Abstract
Geminin is a multifunctional protein previously suggested to both maintain the bone morphogenetic protein inhibition required for neural induction and to control cell-cycle progression and cell fate in the early embryo. Since Geminin is required in the blastocyst on E3.5, we employed shRNA to examine its role during postimplantation development. Geminin knockdown inhibited the epithelial to mesenchymal transition (EMT) required at gastrulation and neural crest delamination, resulting in anterior-posterior axis and patterning defects, while overexpression promoted EMT at both locations. Geminin was negatively correlated with expression of E-cadherin, which is critically involved in controlling epithelial architecture. In addition, Geminin expression level was correlated with Wnt signaling and expression of the Wnt target gene Axin2 and with Msx2, and negatively correlated with the expression of Bmp4 and Neurog1 in quantitative reverse transcriptase–polymerase chain reaction analysis of RNAs from individual embryos. These results suggest that in addition to patterning the early embryo, Geminin plays a previously unrecognized role in EMT via its ability to affect Wnt signaling and E-cadherin expression.
Introduction
The epiblast is patterned by reciprocal signaling between embryonic and extra-embryonic tissues to first position the proximal-distal (PD) axis; the anterior migration of the distal visceral endoderm then breaks the radial symmetry of the embryo and establishes the anterior-posterior (AP) axis [1,2]. At the onset of gastrulation, cells at the posterior pole of the epiblast divide and move medially in the primitive streak [3]. Subsequent epithelial to mesenchymal transition (EMT) and formation of the 3 definitive germ layers depend on the convergence of multiple signal transduction pathways to downregulate E-cadherin [4], and control the ensuing expression of genes involved in cell migration and in lineage differentiation. The ectoderm is then patterned by controlled bone morphogenetic protein (BMP) signaling [5], although additional molecules are likely required in the mammalian embryo [6].
One candidate previously identified, based on its ability to affect the size of the Xenopus neural plate [7], is the 33 kDa protein Geminin. Geminin has been suggested to act as a switch between proliferation and differentiation of many tissues [8–10] and certain cancers [11,12] presumably via its ability to control cell-cycle progression by associating with Cdt1 [13]. Despite its provocative function in Xenopus embryos, the role of Geminin during early mammalian development is largely unknown since deletion is lethal on E3.5 of development [14,15] due to over-replication of DNA.
In the postimplantation embryo, Geminin is expressed initially in the epiblast at E6.5; with neural induction and gastrulation, Geminin is present in the neural plate and primitive streak, while extra-embryonic tissues and the epidermal ectoderm consistently lack Geminin. To examine its role in development, we overexpressed a Geminin cDNA or employed shRNAs to knockdown Geminin expression. As Geminin expression decreased, Wnt signaling and Bmp4 expression were attenuated, while overexpression expanded both. Geminin shRNA embryos also failed to initiate Fgf8 expression in the primitive streak and E-cadherin was strikingly upregulated, abrogating EMT and gastrulation movements. Cell migration through the node and primitive streak was affected, inhibiting AP axis elongation and producing defects of neural tube and body wall closure. Conversely, overexpression attenuated E-cadherin expression, promoting premature EMT at both the primitive streak and neural crest. These results identify a novel role for Geminin in EMT and highlight the central role of this transition in development, metastasis, and recently, in cellular reprogramming.
Materials and Methods
Mice
Time-pregnant ICR strain (Harlan) or “Wnt indicator” mice expressing β-galactosidase via 6 transcription factor/lymphoid enhancer binding factor (TCF/LEF) sites [16] were employed and embryos harvested on E6.5–E17.0. To monitor the health status of pregnant dams after shRNA exposure, blood chemistries were analyzed by Unit for Laboratory Animal Medicine (ULAM) Pathology Core and did not identify significant variations in health profile. All protocols were reviewed and approved by the University of Michigan Committee on the Use and Care of Animals.
DNA delivery
E6.0 time-pregnant mice were injected via the tail-vein with 5 μg each of 2 shRNAs targeting Geminin or scrambled hairpin control shRNA constructs (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/scd) in 150 μL Ringer's solution as previously described [17]. For overexpression, the US2 promoter drove a Geminin cDNA and enhanced green fluorescent protein (EGFP). shRNA targeting Geminin or scrambled control shRNA was also injected into the pronucleus of fertilized zygotes, (C57BL/6×SJL)F2 by the University of Michigan Transgenic Animal Core. Embryos were transferred to pseudopregnant dams and allowed to develop to E6.5–E9.5.
Tissue analysis
Mice were sacrificed, embryos dissected from amnion and chorion, and digital images obtained prior to fixation. For whole-mount immunohistochemistry and whole-mount in situ hybridization (WISH), embryos were fixed for 10 min in 2% PFA. Embryos for WISH were dehydrated in MeOH, then stored at −20°C. For SEM, embryos were fixed in 1% glutaraldehyde, dehydrated, followed by hexamethyldisilazane, and then examined in an Amray 1910 SEM. Individual embryos for quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) were placed in Trizol (ILT), and those for western blot analysis were placed in RIPA/7×complete buffer (Roche) and stored at −20°C.
Quantitative RT-PCR
RNAs were extracted using Trizol. To minimize variation, control embryos were carefully staged: Stage 1: primitive streak (E7.0) to 4–5 somites; Stage 2: 5–10 somites; Stage 3: 10–12 somites; Stage 4: 12–25 somites. RNAs were also collected from blastocyst, E11, E13, E15, and E17 staged embryos, from individual Stage 3 and Stage 4 embryos exposed to Geminin shRNA (n=7 and n=30), scrambled shRNA (n=10 and n=22), or to the Geminin cDNA (Stage 4, n=36) and reverse transcribed using 1 μg RNA and random nonamers. qRT-PCR was performed in triplicate using iQ SYBR green supermix (BioRad) with β-actin as a reference. Primer sequences are provided in Supplementary Table S1. Mean control Ct values were used as the reference for embryos exposed to Geminin shRNA or cDNAs, and fold changes calculated [18].
Western blotting
Individual E9.0 embryos were homogenized, protein quantified using the Bradford reagent (BioRad), subjected to polyacrylamide gel electrophoresis, and exposed to Geminin (1:1000; Santa Cruz sc-13015), β-actin (1:20,000; Sigma #A1978), and E-cadherin (1: 200; Cell Signaling) antibodies. Exposure to rabbit anti-HRP (1:2000) and mouse anti-HRP (1:20,000; Jackson Immunoresearch) was followed by detection in luminol (Supersignal West pico chemiluminescent substrate, Thermo Scientific) and exposure to film for 1 min. Band intensity was determined using a BioRad ChemiDoc.
Statistical analysis
Expression data were analyzed using Student's t-test, with linear regression analyses of western blot and qRT-PCR data using SPSS.
Immunohistochemistry
Embryos were exposed to primary antibody overnight at 4°C followed by appropriate secondary antibodies. Primary antibodies were: Foxa2 (1:1000; Millipore #07-633), Sox3 (1:2000 ; MW Klymkowsky, Denver, CO), E-cadherin (1:500; BD #610181 and S Weiss, University of Michigan), laminin (1:25; Sigma #L9393), TuJ1 (1:200; Covance #MMS-435P), phospho-histone H3 (1:2000; Upstate #09-797), DsRed (1:100; SantaCruz, #SC33353), and EGFP (1:500; Abcam #42560). Secondary antibodies were conjugated to Alexa-488 (1:1000), Texas Red (1:500), FITC (1:500), or HRP (1:200) (Jackson Immunoresearch Laboratories). Hoechst 33258 (1 μM) was used to identify nuclei. X-gal staining was carried out (www.sanger.ac.uk/genetrap/) with 20 μg/μL X-gal (ILT #15520-034). TUNEL staining was carried out following the manufacturer's protocol (Promega). Images were captured using an Olympus BX-51, Leica dissecting scope, Leitz Fluovert, and/or a Zeiss 541 confocal microscope and transferred to Photoshop to assemble plates.
Whole-mount in situ hybridization
Embryos were treated with proteinase K, and WISH was carried out [19]. Probes were generated using T3 or T7 polymerase and DIG-11-UTP labeling kit (Roche). A 412 bp-Geminin probe was generated by linearizing pSK using BsAI and transcribing with T3 polymerase (NM_020567 nt 564 to 976). The 751 bp-Brachyury probe was first linearized from pSK by Xho 1 and transcribed using T3 (NM_009309 nt 476 to 1227). Additional probes used were obtained from Bmp4 [20], Cer1 [21], Engrailed-2 [22], Hesx1 [23], Shh [24], Fgf8 [25], Msx2 [26], Snail1 [4], and Nodal [27]. Detection was carried out using anti-DIG-AP and NBT/BCIP (Roche). After photography as whole mounts, 10 micron frozen sections were cut and photographed.
Results
Geminin expression is initially restricted to the epiblast, primitive streak, and neural ectoderm
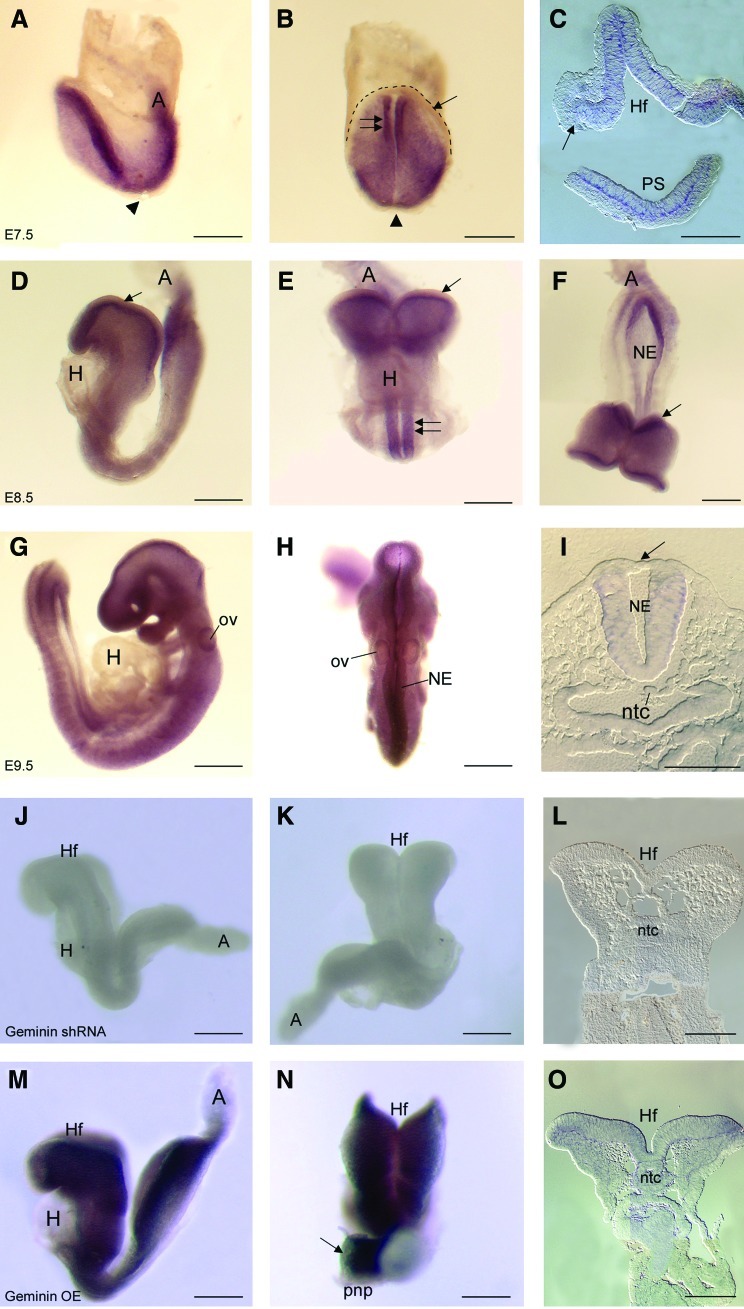
Geminin mRNA was present in the epiblast and forming neural ectoderm (Fig. 1A–C), expression terminating sharply at its boundary with the epidermal ectoderm (Fig. 1B, C arrow). It was expressed in the primitive streak (Fig. 1A, C) and dorsal node (Fig. 1A, B arrowheads) on E7.5, but was not present in extra-embryonic tissues at any stage examined (Fig. 1). By E8.5 and E9.5, Geminin was expressed throughout the neural ectoderm (Fig. 1D–I, E double arrows), but continued to be excluded from the epidermal ectoderm (Fig. 1D–F, I arrows), from the endoderm, and the heart. Overall, mRNA expression peaked at 5–10 somites, declining until E13 then increasing during organogenesis (Supplementary Fig. S3A).
FIG. 1.
Geminin expression. Whole-mount in situ hybridization localization of Geminin mRNA in control embryos on E7.5 (A–C), E8.5 (D–F), and E9.5 (G–I); in E8.5 Geminin shRNA embryos (J–L) and E8.5 Geminin overexpressing embryos (M–O). A, allantois; H, heart; Hf, headfolds; NE, neural ectoderm; ntc, notochord; ov, otic vesicle; pnp, posterior neuropore; PS, primitive streak; arrowhead in A, B, node; arrow in B, C, D, E, F, I, N, epidermal ectoderm; dashed line in B, edge of epidermal ectoderm; double arrows in E, Geminin expression in the neural tube. Anterior is to the left in: A, D, G, J, M. Anterior (coronal) views: B, E. Dorsal views: F, H, K, N. Transverse sections with anterior at top: C, I, L, O. Scale bars=200 μm except (C, L, O) which are 100 μm. Color images available online at www.liebertonline.com/scd
Geminin expression can be modified by exposure to Geminin shRNA or a Geminin cDNA
shRNA to knockdown or a Geminin cDNA to overexpress Geminin were delivered to pregnant dams on E6.0 [28,29]. Additional embryos were injected at the pronuclear stage with either an shRNA or a scrambled control shRNA (Supplementary Fig. S1). Little mRNA was detected in shRNA embryos (Fig. 1J–L), while exposure to the Geminin cDNA (Fig. 1M–O) increased Geminin mRNA in the neural ectoderm, but not heart or epidermal ectoderm (Fig. 1N arrow), possibly regulated by the many microRNAs that may target Geminin. Immunohistochemical localization of DsRed or EGFP indicated that the plasmids were widely expressed in postimplantation embryos, with considerable overlap with Hoechst 33258 (Supplementary Fig. S2).
shRNA exposure significantly reduced Geminin mRNA expression (>6-fold, P<0.001) compared with scrambled shRNA embryos, while the Geminin cDNA increased expression by 6.2-fold (P<0.001; Supplementary Fig. S3B). Geminin protein was 134.1% of control expression after cDNA exposure and 54.4% of control levels (P<0.001) after shRNA-treatment (Supplementary Fig. S3C and Table 1); individual embryos exhibited a range in Geminin protein from control levels to virtually no protein (Supplementary Fig. S3D). This technique can produce graded knockdown similar to electroporation or to an allelic series with a corresponding range of phenotypes within a litter (Supplementary Fig. S4).
Table 1.
Regression Analysis
| Group | E-cadherin | Geminin | Ratio | R2 | f | P< | T |
|---|---|---|---|---|---|---|---|
| Pooled embryos | 1.3 | 0.001 | 0.77 | 0.00001 | 5.48 | ||
| Missense | 99.8+7.7 | 100.2+12.6 | 0.99 | 0.275 | 0.02 | 0.0009 | 4.8 |
| shRNA | 202.3+26.8 | 54.4+4.7 | 3.72 | 0.776 | 0.00 | 0.021 | −2.4 |
| OE | 65.2+5.6 | 134.1+15.0 | 0.49 | 0.15 | 0.02 | 0.00008 | 4.74 |
E-cadherin expression is inversely correlated with Geminin protein levels. Regression analysis of individual embryos after knockdown or overexpression of Geminin in comparison to E-caderin expression.
OE, overexpressing.
Geminin is required during preimplantation development
Pronuclear injection of shRNA produced significant preimplantation loss with surviving embryos (18%) often exhibiting few anomalies (Supplementary Fig. S1G–I). However, by delivering the shRNAs on E6.0 to bypass the early requirement for Geminin, we were able to obtain knockdown in more than 70% of the embryos.
Geminin is required in the epiblast for axis elongation and neurulation
By E7.5, control embryos had well-organized neural folds and an expanded amniotic cavity as a result of normal axis elongation (Supplementary Fig. S1A). In Geminin shRNA-exposed embryos (Supplementary Fig. S1D), the node was positioned anteriorly and AP axis elongation was inhibited, like the rare surviving embryos after pronuclear injection of shRNA (Supplementary Fig. S1G). Geminin overexpression was often lethal; surviving embryos (Supplementary Fig. S1J) were often composed of headfolds and membranes with disorganized posterior tissues.
By E8.5, compared with control embryos (Supplementary Fig. S1B), the anterior- and posterior neuropores of shRNA embryos remained widely open (Supplementary Fig. S1E, H). Overexpression of Geminin resulted in posterior disorganization with relatively well-formed headfolds (Supplementary Fig. S1K). When E9.0 embryos were examined using SEM, the anterior neural folds of controls were approaching the dorsal midline (Supplementary Fig. S5A, D, G) compared with shRNA- and cDNA-exposed embryos, where the neural folds were widely everted (Supplementary Fig. S5B, C, E, F, H, I) and massively overgrown in overexpressing embryos (Supplementary Fig. S5C, F, I). The proepicardium, which also undergoes EMT, failed to cover the heart tube of shRNA embryos (Supplementary Fig. S5H). By E9.5, the anterior neural tube had closed in control embryos (Supplementary Fig. S1C), while shRNA embryos often had open, everted neural folds (Supplementary Fig. S1F arrows). The first branchial arch was also small and abnormally oriented. Surviving embryos carrying the hairpin shRNA express DsRed or EGFP (Supplementary Figs. S1F, I, S2D, G). Embryos that survived Geminin overexpression to E9.5 often failed to undergo the normal turning process and the neural ectoderm was overgrown (Supplementary Fig. S1L arrows).
Geminin is required for morphogenetic cell movements at gastrulation
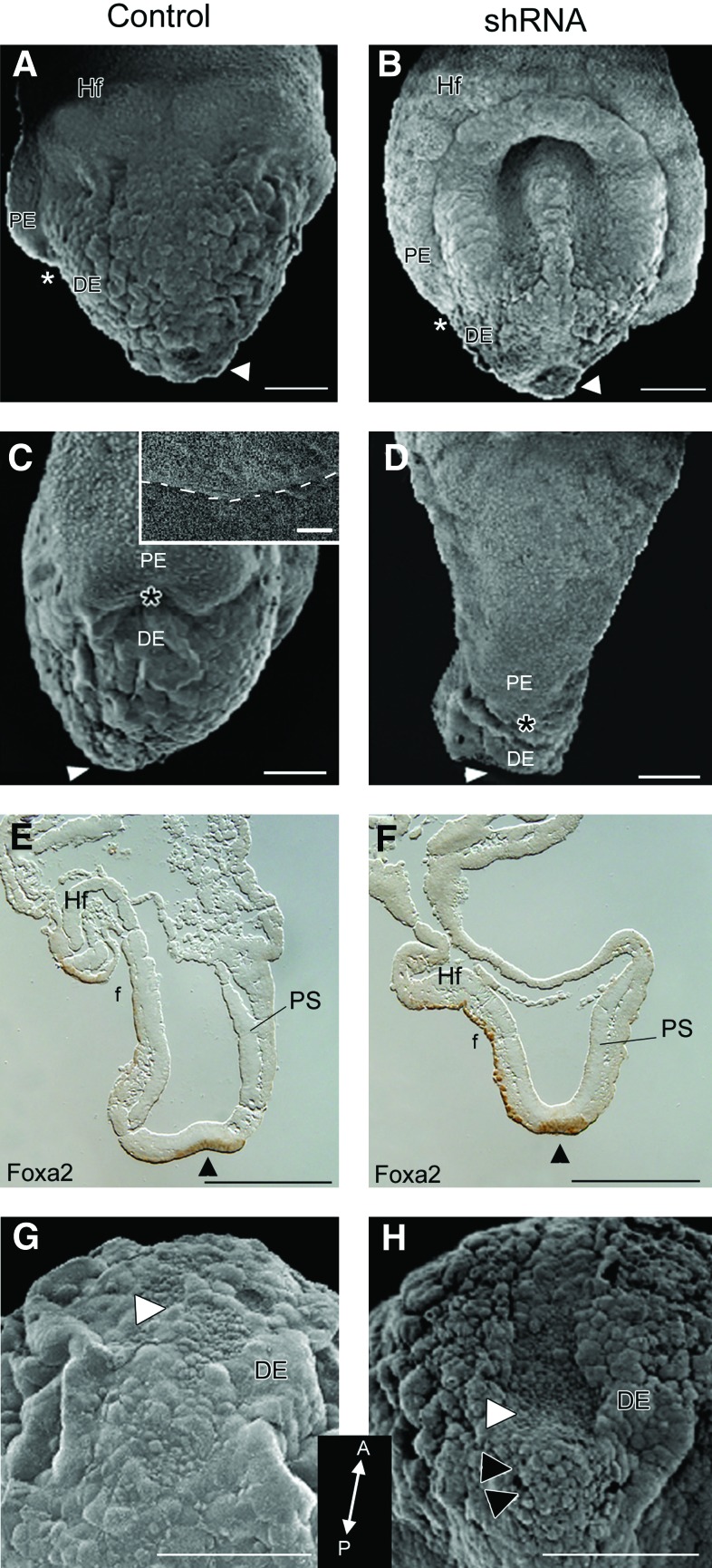
At gastrulation, cells delaminate from the primitive streak to form endoderm by intercalating and replacing the primitive endoderm (PE) with definitive (embryonic) endoderm (DE) [30]. As gastrulation proceeds, the boundary between the PE and DE progresses from distal to proximal, terminating at the extra-embryonic margin (Fig. 2A, C asterisks; 2C dashed line in inset). In Geminin shRNA embryos, this boundary failed to migrate (Fig. 2B, D asterisks) suggesting that DE does not replace PE. PE markers including HRP, illustrate a similar lack of cell movement (not shown) indicating that Geminin is required for differentiation or migration of the DE.
FIG. 2.
Alterations at the node. E7.5 embryos exposed to scrambled shRNA (A, C, E, G) or to Geminin shRNA (B, D, F, H) were analyzed using SEM or Foxa2 expression in sagittal sections (E, F). Arrowheads (A–F), node; *(A–D), border of PE and DE; white arrowhead (G, H), ciliated cells of the node; black arrowheads (H), aggregate of cells at the node; f, foregut pocket; Hf, headfolds; PS, primitive streak. Anterior view in A, B; Anterior is to the left in C–F; Distal tip of the embryo toward the top in G, H (inset is posterior to anterior orientation). Scale bars=100 μm (A–D, G, H), 10 μM (inset in C), and 200 μm (E, F). PE, primitive endoderm; DE, definitive endoderm. Color images available online at www.liebertonline.com/scd
Compared with control embryos (Fig. 2A, C, E), both the foregut and hindgut pockets were widely open in shRNA embryos (Fig. 2B, D, F), likely due to the lack of elongation and morphogenetic movements required for body wall closure. Foxa2 was expressed in the DE that lines the node and foregut pocket (Fig. 2E, F), highlighting the widely open foregut pocket, shortened anterior-posterior axis, and abnormal headfold positioning in shRNA embryos (Fig. 2F).
Loss of Geminin affects cell migration through the node
Normally, the node has a slightly concave appearance (Fig. 2A, E, G); cells at the tip involute as the AP axis elongates and the node moves posteriorly [31]. When Geminin expression was downregulated, the node failed to regress, forming a deep, concave pit (Fig. 2B, H white arrowheads), leaving a cluster of unorganized cells behind (Fig. 2H black arrowheads). However, Nodal expression in the surrounding crown cells was not affected by Geminin dosage (not shown).
Geminin is required for EMT at gastrulation
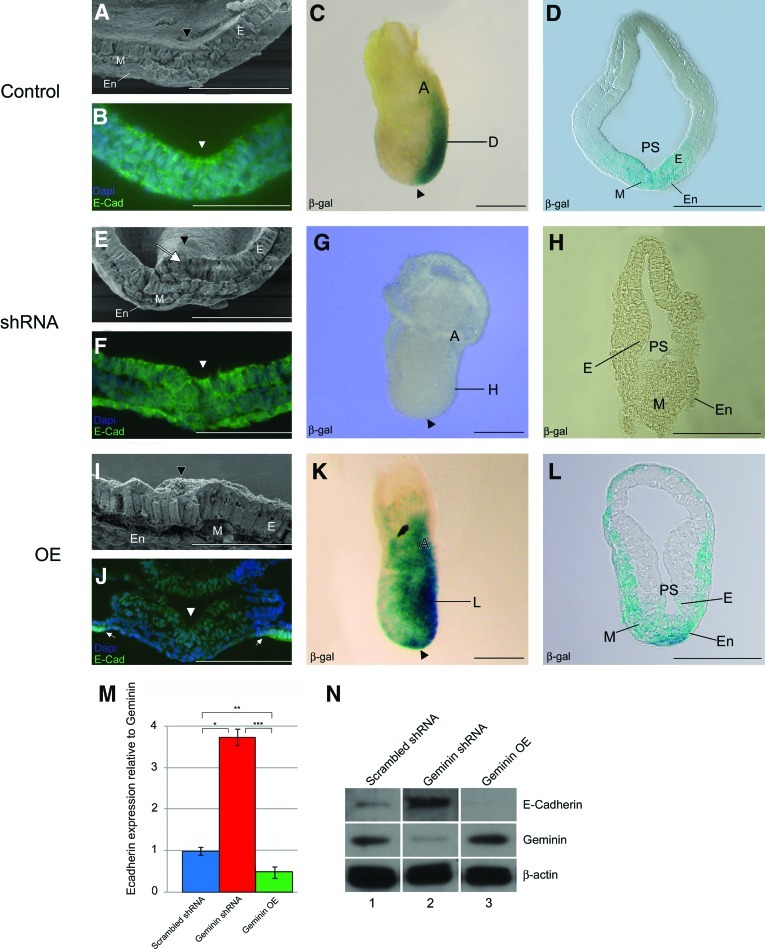
The primitive streak is a pseudostratified epithelium from which mesendoderm cells delaminate at the midline (Fig. 3A arrowhead). In shRNA embryos, cells fail to leave the primitive streak resulting in an overgrown epiblast (Fig. 3E white arrow), while Geminin overexpression resulted in premature EMT and disorganization of the posterior pole of the embryo (Fig. 3I). In control embryos, E-cadherin expression decreased at the midline (Fig. 3B, arrowhead); however, in Geminin shRNA embryos E-cadherin persisted in the primitive streak, mesoderm, and endoderm (Fig. 3F arrowhead). In overexpressing embryos, there was less E-cadherin in the epithelium and the newly formed mesendoderm, whereas it remained at high levels in the PE (Fig. 3J arrows). Laminin, which is expressed in the basement membrane of the ectoderm except the midline of the primitive streak where cells ingress, was also affected by Geminin dose. When Geminin was overexpressed, the normally linear pattern of laminin immunoreactivity was irregular and punctate, while in knockdown embryos it remained in the basement membrane throughout its extent. In immunoblots of protein from individual embryos, Geminin protein was strongly negatively correlated with E-cadherin levels (Fig. 3M, N; discussion below).
FIG. 3.
Organization of the primitive streak. The primitive streak of control embryos (A–D), Geminin shRNA embryos (E–H), and Geminin overexpressing embryos (I–L) was analyzed using SEM of transverse fractures (A, E, I), E-cadherin localization in transverse sections (B, F, J), and β-gal expression in “Wnt-indicator” mice (C, D, G, H, K, L). Anterior is to the left in C, G, K. D, H, L are transverse sections at levels indicated in C, G, and K with anterior at top. A, allantois; E, ectoderm; En, endoderm; M, mesoderm; PS, primitive streak; arrowhead (A–B, E–F, I–J), primitive streak; arrowhead (C, G, K), node; white arrow (E), overgrown epiblast. Scale bars=100 μm (A, B, E, F, I, J), 200 μm (C, D, G, H, K, L). M. Western blot analysis of the relative expression of E-cadherin in scrambled shRNA control, Geminin shRNA-exposed embryos and embryos over expressing (OE) Geminin. E-cadherin expression is inversely related to Geminin. *P<2.1×10−8, **P<0.02, ***P<2.6×10−14, Students t-tests. N. Representative western blots illustrating E-cadherin, Geminin, and β-actin in individual E9.0 embryos exposed to scrambled (lane 1), Geminin shRNA (lane 2), or Geminin overexpression (OE; lane 3) constructs. Color images available online at www.liebertonline.com/scd
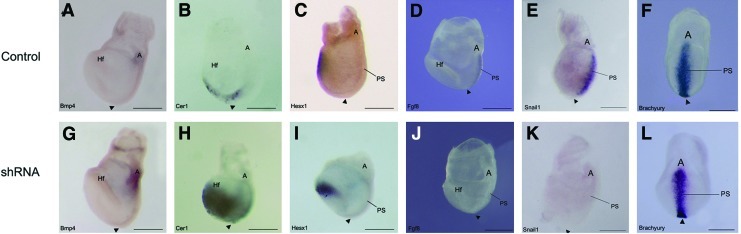
X-gal staining of control “Wnt indicator” embryos [16] indicated that Wnt signaling was normally tightly restricted to the primitive streak (Fig. 3C, D). However, in Geminin shRNA embryos, signaling was reduced (Fig. 3G, H), while overexpression expanded the Wnt-signaling domain into the mesoderm (Fig. 3K, L). The early expression of Bmp4 in the extra-embryonic ectoderm and epidermal ectoderm of control embryos (Fig. 4A) was expanded in shRNA embryos, (Fig. 4G), suggesting that Bmp4 (and Nodal) are expressed at sufficient levels to position the node, but Geminin is required to constrain the Bmp4 expression domain as in Xenopus [7].
FIG. 4.
Geminin is required to pattern the embryo on E7.5. Whole-mount in situ hybridization was used to analyze gene expression in E7.5 control (A–F) and Geminin shRNA (G–L) embryos. Embryos were hybridized with probes to: Bmp4 (A, G), Cer1 (B, H), Hesx1 (C, I), Fgf8 (D, J), Snail1 (E, K), and Brachyury (F, L). Anterior is oriented to the left in all images except (F, L), which are posterior (ventral) views. A, allantois; Hf, headfolds; PS, primitive streak; arrowhead, node. Scale bars=200 μm for all embryos. Color images available online at www.liebertonline.com/scd
In control embryos, Cer1 and Hesx1 were expressed in the anterior endoderm (Fig. 4B, C); Geminin knockdown expanded their expression domains posteriorly (Fig. 4H, I). Fgf8 and Snail1 were present in the primitive streak of control embryos (Fig. 4D, E); but they were down regulated in Geminin shRNA embryos (Fig. 4J, K). Brachyury, present in early mesoderm of the primitive streak and node (Fig. 4F), had a slightly wider expression domain near the allantois of shRNA embryos (Fig. 4L). In transverse section, Brachyury is expressed in the primitive streak and newly forming mesoderm in missense control embryos (Fig. 7J), compared with shRNA embryos, where there is little mesoderm and strong ectodermal expression (Fig. 7N).
FIG. 7.
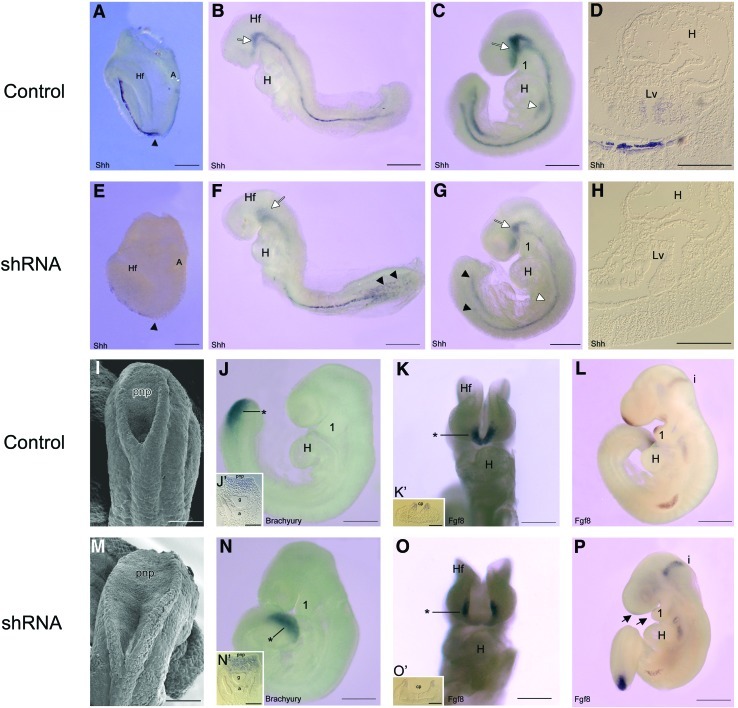
Geminin shRNA embryos are mispatterned along the length of the neuraxis. Whole-mount in situ hybridization was used to analyze the expression of Shh (A–H), Brachyury (J, N), and Fgf8 (K, L, O, P) with SEM employed for high-magnification views of the posterior neuropore (I, M) in control (A–D, I–L) and Geminin shRNA-exposed embryos (E–H, M–P). Sagittal views, with anterior to the left, include: (A–C, E–G, J, L, N, P), (K, O) are coronal (ventral) views, (I, M) are dorsal views of the closing posterior neuropore, (J′, K′, N′, O′) are transverse sections (section level indicated by * in J, K, N, O), and (D, H) are sagittal sections through the heart and liver bud of E9.0 embryos. A, allantois; a, aorta; cp, commissural plate; g, gut; H, heart; Hf, headfold; i (L, P), isthmus; Lv, liver bud; pnp, posterior neuropore; 1, first branchial arch; black arrowhead (A, E), node; black arrowheads (F, G), misexpression of Shh; white arrowhead (C, D), Shh expression in the liver bud; black arrows (P), loss of Fgf8 in the commissural plate and branchial arch; white arrows (B, C, F, G), Shh expression in the cranial region. Scale bars=200 μm (A–C, E–G, J–L, N–P), 100 μm (D, H, I, J′, K′, M, N′, O′). Color images available online at www.liebertonline.com/scd
Geminin dose controls neural crest EMT
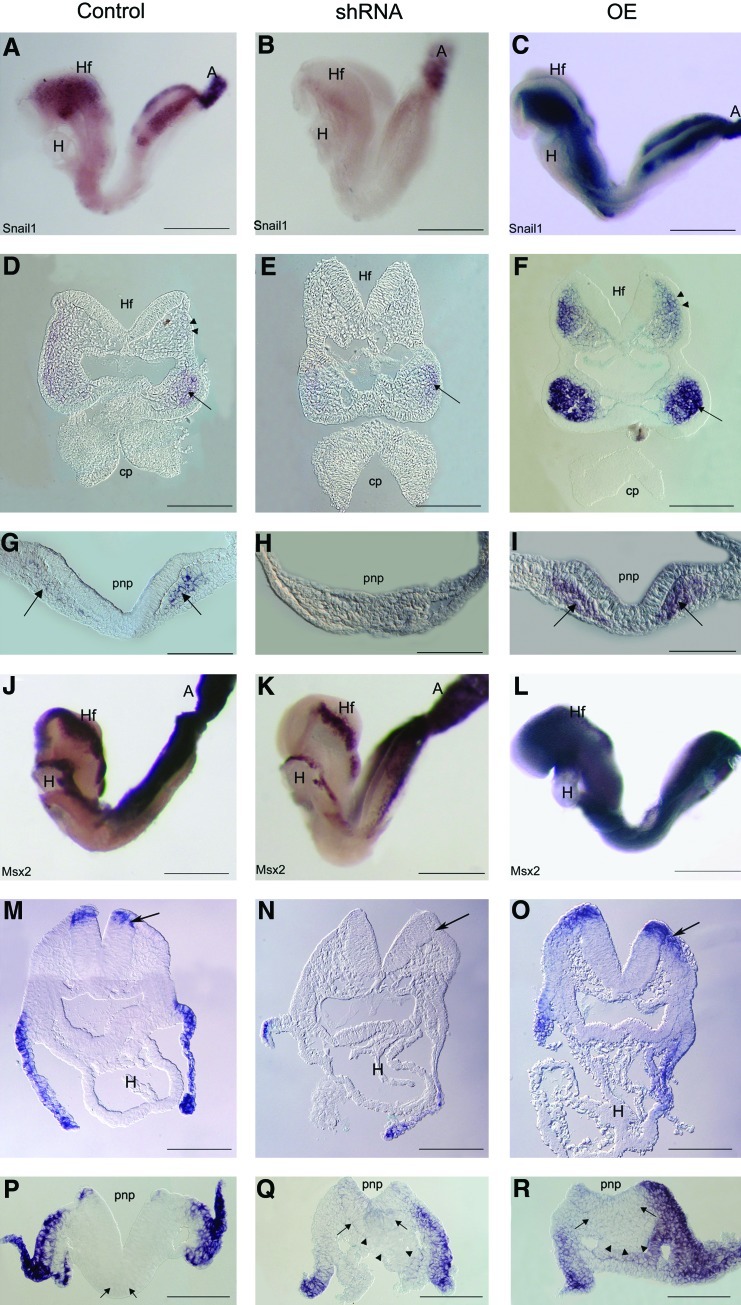
Combinatorial signaling by Wnts, BMPs, and fibroblast growth factors (FGFs) is thought to specify the neural crest [32]. In control embryos, Snail1 and Msx2 were expressed at the margins of the neural folds and by neural crest cells (NCC) migrating into the mesenchyme (Fig. 5A, J) in both the cephalic region (Fig. 5D, M) and the posterior neuropore (Fig. 5G, P). Msx2 was also expressed in the lateral plate mesoderm that will line the body wall and gut tube (Fig. 5M, P), and Snail1 in the extra-embryonic mesoderm of the allantois (not shown). In Geminin shRNA embryos, expression of both genes was strikingly downregulated in the anterior neural crest and mesenchyme (Fig. 5B, E, K, N arrow), although some Snail1 was present in NCC in the branchial arches (Fig. 5E arrow). The posterior neuropore was often flattened and there was little Snail1 expressed in this region (Fig. 5H), or in the allantois (not shown). Msx2 was similarly strongly downregulated in the posterior region although there was ectopic expression of Msx2 in the aggregate of cells at the junction of the posterior neuropore with the primitive streak (Fig. 5Q arrowheads). Overexpression of Geminin increased Snail1 (Fig. 5C, F) and Msx2 (Fig. 5L, O) in NCC. Both were strongly expressed in the mesenchyme of the headfolds and branchial arches (Fig. 5F, O arrows), the posterior neuropore (Fig. 5I, R arrows), and allantois (not shown). Geminin dose did not produce alterations in either proliferation or cell death (not shown), however.
FIG. 5.
Geminin dose controls expression of Snail1 and Msx2. Localization at E8.5 of Snail1 (A–I) and Msx2 (J–R) in control (A, D, G, J, M, P), Geminin shRNA (B, E, H, K, N, Q) and overexpressing embryos (C, F, I, L, O, R) in transverse sections of the anterior neural folds (D–F, M–O), or posterior neuropore (G–I, P–R). A, allantois; cp, commissural plate; H, heart; Hf, headfolds; pnp, posterior neuropore. Arrows indicate NCC in D–I, M–O. In P–R arrowheads identify clusters of cells at the junction of the pnp and primitive streak. White arrows indicate ectopic Msx2 expression. Anterior is to the left in: A–C, J–L. Anterior toward the top in: D–I, M–R. Scale bars=200 μm (A–C, J–L); 100 μm (D–I, M–R). Color images available online at www.liebertonline.com/scd
E-cadherin expression is negatively correlated with Geminin level
Given the striking changes in E-cadherin expression in the embryo in immunohistochemistry, and the central role of E-cadherin in controlling EMT, we quantified E-cadherin protein from individual embryos using western blot. Geminin expression level was normalized to β-actin to control loading, then compared to E-cadherin. Geminin was expressed at highest levels in single overexpressing embryos and significantly decreased in shRNA-treated embryos (Table 1). E-cadherin was reciprocally expressed; E-cadherin was high in shRNA-exposed embryos and significantly decreased when Geminin levels were increased. We examined the relationship between E-cadherin and Geminin first using ratios (Table 1 and Fig. 3M, N); the means were significantly different between the groups when analyzed using t-test. Data were also analyzed using regression analysis. When data from all embryos were pooled, there was a consistent significant correlation in Geminin expression level and the expression of E-cadherin (P≤0.00001, t=5.479). Within treatment groups, the correlations were positive, except in the shRNA embryos where high E-cadherin was associated with low levels of Geminin expression.
Wnt signaling is responsive to Geminin dosage
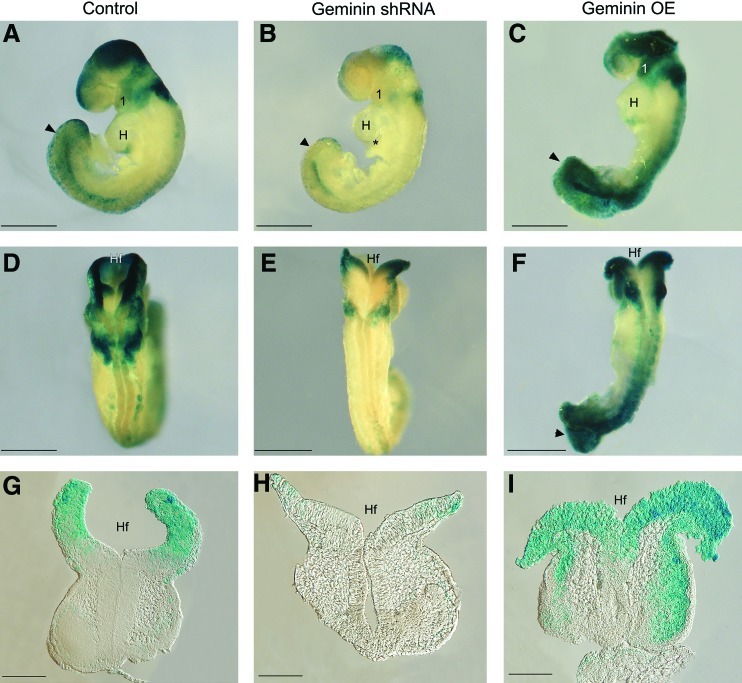
On E9.5, Wnt signaling was active in the dorsal rhombencephalon, midbrain, and forebrain (Fig. 6A, D, G), in NCC migrating to the first branchial arch, differentiating somites, and posterior neuropore (Fig. 6A arrowhead). Geminin shRNA decreased β-gal expression throughout the embryo (Fig. 6B), but particularly in migrating NCC, in the forebrain and midbrain (Fig. 6B, E, H), the forming proepicardium (Fig. 6B, asterisk), and posterior neuropore (Fig. 6B arrowhead). Overexpression strikingly elevated signaling throughout the neuraxis, particularly in the first arch, the midbrain, and the hindbrain (Fig. 6C, F, I) and in somites and the posterior neuropore (Fig. 6C, F).
FIG. 6.
Geminin dose affects Wnt signaling. Embryos were exposed to scrambled (A, D, G), Geminin shRNA (B, E, H), or Geminin overexpression (C, F, I) constructs followed by X-gal staining to identify sites of Wnt signaling. (A–C) Sagittal views of E9.5 embryos with anterior to the left. (D–F) Dorsal views. (G–I) Coronal sections through the midbrain. *Proepicardium; 1, first branchial arch; H, heart; Hf, headfolds; arrowhead, posterior neuropore. Scale bars=200 μm (A–F), 100 μm (G–I). Color images available online at www.liebertonline.com/scd
Anterior and posterior patterning is affected by Geminin knockdown
In control embryos, Shh is expressed in the node and notochord from the posterior pole of the embryo to the commissural plate (Fig. 7A–C). In shRNA embryos, there was patchy expression of Shh in the notochord, (Fig. 7E, F) producing a widened floor plate and neural tube closure abnormalities (Fig. 7F, G, M, O). Unlike controls (Fig. 7D), Shh was not expressed in the liver bud of shRNA embryos (Fig. 7H), which would affect later differentiation of both the liver and gall bladder.
By E9.5, the posterior neuropore (pnp) was closing (Fig. 7I) and Brachyury expression was restricted to this region (Fig. 7J, J′). In contrast, the pnp was consistently widely-open in shRNA embryos (Fig. 7M) and Brachyury expanded into the dorsal wall of the hindgut (Fig. 7N, N′). There was decreased Fgf8 expression in the wide ventral commissural plate, and in the first branchial arch of Geminin shRNA embryos (Fig. 7O, O′, P arrows) compared with controls (Fig. 7K, K′, L). This was specific to the anterior neural folds as Fgf8 expression in the isthmus and limb bud was unaffected by Geminin knockdown. Engrailed-2, which is normally present in the posterior midbrain and in rhombomere 1 of the hindbrain was strikingly reduced in the midbrain but not in rhombomere 1 of shRNA embryos (not shown).
Geminin alters differentiation of neural precursor cells
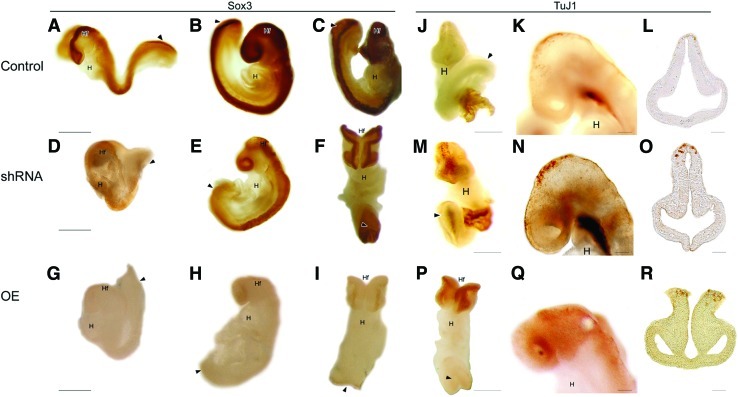
Prior research suggested that Geminin controls the switch between proliferation of neural precursors and differentiation into immature neurons [33,34]. In control embryos, Sox3 identifies proliferative neural precursors (Fig. 8A–C), while TuJ1 appears later in midbrain neurons (Fig. 8J–L). In shRNA embryos, Sox3 was reduced (Fig. 8D–F) and TuJ1 prematurely expressed throughout the neuraxis (Fig. 8M–O). Overexpression of Geminin also decreased Sox3 (Fig. 8G–I), while TuJ1 expression was increased (Fig. 8P–R). These results suggest that Geminin exerts stage-specific and context-dependent control of ectoderm differentiation.
FIG. 8.
Geminin affects the differentiation of neurons from neural precursors. Localization of Sox3 (A–I) and TuJ1 (J–R) in control (A–C, J–L), Geminin shRNA (D–F, M–O), and Geminin overexpressing (G–I, P–R) embryos. E8.5 (A, D, G) and E9.5 (B–C, E–F, H–I, J–R) embryos were examined. (A, B, D, E, G, H, K, N, Q) are sagittal views with anterior to the left. (C, F, I, J, M, P) are coronal/ventral views and (L, O, R) are coronal sections with anterior toward the top. H, heart; Hf, headfolds; arrowhead (A–P), posterior neuropore. Scale bars=200 μm (A–I, J–K, M–N, P–Q), 100 μm (L, O, R). Color images available online at www.liebertonline.com/scd
Gene expression analysis
To correlate Geminin and target gene expression, we carried out qRT-PCR analysis of RNAs from individual embryos exposed to shRNAs (Supplementary Table S2) and from single embryos overexpressing Geminin (Supplementary Table S3), followed by Pearson correlation. Geminin levels were positively correlated with the expression of the Wnt target Axin2, and negatively correlated with Bmp4 and Neurog1 in shRNA embryos. Brachyury expression was also negatively correlated with Geminin, but did not reach statistical significance. Interestingly, Foxa2 was correlated with both Sox3 and Neurog1, likely reflecting overall embryonic development. Axin2 and Neurog1 expression were also highly correlated, markers of mesoderm (Brachyury) and neurons (Neurog1) were negatively correlated, as were Bmp4 and Sox3 expression (Supplementary Table S2). Like our in situ results, Msx2 and Geminin were strongly correlated in overexpressing embryos. Bmp4 was again strongly, negatively correlated with Sox3 expression while Neurog1 was positively correlated with Snail1 and Axin2 levels (Supplementary Table S3). These results are consistent with the observed increase in Snail1 expression in the embryo and the strong stimulation of Wnt signaling in these embryos. Overall, the most consistent effects were the strong negative correlation between Geminin levels with Bmp4 and positive correlation with Axin2, and the negative correlation between Bmp4 and Sox3.
Discussion
Despite its provocative prediction of the neural plate in other species [7,10,35], this is the first investigation to examine the expression and role of Geminin in the early postimplantation-staged mouse embryo. Geminin was originally described as a bi-functional molecule; the N-terminus implicated in neural-cell fate determination [7], the C-terminus and central regions involved in DNA replication and cell cycle progression [13] in the Xenopus embryo. Geminin also binds polycomb and Brahma proteins to regulate gene expression [33,9]; it binds Hox and Six proteins to influence patterning of the embryo [8,9,36]; our in vitro results (Slawny and O'Shea this issue) suggest that it also binds nuclear TLEs to control the Wnt-signaling cascade. Ultimately, competition for partner proteins in a position- or stage-restricted manner may place Geminin in a unique position to influence the differentiation of many tissues.
Geminin dosage controls EMT
The conversion of cells within an epithelial sheet to mesenchyme is essential in transforming the early egg cylinder into the 3-germ layer embryo at gastrulation and is reiterated and reversed multiple times during development [37,38]. EMT involves a series of well-characterized alterations in cell-cell adhesion, cell polarity, expression of integrin receptors, and cell migration [39]. Interestingly, the opposite process—mesenchymal to epithelial transition—also appears to be essential in reprogramming fibroblasts to epithelial induced pluripotent cells [40,41].
Gene targeting has established that members of the Wnt, Fgf, and transforming growth factor β superfamilies are essential for EMT at gastrulation [37]. Lacking Geminin, embryos fail to express Fgf8 in the primitive streak, thereby affecting the cascade of Msx2, Msx1, and E-cadherin expression. E-cadherin is critically involved in maintaining the integrity of the epithelial sheet; downregulation is required for EMT and metastasis [38]. Our data demonstrate that at the level of the individual embryo the amount of Geminin present inversely correlates with the expression of E-cadherin. The posterior disorganization observed with Geminin overexpression is reminiscent of embryos where Wnt pathway members are overexpressed, resulting in premature EMT of the epiblast [42]. The Geminin gastrulation phenotype in KD embryos is also similar to mouse embryos lacking Hira [43], Fgfr1 [44,45], Fgf8 [46], the Nodal target Eomesodermin [47], and Snail1 [48], where cells fail to leave the primitive streak. Exposure of Zebrafish embryos to Geminin morpholinos similarly repressed Snail1a; mesendodermal cells did not delaminate, while overexpression promoted Snail1a production and endodermal differentiation [49], suggesting that Geminin plays a central, critical role in this cascade.
Sequelae of EMT inhibition
Failure of EMT had many sequelae, including the lack of axis elongation after the failure of mesendoderm migration, as in our EB model (Slawny and O'Shea, this volume). Geminin appears to be required downstream of Nodal signaling, since the PD axis was initially specified properly and Nodal expression was not affected by Geminin dosage. Migration through the posterior streak was less affected, although ectopic Snail1 and Brachyury positive cells were present in the dorsal hindgut wall in Geminin mutant embryos, and Geminin dose affected Snail1 expression in the allantoic mesenchyme. The early migrating cardiac mesoderm also appears to be unaffected by Geminin dose, suggesting a cell type-, region- or stage-restricted role for Geminin in lineage allocation at gastrulation.
Geminin mutant embryos exhibit body wall closure defects after failure of morphogenetic movements and endoderm differentiation, as previously reported in Zebrafish [49]. Lack of normal anterior mesendoderm migration also resulted in the failure of DE to replace PE, and persistent signaling in the anterior region, identified by expanded Cer1 and Hesx1 expression, which likely have long-term effects on patterning of the anterior central nervous system (CNS) [50]. Neural tube closure defects were commonly observed in Geminin-targeted embryos, and may result from notochord abnormalities as secreted factors from the notochord are essential for floorplate induction and the resulting elevation of the neural folds [51]. Inhibition of BMP signaling is then required for the formation of the dorsolateral hingepoints and midline approximation of the neural folds [52], suggesting that alterations in Shh and/or BMP expression may play a role in the genesis of the observed neural tube defects.
Geminin and the neural crest
Neural crest emigration has been considered a second gastrulation event [53], and neural crest versus epidermal identity is determined by BMP and Wnt signaling; intermediate levels of BMP signaling position the border [54–56]. A second mesodermal Wnt signal [57] with BMP then activates Msx1,2 gene expression [58], which induce expression of Twist1, Snail1, and Slug [59] that then induce p21, and downregulate E-Cadherin expression to promote EMT. In fact, intermediate levels of Geminin overexpression (insufficient to induce neural markers) similarly expanded the Twist-positive neural crest field [7,33]. In the current investigation, Geminin expression level strongly affected Bmp4, Snail1, and Msx2 gene expression and Wnt signaling by NCC.
In NCC, BMP-dependent Wnt signaling has been shown to control the transition from G1 to S by modulating intracellular levels of cyclins [60]. Premigratory NCC are arrested in G1 and enter S on delamination; repression of the G1 to S transition inhibits the EMT process [61]. The unique ability of Geminin to both modulate cell cycle, while negatively modulating E-cadherin, BMP, and Fgf gene expression and to positively regulate Wnt signaling place it in a unique position to control EMT and thereby tissue organization during development. The observation that decreased Geminin levels inhibit EMT and maintain neural crest stem cells, while overexpression promotes their migration, are similar to results in subventricular zone neural stem cells where deletion of Geminin promoted self-renewal and expanded the progenitor population while overexpression promoted migration and differentiation [62], suggesting that similar mechanisms may maintain these stem cell niches. The observation that Geminin controls EMT may also inform our understanding of metastasis, as Geminin is overexpressed in a number of tumors where it is negatively correlated with outcome [63].
Geminin dose affects Bmp4 and Wnt Signaling
This is the first identification of the strong correlation between Geminin and Wnt signaling, which is required in the epiblast to position and maintain the primitive streak [64]. Overexpression of Wnt8c [65], interference with β-catenin degradation [66], or deletion of negative regulators [67,68] expand or duplicate the primitive streak, as observed with Geminin knockdown. Geminin and Wnts may act reciprocally since the Geminin promoter contains TCF-binding sites [69], Geminin may control Wnt target gene expression via Snail1 [70], or Geminin may bind a Groucho factor to modulate Wnt signaling (Slawny and O'Shea, this volume).
Given the strong negative correlation between Geminin and Bmp4 expression, the requirement for BMP signaling in the formation of the node and primitive streak, and specification of mesoderm [71,72], interference with BMP signaling may also play a role in the gastrulation block observed in Geminin mutant embryos. The effect may be direct as suggested by data from amphibian [7] and chick embryos [35], or Geminin may interact with additional pathway members, for example, Smad proteins in the nucleus.
Neural differentiation
Geminin has been suggested to mark the acquisition by the ectoderm of neural competence [73]. The striking lack of Geminin in the epidermal ectoderm suggests that Geminin may function in the ectoderm like other preneural genes, such as Sox3 and Zic2 that are required for the induction of neural tissue by BMP inhibition, thereby positioning the neural-epidermal ectodermal border [54,73], consistent with observations that overexpression increases and reductions in Geminin reduce the size of the neural plate in Xenopus [7], Zebrafish [49], and Drosophila [10] embryos. Geminin may then maintain proliferation in the neural ectoderm and prevent premature expression of genes involved in fate-restriction [34,35,74]. These effects are both context- and dose-dependent. In the Xenopus embryo, high levels of Geminin are required to induce ectopic expression of neuronal markers, while low levels of suppression did not affect neuronal fate acquisition, but promoted premature neuronal differentiation [8,74]. Our results reflect a similar dosage effect; with knockdown producing premature differentiation of midbrain neurons, consistent with the strong negative correlation between Geminin and Neurog1. High levels of expression also resulted in a loss of Sox gene expression and dysregulated expression of neuronal markers, and it is possible that Sox3 expression was accelerated rather than abrogated. Recent reports indicate that deletion of Geminin in nestin+ cells of the nervous system had little effect on the number or differentiation of precursors in one case [75], while deletion expanded progenitors in the subventricular zone and overexpression promoted migration and premature differentiation [62] in another. These results, and in vitro studies in embryonic stem cells (Slawny and O'Shea, this volume) emphasize the context- and dose-dependent effects of Geminin and suggest that it may play very different roles in lineage-committed precursors versus cells with multilineage differentiation potential such as the neural crest and primitive streak.
Ultimately, Geminin's effects are likely determined by the availability of protein partners in the nucleus, such as the Brahma-related gene 1, Brg1, which regulates both proneural bHLH [76] and Sox gene expression [35]. Brg1 activates Wnt reporters [77], interacts with nuclear β-catenin to control gene expression [78,79], and can be recruited to target gene promoters by Groucho proteins [78]. The striking correlation between Geminin expression, Wnt reporter activity, and target gene expression suggests that in the mouse embryo, Geminin may regulate Wnt signaling via Brg1 or yet unidentified nuclear factors. Geminin is expressed in multiple stem cell populations—embryonic stem cells, neural stem cells, the primitive streak, and gut stem cells, and overexpression in tumors is strongly negatively correlated with survival. Additional study of Geminin's role in tissue-type transitions involved in the morphogenesis of other organs will likely require reconstruction in tissue restricted or inducible systems.
Supplementary Material
Acknowledgments
We are grateful to: MW Klymkowsky (Sox3 antibody), S Weiss (E-cadherin antibody), A Joyner (En-2 probe), SG Gong (Bmp4), V Kaartinen (Snail1), Y Mishina (Cer1 and Hesx1), P Gage (Msx2, Nodal, Fgf8, and Shh probes as well as Wnt reporter mice), and T Saunders, M Van Keuren in the University of Michigan Transgenic Animal Core. Also: Christine Belzyt, Kate Eaton, Amanda Evans-Zacharias, Theresa Gratsch, Maria Morell, John O'Shea, Nicole Slawny, Yao-Chang Tsan, and Derrick Yang for assistance, reagents, and comments on this work. This research was supported by NIH grant RR-023187.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Thomas P. Beddington R. Anterior primitive endoderm may be responsible for patterning the anterior neural plate in the mouse embryo. Curr Biol. 1996;6:1487–1496. doi: 10.1016/s0960-9822(96)00753-1. [DOI] [PubMed] [Google Scholar]
- 2.Arnold SJ. Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 3.Lawson KA. Meneses JJ. Pedersen RA. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- 4.Cano A. Perez-Moreno MA. Rodrigo I. Locascio A. Blanco MJ. del Barrio MG. Portillo F. Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 5.Levine AJ. Brivanlou AH. Proposal of a model of mammalian neural induction. Dev Biol. 2007;15:247–256. doi: 10.1016/j.ydbio.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachiller D. Klingensmith J. Kemp C. Belo JA. Anderson RM. May SR. McMahon JA. McMahon AP. Harland RM. Rossant J. De Robertis EM. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature. 2000;403:658–661. doi: 10.1038/35001072. [DOI] [PubMed] [Google Scholar]
- 7.Kroll KL. Salic AN. Evans LM. Kirschner MW. Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development. 1998;125:3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- 8.Del Bene F. Tessmar-Raible K. Wittbrodt J. Direct interaction of geminin and Six3 in eye development. Nature. 2004;427:745–749. doi: 10.1038/nature02292. [DOI] [PubMed] [Google Scholar]
- 9.Luo L. Yang X. Takihara Y. Knoetgen H. Kessel M. The cell-cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature. 2004;427:749–753. doi: 10.1038/nature02305. [DOI] [PubMed] [Google Scholar]
- 10.Quinn LM. Herr A. McGarry TJ. Richardson H. The Drosophila Geminin homolog: roles for Geminin in limiting DNA replication, in anaphase and in neurogenesis. Genes Dev. 2001;15:2741–2754. doi: 10.1101/gad.916201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudderidge TJ. Stoeber K. Loddo M. Atkinson G. Fanshawe T. Griffiths DF. Williams GH. Mcm2, Geminin, and Ki67 define proliferative state and are prognostic markers in renal cell carcinoma. Clin Cancer Res. 2005;11:2510–2517. doi: 10.1158/1078-0432.CCR-04-1776. [DOI] [PubMed] [Google Scholar]
- 12.Xouri G. Lygerou Z. Nishitani H. Pachnis V. Nurse P. Taraviras S. Cdt1 and geminin are down-regulated upon cell cycle exit and are over-expressed in cancer-derived cell lines. Eur J Biochem. 2004;271:3368–3378. doi: 10.1111/j.1432-1033.2004.04271.x. [DOI] [PubMed] [Google Scholar]
- 13.McGarry TJ. Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez MA. Tachibana KE. Adams DJ. vander Weyden L. Hemberger M. Coleman N. Bradley A. Laskey RA. Geminin is essential to prevent endoreplication and to form pluripotent cells during mammalian development. Genes Dev. 2006;20:1880–1884. doi: 10.1101/gad.379706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara K. Nakayama KI. Nakayama K. Geminin is essential for the development of preimplantation mouse embryos. Genes Cells. 2006;11:1281–1293. doi: 10.1111/j.1365-2443.2006.01019.x. [DOI] [PubMed] [Google Scholar]
- 16.Mohamed OA. Clarke HJ. Dufort D. β-catenin signaling marks the prospective site of primitive streak formation in the mouse embryo. Dev Dyn. 2004;231:416–424. doi: 10.1002/dvdy.20135. [DOI] [PubMed] [Google Scholar]
- 17.Gratsch TE. De Boer LS. O'Shea KS. RNA inhibition of BMP-4 gene expression in postimplantation mouse embryos. Genesis. 2003;37:12–17. doi: 10.1002/gene.10221. [DOI] [PubMed] [Google Scholar]
- 18.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson DG. Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- 20.Gong SG. Guo C. Bmp4 gene is expressed at the putative site of fusion in the midfacial region. Differentiation. 2003;71:228–236. doi: 10.1046/j.1432-0436.2003.710304.x. [DOI] [PubMed] [Google Scholar]
- 21.Shawlot W. Deng JM. Behringer RR. Expression of the mouse cerberus-related gene, Cerr1, suggests a role in anterior neural induction and somitogenesis. Proc Natl Acad Sci USA. 1998;95:6198–6203. doi: 10.1073/pnas.95.11.6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis CA. Noble-Topham SE. Rossant J. Joyner AL. Expression of the homeo box-containing gene En-2 delineates a specific region of the developing mouse brain. Genes Dev. 1988;2:361–371. doi: 10.1101/gad.2.3.361. [DOI] [PubMed] [Google Scholar]
- 23.Hermesz E. Mackem S. Mahon KA. Rpx: a novel anterior-restricted homeobox gene progressively activated in the prechordal plate, anterior neural plate and Rathke's pouch of the mouse embryo. Development. 1996;122:41–52. doi: 10.1242/dev.122.1.41. [DOI] [PubMed] [Google Scholar]
- 24.Echelard Y. Epstein DJ. St-Jacques B. Shen L. Mohler J. McMahon JA. McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 25.Crossley PH. Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 26.Jowett AK. Vainio S. Ferguson MWJ. Sharpe PT. Thesleff I. Epithelial-mesenchymal interactions are required for msx 1 and msx 2 gene expression in the developing murine molar tooth. Development. 1993;117:461–470. doi: 10.1242/dev.117.2.461. [DOI] [PubMed] [Google Scholar]
- 27.Fischer A. Viebahn C. Blum M. FGF8 acts as a right determinant during establishment of the left-right axis in the rabbit. Curr Biol. 2002;12:1807–1816. doi: 10.1016/s0960-9822(02)01222-8. [DOI] [PubMed] [Google Scholar]
- 28.Gratsch TE. O'Shea KS. Noggin and chordin have distinct activities in promoting lineage commitment of mouse embryonic stem (ES) cells. Dev Biol. 2002;245:83–94. doi: 10.1006/dbio.2002.0629. [DOI] [PubMed] [Google Scholar]
- 29.O'Shea KS. De Boer LS. Slawny NA. Gratsch TE. Transplacental RNAi: deciphering gene function in the postimplantation-staged embryo. J Biomed Biotechnol. 2006;4:18657. doi: 10.1155/JBB/2006/18657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon GS. Viotti M. Hadjantonakis AK. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev Cell. 2008;15:509–520. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solnica-Krezel L. Conserved patterns of cell movements during vertebrate gastrulation. Curr Biol. 2005;15:R213–R228. doi: 10.1016/j.cub.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 32.Sauka-Spengler T. Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- 33.Seo S. Herr A. Lim JW. Richardson GA. Richardson H. Kroll KL. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev. 2005;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spella M. Britz O. Kotantaki P. Lygerou Z. Nishitani H. Ramsay RG. Flordellis C. Guillemot F. Mantamadiotis T. Taraviras S. Licensing regulators Geminin and Cdt1 identify progenitor cells of the mouse CNS in a specific phase of the cell cycle. Neuroscience. 2007;147:373–387. doi: 10.1016/j.neuroscience.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 35.Papanayotou C. Mey A. Birot AM. Saka Y. Boast S. Smith JC. Samarut J. Stern CD. A mechanism regulating the onset of Sox2 expression in the embryonic neural plate. PLoS Biol. 2008;6:e2. doi: 10.1371/journal.pbio.0060002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanagi K. Mizuno T. Tsuyama T. Tada S. Lida Y. Sugimoto A. Eki T. Enomoto T. Hanaoka F. Caenorhabditis elegans geminin homologue participates in cell cycle regulation and germ line development. J Biol Chem. 2005;280:19689–19694. doi: 10.1074/jbc.C500070200. [DOI] [PubMed] [Google Scholar]
- 37.Yang J. Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Thiery JP. Acloque H. Huang RY. Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat. 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 40.Li R. Liang J. Ni S. Zhou T. Qing X. Li H. He W. Chen J. Li F, et al. A Mesenchymal-to-Epithelial Transition Initiates and Is Required for the Nuclear Reprogramming of Mouse Fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 41.Samavarchi-Tehrani P. Golipour A. David L. Sung HK. Beyer TA. Datti A. Woltjen K. Nagy A. Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 42.Kemler R. Hierholzer A. Kanzler B. Kuppig S. Hansen K. Taketo MM. de Vries WN. Knowles BB. Solter D. Stabilization of beta-catenin in the mouse zygote leads to premature epithelial-mesenchymal transition in the epiblast. Development. 2004;131:5817–5824. doi: 10.1242/dev.01458. [DOI] [PubMed] [Google Scholar]
- 43.Roberts C. Sutherland HF. Farmer H. Kimber W. Halford S. Carey A. Brickman JM. Wynshaw-Boris A. Scambler PJ. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesendodermal derivatives prior to early embryonic lethality. Mol Cell Biol. 2002;22:2318–2328. doi: 10.1128/MCB.22.7.2318-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng CX. Wynshaw-Boris A. Shen MM. Daugherty C. Ornitz DM. Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 45.Yamaguchi TP. Harpal K. Henkemeyer M. Rossant J J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 46.Sun X. Meyers EN. Lewandoski M. Martin GR. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnold SJ. Hofmann UK. Bikoff EK. Robertson EJ. Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development. 2008;135:501–511. doi: 10.1242/dev.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carver EA. Jiang R. Lan Y. Oram KF. Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X. Huang S. Ma J. Li C. Zhang Y. Luo L. NF-kappa B and Snail1a coordinate the cell cycle with gastrulation. J Cell Biol. 2009;184:805–815. doi: 10.1083/jcb.200806074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tam PP. Loebel DA. Gene function in mouse embryogenesis: get set for gastrulation. Nat Rev Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- 51.Placzek M. Dodd J. Jessell TM. The case for floor plate induction by the notochord. Curr Opin Neurobiol. 2000;10:15–22. doi: 10.1016/s0959-4388(99)00060-4. [DOI] [PubMed] [Google Scholar]
- 52.Ybot-Gonzalez P. Gaston-Massuet C. Girdler G. Klingensmith J. Arkell R. Greene ND. Copp AJ. Neural plate morphogenesis during mouse neurulation is regulated by antagonism of Bmp signaling. Development. 2007;134:3203–3211. doi: 10.1242/dev.008177. [DOI] [PubMed] [Google Scholar]
- 53.Duband JL. Neural crest delamination and migration: integrating regulations of cell interactions, locomotion, survival, and fate. Adv Exp Med Biol. 2006;589:45–77. doi: 10.1007/978-0-387-46954-6_4. [DOI] [PubMed] [Google Scholar]
- 54.Anderson RM. Stottmann RW. Choi M. Klingensmith J. Endogenous bone morphogenetic protein antagonists regulate mammalian neural crest generation and survival. Dev Dyn. 2006;235:2507–2520. doi: 10.1002/dvdy.20891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raible DW. Development of the neural crest: achieving specificity in regulatory pathways. Curr Opin Cell Biol. 2006;18:698–703. doi: 10.1016/j.ceb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 56.Patthey C. Edlund T. Gunhaga L. Wnt-regulated temporal control of BMP exposure directs the choice between neural plate border and epidermal fate. Development. 2009;136:73–83. doi: 10.1242/dev.025890. [DOI] [PubMed] [Google Scholar]
- 57.Steventon B. Araya C. Linker C. Kuriyama S. Mayor R. Differential requirements of BMP and Wnt signalling during gastrulation and neurulation define two steps in neural crest induction. Development. 2009;136:771–779. doi: 10.1242/dev.029017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hussein SM. Duff EK. Sirard C. Smad4 and beta-catenin co-activators functionally interact with lymphoid-enhancing factor to regulate graded expression of Msx2. J Biol Chem. 2003;278:48805–48814. doi: 10.1074/jbc.M305472200. [DOI] [PubMed] [Google Scholar]
- 59.Conacci-Sorrell M. Simcha I. Ben-Yedidia T. Blechman J. Savagner P. Ben-Ze'ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163:847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burstyn-Cohen T. Stanleigh J. Sela-Donenfeld D. Kalcheim C. Canonical Wnt activity regulates trunk neural crest delamination linking BMP/noggin signaling with G1/S transition. Development. 2004;131:5327–5339. doi: 10.1242/dev.01424. [DOI] [PubMed] [Google Scholar]
- 61.Burstyn-Cohen T. Kalcheim C. Association between the cell cycle and neural crest delamination through specific regulation of G1/S transition. Dev Cell. 2002;3:383–395. doi: 10.1016/s1534-5807(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 62.Spella M. Kyrousi C. Kritikou E. Stathopoulou A. Guillemot F. Kioussis D. Pachnis V. Lygerou Z. Taraviras S. Geminin regulates cortical progenitor proliferation and differentiation. Stem Cells. 2011;29:1269–1282. doi: 10.1002/stem.678. [DOI] [PubMed] [Google Scholar]
- 63.Doble BW. Patel S. Wood GA. Kockeritz LK. Woodgett JR. Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev Cell. 2007;12:957–971. doi: 10.1016/j.devcel.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haegel H. Larue L. Ohsugi M. Fedorov L. Herrenknecht K. Kemler R. Lack of β-catenin affects mouse development at gastrulation. Development. 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 65.Popperl H. Schmidt C. Wilson V. Hume CR. Dodd J. Krumlauf R. Beddington RS. Misexpression of Cwnt8c in the mouse induces an ectopic embryonic axis and causes a truncation of the anterior neuroectoderm. Development. 1997;124:2997–3005. doi: 10.1242/dev.124.15.2997. [DOI] [PubMed] [Google Scholar]
- 66.Ishikawa TO. Tamai Y. Li Q. Oshima M. Taketo MM. Requirement for tumor suppressor Apc in the morphogenesis of anterior and ventral mouse embryo. Dev Biol. 2003;253:230–246. doi: 10.1016/s0012-1606(02)00020-9. [DOI] [PubMed] [Google Scholar]
- 67.Zeng L. Fagotto F. Zhang T. Hsu W. Vasicek TJ. Perry WL., 3rd Lee JJ. Tilghman SM. Gumbiner BM. Costantini F. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90:181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 68.Merrill BJ. Pasoli HA. Polak L. Rendl M. Garcia-Garcia MJ. Anderson KV. Fuchs E. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- 69.Taylor JJ. Wang T. Kroll KL. Tcf- and Vent-binding sites regulate neural-specific geminin expression in the gastrula embryo. Dev Biol. 2006;289:494–506. doi: 10.1016/j.ydbio.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 70.Stemmer V. de Craene B. Berx G. Behrens J. Snail promotes Wnt target gene expression and interacts with beta-catenin. Oncogene. 2008;27:5075–5080. doi: 10.1038/onc.2008.140. [DOI] [PubMed] [Google Scholar]
- 71.Mishina Y. Suzuki A. Ueno N. Behringer RR. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Development. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- 72.Fujiwara T. Dehart DB. Sulik KK. Hogan BL. Distinct requirements for extra-embryonic and embryonic bone morphogenetic protein 4 in the formation of the node and primitive streak and coordination of left-right asymmetry in the mouse. Development. 2002;129:4685–4696. doi: 10.1242/dev.129.20.4685. [DOI] [PubMed] [Google Scholar]
- 73.Rogers CD. Harafuji N. Archer T. Cunningham DD. Casey ES. Xenopus Sox3 activates sox2 and geminin and indirectly represses Xvent2 expression to induce neural progenitor formation at the expense of non-neural ectodermal derivatives. Mech Dev. 2009;126:42–55. doi: 10.1016/j.mod.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo L. Kessel M. Geminin coordinates cell cycle and developmental control. Cell Cycle. 2004;3:711–714. [PubMed] [Google Scholar]
- 75.Schultz KM. Banisadr G. Lastra RO. McGuire T. Kessler JA. Miller RJ. McGarry TJ. Geminin-deficient neural stem cells exhibit normal cell division and normal neurogenesis. PLoS One. 2011;6:e17736. doi: 10.1371/journal.pone.0017736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seo S. Richardson GA. Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005b;132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- 77.Park JI. Venteicher AS. Hong JY. Choi J. Jun S. Shkreli M. Chang W. Meng Z. Cheung P, et al. Telomerase modulates Wnt signaling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barker N. Hurlstone A. Musisi H. Miles A. Bienz M. Clevers H. The chromatin remodeling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eroglu B. Wang G. Tu N. Sun X. Mivechi NF. Critical role of Brg1 member of the SWI/SNF chromatin remodeling complex during neurogenesis and neural crest induction in zebrafish. Dev Dyn. 2006;235:2722–2735. doi: 10.1002/dvdy.20911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.