Abstract
Genome-wide structural and gene content variations are hypothesized to drive important phenotypic variation within a species. Structural and gene content variations were assessed among four soybean (Glycine max) genotypes using array hybridization and targeted resequencing. Many chromosomes exhibited relatively low rates of structural variation (SV) among genotypes. However, several regions exhibited both copy number and presence-absence variation, the most prominent found on chromosomes 3, 6, 7, 16, and 18. Interestingly, the regions most enriched for SV were specifically localized to gene-rich regions that harbor clustered multigene families. The most abundant classes of gene families associated with these regions were the nucleotide-binding and receptor-like protein classes, both of which are important for plant biotic defense. The colocalization of SV with plant defense response signal transduction pathways provides insight into the mechanisms of soybean resistance gene evolution and may inform the development of new approaches to resistance gene cloning.
Genetic variation within and between species is most commonly quantified by single nucleotide polymorphisms (SNPs). There has been increased interest in recent years to also resolve genetic differences in terms of structural variation (SV), which includes copy number variation (CNV) caused by large insertions and deletions, and other types of rearrangements such as inversions and translocations. The copy number of a specific gene or gene family has been associated with variation for specific traits, such as the digestion of starchy foods in humans (Perry et al., 2007), boron toxicity tolerance and winter hardiness in barley (Hordeum vulgare; Sutton et al., 2007; Knox et al., 2010), dwarfism and flowering time in wheat (Triticum aestivum; Pearce et al., 2011; Díaz et al., 2012), and insecticide and virus resistance in Drosophila melanogaster (Schmidt et al., 2010; Magwire et al., 2011). Genomic SV is thought to be an important factor in determining phenotypic variation for a wide range of traits (for review, see Stankiewicz and Lupski, 2010).
SV studies have been published in various invertebrate (Dopman and Hartl, 2007; Emerson et al., 2008; Maydan et al., 2010) and mammalian (Graubert et al., 2007; Guryev et al., 2008; Lee et al., 2008; Perry et al., 2008; Gazave et al., 2011; Golzio et al., 2012) systems; however, a high proportion of such studies have been conducted in humans, where there is interest in identifying associations between SV, complex diseases, and neurological disorders (Conrad et al., 2010; Craddock et al., 2010; Sudmant et al., 2010; Girirajan et al., 2011). Domesticated animal species, including dog, cow, and silkworm, have also been the focus of recent investigations of SV (Chen et al., 2009; Nicholas et al., 2009, 2011; Liu et al., 2010; Sakudoh et al., 2011; Clop et al., 2012). CNV have been identified within genes and gene families associated with specific biological functions, such as immunity. Some evidence from these studies suggests that phenotypic variation caused by CNV can rapidly emerge and be driven to fixation by breeders.
Recent studies in maize (Zea mays) have explored the exceptionally high rates of SV between inbred accessions (Springer et al., 2009; Beló et al., 2010; Swanson-Wagner et al., 2010). The maize profile indicates that there are continuously high levels of SV between accessions throughout all 10 chromosomes, interspersed by relatively small regions of conservation thought to be regions of identity by descent. Other than this work in maize and work in Arabidopsis (Arabidopsis thaliana; Santuari et al., 2010; Cao et al., 2011; Gan et al., 2011; Lu et al., 2012), rice (Oryza sativa; Yu et al., 2011; Xu et al., 2012), and sorghum (Sorghum bicolor; Zheng et al., 2011), relatively little is known about the intraspecific structural genomic variation within plant species. Maize is a primarily outcrossing species known to have a remarkably diverse germplasm (Wright et al., 2005; Gore et al., 2009); most other domesticated plant and crop species would be expected to have lower rates of SV as a consequence of their narrower genetic base. Soybean (Glycine max) is an interesting system for comparison. Soybean is a self-pollinating species with a comparatively narrow genetic base that has experienced severe genetic bottlenecks during domestication (Hyten et al., 2006). Soybean SNP rates among accessions are relatively low, typically on the order of one SNP per kb (Hyten et al., 2006; Lam et al., 2010). Given the low polymorphism rate and narrow genetic base, one might surmise that soybean accessions are similarly devoid of SV. However, a recent study using microarray-based comparative genomic hybridization (CGH) analysis found surprisingly high rates of SV between two cultivars (‘Kingwa’ and ‘Williams’; Haun et al., 2011). Additionally, regions of SV were identified within sublineages of the reference cultivar (‘Williams 82’), including several genic loci that exhibited presence-absence variation (PAV) among the different Williams 82 individuals (Haun et al., 2011). PAV is a subclass of CNV in which a specific gene or other sequence is present in some accessions and entirely absent in others (Springer et al., 2009).
In this study, we sought to define the range of SV between four genetically diverse soybean accessions: ‘Archer,’ ‘Minsoy,’ ‘Noir 1,’ and Williams 82. Minsoy and Noir 1 are plant introductions, while Archer is a North American cultivar. These three genotypes were of particular interest because they are the parental lines for the recombinant inbred line populations utilized in the first agronomic quantitative trait loci (QTL) analyses of soybean (Lark et al., 1995; Orf et al., 1999) and represent a wide range of soybean sequence diversity (Zhu et al., 2003). Zhu et al. (2003) reported that approximately two-thirds of the common soybean SNPs found in a set of 25 diverse lines are polymorphic among these three genotypes. Williams 82 is a modern soybean cultivar that provided the reference genome sequence (Schmutz et al., 2010). Coarse structural differences between the four genotypes (i.e. CNV) were resolved using CGH technology, and specific gene content variants (i.e. gene PAV) were identified using exome-resequencing analyses. These data were used to catalog the set of genes located within regions enriched for SV, giving new insight into the mechanisms and forces that may be driving SV in soybean.
RESULTS
SNP Variation among Four Soybean Genotypes
Little is known about the genomic SV among soybean accessions. Furthermore, the relationship between genomic SV and haplotype variation is essentially unstudied. To investigate this relationship, we generated whole-genome CGH data and comprehensive SNP genotype data on the four soybean genotypes Archer, Minsoy, Noir 1, and Williams 82. There is known to be genetic heterogeneity within some soybean cultivars (Fasoula and Boerma, 2007; Haun et al., 2011; Varala et al., 2011; Yates et al., 2012), meaning that there can be differences between individual plants or sublines within an accession. Therefore, the four genotypes in this study were each represented by a single plant. Herein, the four individuals are referred to by their abbreviated or cultivar name (Archer, Minsoy, Noir 1, and Wm82) for brevity and simplicity, but their full subline names are given in the Materials and Methods.
Comprehensive SNP genotyping data were generated using the Illumina Infinium platform for soybean, which consists of approximately 44,000 SNPs spaced at regular intervals across the soybean genome. The SNP profiles of the Archer, Minsoy, and Noir 1 individuals were compared with the Wm82 individual (Fig. 1). The three genotypes all displayed discontinuous patterns of polymorphism along the 20 chromosomes. Archer showed the lowest level of polymorphism relative to Wm82, including several stretches that appeared to be shared haplotypes across long chromosomal regions (e.g. chromosomes 3, 19, and 20 in Fig. 1). These shared haplotypes may be regions of identity by descent, as Williams 82 was the Phytophthora root rot resistance donor (Rps1k) in the Archer pedigree (Cianzio et al., 1991). Minsoy and Noir 1 also appeared to have some haplotype regions shared with Wm82, although to a lesser degree than Archer (Fig. 1).
Figure 1.
SNP genotyping reveals regions of conservation and divergence between Wm82 and Archer (A), Wm82 and Minsoy (B), and Wm82 and Noir 1 (C). Gray spots indicate matching SNPs, and black spots indicate polymorphic SNPs between genotypes. The spots are jittered along the x axis to enhance the resolution of the data points.
Genomic SV among Four Soybean Genotypes
To gauge SV between genotypes, Archer, Minsoy, and Noir 1 were each hybridized with Wm82 as the reference in CGH experiments (Supplemental Fig. S1). Among the three comparisons, the number of genomic segments exhibiting significant CNV ranged from 188 to 267 segments per genotype comparison (Table I). The CNV false discovery rate based on technical variables is likely low, as a control Wm82-Wm82 self-hybridization identified only 13 significant CNV (Table I).
Table I. Summary statistics of soybean CNV number and size.
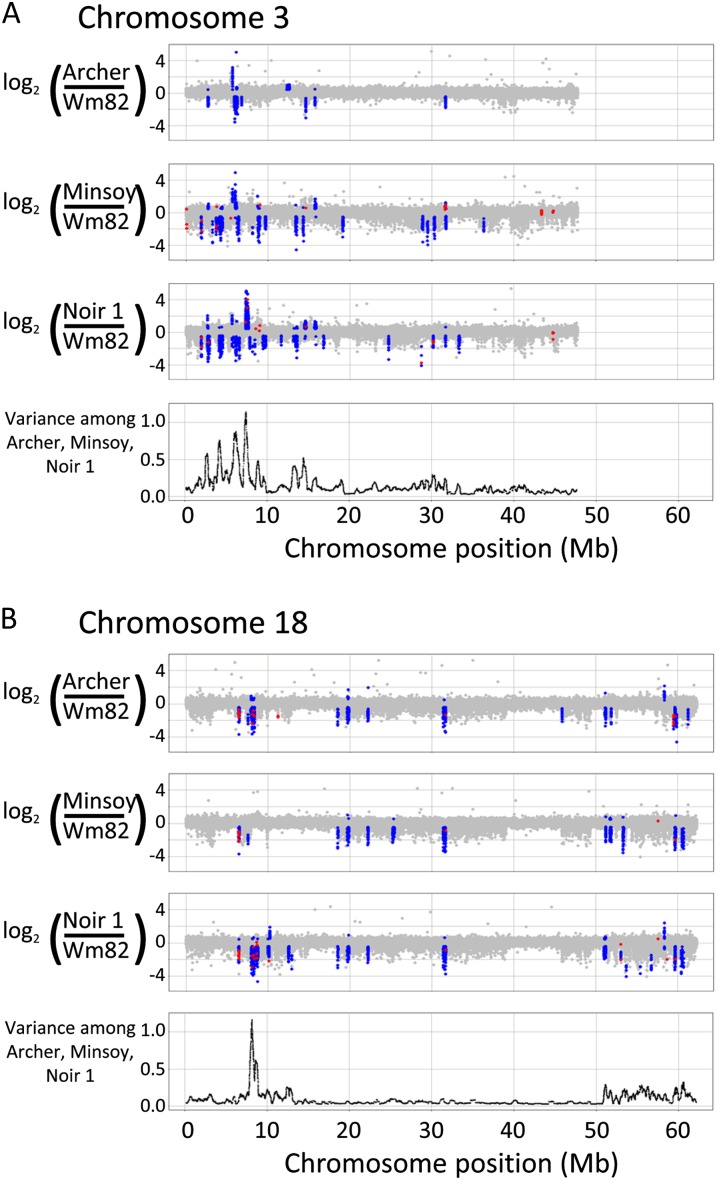
The median size of a CNV segment was approximately 18 to 23 kb for all three comparisons (Table I). The distribution of significant CNV was discontinuous throughout each comparison (Supplemental Fig. S1). The chromosomes exhibited differing levels of SV, including whole chromosomes with little to no evidence of SV (e.g. chromosomes 5 and 11) and chromosomes with extended regions of SV (e.g. chromosomes 3 and 18; Fig. 2).
Figure 2.
CNV among soybean genotypes on chromosomes 3 (A) and 18 (B). Log2 ratios between each genotype relative to the Wm82 reference are shown. Blue spots indicate probes within significant CNV segments with values beyond threshold. Red spots indicate probes within PAV genes as determined by exome-resequencing analysis. The plots at the bottom of both A and B indicate the average variance (along a sliding window of 250 probes) between the log2 ratios of the Archer, Minsoy, and Noir 1 hybridizations.
Hundreds of genes colocalized to regions of SV. The gene models overlapping with significant CNV regions were identified for each genotype comparison (Supplemental Table S1). In total, 672 different gene models overlapped with CNV regions in at least one of the three comparisons (Supplemental Table S1). Over one-third (37%) of the 672 gene models were significant in more than one comparison, and 120 were significant in all three comparisons (Supplemental Fig. S2). The false discovery rate is likely to be low, as the control Wm82-Wm82 self-hybridization identified only one gene model within a significant CNV. Furthermore, CGH technical replications of Minsoy-Wm82 and Noir 1-Wm82 were performed to estimate the reproducibility of these discoveries. While the technical replicate hybridizations had more noise than the original hybridizations, the CNV patterns were essentially the same (Supplemental Fig. S3). Importantly, the vast majority of gene models identified within the CNV regions in the technical replicates were also significant in the original hybridizations (77% for Minsoy-Wm82 and 77% for Noir 1-Wm82; Supplemental Table S1).
Most of the polymorphic loci consist of DownCNV (Table I), which can be interpreted as regions in which the tested genotype has fewer DNA copies than the reference genotype Wm82 or that hybridize less efficiently than Wm82 (presumably due to nucleotide sequence polymorphisms). The DownCNV can be visualized as downward peaks in Figure 2 and Supplemental Figure S1. The high frequency of DownCNV relative to UpCNV is expected, as the microarray was developed based on the Williams 82 reference genome sequence (Schmutz et al., 2010). Significant UpCNV were observed in some instances (approximately 12% of all CNV; Table I), most prominently along chromosomes 3 and 7. These UpCNV occur within regions of known intracultivar heterogeneity, in which the Williams 82 reference genome sequence does not perfectly match the Wm82-ISU-01 haplotype (Haun et al., 2011). The UpCNV likely represent genomic regions that are absent from Wm82-ISU-01, present in the Williams 82 reference sequence, and also present in the respective genotypes (Archer, Minsoy, and/or Noir 1) in which the UpCNV is observed. There are also some examples of UpCNV that do not occur within regions of known heterogeneity (e.g. the UpCNV on chromosome 12) and that possibly represent copy number gains in the respective genotypes relative to the Williams 82 genome.
The relationship between the genomic SV profiles and the Infinium SNP variation profiles is shown in Supplemental Figure S4. The interpretation of this comparison is complicated because the Infinium SNP assays were selected by virtue of being highly polymorphic, while the CGH probes were selected without regard to their potential to underlie CNV. This ascertainment bias leads to a distortion in the fraction of data that the two technologies detect as polymorphic. Despite this complication, some general trends were observed. SNP and SV rates were generally coincident along each chromosome, such that regions of high SV were localized to regions with high rates of nucleotide polymorphism. The colocalization of SNP and CNV suggests that areas rich for both SNP and SV represent divergent haplotypes for a given comparison. However, exceptional instances were observed in which regions with profound SNP polymorphism rates exhibited little SV (e.g. Archer-Wm82 chromosome 1) or regions with abundant CNV exhibited little to no SNP variation (e.g. Archer-Wm82 chromosome 3; Supplemental Fig. S4).
Exome capture and resequencing data were generated for the four soybean genotypes to validate the array CGH data and further evaluate gene content variation between the lines using a fixed exon content measure. A stringent analysis pipeline was developed to identify gene content variation among the four genotypes based on the number of normalized exome read counts mapping to each gene model (for a description of the analysis details, see “Materials and Methods”). This allowed for the identification of a subset of genes that exhibited high read counts (more than 30) in at least one genotype and zero read counts in at least one other genotype. These 133 genes make up the high-confidence list of PAV. The locations of the PAV along each chromosome are shown as red spots in Figure 2 and Supplemental Figure S1. The gene models and presence-absence profiles of these 133 genes are shown in Supplemental Table S2, and the distribution of “absent” genes among the four genotypes is shown in Supplemental Figure S5.
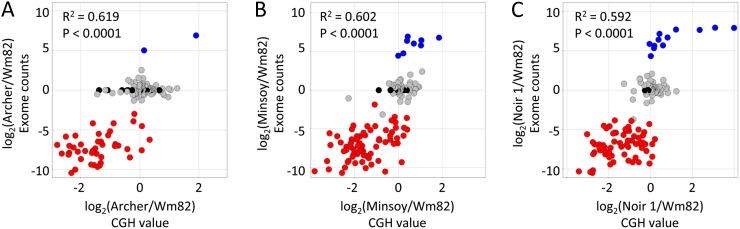
The CGH and exome-resequencing data were compared to cross-validate the PAV calls on the 133 genes. The log2 ratios of the exome-resequencing counts and CGH data for each gene × genotype comparison are shown in Figure 3. There was a strong significant correlation (P < 0.0001) between the CGH and exome-resequencing platforms for all of the genotype comparisons. Furthermore, the majority (58%) of the PAV genes called absent in Archer, Minsoy, or Noir 1 relative to Wm82 were located within significant CNV segments.
Figure 3.
Cross-validation of CGH and exome-resequencing data for the 133 genes identified as PAV in the exome-resequencing data set. Archer/Wm82 (A), Minsoy/Wm82 (B), and Noir 1/Wm82 (C) log2 ratios based on CGH (x axis) and exome-resequencing counts (y axis) each exhibited significant correlations. Each spot represents one of the 133 genes. Spot coloration is based on the gene presence/absence call in the exome-resequencing data. Red spots indicate genes called absent in Archer (A), Minsoy (B), or Noir 1 (C). Blue spots indicate genes called present in the respective genotype and absent in Wm82. Gray spots indicate genes called present in both the respective genotype and Wm82. Black spots indicate genes called absent in both the respective genotype and Wm82.
The 133 PAV genes identified represent a high-confidence list but almost certainly underestimate the number of genes that have full or partial gene content variation among the tested genotypes (for further explanation, see Supplemental Materials and Methods S1). Additionally, any novel gains in gene content exhibited by the lines would be missing from this analysis because the exon capture can only deliver sequences homologous to the probes designed from the Williams 82 reference sequence.
PCR assays were conducted to estimate the presence-absence distribution of high-confidence PAV genes in a sample of 31 germplasm accessions, including 10 modern cultivars, 17 North American ancestors, and four landraces. PCR primers were designed within the predicted coding regions of 13 putative PAV genes (Supplemental Table S3); these 13 genes also showed evidence of CNV in the CGH experiment. The presence-absence state of these 13 genes was tested by PCR on the full set of 31 accessions (Supplemental Table S4; Supplemental Fig. S6). A wide range of gene content variation among the accessions was observed for the genes. Two genes, Glyma05g24800 and Glyma17g18890, showed present amplicons in all of the accessions except for the genotype in which absence was initially observed (Archer and Noir 1, respectively). Thus, these two genes are likely to be rare variants in which the absent state is found in few genotypes. The remaining 11 genes exhibited absent rates ranging from 13% to 65% of the accessions (Supplemental Table S5).
High Levels of SV Associated with Disease Resistance Genes
The full set of genes associated with CNV or PAV regions, along with their Gene Ontology (GO) annotations and protein family prediction, is shown in Supplemental Table S5. GO analyses revealed 24 categories that were significantly enriched and nine categories that were significantly depleted for genes within CNV regions relative to all genes not found within significant CNV regions (Fisher’s exact test; multiple testing adjusted P < 0.05; Supplemental Table S6). Although the level of significance varied for each genotypic comparison, the direction (enrichment or depletion) was consistent among the Archer, Minsoy, and Noir 1 comparisons with Wm82 (Supplemental Table S6).
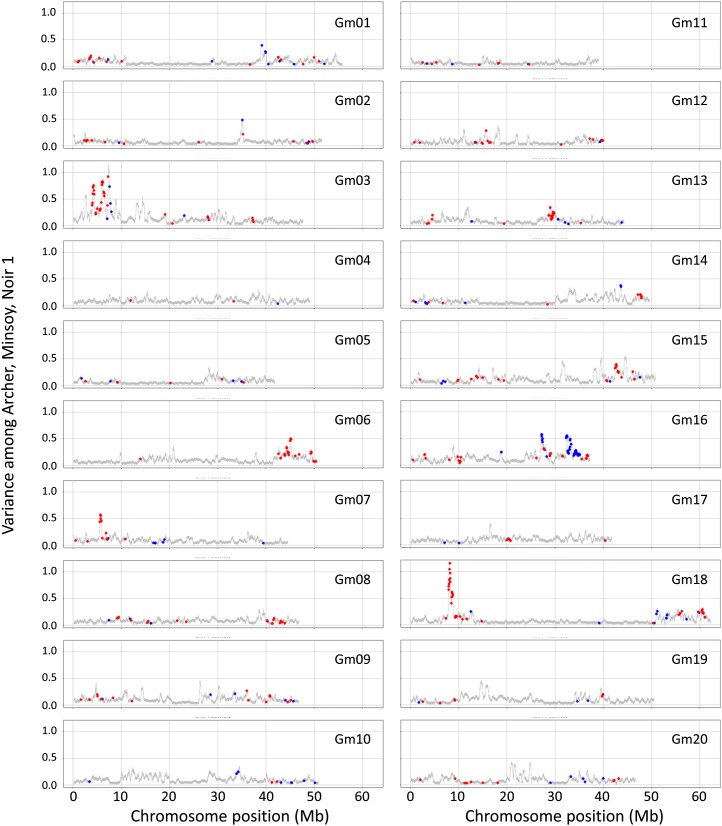
Genes within regions of structural and gene content variation frequently had potential functions in disease resistance and response to biotic stress. GO categories with the greatest enrichment of genes in CNV regions were related to plant-pathogen interactions and included “defense response,” “plant-type hypersensitive response,” “programmed cell death,” and “apoptosis.” A specific enrichment was observed for genes encoding nucleotide-binding (NB) proteins and receptor-like proteins (RLPs; Fisher’s exact test; P = 1.87 × 10−58 and P = 4.32 × 10−122, respectively; Table II), which often function in disease resistance (Kruijt et al., 2005; DeYoung and Innes, 2006). Enrichment of NB- and RLP-encoding genes was also observed in PAV (Fisher’s exact test; P < 0.01; Supplemental Table S6). The full list and genomic positions of soybean gene models identified as NB (392 gene models) and RLP (220 gene models) are shown in Supplemental Tables S7 and S8, respectively. Figure 4 shows the colocalization of these gene classes with CNV spikes. The hybridization variances among the Archer-Wm82, Minsoy-Wm82, and Noir 1-Wm82 comparisons are shown, revealing the approximate locations of structurally conserved regions (shown as relatively flat intervals) as well as structurally diverged regions (shown as peaks or peak clusters) among the Archer, Minsoy, and Noir 1 genomes. The colored spots indicate the locations of all annotated NB (red) and RLP (blue) genes in the genome.
Table II. Enrichment of CNV within subclasses of genes.
For all eligible NB-encoding and RLP-encoding genes, significant differences between the number of genes for a given gene category located in CNV regions in comparison with non-CNV regions were determined by Fisher’s exact test (*P < 0.05, **P < 0.001). na, Not applicable.
| Genes | All Eligible Genes |
NB Encoding |
RLP Encoding |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | CNV | %CNVa | Fold Changeb | Total | CNV | %CNV | Fold Change | Total | CNV | %CNV | Fold Change | |
| Unique genes | 10,096 | 188** | 1.9 | 1.3 | 91 | 10* | 11 | 7.6 | 36 | 10* | 28 | 19 |
| Small multigene family (2–10) | 28,299 | 247** | 0.9 | 0.6 | 98 | 16 | 16 | 11 | 48 | 0** | 0 | 0 |
| Large multigene family (more than 10) | 7,858 | 237** | 3.0 | 2.1 | 201 | 47* | 23 | 16 | 136 | 89** | 65 | 45 |
| Isolated multigene family members | 30,928 | 196** | 0.6 | 0.4 | 93 | 3** | 3.2 | 2.2 | 59 | 4** | 6.8 | 4.7 |
| Clustered multigene family members | 5,229 | 288** | 5.5 | 3.8 | 206 | 60** | 29 | 20 | 125 | 85** | 68 | 47 |
| Genes containing tandem repeats | 13,684 | 217 | 1.6 | 1.1 | 124 | 27 | 22 | 15 | 85 | 54** | 64 | 44 |
| Genes with nearby TEs | 11,347 | 250** | 2.2 | 1.5 | 152 | 22 | 14 | 10 | 65 | 28 | 43 | 30 |
| All CNV-eligible genes | 46,253 | 672 | 1.5 | na | 385 | 73 | 19 | 13 | 220 | 99 | 43 | 31 |
%CNV is calculated as the number of CNV in a particular category divided by the total number of genes in that category.
Fold change is relative to the genome-wide %CNV: the %CNV for a particular category divided by genome-wide %CNV (1.5%).
Figure 4.
Colocalization of genome SV within defense gene clusters. Archer, Minsoy, and Noir 1 were each independently hybridized to the CGH microarray, with Wm82 serving as the constant reference. The variance between the log2 ratios of the Archer, Minsoy, and Noir 1 hybridizations was calculated for each probe on the microarray. The average variance along a sliding window of 250 probes is shown on the y axis. Colored spots indicate the probes nearest to the physical positions of genes defined within the NB (red spots) or RLP (blue spots) classes. All soybean NB and RLP gene positions are shown. Regions with high SV tend to localize to the NB- and/or RLP-encoding gene clusters (note the prominent peaks on chromosomes 3, 6, 7, 16, and 18).
The locations of structurally diverged regions were compared with the results of previous soybean genetic mapping studies, which are publicly available at http://www.soybase.org. The regions with the greatest amplitude of CNV variance account for 94 centimorgans of the soybean composite genetic map, which is less than 4% of the total map. Previous genetic mapping experiments in soybean indicate that multiple QTL and/or genes for disease resistance map to nearly all of the regions with highest CNV variance (genetic mapping data obtained from http://www.soybase.org). Fourteen percent (43 of 311) of QTL for disease resistance map to the regions with highest CNV variance, while only 7% (85 of 1,221) of non-disease-related QTL map to these regions. This suggests that a large portion of the qualitative and quantitative variation in disease resistance may be derived from gene content variation in NB- or RLP-encoding gene clusters.
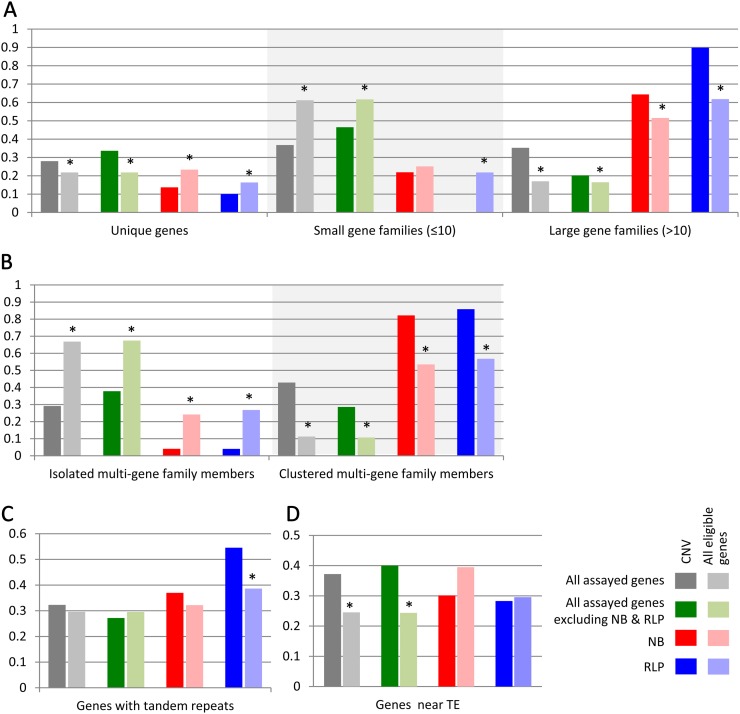
The physical arrangement, copy number, and repetitive nature of genes residing within regions of CNV were assessed to determine whether the enrichment of the NB- and RLP-encoding genes may be influenced by factors other than their functions in disease resistance and the selection pressures potentially associated with those functions (Bishop et al., 2000; Mondragón-Palomino et al., 2002; Chen et al., 2010). Gene family size appeared to be one such factor; CNV regions were associated with large gene families more often than predicted by random expectations (Table II; Supplemental Table S9). Seventeen percent of predicted genes in the soybean genome are members of large multigene families (families with more than 10 members). In comparison, 35% of genes within CNV regions in our study were members of large multigene families (Fig. 5A).
Figure 5.
Enrichment or depletion of CNV in comparison with expectation (all eligible genes) for each gene class. Genes were divided by gene class (e.g. all genes, NB or RLP) as well as groups based on structural characteristics (e.g. size of the gene family). The proportion of genes with a particular structural feature within each gene class is presented. This proportion is presented for genes located within CNV as well as for all genes assayed for CGH comparisons. Significant differences (Fisher’s exact test; P < 0.05) between the proportion of genes within CNV regions and within all eligible genes for each category are indicated by asterisks. A, Proportion of genes that are unique or in small or large gene families. B, Proportion of genes that are clustered or isolated members of multigene families. C, Proportion of genes that contain tandem repeats. D, Proportion of genes within 5 kb of a TE.
The relationship between large gene families and CNV regions appears to be primarily due to the NB- and RLP-encoding gene families (Fig. 5A). When these genes, representing 4% of the large multigene families, 0.5% of the small multigene families, and 1% of the unique genes, are removed from the analysis, the frequency of large gene family members within CNV is near the frequency that would be expected at random. These correlations indicate that CNV occur more frequently within the NB- and RLP-encoding families than other large multigene families. It is evident that not all large gene families are associated with CNV regions and that there are requirements for CNV beyond gene family size.
The physical distribution of the members of multigene families appears to be a key component of the SV-enriched regions. Only members of multigene families that are located within clusters tend to be associated with CNV regions (Fig. 5B); isolated family members that are not clustered do not often associate with CNV regions. This trend is observed across all classes of genes (Fig. 5B); the tightly clustered NB- and/or RLP-encoding families on chromosomes 3, 6, 7, 16, and 18 are notable examples (Fig. 4).
Genes that contain tandem repeats within CNV regions were identified at a slightly higher frequency than expected, with 30% of all genes containing tandem repeats and 32% of genes with CNV containing tandem repeats (Fig. 5C; Supplemental Table S10). In comparison with all genes, there is a slightly higher percentage of tandem repeats in the NB and RLP classes (32% and 39%, respectively). In addition, there is a higher percentage of tandem repeats in both NB- and RLP-encoding genes within CNV regions (37% and 55%, respectively; Fig. 5C). This association of tandem repeats to genes within CNV is specific to the NB and RLP classes and is not observed in the remainder of the genic sequences (Fig. 5C), indicating that the presence of tandem repeats without other factors attributed to NB- and RLP-encoding genes does not influence CNV.
Transposable elements (TEs) can result in duplication, deletion, or transposition of nearby non-TE genes through a variety of mechanisms (Bennetzen, 2000). Possible evidence of TE-mediated transposition and deletion of NB-leucine-rich repeat (LRR)-encoding genes in soybean has been reported previously (Innes et al., 2008; Wawrzynski et al., 2008). We observed an enrichment of nearby TEs (Du et al., 2010) in genes located within CNV regions in comparison with genes not located within CNV regions (Fisher’s exact test; P < 0.05; Fig. 5D). However, NB- and RLP-encoding genes within CNV regions have no significant enrichment for nearby TEs (Table II; Fig. 5D). These observations indicate that while TEs may play an important role in generating CNV, the CNV regions surrounding NB- and RLP-encoding genes are either primarily generated by other mechanisms or nearby TEs were sufficiently fragmented to not be detectable by methods implemented to identify partial TEs for inclusion in SoyTEdb (Du et al., 2010).
Confirmation of CGH Trends on Additional Soybean Accessions
We performed additional CGH experiments with additional soybean accessions to test whether the trends observed in the CGH experiments involving Archer, Minsoy, Noir 1, and Wm82 would also be observed in other genotype comparisons. The first two hybridizations utilized Wm82 as the reference dye and genotypes ‘Essex’ and ‘Richland’ as the experimental dye. Essex is a cultivar release from 1973 (Smith and Camper, 1973), and Richland is a North American ancestor accession. A third hybridization directly compared the cultivars Archer and ‘M92-220’, which is a recently developed cultivar derived from the 2006 Crop Improvement Association seed stock of cv ‘MN1302’ (Orf and Denny, 2004). The last hybridization used for comparison involved accessions Kingwa and Williams. This hybridization was described previously (Haun et al., 2011) but was never analyzed for the patterns of CNV regional abundance.
The CNV frequency and size for each of the hybridizations are shown in Supplemental Table S11. The gene models associated with significant CNV are shown in Supplemental Table S12, and the enriched GO and Pfam categories are shown in Supplemental Table S13. The patterns and trends observed in these additional hybridizations closely paralleled the results of the Archer, Minsoy, Noir 1, and Wm82 experiments. For each of the four additional hybridizations, defense-related GO terms and Pfam domains associated with plant resistance genes had significant overrepresentation within genes in CNV regions (Supplemental Table S13). Combining the Essex-Wm82 and Richland-Wm82 data with the previous experiments on Archer, Noir 1, and Minsoy, 782 different gene models overlapped with CNV regions in at least one of the five comparisons (Tables S1 and S12). Nearly half (46%) of these gene models were significant in more than one comparison (Supplemental Fig. S7).
DISCUSSION
Distribution and Rates of SV in the Soybean Genome
The CNV profiles in this study indicate that soybean has relatively long chromosomal regions (and nearly entire chromosomes) that exhibit virtually no SV among genotypes, interspersed with pockets of high SV ranging from several kb to greater than 10 Mb in length. DownCNV (segment loss relative to Wm82) were much more abundant than UpCNV (segment gain relative to Wm82). This is expected, considering that the reference dye in all of the hybridizations was Wm82, which serves as the reference sequence used to design the microarray platform. The relative abundance of DownCNV is consistent with previous CGH studies of similar design, including those of maize (Springer et al., 2009; Swanson-Wagner et al., 2010) and rice (Yu et al., 2011). The patterning of the statistically significant DownCNV resembles that observed in soybean fast-neutron deletion lines (Bolon et al., 2011), indicating that the most prominent peaks likely represent missing genomic regions within the Archer, Minsoy, and/or Noir 1 genotypes relative to Wm82. The rare UpCNV likely represent heterogeneous regions within the Williams 82 cultivar (Haun et al., 2011) and may represent sequences and gene content variants absent in the particular Wm82 individual used in these hybridization experiments. Collectively, these findings indicate that many of the SV detected in this study are likely caused by DNA segments that are present in some lines and absent in others.
Conversely, nucleotide polymorphism among lines may have also contributed to the microarray hybridization differences. This is particularly likely within large segments, in which true SV may be interspersed with regions of high sequence polymorphism. For example, the largest CNV identified in this study was a nearly 2-Mb DownCNV located in a gene-poor pericentromeric region of chromosome 4. This region, which is approximately three times larger than the next largest CNV identified in this study, was barely beyond the significance threshold in the Archer-Wm82 and Minsoy-Wm82 comparisons. Clearly, this region is not a true 2-Mb deletion in Archer and Minsoy but instead may indicate a combination of SV and SNP polymorphisms throughout the region. In this sense, the CGH analysis represents a scan of genome-wide polymorphisms that is particularly sensitive to identifying strong SV (but the “CNV” terminology is not necessarily an accurate description for all of the polymorphic segments identified in the analyses). However, the low SNP rates in soybean (Hyten et al., 2006) and the application of stringent significance thresholds provide confidence that a sizeable fraction of polymorphic segments identified in this study consists primarily of true SV. A PCR survey of 31 accessions supported this conclusion, indicating that the subset of CNV and PAV identified in this study likely represented a range of both rare and common structural variants throughout the soybean germplasm.
The most extensive studies of crop plant SV to date have been performed in maize (Springer et al., 2009; Swanson-Wagner et al., 2010). In terms of CNV distribution, the soybean comparisons are virtually the opposite of what has been observed in maize. In maize, accessions tend to exhibit high rates of SV throughout their chromosomes, with infrequent regions of structural conservation interspersed. In soybean, chromosomes tend to exhibit long stretches of conservation interspersed with regions enriched for CNV. The relative rates of SV between the two species are consistent with published rates of nucleotide variation, in which domesticated maize lines exhibit much higher SNP rates than domesticated soybean lines (Wright et al., 2005; Hyten et al., 2006; Gore et al., 2009; Lam et al., 2010). Not surprisingly, the rates of SV between soybean genotypes are also an order of magnitude less than the differences observed between the genome sequences of soybean and its nearest wild relative Glycine soja, in which more than 1,000 genes are estimated to have large structural differences caused by deletions, insertions, inversions, transpositions, or translocations (Kim et al., 2010).
Regions of High SV
The soybean CNV data showed elevated SV within clusters of NB- and RLP-encoding genes. These are common classes of disease resistance genes (R genes) that have been shown to frequently have functions in pathogen perception and signaling of plant host defense responses (Kruijt et al., 2005; DeYoung and Innes, 2006). Genes involved in immunity, environmental response, and defense have also been reported to be enriched within CNV regions in human and other mammalian genomes (Feuk et al., 2006; Nguyen et al., 2006; Perry et al., 2006; Nicholas et al., 2009; Liu et al., 2010; Gokcumen et al., 2011; Hou et al., 2012). The co-occurrence of CNV with clusters of NB- and RLP-encoding genes in soybean is intriguing when one considers the biological function of the loci, the potential mechanisms for acquiring the diversity necessary for recognizing new pathogens, and the consequences possibly imposed upon the genome by breeder-assisted positive selection.
It has been proposed that the plant’s first line of defense is through the perception of highly conserved pathogen-associated molecular patterns (PAMPs) by cell surface pattern recognition receptors (Jones and Dangl, 2006; Zipfel, 2009). This perception can lead to a non-race-specific resistance termed PAMP-triggered immunity. To defeat or suppress PAMP-triggered immunity, pathogens have evolved effector proteins. In turn, plants have evolved R genes that act to directly or indirectly perceive the pathogen effector protein and signal a defense response (Jones and Dangl, 2006). Unlike the PAMP/pattern recognition receptor relationship, which may be highly conserved (Boller and Felix, 2009), the effector/R gene relationship exists in flux and results in race-specific resistance as the pathogen evolves to escape perception and R genes coevolve to adapt to the evolved pathogen (de Wit et al., 2002; Takken and Rep, 2010; Ravensdale et al., 2011). Therefore, the importance of any given R gene may change depending on environmental circumstances. The R gene may be essential for survival in the presence of a pathogen harboring the cognate effector protein. However, in the absence of the pathogen, the R gene may become dispensable.
Structural changes, particularly gene loss or gene gain, may occur within R gene clusters as a consequence of natural and/or artificial selection. Unequal crossing over within existing gene parts or recombination between diverged haplotypes may give rise to new R genes with novel functions, some of which may be beneficial in combating newly arisen pathogen strains. Thus, there may be a gain in fitness for the creation of an R gene with a new specificity. Conversely, in the absence of a pathogen that can be recognized by a given R gene, there is no fitness cost to the loss of the unutilized R gene. In fact, there may be a fitness cost to maintaining R genes in the absence of a pathogen harboring the cognate effector protein (Tian et al., 2003; Bomblies and Weigel, 2007), driving selection to favor recombination and unequal sequence-exchange events that purge R gene copies that are no longer needed in a given environment. These factors lend themselves to the rapid evolution of R genes (Mondragón-Palomino and Gaut, 2005) and predict that R gene clusters may be hotspots for SV. Indeed, rapid evolution of NB-LRR-encoding genes was observed within homeologous regions of the Rpg1b locus of soybean and related species (Ashfield et al., 2012). NB-LRR-encoding genes in these regions experienced a higher rate of duplication and deletion than non-NB-LRR-encoding genes interspersed within the cluster. One might expect that other gene classes that tend to be “environmentally specific” or “conditionally necessary” may also be hotspots for SV, although perhaps undetectable with our current sample size.
The findings of this study are consistent with previous findings in soybean and other plant species. Resequencing of 80 Arabidopsis genomes revealed predicted PAV in 33% of the NB-encoding genes, 2.6-fold greater than the genome average (Guo et al., 2011). A recent CGH study identified 20 NB-encoding genes that exhibited CNV between two rice cultivars (Yu et al., 2011). An association between CNV and disease resistance genes has also been reported in a recent resequencing study of 50 rice accessions (Xu et al., 2012). Resequencing and CGH studies in maize identified a total of 20 NB-encoding genes that exhibited gene content variation among 24 different inbred lines (Springer et al., 2009; Lai et al., 2010; Swanson-Wagner et al., 2010), leading the authors to speculate that these genes may be involved in strain-specific disease resistance (Lai et al., 2010). A membrane array study conducted on multiple accessions within Oryza, Glycine, and Gossypium genera found that the number of NB-encoding genes varied widely both within and among the respective species (Zhang et al., 2010). Resequencing studies in maize and soybean each reported an enrichment of amino acid substitutions with a predicted large effect within NB-encoding genes (Lai et al., 2010; Lam et al., 2010). In concordance with this observation, a genome-wide analysis of segmentally duplicated NB-encoding genes in soybean reported that these genes are evolving at a higher evolutionary rate than other genes (Zhang et al., 2011).
Prospects for Cloning Soybean R Genes
The enrichment of CNV within NB and RLP gene clusters may complicate the detection of direct orthologous relationships between these gene family members, even within intraspecific comparisons. The variation in gene content has often necessitated the construction of bacterial artificial chromosome libraries specific to the resistant accession in order to clone specific R genes (Ashfield et al., 2003, 2004; Bhattacharyya et al., 2005), even from species with a sequenced reference genome (Ashikawa et al., 2008; Lee et al., 2009). A number of previous studies on specific resistance loci have shown that the genes conferring disease resistance are often completely absent in susceptible genotypes. In soybean, a study of the Rps4 gene for resistance to Phytophthora sojae reported gene content variation associated with disease resistance; a specific gene deletion was associated with susceptibility in the Williams-derived genotype L89-1581 (Sandhu et al., 2004). Similarly, analysis of recombinant haplotypes within the cluster of NB-encoding genes at the Rsv1 locus for resistance to Soybean mosaic virus revealed that distinct resistant and susceptible interactions were associated with the presence or absence of the members of this cluster (Hayes et al., 2004). Additionally, bacterial artificial chromosome-based comparative sequencing of candidate R gene regions in soybean have identified several regions in which the content of NB-encoding gene clusters is highly dynamic, including many gene models that exhibit PAV among accessions (M. Graham, personal communication). The dynamic nature of NB- and RLP-encoding gene clusters decreases the utility of a single reference genome sequence for the identification and cloning of resistance genes. Diagnostic platforms capable of assessing genome-wide gene content variation among the wider soybean germplasm (beyond the Williams 82 genome sequence) may be a valuable tool for identifying candidate R genes in the future.
CONCLUSION
This report provides, to our knowledge, the first genome-wide analysis of soybean copy number and PAV among a limited sample of accessions. The CNV dynamics along the individual chromosomes were described, providing insight into the regions and chromosomes with relatively high or low rates of SV. A notable enrichment of significant CNV was identified within known R gene clusters. Furthermore, this study provides the groundwork for a deeper sampling of the germplasm that will allow for a more thorough assessment of soybean SV within the context of population and evolutionary genetics.
MATERIALS AND METHODS
Plant Material and Nucleic Acid Extraction
Seeds for soybean (Glycine max) ‘Williams 82’ were obtained from Dr. Randy Shoemaker at Iowa State University. The individual used for this study was named Wm82-ISU-01. Seeds for accessions Archer, Minsoy, and Noir 1 were obtained from the U.S. Department of Agriculture Soybean Germplasm Collection in Urbana, Illinois. The individuals used for this study were named Archer-SGC-01, Minsoy-SGC-01, and Noir 1-SGC-01, respectively. These individuals are referred to by their abbreviated or cultivar names (Archer, Minsoy, Noir 1, and Wm82) for simplicity.
Seeds were planted in individual 4-inch pots containing a 50:50 mix of sterilized soil and Metro Mix and grown under standard greenhouse conditions. Young trifoliate leaves from 3-week-old plants were harvested and immediately frozen in liquid nitrogen. Frozen leaf tissue was ground with a mortar and pestle in liquid nitrogen. DNA was extracted using the Qiagen Plant DNeasy Mini Kit according to the manufacturer’s protocol. DNA was quantified on a NanoDrop spectrophotometer. These DNA samples were used for SNP genotyping, CGH, and exome-resequencing analyses.
Four more soybean accessions were used for additional CGH experimentation. Seeds for accessions Kingwa and Williams were obtained from the Soybean Germplasm Collection in Urbana, Illinois. Seeds for accessions Richland and M92-220 were obtained from Dr. James Orf at the University of Minnesota. Plants for these accessions were grown and DNA was extracted as described above.
Illumina Infinium Genotyping
The Illumina Infinium iSelect SoySNP50 chip (Q. Song, C.V. Quigley, G. Jia, P.B. Cregan, and D.L. Hyten, unpublished data) was used to obtain genotyping data for the three individual plants: Archer, Minsoy, and Noir 1. The Wm82-ISU-01 Infinium genotyping data from a previous study (Haun et al., 2011) were used for comparisons. SNP calls were made with Illumina GenomeStudioV2010.2 software. Heterozygous, ambiguous, or otherwise uninformative data points were treated as missing data. Visual displays showing the SNP profiles for the three genotypes relative to Wm82 were generated using Spotfire DecisionSite software.
CGH
The microarray used for the CGH experiments is described in detail in a previous publication (Haun et al., 2011). Briefly, the microarray consists of 696,139 unique oligonucleotide probes ranging from 50 to 75 bp in length. The probes tile the assembled soybean genome sequence at a median interval length of 1,120 bp between adjacent probes. This platform may be ordered from Roche NimbleGen by requesting the design 091113_Gmax_RS_CGH_HX3.
CGH protocols, including DNA labeling, microarray hybridization, and scanning, were performed as described (Haun et al., 2011). Genotype Wm82 was used as the Cy5 reference in all hybridizations. Data analyses followed previously described methodologies (Haun et al., 2011). Briefly, the segMNT algorithm in the Nimblescan software (version 2.5) was used to extract the raw data and make segmentation calls. The parameters of the algorithm were as follows: minimum segment difference = 0.1, minimum segment length (number of probes) = 2, acceptance percentile = 0.99, number of permutations = 10; nonunique probes were included, and spatial correction and qspline normalization were applied. The list of resulting segments was processed to identify significant segments. Segments were significant if the log2 ratio mean of the probes within the segment was beyond the threshold level for that genotype. The upper threshold was the log2 ratio value of the 95th percentile of all data points for each individual genotype. The lower threshold was the reciprocal negative value of the upper threshold. The thresholds for Archer, Minsoy, and Noir 1 were ±0.437, ±0.449, and ±0.421, respectively. Following manual inspection, each segment was compared with the soybean high-confidence gene list to identify genes and repetitive elements within significant segments.
To compare the relative SV among Archer, Minsoy, and Noir 1, we calculated the variance between the Archer-Wm82, Minsoy-Wm82, and Noir 1-Wm82 hybridization log2 ratios for each probe on the microarray. The mean variance along a sliding window of 250 probes was calculated and plotted for each chromosome. Visual displays of the CGH data with respect to the significant CNV probes and PAV genes were rendered using Spotfire DecisionSite software.
Additional CGH experiments using genotypes Kingwa, Williams, Richland, and M92-220 were also performed as described above.
Exome-Resequencing and Data Analysis
Exon DNA was captured from the Archer, Minsoy, Noir 1, and Wm82 samples using the NimbleGen soybean exome chip (design 100310_Gmax_public_exome_cap_HX3) and was resequenced using the Illumina Genome Analyzer IIX platform. The exome capture and 76-bp paired-end sequencing were performed as described previously (Haun et al., 2011).
Sequence reads were aligned to the soybean reference genome sequence (Schmutz et al., 2010) using SOAP2 (Li et al., 2009), as described previously (Haun et al., 2011). The number of reads per gene model exon was calculated as described (Haun et al., 2011). The total number of reads mapping to exons for the Wm82 sample was 29,054,888. The respective numbers of reads mapping to exons for the Archer, Minsoy, and Noir 1 samples were 25,915,052, 19,878,546, and 22,480,515, respectively.
The exon read count for each Glyma gene model was used to detect gene content differences among the Wm82, Archer, Minsoy, and Noir 1 samples. Read counts were normalized by applying a correction factor to each sample that adjusted the total number of read counts to 19,878,546 (the number of Minsoy read counts). Gene content variants were defined as any gene model that had a minimum of 30 read counts in at least one genotype and zero read counts in at least one genotype; 133 such gene models were identified. To subclassify the gene content profiles, a cutoff of six reads was set as the standard for calling a gene as “present” or “absent” within a genotype. Genotypes with six or more reads were considered present for the gene; genotypes with five or fewer reads were considered absent. Based on these presence-absence calls, the 133 genes were subclassified into 11 different groups according to their presence-absence profile among genotypes.
To compare the exome-resequencing read counts of the 133 PAV genes with the CGH data, we computed the log2 ratio for each gene in the four genotypes. Counts of zero were converted to a value of 1 to allow for the calculation of count ratios between the genotypes. Calculations and statistical analyses of the exome-resequencing and CGH data log2 ratios were performed using Excel software.
PCR Validation of PAV
A subset of genes exhibiting CNV and PAV among the four genotypes were examined for PAV among a diverse set of 31 soybean accessions. PCR primers were designed for 13 soybean gene models based on genomic DNA sequences available at www.phytozome.net (version 7.0). Primer3 software (version 0.4.0) was used for primer design, targeting a product size range between 300 and 400 bp per gene model. A standard PCR protocol was executed using HotStar Taq DNA Polymerase (Qiagen), with 36 cycles of heat denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 40 s after an initial denaturation at 95°C for 15 min. PCR products were run on 1.4% agarose gels and stained with ethidium bromide. PCR bands were visually scored as either present or absent for each genotype template.
Comparison of Regions of SV with Known QTL
For comparisons of SV and known QTL, we defined the regions with the greatest amplitude of CNV variance as regions exhibiting over 0.5 variance between the log2 ratios of the Archer, Minsoy, and Noir 1 hybridizations (Fig. 4). The genome sequence coordinates of markers from the soybean consensus map 4.0 (Hyten et al., 2010; available at http://www.soybase.org) were used to estimate genetic positions associated with these high-amplitude CNV regions. Genetic locations of QTL and genes for disease resistance from previous mapping studies were estimated from the soybean consensus map 4.0 (www.soybase.org).
Identification and Enrichment Analysis of Gene Classes Associated with Genomic SV
The genes associated with PAV and significant CNV regions were subjected to GO and other analyses to identify gene classes enriched within SV regions. The subset of 133 PAV gene models identified in the exome-resequencing data was selected for this analysis. Additionally, we used the CGH data to select the subsets of gene models in which any part of the predicted coding region overlapped with or resided within a significant CNV segment.
All gene annotations were estimated from the longest open reading frame of the soybean 46,430 high-confidence predicted protein-coding genes (Schmutz et al., 2010). GO designations (Berardini et al., 2004) were assigned based on the highest BLASTp (Altschul et al., 1997) hit of soybean predicted peptides to the Arabidopsis (Arabidopsis thaliana) protein database with an expectation threshold of 1 × e−20. Soybean sequences were classified according to Arabidopsis biological process GO designations. Protein domains were predicted by Pfam, with the cutoff defined by gathering thresholds (Finn et al., 2010).
To analyze the relative effects of structural features versus selection pressures, two large protein families involved in disease resistance, the NB and RLP families, were identified from the total soybean protein set. NB family members were defined by the presence of an NB-ARC (nucleotide-binding adaptor shared by APAF-1, R proteins, and CED-4) domain (Pfam: PF00931). RLPs were defined by the presence of the Pfam LRR domain, the conserved LRR-containing C3 domain, and a transmembrane domain. A hidden Markov model was built from an alignment of the C3 domain from Arabidopsis RLPs (HMMER3; Eddy, 2009). Proteins with predicted LRR and C3 domains, with no other predicted Pfam domain with an e-value less than 0.5 (Tör et al., 2004), and with an identifiable C-terminal transmembrane domain were categorized as RLPs. Transmembrane topology was predicted by hidden Markov models using TMMOD version 2.0 and TMHMM (Krogh et al., 2001; Kahsay et al., 2005). Additionally, RLP-like proteins were identified as gene models with the following characteristics: bidirectional best BLASTp (threshold 1 × e-20) hits to known functional RLPs or RLPs characterized in Arabidopsis (Tör et al., 2004), containing a predicted LRR and C3 domain, and not containing other predicted Pfam domains (but lacking a predicted C-terminal transmembrane domain). The RLP and RLP-like gene models were grouped together into an “RLP-encoding” family for downstream analyses.
Enrichment or depletion of a GO category or protein domain was determined by a hypergeometric distribution (Fisher’s exact test) with adjustment for multiple hypothesis testing achieved by resampling methods implemented by the FuncAssociate 2.0 program using 10,000 simulations (Berriz et al., 2009). All genes eligible to be called within a CNV region or as a PAV were used as reference, 46,275 and 43,530 genes, respectively. Adjusted P values were doubled to account for the two-sided Fisher’s exact test.
Gene families were gathered via BLASTCLUST (Altschul et al., 1997) with greater than 50% amino acid identity over more than 70% of the sequence length. Gene families were defined as large (more than 10 members) or small (two to 10 members). Based on their genomic distribution, gene family members were categorized as isolated or clustered. A cluster was defined as two or more members of a family with a maximum of eight intervening genes (Richly et al., 2002; Meyers et al., 2003). Tandem repeats within the genomic sequence of individual genes were predicted de novo with the Tandem Repeat Finder (Benson, 1999). Settings were modified from defaults to include a maximum repeat period of 2 kb, and repeats were filtered to a minimum length of 30 bp. Tandem repeats and gene family membership and distribution are available in Supplemental Tables S9 and S10. The coordinates of TEs were downloaded from SoyTEdb (Du et al., 2010). For each gene, it was determined whether the gene start or end coordinates were within 5 kb of the start or end coordinates of a TE.
All CGH data from this study are freely available from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/; accession no. GSE28905). All exome-resequencing data are freely available on the National Center for Biotechnology Information Short Read Archive database (http://www.ncbi.nlm.nih.gov/; accession no. SRA039969).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. CNV among soybean genotypes.
Supplemental Figure S2. Frequency of shared and unique CNV associated with soybean gene models.
Supplemental Figure S3. CGH profiles among technical replications of the Minsoy-Wm82 and Noir 1-Wm82 hybridizations.
Supplemental Figure S4. Relationship between genomic SV and nucleotide SNP polymorphism among soybean genotypes.
Supplemental Figure S5. Distribution of presence-absence gene content variants among the four soybean genotypes.
Supplemental Figure S6. Distribution of presence-absence gene content variants among 31 diverse accessions.
Supplemental Figure S7. Frequency of shared and unique CNV associated with soybean gene models across five comparisons.
Supplemental Table S1. The gene models within regions of significant CNV between Wm82 and each of the three other genotypes.
Supplemental Table S2. The gene models and presence-absence profiles of the 133 PAV genes.
Supplemental Table S3. Primers used for presence-absence analysis.
Supplemental Table S4. Presence-absence results for 13 genes, as measured by PCR.
Supplemental Table S5. Annotations and classifications of genes identified as PAV or within CNV.
Supplemental Table S6. Pfam domains and GO terms significantly enriched or depleted within genes in CNV regions or PAV.
Supplemental Table S7. Predicted NB-encoding genes.
Supplemental Table S8. Predicted RLP-encoding genes.
Supplemental Table S9. Genomic distribution of multigene families within the Williams 82 genome.
Supplemental Table S10. Tandem repeats identified in unspliced gene sequences.
Supplemental Table S11. Summary statistics of soybean CNV number and size for additional CGH experiments.
Supplemental Table S12. The gene models within regions of significant CNV for additional CGH experiments.
Supplemental Table S13. Pfam domains and GO terms significantly enriched or depleted within genes in CNV regions for additional CGH experiments.
Supplemental Materials and Methods S1. Details on exome-resequencing PAV genes.
Supplementary Material
Acknowledgments
We are grateful to Carroll Vance and Gary Muehlbauer for contributing toward the development of the CGH platform and offering helpful suggestions throughout this project. We thank Nathan Springer, Michelle Graham, and Peter Morrell for reviewing the manuscript and contributing many excellent suggestions. We thank Randy Nelson, Jim Orf, and Randy Shoemaker for providing the seeds used in this study.
Glossary
- SV
structural variation
- SNP
single nucleotide polymorphism
- CNV
copy number variation
- CGH
comparative genomic hybridization
- PAV
presence-absence variation
- QTL
quantitative trait loci
- Wm82
Williams 82
- GO
Gene Ontology
- NB
nucleotide-binding
- RLP
receptor-like protein
- TE
transposable element
- LRR
leucine-rich repeat
- PAMP
pathogen-associated molecular pattern
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Bocian A, Held D, Henk AD, Marek LF, Danesh D, Peñuela S, Meksem K, Lightfoot DA, Young ND, et al. (2003) Genetic and physical localization of the soybean Rpg1-b disease resistance gene reveals a complex locus containing several tightly linked families of NBS-LRR genes. Mol Plant Microbe Interact 16: 817–826 [DOI] [PubMed] [Google Scholar]
- Ashfield T, Egan AN, Pfeil BE, Chen NW, Podicheti R, Ratnaparkhe MB, Ameline-Torregrosa C, Denny R, Cannon S, Doyle JJ, et al. (2012) Evolution of a complex disease resistance gene cluster in diploid Phaseolus and tetraploid Glycine. Plant Physiol 159: 336–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. (2004) Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 16: 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J, Matsumoto T, Ono K, Yano M. (2008) Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 180: 2267–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beló A, Beatty MK, Hondred D, Fengler KA, Li B, Rafalski A. (2010) Allelic genome structural variations in maize detected by array comparative genome hybridization. Theor Appl Genet 120: 355–367 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42: 251–269 [PubMed] [Google Scholar]
- Benson G. (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al. (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. (2009) Next generation software for functional trend analysis. Bioinformatics 25: 3043–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya MK, Narayanan NN, Gao H, Santra DK, Salimath SS, Kasuga T, Liu Y, Espinosa B, Ellison L, Marek L, et al. (2005) Identification of a large cluster of coiled coil-nucleotide binding site-leucine rich repeat-type genes from the Rps1 region containing Phytophthora resistance genes in soybean. Theor Appl Genet 111: 75–86 [DOI] [PubMed] [Google Scholar]
- Bishop JG, Dean AM, Mitchell-Olds T. (2000) Rapid evolution in plant chitinases: molecular targets of selection in plant-pathogen coevolution. Proc Natl Acad Sci USA 97: 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bolon YT, Haun WJ, Xu WW, Grant D, Stacey MG, Nelson RT, Gerhardt DJ, Jeddeloh JA, Stacey G, Muehlbauer GJ, et al. (2011) Phenotypic and genomic analyses of a fast neutron mutant population resource in soybean. Plant Physiol 156: 240–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Weigel D. (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet 8: 382–393 [DOI] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Chen Q, Han Z, Jiang H, Tian D, Yang S. (2010) Strong positive selection drives rapid diversification of R-genes in Arabidopsis relatives. J Mol Evol 70: 137–148 [DOI] [PubMed] [Google Scholar]
- Chen WK, Swartz JD, Rush LJ, Alvarez CE. (2009) Mapping DNA structural variation in dogs. Genome Res 19: 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianzio SR, Shultz SP, Fehr WR, Tachibana H. (1991) Registration of ‘Archer’ soybean. Crop Sci 31: 1707 [Google Scholar]
- Clop A, Vidal O, Amills M. (2012) Copy number variation in the genomes of domestic animals. Anim Genet (in press) [DOI] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. (2010) Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF, Giannoulatou E, et al. (2010) Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit PJ, Brandwagt BF, van den Burg HA, Cai X, van der Hoorn RA, de Jong CF, van Klooster J, de Kock MJ, Kruijt M, Lindhout WH, et al. (2002) The molecular basis of co-evolution between Cladosporium fulvum and tomato. Antonie van Leeuwenhoek 81: 409–412 [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Innes RW. (2006) Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol 7: 1243–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. (2012) Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS One 7: e33234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopman EB, Hartl DL. (2007) A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci USA 104: 19920–19925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Grant D, Tian Z, Nelson RT, Zhu L, Shoemaker RC, Ma J. (2010) SoyTEdb: a comprehensive database of transposable elements in the soybean genome. BMC Genomics 11: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (2009) A new generation of homology search tools based on probabilistic inference. Genome Inform 23: 205–211 [PubMed] [Google Scholar]
- Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M. (2008) Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320: 1629–1631 [DOI] [PubMed] [Google Scholar]
- Fasoula VA, Boerma HR. (2007) Intra-cultivar variation for seed weight and other agronomic traits within three elite soybean cultivars. Crop Sci 47: 367–373 [Google Scholar]
- Feuk L, Carson AR, Scherer SW. (2006) Structural variation in the human genome. Nat Rev Genet 7: 85–97 [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E, Darré F, Morcillo-Suarez C, Petit-Marty N, Carreño A, Marigorta UM, Ryder OA, Blancher A, Rocchi M, Bosch E, et al. (2011) Copy number variation analysis in the great apes reveals species-specific patterns of structural variation. Genome Res 21: 1626–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Campbell CD, Eichler EE. (2011) Human copy number variation and complex genetic disease. Annu Rev Genet 45: 203–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokcumen O, Babb PL, Iskow R, Zhu Q, Shi X, Mills RE, Ionita-Laza I, Vallender EJ, Clark AG, Johnson WE, et al. (2011) Refinement of primate copy number variation hotspots identifies candidate genomic regions evolving under positive selection. Genome Biol 12: R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, Jacquemont S, Reymond A, Sun M, Sawa A, Gusella JF, et al. (2012) KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature 485: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, et al. (2009) A first-generation haplotype map of maize. Science 326: 1115–1117 [DOI] [PubMed] [Google Scholar]
- Graubert TA, Cahan P, Edwin D, Selzer RR, Richmond TA, Eis PS, Shannon WD, Li X, McLeod HL, Cheverud JM, et al. (2007) A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet 3: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YL, Fitz J, Schneeberger K, Ossowski S, Cao J, Weigel D. (2011) Genome-wide comparison of nucleotide-binding site-leucine-rich repeat-encoding genes in Arabidopsis. Plant Physiol 157: 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryev V, Saar K, Adamovic T, Verheul M, van Heesch SA, Cook S, Pravenec M, Aitman T, Jacob H, Shull JD, et al. (2008) Distribution and functional impact of DNA copy number variation in the rat. Nat Genet 40: 538–545 [DOI] [PubMed] [Google Scholar]
- Haun WJ, Hyten DL, Xu WW, Gerhardt DJ, Albert TJ, Richmond T, Jeddeloh JA, Jia G, Springer NM, Vance CP, et al. (2011) The composition and origins of genomic variation among individuals of the soybean reference cultivar Williams 82. Plant Physiol 155: 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes AJ, Jeong SC, Gore MA, Yu YG, Buss GR, Tolin SA, Maroof MA. (2004) Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics 166: 493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Liu GE, Bickhart DM, Matukumalli LK, Li C, Song J, Gasbarre LC, Van Tassell CP, Sonstegard TS. (2012) Genomic regions showing copy number variations associate with resistance or susceptibility to gastrointestinal nematodes in Angus cattle. Funct Integr Genomics 12: 81–92 [DOI] [PubMed] [Google Scholar]
- Hyten DL, Choi IY, Song QJ, Specht JE, Carter TE, Shoemaker RC, Hwang EY, Matukumalli LK, Cregan PB. (2010) A high density integrated genetic linkage map of soybean and the development of a 1536 universal soy linkage panel for quantitative trait locus mapping. Crop Sci 50: 960–968 [Google Scholar]
- Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB. (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA 103: 16666–16671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes RW, Ameline-Torregrosa C, Ashfield T, Cannon E, Cannon SB, Chacko B, Chen NW, Couloux A, Dalwani A, Denny R, et al. (2008) Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol 148: 1740–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kahsay RY, Gao G, Liao L. (2005) An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 21: 1853–1858 [DOI] [PubMed] [Google Scholar]
- Kim MY, Lee S, Van K, Kim TH, Jeong SC, Choi IY, Kim DS, Lee YS, Park D, Ma J, et al. (2010) Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc Natl Acad Sci USA 107: 22032–22037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox AK, Dhillon T, Cheng H, Tondelli A, Pecchioni N, Stockinger EJ. (2010) CBF gene copy number variation at frost resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet 121: 21–35 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kruijt M, DE Kock MJ, de Wit PJ. (2005) Receptor-like proteins involved in plant disease resistance. Mol Plant Pathol 6: 85–97 [DOI] [PubMed] [Google Scholar]
- Lai J, Li R, Xu X, Jin W, Xu M, Zhao H, Xiang Z, Song W, Ying K, Zhang M, et al. (2010) Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet 42: 1027–1030 [DOI] [PubMed] [Google Scholar]
- Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, Li MW, He W, Qin N, Wang B, et al. (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet 42: 1053–1059 [DOI] [PubMed] [Google Scholar]
- Lark KG, Chase K, Adler F, Mansur LM, Orf JH. (1995) Interactions between quantitative trait loci in soybean in which trait variation at one locus is conditional upon a specific allele at another. Proc Natl Acad Sci USA 92: 4656–4660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Gutiérrez-Arcelus M, Perry GH, Vallender EJ, Johnson WE, Miller GM, Korbel JO, Lee C. (2008) Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum Mol Genet 17: 1127–1136 [DOI] [PubMed] [Google Scholar]
- Lee SK, Song MY, Seo YS, Kim HK, Ko S, Cao PJ, Suh JP, Yi G, Roh JH, Lee S, et al. (2009) Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics 181: 1627–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Liu GE, Hou Y, Zhu B, Cardone MF, Jiang L, Cellamare A, Mitra A, Alexander LJ, Coutinho LL, Dell’Aquila ME, et al. (2010) Analysis of copy number variations among diverse cattle breeds. Genome Res 20: 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Han X, Qi J, Yang J, Wijeratne AJ, Li T, Ma H. (2012) Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res 22: 508–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwire MM, Bayer F, Webster CL, Cao C, Jiggins FM. (2011) Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a duplication. PLoS Genet 7: e1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maydan JS, Lorch A, Edgley ML, Flibotte S, Moerman DG. (2010) Copy number variation in the genomes of twelve natural isolates of Caenorhabditis elegans. BMC Genomics 11: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón-Palomino M, Gaut BS. (2005) Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol Biol Evol 22: 2444–2456 [DOI] [PubMed] [Google Scholar]
- Mondragón-Palomino M, Meyers BC, Michelmore RW, Gaut BS. (2002) Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res 12: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DQ, Webber C, Ponting CP. (2006) Bias of selection on human copy-number variants. PLoS Genet 2: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas TJ, Baker C, Eichler EE, Akey JM. (2011) A high-resolution integrated map of copy number polymorphisms within and between breeds of the modern domesticated dog. BMC Genomics 12: 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas TJ, Cheng Z, Ventura M, Mealey K, Eichler EE, Akey JM. (2009) The genomic architecture of segmental duplications and associated copy number variants in dogs. Genome Res 19: 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orf JH, Chase K, Jarvik T, Mansur LM, Cregan PB, Adler FR, Lark KG. (1999) Genetics of soybean agronomic traits. I. Comparison of three related recombinant inbred populations. Crop Sci 39: 1642–1651 [Google Scholar]
- Orf JH, Denny RL. (2004) Registration of ‘MN1302’ soybean. Crop Sci 44: 693 [Google Scholar]
- Pearce S, Saville R, Vaughan SP, Chandler PM, Wilhelm EP, Sparks CA, Korolev A, Al-Kaff N, Boulton MI, Phillips AL, et al. (2011) Molecular characterisation of Rht-1 dwarfing genes in hexaploid wheat. Plant Physiol 157: 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. (2007) Diet and the evolution of human amylase gene copy number variation. Nat Genet 39: 1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Tchinda J, McGrath SD, Zhang J, Picker SR, Cáceres AM, Iafrate AJ, Tyler-Smith C, Scherer SW, Eichler EE, et al. (2006) Hotspots for copy number variation in chimpanzees and humans. Proc Natl Acad Sci USA 103: 8006–8011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Yang F, Marques-Bonet T, Murphy C, Fitzgerald T, Lee AS, Hyland C, Stone AC, Hurles ME, Tyler-Smith C, et al. (2008) Copy number variation and evolution in humans and chimpanzees. Genome Res 18: 1698–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravensdale M, Nemri A, Thrall PH, Ellis JG, Dodds PN. (2011) Co-evolutionary interactions between host resistance and pathogen effector genes in flax rust disease. Mol Plant Pathol 12: 93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Kurth J, Leister D. (2002) Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol 19: 76–84 [DOI] [PubMed] [Google Scholar]
- Sakudoh T, Nakashima T, Kuroki Y, Fujiyama A, Kohara Y, Honda N, Fujimoto H, Shimada T, Nakagaki M, Banno Y, et al. (2011) Diversity in copy number and structure of a silkworm morphogenetic gene as a result of domestication. Genetics 187: 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu D, Gao H, Cianzio S, Bhattacharyya MK. (2004) Deletion of a disease resistance nucleotide-binding-site leucine-rich-repeat-like sequence is associated with the loss of the Phytophthora resistance gene Rps4 in soybean. Genetics 168: 2157–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santuari L, Pradervand S, Amiguet-Vercher AM, Thomas J, Dorcey E, Harshman K, Xenarios I, Juenger TE, Hardtke CS. (2010) Substantial deletion overlap among divergent Arabidopsis genomes revealed by intersection of short reads and tiling arrays. Genome Biol 11: R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, Good RT, Appleton B, Sherrard J, Raymant GC, Bogwitz MR, Martin J, Daborn PJ, Goddard ME, Batterham P, et al. (2010) Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet 6: e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Smith TJ, Camper HM. (1973) Registration of Essex soybean. Crop Sci 13: 495 [Google Scholar]
- Springer NM, Ying K, Fu Y, Ji T, Yeh CT, Jia Y, Wu W, Richmond T, Kitzman J, Rosenbaum H, et al. (2009) Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet 5: e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. (2010) Structural variation in the human genome and its role in disease. Annu Rev Med 61: 437–455 [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE. (2010) Diversity of human copy number variation and multicopy genes. Science 330: 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton T, Baumann U, Hayes J, Collins NC, Shi BJ, Schnurbusch T, Hay A, Mayo G, Pallotta M, Tester M, et al. (2007) Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 318: 1446–1449 [DOI] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Eichten SR, Kumari S, Tiffin P, Stein JC, Ware D, Springer NM. (2010) Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res 20: 1689–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken F, Rep M. (2010) The arms race between tomato and Fusarium oxysporum. Mol Plant Pathol 11: 309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77 [DOI] [PubMed] [Google Scholar]
- Tör M, Brown D, Cooper A, Woods-Tör A, Sjölander K, Jones JD, Holub EB. (2004) Arabidopsis downy mildew resistance gene RPP27 encodes a receptor-like protein similar to CLAVATA2 and tomato Cf-9. Plant Physiol 135: 1100–1112 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Varala K, Swaminathan K, Li Y, Hudson ME. (2011) Rapid genotyping of soybean cultivars using high throughput sequencing. PLoS ONE 6: e24811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzynski A, Ashfield T, Chen NW, Mammadov J, Nguyen A, Podicheti R, Cannon SB, Thareau V, Ameline-Torregrosa C, Cannon E, et al. (2008) Replication of nonautonomous retroelements in soybean appears to be both recent and common. Plant Physiol 148: 1760–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS. (2005) The effects of artificial selection on the maize genome. Science 308: 1310–1314 [DOI] [PubMed] [Google Scholar]
- Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X, Dong Y, Gutenkunst RN, Fang L, Huang L, et al. (2012) Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol 30: 105–111 [DOI] [PubMed] [Google Scholar]
- Yates JL, Boerma HR, Fasoula VA. (2012) SSR-marker analysis of the intracultivar phenotypic variation discovered within 3 soybean cultivars. J Hered 103: 570–578 [DOI] [PubMed] [Google Scholar]
- Yu P, Wang C, Xu Q, Feng Y, Yuan X, Yu H, Wang Y, Tang S, Wei X. (2011) Detection of copy number variations in rice using array-based comparative genomic hybridization. BMC Genomics 12: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wu YH, Lee MK, Liu YH, Rong Y, Santos TS, Wu C, Xie F, Nelson RL, Zhang HB. (2010) Numbers of genes in the NBS and RLK families vary by more than four-fold within a plant species and are regulated by multiple factors. Nucleic Acids Res 38: 6513–6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Feng Y, Cheng H, Tian D, Yang S, Chen JQ. (2011) Relative evolutionary rates of NBS-encoding genes revealed by soybean segmental duplication. Mol Genet Genomics 285: 79–90 [DOI] [PubMed] [Google Scholar]
- Zheng LY, Guo XS, He B, Sun LJ, Peng Y, Dong SS, Liu TF, Jiang S, Ramachandran S, Liu CM, et al. (2011) Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor). Genome Biol 12: R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, Hyatt SM, Fickus EW, Young ND, Cregan PB. (2003) Single-nucleotide polymorphisms in soybean. Genetics 163: 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2009) Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol 12: 414–420 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.