Summary
Tissue-specific transcription patterns are preserved throughout cell divisions to maintain lineage fidelity. We investigated whether transcription factor GATA1 plays a role in transmitting hematopoietic gene expression programs through mitosis when transcription is transiently silenced. Live cell imaging revealed that a fraction of GATA1 is retained focally within mitotic chromatin. ChIP-seq of highly purified mitotic cells uncovered that key hematopoietic regulatory genes are occupied by GATA1 in mitosis. The GATA1 co-regulators FOG1 and TAL1 dissociate from mitotic chromatin, suggesting that GATA1 functions as platform for their postmitotic recruitment. Mitotic GATA1 target genes tend to re-activate more rapidly upon entry into G1 than genes from which GATA1 dissociates. Mitosis-specific destruction of GATA1 delays reactivation selectively of genes that retain GATA1 during mitosis. These studies suggest a requirement of mitotic “bookmarking” by GATA1 for the faithful propagation of cell type-specific transcription programs through cell division.
Keywords: GATA1, mitosis, bookmarking, hematopoiesis, chromatin
Introduction
During development lineage committed cells undergo numerous cell division cycles. For example, erythroid cells not only traverse several divisions as they mature, but cell cycle progression is required for their differentiation and onset of high level erythroid-specific gene expression (Pop et al., 2010). The mitotic phase of each cell division cycle is characterized by a global silencing of transcription associated with the separation from mitotic chromosomes of RNA polymerase II (pol2) and the majority of general and tissue/gene-specific transcription factors (Prescott and Bender, 1962; Taylor, 1960) for review see (Delcuve et al., 2008; Gottesfeld and Forbes, 1997; Sarge and Park-Sarge, 2009; Zaidi et al., 2010). Mitosis is therefore thought to present a challenge to the maintenance of transcriptional states, and by extension, to the normal maturation of committed progenitor cells and the preservation of lineage fidelity. On the other hand, it has been argued that mitosis might afford an opportunity for directed changes in cell type or developmental stage (Egli et al., 2008).
How transcriptional programs are re-established in newborn cells has become the focus of numerous recent studies. What has become clear is that not all signatures of active gene expression are erased during mitosis. The acetylation and methylation of histones is partially or completely maintained during mitosis (Aoto et al., 2008; Blobel et al., 2009; Bonenfant et al., 2007; Kouskouti and Talianidis, 2005; Kruhlak et al., 2001; McManus et al., 2006; Sasaki et al., 2009; Valls et al., 2005; Zaidi et al., 2003). Histone variants that mark active genes remain localized at the promoter regions (Bruce et al., 2005; Kelly et al., 2010), and in some cases (Kuo et al., 1982; Martinez-Balbas et al., 1995) but not others (Komura et al., 2007) regions of accessible chromatin, as reflected in enhanced DNaseI sensitivity, remain in their un-compacted state during mitosis. There even seems to exist mitosis specific unwinding of promoter DNA at genes that are active during interphase (Michelotti et al., 1997). General factors, such as members of the BET family, the histone methyltransferase MLL and, albeit variably, the acetyltransferase p300, have been reported to remain bound to subsets of genes during mitosis (Blobel et al., 2009; Dey et al., 2000; Kanno et al., 2004; Kouskouti and Talianidis, 2005; Kruhlak et al., 2001; Zaidi et al., 2003). Moreover, the basal transcription factor TBP (TATA binding protein) has been reported to mark active genes during mitosis (Chen et al., 2002; Christova and Oelgeschlager, 2002; Xing et al., 2008) although other studies found TBP to largely depart from mitotic chromatin (Blobel et al., 2009; Komura et al., 2007; Segil et al., 1996; Varier et al., 2010). Finally, a select few sequence-specific DNA binding proteins remain at least partially associated with chromatin during mitosis, including RUNX2, AP2, HSF2, FoxI1 (Martinez-Balbas et al., 1995; Xing et al., 2005; Yan et al., 2006; Young et al., 2007a; Young et al., 2007b), (for review see (Egli et al., 2008).
The presence of one or more of these features at a given gene during mitosis has been invoked to account for a mitotic memory function that ensures the accurate and timely re-activation of gene expression upon G1 entry. Despite these important efforts in defining mitosis-associated factors, a role for transcriptional bookmarking has largely been inferred based simply on their physical association with mitotic chromatin. In particular, the hierarchy of events, such as the mitotic binding of transcription factors and their co-factors, the mitotic preservation of histone modifications, as well as their respective contributions to the reassembly of the transcription apparatus upon mitotic exit, remains mostly speculative. Thus, in our view, a major challenge that has yet to be met is the demonstration that a given factor or histone modification directly bestows a bookmarking function on an associated gene. Past efforts to address this question include the knockdown of potential mitotic bookmarking factors. For instance, the depletion of Brd4 delays the onset of expression of select genes during entry into G1 (Dey et al., 2009; Yang et al., 2008). Knock-down of MLL likewise impairs the timely re-activation of genes that are selectively occupied by MLL during mitosis (Blobel et al., 2009). However, the interpretation of such experiments is confounded by possible effects of the knockdowns during non-mitotic phases of the cell cycle.
Here we examined whether tissue-specific transcription factors might be involved in the mitotic bookmarking of select lineage specific genes. The hematopoietic transcription factor GATA1 seemed an ideal candidate as a bookmarking factor since it controls the expression of virtually all erythroid-specific genes (for review see (Ferreira et al., 2005). GATA1, a zinc finger protein, is essential for normal erythroid development. Mice lacking GATA1 succumb to lethal anemia due to the failure to produce mature viable erythroid cells (Fujiwara et al., 1996). Mutations in GATA1 underlie certain forms of congenital anemias and are invariably found in Down syndrome patients with acute megakaryoblastic leukemias (Nichols et al., 2000; Wechsler et al., 2002).
Here we report a unique pattern of GATA1 chromatin occupancy during mitosis and reveal an essential mitosis-specific role of GATA1 for post-mitotic transcription reactivation. The results suggest that GATA1 is required for the normal propagation of a hematopoietic transcriptional program through mitosis, thus contributing to the stable maintenance of lineage- and stage-appropriate gene expression patterns during development.
Results
GATA1 focally associates with mitotic chromatin in erythroid cells
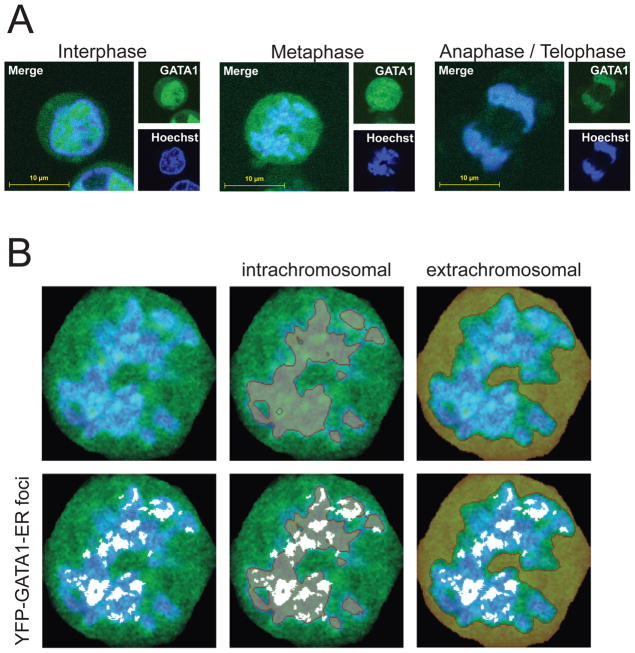
To monitor GATA1 localization on a global scale in living, unsynchronized erythroid cells, GATA1-YFP fusion constructs (Figure S1A) were stably introduced into G1E cells. G1E cells are erythroid precursors that lack GATA1 and consequently fail to mature (Weiss et al., 1997). Introduction of a conditional form of GATA1 (GATA1 fused to the ligand binding domain of the estrogen receptor, ER) conveys estradiol (E2)-dependent erythroid maturation in a manner faithfully reproducing that of normal erythroid cells. GATA1-ER target gene occupancy and expression closely match that of endogenous GATA1 in primary erythroblasts (Pilon et al., 2011; Welch et al., 2004), providing a physiological assay for GATA1 function. Both N-terminal and C-terminal YFP fusions of GATA1-ER were generated to account for potential effects of YFP on GATA1-ER function. YFP-GATA1-ER and GATA1-ER-YFP were expressed at levels similar to endogenous GATA1 (Figure S1B) and were equally capable of inducing erythroid differentiation when compared to wild-type GATA1 (Figure S1C–D). The localization of E2-activated YFP-GATA1-ER was monitored in interphase and mitosis using confocal microscopy (Figure 1A) in live cells stained with the Hoechst 33342 DNA stain. Although G1E cells are non-adherent, we obtained sufficiently clear images allowing conclusions regarding the subnuclear localization of YFP fusion proteins. During interphase, YFP-GATA1-ER showed strong nuclear accumulation. In metaphase, most of YFP-GATA1-ER was dispersed throughout the cytoplasm. However, strong focal accumulations of YFP-GATA1-ER appeared to co-localize with chromatin. Similar results were observed with GATA1-ER-YFP, and constructs lacking the ER moiety (YFP-GATA1, Cherry-GATA1), but not YFP alone (Figure S1A). Towards the end of mitosis, GATA1 fully re-localized with DNA-dense regions, as expected.
Figure 1. Focal Co-localization of GATA1 with Mitotic Chromosomes in Erythroid Cells.
A. Confocal microscopy of live, unsynchronized cells expressing YFP-GATA1-ER exposed to Hoechst live stain. Images of single cells were cropped to 300 × 300 pixels. B. Quantitative analysis of YFP-GATA1-ER localization of a representative cell in metaphase as in the middle of panel A. Intra- and extrachromosomal territories (brown, overlaid in middle and right panels, respectively) and GATA1 foci (white, overlaid in lower panels) were defined as described in the text. Note that GATA1 foci colocalize with chromatin.
Quantitative image analysis confirmed the co-localization of YFP-GATA1-ER foci with metaphase chromatin (Figure 1B). Hoechst staining intensities were used to define intra- and extra-chromosomal territories. Subsequently, we examined the partitioning with regard to these territories of the most intense GATA1 foci, defined as contiguous patches of bright (top 10th percentile) pixels. This revealed unequivocal co-localization of YFP-GATA1-ER foci with Hoechst positive mitotic chromatin (Pearson correlation coefficient of 0.98). These foci were virtually excluded from the extrachromosomal space.
Prior work did not identify mitotic chromatin-associated GATA1 (Xin et al., 2007), perhaps because it represents a relatively small fraction of total GATA1. However, ChIP analysis unequivocally demonstrates that GATA1 selectively occupies a subset of its targets in mitosis (below).
Purification of mitotic hematopoietic cells
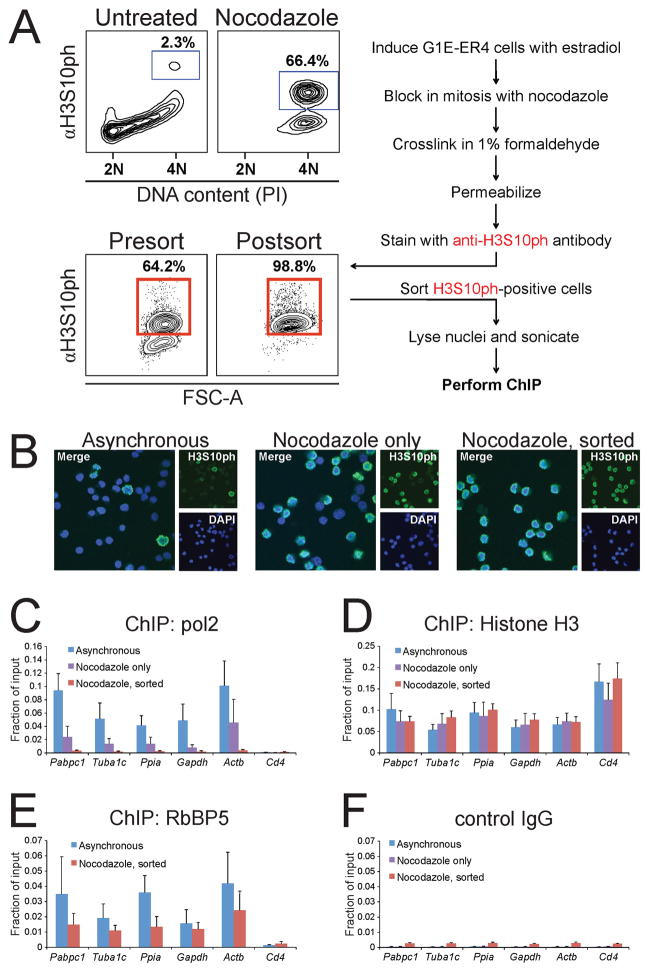
Determining the genomic occupancy of GATA1 specifically during mitosis by ChIP requires the generation of pure preparations of mitotic cells. We developed a protocol that is based on fluorescence-activated cell sorting (FACS) using the mitosis-specific marker histone H3 phosphorylated at serine 10 (H3S10ph) (Hendzel et al., 1997) under conditions allowing for subsequent ChIP (Figure 2A). This method consistently yielded ≥98% mitotic cells as determined by post-sort flow cytometry, IF visualizing anti-H3S10ph staining, or DAPI staining to detect cells with condensed prometaphase chromatin (Figure 2A, B). We confirmed that chromatin fragmentation of asynchronous and mitotic material was comparable (Figure S2A). Importantly, ChIP showed that pol2 occupancy at the 5′ transcribed regions of highly expressed housekeeping genes was reduced to undetectable levels in purified mitotic cells (Figure 2C), consistent with their transcriptional silencing, whereas in cells that were not FACS-purified, pol2 occupancy was only partially reduced commensurate with the incomplete degree of mitotic enrichment. Anti-histone H3 ChIP produced equal signals in mitotic and asynchronous cells (Figure 2D). Moreover, RbBP5, a MLL-associated protein that is retained at highly transcribed housekeeping genes during mitosis (Blobel et al., 2009), remained bound at target sites (Figure 2E). Control ChIP with isotype-matched IgG showed a very small but consistent increase in background signal in the sorted samples (Figure 2F), likely due to a small amount of residual anti-H3S10ph antibody bound to mitotic chromatin after sonication. In concert, these results demonstrate the usefulness of this protocol to purify mitotic cells for ChIP analysis.
Figure 2. Purification of Mitotic Erythroid Cells.
A. Purification protocol (right). DNA content and H3S10ph were measured in untreated or nocodazole-arrested G1E cells (upper left). Note incomplete enrichment (~66%) of mitotic cells in the nocodazole treated population. Purification of H3S10ph enriched mitotic cells to >98% (lower left). Propidium iodide (PI) was omitted from sorting experiments since the H3S10ph mark was sufficient to detect mitotic cells. Percentages indicate mitotic indices of representative experiments. FSC-A, forward scatter (cell size). B. IF of anti-H3S10ph and DAPI stained cells confirms high purity of mitotic cells following nocodazole treatment only after cell sorting. C. ChIP experiments showing complete pol2 dissociation from highly transcribed housekeeping genes in sorted but not unsorted nocodazole-treated cells. Cd4, negative control. D–F, ChIP using control IgG, anti-histone H3, and anti-RbBP5 served as controls. Error bars denote SEM (n = 3).
Mitotic GATA1 binding sites tend to mark key regulators of hematopoiesis
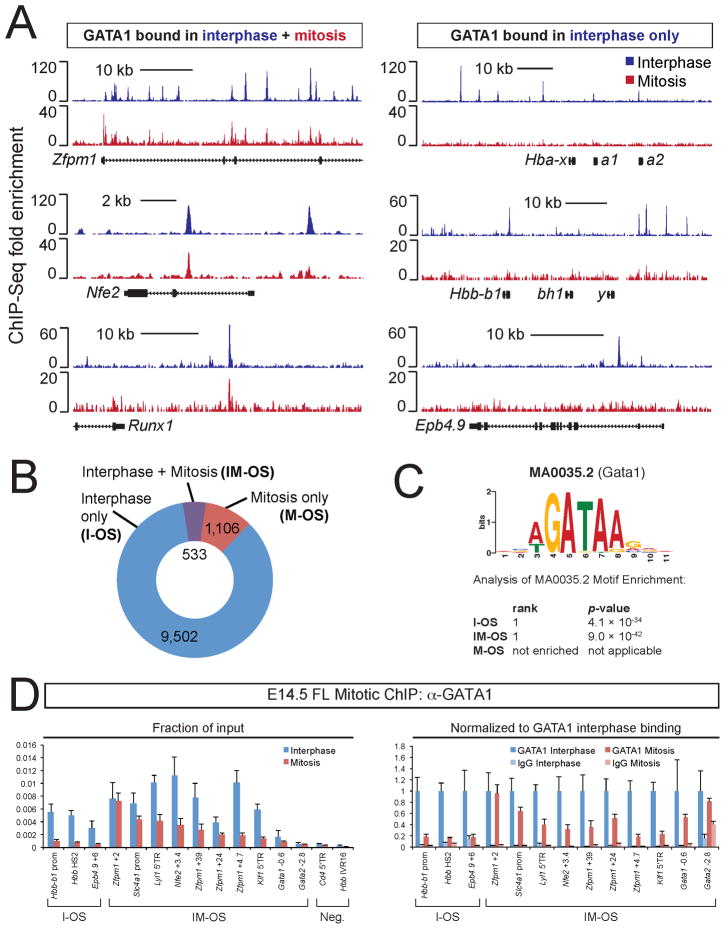
Using ChIP-seq we determined the genome-wide occupancy of GATA1 in purified mitotic G1E cells expressing GATA1-ER (G1E-ER4). Data sets were aligned with those from asynchronous G1E-ER4 cells (Cheng et al., 2009), which consist to >97% of interphase cells (Figure 2A). The results show that GATA1 binding profiles in interphase and mitosis are overlapping but distinct (Figure 3A). We observed a substantial reduction of GATA1 occupancy during mitosis (Figure 3B), consistent with the imaging studies. 533 out of 10,035 (5.3%) interphase GATA1 occupied sites (OS), mapping near ~4% of all genes, were retained on chromatin during mitosis (IM-OS). In addition, at most IM-OS the signal was lower during mitosis when compared to interphase (Figure 3A, 4A). It is possible that this is due to reduced binding of GATA1 co-factors during mitosis (see below) that might contribute to stable chromatin association (Letting et al., 2004; Pal et al., 2004). Surprisingly, a considerable number of sites (1,106) appeared to bind GATA1 preferentially during mitosis (M-OS), which might account for part of the signal seen in Figure 1. ChIP-qPCR showed substantial GATA1 enrichment at 17 out of 18 examined M-OS and IM-OS and the loss of binding at I-OS, thus validating the ChIP-seq data (Figure S2B, 3A). Results were further validated with antibodies against the ER moiety of GATA1-ER (Figure S2C). ChIP-qPCR also showed that a few regions identified as M-OS also bind GATA1 in interphase, albeit at lower levels (data not shown), which is likely due to stringent peak calling criteria.
Figure 3. Genome-wide Distribution of GATA1 in Interphase and Mitosis.
A. GATA1 ChIP-seq profiles in interphase and mitosis at indicated representative loci with peaks in interphase and mitosis (left) or interphase only (right). B. Proportion of sites with GATA1 binding only in interphase (I-OS, blue), mitosis (M-OS, red), or both (IM-OS, purple). C. Analysis of motif enrichment of I-OS, IM-OS, or M-OS peaks. The top-ranked motif in both I-OS and IM-OS was the GATA1 consensus motif MA0035.2. Note: M-OS did not show significant enrichment for GATA1 consensus (WGATAR) motifs. D. Anti-GATA1 ChIP-qPCR of E14.5 primary fetal liver erythroblasts at key I-OS and IM-OS. Left panel, raw data plotted as fraction of input DNA. Right panel, the same data (except negative sites) re-plotted as normalized to GATA1 interphase binding, and control IgG added for comparison. Please note that several sites (Gata1 −0.6, Gata2 −2.8) with relatively low signals still display significant enrichment over IgG. Error bars denote SEM (n = 3).
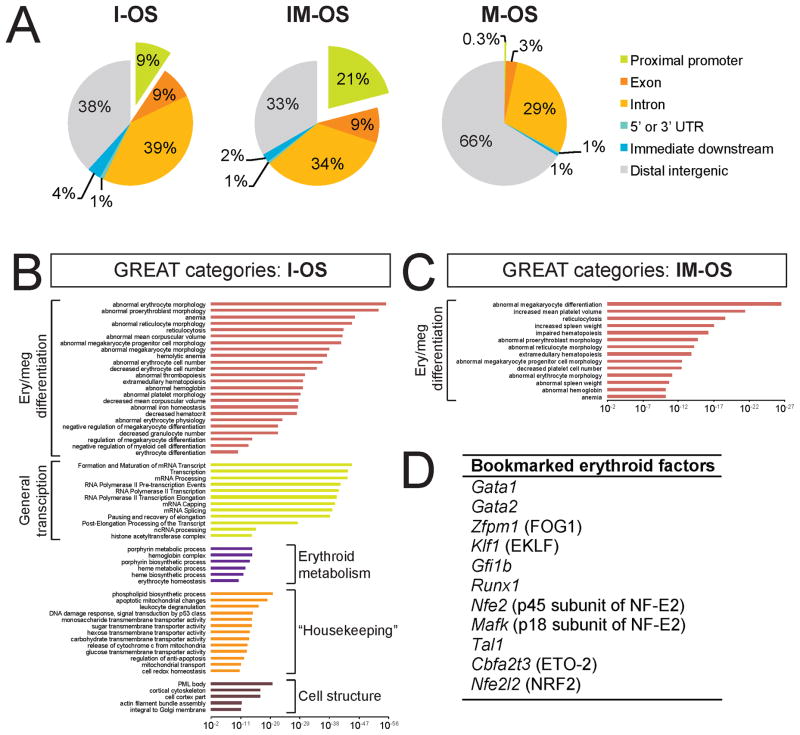
Figure 4. Functional Annotation of Mitotic GATA1 Occupancy Reveals Preference for Sites near Tissue-Specific Regulatory Genes.
A. Distribution of peaks relative to the nearest annotated transcription start site (TSS). I-OS and IM-OS but not M-OS are frequently found at proximal promoters (green, exploded away from pie chart). “Proximal promoter”, ≤ 3 kb upstream of TSS; “immediate downstream”, ≤ 3 kb from transcription termination site. B–C. Functional analysis of I-OS (B) and IM-OS (C) using GREAT. Significantly enriched gene ontology categories were manually grouped by their overarching functional theme. The x axes show binomial raw (uncorrected) p values (log scale). Note that I-OS are associated with ontologies relating to a broad variety of processes while IM-OS are associated with specific defects in erythropoiesis and megakaryopoiesis. Also note that erythroid and megakaryocytic cells share a large fraction of tissue-specific nuclear factors, likely accounting for the admixture of megakaryocyte-themed categories (see Table S1). D. GATA1 bookmarks key erythroid transcription factors.
To test whether endogenous GATA1 associates with mitotic chromatin in primary cells, we performed ChIP in mitotic E14.5 fetal liver erythroblasts (Figure S2D). In spite of subtle differences between those cells and G1E-ER4 cells, likely due to the heterogeneity of fetal liver erythroblasts with regards to their differentiation, the results essentially confirmed the occupancy patterns observed in G1E-ER4 cells (Figure 3D). These experiments, together with mitotic ChIP experiments performed in MEL cells, also allowed us to confirm that GATA1 bookmarks the Gata1 locus, which is altered in G1E-ER4 cells due to the presence of the targeting construct (Figure S2E–G).
Bioinformatic analyses revealed that I-OS and IM-OS are strongly enriched for the GATA1 consensus motif (WGATAR) (Figure 3C) whereas M-OS lack enrichment of the WGATAR motif over random occurrence (Figure 3C), and frequently map to simple (GATA)n repeats distal to annotated gene loci (Figure 4A and data not shown). I-OS and IM-OS preferentially localize to promoters and bodies of genes with IM-OS being more enriched at promoters (21%) when compared to I-OS (9%, Figure 4A). In contrast, M-OS are rarely found near promoters (Figure 4A).
To examine whether I-OS, IM-OS, and M-OS map to genes with distinct functional annotations, we used the Genomic Regions Enrichment of Annotations Tool (GREAT) (McLean et al., 2010). As might be expected, I-OS were highly associated with genes that are essential for differentiation of erythroid and the closely related megakaryocytic lineages (Figure 4B). Additional categories of GATA1 occupied genes include the general transcription machinery, genes involved in erythroid metabolism and “housekeeping” processes. Remarkably, genes associated with IM-OS showed a much-constricted range of ontologies, strongly favoring genes that are essential for the development of the erythro-megakaryocytic lineage (Figure 4C). To ensure that the contraction of ontologies reflects a true redistribution of GATA1 occupancy in mitosis, we performed a discriminatory analysis in which IM-OS were tested against all interphase sites (i.e. the union of I-OS and IM-OS), which produced a very similar set of enriched categories (Figure S3J). In contrast, GREAT analysis of M-OS showed no significantly associated categories, even when only the minor fraction (17%) of M-OS containing GATA1 consensus motifs was analyzed. Gene-distal binding sites (>3 kb from annotated genes) are overrepresented in the M-OS fraction (Figure 4A), suggesting that they might represent distal cis-regulatory elements regulating erythroid-important genes. We therefore reanalyzed just these gene-distal M-OS with GATA1 consensus motifs and found no enrichment for any specific gene ontologies.
When we inspected the gene “hits” the IM-OS associated ontologies we noticed that, on average, 40.5% correspond to nuclear regulators (Table S1). Notably, these include many of the key DNA binding factors known to regulate erythroid development such as Gata1, Gata2, Zfpm1, Klf1, and others highlighted in Figure 4D. Examination of ChIP-seq tracks revealed that additional erythropoietic factors (e.g. Ldb1, Bcl11a, Zbtb7a/LRF) showed mitotic GATA1 signals clearly above background that were barely below the highly stringent peak calling thresholds. Interestingly, prototypical GATA1 targets not encoding nuclear factors, such as α-(Hba) and β-(Hbb) globins as well as many known GATA1 targets not directly involved in hematopoietic development did not exhibit mitotic occupancy. Examples of contrasting occupancy patterns are shown in Figure 3A.
In conclusion, ChIP-seq of GATA1 in highly pure mitotic erythroid cells revealed a mitotic contraction of GATA1 chromatin occupancy, concentrating on genes that encode nuclear factors essential for erythroid differentiation.
Histone marks at I-OS, IM-OS, and M-OS
We next enquired whether select histone modifications are associated specifically with I-OS, IM-OS, or M-OS. Using genome-wide data sets from G1E-ER4 cells (Wu et al., 2011), we found that I-OS and IM-OS bear histone marks associated with transcriptional enhancers (H3K4me1) or active/poised promoters (H3K4me3) (Figure S3A, B). In contrast, M-OS are enriched for histone marks typically associated with transcriptional repression (H3K27me3 and H3K9me3, Figure S3C, D). We next measured by ChIP-qPCR the enrichment of H3 di-ac, H4 tetra-ac, H3K4me1, H3K4me3, and total histone H3 at multiple I-OS and IM-OS in interphase and mitosis (Figure S3E–I). H3 di-ac, but not H4 tetra-ac appeared high and mitotically stable at select IM-OS. Assessing the discriminatory power of H3 di-ac between I-OS and IM-OS will require genome-wide measurements of this mark. We also examined repressive marks (H3K9me3 and H3K27me3), which were expectedly low at GATA1 target genes (not shown). None of these revealed clear distinctions between I-OS and IM-OS, suggesting that the tendency of I-OS for higher H3K4me1 but lower H3K4me3 levels when compared to IM-OS reflects that the latter have a higher tendency to reside near promoters (Figure 3A).
Tissue-specific GATA1 co-factors vacate mitotic GATA1 binding sites
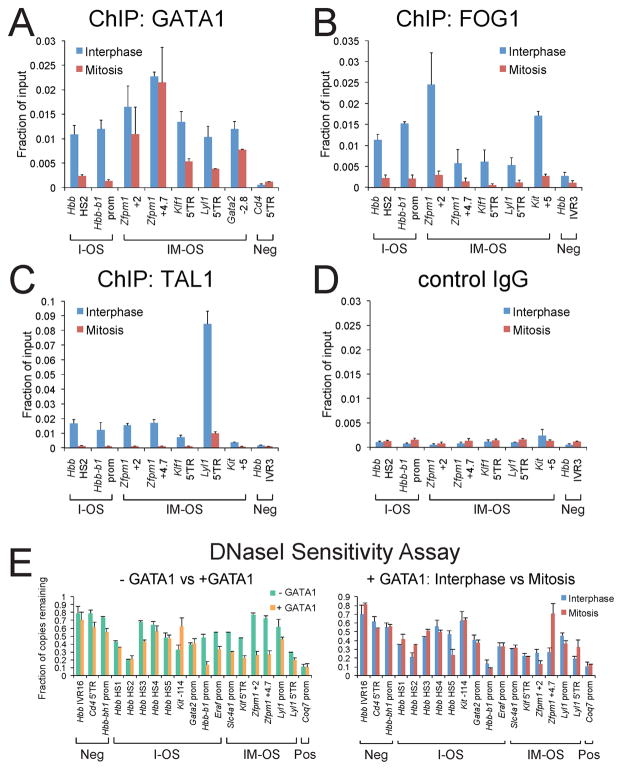
We next examined whether GATA1 retains essential co-factors at mitotically occupied sites. FOG1 is a tissue-specific co-factor whose association with target genes depends entirely on GATA factors (Chang et al., 2002; Tsang et al., 1997). ChIP revealed that during mitosis FOG1 is depleted from all examined GATA1 occupied sites, regardless of whether GATA1 is retained at these sites or not (Figure 5A, B).
Figure 5. GATA1 Co-factor Occupancy and DNaseI Sensitivity at GATA1 Binding Sites during Mitosis.
A–D. ChIP-qPCR with indicated antibodies of asynchronous cells of which >97% are in interphase (blue) or purified mitotic (red) induced G1E-ER4 cells. Primers interrogated I-OS, IM-OS, and negative control sites (Neg). E. DNaseI sensitivity analysis of G1E cells during interphase and mitosis at I-OS and IM-OS or constitutively sensitive (Pos) and insensitive (Neg) control sites. Left panel: asynchronous G1E (− GATA1) cells are compared with E2-induced G1E-ER4 cells (+ GATA1). Note marked increase of DNaseI sensitivity in response to GATA1 activation at the Hbb-b1, Eraf, Slc4a1, Zfpm1, and Klf1 genes. Right panel: Asynchronous (blue) and purified mitotic (red) E2-induced G1E-ER4 cells (+ GATA1). Note that sensitivity is largely maintained in mitosis even at I-OS where GATA1 dissociates during mitosis. All error bars denote SEM (n = 3).
SCL/TAL1 and its associated proteins LMO2, Ldb1/Nli1 are essential for hematopoietic stem cell homeostasis as well as erythroid and megakaryocyte development (Lecuyer and Hoang, 2004). Despite the ability to bind DNA directly, recruitment of the TAL1 complex is to a significant extent independent of direct DNA binding (Porcher et al., 1999) and is instead mediated via interactions with other factors such as GATA1 (Tripic et al., 2009). Similar to FOG1, TAL1 was depleted from all examined sites, including the IM-OS (Figure 5C). We also examined Ldb1 and LMO2 and expectedly, found that both of these factors vacate all tested sites (Figure S4A, B). Notably, the TAL1 complex was lost also from sites where it binds DNA directly, such as the DNaseI hypersensitive site 2 (HS2) of the β-globin locus control region (LCR) (Elnitski et al., 1997) (Figure 5C) and the Gata2 locus (not shown), suggesting that it does not play a direct role in marking mitotic genes. Together, these results indicate that essential co-factor complexes surrounding GATA1 separate from mitotic chromatin.
Maintenance of DNaseI hypersensitivity during mitosis is independent of GATA1
It is possible that changes in chromatin structure that are exerted by GATA1 contribute to the epigenetic propagation through mitosis of GATA1-dependent transcription states. It is also possible that compaction of chromatin displaces GATA1 from some of its targets, although generally, mitotic chromosomes are accessible to transcription factors (Chen et al., 2005). Since GATA elements are required for the establishment of HSs at the β-globin LCR (Stamatoyannopoulos et al., 1995), we measured HS formation by qPCR in G1E and E2-treated G1E-ER4 cells at several GATA1 occupied sites. GATA1 increased DNaseI sensitivity at some but not all of its binding sites (Figure 5E). Similar results were obtained over a range of DNaseI concentrations (Figure S4C). To determine whether GATA1-occupied HS are stable during mitosis and whether their maintenance depends on GATA1, we compared DNaseI sensitivity of I-OS and IM-OS in mitotic and asynchronous E2-treated G1E-ER4 cells. We found that almost all HSs persisted during mitosis, regardless of their degree of hypersensitivity and independently of GATA1 occupancy (Figure 5E). This suggests that although GATA1 participates in HS formation, HS propagation through mitosis is mediated by a GATA1-independent mechanism.
Mitotic GATA1 occupancy correlates with rapid transcriptional reactivation after mitosis
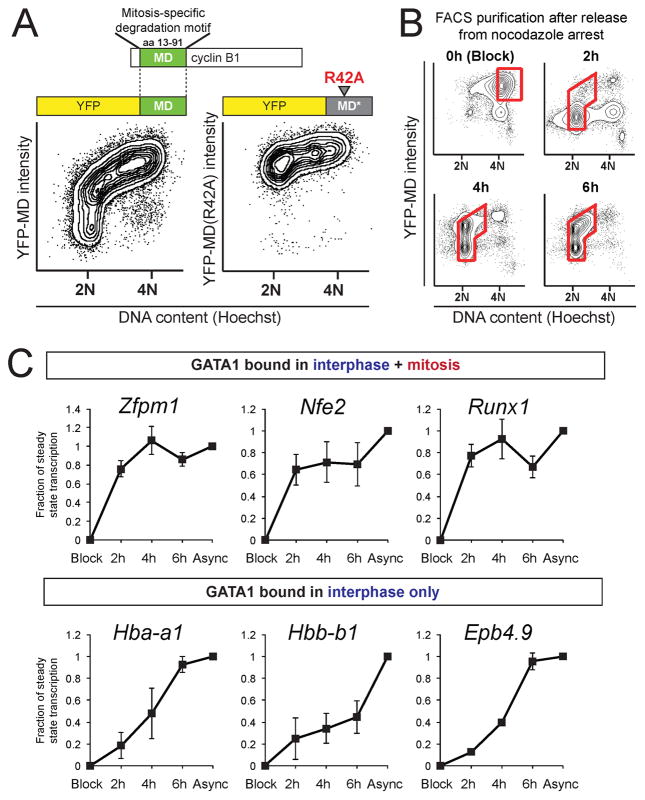
Maintenance of GATA1 occupancy through mitosis might aid timely reactivation of transcription in newly divided cells. We therefore compared gene reactivation kinetics at bookmarked and vacated GATA1 target genes following mitosis. However, as discussed above, incomplete cell synchronization by conventional treatments hampered mitotic arrest-release experiments. The mitosis-specific degradation domain (MD) of cyclin B1 (amino acids 13–91) is sufficient for APCCdc20-dependent protein destruction at the metaphase-anaphase transition (Glotzer et al., 1991; Holloway et al., 1993). We reasoned that fusion of YFP to the cyclin B1 MD would allow tracing of live cells through the cell cycle when combined with Hoechst 33342 staining for DNA content. When asynchronous G1E stably expressing YFP-MD were exposed to Hoechst 33342, and analyzed by flow cytometry (Figure 6A), the highest YFP fluorescence was evident in cells with 4N DNA content, representing late G2 and early mitotic cells. Progression through anaphase was accompanied by YFP-MD destruction. Subsequent cytokinesis generated 2N cells that gradually resynthesized YFP-MD after APCCdc20 inactivation. Mutation of a single arginine (R42A) within the destruction box inactivates the MD (Glotzer et al., 1991) leading to constitutive expression of YFP-MD (R42A). Expression of YFP-MD or YFP-MD (R42A) did not perturb the cell cycle profile, growth rate, or viability of G1E cells (Figure S5A and not shown). These findings establish the usefulness of YFP-MD in combination with Hoechst stain to purify live cells at distinct cell cycle stages.
Figure 6. Rapid Transcriptional Reactivation of Genes Occupied by GATA1 during Mitosis.
A. Flow cytometry of YFP-MD (left) or YFP-MD(R42A) (right) intensity as a function of DNA content. Note that MD but not MD (R42A) confers mitotic destruction. B. Flow cytometry as in A) of cells treated with nocodazole and released for 0h, 2h, 4h, or 6h shows gradual accumulation of cells in G1. Red boxes indicate gates that were used to collect cells for pre-mRNA (PT) RT-qPCR analysis. C. PT RT-qPCR in G1E-ER4 cells of genes bound by GATA1 in interphase and mitosis (upper panels) or in interphase only (lower panels). Data are plotted as a fraction of steady state levels in asynchronous cells (Async) that were set to one. Error bars show SEM (n = 3).
To enrich for cells exiting mitosis, cells were treated with nocodazole for 6 hours and sorted by FACS at various time points following nocodazole wash-out (Figure 6B). As expected, nocodazole-induced metaphase block enriched for YFP-high/4N populations but also produced YFP-low/4N cells. The latter population, which represents cells that “slipped” through the mitotic checkpoint, was efficiently eliminated by FACS. At 2h post release, the majority of cells was YFP-low/2N, consistent with the emergence of early G1 cells. At 4h and 6h post release, cells resumed synthesis of YFP and progressed into early S phase. The validity of this method to examine transcription reactivation as cells exit mitosis was confirmed by measuring pre-mRNA (primary transcript, PT) and mature mRNA of the MLL/RbBP5-bookmarked gene Pabpc1 (Figure 2E and (Blobel et al., 2009)) in G1E-ER4 cells (Figure S5B, C), and by demonstrating that Pol2 occupancy at 5′ transcribed regions closely mirrors PT levels (Figure S5C–H).
To determine whether mitotic GATA1 occupancy correlates with rapid reactivation of transcription, we examined three GATA1 target genes that display mitotic GATA1 binding sites (Zfpm1, Nfe2, and Runx1) and three that are bound by GATA1 only in interphase (Hba-a1, Hbb-b1, Epb4.9). Primary transcript RT-qPCR of induced G1E-ER4 cells showed that the latter group reached steady state levels in a gradual fashion (Figure 6C). In contrast, and analogous to the MLL-bookmarked Pabpc1 gene (Figure S5C), genes marked by GATA1 in mitosis reactivated more rapidly, reaching near-steady state levels as early as 2 hours after release. Mature mRNA levels of these genes were not significantly altered throughout these time points, as expected (not shown). These results provide correlative evidence that GATA1 acts on its mitotic target genes to facilitate rapid transcription reactivation after mitosis.
Mitosis-specific degradation of GATA1 delays re-activation of bookmarked genes
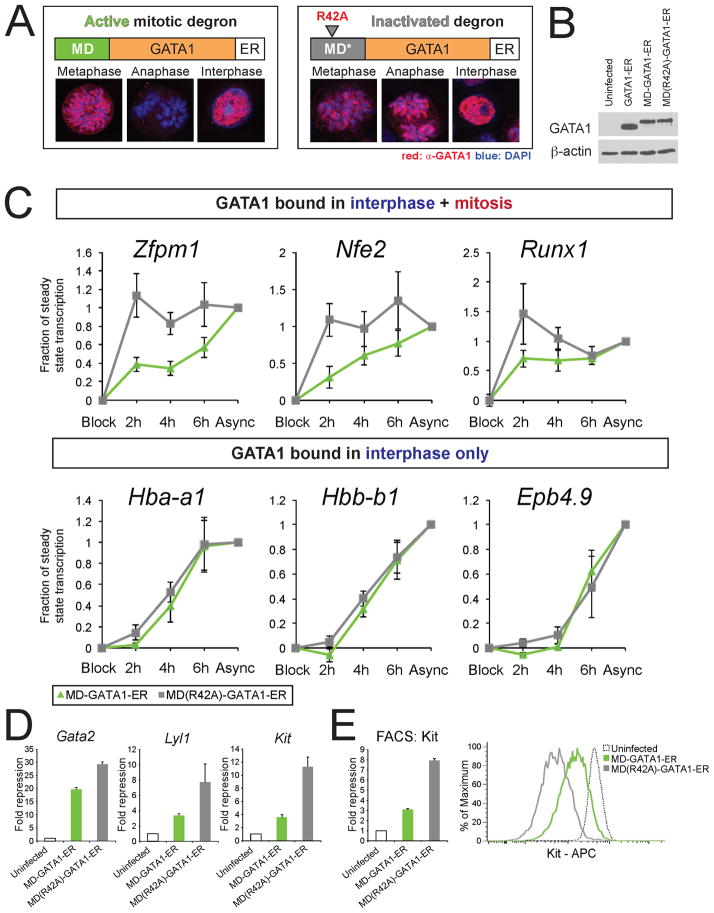
Mitotic retention of select nuclear factors has led to the speculation that they serve to faithfully preserve transcription patterns or the rapid onset of transcription in early G1. Although this idea is attractive, there is scarce direct experimental evidence to support it (see Introduction). To inhibit GATA1 activity during mitosis while minimizing the impact on GATA1 activity in interphase, we set out to degrade GATA1 specifically in mitosis. To this end we fused GATA1-ER to the cyclin B MD or MD(R42A), stably introduced fusion constructs into G1E cells, and analyzed their abundance by IF microscopy. Notably, MD-GATA1-ER but not MD (R42A)-GATA1-ER was depleted in early anaphase (Figure 7A). Please note that steady state levels of both proteins were similar (Figure 7B). In addition, MD-GATA1-ER is restored to the same levels as MD(R42A)-GATA1-ER at 4 hours after mitosis (Figure S6A).
Figure 7. Mitosis-specific Destruction of GATA1 Delays Reactivation of Bookmarked Genes.
A. IF images demonstrate destruction of MD- but not MD(R42A)-tagged GATA1 constructs at the metaphase-anaphase transition. All cells shown had similar expression of GFP, indicative of similar steady-state MD-GATA1-ER levels. B. Western blots of whole cell lysates show that MD-GATA1-ER and MD(R42A)-GATA1-ER expression is comparable to endogenous GATA1 levels (compare with Figure S1B). C. PT-RT-qPCR analysis in G1E cells expressing MD-GATA1-ER (green) and MD(R42A)-GATA1-ER (gray). MD(R42A)-GATA1-ER reactivates bookmarked genes similarly to GATA1-ER (compare with Figure 6C). Mitotic destruction of GATA1 delays reactivation of genes occupied by GATA1 in mitosis, but does not affect reactivation of genes that are vacated during mitosis. Data are plotted as in Figure 6C. Error bars denote SEM and n = 6. D. Gene expression levels in G1E cells expressing indicated constructs plotted as fold repression compared to uninfected G1E cells. E. Flow cytometry of Kit surface expression reveals failure to fully repress Kit expression by MD-GATA1-ER. Left: Mean fluorescent intensity; right: histogram of Kit detection in a representative experiment. All error bars denote SEM and n = 3 for RT-qPCR and FACS.
To examine the effect of mitosis-specific GATA1 degradation on transcription reactivation we combined the MD-GATA1-ER and the YFP-MD systems. MD-GATA1-ER or MD (R42A)-GATA1-ER was co-expressed with YFP-MD in G1E cells, followed by exposure to E2 and nocodazole treatment. At indicated time points following release, cells were FACS-purified and GATA1 target gene transcription rates measured by PT-RT-qPCR. Pabpc1, which is not a GATA1 target gene and is bookmarked independently of GATA1, showed rapid reactivation independently of the mitotic presence of GATA1 (Figure S6B). Cells expressing MD (R42A)-GATA1-ER activated the mitotically marked genes (Zfpm1, Nfe2, and Runx1) with rapid kinetics while the Hba-a1, Hbb-b1, and Epb4.9 genes activated more slowly (Figure 7C), similar to cells expressing GATA1-ER. In contrast, in cells expressing MD-GATA1-ER, the time for bookmarked genes to reach steady state transcription levels was significantly prolonged. However, the activation of the non-bookmarked genes Hba-a1, Hbb-b1, and Epb4.9 was remarkably similar in cells expressing either construct. These results indicate that genes at which GATA1 is retained during mitosis require GATA1 for their timely reactivation.
GATA1 also functions as a direct repressor of genes that mark the immature proliferative state, including Kit, Gata2 and Lyl1 (Grass et al., 2003; Jing et al., 2008; Johnson et al., 2007; Munugalavadla et al., 2005; Weiss et al., 1994; Weiss et al., 1997), all three of which retain GATA1 at some sites during mitosis. We asked whether mitotic retention of GATA1 is required to maintain full repression. Notably, MD-GATA1-ER was significantly less effective in repressing these genes when compared to MD (R42A)-GATA1-ER (Figure 7D). Since Kit is expressed on the cell surface, results were confirmed by flow cytometry (Figure 7E). In summary, mitosis-specific destruction of GATA1 leads to delayed activation and impaired repression of mitotically marked GATA1 target genes.
Discussion
Here, we investigated mechanisms by which cells transmit transcriptional information through mitosis. We found that in contrast to most other DNA-binding factors, GATA1 remains bound during mitosis to a subset of its target genes. Notably, mitotic GATA1 preferentially occupies genes encoding lineage-specific transcription factors. Using a version of GATA1 that is selectively destroyed during mitosis, we show that mitotic GATA1 occupancy is required for the rapid reactivation of these genes in newborn cells. We suggest that mitotic GATA1 occupancy of hematopoietic regulatory genes ensures their rapid expression to faithfully maintain lineage-specific gene expression patterns.
In contrast to MLL, which preferentially marks highly expressed genes during mitosis (Blobel et al., 2009), GATA1’s mitotic binding is not correlated with the level of transcription. In addition, MLL tends to be retained at housekeeping genes, while GATA1 favors association with lineage-specific regulatory genes. Thus, distinct categories of genes appear to rely on distinct classes of bookmarking factors.
An important technical aspect of the present work concerns the high degree of purification of mitotic cells using FACS-based method. Omission of a mitosis-specific marker can lead to an underestimate of interphase cells contaminating a mitotic cell preparation. Indeed, it is possible that disparate results regarding the degree of maintenance of certain histone marks and nuclear factors on chromatin (Delcuve et al., 2008) might be attributed to variable contributions by contaminating interphase cells. Since histone H3S10 is globally phosphorylated in mitosis in all eukaryotes (Prigent and Dimitrov, 2003) FACS-based purification of mitotic cells should be broadly applicable.
How is GATA1 retained at some sites but not others? No differences in the GATA consensus sequence were found between I-OS and IM-OS. GATA1 associates with tissue-specific co-factors some of which (e.g. FOG1) facilitate GATA1 chromatin occupancy (Letting et al., 2004; Pal et al., 2004). We found that all examined GATA1 cofactors (FOG1, TAL1, Ldb1 and LMO2) vacate mitotic chromatin regardless of whether GATA1 is retained, suggesting that they do not influence GATA1 binding to mitotic chromatin. Mitotic removal of nuclear factors often occurs via phosphorylation by mitotic kinases (for review see Delcuve et al., 2008). It will be interesting to examine phosphorylation states of the FOG1 zinc fingers that bind GATA1, as well as GATA1 binding modules of the other co-factors. Regardless of the mechanism, these results are consistent with a function for GATA1 as mitotically stable platform upon which co-regulator complexes are re-assembled at the appropriate genomic locations to restore cell specific transcription programs.
Surprisingly, DNaseI hypersensitivity is preserved during mitosis at all examined GATA sites independently of the mitotic presence of GATA1, reminiscent of HSs at the Hsp70 promoter (Martinez-Balbas et al., 1995). These results suggest that occlusion of GATA elements by nucleosomes is not a general mechanism by which GATA1 is ejected from its binding sites during mitosis. At present it remains unclear what structurally distinguishes IM-OS from I-OS. Neither de novo computational motif discovery nor the directed search for elements known to frequently locate close to GATA1 elements, including E-boxes, Ets and CACCC elements revealed sequences that reliably discriminate between I-OS and IM-OS. However, select features showed moderate differences among IM-OS when compared to I-OS (summarized in Table S2). These include promoter proximity, enrichment for H3K4me3, lower levels of H3K4me1, and the more frequent occurrence of multiple GATA1 elements per GATA1 OS. The identification of patterns that discriminate between I-OS and IM-OS will require a comprehensive characterization in interphase and mitosis and subsequent functional analysis of chromatin modifying factors, histone modifications, transcription factors and their modifications.
In contrast to I-OS and IM-OS, M-OS frequently contain (GATA)n repeats and repressive H3K9me3 and H3K27me3 marks. Moreover, M-OS were not found near annotated promoters, and GREAT analysis did not reveal any significantly enriched gene ontologies of M-OS associated genes, calling into question whether these sites play a direct role in transcriptional regulation. It is possible that they serve as temporary “storage” sites for GATA1 that has been displaced from its interphase genomic locations.
By comparing post-mitotic reactivation kinetics of GATA1 bookmarked versus non-bookmarked genes, we found that the former reached steady state levels more rapidly than the latter. In cells expressing mitotically unstable GATA1, post-mitotic reactivation of bookmarked target genes is delayed. While we cannot fully exclude the possibility that the need to resynthesize the mitotically degraded GATA1 protein in the earliest stage of G1 contributed to the delay in transcriptional reactivation, GATA1 target genes that are not bookmarked reached steady state levels with normal kinetics. In concert, these experiments demonstrate a mitosis/early G1-specific requirement of a DNA binding factor.
Since a tissue-determining transcription factor such as GATA1 assumes a bookmarking function during mitosis, it is likely that additional nuclear factors that regulate lineage choice and cellular maturation will emerge to carry out similar functions. In this context it is worth noting that the mitotic degradation domain is modular and the mitosis-specific protein destruction functions through ubiquitous intracellular machinery requiring no exogenous factors. These features should make it possible to examine any factor in any given tissue for a potential role in mitotic bookmarking.
Experimental Procedures
Mitotic ChIP
ChIP assays were performed as described (Letting et al., 2003) with the following modifications. G1E-ER4 cells were induced with 100 nM estradiol for 18 hours. Nocodazole (Sigma) was added (200 ng/mL) for 6–7 hours before harvest. Cells were crosslinked with 1% (0.33 M) formaldehyde in PBS at room temperature for 10 min and quenched with 1 M glycine for 5 min to avoid formation of clogs during sorting. Cells were stained with anti-H3S10ph antibodies and Dy649-conjugated anti-rabbit IgG (Jackson Immuno) F(ab′)2 antibody fragments in PBS supplemented with 2% FBS, 2 mM EDTA, 1 mM PMSF, and protease inhibitor cocktail (Sigma). The use of F(ab′)2 minimized nonspecific capture by protein A/G beads used for subsequent ChIP. 107 H3S10ph-positive cells per IP were sorted on a FACSAria II (Beckton Dickinson). ChIP material from 6 × 107 cells was analyzed by high-throughput sequencing (Supplemental Experimental Procedures).
Mitotic Arrest-Release Experiments
YFP-MD constructs were introduced into G1E-ER4 cells. YFP-high cells were enriched by FACS. For mitosis-release experiments, cells were treated with nocodazole for 6–12 hours and replated into fresh medium (37°C). After staining with 32 μM Hoechst 33342 for 45 min at 37°C, cells were sorted into TRIZOL LS (Invitrogen). RNA was purified using RNeasy mini columns (Qiagen), reverse transcribed, and cDNA was quantitated by qPCR (SYBR green, Applied Biosystems). For primer sequences see Supplemental Experimental Procedures.
Mitotic Destruction of GATA1
MD-GATA1-ER and MD (R42A)-GATA1-ER were introduced into G1E cells. Expression was adjusted by monitoring GFP levels. Whole cell lysates were prepared by boiling 106 cells in Laemmli buffer (63 mM Tris HCl, 10% glycerol, 2% SDS, 0.0025% bromophenol blue) followed by sonication, SDS-PAGE, and Western blotting. IF was performed as described in the supplement using GATA1 antibodies. Surface expression of Kit on G1E cells was measured using a FACSCanto (BD) after staining with APC-conjugated anti-Kit antibodies.
Supplementary Material
Highlights.
GATA1 remains focally bound to chromatin during mitosis
Mitotic GATA1 sites mark key hematopoietic transcription factor genes
Mitotic retention of GATA1 is required for normal transcription reactivation
Mitotic retention of GATA1 is required for repression of immature cell markers
Acknowledgments
We thank Alan Lau, Margaret Chou, Jordan Orange, Aline Disimone, Paul Gadue, Rena Zheng, Joanna Balcerek, Chris Thom, Ariel Yang, Merav Socolovsky for technical advice, and Christopher Vakoc, Mitchell Weiss, Mitchell Lazar, and members of the laboratory for comments on the manuscript. This work was supported by an AHA predoctoral fellowship (90PRE2300139, SK), NIH grants DK058044 and DK054937 (GAB), and DK065806 and HG005573 (RCH).
Footnotes
Author contributions: SK, MU, JMP, JCA performed experiments; SK, DPJ, YC performed computational analyses; SK, RCH and GAB conceived the study and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoto T, Saitoh N, Sakamoto Y, Watanabe S, Nakao M. Polycomb group protein-associated chromatin is reproduced in post-mitotic G1 phase and is required for S phase progression. J Biol Chem. 2008;283:18905–18915. doi: 10.1074/jbc.M709322200. [DOI] [PubMed] [Google Scholar]
- Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36:970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonenfant D, Towbin H, Coulot M, Schindler P, Mueller DR, van Oostrum J. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol Cell Proteomics. 2007;6:1917–1932. doi: 10.1074/mcp.M700070-MCP200. [DOI] [PubMed] [Google Scholar]
- Bruce K, Myers FA, Mantouvalou E, Lefevre P, Greaves I, Bonifer C, Tremethick DJ, Thorne AW, Crane-Robinson C. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 2005;33:5633–5639. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AN, Cantor AB, Fujiwara Y, Lodish MB, Droho S, Crispino JD, Orkin SH. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc Natl Acad Sci U S A. 2002;99:9237–9242. doi: 10.1073/pnas.142302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Dundr M, Wang C, Leung A, Lamond A, Misteli T, Huang S. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Hinkley CS, Henry RW, Huang S. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol Biol Cell. 2002;13:276–284. doi: 10.1091/mbc.01-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen KB, Zhang Y, Drautz D, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova R, Oelgeschlager T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- Delcuve GP, He S, Davie JR. Mitotic partitioning of transcription factors. J Cell Biochem. 2008;105:1–8. doi: 10.1002/jcb.21806. [DOI] [PubMed] [Google Scholar]
- Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell. 2009;20:4899–4909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D, Birkhoff G, Eggan K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat Rev Mol Cell Biol. 2008;9:505–516. doi: 10.1038/nrm2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L, Miller W, Hardison R. Conserved E boxes function as part of the enhancer in hypersensitive site 2 of the beta-globin locus control region. Role of basic helix-loop-helix proteins. Journal of Biological Chemistry. 1997;272:369–378. doi: 10.1074/jbc.272.1.369. [DOI] [PubMed] [Google Scholar]
- Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 Function, a Paradigm for Transcription Factors in Hematopoiesis. Mol Cell Biol. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Holloway SL, Glotzer M, King RW, Murray AW. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993;73:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Boyer ME, Kang JA, Wickrema A, Cantor AB, Bresnick EH. Friend of GATA-1-independent transcriptional repression: a novel mode of GATA-1 function. Blood. 2007;109:5230–5233. doi: 10.1182/blood-2007-02-072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- Kelly TK, Miranda TB, Liang G, Berman BP, Lin JC, Tanay A, Jones PA. H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol Cell. 2010;39:901–911. doi: 10.1016/j.molcel.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komura J, Ikehata H, Ono T. Chromatin fine structure of the c-MYC insulator element/DNase I-hypersensitive site I is not preserved during mitosis. Proc Natl Acad Sci U S A. 2007;104:15741–15746. doi: 10.1073/pnas.0702363104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskouti A, Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. Embo J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Hendzel MJ, Fischle W, Bertos NR, Hameed S, Yang XJ, Verdin E, Bazett-Jones DP. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J Biol Chem. 2001;276:38307–38319. doi: 10.1074/jbc.M100290200. [DOI] [PubMed] [Google Scholar]
- Kuo MT, Iyer B, Schwarz RJ. Condensation of chromatin into chromosomes preserves an open configuration but alters the DNase I hypersensitive cleavage sites of the transcribed gene. Nucleic Acids Res. 1982;10:4565–4579. doi: 10.1093/nar/10.15.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Hoang T. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp Hematol. 2004;32:11–24. doi: 10.1016/j.exphem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Letting DL, Chen YY, Rakowski C, Reedy S, Blobel GA. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci U S A. 2004;101:476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letting DL, Rakowski C, Weiss MJ, Blobel GA. Formation of a Tissue-Specific Histone Acetylation Pattern by the Hematopoietic Transcription Factor GATA-1. Mol Cell Biol. 2003;23:1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus KJ, Biron VL, Heit R, Underhill DA, Hendzel MJ. Dynamic changes in histone H3 lysine 9 methylations: identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. J Biol Chem. 2006;281:8888–8897. doi: 10.1074/jbc.M505323200. [DOI] [PubMed] [Google Scholar]
- Michelotti EF, Sanford S, Levens D. Marking of active genes on mitotic chromosomes. Nature. 1997;388:895–899. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- Munugalavadla V, Dore LC, Tan BL, Hong L, Vishnu M, Weiss MJ, Kapur R. Repression of c-kit and its downstream substrates by GATA-1 inhibits cell proliferation during erythroid maturation. Mol Cell Biol. 2005;25:6747–6759. doi: 10.1128/MCB.25.15.6747-6759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, Weiss MJ. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24:266–270. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Cantor AB, Johnson KD, Moran TB, Boyer ME, Orkin SH, Bresnick EH. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci U S A. 2004;101:980–985. doi: 10.1073/pnas.0307612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon AM, Ajay SS, Kumar SA, Steiner LA, Cherukuri PF, Wincovitch S, Anderson SM, Mullikin JC, Gallagher PG, et al. Comparative Sequencing Program, N. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood. 2011 doi: 10.1182/blood-2011-05-355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop R, Shearstone JR, Shen Q, Liu Y, Hallstrom K, Koulnis M, Gribnau J, Socolovsky M. A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU.1 and S-phase progression. PLoS Biol. 2010:8. doi: 10.1371/journal.pbio.1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher C, Liao EC, Fujiwara Y, Zon LI, Orkin SH. Specification of hematopoietic and vascular development by the bHLH transcription factor SCL without direct DNA binding. Development. 1999;126:4603–4615. doi: 10.1242/dev.126.20.4603. [DOI] [PubMed] [Google Scholar]
- Prescott DM, Bender MA. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- Prigent C, Dimitrov S. Phosphorylation of serine 10 in histone H3, what for? J Cell Sci. 2003;116:3677–3685. doi: 10.1242/jcs.00735. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK. Mitotic bookmarking of formerly active genes: keeping epigenetic memories from fading. Cell Cycle. 2009;8:818–823. doi: 10.4161/cc.8.6.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Ito T, Nishino N, Khochbin S, Yoshida M. Real-time imaging of histone H4 hyperacetylation in living cells. Proc Natl Acad Sci U S A. 2009;106:16257–16262. doi: 10.1073/pnas.0902150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segil N, Guermah M, Hoffmann A, Roeder RG, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Goodwin A, Joyce T, Lowrey CH. NF-E2 and GATA binding motifs are required for the formation of DNase I hypersensitive site 4 of the human β-globin locus control region. EMBO J. 1995;14:106–116. doi: 10.1002/j.1460-2075.1995.tb06980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JH. Nucleic acid synthesis in relation to the cell division cycle. Ann N Y Acad Sci. 1960;90:409–421. doi: 10.1111/j.1749-6632.1960.tb23259.x. [DOI] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AP, EVJ, ATC, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Valls E, Sanchez-Molina S, Martinez-Balbas MA. Role of histone modifications in marking and activating genes through mitosis. J Biol Chem. 2005;280:42592–42600. doi: 10.1074/jbc.M507407200. [DOI] [PubMed] [Google Scholar]
- Varier RA, Outchkourov NS, de Graaf P, van Schaik FM, Ensing HJ, Wang F, Higgins JM, Kops GJ, Timmers HT. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. EMBO J. 2010;29:3967–3978. doi: 10.1038/emboj.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, Crispino JD. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Keller G, Orkin SH. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1_ embryonic stem cells. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properities of transcription factor GATA-1 revealed by phenotypic rescue of a gene targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Wu W, Cheng Y, Keller CA, Ernst J, Kumar SA, Mishra T, Morrissey C, Dorman CM, Chen KB, Drautz D, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 2011 doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L, Zhou GL, Song W, Wu XS, Wei GH, Hao DL, Lv X, Liu DP, Liang CC. Exploring cellular memory molecules marking competent and active transcriptions. BMC Mol Biol. 2007;8:31. doi: 10.1186/1471-2199-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Vanderford NL, Sarge KD. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat Cell Biol. 2008;10:1318–1323. doi: 10.1038/ncb1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, Hong Y, Park-Sarge OK, Sarge KD. Mechanism of hsp70i gene bookmarking. Science. 2005;307:421–423. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- Yan J, Xu L, Crawford G, Wang Z, Burgess SM. The forkhead transcription factor FoxI1 remains bound to condensed mitotic chromosomes and stably remodels chromatin structure. Mol Cell Biol. 2006;26:155–168. doi: 10.1128/MCB.26.1.155-168.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007a;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, et al. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci U S A. 2007b;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet. 2010;11:583–589. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Pockwinse SM, Javed A, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic partitioning and selective reorganization of tissue-specific transcription factors in progeny cells. Proc Natl Acad Sci U S A. 2003;100:14852–14857. doi: 10.1073/pnas.2533076100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.