Abstract
Superhelical PM2 DNA I can be modified with N-cyclohexyl-N'-beta-(4-methylmorpholinium)ethyl carbodiimide (CMC). The transition of the sedimentation coefficient uncorrected for buoyant density change (S20,*) vs. % reactivity in terms of base pairs shows the following characteristics. The S20,* increases by 4.5 S units upon 1% modification. There is a plateau in S20,* between 1 and 4% reactivity. The extent of reactivity was determined by buoyant density and 14C radioactive CMC binding measurements. Further reactivity was not explored since Pulleyblank and Morgan's (22) data of S20,* vs. % reactivity from 6 to 34% was previously published. The initial results obtained in this study are complementary to the cited results of the above authors. Consequently, both sets of data taken together represent a complete description of S20,* vs. % reactivity with CMC. It is shown that the model in which superhelical DNA is proposed to contain small intrastrand hairpin regions can be extended to account for the observed transitions in S20,* vs. reactivity.
Full text
PDF




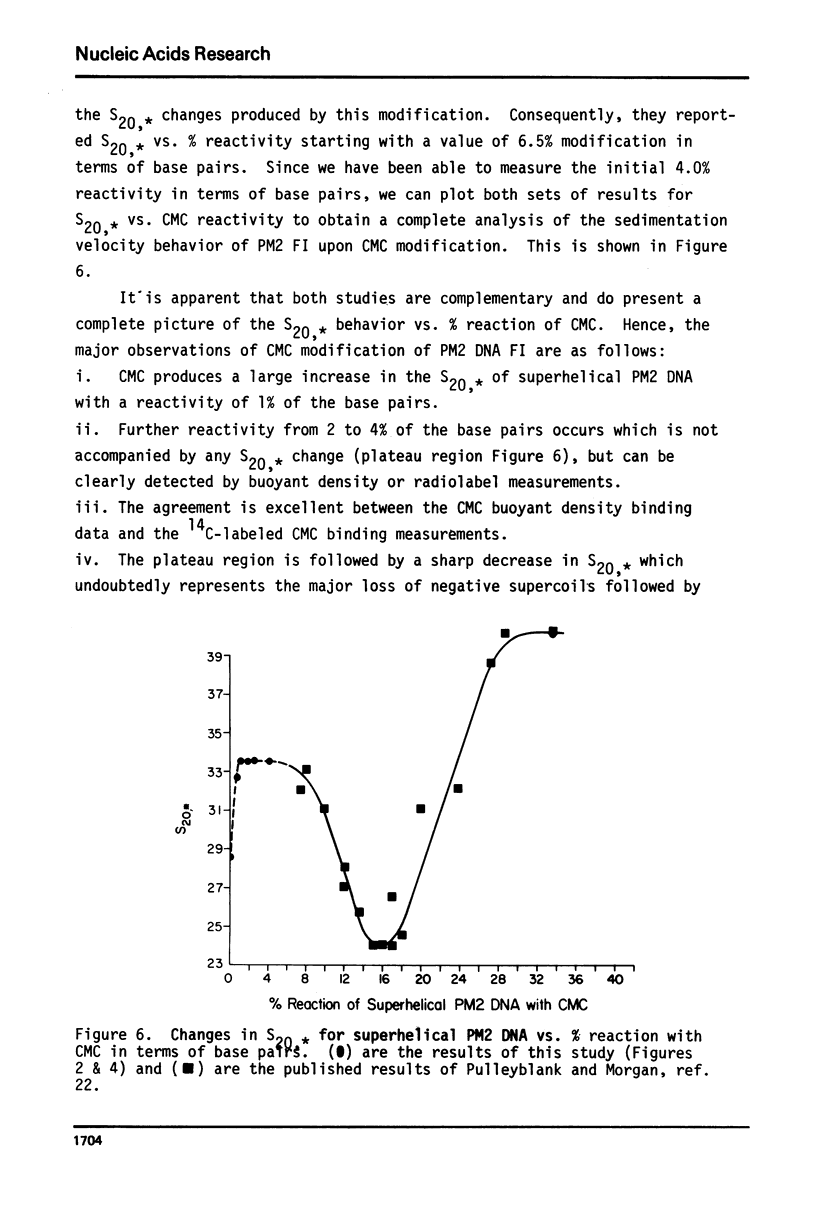



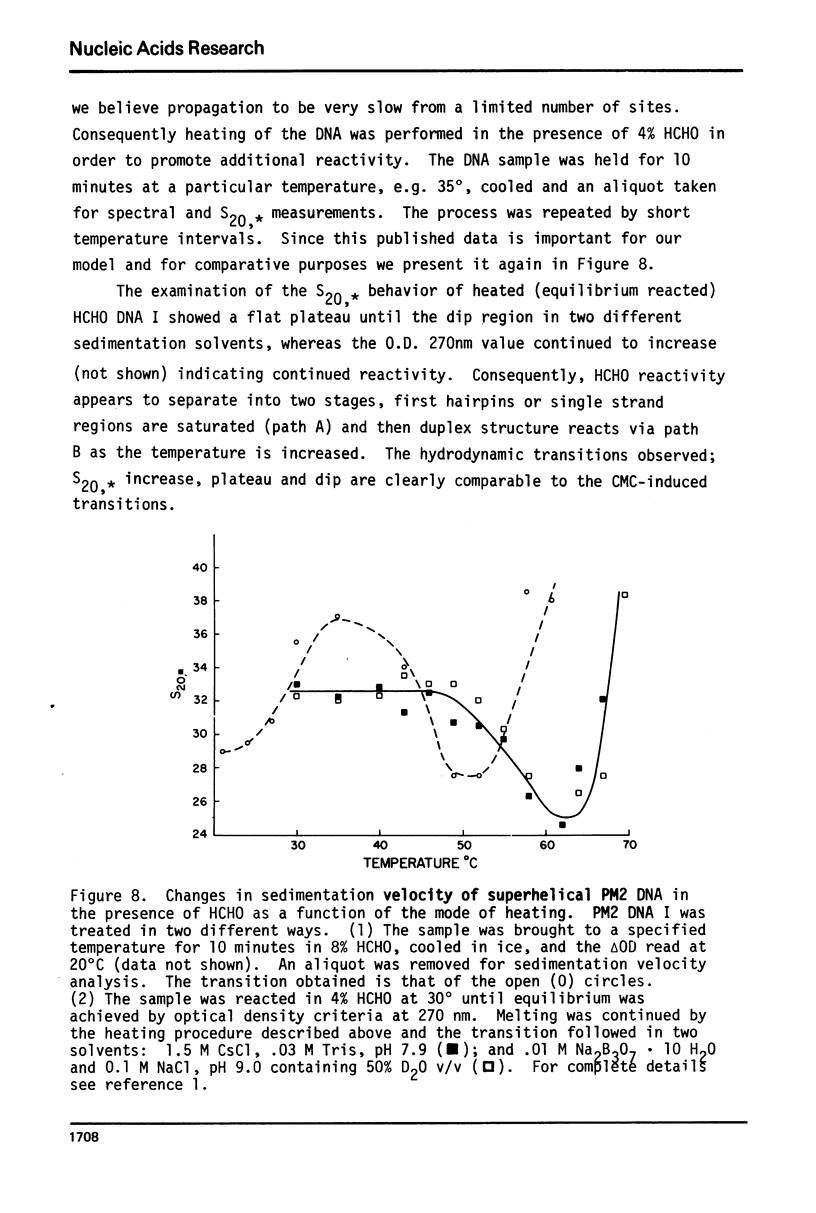



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartok K., Denhardt D. T. Site of cleavage of superhelical phiX174 replicative form DNA by the single strand-specific Neurospora crassa endonuclease. J Biol Chem. 1976 Jan 25;251(2):530–535. [PubMed] [Google Scholar]
- Beard P., Morrow J. F., Berg P. Cleavage of circular, superhelical simian virus 40 DNA to a linear duplex by S1 nuclease. J Virol. 1973 Dec;12(6):1303–1313. doi: 10.1128/jvi.12.6.1303-1313.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman T. A., Lebowitz J. Further analysis of the altered secondary structure of superhelical DNA. Sensitivity to methylmercuric hydroxide a chemical probe for unpaired bases. J Mol Biol. 1973 Sep 25;79(3):451–470. doi: 10.1016/0022-2836(73)90398-7. [DOI] [PubMed] [Google Scholar]
- Brack C., Bickle T. A., Yuan R. The relation of single-stranded regions in bacteriophage PM2 supercoiled DNA to the early melting sequences. J Mol Biol. 1975 Aug 25;96(4):693–702. doi: 10.1016/0022-2836(75)90146-1. [DOI] [PubMed] [Google Scholar]
- Bruner R., Vinograd J. The evaluation of standard sedimentation coefficients of sodium RNA and sodium DNA from sedimentation velocity data in concentrated NaCl and CsCl solutions. Biochim Biophys Acta. 1965 Sep 6;108(1):18–29. doi: 10.1016/0005-2787(65)90104-8. [DOI] [PubMed] [Google Scholar]
- Campbell A., Eason R. Effects of DNA primary structure on tertiary structure. FEBS Lett. 1975 Jul 15;55(1):212–215. doi: 10.1016/0014-5793(75)80994-x. [DOI] [PubMed] [Google Scholar]
- Chen M., Lebowitz J., Salzman N. P. Hin D restriction mapping of upaired regions in simian virus 40 superhelical DNA I: considerations regarding structure-function relationships. J Virol. 1976 Apr;18(1):211–217. doi: 10.1128/jvi.18.1.211-217.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer F. Three-dimensional structure of tRNA. Prog Nucleic Acid Res Mol Biol. 1971;11:391–421. doi: 10.1016/s0079-6603(08)60333-5. [DOI] [PubMed] [Google Scholar]
- Dean W. W., Lebowitz J. Partial alteration of secondary structure in native superhelical DNA. Nat New Biol. 1971 May 5;231(18):5–8. [PubMed] [Google Scholar]
- Espejo R. T., Canelo E. S., Sinsheimer R. L. DNA of bacteriophage PM2: a closed circular double-stranded molecule. Proc Natl Acad Sci U S A. 1969 Aug;63(4):1164–1168. doi: 10.1073/pnas.63.4.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Kamenetskii M. D., Lazurkin Y. S. Conformational changes in DNA molecules. Annu Rev Biophys Bioeng. 1974;3(0):127–150. doi: 10.1146/annurev.bb.03.060174.001015. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Vogt V. M., Hirt B. Characterization of the single-strand-specific nuclease S1 activity on double-stranded supercoiled polyoma DNA. Eur J Biochem. 1974 Apr 16;43(3):591–600. doi: 10.1111/j.1432-1033.1974.tb03446.x. [DOI] [PubMed] [Google Scholar]
- Goad W. B., Cann J. R. V. Chemically interacting systems. I. Theory of sedimentation of interacting systems. Ann N Y Acad Sci. 1969 Nov 7;164(1):192–225. doi: 10.1111/j.1749-6632.1969.tb14039.x. [DOI] [PubMed] [Google Scholar]
- Holloman W. K., Wiegand R., Hoessli C., Radding C. M. Uptake of homologous single-stranded fragments by superhelical DNA: a possible mechanism for initiation of genetic recombination. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2394–2398. doi: 10.1073/pnas.72.6.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob R. J., Lebowitz J., Printz M. P. Unpaired bases in superhelical DNA: kinetic evidence. Nucleic Acids Res. 1974 Apr;1(4):549–558. doi: 10.1093/nar/1.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A. C., Bartok K., Fraser M. J., Denhardt D. T. Sensitivity of superhelical DNA to a single-strand specific endonuclease. Biochim Biophys Acta. 1973 Apr 21;308(7):68–78. doi: 10.1016/0005-2787(73)90123-8. [DOI] [PubMed] [Google Scholar]
- McConnell B., von Hippel P. H. Hydrogen exchange as a probe of the dynamic structure of DNA. I. General acid-base catalysis. J Mol Biol. 1970 Jun 14;50(2):297–316. doi: 10.1016/0022-2836(70)90194-4. [DOI] [PubMed] [Google Scholar]
- Metz D. H., Brown G. L. The investigation of nucleic acid secondary structure by means of chemical modification with a carbodiimide reagent. I. The reaction between N-cyclohexyl-N'-beta-(4-methylmorpholinium)ethylcarbodiimide and model nucleotides. Biochemistry. 1969 Jun;8(6):2312–2328. doi: 10.1021/bi00834a012. [DOI] [PubMed] [Google Scholar]
- Morrow J. F., Berg P. Location of the T4 gene 32 protein binding site on simian virus 40 DNA. J Virol. 1973 Dec;12(6):1631–1632. doi: 10.1128/jvi.12.6.1631-1632.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulleyblank D. E., Morgan A. R. The sense of naturally occurring superhelices and the unwinding angle of intercalated ethidium. J Mol Biol. 1975 Jan 5;91(1):1–13. doi: 10.1016/0022-2836(75)90368-x. [DOI] [PubMed] [Google Scholar]
- Reed S. I., Ferguson J., Davis R. W., Stark G. R. T antigen binds to simian virus 40 DNA at the origin of replication. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1605–1609. doi: 10.1073/pnas.72.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman N. P., Lebowitz J., Chen M., Sebring E., Garon C. F. Properties of replicating SV40 DNA molecules and mapping unpaired regions in SV40 DNA I. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):209–218. doi: 10.1101/sqb.1974.039.01.027. [DOI] [PubMed] [Google Scholar]
- Upholt W. B., Gray H. B., Jr, Vinograd J. Sedimentation velocity behavior of closed circular SV40 DNA as a function of superhelix density, ionic strength, counterion and temperature. J Mol Biol. 1971 Nov 28;62(1):21–38. doi: 10.1016/0022-2836(71)90128-8. [DOI] [PubMed] [Google Scholar]
- VINOGRAD J., HEARST J. E. Equilibrium sedimentation of macromolecules and viruses in a density gradient. Fortschr Chem Org Naturst. 1962;20:373–422. [PubMed] [Google Scholar]
- Wang J. C. Interactions between twisted DNAs and enzymes: the effects of superhelical turns. J Mol Biol. 1974 Aug 25;87(4):797–816. doi: 10.1016/0022-2836(74)90085-0. [DOI] [PubMed] [Google Scholar]
- Wiegand R. C., Godson G. N., Radding C. M. Specificity of the S1 nuclease from Aspergillus oryzae. J Biol Chem. 1975 Nov 25;250(22):8848–8855. [PubMed] [Google Scholar]
- Woodworth-Gutai M., Lebowitz J. Introduction of interrupted secondary structure in supercoiled DNA as a function of superhelix density: consideration of hairpin structures in superhelical DNA. J Virol. 1976 Apr;18(1):195–204. doi: 10.1128/jvi.18.1.195-204.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]


