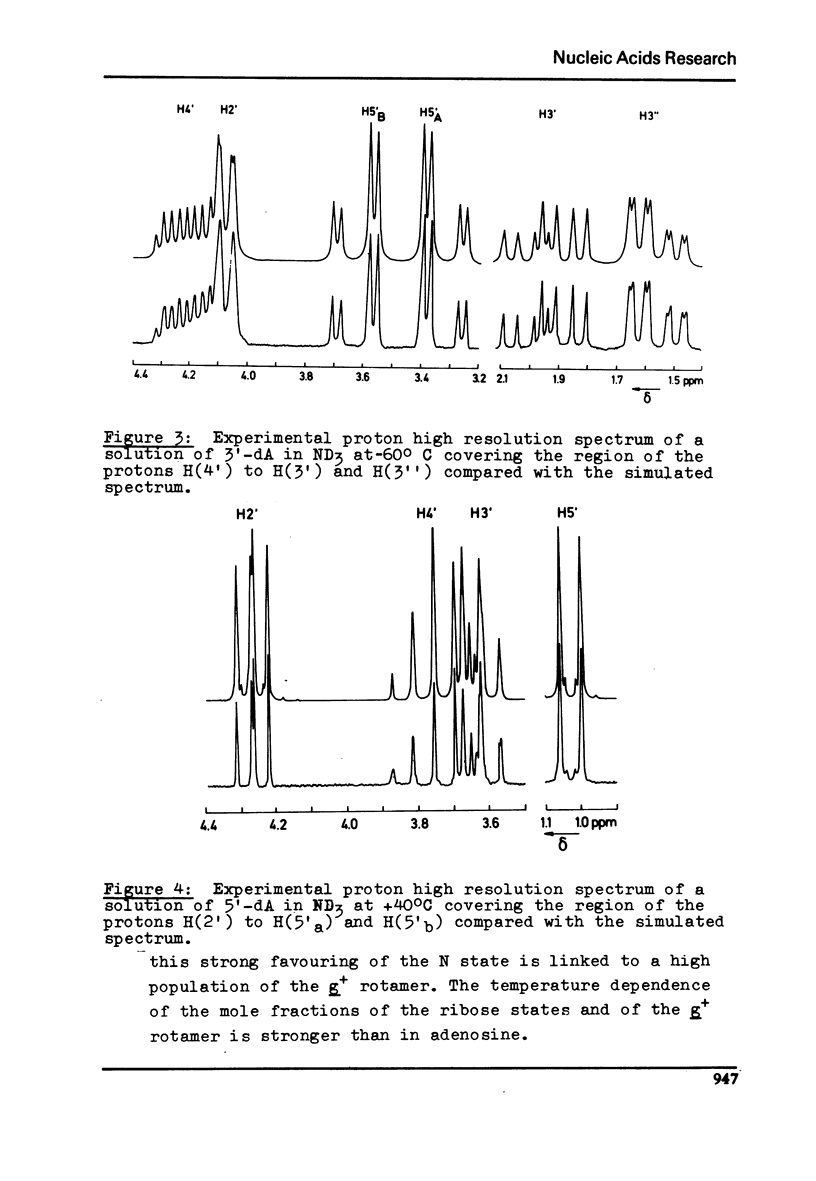
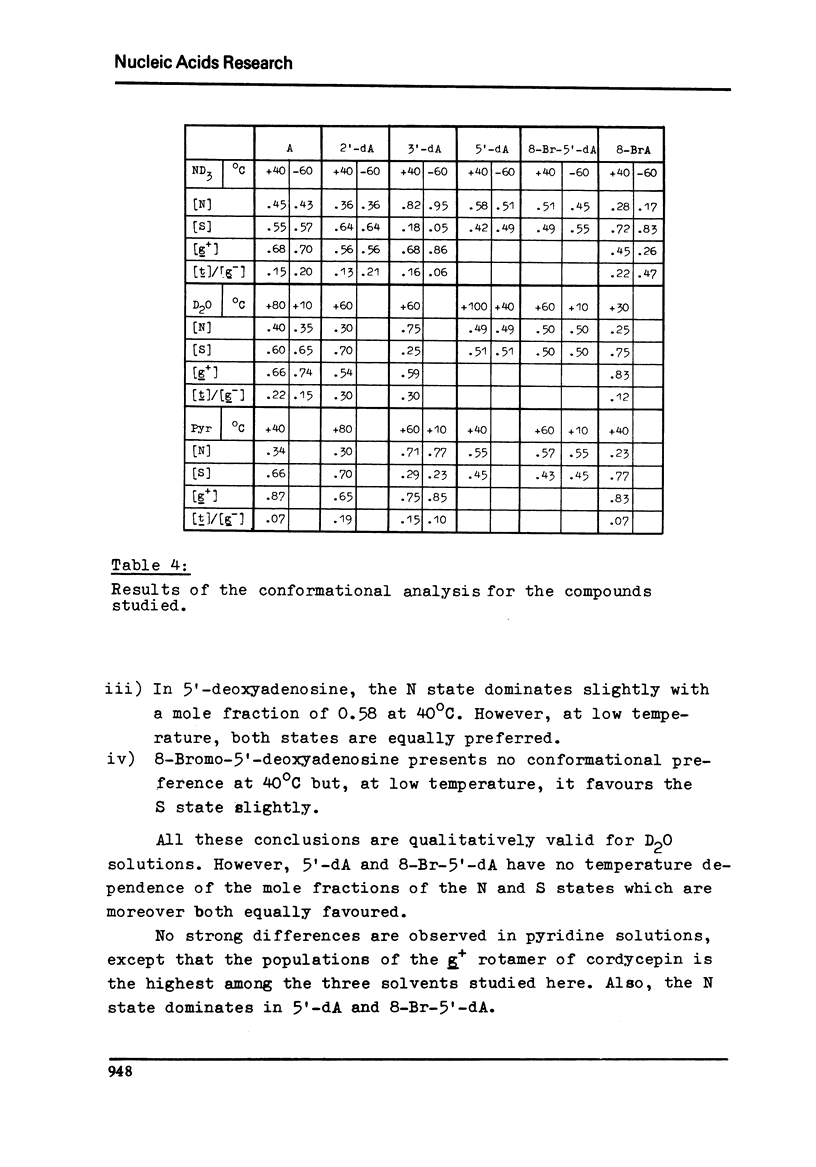
Proton magnetic resonance studies of 2'-deoxyadenosine (2'-dA), 3'-deoxyadenosine (3'-dA), 5'-deoxyadenosine (5'-dA) and 8-bromo-5'-deoxyadenosine (8-Br-5'-dA) have been carried out in the temperature range between -60 degrees and +40 degrees C for ND3 solutios, +40 degrees and +100 degrees C for D2O solutions, and finally +10 degrees and +60 degrees C for pyridine solutions. The analysis is based on the two-state S in equilibrium N model of the ribose moiety proposed by Altona and Sundaralingam. In all solvents, 2'-dA favours slightly the S state of the ribose and the g+ conformer of the exocyclic CH2OH group. However, 3'-dA prefers strongly the N state of the ribose and the g+ conformation. Both the S and N states of the ribose are equally favoured by 5'-DA and 8-Br-5'-dA. Evidence for the existence of an intramolecular hydrogen bond between 0(5') and N3 in purine (beta)-nucleosides is presented. It is also concluded that cordycepin (3'-dA) exists in solution mainly in the anti conformation of the base relative to the ribose.
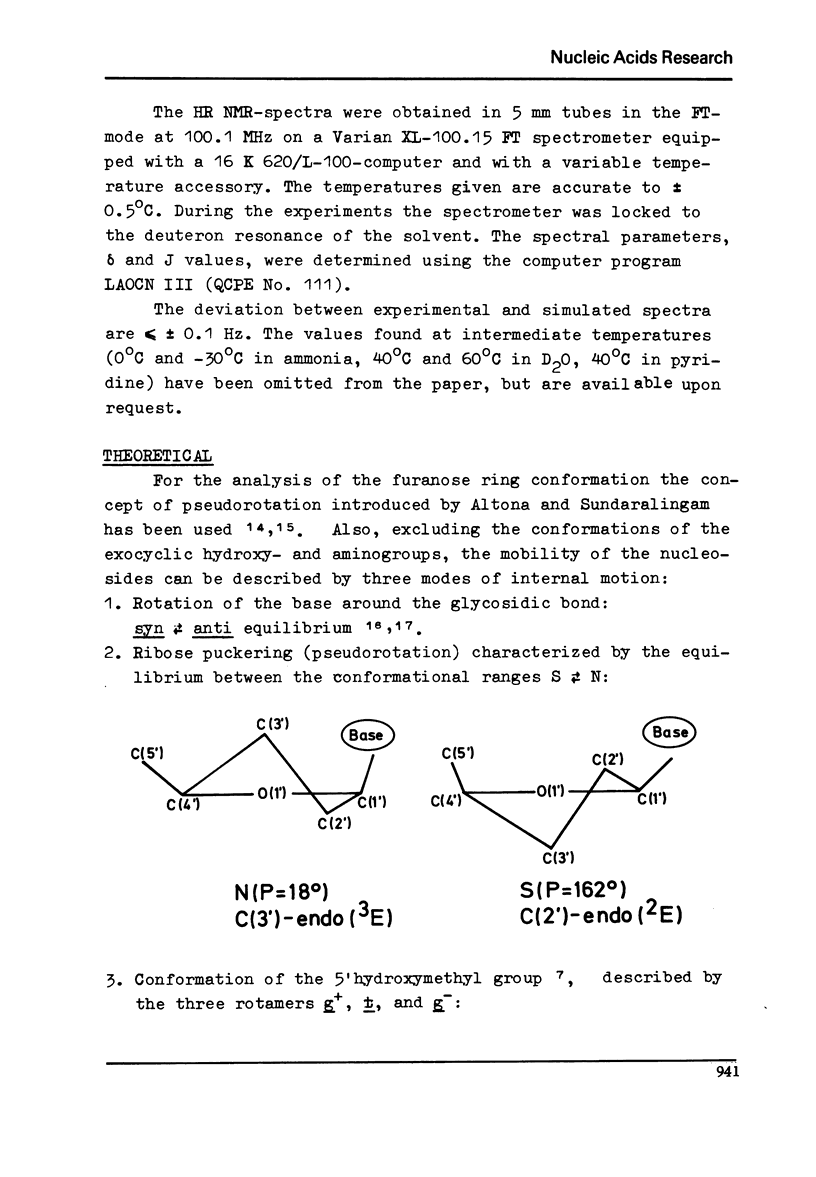
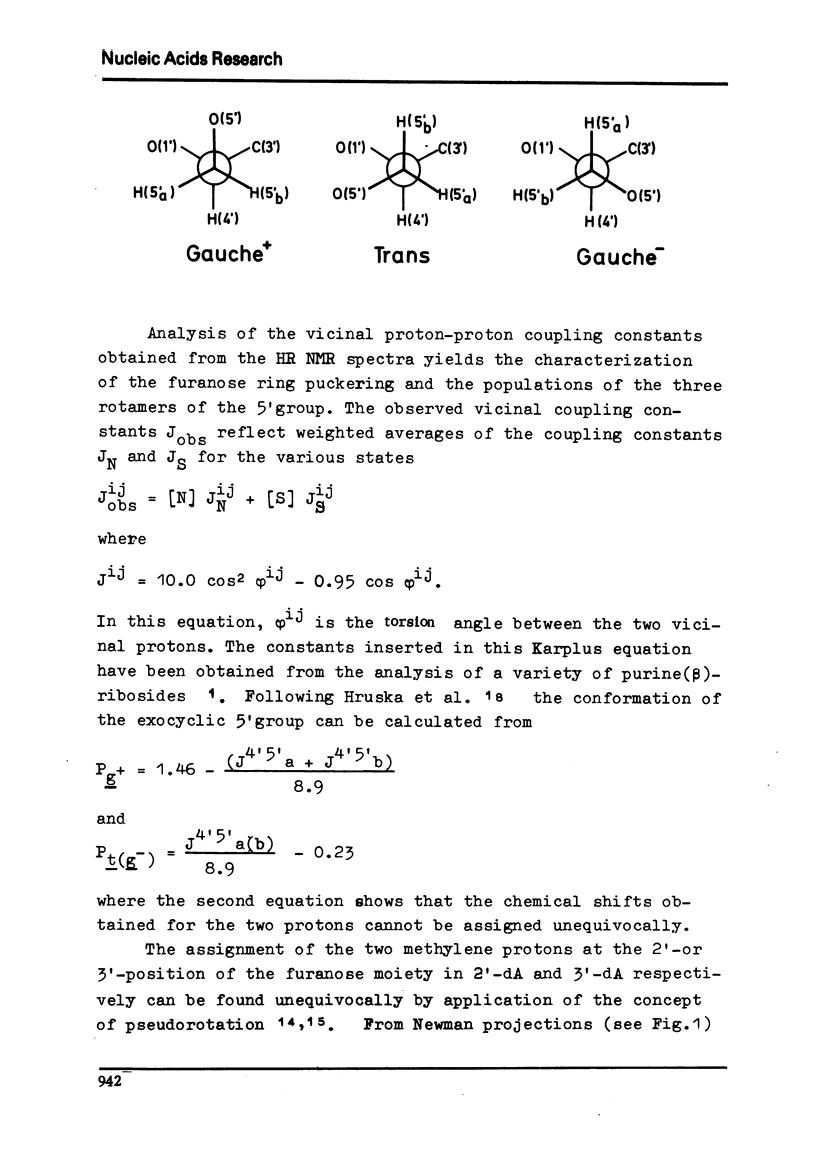
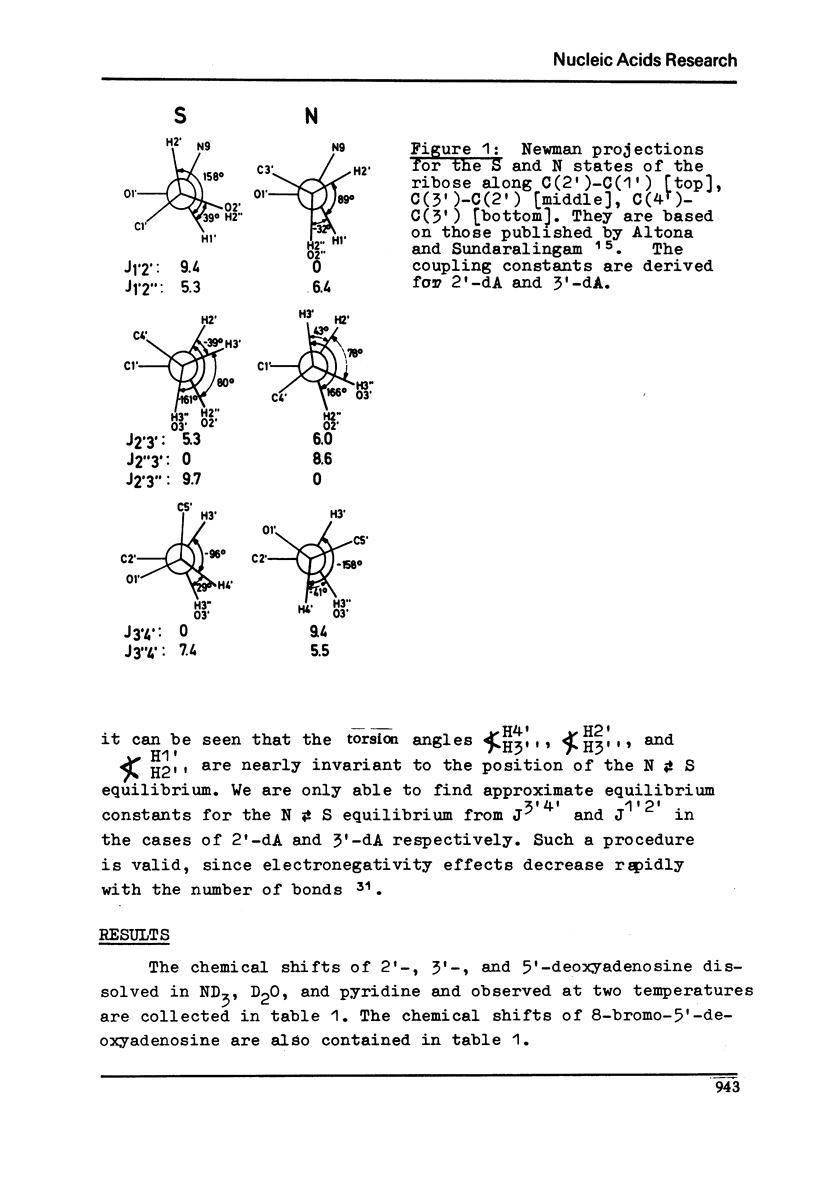
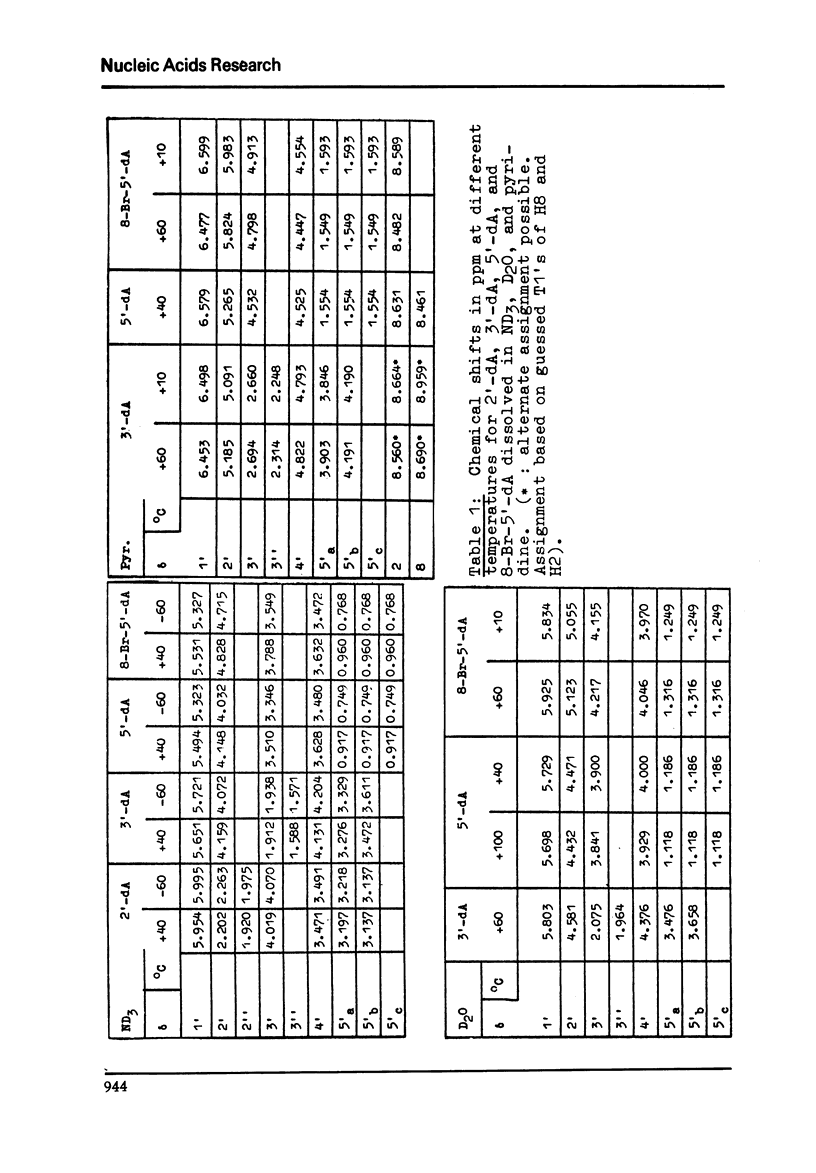
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. Improved method for the interpretation of proton magnetic resonance coupling constants. J Am Chem Soc. 1973 Apr 4;95(7):2333–2344. doi: 10.1021/ja00788a038. [DOI] [PubMed] [Google Scholar]
- Berthod H., Pullman B. Molecular orbital calculations on the conformation of nucleic acids and their constituents. I. Conformational energies of beta-nucleosides with C(3')-and C(2')-endo sugars. Biochim Biophys Acta. 1971 Apr 8;232(4):595–606. doi: 10.1016/0005-2787(71)90750-7. [DOI] [PubMed] [Google Scholar]
- DONOHUE J., TRUEBLOOD K. N. Base pairing in DNA. J Mol Biol. 1960 Dec;2:363–371. doi: 10.1016/s0022-2836(60)80047-2. [DOI] [PubMed] [Google Scholar]
- Davies D. B., Danyluk S. S. Nuclear magnetic resonance studies of 2'- and 3'-ribonucleotide structures in solution. Biochemistry. 1975 Feb 11;14(3):543–554. doi: 10.1021/bi00674a013. [DOI] [PubMed] [Google Scholar]
- Davies D. B., Danyluk S. S. Nuclear magnetic resonance studies of 5'-ribo- and deoxyribonucleotide structures in solution. Biochemistry. 1974 Oct 8;13(21):4417–4434. doi: 10.1021/bi00718a027. [DOI] [PubMed] [Google Scholar]
- Evans F. E., Sarma R. H. Comparative study of the structure and conformation in aqueous solution of the antileukemic agent 6-thiopurine ribonucleoside 5'-phosphate to that of common purine 5'-nucleotides. J Am Chem Soc. 1975 May 28;97(11):3215–3218. doi: 10.1021/ja00844a049. [DOI] [PubMed] [Google Scholar]
- Fox J. J., Watanabe K. A., Bloch A. Nucleoside antibiotics. Prog Nucleic Acid Res Mol Biol. 1966;5:251–313. doi: 10.1016/s0079-6603(08)60236-6. [DOI] [PubMed] [Google Scholar]
- Haschemeyer A. E., Rich A. Nucleoside conformations: an analysis of steric barriers to rotation about the glycosidic bond. J Mol Biol. 1967 Jul 28;27(2):369–384. doi: 10.1016/0022-2836(67)90026-5. [DOI] [PubMed] [Google Scholar]
- Hruska F. E., Wood D. J., Mynott R. J., Sarma R. H. H NMR study of the conformation of the ribose phosphate moiety of 6-azauridine-5'-monophosphate--a nucleotide with an unusual conformation. FEBS Lett. 1973 Apr 1;31(1):153–155. doi: 10.1016/0014-5793(73)80094-8. [DOI] [PubMed] [Google Scholar]
- Hruska F. E., Wood D. J., Singh H. Effect of temperature and protonation upon the conformation of 2'-o-methyladenosine. Correlation of conformational parameters in purine nucleosides. Biochim Biophys Acta. 1977 Jan 3;474(1):129–140. doi: 10.1016/0005-2787(77)90220-9. [DOI] [PubMed] [Google Scholar]
- Lüdemann H. D., Röder O., Westhof E., Goldammer E., Müller A. Conformation of the common purine (beta) ribosides in solution: further evidence for a correlation between N-S state of the ribose moiety and syn-anti equilibrium. Biophys Struct Mech. 1975 Feb 19;1(2):121–137. doi: 10.1007/BF00539774. [DOI] [PubMed] [Google Scholar]
- Lüdemann H. D., Westhof E., Cuno I. Ribose conformations of 8-azapurine nucleosides in solution. Z Naturforsch C. 1976 Mar-Apr;31(3-4):135–140. doi: 10.1515/znc-1976-3-407. [DOI] [PubMed] [Google Scholar]
- Lüdemann H. D., Westhof E., Röder O. A dynamic correlation between ribose conformation and glycosyl torsion angle of dissolved xanthosine studies by continuous-wave-mode and pulsed nuclear-magnetic-resonance methods. Eur J Biochem. 1974 Nov 1;49(1):143–150. doi: 10.1111/j.1432-1033.1974.tb03819.x. [DOI] [PubMed] [Google Scholar]
- Prusiner P., Yathindra N., Sundaralingam M. Effect of ribose O(2')-methylation on the conformation of nucleosides and nucleotides. Biochim Biophys Acta. 1974 Oct 11;366(2):115–123. doi: 10.1016/0005-2787(74)90325-6. [DOI] [PubMed] [Google Scholar]
- Remin M., Shugar D. Conformational analysis of cytidine, 1-beta-D-(arabinofuranosyl)cytosine and their O'-methyl derivatives by proton magnetic resonance spectroscopy. J Am Chem Soc. 1973 Nov 28;95(24):8146–8156. doi: 10.1021/ja00805a031. [DOI] [PubMed] [Google Scholar]
- Sarma R. H., Lee C. H., Evans F. E., Yathindra N., Sundaralingam M. Probing the interrelation between the glycosyl torsion, sugar pucker, and the backbone conformation in C(8) substituted adenine nucleotides by 1H and 1H-(31P) fast Fourier transform nuclear magnetic resonance methods and conformational energy calculations. J Am Chem Soc. 1974 Nov 13;96(23):7337–7348. doi: 10.1021/ja00830a028. [DOI] [PubMed] [Google Scholar]
- Son T. D., Guschlbauer W. Necleoside conformations. 19. Temperature and pH effects on the conformation of guanosine phosphates. Nucleic Acids Res. 1975 Jun;2(6):873–886. doi: 10.1093/nar/2.6.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaralingam M., Arora S. K. Crystal structure of the aminoglycosyl antibiotic puromycin dihydrochloride pentahydrate. Models for the terminal 3'-aminoacyladenosine moieties of transfer RNA's and protein-nucleic acid interactions. J Mol Biol. 1972 Oct 28;71(1):49–70. doi: 10.1016/0022-2836(72)90400-7. [DOI] [PubMed] [Google Scholar]
- Westhof E., Röder O., Croneiss I., Lüdemann H. D. Ribose conformations in the common purine(beta)ribosides, in some antibiotic nucleosides, and in some isopropylidene derivatives: a comparison. Z Naturforsch C. 1975 Mar-Apr;30(2):131–140. doi: 10.1515/znc-1975-3-401. [DOI] [PubMed] [Google Scholar]


