Abstract
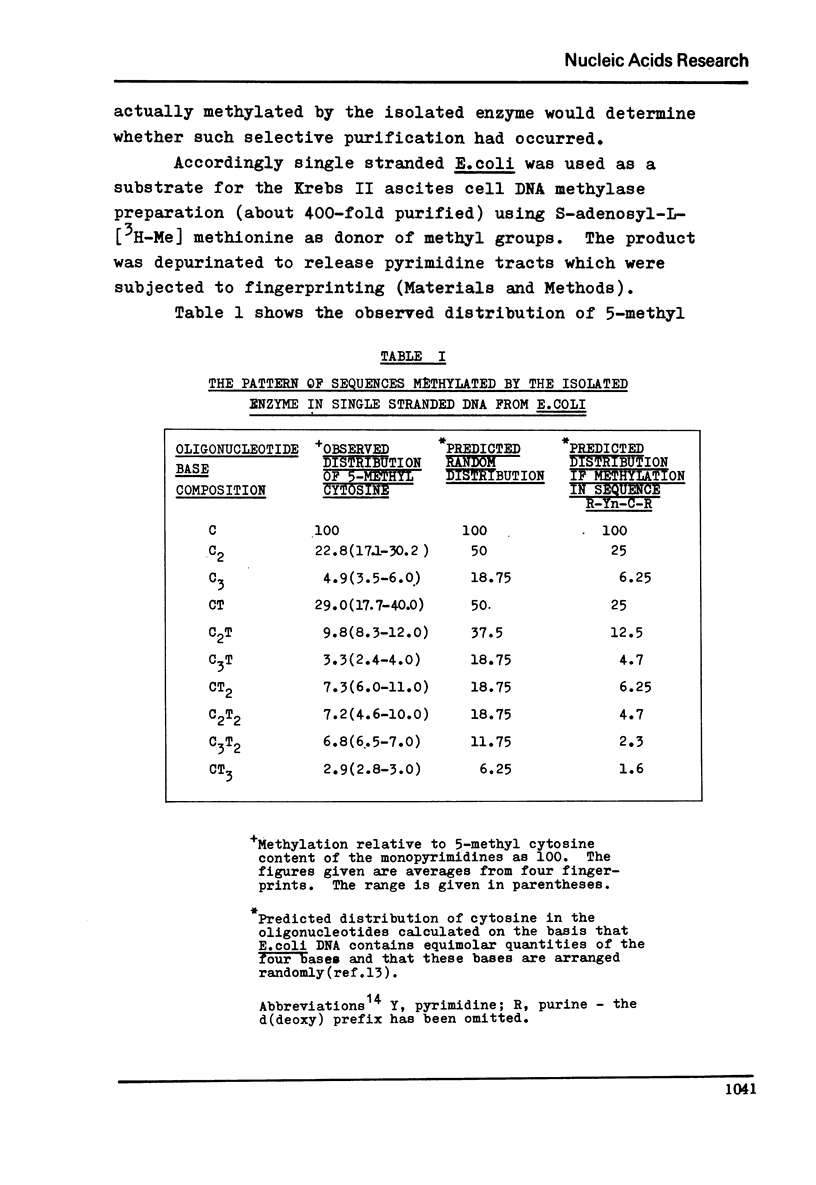
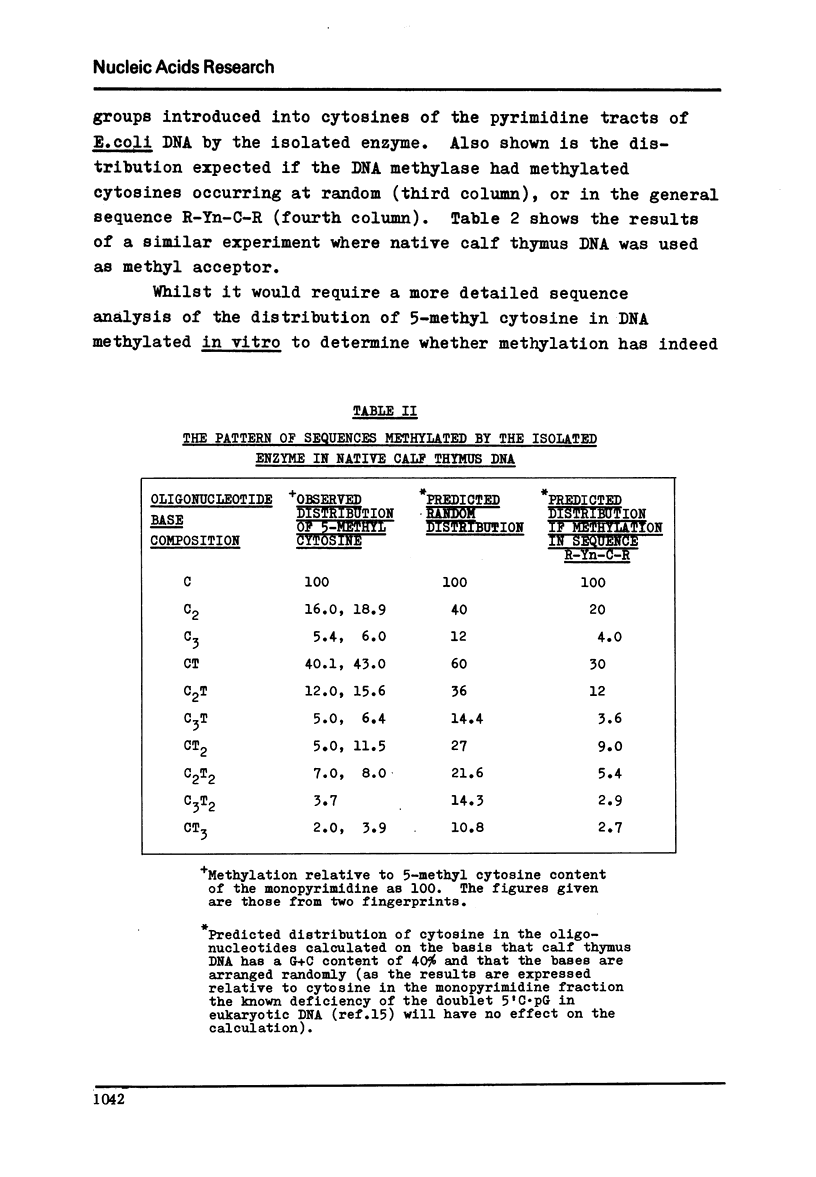
The sequence specificity of an extensively purified DNA methylase preparation from Krebs II mouse ascites cells has been examined. The enzyme appears to be highly sequence dependent. Moreover the sequence distribution of cytosine residues that are methylated, bears a very close resemblance to the sequence distribution of 5'-methyl cytosine found in vivo in a wide range of vertebrate cells and is consistent with methylation of cytosines in the sequence R-Yn-C-R.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arber W. DNA modification and restriction. Prog Nucleic Acid Res Mol Biol. 1974;14(0):1–37. doi: 10.1016/s0079-6603(08)60204-4. [DOI] [PubMed] [Google Scholar]
- Browne M. J., Burdon R. H. The sequence specificity of vertebrate DNA methylation. Nucleic Acids Res. 1977 Apr;4(4):1025–1037. doi: 10.1093/nar/4.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Drahovsky D., Morris N. R. The mechanism of action of rat liver DNA methylase. 3. Nucleotide requirements for binding and methylation. Biochim Biophys Acta. 1972 Aug 25;277(2):245–250. doi: 10.1016/0005-2787(72)90404-2. [DOI] [PubMed] [Google Scholar]
- Drahovský D., Morris N. R. Mechanism of action of rat liver DNA methylase. I. Interaction with double-stranded methyl-acceptor DNA. J Mol Biol. 1971 May 14;57(3):475–489. doi: 10.1016/0022-2836(71)90104-5. [DOI] [PubMed] [Google Scholar]
- Drahovský D., Morris N. R. Mechanism of action of rat liver DNA methylase. II. Interaction with single-stranded methyl-acceptor DNA. J Mol Biol. 1971 Oct 28;61(2):343–356. doi: 10.1016/0022-2836(71)90384-6. [DOI] [PubMed] [Google Scholar]
- Grippo P., Iaccarino M., Parisi E., Scarano E. Methylation of DNA in developing sea urchin embryos. J Mol Biol. 1968 Sep 14;36(2):195–208. doi: 10.1016/0022-2836(68)90375-6. [DOI] [PubMed] [Google Scholar]
- Kalousek F., Morris N. R. Deoxyribonucleic acid methylase activity in pea seedlings. Science. 1969 May 9;164(3880):721–722. doi: 10.1126/science.164.3880.721. [DOI] [PubMed] [Google Scholar]
- Morris N. R., Pih K. D. The preparation of soluble DNA methylase from normal and regenerating rat liver. Cancer Res. 1971 Apr;31(4):433–440. [PubMed] [Google Scholar]
- Roy P. H., Weissbach A. DNA methylase from HeLa cell nuclei. Nucleic Acids Res. 1975 Oct;2(10):1669–1684. doi: 10.1093/nar/2.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell G. J., Walker P. M., Elton R. A., Subak-Sharpe J. H. Doublet frequency analysis of fractionated vertebrate nuclear DNA. J Mol Biol. 1976 Nov;108(1):1–23. doi: 10.1016/s0022-2836(76)80090-3. [DOI] [PubMed] [Google Scholar]
- Sneider T. W. Methylation of mammalian deoxyribonucleic acid. II. The distribution of 5-methylcytosine in pyrimidine deoxyribonucleotide clusters in Novikoff hepatoma cell deoxyribonucleic acid. J Biol Chem. 1971 Aug 10;246(15):4774–4783. [PubMed] [Google Scholar]


