Abstract
OBJECTIVE
The C allele at the rs11212617 polymorphism in the ataxia-telangiectasia–mutated (ATM) gene has been associated with greater clinical response to metformin in people with type 2 diabetes. We tested whether this variant modified the effect of metformin in the Diabetes Prevention Program (DPP), in which metformin reduced diabetes incidence by 31% in volunteers with impaired glucose tolerance.
RESEARCH DESIGN AND METHODS
We genotyped rs11212617 in 2,994 DPP participants and analyzed its effects on diabetes incidence and related traits.
RESULTS
Contrary to expectations, C carriers enjoyed no preventive advantage on metformin; their hazard ratio, compared with A carriers, was 1.17 ([95% CI 0.96–1.42], P = 0.13) under metformin. There were no significant differences by genotype in metformin’s effects on insulin sensitivity, fasting glucose, glycated hemoglobin, or disposition index.
CONCLUSIONS
The reported association of rs11212617 with metformin response was not confirmed for diabetes prevention or for effects on relevant physiologic parameters in the DPP.
Metformin is an effective, cheap, and safe drug used as the first-line agent for treating type 2 diabetes (T2D) (1–3). Nevertheless, hyperglycemia eventually progresses in many patients, causing escalation of therapy (4). The reasons for such failure, which takes some time to become apparent, are unknown. Genetic factors may contribute to this process.
Recently, a genome-wide association study (GWAS) for metformin response has been published (5). The Genetics of Diabetes Audit and Research Tayside (GoDARTS) investigators analyzed 705,125 single nucleotide polymorphisms (SNPs) in 1,024 individuals who had a definable metformin response in a retrospective clinical database. Treatment success (analyzed as a categorical trait) was declared if glycated hemoglobin (A1C) became ≤7% within 18 months of starting therapy; change in A1C was also analyzed as a quantitative trait. Fourteen SNPs, concentrated around the ataxia-telangiectasia–mutated (ATM) gene, were associated with categorical metformin response at a suggestive level of P < 10−6. Consistent results were obtained for metformin response as a quantitative trait. The top SNP (rs11212617) was genotyped in 1,783 additional GoDARTS participants and 1,113 participants in the UK Prospective Diabetes Study (UKPDS) clinical trial (6). Again, in both cohorts, its minor C allele (frequency 44%) was nominally associated with greater metformin response, either as a categorical or quantitative trait. Joint analysis exceeded conventional genome-wide statistical significance (P = 2.9 × 10−9), i.e., this P value withstands correction for multiple comparisons. We therefore tested this SNP for metformin response in the Diabetes Prevention Program (DPP), a clinical trial with participants from five U.S. ethnic groups who were at high risk of T2D and were treated with an intensive lifestyle intervention or metformin for diabetes prevention.
RESEARCH DESIGN AND METHODS
Participants
The DPP enrolled 3,234 overweight or obese, nondiabetic people with impaired glucose tolerance. They were randomized to placebo, metformin (850 mg twice daily), or a lifestyle intervention. The participants’ mean age was 51 years and mean BMI was 34.0 kg/m2, 68% were women, and 45% belonged to U.S. ethnic minority groups; their demographic characteristics are shown in Table 1. The lifestyle and metformin interventions reduced the incidence of diabetes by 58 and 31%, respectively, versus placebo (7). In total, 2,994 participants (988 on metformin) consented to genetic investigation. All procedures were approved by institutional review boards at the 27 study sites.
Table 1.
Demographic characteristics of the DPP cohort by treatment arm and genotype at rs11212617
Genotyping
We genotyped rs11212617 on a Sequenom iPLEX platform as previously described (8,9). Genotyping success was 99.9%.
Measurements
Diabetes incidence was determined by semiannual measurements of fasting glucose and an annual oral glucose tolerance test. The principal study outcome was the development of diabetes by American Diabetes Association criteria, including confirmation. Besides diabetes incidence as a categorical outcome, we selected the insulin sensitivity index (ISI), fasting glucose, A1C, weight, and oral disposition index (DIo) as indices of metformin response. We calculated the ISI as 22.5/([fasting insulin × fasting glucose]/18.01) (or 1/homeostasis model assessment of insulin resistance) (10), and the DIo as 1/fasting insulin × insulinogenic index (Δinsulin/Δglucose over the first 30 min of the oral glucose tolerance test) (11). Quantitative traits were natural log–transformed.
Statistical analysis
We tested the additive effect of genotype at rs11212617 on diabetes incidence by Cox proportional hazards regression models with genotype and intervention and their interactions as the independent variables predicting time to diabetes over mean 3.2 years follow-up, adjusted for sex, ethnicity, treatment arm, baseline age, and waist circumference. We included all three treatment arms and an interaction test to simultaneously rule out a main effect of this variant on diabetes incidence independent of metformin, or under the action of a lifestyle intervention. We used generalized mixed models to test additive effect of genotype on baseline log-transformed traits and, to model change under metformin action, on the same traits after 1 year of intervention adjusted for the baseline value of the respective trait, age, sex, ethnicity, treatment arm, and waist circumference. Post hoc power calculations (which should be interpreted with caution) show that the sample size in the DPP metformin arm has >99% power to detect the change in A1C of 0.61% that was reported in the UKPDS (5). To control for the potential effect of ethnicity, we performed sensitivity analyses in the largest race/ethnic group (white participants), which is most closely related to the populations examined in the original report and whom we have previously shown to be essentially free of non-European admixture (12).
RESULTS
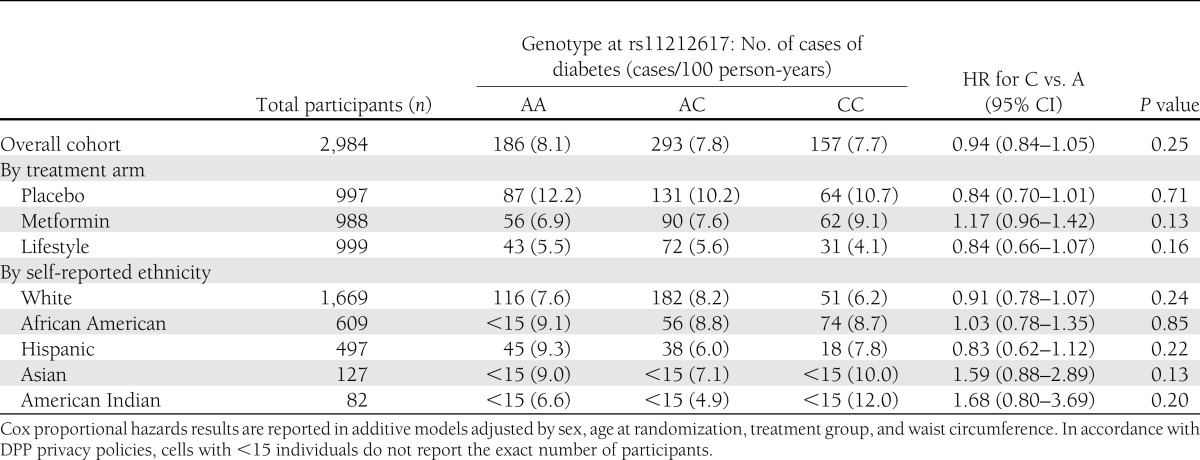
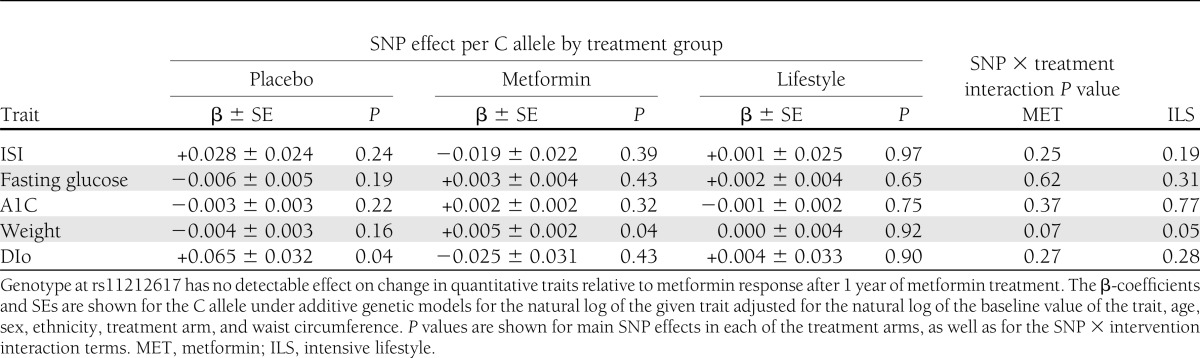
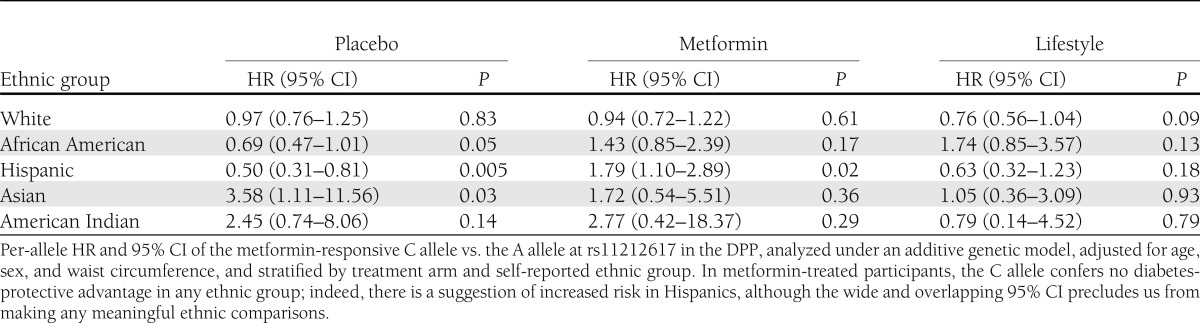
The frequency of the C (metformin-responsive) allele was 42.4, 72.4, 40.1, 51.6, and 41.5% in 1,669 white, 609 African American, 497 Hispanic, 127 Asian/Pacific Islander, and 82 American Indian participants, respectively (Table 1). We found no association of genotype with diabetes incidence in all arms combined, either in unadjusted analyses (hazard ratio [HR] per copy of the C allele 0.98 [95% CI 0.88–1.10], P = 0.76) or after adjusting for age, sex, ethnicity, and treatment arm (HR per copy of the C allele 0.95 [0.85–1.07], P = 0.42); further adjustment for waist circumference produced indistinguishable results (Table 2). There was no significant genotype × metformin interaction in the unadjusted model. Though there was a nominal SNP × metformin interaction in the fully adjusted model (P = 0.04), the observed trend was in the opposite direction from the expected prevention effect; the C allele conferred no detectable advantage on metformin recipients in diabetes prevention but was associated with a nonsignificant trend toward increased risk of diabetes (HR per copy of the C allele 1.17 [0.96–1.42], P = 0.13). We found no significant associations of genotype with relevant quantitative glycemic traits at baseline; similarly, there were no significant differences across genotype groups in change in ISI, fasting glucose, A1C, or DIo after 1 year of metformin (Table 3). The C allele was associated with greater weight gain in the metformin arm. In this arm, there were no statistically significant interactions between the C allele and BMI or waist circumference on diabetes incidence. Analyses stratified by ethnic group failed to show any ethnic-specific beneficial effects of the C allele with regard to diabetes incidence on metformin-treated participants (Table 4).
Table 2.
Diabetes incidence in the DPP by genotype at rs11212617, treatment arm, and self-reported ethnicity
Table 3.
Association of rs11212617 with quantitative glycemic traits at 1 year
Table 4.
Ethnic-specific effects of rs11212617 on diabetes incidence in the DPP
CONCLUSIONS
In the DPP, the effect of metformin to prevent diabetes or improve relevant glycemic traits was not magnified among carriers of the C allele at rs11212617 in the ATM gene. Our findings do not support the previously reported association of this allele with improved metformin action on glycemic control. The original association was consistent in three different datasets (the discovery sample and two follow-up cohorts) and has been reported recently in other clinical cohorts similarly ascertained (13). Inconsistent results in the DPP could be due to multiple reasons. First, metformin response is defined differently in a prediabetic cohort (impact on diabetes incidence or quantitative glycemic traits) than it is in a disease cohort (ability to reach A1C ≤7% under treatment). Second, metformin may be more effective in individuals with a higher A1C at baseline, and thereby the effects of genotype on response might be easier to detect in the disease setting. Third, the reported effect might be confined to populations of European descent, e.g., if rs11212617 tags a low-frequency variant unique to white populations, further diminishing statistical power in the DPP multiethnic cohort. And fourth, the previously reported GWAS was based on a retrospective evaluation of clinical records, where potential confounders (e.g., if genotype were to influence comorbidities that affect patient adherence, continuity of care, or frequency of A1C measurements) are harder to control than in a clinical trial.
To address potential ethnic differences in the genomic architecture of this region that might explain our negative results, we examined the haplotype structure around this locus in the HapMap European (CEU) and West African (YRI) datasets. The full ATM gene and the rs11212617 variant share a segment of tight linkage disequilibrium in both the CEU and YRI populations; there is a recombination hot spot downstream from rs11212617, beyond which SNPs display equally low correlations with rs11212617 in CEU and YRI, indicating that major differences in linkage disequilibrium patterns would be unlikely to account for potentially discrepant findings in Europeans and Africans. Furthermore, the region distal to this hot spot was well captured by the original GWAS array, suggesting that a true signal emerging from this region (and which might have explained a stronger association in Europeans than in Africans) would have also been detected by the original GWAS.
Nevertheless, this previously reported association merits additional follow-up in independent cohorts. More generally, a better-powered genome-wide assessment of pharmacogenetic responses in T2D is needed; whether genetic information will prove useful in diabetes prevention or therapeutics must be tested in prospective clinical trials.
Acknowledgments
The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health provided funding to the clinical centers and the coordinating center for the design and conduct of the study, including collection, management, analysis, and interpretation of the data. The Southwestern American Indian Centers were supported directly by the NIDDK and the Indian Health Service. The General Clinical Research Center Program, National Center for Research Resources, and the Department of Veterans Affairs supported data collection at many of the clinical centers. Funding for data collection and participant support was also provided by the Office of Research on Minority Health, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the National Institute on Aging, the Office of Research on Women’s Health, the Centers for Disease Control and Prevention, and the American Diabetes Association. This research was also supported, in part, by the intramural research program of the NIDDK and Grant R01-DK-072041 to J.C.F., K.A.J., A.R.S., and T.I.P. McKesson BioServices Corp., Matthews Media Group, Inc., and the Henry M. Jackson Foundation provided support services under subcontract with the coordinating center.
Bristol-Myers Squibb and Parke-Davis provided medication. LifeScan, Inc., Health O Meter, Hoechst Marion Roussel, Inc., Merck-Medco Managed Care, Inc., Merck and Co., Inc., Nike Sports Marketing, Slim Fast Foods Co., and Quaker Oats Co. donated materials, equipment, or medicines for concomitant conditions. J.C.F. has received consulting honoraria from Novartis, Eli Lilly and Co., and Pfizer. N.H.W. has received consulting honoraria from NovoNordisk and Daiichi-Sankyo. A.R.S. has received consulting honoraria from Merck. No other potential conflicts of interest relevant to this article were reported.
The opinions expressed are those of the investigators and do not necessarily reflect the views of the Indian Health Service or other funding agencies.
J.C.F. researched data, contributed to discussion, wrote the manuscript, and reviewed and edited the manuscript. K.A.J., A.T., W.C.K., A.R.S., and T.I.P. researched data, contributed to discussion, and reviewed and edited the manuscript. K.M., E.H., N.H.W., and E.B.-C. contributed to discussion and reviewed and edited the manuscript. J.C.F. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Parts of this study were presented at the 71st Scientific Sessions of the American Diabetes Association, San Diego, California, 24–28 June 2011.
The authors gratefully acknowledge the commitment and dedication of the participants of the DPP.
Footnotes
Clinical trial reg. no. NCT00004992, clinicaltrials.gov.
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc11-2301/-/DC1.
*A complete list of the Diabetes Prevention Program Research Group investigators is provided in the Supplementary Data online.
References
- 1.Nathan DM, Buse JB, Davidson MB, et al. American Diabetes Association. European Association for Study of Diabetes Medical management of hyperglycemia in type 2 diabetes: a consensus algorithm for the initiation and adjustment of therapy: a consensus statement of the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care 2009;32:193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathan DM, Buse JB, Davidson MB, et al. American Diabetes Association. European Association for the Study of Diabetes Medical management of hyperglycaemia in type 2 diabetes mellitus: a consensus algorithm for the initiation and adjustment of therapy: a consensus statement from the American Diabetes Association and the European Association for the Study of Diabetes. Diabetologia 2009;52:17–30 [DOI] [PubMed] [Google Scholar]
- 3.Rodbard HW, Jellinger PS, Davidson JA, et al. Statement by an American Association of Clinical Endocrinologists/American College of Endocrinology consensus panel on type 2 diabetes mellitus: an algorithm for glycemic control. Endocr Pract 2009;15:540–559 [DOI] [PubMed] [Google Scholar]
- 4.Kahn SE, Haffner SM, Heise MA, et al. ADOPT Study Group Glycemic durability of rosiglitazone, metformin, or glyburide monotherapy. N Engl J Med 2006;355:2427–2443 [DOI] [PubMed] [Google Scholar]
- 5.Zhou K, Bellenguez C, Spencer CC, et al. GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group. Wellcome Trust Case Control Consortium 2. MAGIC investigators Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet 2011;43:117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.UK Prospective Diabetes Study (UKPDS) Group Effect of intensive blood-glucose control with metformin on complications in overweight patients with type 2 diabetes (UKPDS 34). Lancet 1998;352:854–865 [PubMed] [Google Scholar]
- 7.Knowler WC, Barrett-Connor E, Fowler SE, et al. Diabetes Prevention Program Research Group Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002;346:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hivert MF, Jablonski KA, Perreault L, et al. DIAGRAM Consortium. Diabetes Prevention Program Research Group Updated genetic score based on 34 confirmed type 2 diabetes loci is associated with diabetes incidence and regression to normoglycemia in the diabetes prevention program. Diabetes 2011;60:1340–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang K, Fu DJ, Julien D, Braun A, Cantor CR, Köster H. Chip-based genotyping by mass spectrometry. Proc Natl Acad Sci USA 1999;96:10016–10020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–419 [DOI] [PubMed] [Google Scholar]
- 11.Utzschneider KM, Prigeon RL, Faulenbach MV, et al. Oral disposition index predicts the development of future diabetes above and beyond fasting and 2-h glucose levels. Diabetes Care 2009;32:335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jablonski KA, McAteer JB, de Bakker PI, et al. Diabetes Prevention Program Research Group Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes 2010;59:2672–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Leeuwen N, Nijpels G, Becker ML, et al. A gene variant near ATM is significantly associated with metformin treatment response in type 2 diabetes: a replication and meta-analysis of five cohorts. Diabetologia. 28 March 2012 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]