Abstract
Pollen elimination provides an effective containment method to reduce direct gene flow from transgenic trees to their wild relatives. Until now, only limited success has been achieved in controlling pollen production in trees. A pine (Pinus radiata) male cone-specific promoter, PrMC2, was used to drive modified barnase coding sequences (barnaseH102E, barnaseK27A, and barnaseE73G) in order to determine their effectiveness in pollen ablation. The expression cassette PrMC2-barnaseH102E was found to efficiently ablate pollen in tobacco (Nicotiana tabacum), pine, and Eucalyptus (spp.). Large-scale and multiple-year field tests demonstrated that complete prevention of pollen production was achieved in greater than 95% of independently transformed lines of pine and Eucalyptus (spp.) that contained the PrMC2-barnaseH102E expression cassette. A complete pollen control phenotype was achieved in transgenic lines and expressed stably over multiple years, multiple test locations, and when the PrMC2-barnaseH102E cassette was flanked by different genes. The PrMC2-barnaseH102E transgenic pine and Eucalyptus (spp.) trees grew similarly to control trees in all observed attributes except the pollenless phenotype. The ability to achieve the complete control of pollen production in field-grown trees is likely the result of a unique combination of three factors: the male cone/anther specificity of the PrMC2 promoter, the reduced RNase activity of barnaseH102E, and unique features associated with a polyploid tapetum. The field performance of the PrMC2-barnaseH102E in representative angiosperm and gymnosperm trees indicates that this gene can be used to mitigate pollen-mediated gene flow associated with large-scale deployment of transgenic trees.
One possible concern about field trials and commercial deployment of transgenic trees is the potential for transgene introgression into the gene pool of wild relatives via cross-pollination (Hoenicka and Fladung, 2006). Genetic engineering to eliminate pollen production to generate male-sterile plants to facilitate breeding, hybrid seed production, and reducing out-crossing has been widely studied in targeted crops (Goldberg et al., 1993; Strauss et al., 1995; Brunner et al., 2007), and pollen ablation, using an anther-specific promoter driving a cytotoxic gene, has been proven to be very effective (Mariani et al., 1990).
Pollen formation occurs in anthers in angiosperms and in male cones (pollen cones) in conifers. Both of these male reproductive organs produce microsporangia, in which the tapetum, the innermost layer of the pollen sac, plays a crucial role in the development of microspores into mature pollen grains (Shivanna et al., 1997). In both gymnosperms and angiosperms, one of the contributions of the tapetum to the development of mature microspores is the release of enzymes that degrade the callose and pectic molecules to free individual microspores from the tetrad (Pacini et al., 1984; Rhee and Somerville, 1998). Soon after the release of individual microspores, tapetal cells undergo programmed cell death and release sugars, starch, crystallized proteins, and lipids that are utilized by the developing microspores. A number of male-sterile mutants have been found to have defects in the structure of the tapetum (Raghavan, 1997). The indispensability of the tapetum during microspore development was further demonstrated by the specific ablation of tapetal cells in tobacco (Nicotiana tabacum), leading to the premature abortion of microspores (Goldberg et al., 1993). However, the tapetum is not considered to play an active role in the nutrition of pollen mother cells before and during meiosis (Raghavan, 1989).
Genetic ablation refers to the process or methodology of expressing a cytotoxic gene under the control of a tightly regulated promoter. The result of genetic ablation is the targeted elimination of specific cells or tissues of living organisms without lethal effects. The PrMC2 gene of radiata pine (Pinus radiata) encodes a protein similar to the anther-specific A9 protein in Brassica napus and Arabidopsis (Arabidopsis thaliana). In situ hybridization analysis indicated that PrMC2 was expressed only in the tapetum of male cones of radiata pine (Walden et al., 1999). The PrMC2 promoter contains a 16-bp sequence similar to the anther box previously identified in Petunia (van Tunen et al., 1990; van der Meer et al., 1992). The tissue specificity of the PrMC2 promoter was analyzed in the heterologous host Arabidopsis, and the results showed that the promoter had high activities in the tapetum, microspores, as well as other anther tissues, including epidermis, vascular bundles, and parietal cells, but no promoter activity was found in pollen mother cells (Höfig et al., 2003).
The most commonly used cytotoxic genes in genetic ablation are diphtheria toxin A chain and barnase. The use of the diphtheria toxin A chain, as it is derived from a human pathogen, may face negative public perception and health concerns. Barnase is an RNase from Bacillus amyloliquefaciens that has been extensively studied in protein denaturation and refolding research, since it is a small protein with no disulfide bonds (Serrano et al., 1992). It was found that the barnase protein had a very strong RNase activity and was stable in different conditions. The full-length barnase protein has 149 amino acids, which contains a leader peptide (the first 39 amino acids from the N terminus) for secretion.
In this paper, the coding sequence of barnase corresponds to the sequence that encodes a mature barnase protein of 110 amino acids without the leader peptide. Almost every one of the 110 amino acids of the barnase protein has been mutated by single substitution with multiple amino acids, and the mutagenesis studies identified that the Lys at position 27, the Glu at position 73, and the His at position 102 were involved in the catalytic activity for RNA hydrolysis (Mossakowska et al., 1989; Meiering et al., 1992; Axe et al., 1998).
The first demonstration of pollen ablation using a tapetum-specific promoter driving barnase was reported by Mariani et al. (1990). In their study, transgenic tobacco lines containing the tobacco TA29 promoter driving barnase were analyzed, and 92% of the transgenic lines produced no pollen, lacked a detectable tapetum, and had a collapsed pollen sac. No deleterious vegetative effects on plant regeneration were reported during the transformation of TA29-barnase into tobacco. However, other studies have reported negative effects imposed by the barnase gene on plant regeneration and vegetative growth associated with low levels of promoter activity in nontarget tissues (Brunner et al., 2007; Wei et al., 2007). It has been shown that many floral promoters are not expressed exclusively in flowers (Brunner et al., 2000; Rottmann et al., 2000), and even low levels of unintended expression of a cytotoxic gene may impose negative effects on tree growth (Skinner et al., 2000). Lännenpää et al. (2005) reported that the birch tree (Betula pendula) promoter of BpFULL, a homolog of Arabidopsis FRUITFULL (formerly AGL8), was isolated and used to drive barnase, and the resulting construct was introduced into Arabidopsis, tobacco, and birch. In Arabidopsis, all of the 29 BpFULL-barnase lines showed serious defects in vegetative growth and did not produce floral inflorescences. In 13 BpFULL-barnase transgenic tobacco lines, three showed normal flowers and normal growth, four showed dramatic defects in vegetative growth and no flowers, and the remaining six showed relatively normal growth and no flower formation. When BpFULL-baranse was tested in birch, nine of the 21 transgenic lines obtained (43%) had normal flowers and the remaining 12 lines (57%) had no flowers. Of the nonflowering lines, four showed severe defects in vegetative growth and had a bush-like phenotype. The other eight nonflowering lines grew normally initially, but later in development, they developed a bush-like phenotype. The authors speculated that this phenotype was possibly due to the expression of the barnase gene in the early stage of shoot development. The above results strongly suggest that when barnase is used as the ablation gene, slight promoter activity in nontarget tissues could result in negative effects on vegetative growth, especially in plant species with long regeneration times and extended periods of vegetative growth, such as woody plants. For a pollen ablation cassette for commercial applications, the ablation efficiency should be high and stable without any negative effects on vegetative growth.
In this paper, we report a highly effective pollen ablation cassette PrMC2-barnaseH102E for both gymnosperm and angiosperm tree species. This effective pollen ablation cassette is likely the result of a unique combination of the tapetal cell specificity and the activity of the PrMC2 promoter, the reduced RNase activity of barnase mutants, and the polyploidy of tapetal cells. The stable and efficient pollen ablation performance of PrMC2-barnaseH102E under field conditions, achieved without any concomitant detrimental effects on vegetative growth, makes this expression cassette useful in field plantings of transgenic trees when mitigation of pollen-mediated transgene flow is desired.
RESULTS
GUS Staining Indicates That the PrMC2 Promoter Is Expressed Only in Anthers of Transgenic Tobacco
Two versions of the PrMC2 promoter isolated from radiata pine were investigated. The first version contains 351 nucleotides at the 5′ end of the translation initiation ATG and 39 nucleotides of the coding region including the translation initiation ATG. This 390-nucleotide version of the promoter was designated as PrMC2 promoter version I (PrMC2pro-I). PrMC2pro-I, when linked to the barnase coding sequence, is expected to result in a transcript encoding a chimeric protein with the 13 amino acids of the PrMC2 protein at the N terminus of the expressed barnase protein. A second version of the PrMC2 promoter was generated, in which the 36 nucleotides of the coding region, except the translation initiation ATG, were deleted, creating PrMC2 promoter version II (PrMC2pro-II). PrMC2pro-II gives a transcript encoding a “clean” barnase protein with no additional amino acids from PrMC2. In this paper, we refer to PrMC2pro-I and PrMC2pro-II collectively as the PrMC2 promoter (PrMC2pro).
We fused PrMC2pro-I with the Escherichia coli GUS gene, and the resulting construct, PrMC2pro-I-GUS, was introduced into tobacco. GUS staining of young floral buds revealed anther-specific expression in 15 of 17 transgenic lines (Fig. 1), with no GUS staining found in sepals, petals, carpels, young shoot tips, young leaves, and roots. To compare the anther specificity of PrMC2pro-I and PrMC2pro-II, we obtained 18 transgenic tobacco lines carrying PrMC2pro-II-GUS. GUS staining intensity and patterns of floral buds from PrMC2pro-II-GUS tobacco closely matched those of PrMC2pro-I-GUS tobacco in 17 of the 18 transgenic lines, suggesting that removal of the 36 nucleotides of the coding sequence from PrMC2pro-I had little or no impact on the activity and anther specificity of the PrMC2pro.
Figure 1.
GUS staining of a young floral bud of tobacco carrying PrMC2pro-I-GUS. The unopened floral bud was cut longitudinally in the middle using a blade, with the surface of the two anthers being partially cut, and stained for GUS activity overnight at 37°C. The photograph was taken after briefly destaining in ethanol to remove chlorophyll. The bright spots in the photograph are reflections in the ethanol left in the specimen during photographing. The GUS staining was found to be present only in the anthers.
In Vivo Comparison of RNase Activities of Four Barnase Mutants
It is hypothesized that the strong RNase activity of native barnase, when used for flower ablation, often damages nontargeted tissues due to low but non-flower-specific activities in many floral promoters. We reasoned that a modified barnase with reduced RNase activity may be more suitable for flower ablation, since the reduced RNase activity would cause less damage in nonfloral tissues during plant transformation and vegetative growth. Four barnase mutants carrying single amino acid substitutions were generated: barnaseH102E, barnaseK27A, barnaseE73G, and barnaseF106S. BarnaseH102E, in which the His at position 102 of the barnase protein was replaced by a Glu, and barnaseK27A, in which the Lys at position 27 of the barnase protein was replaced by an Ala, were generated using site-directed mutagenesis. BarnaseE73G, where the Glu at position 73 was replaced by a Gly, and barnaseF106S, where the Phe at position 106 was replaced by a Ser, were obtained unexpectedly while cloning the barnase coding sequence under the control of the radiata pine AGAMOUS (PrAG) promoter, a male and female cone-specific promoter (Tandre et al., 1998), without the presence of barstar in the cloning vector of pUC19. The barstar protein is a specific inhibitor of barnase produced by the same B. amyloliquefaciens bacterium (Hartley, 1988). We hypothesized that the two barnase mutants obtained from this cloning experiment were the result of reduced barnase activities and E. coli tolerance to low levels of barnase activities. Based on this observation, we developed an in vivo system for comparison of low levels of barnase activities in E. coli by measuring the diameters of single colonies growing on agar medium (see “Materials and Methods”). We used this system to rank the RNase activities of the four barnase mutants and a control mutant of barnaseH102Y, which was reported to have no biological activity (Axe et al., 1998). No E. coli colonies were obtained upon transformation of the PrAG-barnaseK27A construct, suggesting that the barnaseK27A had strong RNase activity and prevented E. coli growth. However, hundreds of single colonies were obtained with the other three barnase mutants as well as the control of barnaseH102Y driven by the same PrAG promoter. The growth rate of E. coli on agar medium was affected by barnase activity, and the relative RNase activities of the barnase mutants could be assessed based on the average diameter of single colonies of E. coli bearing each of the barnase mutants. The activity rankings based on colony size on agar medium, from strongest to weakest, were barnaseK27A > barnaseF106S > barnaseE73G > barnaseH102E = barnaseH102Y (Supplemental Table S1).
PrMC2pro-I-barnaseH102E Effectively Prevented Pollen Formation in Tobacco with No Negative Effects on Vegetative Growth, While the Same Promoter Driving barnaseE73G or barnaseK27A Imposed Detrimental Effects on Tobacco Regeneration and Growth
To facilitate the presentation of the results associated with both glasshouse and field tests, Table I shows the information of experimental materials and establishment time, DNA constructs, plant species, and test locations. Three pollen ablation constructs, PrMC2pro-I-barnaseH102E, PrMC2pro-I-barnaseE73G, and PrMC2pro-I-barnaseK27A, and a control construct, PrMC2pro-I-GUS, were introduced into tobacco ‘Xanthi’ via Agrobacterium tumefaciens-mediated transformation. We found that the transformation efficiency varied depending on the barnase mutants. We obtained 18 barnaseH102E lines and 12 GUS lines, and the transformation efficiency of barnaseH102E tobacco was similar to that of the GUS control. By contrast, we obtained only six barnaseE73G lines and five barnaseK27A lines in two transformation attempts. In our tobacco transformation protocol, we targeted one transgenic event per leaf disc, so the transformation efficiency was calculated by the number of transgenic events obtained from 100 leaf discs. The transformation efficiency for the constructs PrMC2pro-I-barnaseE73G and PrMC2pro-I-barnaseK27A was approximately 5%, while the transformation efficiency for PrMC2pro-I-barnaseH102E and PrMC2pro-I-GUS was approximately 80%. All transgenic lines were transplanted into pots and grown in the glasshouse until they flowered.
Table I. Information for glasshouse experiments and field tests evaluating the pollen ablation effect in plants.
The glasshouse experiments and field tests are listed in sequential order as they appear in the text.
| Experiment Materialsa and Establishment Year | Construct Name | Cassette(s) Carried by the Indicated Constructsbc | Experiment Location |
|---|---|---|---|
| Tobacco, 2003 | pWVR234 | PrMC2pro-I-GUS | Summerville, South Carolina |
| Tobacco, 2004 | pAGF245 | PrMC2pro-II-GUS | Summerville, South Carolina |
| Tobacco, 2003 | pWVR220 | PrMC2pro-I-barnaseH102E | Summerville, South Carolina |
| Tobacco, 2003 | pWVR216 | PrMC2pro-I-barnaseK27A | Summerville, South Carolina |
| Tobacco, 2003 | pWVR232 | PrMC2pro-I-barnaseE73G | Summerville, South Carolina |
| Tobacco, 2004 | pAGF243 | PrMC2pro-II-barnaseH102E | Summerville, South Carolina |
| PxL grafts, 2005 | pWVR220 | PrMC2pro-I-barnaseH102E | Southern Georgia |
| PxL grafts, 2005 | pWVR234 | PrMC2pro-I-GUS | Southern Georgia |
| Field-grown PxL trees, 2004 | pWVR220 | PrMC2pro-I-barnaseH102E | Eastern South Carolina |
| Loblolly grafts, 2007 | pWVK310 | PrMC2pro-II-barnaseH102E and PrUBQ-AtGOGAT | Southern Georgia |
| Loblolly grafts, 2007 | pWVK312 | PrMC2pro-II-barnaseH102E and PrUBQ-PsGS | Southern Georgia |
| Loblolly grafts, 2007 | pAGK316 | PrMC2pro-II-barnaseH102E and Pt4CL-PtUDP-GBD | Southern Georgia |
| Loblolly grafts, 2007 | pAGK320 | PrMC2pro-II-barnaseH102E and PrQUB-AtGOGAT and Pt4CL-PtUDP-GBD | Southern Georgia |
| Loblolly grafts, 2007 | pAGK321 | PrMC2pro-II-barnaseH102E and PrUBQ-PsGS and Pt4CL-PtUDP-GBD | Southern Georgia |
| E. occidentalis, 2005 | pARB598 | PrMC2pro-I-barnaseH102E and Pt4CL-Eg4CL antisense | Summerville, South Carolina |
| E. occidentalis, 2006 | pAGF243 | PrMC2pro-II-barnaseH102E | Summerville, South Carolina |
| Field-grown EH1, 2005 | pARB598 | PrMC2pro-I-barnaseH102E and Pt4CL-Eg4CL antisense | Central Florida |
| Field-grown EH1, 2005 | pARB599 | PrMC2pro-I-barnaseH102E and Pt4CL- Eg4CL RNA interference | Central Florida |
| Field-grown EH1, 2005 | pABCTE01 | PrMC2pro-II-barnaseH102E and AtRd29A-AtCBF2 | Southern Alabama |
Tobacco and E. occidentalis were grown in the glasshouse. bPromoter and gene symbols are as follows: PrMC2pro-I, PrMC2 promoter version I from P. radiata; PrMC2pro-II, PrMC2 promoter version II from P. radiata; PrUBQ, polyubiquitin promoter from P. radiata; AtGOGAT, Arabidopsis Glu synthase; PsGS, Gln synthetase from pea (Pisum sativum); Pt4CL, 4-coumarate CoA ligase gene promoter from P. taeda; PtUDP-GBD, the UDP-Glc-binding domain protein gene from Populus tremuloides; Eg4CL, 4-coumarate CoA ligase gene from E. grandis; AtRd29A, Arabidopsis Rd29A gene promoter; AtCBF2, Arabidopsis C-repeat binding factor 2. cEach construct also carries the same selectable marker cassette consisting of the E. coli nptII gene driven by an Arabidopsis ubiquitin promoter.
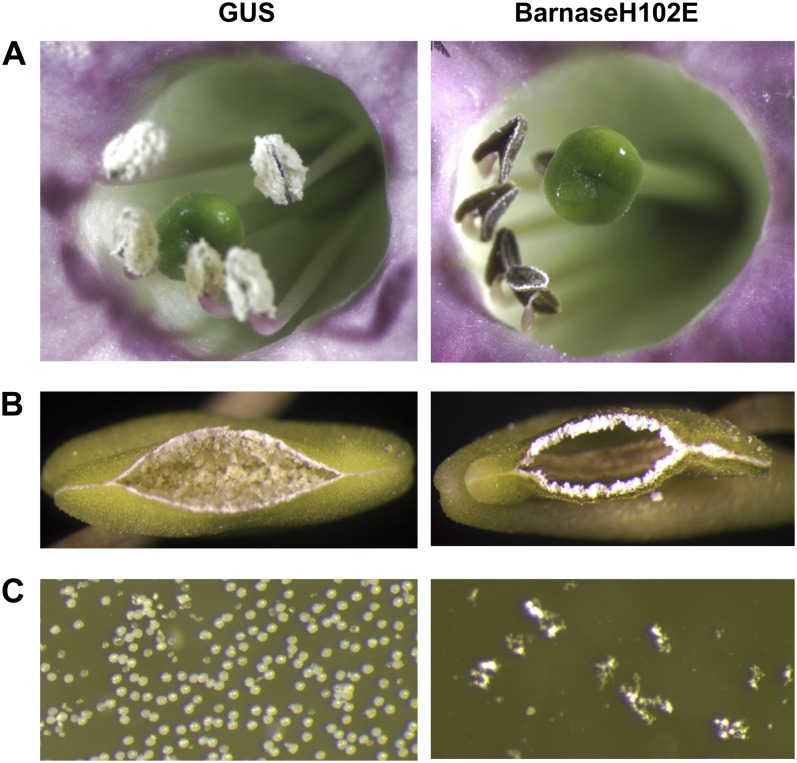
Different pollen ablation responses in tobacco were obtained from the three barnase mutants. A complete pollen ablation phenotype was observed in barnaseH102E tobacco, despite the result of barnaseH102E lacking of any significant biological activity in the E. coli colony assay (see “Discussion”). Visual observation of opened barnaseH102E flowers revealed shriveled and dark brown anthers that were devoid of pollen grains (Fig. 2A). By contrast, the anthers of GUS tobacco were able to produce abundant pollen (Fig. 2A). Undehisced anthers were cut partially open along their long axis and observed with a dissecting microscope. The barnaseH102E anthers appeared empty, while the GUS anthers were filled with white-colored and powder-like material (Fig. 2B). Under higher magnification, observation of the internal contents sampled from dissected anthers revealed that no pollen grains were present inside barnaseH102E anthers (Fig. 2C), while the white material inside GUS anthers was made up of pollen grains (Fig. 2C). None of the 18 barnaseH102E tobacco lines produced detectable pollen. Visual observation revealed that the height, stem diameter, morphology of vegetative and floral organ systems, time of flowering, and flower color patterns were similar between the barnaseH102E and GUS tobacco growing in the glasshouse (Supplemental Fig. S1). Six barnaseH102E tobacco lines were hand pollinated by placing GUS or wild-type pollen on their stigmas, and all of pollinated flowers were able to set seeds, suggesting that the female organs of barnaseH102E tobacco were physiologically normal and not affected by the presence of PrMC2pro-I-barnaseH102E.
Figure 2.
Visual and microscopic observations of tobacco flowers carrying PrMC2pro-I-GUS or PrMC2pro-I-barnaseH102E. A, Images of a single flower of GUS tobacco (left panel) and barnaseH102E tobacco (right panel) taken using a conventional camera. B, Images of a partially opened anther of GUS tobacco (left panel) and barnaseH102E tobacco (right panel) taken using a dissecting microscope with 9-fold magnification. The anthers were cut longitudinally with a blade. C, Images of sampled internal contents from a GUS anther (left panel) and a barnaseH102E anther (right panel) taken using a compound microscope with 200-fold magnification.
Six independent tobacco transgenic lines carrying PrMC2pro-I-barnaseE73G provided a less consistent pollen control response. Three lines produced no pollen; two lines produced pollen; and one line did not flower after 90 d in the glasshouse. Two of the three non-pollen-producing lines and the one line that did not produce flowers exhibited stunted growth. Of the five tobacco lines carrying PrMC2pro-I-barnaseK27A, four lines produced no pollen, while one did produce pollen. All five barnaseK27A lines demonstrated a stunted growth phenotype. These results indicate that PrMC2pro-I was active, at some levels, in tobacco vegetative tissues, and this nonfloral expression caused vegetative damage when barnase mutants with stronger RNase activities were employed in pollen ablation.
Version II of the PrMC2 promoter driving barnaseH102E (PrMC2pro-II-barnaseH102E) has similar pollen ablation efficiency as PrMC2pro-I-barnaseH102E in tobacco. PrMC2pro-II-barnaseH102E was introduced into tobacco, and 12 transgenic lines were obtained (Table I). At the time of flowering, none of the 12 lines produced pollen, a result similar to that obtained from PrMC2pro-I-barnaseH102E tobacco. No negative vegetative effects were observed during tobacco transformation and the growth of transgenic plants, suggesting that PrMC2pro-II had comparable effectiveness and anther specificity to PrMC2pro-I.
Male Cones Growing on Transgenic Pine Grafts Carrying PrMC2pro-I-barnaseH102E Did Not Produce Pollen for Four Consecutive Years in the Field
The same three pollen ablation constructs tested in tobacco, PrMC2pro-I-barnaseH102E, PrMC2pro-I-barnaseE73G, and PrMC2pro-I-barnaseK27A, and the control construct PrMC2pro-I-GUS were introduced into a hybrid pine genotype, “PxL,” resulting from the cross of pitch pine (Pinus rigida) with loblolly pine (Pinus taeda). Transformations resulted in 17 barnaseH102E PxL lines and 13 GUS PxL lines, and the presence of the transgenes was confirmed by PCR. Efforts to produce PxL lines containing either the barnaseE73G or barnaseK27A expression cassette proved difficult, with only two to three transgenic lines being produced for each of the two constructs after multiple transformation attempts. These few barnaseE73G or barnaseK27A lines were not evaluated further.
Usually, it takes 3 years for a PxL pine and 5 years for a loblolly pine to begin to bear male and female cones in the southeastern United States. However, it has been documented that grafting physiologically immature scions onto a tree that has already entered reproductive growth can accelerate reproductive growth on grafts, and pine grafting can be utilized to produce male cones and female cones earlier than it would be possible on trees grown from seedlings (Bramlett et al., 1995). In order to stimulate flowering, 10 scions of each of the 17 barnaseH102E and each of six GUS PxL lines were collected from 1-year-old potted trees and then grafted on 7-year-old loblolly pine trees in southern Georgia in January 2005 (Table I). We also grafted 40 untransformed loblolly pine scions, collected from 4-year-old field-grown trees, at the grafting site. Several months later (May 2005), the survival rate of the grafts was checked and the overall survival rate of transgenic grafts, including barnaseH102E and GUS grafts, was 38%. This resulted in three or four surviving grafts of each transgenic line on the rootstock trees. The survival rate of untransformed loblolly grafts was 60%, likely due to a better size match between the older scion material and the rootstock at the grafting site. One year after grafting (January 2006), 15 barnaseH102E grafts, four GUS grafts, and two untransformed loblolly grafts bore male cones and some grafts also produced female cones. The 15 barnaseH102E grafts with male cones represented 12 of the 17 grafted barnaseH102E lines. Four GUS grafts bearing male cones belong to three of the six grafted GUS lines. No differences were observed between barnaseH102E and GUS PxL grafts with respect to growth rate, the color of needles, the branching patterns, and the timing of male cone and female cone initiation. The untransformed loblolly grafts grew more than any of the transgenic PxL grafts, including GUS grafts, and this was expected because loblolly pine typically has a faster growth rate than PxL pine. Usually, individual loblolly or PxL male cones grow closely together to form male cone clusters, which contain from two to approximately 20 individual male cones, although occasionally single male cones can be found. To prevent transgenic pollen release into the environment, branches bearing male cone clusters were bagged approximately 2 weeks before pollen release, with each bag containing only one male cone cluster (see “Materials and Methods”).
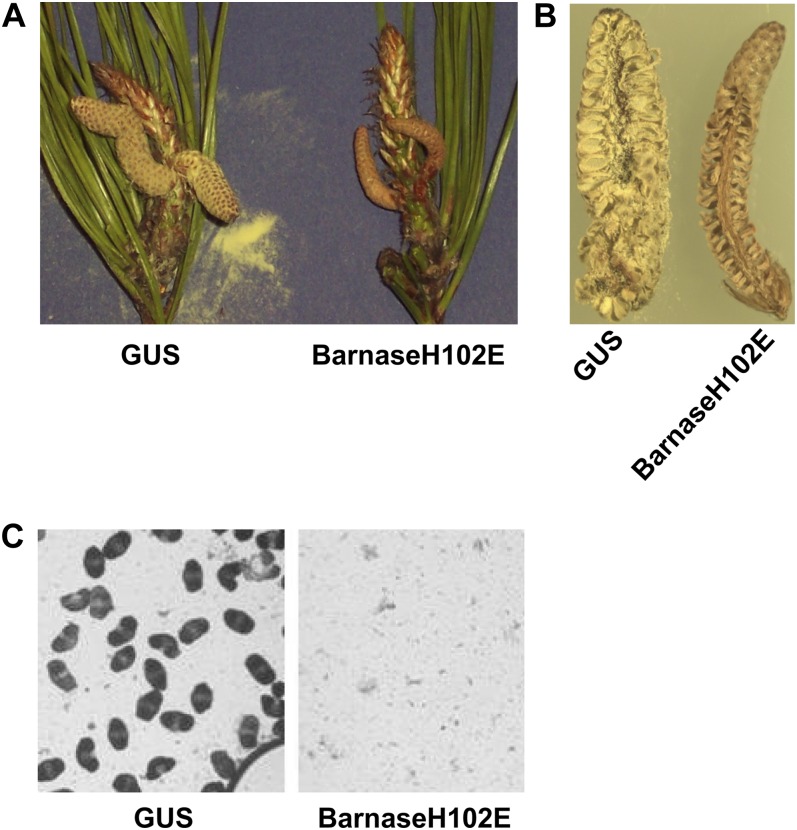
Approximately 15 months after grafting (April 2006), we harvested 19 bagged male cone clusters from the 15 barnaseH102E grafts, four bagged male cone clusters from the four GUS grafts, and two bagged male cone clusters from the untransformed loblolly grafts. The bagged barnaseH102E male cones were harvested after observing pollen release inside the bags containing GUS male cone clusters or the untransformed male cone clusters. Visual observation of harvested bags revealed that all 19 barnaseH102E male cone clusters did not release pollen inside the bags, while the four GUS and the two untransformed male cone clusters released copious amounts of pollen inside the bags (Fig. 3; Table II). Visual observation of individual male cones revealed that the GUS male cones were elongated, swollen, and light yellow in color; the cone scales had opened and yellow pollen was released (Fig. 4A). In contrast to the GUS male cones, the barnaseH102E male cones were elongated but not swollen and were brown or dark brown in color, with closed cone scales and no evidence of pollen release (Fig. 4A). Individual male cones were cut in the middle along their long axis and observed with a dissecting microscope. In the GUS male cone, the microsporangia between two scales were filled with pollen grains (Fig. 4B). By contrast, in the barnaseH102E male cone, the microsporangia were empty (Fig. 4B). To investigate if small quantities of pollen grains, which may not be visible, were present inside the male cones, procedures for pollen extraction were performed with individual male cones, and the resulting samples were observed with a compound microscope. The results showed that in most cases, no pollen grains were observed in multiple views of a slide prepared from an individual barnaseH102E male cone (Fig. 4C), but in rare cases, a few pollen grains were observed that were attributed to contamination during sample preparation from bagged material that had been exposed to pollen from surrounding pine trees. In general, untransformed loblolly male cones released pollen 1 or 2 weeks earlier than the PxL GUS male cones at the grafting site. Having verified that the GUS PxL grafts produced normal pollen, all GUS grafts were cut down after data collection was completed in May 2006.
Figure 3.
Visual observation of bagged male cone clusters at 15 months after grafting. The GUS-labeled bag contains a PxL pine male cone cluster carrying PrMC2pro-I-GUS, while the barnaseH102E bag contains a PxL male cone cluster carrying PrMC2pro-I-barnaseH102E. The clear bag was made of cellulose membrane, which allows air exchange between inside and outside of the bag. The male cone clusters and vegetative shoots were able to grow and develop normally inside the bags without the possibility of pollen release into the environment. The aluminum rings inside the bags were used to anchor and support the bags on the branches of grafts. The open end of the bag was toward the bottom, and it was sealed with a sponge and a cable tie. Massive amounts of yellow-colored pollen grains were readily visible inside the GUS bag, while no pollen grains were visually observed inside the barnaseH102E bag.
Table II. Results of visual and microscopic observation of male cones collected from barnaseH102E grafts and untransformed grafts at the grafting site in southern Georgia for 4 consecutive years.
The barnaseH102E grafts, which were grafted in January 2005, are the pine PxL hybrid transformed with PrMC2pro-I-barnaseH102E. The control grafts were untransformed loblolly pine. Each bag contained one male cone cluster. Wild type refers to untransformed loblolly grafts.
| Year | No. of Grafts Bearing Male Cones |
No. of Lines Bearing Male Cones |
No. of Bags Harvested |
No. of Bags with Pollen Inside by Visual Observation |
No. of Bags of barnaseH102E Male Cone Cluster with Abnormal Pollen Identified by Microscopic Observation | ||||
|---|---|---|---|---|---|---|---|---|---|
| barnaseH102E | Wild Type | barnaseH102E | Wild Type | barnaseH102E | Wild Type | barnaseH102E | Wild Type | ||
| 2006 | 15 | 2 | 12 | 1 | 19 | 2 | 0 | 2 | 0 |
| 2007 | 35 | 24 | 17 | 1 | 103 | 61 | 0 | 61 | 1 |
| 2008 | 31 | 20 | 17 | 1 | 93 | 22 | 0 | 22 | 1 |
| 2009 | 30 | 20 | 17 | 1 | 88 | 60 | 0 | 60 | 1 |
Figure 4.
Visual and microscopic observations of bagged male cone clusters from PxL pine grafts carrying either PrMC2pro-I-GUS or PrMC2pro-I-barnaseH102E. A, The bagged male cone clusters were taken out of the bags, and the needles were trimmed to expose individual male cones. The GUS male cones were releasing pollen, while the barnaseH102E male cones contained no pollen. B, One GUS male cone and one barnaseH102E male cone in A were cut longitudinally in the middle using a blade and observed using a dissecting microscope. C, The pollen extraction procedure was performed with GUS or barnaseH102E male cones, and the resulting samples were observed using a compound microscope. A normal pine pollen grain has two wing-like appendages as seen in C from a GUS male cone.
At 27 months after grafting (April 2007), multiple grafts of all 17 barnaseH102E lines and 24 untransformed loblolly grafts produced male cones. In total, 103 bagged barnaseH102E male cone clusters and 61 bagged untransformed loblolly male cone clusters were harvested. Visual observation of bagged untransformed male cone clusters revealed that all of them had released readily visible yellow pollen inside the bags. By contrast, visual observation of all bags of barnaseH102E male cone clusters collected from the 17 transgenic lines showed that none of them released visually detectable pollen inside the bags (Table II). However, dissection and microscopic observation of individual male cones sampled from the bags showed that one graft of a barnaseH102E line, TRT001343-2, produced a small quantity of unreleased and underdeveloped pollen grains, while the remaining 16 barnaseH102E lines produced no pollen. The TRT001343 barnaseH102E line was represented by three grafts, TRT001343-1, TRT001343-2, and TRT001343-3, on different rootstock trees. We found that the underdeveloped pollen was present only in TRT001343-2 but not in TRT001343-1 and TRT001343-3. Furthermore, three male cone clusters were bagged and harvested from the graft of TRT001343-2. The microscopic observation revealed that only one of the three clusters produced the underdeveloped pollen while the other two did not. Furthermore, the pollen-producing cluster had 12 individual male cones, and only two of the 12 showed the presence of underdeveloped pollen while the other 10 male cones did not. The pollen grains produced by TRT001342-2 appeared to be abnormal, underdeveloped, and smaller in size compared with untransformed pollen grains (Supplemental Fig. S2). We believe that none of the underdeveloped pollen grains were released from the male cones, since no visible yellow pollen was observed inside the bag and the male cones that produced the underdeveloped pollen were degenerated in a similar way as the male cones with no pollen development. We concluded, therefore, that none of the 103 barnaseH102E male cone clusters produced or shed any normal or mature pollen grains.
At 39 months and 51 months after grafting (April 2008 and April 2009), the visual and microscopic observation of bagged male cone clusters yielded similar results as the previous 2 years (Table II). The male cones harvested from 16 of the 17 barnaseH102E lines did not produce any pollen, determined by both visual and microscopic observation, while all untransformed male cones released pollen inside bags. Pollen development was found again in a few male cones collected from the graft TRT001343-2 in 2008 and 2009. In 2008, two of the 20 individual male cones collected from TRT001343-2 produced a small quantity of underdeveloped pollen grains, which were irregular in shape and smaller in size compared with the untransformed pollen grains. In 2009, one of the 22 individual male cones harvested from TRT001343-2 produced a small quantity of underdeveloped pollen of a smaller size than pollen from untransformed male cones.
It was observed that female cone (seed cone) initiation and development on barnaseH102E PxL grafts were similar to those on GUS PxL grafts and untransformed loblolly grafts. Female cones were left on the grafts for observation and later harvest. Immature female cones were collected prior to complete seed development from nine barnaseH102E lines and from untransformed loblolly grafts approximately 31 months after grafting (August 2007). The successful development of female cones on the barnaseH102E grafts was the result of natural pollination by wild-type pollen from surrounding loblolly trees. The immature barnaseH102E female cones appeared normal and were green in color. Immature embryos were dissected from the female cones and observed with a dissecting microscope. The comparison revealed that the dissected barnaseH102E embryos were morphologically and developmentally similar to embryos from untransformed female cones, indicating that the presence of PrMC2pro-I-barnaseH102E did not affect embryo development.
The Field-Grown PxL Trees Carrying PrMC2pro-I-barnaseH102E Showed the Same Pollen Ablation Results as the PxL Grafts
To validate the pollen ablation results obtained from grafts, multiple trees of 17 transgenic PxL lines carrying PrMC2pro-I-barnaseH102E and untransformed PxL trees were planted in a field trial in eastern South Carolina in June 2004 (Table I). These 17 barnaseH102E lines were the same lines grafted in southern Georgia described above. Approximately 31 months after planting (January 2007), we observed the initiation of male cones and female cones on some of the barnaseH102E PxL trees as well as on untransformed PxL trees. Visual observation revealed that the barnaseH102E PxL trees were similar to the untransformed PxL trees with respect to height, stem diameter, needle color, crown shape, and male and female cone initiation time and pattern. In order to facilitate the monitoring and management of male cones on these trees, the number of trees in this field test was reduced. A single tree of each of seven randomly selected barnaseH102E lines and an untransformed PxL tree were kept in the field while the rest of the trees were cut down. Unfortunately, the barnaseH102E line TRT001343, in which one of the three grafts showed pollen development in a few pollen cones in southern Georgia, was not selected and all ramets of this line were cut down. Therefore, we were unable to verify this phenomenon in field-grown trees. In March 2007, four male cone clusters were bagged on each tree remaining in the field trial. A total of 28 bagged barnaseH102E male cone clusters and four bagged untransformed male cone clusters were harvested at the time when pollen shed was evident in the untransformed male cones. Visual observation of the bags revealed that the four untransformed male cone clusters released pollen inside bags while all barnaseH102E male cone clusters did not. Three individual male cones from each barnaseH102E male cone cluster were dissected and microscopically observed, confirming that none of them produced pollen. The pollen ablation results obtained over the next 2 years (2008 and 2009) for the same seven barnaseH102E trees and a single untransformed tree were identical to the results obtained in 2007. In addition, these pollen ablation results obtained from the field-grown trees in South Carolina were consistent with the results obtained from grafts in Georgia, indicating that the reproductive growth and development in the grafts and field-grown trees were the same or very similar over the two different geographic locations.
PrMC2pro-II-barnaseH102E Has Similar Pollen Ablation Efficacy to PrMC2pro-I-barnaseH102E, and a Single Copy Is Sufficient to Abolish Pollen Formation in Pine
In January 2007, 460 loblolly pine scions, each of which carried one of five stacking constructs, pWVK310, pWVK312, pAGK316, pAGK320, and pAGK321 (for construct information, see Table I), were grafted on 7-year-old loblolly trees in southern Georgia. All five stacking constructs contained the same pollen ablation cassette, PrMC2pro-II-barnaseH102E, as well as a different second expression cassette for a putative growth gene. Since the promoters used to drive the growth genes in the five stacking constructs are either constitutive, such as radiata pine ubiquitin (PrUBQ) promoter (Perera et al., 2000), or xylem-specific, such as loblolly pine 4-coumarate CoA ligase (Pt4CL) promoter (Osakabe et al., 2009), the performance of the PrMC2pro-II-barnaseH102E could be evaluated to determine if there are any interactions between the pollen ablation cassette and the growth gene cassettes that could potentially interfere with pollen ablation efficacy. At 27 months after grafting (April 2009), 19 barnaseH102E grafts and two untransformed grafts bore male cones. These 19 grafts represented 10 transgenic lines distributed across four of the five stacking constructs mentioned above. Visual observation of the grafts did not identify any difference between the barnaseH102E and untransformed grafts with respect to growth rate, the color of needles, the branching patterns, and the timing of male cone and female cone initiation (for additional information, see Supplemental Fig. S3). Visual observation of bagged male cone clusters and microscopic observation of individual male cones showed that all male cones collected from the 19 barnaseH102E grafts did not produce pollen, while the bagged male cones collected from the two untransformed grafts released pollen inside the bags. At 39 months after grafting (April 2010), more grafts bore male cones. In total, we harvested 138 barnaseH102E and 35 untransformed bagged male cone clusters. The 138 barnaseH102E male cone clusters represented 25 transgenic lines across all five stacking constructs. Visual observation of barnaseH102E male cone clusters and microscopic observation of barnaseH102E individual male cones did not find pollen, while all untransformed and bagged male cones released pollen inside bags. These results are identical to the results obtained 1 year earlier in April 2009.
The pollen ablation phenotype is conveyed with either one or two copies of the PrMC2pro-II-barnaseH102E cassette present. The transgene copy number of the 25 barnaseH102E lines, which did not produce pollen, had been determined using real-time quantitative PCR before grafting in the field, and the results indicated that 16 of those 25 barnaseH102E lines contain a single copy, while the remaining nine lines carry two copies. The pollen ablation efficacy of PrMC2pro-II-barnaseH102E carried in the five stacking constructs in this experiment was similar to the constructs that carried just the pollen ablation gene PrMC2pro-I-barnaseH102E or PrMC2pro-II-barnaseH102E, indicating that the pollen ablation cassette in the stacking constructs was not affected by flanking cassettes with either a strong constitutive promoter (PrUBQ) or a strong xylem-preferred promoter (Pt4CL). The above results also confirm that PrMC2pro-II-barnaseH102E has the same high efficiency of pollen ablation as PrMC2pro-I-barnaseH102E in pine.
The PrMC2 Promoter Driving barnaseH102E Abolished Pollen Formation Effectively in Eucalyptus (spp.) Trees Grown in a Glasshouse as Well as in the Field
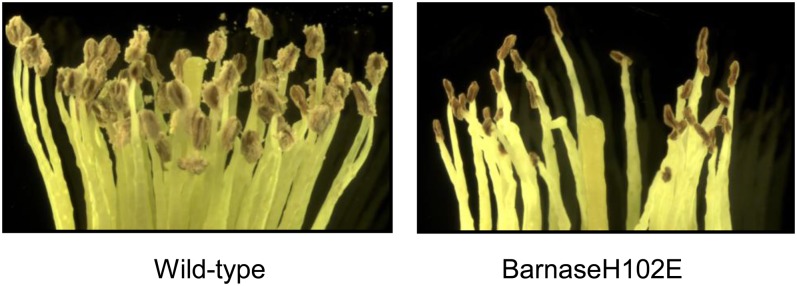
A stacking construct, pARB598, a lignin modification construct containing a cassette of an antisense Eucalyptus grandis 4CL coding sequence driven by the pine Pt4CL promoter and the pollen ablation cassette PrMC2pro-I-barnaseH102E, was introduced into Eucalyptus occidentalis (Table I). E. occidentalis is an ideal model hardwood tree for testing pollen ablation genes in a glasshouse environment due to its characteristics of early flowering and relatively small size (Southerton, 2007). Replicate trees of 23 pARB598 E. occidentalis lines and untransformed trees were transplanted into soil in 1-gallon pots and grown in the glasshouse in 2005 (Table I). Four months after transplanting, the potted trees flowered. The pARB598 E. occidentalis was similar to the untransformed trees with respect to height, stem diameter, leaf color, flowering time, and flowering patterns. E. occidentalis flowers have numerous stamens that are enclosed in a cap known as the operculum, which is composed of fused sepals or petals or both. During flowering time, flowers at different developmental stages, from young floral buds to opened flowers, are typically present on a single tree. Visual observation of opened flowers revealed that the untransformed anthers were covered with yellow-colored pollen grains. In comparison, the pARB598 anthers were shriveled, pollenless, and smaller in size (Fig. 5). Visual observation of 50 individual flowers on each tree indicated that none of the 23 pARB598 lines released pollen. Twenty unopened flowers with brown-colored operculum, which occurs 10 to 15 d before anthesis, were dissected and observed microscopically for each transgenic line and untransformed trees. Microscopic observation of the anthers of these flowers revealed that one of the 23 pARB598 lines, TEO500014, produced a small quantity of normal-appearing immature pollen grains. When the pollen production of individual flowers of TEO500014 and untransformed trees was compared, we estimated that the amount of immature pollen produced by a single flower of TEO500014 was approximately 2% of the amount of pollen produced by a single untransformed flower.
Figure 5.
Microscopic observation of E. occidentalis single flowers. The barnaseH102E flower carries the construct pARB598 (Table I), which contains the pollen ablation cassette PrMC2pro-I-barnaseH102E and an antisense 4CL fragment driven by the loblolly pine 4CL promoter. The images were taken using a dissecting microscope with 9-fold magnification.
We also investigated if the PrMC2pro-II driving barnaseH102E has similar pollen ablation efficacy as PrMC2pro-I in E. occidentalis. Replicate trees of seven E. occidentalis lines carrying PrMC2pro-II-barnaseH102E were obtained, transplanted into soil in 1-gallon pots, and grown in the glasshouse in 2006 (Table I). At the time of flowering, visual observation of opened flowers failed to find pollen on the anthers of the seven barnaseH102E lines, while pollen was readily visible on the anthers of untransformed E. occidentalis. Dissection and microscopic observation showed that no pollen was evident in the unopened flowers sampled from the seven barnaseH102E lines. The phenotypes of pollenless anthers carrying PrMC2pro-II-barnaseH102E were identical to the anthers containing PrMC2pro-I-barnaseH102E, as shown in Figure 5.
To validate the pollen ablation results obtained in the glasshouse, we planted transgenic Eucalyptus (spp.) in field trials in May 2005. Construct pARB598 described above and a second stacking construct, pARB599, were introduced into a Eucalyptus (spp.) hybrid, EH1 (Eucalyptus grandis × Eucalyptus urophylla). pARB599 is similar to pARB598 and contains an RNA interference version of the E. grandis 4CL coding sequence driven by the loblolly pine 4CL promoter together with PrMC2pro-I-barnaseH102E (Table I). Replicate trees of 29 independent transgenic lines carrying either pARB598 or pARB599 were planted in a field trial in central Florida in 2005 (Table I). At 24 months after the field planting (May 2007), these trees produced floral buds that did not open until 4 months after formation. As the size of the EH1 flower is smaller than that of E. occidentalis, it is difficult to determine the presence or absence of pollen on the EH1 anthers with the naked eye. Therefore, unopened flowers with a brown-colored operculum (approximately 10–15 d prior to opening) were dissected to assess the presence of pollen. The dissection and microscopic observation showed that 27 of the 29 lines containing either pARB598 or pARB599 failed to produce pollen while two lines produced pollen, which appeared similar in shape, size, and quantity compared with untransformed pollen. All trees of the two pollen-producing lines were cut down after the completion of data collection and before the maturation or opening of the flowers in 2007. At 40 months after field planting (September 2008), nine of the 27 transgenic lines, which did not produce pollen in the previous year, were reexamined, and again, none of them produced pollen. The rest of the 27 lines were not examined, as they had been extensively damaged due to inclement weather experienced in the year.
In a second field site in southern Alabama, replicate trees of 44 independently transformed EH1 lines, carrying the stacking construct pABCTE01, were planted in November 2005 (Table I). The pABCTE01 contains two target gene cassettes: a freeze tolerance expression cassette composed of the Arabidopsis rd29A promoter driving the Arabidopsis CBF2 coding sequence (Cook et al., 2004), and the pollen ablation cassette PrMC2pro-II-barnaseH102E. Twelve pABCTE01 lines were selected for the investigation of pollen ablation (Table III). The transgene copy number of these 12 lines was determined by either Southern blots or real-time quantitative PCR or both before field planting, and the results showed that nine of the 12 lines had a single copy, two lines had two copies, and the remaining one line had more than two copies. The field trial included eight trees of each of the 12 lines and eight trees of the untransformed EH1 control. Opened and unopened flowers were collected from the 12 transgenic lines as well as from the untransformed trees. Forty-four unopened flowers from each tree, including transgenic and untransformed trees, were dissected and microscopically observed, with a total of over 4,500 flowers being observed. The results revealed that none of the unopened flowers collected from the 12 pABCTE01 lines produced pollen, while pollen was found in all untransformed flowers (Table III).
Table III. Pollen ablation data from microscopic observation of more than 4,500 flowers collected from pABCTE01 EH1 and untransformed EH1 trees in two field tests.
The stacking construct pABCTE01contains PrMC2pro-II-barnaseH102E for pollen ablation and AtRd29A-AtCBF2 for improving freeze tolerance.
| Field Location (County/State) | Flower Collection Period | Tree Age at Flower Collection | Transgenic Line Identifier | No. of Trees Sampled | Pollen Production (Yes/No) |
|---|---|---|---|---|---|
| Baldwin/Alabama | September 2007 | Approximately 2 Years | Control | 8 | Yes |
| TUH000427 | 8 | No | |||
| TUH000434 | 8 | No | |||
| TUH000435 | 8 | No | |||
| TUH000447 | 8 | No | |||
| TUH000456 | 8 | No | |||
| TUH000470 | 8 | No | |||
| TUH000489 | 8 | No | |||
| TUH000490 | 8 | No | |||
| TUH000493 | 8 | No | |||
| TUH000494 | 8 | No | |||
| TUH000495 | 8 | No | |||
| TUH000517 | 8 | No | |||
| Highlands/Florida | September 2010 | Approximately 4 Years | Control | 3 | Yes |
| TUH000427 | 4 | No | |||
| TUH000435 | 2 | No |
In a third field test in central Florida, flowers were collected from two selected lines of pABCTE01 EH1, which were also included in the Alabama test, as well as from untransformed EH1 trees (Table III). In total, 264 pABCTE01 flowers and 132 untransformed flowers were dissected and microscopically observed. The results show that none of the flowers from these two pABCTE01 lines produced pollen while all flowers from untransformed trees did (Table III).
Pollen Development in Pine and Eucalyptus (spp.) Carrying the PrMC2 Promoter Driving barnaseH102E Ceases at the Stage of Tetrad Formation
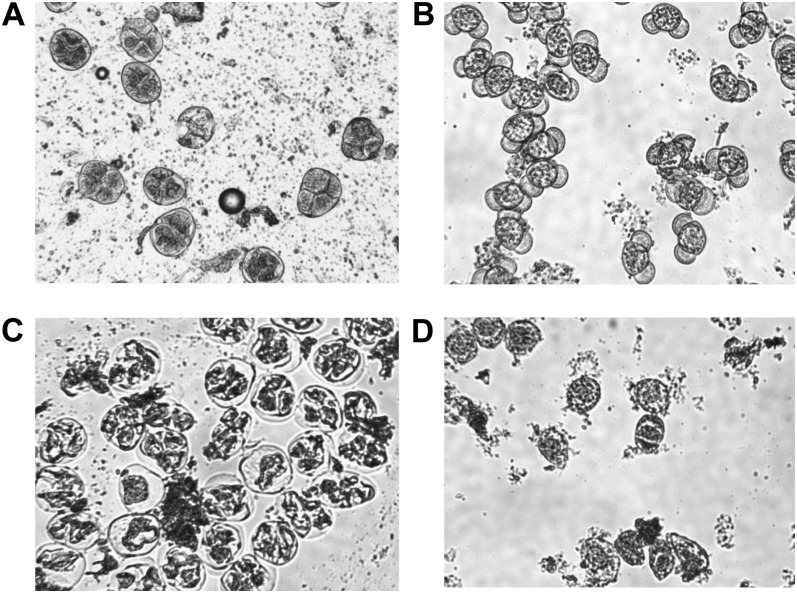
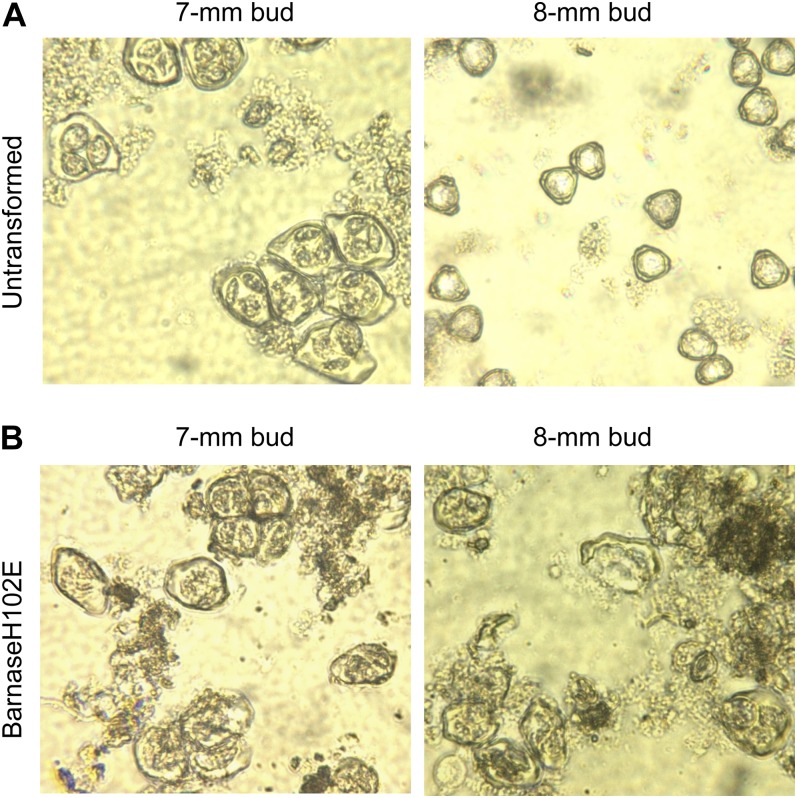
We compared male reproduction development in both transgenic pine and Eucalyptus (spp.). Male cones of transgenic PxL grafts containing PrMC2pro-I-GUS or PrMC2pro-I-barnaseH102E were assessed. The developmental stages of male cones were classified based on the Pollen Development Classification System described by Bramlett and Bridgwater (1989). No differences between GUS and barnaseH102E male cones were observed with respect to the timing of male cone initiation, male cone color, and male cone elongation from stage 1.0 (male cone buds initiate on vegetative shoots and are enclosed by bud scales) to stage 3.0 (male cones increase in length over an extended period and exude clear fluid when pressed between the fingers). However, major morphological differences between barnaseH102E male cones and GUS male cones appeared at stage 3.3 (male cones exude a yellow fluid when pressed between the fingers, approximately 7 to 10 d before pollen release). At this stage, the GUS male cones were swollen and light yellow in color. By contrast, the barnaseH102E male cones did not appear swollen and were brown in color. At stage 4.0 (the beginning of pollen release), the GUS male cones were yellow with open scales and released pollen. By contrast, the barnaseH102E male cones were dark brown with closed scales and with no pollen shed (Fig. 4, A and B). We sampled internal tissues of male cones between stage 2.0 and stage 3.3 and observed these with a microscope. At late stage 2.0 and/or early stage 3.0, the pollen mother cells looked similar in both GUS and barnaseH102E male cones, suggesting that the pollen mother cells were not affected by the presence of PrMC2pro-I-barnaseH102E. Pollen tetrads were also found in some GUS male cones (Fig. 6A) but not in barnaseH102E male cones at this stage. One week later, when male cones on grafts developed to stage 3.3, microscopic observation of sampled GUS male cones revealed that the pollen development had already passed the tetrad stage and individual pollen grains were formed (Fig. 6B). By contrast, pollen development in barnaseH102E male cones was at the tetrad stage. In four of the 12 barnaseH102E lines studied, many tetrads appeared large with ill-defined quadrant boundaries (Fig. 6C) relative to the tetrads observed 1 week earlier in GUS male cones. In five of the 12 barnaseH102E lines, it appeared that many tetrads were disrupted and irregular in shape, with the microspore nuclei appearing degraded (Fig. 6D). In the remaining three lines, no tetrads were found. At stage 4.0, all male cones of the 12 barnaseH102E lines degenerated without releasing pollen while the GUS male cones released pollen inside bags, as shown in Figures 3 and 4A.
Figure 6.
Microscopic observation of internal tissues sampled from different stages of male cones of transgenic PxL pine grafts carrying PrMC2pro-I-GUS or PrMC2pro-I-barnaseH102E. A, Image of tetrads from a GUS male cone at late stage 2.0 or early stage 3.0. The male cones were sampled on February 27, 2006, which is 1 week earlier than in B to D. B, Image of immature pollen grains of a GUS male cone at stage 3.3. C and D, Abnormal pollen development in barnaseH102E male cones at stage 3.3, with C showing enlarged tetrads with disorganized nuclear DNA and D showing degenerated tetrads. The magnification is 200-fold for all images.
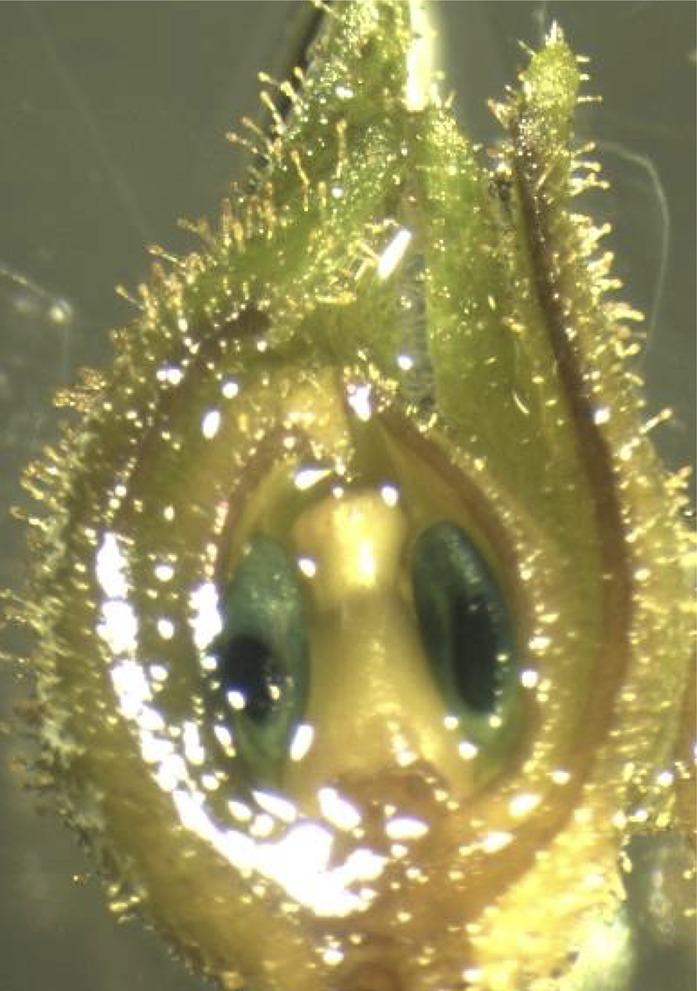
Similar abnormal tetrad and microspore development was observed in flowers sampled from barnaseH102E Eucalyptus (spp.). As mentioned earlier, pollen ablation was studied in seven glasshouse-grown transgenic lines of E. occidentalis containing PrMC2pro-II-barnaseH102E. These E. occidentalis trees continued flowering after the onset of the first flower, and flowers at different developmental stages could be found on a single tree. Floral buds of different maturity identified by operculum length were dissected and observed with a microscope. Pollen tetrads were found in 7-mm floral buds of both untransformed and barnaseH102E E. occidentalis, but their appearance differed from one another. Clear quadrant boundaries and individual microspores were visible in tetrads from untransformed E. occidentalis (Fig. 7A). By contrast, quadrant boundaries and individual microspores were ill defined and the microspore nuclei were disorganized in tetrads from barnaseH102E E. occidentalis (Fig. 7B). In 8-mm untransformed floral buds, immature individual pollen grains can be clearly seen (Fig. 7A), while in the barnaseH102E floral buds of the same size, pollen development was arrested at the tetrad stage, with the tetrads appearing disrupted and degenerated (Fig. 7B). No individual pollen grains were observed in the 8-mm barnaseH102 floral buds, and later on, no pollen was produced in barnaseH102E flowers, as shown in Figure 5.
Figure 7.
Microscopic observation of internal tissues sampled from 7- and 8-mm untransformed (A) and barnaseH102E (B) floral buds of E. occidentalis carrying PrMC2pro-II-barnaseH102E. The size of the floral buds was classified by the length of the operculum. The floral buds were cut longitudinally in the middle, and internal tissues were sampled and observed using a compound microscope. The magnification was 468-fold for A and B.
DISCUSSION
In this paper, we report the development of an in vivo system of E. coli to detect low levels of barnase activities. It has been previously reported that E. coli cells are able to tolerate low levels of barnase activity and have been used for in vivo assays of barnase mutant activities (Jucovic and Hartley, 1995; Axe et al., 1996; Yazynin et al., 1999). We demonstrated that the in vivo system reported in this paper was able to detect low levels of RNase activity of barnase mutants, and this system helped us identify mutants with sufficient activity to achieve high and consistent pollen ablation efficacy while minimizing any vegetative damage. In this in vivo system, the modified barnase sequences were driven by the radiata pine PrAG promoter that is not expected to be functional in a prokaryotic environment. Clearly, however, some low levels of the mutant barnase protein were made in the E. coli cells, which affected colony growth.
We have shown that the expression cassette PrMC2pro-barnaseH102E confers complete pollen ablation at high efficacy across independent transgenic events in widely divergent species (tobacco, Eucalyptus (spp.), and pine). Utilization of barnaseH102E with very low RNase activity was critical to this success. The selection of barnaseH102E for pollen ablation was based on a similar mutant in binase, an RNase produced by Bacillus intermedius and very similar to barnase. The binaseH101E mutant had 2.0% to 2.7% of the wild-type binase activity based on enzyme assay (Yakovlev et al., 1994). However, the in vivo comparison of barnase mutant activities in this study revealed that barnaseH102E had no detectable activity in the E. coli colony assay. Many single amino acid substitutions of the His at position 102 of the barnase protein have been reported, including substitution with Ala, Gln, Tyr, Asn, Asp, Arg, Lys, Gly, Leu, and Pro (Mossakowska et al., 1989; Meiering et al., 1992; Jucovic and Hartley, 1995; Axe et al., 1998). Among all the single substitution mutants listed above, only mutants carrying a substitution with Leu or Pro had detectable activities, while all others were found to render the enzyme inactive, including barnaseH102D. The barnaseH102D mutant is notably similar to barnaseH102E, since Asp is structurally similar to Glu and both are negatively charged amino acids. Since pollen formation was completely abolished in plants carrying PrMC2pro-barnaseH102E, it is clear that the in vivo system of using the transgenic plants in this study is able to reveal the low level of RNase activity of barnaseH102E, which is likely below detectable levels by conventional enzyme assays or bacterium-based in vivo assay systems.
During the course of this study, an analysis of the radiata pine PrMC2 promoter in Arabidopsis was published by Höfig et al. (2003), who showed that the major activity of the PrMC2 promoter was found in tapetal cells. The promoter used by Höfig et al. (2003) is 1.3 kb in length, while the PrMC2 promoter used in this paper is 0.4 kb. The GUS staining and pollen ablation results in tobacco, pine, and Eucalyptus (spp.) presented in this paper clearly showed that the 0.4-kb PrMC2 promoter was sufficient to drive the male reproduction-specific expression for pollen ablation. By comparing the 1.3- and 0.4-kb PrMC2 promoters and the PrMC2 complementary DNA sequence in GenBank, we determined that PrMC2pro-I contained part of the coding sequence and added 13 amino acids to the N terminus of barnaseH102E. We removed the coding sequences and tested the modified PrMC2 promoter, PrMC2pro-II, in plants. The results obtained from transgenic plants indicated that PrMC2pro-II was essentially identical to PrMC2pro-I with respect to promoter activity and anther specificity. PrMC2pro-II may be more desirable for use in future commercial applications since it expresses a “clean” barnaseH102E.
The results presented here showed that the same pollen ablation gene PrMC2pro-barnaseH102E was able to prevent pollen formation in the annual angiosperm tobacco and the perennial angiosperm Eucalyptus (spp.) and in the evolutionarily more primitive perennial gymnosperm pine. The PrMC2 gene was demonstrated to be expressed predominantly in the tapetum of the pollen sac (Walden et al., 1999; Höfig et al., 2003). In pollen development, the first known contribution of the tapetum to the developing microspores is the release of β-1,3-glucanase, which degrades callose to free individual microspores from the tetrad (Rhee and Somerville, 1998). Soon after the release of individual pollen from tetrads, the tapetal cells begin to undergo programmed cell death and release sugars, starch, proteins, and lipids, which are utilized by the developing pollen. The barnaseH102E tetrads, observed in male cones of barnaseH102E pine and flowers of barnaseH102E Eucalyptus (spp.), were enlarged, developed slowly, and degenerated without forming individual pollen grains. These observations strongly suggest that the tapetal cells were not functioning normally or were absent in barnaseH102E male cones and barnaseH102E Eucalyptus (spp.) flowers. The enlarged size of tetrads and longer duration at the tetrad stage are likely the result of lacking glucanase secreted by tapetal cells to degrade the tetrad wall and free pollen grains. Normal tetrad development may also have been affected by the lack of nutrients supplied by the tapetum. The results presented here are consistent with the results obtained by Mariani et al. (1990), who demonstrated that the lack of tapetal cells in TA29-barnase tobacco resulted in the cessation of pollen formation.
The PrMC2pro driving barnaseH102E showed high efficiency of pollen ablation at the transgenic line level in tobacco, Eucalyptus (spp.), and pine. The total number of tested transgenic lines in the three species is summarized in Supplemental Table S2. In tobacco, a total of 30 transgenic lines carrying PrMC2pro-barnaseH102E were studied, and none of them were capable of shedding pollen (Supplemental Table S2). In E. occidentalis and the EH1 hybrid, a total of 71 independently transformed lines carrying PrMC2pro-barnaseH102E either alone or in stacking constructs were tested in a glasshouse or in the field. The results showed that 68 of the 71 lines (95%) did not produce pollen (Supplemental Table S2). In the field test of EH1 carrying pARB598, complete pollen ablation was demonstrated in nine transgenic lines for 2 consecutive years. In addition, two EH1 lines carrying pABCTE01, which were examined at two ages, also showed complete pollen ablation regardless of age (Table III). These results suggest that the expression of the pollen ablation gene was stable in field-grown Eucalyptus (spp.) trees over time. In pine, a total of 42 lines (across both the PxL and loblolly pine genotypes) containing PrMC2pro-barnaseH102E, either alone or stacked with other genes of interest, were assessed, and 41 of the 42 lines (97%) did not release pollen (Supplemental Table S2). Sixteen of the 17 PxL barnaseH102E lines did not release pollen consistently for 4 consecutive years in the field. One or two individual male cones in a male cone cluster of the transgenic PxL graft TRT001343-2, carrying PrMC2pro-I-barnaseH102E, produced a small quantity of underdeveloped pollen grains, while other male cones in the same cluster did not produce pollen. This phenomenon was observed repeatedly on the same graft for 3 consecutive years. At this time, we do not know the biological basis behind this result. Since this phenomenon was observed when TRT001343-2 bore male cones the first time and consistently on the same graft for 3 years, it is unlikely that the failure of 100% pollen ablation in graft TRT001343-2 is due to random mutations of the PrMC2 promoter or barnaseH102E or to the instability of the barnaseH102E protein in individual male cones. The majority of the male cones collected from graft TRT001343-2 did not produce pollen, and all male cones collected from two additional grafts of this transgenic line did not produce pollen, suggesting that this phenomenon was highly graft specific.
The prevention of pollen formation affected the flowering response of the tested plant species in different ways. As tobacco plants can be self-pollinated, many barnaseH102E tobacco flowers did not set seeds due to the lack of pollen, and flowers simply senesced and abscised. As a result, barnaseH102E tobacco plants continued to produce flowers and therefore produced more flowers than the GUS or untransformed plants that set seed normally and stopped flower development. In pine, the barnaseH102E male cones senesced early, suggesting that the lack of pollen development and associated pollen sac collapse signaled the end of male reproductive growth. In Eucalyptus (spp.), we did not see premature senescence of flowers in barnaseH102E plants compared with the untransformed plants. This observation can be explained by the fact that the hybrid EH1, as has been reported for E. grandis, can produce mature fruits without seed development (Horsley and Johnson, 2007; McGowen et al., 2010).
We considered how barnaseH102E with such a low-level activity was able to ablate the tapetum in the transgenic plants. One possible explanation relates to the polyploidy of the tapetum. In many plant species, tapetal cells are polyploid and contain duplicated genomes (Weiss and Maluszynska, 2001). One of the proposed functions of duplicated genomes is to increase the copy number of gene transcription. It has been shown that many homologous gene pairs are expressed at equal levels after polyploid formation (Adams et al., 2003), leading to a higher level of gene transcripts. Tapetal cells are known to have higher metabolic activities than other anther cells and to accumulate proteins, lipids, and carbohydrates, which are released later in supporting pollen development (Pacini et al., 1984; Rhee and Sommerville, 1998). We hypothesize that in the barnaseH102E male cones or flowers, the protein level of barnaseH102E is relatively high in the tapetum compared with other cells and that this high level of barnaseH102E protein compensates the low level of its RNase activity, leading to ablation of the tapetal cells.
PrMC2pro-barnaseH102E showed a high efficacy of pollen ablation in tobacco, pine, and Eucalyptus (spp.) without detrimental effects on vegetative growth. We believe that this highly significant outcome is the result of a unique combination of the PrMC2 promoter, barnaseH102E activity, and the polyploid characteristics of tapetal cells. First, if the gene is expressed at other developmental stages, the presence of barnaseH102E could disrupt the normal cycle of vegetative and floral development. The GUS staining of tobacco floral buds carrying PrMC2pro-I-GUS indicated that the activity and anther specificity of the PrMC2 promoter were relatively high, and we did not see staining in any other tissues (Fig. 1). But when the same promoter was driving barnaseE73G or barnaseK27A, we observed a reduction of transformation efficiency, likely due to promoter leakage in vegetative tissues and the stronger RNase activities of these two barnase mutants. When the PrMC2 promoter was driving barnaseH102E, the transformation efficiency of tobacco and pine were similar to that of the GUS construct. This observation strongly suggested that the low level of RNase activity of barnaseH102E mitigated the low levels of vegetative activity of the PrMC2 promoter.
The phenotypes of barnaseH102E tobacco, pine, and Eucalyptus (spp.) were normal in all respects except failure to shed pollen, and this observation is consistent with the lack of tapetal function. It appears that the presence of PrMC2pro-barnaseH102E did not interfere with vegetative growth and the development of anthers, with the exception of pollen development, in flowering plants and male cones in pine. Our results also showed that the regulatory sequences for tapetum-specific expression of the pine PrMC2 promoter are functional in flowering tobacco and Eucalyptus (spp.), suggesting that the PrMC2 protein homolog may play the same roles during pollen formation in both pine and angiosperms. The observations of similar mechanisms leading to the pollenless phenotype in pine and Eucalyptus (spp.) strongly suggest that the functions of the tapetum during pollen formation are conserved in both angiosperm and gymnosperm plants.
Multiple-year field tests in different geographic locations yielded similar results of pollen ablation in pine and Eucalyptus (spp.), strongly suggesting that the expression and effectiveness of PrMC2pro-barnaseH102E are stable in perennial trees. We did not see any altered or diminished activity of PrMC2pro-barnaseH102E as a consequence of the potential interaction with flanking gene cassettes containing either a strong constitutive or xylem-preferred promoter. The tobacco, pine, and Eucalyptus (spp.) plants containing PrMC2pro-barnaseH102E were phenotypically identical to their corresponding control plants in all respects except pollenless anthers or pollenless male cones. The ability of PrMC2pro-barnaseH102 to induce male sterility without altering female fertility still allows breeding to produce hybrid trees. The very high and stable pollen ablation efficacy in three diverse species indicates that the PrMC2pro-barnaseH102E gene is likely to function in most, if not all, pollen-producing plants. Our results demonstrate that PrMC2pro-barnaseH102E is a highly effective pollen ablation gene for broad-scale applications of transgenic trees in industrial forestry in cases where the prevention of pollen-mediated transgene flow is desired.
MATERIALS AND METHODS
Isolation of the PrMC2 Promoter
Genomic DNA was isolated from Pinus radiata, and sequences upstream of the translation initiation ATG of the PrMC2 gene were isolated using the Genomic Walker Kit (Clontech). The experimental procedures were carried out according to the manufacturer’s instructions. The general strategy of the Genomic Walker Kit was to cut the genomic DNA with enzymes to generate blunt ends and then ligate the resulting DNA fragments to an adaptor with known sequences. In the first round of PCR amplification, the upstream primer (forward primer) annealed to the known sequences of the adaptor while the downstream primer (reverse primer) annealed to the known sequence of PrMC2 complementary DNA 3′ to the translation initiation ATG but close to the ATG. The amplified product from the first PCR was used as the template in the second PCR (nested PCR), with an outside primer annealing to the adaptor and a nested gene-specific primer. The cloned PrMC2 genomic DNA had 390 bp including 354 bp 5′ of the translation initiation ATG and 36 bp of the coding region. The 390-bp sequence is identical to the same sequence (accession no. AF337657) reported in GenBank except three nucleotides 5′ of the translation initiation ATG.
Site-Specific Mutagenesis to Generate barnaseH102E and barnaseK27A
The barnaseH102E and barnaseK27A coding sequences were generated using oligonucleotide-mediated mutagenesis described by Higuchi et al. (1988). The overall strategy was to use four DNA primers, two mutagenesis primers and two flanking primers, to introduce the mutations. The two mutagenesis primers introduced the mutation at a selected site, while the two flanking primers amplified the whole coding region of barnase. The first round of PCR was composed of two independent PCRs; one reaction contained the 5′ flanking primer (forward) and the reverse mutagenesis primer, while the other reaction contained the forward mutagenesis primer and the 3′ flanking primer. The outcome of these two PCRs was the two partially overlapping fragments, and the overlapping portion of the two fragments contained the mutated sequence. In the second round of PCR, the two partially overlapping fragments were used as the template with the two flanking primers to amply the full length of the barnase coding sequence.
Obtaining barnaseE73G and barnaseF106S and in Vivo Comparison of RNase Activities of Barnase Mutants in Escherichia coli
The entire barnase coding region was amplified in a PCR using Taq polymerase from New England Biolabs. The amplified fragment was ligated downstream of a 1.5-kb fragment of the pine AG gene promoter carried in pUC19, which does not contain the barstar sequence. The resulting ligation mixture was introduced into E. coli strain DH5α, and single colonies were obtained on Luria-Bertani (LB) plates containing ampicillin (75 μg mL−1) after incubation at 37°C overnight (approximately 16 h). The diameters of the single colonies were classified into three groups, large, medium, and small. Two hundred single colonies with medium or small diameters were selected and inoculated into 1 mL of LB liquid supplemented with ampicillin. After overnight growth at 37°C, the cell density (cell growth rate) of the bacterial cultures was determined visually, and the cultures growing far behind others (slow-growing cultures) were selected. The plasmid DNA was extracted from the slow-growing cultures and reintroduced into E. coli DH5α to confirm that the single colonies harboring the plasmid DNAs had smaller diameters compared with the colonies harboring a control construct of the same pine AG promoter driving barnaseY102E on the ampicillin LB plates. Then, the plasmid DNA extracted from slow-growing colonies was sequenced.
To compare the RNase activities of the barnase mutants in vivo, an equal amount of the plasmid DNA from each of the mutants was mixed with 50 μL of E. coli DH5α. After incubation on ice and heat shock, the cells were transferred to 1 mL of LB liquid medium and incubated at 37°C for 45 min with slow agitation. After incubation, series dilutions of the cultures were made, and 50 μL from each dilution was placed on a LB agar petri dish (100 × 15 mm) supplemented with ampicillin. After overnight (approximately 16 h) incubation at 37°C, the petri dishes with similar numbers of single colonies (between 180 and 320) were selected for comparison, and the diameters of the single colonies were measured with a dissecting microscope. The stronger the barnase activity is, the smaller the colony diameter.
Plant Transformation and Growth Conditions
Tobacco (Nicotiana tabacum) transformation was performed by following the protocol of leaf disc transformation (Horsch et al., 1988). The Eucalyptus (spp.) transformation procedures were developed at ArborGen Inc., and detailed descriptions can be found in two patent applications (Chang et al., 2006a, 2006b). In general, tissues from young leaves grown in vitro were infected with Agrobacterium tumefaciens on cocultivation medium for 4 d. After cocultivation, the leaf explants were transferred to medium containing kanamycin for the selection of transgenic tissue and Timentin to eradicate the Agrobacterium. After adventitious shoot development, the tissue was sampled for DNA extraction and PCR analysis. The kanamycin-resistant shoots containing the gene of interest were subcultured in rooting medium and then transferred into soil. Pine transformation was performed by following the procedure described by Connett-Porceddu and Gulledge (2005) and Connett-Porceddu et al. (2007). In general, embryogenic cell cultures were infected by Agrobacterium. After infection, the infected cell culture was washed several times with washing medium containing Timentin, and then the cell cultures were placed on medium containing geneticin for the selection of transgenic embryogenic calli. Somatic embryos were induced from the transgenic embryogenic calli, and rooted seedlings were produced from the embryos.
Potted tobacco and Eucalyptus occidentalis were grown in soil in 1-gallon pots in a glasshouse. The temperature inside the glasshouse ranged from 16°C to 24°C in winter and from 27°C to 35°C in summer. A 16-h photoperiod was maintained constantly inside the glasshouse using natural light and supplemental light at an intensity of 65 μmol m−2 s−1.
Pine Grafting
Scions were collected either from 1-year-old potted trees or from 2-year-old field-grown trees in winter and then stored at 4°C for 1 or 2 months before grafting. The grafting was performed by following the standard cleft-and-paraffin grafting technique (White et al., 1983). In general, the scions, with diameters similar to a No. 2 pencil and 10 to 15 cm in length, were completely covered with paraffin and then cut into a V shape at the base. The V shape end was inserted into a rootstock tree branch with similar diameters, and careful matching of cambium was conducted. The graft unions were sealed with Parafilm and wrapped completely with degradable rubber tape. Two months after grafting, the survival rate was checked by visually inspecting the newly growing needles on the grafted scions.
Pollen Ablation Data Collection in Tobacco, Pine, and Eucalyptus (spp.)
In tobacco, the presence of pollen was determined by three methods. First, 10 opened flowers of each plant were observed visually for the presence of pollen. Second, four anthers collected from each plant were cut open along their long axis and observed with a dissecting microscope with 9-fold magnification. Third, the internal tissues of anthers were sampled and observed with a compound microscope from selected transgenic lines and untransformed tobacco.
In pine, the initiation and development of male cone and female cone on grafts were monitored once per week from January to April each year. The developmental stages of male cones were classified based on the Pollen Development Classification System described by Bramlett and Bridgwater (1989). Approximately 2 weeks before pollen release, three randomly selected male cone clusters on each graft or four male cone clusters on each field-grown tree were bagged, and then the rest of the male cone clusters on the same grafts or field-grown trees were removed. Each bag contained one male cone cluster. The bagging practice is for the purpose of preventing the release of transgenic pollen into the environment. The bag was manufactured by Devro-Teepak, a producer of manufactured casings for the food industry. The bag was clear, and the bagged pollen clusters were clearly visible from the outside. The bag is made of cellulose membrane, and air exchange occurs between inside and outside of the bag while the pollen grains are contained inside. The bagged male cones and vegetative shoots were able to develop normally inside the bags. This bag has been used for pollen collection in pine breeding practice for many years. It takes 18 month for a loblolly female cone to release seeds after initiation. The immature female cones on grafts were collected 2 months before seed release.
Two sets of pollen ablation data were collected from the same transgenic and untransformed pine grafts or field-grown trees. The first set was the visual observation of bags containing male cone clusters collected either from grafts or from field-grown trees. It is easy to determine the massive pollen release inside bags by visual observation. The second set of pollen ablation data in pine was collected by extracting pollen grains from individual male cones sampled from the bagged male clusters and microscopic observation of the extracted pollen grains. Three to six individual male cones from each bagged male cone cluster were randomly selected, and each male cone was placed into a microcentrifuge tube containing 0.5 mL of water. Pollen grains inside the male cone were released into the water by gently pressing the male cone five to six times with a plastic pestle. A small aliquot (10 μL) of the resulting liquid sample was loaded on a glass slide, coverslipped, and observed with a compound microscope with 200-fold magnification.
In Eucalyptus (spp.), the pollen ablation data were collected in a manner similar to the data collection in pine, but with some differences. First, two sets of pollen ablation data were collected from E. occidentalis growing in the glasshouse. For each E. occidentalis tree, 50 opened flowers were visually observed for the presence of pollen. Second, we dissected anthers of unopened flowers to determine the presence of pollen in the flowers. In the dissection, the flowers with brown-colored operculum were selected to ensure the presence of immature pollen grains in the sampled flowers. The operculum of an unopened flower was removed by hand, and all anthers were cut off and placed into a microcentrifuge tube containing 0.2 mL of water. Immature pollen grains inside the anthers were released into the water by gently pressing the anthers five to six times with a plastic pestle. A small aliquot (10 μL) of the resulting liquid sample was loaded on a glass slide, coverslipped, and observed with a compound microscope with 468-fold magnification. To compare the pollen production between a barnaseH102E flower and an untransformed flower, pollen grains were extracted from 10 anthers of each individual flower and microscopically observed in the same way as described above except for the reduced volume of water inside the tube (0.1 mL). The pollen production for an individual flower was estimated by counting pollen grains from six randomly selected views and then averaged.
For the field-grown trees of Eucalyptus (spp.) hybrid EH1 (Eucalyptus grandis × Eucalyptus urophylla), the visual observation of opened flowers is difficult, since the flowers are smaller than E. occidentalis flowers. As a result, only the microscopic observation of pollen ablation data were collected, which involved the dissection and microscopic observation of unopened flowers. Two types of dissection samples were prepared. One was called single flower dissection, in which all anthers of a single flower were collected. The other was called pooled flower dissection, in which anthers collected from 10 individual flowers were pooled together to form one sample. The pollen grain extraction and microscopic observation of EH1 were similar to the practice in E. occidentalis except for the water volume in a microcentrifuge tube for the pooled sample, which was increased to 0.5 mL.
TaqMan Real-Time PCR to Determine Transgene Copy Number in Pine and Eucalyptus (spp.)
Genomic DNA was isolated from pine needles or Eucalyptus (spp.) leaves using the cetyl-trimethyl-ammonium bromide method described by Doyle and Doyle (1990) and then quantified by a SpectraMAX Gemini Fluorescence microplate reader (Molecular Devices). The TaqMan quantitative PCR technology and the δδCt method (Livak and Schmittgen, 2001) were employed to determine transgene copy number. Primers and probes for the targeted gene of bacterial NPTII (for neomycin phosphotransferase II) and a normalizing gene were designed using Beacon Designer 6 software (Premier Biosoft) and optimized in test runs. The sequence of the forward primer for NPTII was 5′-CACGACGGGCGTTCCTTGC-3′, while the reverse primer sequence was 5′-GGTGGTCGAATGGGCAGGTAGC-3′. The TaqMan probe sequence for NPTII was 5′-[FAM]ACTGAAGCGGGAAGGGACTGGCT[TAMRA]-3′. Prior to the PCR, a few transgenic lines of pine or Eucalyptus (spp.) with one, two, or three copies were identified using Southern blots. The TaqMan PCR was run on a Mx4000 QPCR machine (Stratagene). In every run for copy number determination, the genomic DNA isolated from the transgenic plants of Southern-confirmed one-, two-, and three-copy lines were included for calibrating the results. The copy number was determined by comparing the results obtained from the transgenic lines under investigation with the results from calibrating transgenic lines using the method described by Livak and Schmittgen (2001).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypes of transgenic tobacco carrying PrMC2pro-I-barnaseH102E.
Supplemental Figure S2. Microscopic observation of normal and underdeveloped pine pollen grains.
Supplemental Figure S3. Phenotypic comparison of barnaseH102E and untransformed pine grafts on potted rootstock trees 8 months after grafting.
Supplemental Table S1. In vivo comparison of RNase activities of four barnase mutants in E. coli.
Supplemental Table S2. Number of transgenic lines of tobacco, pine, and Eucalyptus (spp.) plants that contained either PrMC2pro-I-barnaseH102E or PrMC2pro-II-barnaseH102E and were tested in the glasshouse or in field trials.
Supplementary Material
Glossary
- LB
Luria-Bertani
References
- Adams KL, Cronn R, Percifield R, Wendel JF. (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100: 4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe DD, Foster NW, Fersht AR. (1996) Active barnase variants with completely random hydrophobic cores. Proc Natl Acad Sci USA 93: 5590–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe DD, Foster NW, Fersht AR. (1998) A search for single substitutions that eliminate enzymatic function in a bacterial ribonuclease. Biochemistry 37: 7157–7166 [DOI] [PubMed] [Google Scholar]
- Bramlett DL, Bridgwater FE. (1989) Pollen development classification system for loblolly pine. In Proceedings of the 20th Southern Forest Tree Improvement Conference. National Reforestation, Nurseries, and Genetic Resources (RNGR) Program, USDA Forest Service, Washington, DC, pp 116–121
- Bramlett DL, Williams CG, Burris LC. (1995) Surrogate pollen induction shortens the breeding cycle in loblolly pine. Tree Physiol 15: 531–535 [DOI] [PubMed] [Google Scholar]
- Brunner AM, Li J, DiFazio S, Shevchenko O, Mohamed R, Montgomery B, Elias A, Van Wormer K, DiFazio SP, Strauss SH. (2007) Genetic containment of forest plantations. Tree Genet Genomes 3: 75–100 [Google Scholar]
- Brunner AM, Rottmann WH, Sheppard LA, Krutovskii K, DiFazio SP, Leonardi S, Strauss SH. (2000) Structure and expression of duplicate AGAMOUS orthologues in poplar. Plant Mol Biol 44: 619–634 [DOI] [PubMed] [Google Scholar]
- Chang S, Thomas RD, Handley LW, Connett MB, Hamilton RL, inventors. May 11, 2006a. Eucalyptus urophylla transformation and regeneration. US Patent Application No. US 2006/0101536 A1
- Chang S, Thomas RD, Handley LW, Connett MB, Hamilton RL, inventors. May 11, 2006b. Eucalyptus urophylla transformation and selection. US Patent Application No. US 2006/0101537 A1
- Connett-Porceddu MB, Gladfelter HJ, Gulledge JE, McCormack RR, inventors. January 2, 2007. Enhanced transformation and regeneration of transformed embryogenic pine tissue. US Patent No. US 7,157,620 B2
- Connett-Porceddu MB, Gulledge JE, inventors. November 15, 2005. Enhanced selection of genetically modified pine embryogenic tissue. US Patent No. US 6,964,870 B2
- Cook D, Fowler S, Fiehn O, Thomashow MF. (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 101: 15243–15248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15 [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM. (1993) Anther development: basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley RW. (1988) Barnase and barstar: expression of its cloned inhibitor permits expression of a cloned ribonuclease. J Mol Biol 202: 913–915 [DOI] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res 16: 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenicka H, Fladung M. (2006) Biosafety in Populus spp. and other forest trees: from non-native species to taxa derived from traditional breeding and genetic engineering. Trees (Berl) 20: 131–144 [Google Scholar]
- Höfig KP, Moyle RL, Putterill J, Walter C. (2003) Expression analysis of four Pinus radiata male cone promoters in the heterologous host Arabidopsis. Planta 217: 858–867 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry J, Hoffmann N, Neidermeyer J, Rogers SG, Fraley RT. (1988) Leaf disc transformation. In RA Schilperoort, SB Gelvin, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp A5 1–9 [Google Scholar]
- Horsley TN, Johnson SD. (2007) Is eucalyptus cryptically self-incompatible? Ann Bot (Lond) 100: 1373–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucovic M, Hartley RW. (1995) In vivo system for the detection of low level activity barnase mutants. Protein Eng 8: 497–499 [DOI] [PubMed] [Google Scholar]
- Lännenpää M, Hassinen M, Ranki A, Hölttä-Vuori M, Lemmetyinen J, Keinonen K, Sopanen T. (2005) Prevention of flower development in birch and other plants using a BpFULL1:BARNASE construct. Plant Cell Rep 24: 69–78 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mariani C, De Beuckeleer M, Truettner J, Leemans J, Goldberg RB. (1990) Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature 347: 737–741 [Google Scholar]
- McGowen MH, Vaillancourt RE, Pilbeam DJ, Potts BM. (2010) Sources of variation in self-incompatibility in the Australian forest tree, Eucalyptus globulus. Ann Bot (Lond) 105: 737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiering EM, Serrano L, Fersht AR. (1992) Effect of active site residues in barnase on activity and stability. J Mol Biol 225: 585–589 [DOI] [PubMed] [Google Scholar]
- Mossakowska DE, Nyberg K, Fersht AR. (1989) Kinetic characterization of the recombinant ribonuclease from Bacillus amyloliquefaciens (barnase) and investigation of key residues in catalysis by site-directed mutagenesis. Biochemistry 28: 3843–3850 [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Osakabe K, Chiang VL. (2009) Isolation of 4-coumarate Co-A ligase gene promoter from loblolly pine (Pinus taeda) and characterization of tissue-specific activity in transgenic tobacco. Plant Physiol Biochem 47: 1031–1036 [DOI] [PubMed] [Google Scholar]
- Pacini E, Franchi GG, Hesse M. (1984) The tapetum: its form, function, and possible phylogeny in Embryophyta. Plant Syst Evol 149: 155–185 [Google Scholar]
- Perera RJ, Rice S, Eagleton C, De Ath A, Shenk A. (2000) The Pinus ubiquitin promoter: a strong constitutive promoter for gene expression in gymnosperms and angiosperms. Plant and Animal Genome VIII Conference at Town & Country Hotel, San Diego, January 9–12, 2000. http://www.intl-pag.org/8/abstracts/pag8138.html
- Raghavan V. (1989) mRNAs and a cloned histone gene are differentially expressed during anther and pollen development in rice (Oryza sativa L.). J Cell Sci 92: 217–229 [DOI] [PubMed] [Google Scholar]
- Raghavan V. (1997) Pollen abortion and male sterility. In V Raghavan, ed, Molecular Embryology of Flowering Plants. Cambridge University Press, Cambridge, UK, pp 120–151
- Rhee SY, Somerville CR. (1998) Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J 15: 79–88 [DOI] [PubMed] [Google Scholar]
- Rottmann WH, Meilan R, Sheppard LA, Brunner AM, Skinner JS, Ma C, Cheng S, Jouanin L, Pilate G, Strauss SH. (2000) Diverse effects of overexpression of LEAFY and PTLF, a poplar (Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant J 22: 235–245 [DOI] [PubMed] [Google Scholar]
- Serrano L, Kellis JT, Jr, Cann P, Matouschek A, Fersht AR. (1992) The folding of an enzyme. II. Substructure of barnase and the contribution of different interactions to protein stability. J Mol Biol 224: 783–804 [DOI] [PubMed] [Google Scholar]
- Shivanna KR, Cresti M, Ciampolini F. (1997) Pollen development and pollen-pistil interaction. In KR Shivanna, VK Sawhney, eds, Pollen Biotechnology for Crop Production and Improvement. Cambridge University Press, Cambridge, UK, pp 15–39
- Skinner JS, Meilan R, Brunner AM, Strauss SH. (2000) Options for genetic engineering of floral sterility in forest trees. In SM Jain, SC Minocha, eds, Molecular Biology of Woody Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 135–153
- Southerton SG. (2007) Early flowering induction and Agrobacterium transformation of the hardwood tree species Eucalyptus occidentalis. Funct Plant Biol 34: 707–713 [DOI] [PubMed] [Google Scholar]
- Strauss SH, Rottmann WH, Brunner AM, Sheppard AL. (1995) Genetic engineering of reproductive sterility in forest trees. Mol Breed 1: 5–26 [Google Scholar]
- Tandre K, Svenson M, Svensson ME, Engström P. (1998) Conservation of gene structure and activity in the regulation of reproductive organ development of conifers and angiosperms. Plant J 15: 615–623 [DOI] [PubMed] [Google Scholar]
- van der Meer IM, Stam ME, van Tunen AJ, Mol JN, Stuitje AR. (1992) Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tunen AJ, Mur LA, Brouns GS, Rienstra JD, Koes RE, Mol JN. (1990) Pollen- and anther-specific chi promoters from petunia: tandem promoter regulation of the chiA gene. Plant Cell 2: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden AR, Walter C, Gardner RC. (1999) Genes expressed in Pinus radiata male cones include homologs to anther-specific and pathogenesis response genes. Plant Physiol 121: 1103–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Meilan R, Brunner AM, Skinner JS, Ma C, Gandhi HT, Strauss SH. (2007) Field trial detects incomplete barstar attenuation of vegetative cytotoxicity in Populus trees containing a poplar LEAFY promoter:barnase sterility transgene. Mol Breed 19: 69–85 [Google Scholar]
- Weiss H, Maluszynska J. (2001) Molecular cytogenetic analysis of polyploidization in the anther tapetum of diploid and autotetraploid Arabidopsis thaliana plants. Ann Bot (Lond) 87: 729–735 [Google Scholar]
- White G, Lowe WJ, Wright J. (1983) Paraffin grafting techniques for loblolly pine. South J Appl For 7: 116–118 [Google Scholar]
- Yakovlev GI, Moiseyev GP, Struminskaya NK, Borzykh OA, Kipenskaya LV, Znamenskaya LV, Leschinskaya IB, Chernokalskaya EB, Hartley RW. (1994) Mutational analysis of the active site of RNase of Bacillus intermedius (BINASE). FEBS Lett 354: 305–306 [DOI] [PubMed] [Google Scholar]
- Yazynin S, Lange H, Mokros T, Deyev S, Lemke H. (1999) A new phagemid vector for positive selection of recombinants based on a conditionally lethal barnase gene. FEBS Lett 452: 351–354 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.