Abstract
Xyloglucan is the major hemicellulosic polysaccharide in the primary cell walls of most vascular dicotyledonous plants and has important structural and physiological functions in plant growth and development. In Arabidopsis (Arabidopsis thaliana), the 1,4-β-glucan synthase, Cellulose Synthase-Like C4 (CSLC4), and three xylosyltransferases, XXT1, XXT2, and XXT5, act in the Golgi to form the xylosylated glucan backbone during xyloglucan biosynthesis. However, the functional organization of these enzymes in the Golgi membrane is currently unknown. In this study, we used bimolecular fluorescence complementation and in vitro pull-down assays to investigate the supramolecular organization of the CSLC4, XXT1, XXT2, and XXT5 proteins in Arabidopsis protoplasts. Quantification of bimolecular fluorescence complementation fluorescence by flow cytometry allowed us to perform competition assays that demonstrated the high probability of protein-protein complex formation in vivo and revealed differences in the abilities of these proteins to form multiprotein complexes. Results of in vitro pull-down assays using recombinant proteins confirmed that the physical interactions among XXTs occur through their catalytic domains. Additionally, coimmunoprecipitation of XXT2YFP and XXT5HA proteins from Arabidopsis protoplasts indicated that while the formation of the XXT2-XXT2 homocomplex involves disulfide bonds, the formation of the XXT2-XXT5 heterocomplex does not involve covalent interactions. The combined data allow us to propose that the proteins involved in xyloglucan biosynthesis function in a multiprotein complex composed of at least two homocomplexes, CSLC4-CSLC4 and XXT2-XXT2, and three heterocomplexes, XXT2-XXT5, XXT1-XXT2, and XXT5-CSLC4.
The major structural components of plant cell walls are polysaccharide networks composed of pectins, hemicelluloses, and cellulose. The precise arrangement and composition of these components exert important influences on plant growth and development. Moreover, understanding (and possibly manipulating) cell wall formation and structure is key for industrial applications such as the production of biofuels and biomaterials. In dicotyledons and nongraminaceous monocotyledons, xyloglucan (XyG) is the major hemicellulosic polysaccharide (Scheller and Ulvskov, 2010). XyG has a backbone made of β-1,4-linked glucosyl residues and is branched, with short side chains made of Xyl, Gal, and Fuc. For example, in Arabidopsis (Arabidopsis thaliana), the basic repeating XyG subunit is XXXG, which is composed of a β-1,4-glucan, where three of four glucosyl residues are linked to α-d-xylosyl residues at the O-6 position. These basic XXXG repeating subunits can be further substituted at the O-2 position of the xylosyl residues by either a β-d-galactosyl residue (L) or a dimer, α-l-fucosyl-(1,2)-β-d-galactosyl (F), forming XXLG, XLLG, and XLFG subunits, respectively (Fry et al., 1993).
All polysaccharides in plants, except cellulose and callose, are assembled by Golgi membrane-bound glycan synthases and glycosyltransferases (Keegstra and Raikhel, 2001) from various nucleotide diphosphate sugars synthesized mainly in the cytosol (Reyes and Orellana, 2008). Glycosyltransferases transfer sugar residues from a nucleotide diphosphate sugar onto the polysaccharide backbone, such as glucan in XyG. Most glycosyltransferases are localized in the Golgi membrane and have a type II membrane protein topology (Perrin et al., 2001), with a short N-terminal fragment most likely protruding into the cytosol, one helical transmembrane domain, and a catalytic domain containing a DXD motif, which is attached to a flexible stem region; both the catalytic domain and the stem region are localized in the Golgi lumen (Albersheim et al., 2010). These glycosyltransferases are highly specific; it is postulated that a distinct enzyme is required to create each type of linkage (Keegstra and Raikhel, 2001). The other Golgi-resident enzymes involved in polysaccharide biosynthesis are glycan synthases belonging to the Cellulose Synthase-Like (CSL) superfamily. Like cellulose synthases, CSLs have several transmembrane domains and the catalytic domain synthesizing glycan polymeric chains, which is predicted to have the D,D,D,Q/RXXRW motif (Saxena et al., 1995; Charnock et al., 2001; Doblin et al., 2002).
It has been proposed that glycosyltransferases form multienzyme complexes to synthesize complex polysaccharide structures (Keegstra, 2010). Recently, several pieces of evidence supporting this hypothesis have emerged. First, several putative protein complexes involved in polysaccharide biosynthesis have been identified. Using a wheat (Triticum aestivum) microsomal fraction and a coimmunoprecipitation approach, Zeng et al. (2010) demonstrated that three Golgi-resident glycosyltransferases, members of the GT43, GT47, and GT75 families, are involved in arabinoxylan biosynthesis and can be coimmunoprecipitated with antibodies specific to GT43. Atmodjo et al. (2011) reported that GAUT1, a galacturonosyltransferase involved in pectin biosynthesis, interacts with another homologous protein, GAUT7, forming an active multiprotein complex localized in the Golgi membrane. Two other putative glycosyltransferases involved in pectin biosynthesis, ARABINAN DEFICIENT1 (ARAD1) and ARAD2 were shown to form homocomplexes and heterocomplexes in the Golgi when transiently coexpressed in Nicotiana benthamiana (Sakuragi et al., 2011; Harholt et al., 2012). In yeast, Stolz and Munro (2002) demonstrated that several mannosyltransferases involved in cell wall mannan synthesis form two types of protein complexes, M-Pol I and II.
The second line of evidence supporting the involvement of multiprotein complexes in glycan biosynthesis is the finding that glycosyltransferases either tend to form or are even required to form homooligomers and heterooligomers or complexes in order to be targeted to and retained in the Golgi. Moreover, some glycosyltransferases function as homooligomers or heterooligomers rather than in monomeric form (McCormick et al., 2000; van Meer, 2001; Hassinen et al., 2010). For example, for cellulose biosynthesis, cellulose synthases must form homodimers and heterodimers organized into a rosette-like multisubunit complex (Kurek et al., 2002; Taylor et al., 2003; Desprez et al., 2007; Carpita, 2011). Studying Cellulose Synthase-Like C4 (CSLC4; Cocuron et al., 2007) demonstrated that coexpression of XXT1 and CSLC4 in Pichia cells increases the length of the synthesized glucan polymer more than the expression of CSLC4 alone, despite the fact that the glucan chain cannot be xylosylated because yeast do not produce UDP-Xyl. It was proposed that XXT1 may assist CSLC4 function through protein-protein interactions.
In Arabidopsis, at least seven enzymes are involved in XyG biosynthesis: a β-glucan synthase, CSLC4 (Cocuron et al., 2007); three α-1,6-xylosyltransferases, XXT1, XXT2, and XXT5 (Cavalier and Keegstra, 2006; Cavalier et al., 2008; Zabotina et al., 2008); two β-1,2-galactosyltransferases, MUR3 (Madson et al., 2003) and XTLT2 (Jensen et al., 2012); and an α-fucosyltransferase, FUT1 (Perrin et al., 1999; Vanzin et al., 2002). Although the structure of XyG is well characterized and most of the enzymes involved in XyG biosynthesis have been identified, knowledge about the mechanism of XyG biosynthesis is still limited.
In order to understand the function of Arabidopsis XyG-synthesizing enzymes, we first investigated complex formation in vivo and potential physical interactions among three xylosyltransferases, XXT1, XXT2, and XXT5, and a glucan synthase, CSLC4. We used two independent approaches: (1) bimolecular fluorescence complementation (BiFC) assays in Arabidopsis protoplasts, visualizing the signal by fluorescence microscopy and quantifying its intensity by flow cytometry (Walter et al., 2004; Li et al., 2010); and (2) in vitro pull-down assays using recombinant proteins expressed in Escherichia coli. Here, we report results that, to our knowledge for the first time, directly demonstrate protein-protein complex formation among the xylosyltransferases and glucan synthase in the Golgi membrane in Arabidopsis cells.
RESULTS
To study the in vivo colocalization of glycosyltransferases involved in XyG biosynthesis into putative protein-protein complexes, we conducted BiFC assays using proteins transiently expressed in Arabidopsis protoplasts (Hu et al., 2002). For the assay, we made two constructs for each enzyme (three xyloglucan xylosyltransferases, XXT1, XXT2, and XXT5, and the glucan synthase CSLC4); each enzyme was fused to either the N- or C-terminal fragment of yellow fluorescent protein (YFP; N-YFP and C-YFP) in the pSAT vectors (Citovsky et al., 2008) to form NC and CC forms, respectively (Fig. 1A). We fused the YFP fragment to the N terminus of the enzyme for all proteins, presuming that the short cytosolic N terminus is less likely to affect YFP reconstitution than the big bulky transferase catalytic domain at the C terminus.
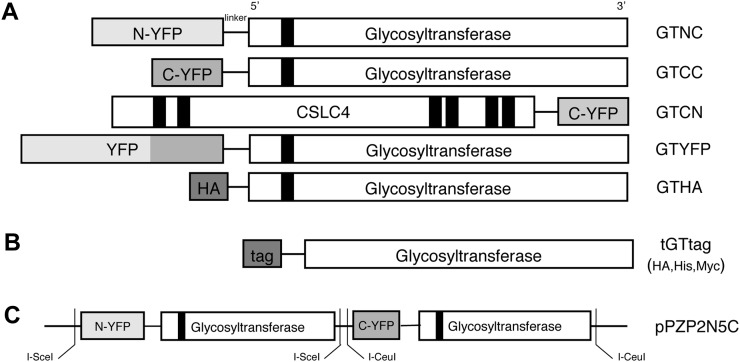
Figure 1.
Plasmid constructs used for BiFC. A, Each gene was cloned into the pSAT vector with the N- or C-terminal fragment of YFP (N-YFP and C-YFP), HA-tagged vector, and full-length YFP-tagged vector. Genes were fused to the C terminus of each YFP fragment (GTNC and GTCC), full-length YFP tag (GTYFP), and HA tag (GTHA). One additional construct was made that fused C-terminal fragments of YFP to the C terminus of CSLC4 (GTCN). B, The truncated versions of the xylosyltransferases with His, HA, or Myc tags cloned into the pET-15b backbone vector. C, Two expression cassettes, GTNC and GTCC, were cloned into the coexpression vector (pPZP2N5C).
BiFC Assays in Arabidopsis Protoplasts
BiFC constructs for each pair of xylosyltransferases, XXT1-XXT2, XXT1-XXT5, and XXT2-XXT5, were transiently coexpressed in Arabidopsis protoplasts using the polyethylene glycol (PEG) method (Jin et al., 2001), and the signal from reconstituted YFP was examined by fluorescence microscopy. All three pairs of XXTs show detectable fluorescence, which forms punctate structures (Fig. 2, A, E, and F). Xylosyltransferases are predicted to localize to the Golgi; to test the localization of the expressed fusion proteins, Golgi were visualized using the marker G-CK (Nelson et al., 2007), which is composed of cyan fluorescent protein (CFP) fused with a Golgi membrane signal peptide. The protoplasts were cotransfected with three constructs simultaneously, XXT2NC, XXT5CC, and G-CK, and examined by fluorescence microscopy (Fig. 2B). The merged image (Fig. 2C) confirms colocalization of the punctate YFP signal from the BiFC pair and the CFP signal marking the Golgi.
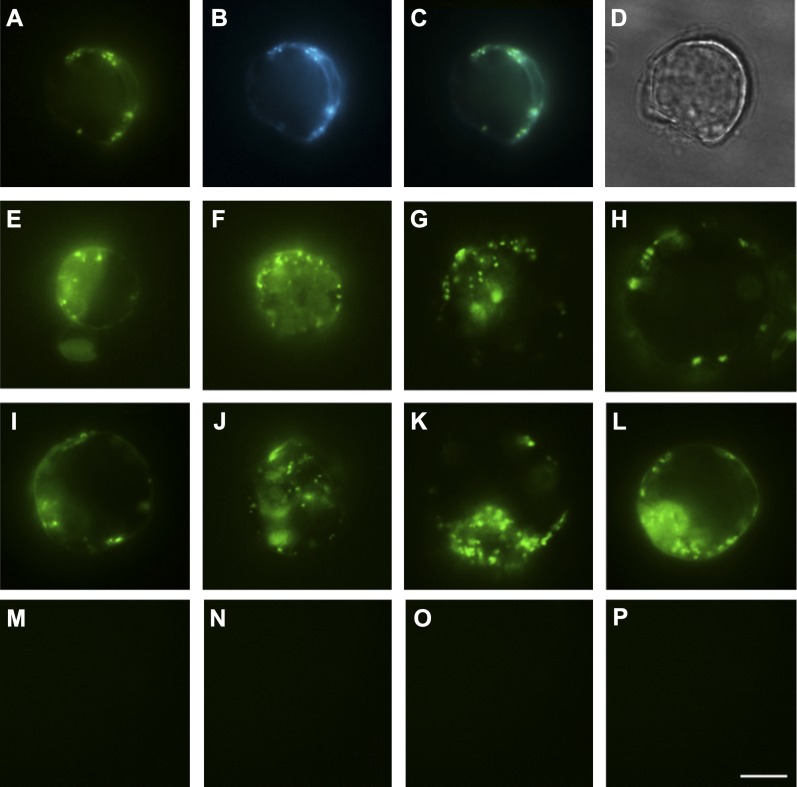
Figure 2.
Fluorescence images of BiFC signal for homodimers and heterodimers in Arabidopsis protoplasts. A and B, Arabidopsis root protoplasts coexpressing XXT2NC and XXT5CC (A) and Golgi marker G-CK (B). C, Merged image of A and B to confirm BiFC signal localization in the Golgi. D, Bright-field image of the same protoplast. E to P, Arabidopsis leaf protoplasts coexpressing XXT1CC and XXT2NC (E), XXT1NC and XXT5CC (F), CSLC4NC and XXT1CC (G), CSLC4NC and XXT5CC (H), XXT2NC and XXT2CC (I), XXT5NC and XXT5CC (J), CSLC4NC and CSLC4CC (K), CSLC4NC and CSLC4CN (L), XXT2NN and XXT5CC (M), XXT2NC and MNS1CC (N), XXT5CC and MNS1NC (O), and CSLC4CC and MNS1NC (P). Representative images from three independent experiments are shown. Bar = 10 μm for all images.
Three different negative controls confirmed that the observed BiFC signals result from specific protein-protein colocalization of glycosyltransferases and not from incidental nonspecific YFP reconstitution. The first negative control was coexpression of two BiFC constructs, where the two YFP complementary fragments were fused to the opposite termini of the two glycosyltransferases: one YFP fragment was fused to the N terminus of XXT2 and the other YFP fragment was fused to the C terminus of XXT5. Another negative control was a transfection of protoplasts with two BiFC constructs, one of which carried an XXT protein fused with one YFP fragment and another construct that carried only the complementary YFP fragment without a fused protein. As a third negative control, we chose an Arabidopsis class I α-mannosidase (MNS1), which is a type II membrane protein with hydrolytic activity and is involved in protein glycosylation in the Golgi (Liebminger et al., 2009). MNS1 fused with the YFP fragment was coexpressed with each of the XXT and CSLC4 proteins fused with the complementary YFP fragment. For all negative controls, we did not observe any BiFC signal by fluorescence microscopy (Fig. 2, M–P). The BiFC results confirm that XXT1, XXT2, and XXT5 have the same topology, with their N termini on the same side of the Golgi membrane forming heterocomplexes, and that the YFP complementary fragments fused to the N termini of XXTs do not affect their localization.
To confirm whether the fusion proteins used in BiFC assays preserved their functions in vivo, XXT5NC and XXT2CC were constitutively coexpressed in Arabidopsis xxt2 xxt5 double mutant plants. The observed complementation of the mutant root hair phenotype (Supplemental Fig. S4) suggests that XXT5NC and XXT2CC are functional proteins localized in the Golgi.
Fluorescence Quantification by Flow Cytometry
To evaluate the probability of complex formation, BiFC fluorescence intensities of XXT pairs coexpressed in Arabidopsis protoplasts were quantified by flow cytometry (Fig. 3, A and B). The combination of BiFC with flow cytometry provides a straightforward and sensitive estimation of the number of positive events (Morell et al., 2008). Fluorescence intensity was expressed as total fluorescence, which was calculated as the percentage of fluorescent cells (events) multiplied by the mean fluorescence of each fluorescent cell (Li et al., 2010). Both values were measured by flow cytometry, where the number of events is equal to the number of protoplasts in the P3 area (Fig. 3A) and the mean fluorescence is the average fluorescence intensity of these protoplasts. The results presented here are expressed as the fluorescence index, the ratio between the total fluorescence of transfected protoplasts and the total fluorescence of nontransfected protoplasts. Nontransfected protoplasts treated with PEG show a low level of fluorescence that was not visible by fluorescence microscopy but was detectable by flow cytometry (Fig. 3A). Therefore, to account for this background fluorescence, the fluorescence index of nontransfected protoplasts was set to 1.0. Two control experiments were performed to investigate the dependence of the BiFC fluorescence signal on the amount of plasmid used for the transfection and on the duration of protoplast incubation with plasmids (Supplemental Fig. S1, A and B). From these experiments, the optimum conditions, 24 h of incubation with 5 or 10 μg of plasmids, were chosen to ensure close to maximum intensity of fluorescence and to avoid overloading the cells with overexpressed proteins.
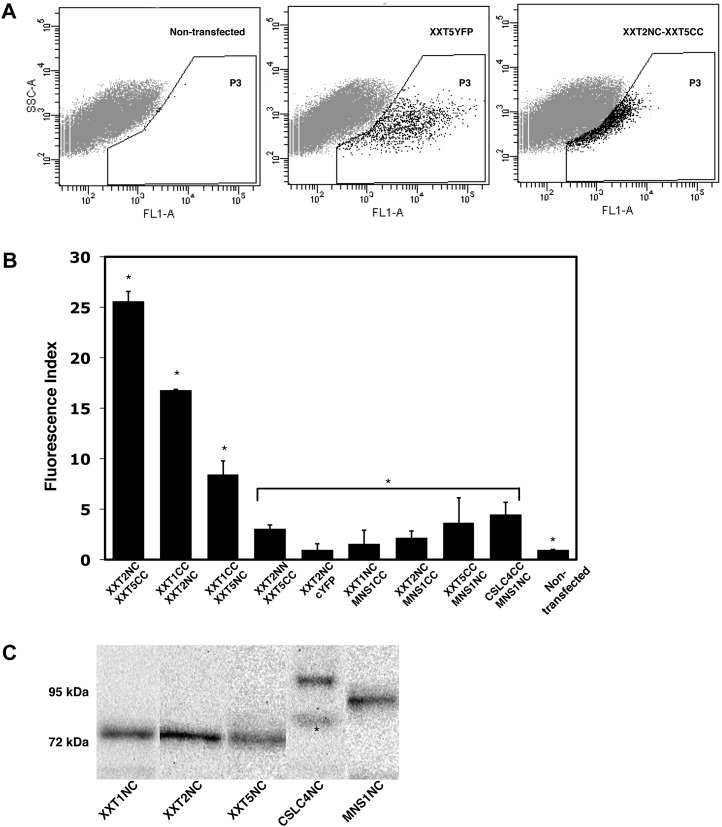
Figure 3.
Heterodimerization of XXTs. A, Flow cytometry quantification of BiFC signal. The plots show the results obtained after protoplasts were analyzed by flow cytometry. A Negative control (nontransfected protoplasts) and a positive control (XXT5YFP) were used to determine the gated area (P3) by the brightness of each protoplast (FL1-A). Each protoplast (a dot) localized in the P3 area represents a fluorescent event (an example is shown for XXT2NC and XXT5CC coexpression). B, BiFC signal intensities determined by flow cytometry for different XXT pairs and all negative controls. The calculation of fluorescence index is described in “Materials and Methods.” Values shown are means ± se. Asterisks indicate significant differences (t test, P < 0.05; n = 5) among experiments, negative controls, and nontransfected protoplasts. C, Estimation of protein expression in Arabidopsis protoplasts. The protein expression of GTNC constructs (all three XXTNCs, approximately 75 kD; CSLC4NC, approximately 99 kD; MNS1NC, approximately 80 kD) was detected using monoclonal GFP antibody. Asterisks indicate nonspecific binding.
The results of quantification by flow cytometry demonstrate that the XXT2NC and XXT5CC pair produced the strongest BiFC signal (Fig. 3B), with a fluorescence index of 25.6 ± 1. The fluorescence index for the XXT1CC and XXT2NC pair was 16.8 ± 0.1, but for XXT1 and XXT5, it was significantly lower, 8.4 ± 1.4. The values of fluorescence indices for all negative controls were in the range from 1.5 ± 1.3 to 4.5 ± 1.2, which is close to the index for nontransfected protoplasts (Fig. 3B).
It has been proposed (Cocuron et al., 2007) that xylosyltransferases can interact with the glucan synthase CSLC4 to xylosylate the glucan backbone during XyG biosynthesis. To investigate this idea, we examined whether XXTs form heterocomplexes with CSLC4 in Arabidopsis protoplasts. The CSLC4NC plasmid was coexpressed with CC constructs for each XXT. The fluorescence of reconstituted YFP was observed by fluorescence microscopy (Fig. 2, G and H) and measured by flow cytometry. The CSLC4-XXT5 pair showed the highest fluorescence index, significantly higher than the indices for the other two pairs, CSLC4-XXT2 and CSLC4-XXT1 (Fig. 4B). This suggests that all XXTs are colocalized in close proximity with CSLC4 and, most likely, that XXT5 occupies the closest position to CLC4 in comparison with the two other XXTs.
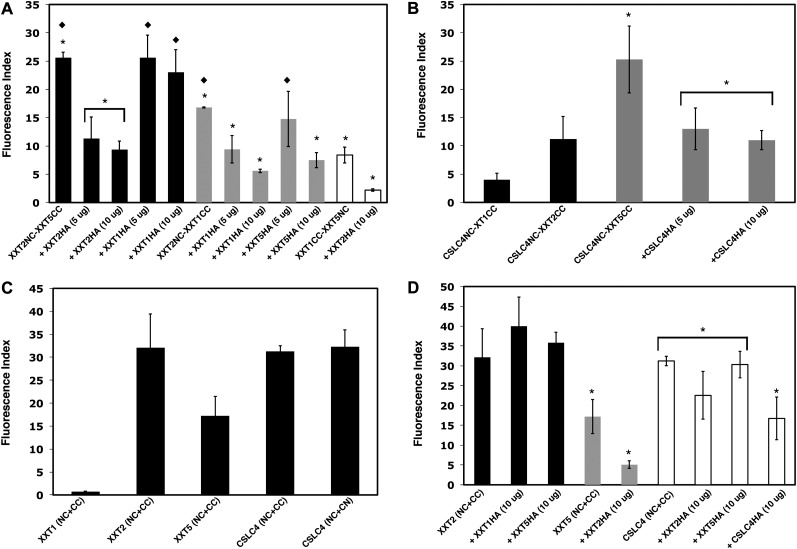
Figure 4.
BiFC competition assays in Arabidopsis protoplasts quantified by flow cytometry. A, BiFC competition assays among XXTs. Coexpression is shown as follows: the XXT2-XXT5 pair with XXT2HA (5 and 10 µg of DNA) or with XXT1HA (5 and 10 µg of DNA; black bars); the XXT1-XXT2 pair with XXT1HA (5 and 10 µg of DNA) or with XXT5HA (5 and 10µg of DNA; gray bars); the XXT1-XXT5 pair with XXT2HA (white bars). B, Coexpression is shown as follows: CSLC4NC with XXT1CC and XXT2CC (black bars); CSLC4NC with XXT5CC and the CSLC4NC-XXT5CC pair with CSLC4HA (5 and 10 µg of DNA; gray bars). C, Homodimerization of XXTs and CSLC4. D, BiFC competition assays among XXTs and the CSLC4 dimer. The XXT2-XXT2 pair was coexpressed with 10 μg of plasmid DNA of XXT1HA or XXT5HA (black bars). The XXT5-XXT5 pair was coexpressed with XXT2HA (gray bars). The CSLC4-CSLC4 pair was coexpressed with 10 μg of plasmid DNA of XXT2HA, XXT5HA, and CSLC4HA (white bars). Values shown are means ± se (t test, P < 0.05; n = 5). Asterisks indicate significant differences and diamonds indicate nonsignificant differences among bars having the same color.
To examine whether XXTs and CSLC4 can form homocomplexes, Arabidopsis protoplasts were transfected with pairs of BiFC constructs that express the same protein fused with N- and C-terminal YFP complementary fragments: XXT1NC and XXT1CC, XXT2NC and XXT2CC, XXT5NC and XXT5CC, CSLC4NC and CSLC4CC (Fig. 1A). The signal was visualized by fluorescence microscopy (Fig. 2, I–L) and measured by flow cytometry (Fig. 4C). The results show high fluorescence signals for the XXT2NC-XXT2CC and CSLC4NC-CSLC4CC pairs, confirming the high probability of homocomplex formation in transfected protoplasts. In the case of the XXT5NC-XXT5CC pair, fluorescence is lower than with XXT2 and CSLC4, but it is high enough to suggest that XXT5 can also form homocomplexes. In contrast, coexpression of the XXT1NC and XXT1CC constructs does not produce a detectable signal under the fluorescence microscope, and measurement by flow cytometry also shows very low total fluorescence (Fig. 4C), suggesting that XXT1 is unable to form stable homocomplexes.
CSLC4 has its N- and C-terminal regions localized in the cytosol (Davis et al., 2010); therefore, the formation of its homocomplex was tested using one CSLC4 fused with a YFP complementary fragment at the N terminus and another CSLC4 fused with YFP at the C terminus (CSLC4NC and CSLC4CN pair; Fig. 1A). The results show that in the CSLC4 homocomplex, both N- and C-terminal regions are close enough to reconstitute YFP fragments fused to the opposite ends of two proteins (Fig. 4C).
Immunoblot Analyses of Fusion Protein Expression
To clarify that the differences in fluorescence intensities reflect differences in the probability of forming a protein-protein complex and not differences in fusion protein expression in protoplasts, we tested protein levels by immunoblot analyses with anti-GFP monoclonal antibodies. Immunoblots were used for each combination of expression constructs, and no significant differences were observed in the expression of proteins fused with the N-terminal YFP complementary fragment (Fig. 3C). However, the GFP monoclonal antibody recognizes an epitope that is close to the N terminus of GFP and its derivatives, such as YFP and CFP; therefore, the monoclonal antibody cannot be used for immunodetection of proteins fused with the C terminus of YFP. To overcome this problem, a coexpression plasmid, pPZP2N5C, containing both XXT2NC and XXT5CC expression cassettes was used (Fig. 1C) to minimize potentially unequal transfection efficiencies of separate BiFC plasmids (Fig. 4A). We found no differences in fluorescence indices between protoplasts transfected with 10 μg of the coexpression plasmid and protoplasts transfected with 10 μg of each separate expression plasmid for XXT2NC and XXT5CC (Supplemental Fig. S2). This demonstrates that XXT2NC and XXT5CC are expressed at similar levels when cotransfected either as separate constructs or as a coexpression plasmid, which suggests that in the same protoplast, the expression levels of proteins fused with either the C- or N-terminal YFP complementary fragments are comparable.
Competition BiFC Assays
To confirm that the observed BiFC signals are the result of protein-protein complex formation and not a consequence of incidental nonspecific binding of two YFP complementary fragments, BiFC competition assays were carried out in addition to the negative controls described above. Arabidopsis protoplasts were cotransfected with the pPZP2N5C plasmid harboring BiFC pair XXT2NC and XXT5CC together with different amounts of XXT2HA plasmid (5 and 10 μg of plasmid DNA; Fig. 1A). In all competition experiments described in this work, the expression levels of all coexpressed proteins were monitored by immunoblot analysis to ensure that the competing hemagglutinin(HA)-tagged proteins and the YFP-tagged proteins were present in comparable amounts (Fig. 3C; Supplemental Fig. S3), so that the competing protein does not significantly exceed the amount of proteins fused with YFP fragments. Measurement of fluorescence signals by flow cytometry demonstrated a gradual reduction of the fluorescence index by 55% and 63% compared with the control, as the amount of XXT2HA was increased (Fig. 4A). These results show that XXT2HA can compete with XXT2NC for a position close to XXT5CC, confirming that the origin of the observed BiFC signal is a protein-protein complex and not an incidental YFP reconstitution. The BiFC competition assay was also performed to confirm the protein-protein complexes formed between XXT2NC and XXT1CC. Different amounts of XXT1HA (5 and 10 μg) were coexpressed with the XXT2NC and XXT1CC pair. As for the other interacting pair, there was a gradual reduction of XXT2NC-XXT1CC fluorescence as the amount of coexpressed XXT1HA was increased (Fig. 4A; Supplemental Fig. S3).
The same approach was used to further investigate the heterocomplexes among XXT1, XXT2, and XXT5. When XXT1HA was coexpressed with the XXT2NC and XXT5CC BiFC pair, the fluorescence signal did not change, indicating that XXT1HA cannot affect XXT2-XXT5 complex formation (Fig. 4A). Coexpression of XXT2HA with the XXT1CC and XXT5NC BiFC pair significantly depressed the total fluorescence signal (Fig. 4A). Similarly, coexpression of XXT5HA with the XXT1NC and XXT2CC pair decreased the total fluorescence signal, although to a lesser extent than XXT2 depresses the fluorescence of the XXT1-XXT5 pair (Fig. 4A). The results of these two experiments suggest that XXT2 and XXT5 have a stronger preference in close colocalization than XXT1 with XXT5 or even XXT1 with XXT2.
The BiFC competition assay was also used to investigate protein complex formation among XXT2, XXT5, and CSLC4. Neither XXT5HA nor XXT1HA coexpressed with the XXT2NC-XXT2CC pair affected its fluorescence, but XXT2HA coexpressed with the XXT5NC-XXT5CC pair decreased its fluorescence intensity (Fig. 4D). Also, the fluorescence of the CSLC4NC-CSLC4CC pair was not affected by coexpression of XXT5HA or by coexpression of XXT2HA (Fig. 4D), but it was depressed by coexpression of CSLC4HA. The latter confirms that the BiFC signal observed for the CSLC4-CSLC4 pair was due to protein-protein homocomplex formation and not due to accidental YFP reconstitution. Similarly, coexpression of CSLC4HA with the CSLC4NC and XXT5CC BiFC pair resulted in decreased fluorescence signal, supporting CSLC4-XXT5 heterocomplex formation (Fig. 4B).
Coimmunoprecipitation of XXT2 and XXT5 from Arabidopsis Protoplasts
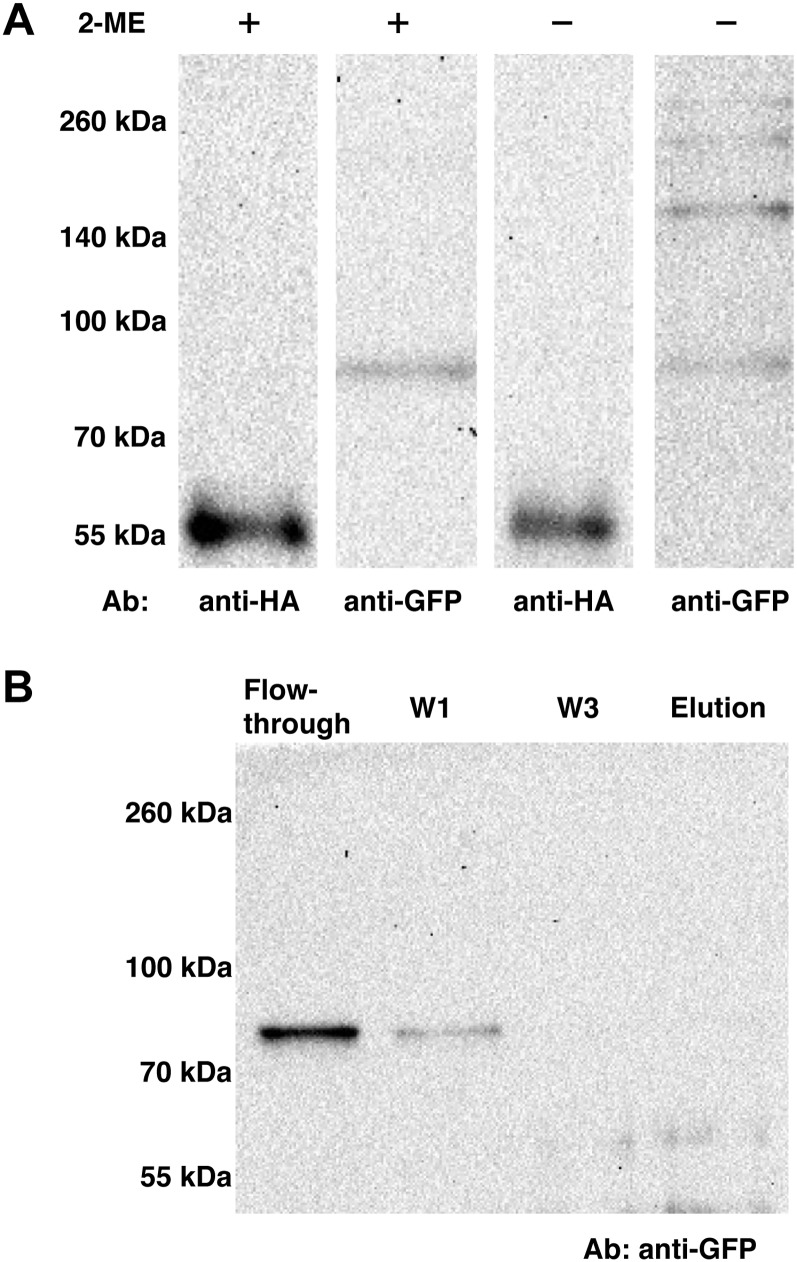
To further our investigation of XXT2-XXT5 heterocomplex formation in vivo, the coimmunoprecipitation assay was performed using protoplasts prepared from Arabidopsis plants expressing XXT5HA (Zabotina et al., 2008) and transfected with the XXT2YFP construct to introduce tagged XXT2 protein. A total protein extract (prepared as described in “Materials and Methods”) was applied to the anti-HA agarose column, and the collected fractions were analyzed by SDS-PAGE under reducing and nonreducing conditions. The XXT2YFP and XXT5HA proteins were detected by western blot. The results show that both proteins can be detected in the elution fraction (Fig. 5A) and not in the flow through or washes. Under nonreducing conditions, XXT2YFP was detected in the elution fraction as a monomer (approximately 83 kD) and also in a set of larger bands (approximately 166, 250, and 300 kD); by contrast, XXT5HA was detected only in monomeric form (approximately 53 kD) under both reducing and nonreducing conditions. To rule out the possibility of XXT2YFP nonspecifically interacting with anti-HA agarose, XXT2YFP was expressed in protoplasts prepared from wild-type Arabidopsis plants, and extracted proteins were applied to anti-HA agarose. XXT2YFP was detected only in the flow-through and first wash fractions but not in the elution fraction (Fig. 5B), confirming that XXT2YFP was retained on the column because of its interaction with XXT5HA and not because of a nonspecific interaction with anti-HA agarose. The results obtained in reducing and nonreducing conditions suggest that formation of the XXT2-XXT2 homocomplex most likely involves disulfide bonds, but the XXT2-XXT5 heterocomplex is formed through noncovalent interactions.
Figure 5.
Coimmunoprecipitation of XXT5HA and XXT2YFP from Arabidopsis protoplasts. A, XXT2YFP was transiently expressed in protoplasts prepared from XXT5HA-expressing plants. Total protein extracts from protoplasts treated with Triton X-100 were applied to an anti-HA agarose column. The elution fractions were treated with or without β-mercaptoethanol (2-ME) SDS-PAGE under reducing and nonreducing conditions, respectively. Proteins were detected by either polyclonal anti-HA or monoclonal anti-GFP antibody (Ab). B, Negative control for immunoprecipitation of XXT5HA and XXT2YFP. Protein extract from wild-type protoplast transiently expressed XXT2YFP was applied to an anti-HA agarose column. The flow-through, wash (W1 and W3), and elution fractions were separated by SDS-PAGE and detected by monoclonal anti-GFP antibodies.
In Vitro Pull-Down Assays
To confirm the formation of protein-protein complexes and investigate possible physical interactions among XXT proteins, in vitro pull-down assays were performed using recombinant proteins expressed in E. coli. To obtain soluble proteins, truncated XXT1, XXT2, and XXT5 mutant proteins, lacking their N termini and transmembrane domains, were fused with HA, His, and Myc tags, respectively (tGTtag; Fig. 1B) and expressed in E. coli (BL21) under the control of an inducible promoter in the pET-15b vector (www.novagen.com). The transmembrane domains of the XXTs were predicted using the program HMMTOP (http://www.enzim.hu/hmmtop), and 45-, 41, and 71-amino acid coding sequences were truncated from the 5′ ends of XXT1, XXT2, and XXT5, respectively. The proteins were designated with a “t” for truncated. The in vitro pull-down assays were performed two different ways: (1) two lysates prepared from cells expressing two different truncated proteins were mixed and then applied to an affinity column; (2) one of the lysates was first applied to the affinity column, and after washing, the second lysate was applied to the same column. After washing, the proteins bound to the column were eluted and examined by immunoblotting with antibodies specific for the tag on each protein. Both assays showed very similar results; therefore, only results from variant 1 are presented here.
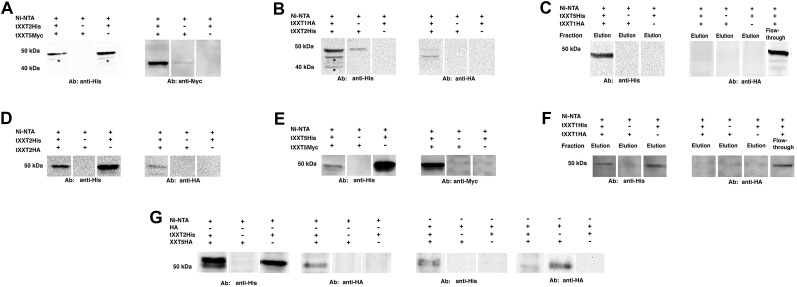
After lysates from tXXT2His- and tXXT5Myc-expressing cells were applied to a nickel-nitrilotriacetic acid agarose (Ni-NTA) affinity column, both tXXT2His (approximately 50 kD) and tXXT5Myc (approximately 46 kD) were detected in the elution fraction (Fig. 6A). When either tXXT2His or tXXT5Myc alone was applied to the Ni-NTA affinity column, only tXXT2His was detected in the elution fraction (Fig. 6A). Similarly, both tXXT1HA and tXXT2His could be pulled down and detected by both anti-HA and anti-His antibodies when lysates from tXXT1HA- (approximately 48 kD) and tXXT2His-expressing cells were applied to the Ni-NTA column (Fig. 6B). When tXXT1HA was applied to the column alone, it was not detected in the elution fraction. We were unable to pull down tXXT1HA and tXXT5His together; tXXT1HA was detected in the flow-through fraction and not in the elution fraction (Fig. 6C). These results are in agreement with the BiFC results, where very weak fluorescence was observed for the XXT1-XXT5 pair. Next, the ability of XXT2 and XXT5 to form homodimers was confirmed using a mixture of tXXT2His and tXXT2HA and a mixture of tXXT5Myc and tXXT5His, respectively (Fig. 6, D and E). By contrast, a pull-down assay using tXXT1HA and tXXT1His did not show an interaction (Fig. 6F), confirming the BiFC results and demonstrating that XXT1 does not homodimerize.
Figure 6.
Interactions between XXTs confirmed by in vitro pull-down assay. A, In vitro Ni-NTA pull-down assay for interaction between tXXT2His and tXXT5Myc. tXXT2His and tXXT5Myc lysates and a mixture of the two lysates were applied to Ni-NTA affinity columns. The elution fractions were detected by either anti-His or anti-Myc antibody (Ab). Asterisks indicate nonspecific bound signal. B, In vitro Ni-NTA pull-down assay for the interaction of tXXT1HA and tXXT2His. tXXT1HA and tXXT2His lysate mixture was applied onto a Ni-NTA affinity column. The elution fractions were detected by either anti-His or anti-HA antibody. Asterisks indicate nonspecific bound signal. C, In vitro Ni-NTA pull-down assay for the interaction of tXXT1HA and tXXT5His. tXXT1HA and tXXT5His lysate mixture was applied onto a Ni-NTA affinity column. The elution fractions were detected by either anti-His or anti-HA antibody. The flow-through fraction was detected by anti-HA antibodies to confirm the presence of nonbound tXXT1HA. D, In vitro Ni-NTA pull-down assay for interaction between tXXT2His and tXXT2HA. tXXT2His and tXXT2HA lysate mixture was applied onto a Ni-NTA affinity column. The elution fractions were detected by either anti-His or anti-HA antibody. E, In vitro Ni-NTA pull-down assay for interactions between tXXT5His and tXXT5Myc. tXXT5His and tXXT5Myc lysate mixture was applied onto a Ni-NTA affinity column. The elution fractions were detected by either anti-His or anti-Myc antibody. F, In vitro Ni-NTA pull-down assay for interactions between tXXT1His and tXXT1HA. tXXT1His and tXXT1HA lysate mixture was applied onto a Ni-NTA affinity column. The elution fractions were detected by either anti-His or anti-HA antibody. The flow through fraction was detected by anti-HA antibodies to confirm the presence of nonbound tXXT1HA. G, In vitro Ni-NTA and anti-HA agarose pull-down assay for interactions between tXXT2His and HA tag-fused full-length XXT5. tXXT2His lysate was mixed with protein extract from XXT5HA-expressing transgenic plants. The mixture was pulled down by either a Ni-NTA or HA agarose column. Both elution fractions were detected by anti-HA and anti-His antibodies.
To verify that the observed in vitro interactions among truncated XXTs were specific and not due to nonspecific attraction between soluble truncated proteins, we performed pull-down assays using the native full-length XXT5HA (approximately 53 kD) and tXXT2His proteins. Total protein extract from protoplasts prepared from XXT5HA-expressing plants (Zabotina et al., 2008) treated with detergent (as described in “Materials and Methods”) was mixed with lysate from tXXT2His-expressing E. coli. The mixture was applied either to a Ni-NTA affinity column or an anti-HA agarose column. Both XXT5HA and tXXT2His were detected in elution fractions obtained from either Ni-NTA or HA columns (Fig. 6G).
Since CSLC4 has six predicted transmembrane domains distributed through its protein sequence, it is not feasible to obtain this protein in soluble form. Therefore, in this study, we were unable to perform pull-down experiments using CSLC4.
DISCUSSION
Previous studies revealed that three xylosyltransferases, XXT1, XXT2, and XXT5, and a glucan synthase, CLSC4, are involved in the synthesis of the xylosylated glucan backbone of XyG in Arabidopsis (Cavalier and Keegstra, 2006; Cocuron et al., 2007; Cavalier et al., 2008; Zabotina et al., 2008). Here, we investigated the protein-protein interactions among these enzymes to shed light on their functional organization and to explore the putative multiprotein complex involved in XyG formation in the Golgi. The idea that XyG assembly involves a multienzyme complex localized in the Golgi membrane first emerged from biochemical studies demonstrating the cooperativity of glucan synthase and xylosyltransferase activities in vitro (Zhang and Staehelin, 1992; Cocuron et al., 2007). In addition, recent reverse-genetic studies indicated that three xylosyltransferases, XXT1, XXT2, and XXT5, play different roles in XyG biosynthesis (Cavalier et al., 2008; Zabotina et al., 2008), and the presence of all three proteins is essential for the formation of the wild-type XyG (Zabotina et al., 2012). In this study, we investigated only one glucan synthase, CSLC4, as a potential member of the XyG synthetic complex, although other CSLC proteins, such as CSLC5 and CSLC6, have been implicated in XyG formation also (Cavalier and Keegstra, 2010). Our selection is based on the facts that CSLC4 was shown to have β-glucan synthase activity when expressed heterologously and that this activity was enhanced by coexpression of CSLC4 and XXT1 (Cocuron et al., 2007). CSLC4 is also expressed in all Arabidopsis tissues and has the highest expression level of all AtCSLCs (http://wardlab.cbs.umn.edu/arabidopsis/). This does not exclude the participation of other CSLCs in the multienzyme complex involved in XyG biosynthesis, and this needs to be investigated in the future. Available transcription data demonstrate that all three XXTs and CSLC4 are coexpressed in all Arabidopsis tissues studied, and XXT2 and CSLC4 have levels of expression about two times higher than XXT1 and XXT5 (Supplemental Fig. S5; Schmid et al., 2005). Higher expression levels of XXT2 and CSLC4 fit with our observation of two homocomplexes, XXT2-XXT2 and CSLC4-CSLC4, while XXT1 and XXT5 are most likely present only in heterocomplexes.
The absence of a fluorescence signal from coexpression of XXT proteins with YFP complementary fragments fused to their opposite termini confirms that all three xylosyltransferases are localized in the Golgi membrane in the same orientation, which brings their N termini into close proximity. It was suggested that XXT1, XXT2, and XXT5 proteins are positioned in the Golgi membrane with their N termini on the cytosolic side and their C termini inside the Golgi lumen (Søgaard et al., 2012). Unlike XXTs, which have only one predicted transmembrane domain and span the Golgi membrane only once, CSLC4 has six predicted transmembrane domains spanning the membrane six times, with the catalytic site and both termini located on the cytosolic side of the Golgi membrane (Davis et al., 2010). This information about the topologies of XXTs and CSLC4 prompted us to fuse the YFP fragments to the N terminus of each protein in the BiFC experiments, because the C termini of XXT and CSLC4 proteins are localized on opposite sides of the Golgi membrane, which would prevent YFP reconstitution in XXT-CSLC4 pairs. The results obtained in this study support the earlier assumptions about the XXTs and CSLC4 topologies.
The competition BiFC assays, together with all the negative controls used in our experiments, confirm that the observed fluorescence signals are the result of specific protein-protein complex formation. Additionally, the competition assay allowed some insights into the putative composition of the protein-protein complexes. For example, it was demonstrated that XXT2 and XXT5 prefer to form a heterocomplex and that the potential interaction between XXT2 and XXT1 occurs most likely via different residues in XXT2. In addition, XXT2 can simultaneously form homocomplexes and heterocomplexes, most likely involving different interacting surfaces. The formation of the XXT2-XXT2 homocomplex and the XXT5-XXT2 heterocomplex is also supported by coimmunoprecipitation of the XXT2YFP homocomplex and the XXT2YFP-XXT5HA heterocomplex from Arabidopsis protoplasts (Fig. 5). Additionally, coimmunoprecipitation demonstrated that the XXT2-XXT2 homocomplex is linked by disulfide bonds, but formation of the XXT2-XXT5 heterocomplex does not involve covalent interactions. Currently, we cannot explain the presence of two bigger bands (approximately 250 and 300 kD) detected with GFP antibodies under nonreducing conditions (Fig. 5). It is possible that XXT2 can form larger homocomplexes or can pull down other components of the multiprotein complex, interacting with them through disulfide bonds.
The weaker ability of XXT5 to compete with the XXT1-XXT2 complex suggests that the XXT1 and XXT2 interaction is stronger than the interaction between XXT1 and XXT5 and, most likely, that XXT2 can form a complex with XXT1 and XXT5 simultaneously. This conclusion is supported by in vitro pull-down assays. XXT2 can be pulled down with XXT5 and XXT1, but XXT1 and XXT5 were not pulled down together, which demonstrates weak or no interaction between XXT1 and XXT5. Perhaps XXT1 and XXT5 do not directly interact in vivo but colocalize in the Golgi close enough to reconstitute YFP in the BiFC assay.
The BiFC assay using complementary YFP fragments fused to the N termini of two CSLC4 proteins or to the C terminus of one protein and the N terminus of another showed comparable fluorescence indices, which is in accordance with the hexagonal rosette-like structure of glucan synthase, similar to cellulose synthases. In addition, this suggests that the termini of two CSLC4 molecules are in closest proximity while their catalytic loops are positioned on opposite sides. The presence of at least two CSLC4s in the complex is also supported by an earlier suggestion that the synthesis of a glucan chain with alternating Glc residues in opposite orientations requires two glucan synthases working alternately (Sandhu et al., 2009; Carpita, 2011). Davis et al. (2010) proposed that CSLC4 has a topology similar to that of cellulose synthases and therefore operates in a similar manner (i.e. by coupling the addition of Glc to the elongating glucan chain with translocation of the chain across the Golgi membrane). BiFC assays demonstrate that expression of XXT5 or XXT2 with CSLC4 gives a strong fluorescence signal, but coexpression of XXT5HA or XXT2HA with the CSLC4-CSLC4 BiFC pair does not depress the fluorescence signal of the latter. This implies that XXT proteins colocalize with two CSLC4 proteins, most likely in the same complex, to xylosylate the elongating glucan chain. Interactions of XXTs, most likely through their catalytic domains, may support complex integrity, holding the xylosyltransferases around the synthesized glucan backbone. Localization of xylosyltransferases close to the glucan synthases is the most plausible complex organization, assuming that xylosylation of the glucan backbone occurs upon its elongation in a processive manner. There could be a few reasons for the lower fluorescence signal observed for interactions between CSLC4 and XXT1 and XXT2, in comparison with the signal observed for the interaction between CSLC4 and XXT5. First, the distance between the interaction side of CSLC4 and its N terminus fused with the YFP complementary fragment is longer than the distance between each XXT’s interaction side and N terminus. This different spatial proximity likely affects YFP reconstitution. XXT5 has a longer N-terminal cytosolic tail than the other two XXTs, which makes it easier for the YFP fragment fused to the XXT5 N terminus to reach the YFP complementary fragment fused to the CSLC4 N terminus. Second, it is possible that in the CSLC4-XXT heterocomplex, the N termini are separated by CSLC4’s active site loop, which can disturb YFP reconstitution. Finally, other complex components may be localized close to the interaction side between CSLC4 and XXTs, increasing the distance between two YFP complementary fragments; these components may include, for example, other glycosyltransferases or nucleotide sugar transporters (Zhang et al., 2011).
In conclusion, we propose that the putative XyG synthase complex contains at least two glucan synthases colocalized with their N and C termini in close proximity to each other, two XXT2 proteins interacting with each other through disulfide bonds, and XXT5 and XXT1, which interact with XXT2-XXT2 and CSLC4-CSLC4 homocomplexes. XXT2, XXT5, and XXT1 physically interact with each other through their catalytic domains localized in the Golgi lumen, and most likely, these interactions do not involve covalent bonds. By contrast, since the catalytic domains of CSLC4 and the XXTs are localized on opposite sides of the Golgi membrane, most likely CSLC4 and the XXTs interact through their transmembrane domains, or the catalytic domains or stem regions of XXTs interact with CSLC4’s loops that protrude into the Golgi lumen. Further investigation will be required to address this question.
Future research will also include other glycosyltransferases known to be involved in XyG decoration, to define the full composition and structure of the XyG synthase complex in Arabidopsis. Demonstration of these complexes in vivo further extends our understanding of XyG biosynthesis; this knowledge will shed light on polysaccharide formation and offer opportunities for the direct manipulation of polysaccharide formation to modify plant cell walls for various industrial applications.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana) ecotype Columbia were sterilized in 70% (v/v) NaOCl solution with 0.1% (v/v) Triton X-100. Sterilized seeds were germinated, and seedlings were grown for 10 d on plates with one-half-strength Murashige and Skoog medium under 16-h-light/8-h-dark photoperiod conditions in a growth incubator at 22°C.
DNA Constructs
Constructs for BiFC Assays
All genes, XXT1, XXT2, XXT5, and CSLC4, were amplified using gene-specific primers from full-length complementary (cDNA) clones (XXT1 and XXT2 cDNA constructs were obtained from Prof. K. Keegstra). CSLC4 was amplified from Arabidopsis cDNA directly. After PCR amplification, PCR products were inserted into the Gateway entry vector pCR8⁄GW⁄TOPO TA Cloning vector (Invitrogen). Gene-TOPO DNAs were recombined into both destination Gateway pSAT-BiFC vectors, pSAT4-DEST-nEYFP-C1 (N-terminal YFP fragment) and pSAT5-DEST-cEYFP-C1(B) (C-terminal YFP fragment; Arabidopsis Biological Resource Center [ABRC]) and into pEarleyGate104 (ABRC). For cloning coexpression plasmids, the expression cassette with the N- or C-terminal YFP fragment was digested with a homing endonuclease, I-SceI or I-CeuI, respectively, and cloned into the pPZP-RCS2-ocs-bar-R1 (ABRC) coexpression binary vector.
Constructs for Pull-Down Assays
The N-terminal His-tagged truncated tXXT1 and tXXT2 were prepared by amplification of XXT1 and XXT2 from the corresponding cDNAs and cloning into the pET-15b vector (Novagen) containing an N-terminal 6xHis tag using gene-specific forward and reverse primers containing NdeI and BamHI sites, respectively. The N-terminal HA-tagged truncated tXXT1 was made by removing the N-terminal 6xHis tag sequence from the pET-15b vector and replacing it with the HA-tXXT1 sequence, which was made by amplification from XXT1 cDNA using a gene-specific forward primer containing both the HA sequence (5′-ACTCATGAATGTACGACGTACCAGATTACGCTACGCCGGAGAAAGATATCGAGG-3′) and a BspHI site as well as a gene-specific reverse primer with a BamHI site (5′-ACGGATCCTCACGTCGTCGTCGTACTAAGCT-3′). The N-terminal Myc-tagged truncated tXXT5 was made by amplification from the cDNA using a gene-specific forward primer containing both the Myc tag and an NcoI site (5′-AGCCATGGATGGAACAAAAACTCATCTCAGAAGAGGATCTGAACCTAGGAAGCTCAAGCGCCG-3′) as well as a gene-specific reverse primer containing an NdeI site (5′-ACCATATGCTAGTTCTGTGGTTTGGTTTCCAC-3′). The amplified Myc-tXXT5 PCR fragment was then cloned into pET-15b digested with NcoI and NdeI to generate the plasmid with the Myc-tXXT5 sequence.
Preparation of Protoplasts
Forty Arabidopsis seedlings grown on plates for 10 d were harvested and incubated in 5 mL of enzyme solution (0.25% [w/v] Macerozyme, 1.0% Cellulase, 0.4 mm mannitol, 8 mm CaCl2, 5 mm MES-KOH, pH 5.6, and 0.1% bovine serum albumin) for 10 to 12 h in the dark with gentle agitation at 50 rpm. After incubation, suspended protoplasts were filtered through a 100-μm cell strainer, laid onto 10 mL of 21% (w/v) Suc solution, and centrifuged at 300g for 5 min. The supernatant, which contained the protoplasts, was collected and diluted with 10 mL of W5 solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, 5 mm Glc, and 1.5 mm MES-KOH, pH 5.6). Protoplasts were collected by centrifugation at 300g for 5 min and resuspended in 1 mL of W5 solution. The amount of protoplasts was measured using a hemocytometer (0.1 mm deep).
Transient Expression in Protoplasts
The protocol for protoplast transfection was adapted from Jin et al. (2001). After protoplast suspensions were counted, protoplasts were pelleted again by centrifugation at 300g for 5 min and resuspended to a density of 2 × 105 mL−1 in a solution of 400 mm mannitol, 15 mm MgCl2, and 5 mm MES-KOH, pH 5.6. Each plasmid (5 or 10 μg) was added into 100 μL of protoplast suspension followed by the addition of 120 μL of PEG solution [30% (w/v) PEG-4000, 400 mm mannitol, and 15 mm Ca(NO3)2]. The transfection mixture was incubated at room temperature for 30 min. After incubation, the transfection mixture was diluted with 4 mL of W5 solution to terminate the transfection process. The transfected protoplasts were collected by centrifugation at 300g for 5 min and resuspended in 1 mL of W5 solution. The transfected protoplast suspension was incubated at room temperature for 8 h in the dark and then moved to 4°C for 10 h. The BiFC fluorescence signal was visualized using a fluorescence microscope (DMIRE; Leica) with distinct filter cubes for YFP (filter set: excitation, 485/20; emission, 460/20) and for CFP (filter set: excitation, 436/20; emission, 480/40), and examined with the attached digital camera.
Western Blot of Expressed Fusion Proteins
Transfected protoplasts expressing fusion proteins (approximately 60,000 per sample) were pelleted by centrifugation at 300g for 5 min and resuspended in 300 μL of protein extraction buffer (40 mm HEPES, 0.45 m Suc, 1 mm EDTA, 1 mm MgCl2, 1 mm KCl, 1 mm dithiothreitol, and protease inhibitor cocktail [Roche], pH 8.0) by vortexing three times for 10 s each. The protein extract was treated with 1% Triton X-100 for 30 min at 4°C to solubilize membrane-bound proteins. After solubilization, proteins were precipitated with 10% (v/v) TCA. Precipitated proteins were resuspended in loading buffer (30 mg mL−1) with or without β-mercaptoethanol for reducing or nonreducing SDS-PAGE, respectively. After SDS-PAGE separation, the proteins were electrophoretically transferred to a nitrocellulose membrane (0.2 μm; Bio-Rad) for immunodetection. Monoclonal anti-GFP antibodies (MMS-118P; Covance) were used (1:5,000 dilution) for the detection of YFP. Monoclonal anti-HA antibodies (LT0422; LifeTein) were used (1:500 dilution) to detect HA-fused proteins. Polyclonal His antibodies (sc-803; Santa Cruz Biotechnology) were used (1:10,000 dilution) to detect His-fused proteins, and monoclonal Myc antibodies (MA1-21316; Thermo) were used (1:2,000 dilution) to detect Myc-fused proteins. Membranes were treated with the reagents to detect peroxidase activity and immediately visualized by ChemiDoc XRS+ (Bio-Rad). Prestained size markers were visualized on the same membrane using visible light. Protein concentration was measured using the Bio-Rad kit (Quick Start Bradford Dye reagent 1X; catalog no. 500-0205) following the manufacturer’s instructions.
Flow Cytometry
Fluorescence intensities of the BiFC signal in transfected protoplasts were quantified by flow cytometry (FACSCanto; BD Biosciences). Approximately 20,000 to 25,000 protoplasts (counted by hemocytometer) were suspended in 500 μL of W5 solution. The YFP was excited with a laser at 488 nm and captured with a FL1-A sensor (the emission wavelength was 505–554 nm). The fluorescence intensity was calculated as described by Li et al. (2010) with the following equation: total fluorescence = mean fluorescence level × the percentage of fluorescent events. The fluorescence intensity index was determined as total fluorescence of transfected protoplasts divided by total fluorescence of nontransfected but PEG-treated protoplasts. For each BiFC pair described here, five independent protoplast transfections were performed, and fluorescence was measured two times for each of five transfections.
Coimmunoprecipitation Assay
Protoplasts from XXT5HA-expressing or wild-type plants (around 80,000 protoplasts) were transfected with the XXT2YFP construct (10 µg), and protein extraction and solubilization with Triton X-100 were performed as described above. Total protein extract was diluted with extraction buffer to reduce the final concentration of detergent to 0.2%, and the extract was applied to an affinity column with anti-HA-conjugated agarose. After 1.5 h of incubation at 4°C, the anti-HA agarose column was washed with 500 μL of wash buffer (25 mm Tris and 150 mm sodium chloride, pH 7.2) three times and eluted with 200 μL of elution buffer (200 mm Gly, pH 2.8). Collected flow-through, wash, and elution fractions were mixed with loading buffer with and without β-mercaptoethanol for reducing and nonreducing conditions, respectively. Proteins were separated by SDS-PAGE and detected by western blotting as described above.
In Vitro Pull-Down Assay
All prepared plasmids with truncated XXT-tagged proteins were transformed into Escherichia coli (BL21) using the heat shock method. Transformed E. coli cells were incubated in 4 mL of lysogeny broth medium at 37°C for 2.5 h at 150 rpm. When the cells reached an optical density at 600 nm of 0.5 to 0.6, the culture was moved to 16°C for 1 h with continuous shaking. After the 16°C treatment, cells were induced with 0.5 mm isopropyl-β-d-thiogalactopyranoside and incubated at 37°C for an additional 3 h. Isopropyl-β-d-thiogalactopyranoside-induced cells were pelleted and lysed by incubation in lysis buffer (1 mg mL−1 lysozyme, 20 mm HEPES, 100 mm NaCl, 10 μg mL−1 DNaseI, and 5 mm MgCl2, pH 7.0) for 1 h at room temperature followed by five freeze/thaw cycles in liquid nitrogen. Lysate containing soluble proteins including tXXT was separated from the insoluble pellet by centrifugation at 13,000 rpm for 10 min.
In vitro pull-down assays were performed using Ni-NTA affinity resin (Thermo Scientific; 88221) or HA agarose (Pierce) following each manufacturer’s instructions. For Ni-NTA, two truncated protein lysates (1 mg of total crude protein) were mixed with 300 μL of equilibration buffer (20 mm sodium phosphate, 300 mm sodium chloride, and 10 mm imidazole, pH 7.4). The mixture was then added to the affinity resin and incubated at 4°C overnight with end-to-end shaking. The column was washed with 600 μL of wash buffer (20 mm sodium phosphate, 300 mm sodium chloride, and 25 mm imidazole, pH 7.4) two times and eluted with 300 μL of elution buffer (20 mm sodium phosphate, 300 mm sodium chloride, and 250 mm imidazole, pH 7.4). All fractions were analyzed by western blotting with tag-specific antibodies. For anti-HA agarose, two truncated protein lysates (1 mg of total crude protein) were mixed with 300 μL of equilibration buffer, applied to the column with anti-HA-conjugated agarose, and then washed and eluted as described above for the coimmunoprecipitation assay.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers 2081625 (XXT1), 2132293 (XXT2), 2019090 (XXT5), and 2089730 (CSLC4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. BiFC signal dependence on the amount of plasmid DNA and time of incubation.
Supplemental Figure S2. BiFC signal intensity for XXT2NC and XXT5CC coexpressed using two approaches.
Supplemental Figure S3. Expression level of HA-tagged proteins used in competition BiFC assays.
Supplemental Figure S4. Expression of XXT2NC and XXT5CC in the Arabidopsis xxt2 xxt5 double mutant.
Supplemental Figure S5. Expression of XXT1, XXT2, XXT5, and CSLC4 in Arabidopsis organs.
Supplementary Material
Acknowledgments
We thank Prof. Kenneth Keegstra from Plant Research Laboratory, Michigan State University for the cDNA clones of XXT1 and XXT2 he kindly provided for this work.
Glossary
- BiFC
bimolecular fluorescence complementation
- XyG
xyloglucan
- YFP
yellow fluorescent protein
- CFP
cyan fluorescent protein
- PEG
polyethylene glycol
- HA
hemagglutinin
- Ni-NTA
nickel-nitrilotriacetic acid agarose
- ABRC
Arabidopsis Biological Resource Center
- cDNA
complementary DNA
References
- Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A. (2010) Plant Cell Walls. Garland Science, New York.
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, III, Orlando R, Scheller HV, Mohnen D. (2011) Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci USA 108: 20225–20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC. (2011) Update on mechanisms of plant cell wall biosynthesis: how plants make cellulose and other (1→4)-β-d-glycans. Plant Physiol 155: 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Keegstra K. (2006) Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. J Biol Chem 281: 34197–34207 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Keegstra K. (2010) Members of Arabidopsis cellulose synthase-like C (CSLC) family are involved in xyloglucan biosynthesis. In S Coimbra, LG Pereira, eds, Abstract Book: XII Cell Wall Meeting, Porto-Portugal, July 25-30. Invulgar, Penafiel, Portugal, p 84.
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnock SJ, Henrissat B, Davies GJ. (2001) Three-dimensional structures of UDP-sugar glycosyltransferases illuminate the biosynthesis of plant polysaccharides. Plant Physiol 125: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Gafni Y, Tzfira T. (2008) Localizing protein-protein interactions by bimolecular fluorescence complementation in planta. Methods 45: 196–206 [DOI] [PubMed] [Google Scholar]
- Cocuron J-C, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel NV, Wilkerson CG. (2007) A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc Natl Acad Sci USA 104: 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J, Brandizzi F, Liepman AH, Keegstra K. (2010) Arabidopsis mannan synthase CSLA9 and glucan synthase CSLC4 have opposite orientations in the Golgi membrane. Plant J 64: 1028–1037 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S. (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP. (2002) Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 43: 1407–1420 [DOI] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau J-P, Kato Y, Lorences EP, Maclachlan GA, McNeil M, et al. (1993) An unambiguous nomenclature for xyloglucan derived oligosaccharides. Physiol Plant 89: 1–3 [Google Scholar]
- Harholt J, Jensen JK, Verhertbruggen Y, Søgaard C, Bernard S, Nafisi M, Poulsen CP, Geshi N, Sakuragi Y, Driouich A, et al. (2012) ARAD proteins associated with pectic arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta 236: 115–128 [DOI] [PubMed] [Google Scholar]
- Hassinen A, Rivinoja A, Kauppila A, Kellokumpu S. (2010) Golgi N-glycosyltransferases form both homo- and heterodimeric enzyme complexes in live cells. J Biol Chem 285: 17771–17777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C-D, Chinenov Y, Kerppola TK. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Jensen JK, Schultink A, Keegstra K, Wilkerson CG, Pauly M. (April 2 2012) RNA-Seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Mol Plant http://dx.doi.org/10.1093/mp/sss032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong G-W, Hwang I. (2001) new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K. (2010) Plant cell walls. Plant Physiol 154: 483–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K, Raikhel NV. (2001) Plant glycosyltransferases. Curr Opin Plant Biol 4: 219–224 [DOI] [PubMed] [Google Scholar]
- Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D. (2002) Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci USA 99: 11109–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Doll J, Weckermann K, Oecking C, Berendzen KW, Schöffl F. (2010) Detection of in vivo interactions between Arabidopsis class A-HSFs, using a novel BiFC fragment, and identification of novel class B-HSF interacting proteins. Eur J Cell Biol 89: 126–132 [DOI] [PubMed] [Google Scholar]
- Liebminger E, Hüttner S, Vavra U, Fischl R, Schoberer J, Grass J, Blaukopf C, Seifert GJ, Altmann F, Mach L, et al. (2009) Class I α-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21: 3850–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter W-D. (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C, Duncan G, Goutsos KT, Tufaro F. (2000) The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci USA 97: 668–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell M, Espargaro A, Aviles FX, Ventura S. (2008) Study and selection of in vivo protein interactions by coupling bimolecular fluorescence complementation and flow cytometry. Nat Protoc 3: 22–33 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K. (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976–1979 [DOI] [PubMed] [Google Scholar]
- Perrin RM, Wilkerson C, Keegstra K. (2001) Golgi enzymes that synthesize plant cell wall polysaccharides: finding and evaluating candidates in the genomic era. Plant Mol Biol 47: 115–130 [PubMed] [Google Scholar]
- Reyes F, Orellana A. (2008) Golgi transporters: opening the gate to cell wall polysaccharide biosynthesis. Curr Opin Plant Biol 11: 244–251 [DOI] [PubMed] [Google Scholar]
- Sakuragi Y, Norholm MHH, Scheller HV. (2011) Visual mapping of cell wall biosynthesis. In ZA Popper, ed, The Plant Cell Wall: Methods and Protocols. Springer Science-Business Media, Humana Press, New York pp 153–167. [DOI] [PubMed]
- Sandhu APS, Randhawa GS, Dhugga KS. (2009) Plant cell wall matrix polysaccharide biosynthesis. Mol Plant 2: 840–850 [DOI] [PubMed] [Google Scholar]
- Saxena IM, Brown RM, Jr, Fevre M, Geremia RA, Henrissat B. (1995) Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol 177: 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Søgaard C, Stenbæk A, Bernard S, Hadi M, Driouich A, Scheller HV, Sakuragi Y. (2012) GO-PROMTO illuminates protein membrane topologies of glycan biosynthetic enzymes in the Golgi apparatus of living tissues. PLoS ONE 7: e31324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz J, Munro S. (2002) The components of the Saccharomyces cerevisiae mannosyltransferase complex M-Pol I have distinct functions in mannan synthesis. J Biol Chem 277: 44801–44808 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G. (2001) What sugar next? Dimerization of sphingolipid glycosyltransferases. Proc Natl Acad Sci USA 98: 1321–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter W-D. (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 99: 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Zabotina OA, Avci U, Cavalier D, Pattathil S, Chou YH, Eberhard S, Danhof L, Keegstra K, Hahn MG. (2012) Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol 159: 1367–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, van de Ven WTG, Freshour G, Drakakaki G, Cavalier D, Mouille G, Hahn MG, Keegstra K, Raikhel NV. (2008) Arabidopsis XXT5 gene encodes a putative α-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. Plant J 56: 101–115 [DOI] [PubMed] [Google Scholar]
- Zeng W, Jiang N, Nadella R, Killen TL, Nadella V, Faik A. (2010) A glucurono(arabino)xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Physiol 154: 78–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Liu X, Qian Q, Liu L, Dong G, Xiong G, Zeng D, Zhou Y. (2011) Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc Natl Acad Sci USA 108: 5110–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GF, Staehelin LA. (1992) Functional compartmentation of the Golgi apparatus of plant cells: immunocytochemical analysis of high-pressure frozen- and freeze-substituted sycamore maple suspension culture cells. Plant Physiol 99: 1070–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.