Abstract
Xyloglucan is an important hemicellulosic polysaccharide in dicot primary cell walls. Most of the enzymes involved in xyloglucan synthesis have been identified. However, many important details of its synthesis in vivo remain unknown. The roles of three genes encoding xylosyltransferases participating in xyloglucan biosynthesis in Arabidopsis (Arabidopsis thaliana) were further investigated using reverse genetic, biochemical, and immunological approaches. New double mutants (xxt1 xxt5 and xxt2 xxt5) and a triple mutant (xxt1 xxt2 xxt5) were generated, characterized, and compared with three single mutants and the xxt1 xxt2 double mutant that had been isolated previously. Antibody-based glycome profiling was applied in combination with chemical and immunohistochemical analyses for these characterizations. From the combined data, we conclude that XXT1 and XXT2 are responsible for the bulk of the xylosylation of the glucan backbone, and at least one of these proteins must be present and active for xyloglucan to be made. XXT5 plays a significant but as yet uncharacterized role in this process. The glycome profiling data demonstrate that the lack of detectable xyloglucan does not cause significant compensatory changes in other polysaccharides, although changes in nonxyloglucan polysaccharide amounts cannot be ruled out. Structural rearrangements of the polysaccharide network appear responsible for maintaining wall integrity in the absence of xyloglucan, thereby allowing nearly normal plant growth in plants lacking xyloglucan. Finally, results from immunohistochemical studies, combined with known information about expression patterns of the three genes, suggest that different combinations of xylosyltransferases contribute differently to xyloglucan biosynthesis in the various cell types found in stems, roots, and hypocotyls.
The primary cell walls of plants largely consist of interacting networks of polysaccharides, together with some structural (glyco)proteins. The major polysaccharide components are pectins, hemicelluloses, and cellulose. The most abundant hemicellulosic polysaccharide in the primary cell walls of most vascular dicot species is xyloglucan, which is thought to bind tightly to cellulose to form a xyloglucan/cellulose network (Hayashi and Maclachlan, 1984; Pauly et al., 1999a). Most models of primary cell walls depict xyloglucan as a polymer that cross-links adjacent cellulose microfibrils, thereby forming a load-bearing structure in the wall whose loosening must be carefully controlled during cell wall expansion (Hayashi, 1989; McCann and Roberts, 1991; Carpita and Gibeaut, 1993; Somerville et al., 2004). An alternative model is that by directly interacting with cellulosic molecules, xyloglucan serves as a spacer that prevents the cellulose microfibrils from aggregating (Thompson, 2005) and thereby supports the interaction between cellulose and other cell wall components (Somerville et al., 2004). Apart from cellulose, xyloglucan is the most characterized cell wall polysaccharide, and considerable progress has been made in the identification and characterization of the enzymes involved in its synthesis. Nevertheless, a full understanding of the details of xyloglucan assembly and its regulation in vivo is far from complete.
The xyloglucan backbone in most dicotyledonous plants is composed of a repeating pattern of 1,4-β-d-glucosyl residues, of which two or three out of four residues carry an α-d-Xyl residue substitution at O-6. Major structural features of xyloglucan are generally conserved within a single plant species, and although variations exist in the substitution patterns (Hoffman et al., 2005), most xyloglucans are classified either as “XXXG type” or “XXGG type” depending on the number of substituted glucosyl residues (Fry et al., 1993; Vincken et al., 1997). The side chain xylosyl residues can be further substituted at the O-2 position with either a single β-d-Gal or an α-l-Fuc-(1,2)-β-d-Gal disaccharide, depending on the plant species. For example, Arabidopsis (Arabidopsis thaliana) plants have an XXXG-type xyloglucan that is composed mainly of XXXG, XLXG, XXFG, XLLG, and XLFG subunits (for nomenclature of subunits, see Fry et al., 1993). In other plants, for example the Poaeceae and Solanaceae, the Xyl units carry α-l-Ara instead of β-d-Gal residues. Variations in the substitution of xylosyl residues are observed not only in different plant species but also in different plant tissues (Freshour et al., 1996; O’Neill and York, 2003).
Arabidopsis xyloglucan contains seven different glycosidic linkages. If one makes the reasonable assumption that a different enzyme is required for the formation of each linkage, the biosynthesis of xyloglucan would theoretically require a combination of at least one (1,4)-β-glucan synthase, three xylosyltransferases, two galactosyltransferases, and one fucosyltransferase. Candidate genes encoding most of these enzymes have been identified and partially characterized using a combination of biochemical and genetic approaches. Thus, a candidate gene for the (1,4)-β-glucan synthase, CSLC4, has been identified in Nasturtium and Arabidopsis (Cocuron et al., 2007). A fucosyltransferase (FUT1) that catalyzes the addition of the terminal Fuc residue to the xyloglucan side chain was biochemically purified from pea (Pisum sativum), and its amino acid sequence was used to identify an Arabidopsis gene, FUT1 (Perrin et al., 1999). Heterologous expression of the pea and Arabidopsis FUT1 genes demonstrated that these encode a protein having xyloglucan fucosyltransferase activity (Perrin et al., 1999; Faik et al., 2000). Screening of ethyl methanesulfonate-mutagenized plants for aberrant cell wall formation led to the isolation of the Arabidopsis mur3 mutant (Reiter et al., 1997), and subsequently, MUR3 was determined to be a xyloglucan galactosyltransferase that adds Gal specifically to the third xylosyl residue, forming XXLG from XXXG (Madson et al., 2003). More recently, Jensen et al. (2012) identified a second galactosyltransferase, XLT2, that specifically adds Gal to the middle xylosyl residue, forming XLXG from XXXG or XLLG from XXLG. A gene family (CAZy family GT34) containing seven genes spread among three clades in Arabidopsis was identified and proposed to encode putative xyloglucan xylosyltransferases (Faik et al., 2002). Heterologous expression of two of the genes in this family (At3g62720 and At4g02500) demonstrated that the encoded proteins have xylosyltransferase activity and were thus named XXT1 and XXT2, respectively (Cavalier and Keegstra, 2006). These biochemical studies demonstrated that the xyloglucan xylosyltransferases XXT1 and XXT2 have the same substrate-acceptor specificity (i.e. transferring Xyl from UDP-Xyl to cellohexaose) and are able to catalyze the substitution of two glycosidic residues in adjacent positions, thereby generating GGXXGG structures.
Using a reverse genetic approach, it was demonstrated that xxt1 xxt2 double mutant plants lack detectable xyloglucan in their cell walls (Cavalier et al., 2008), confirming that XXT1 and XXT2 are xyloglucan xylosyltransferases and are essential for xyloglucan formation. The lack of detectable xyloglucan in the xxt1 xxt2 double mutant plants challenges conventional models for the functional organization of components in the primary cell wall (Cavalier et al., 2008). The same approach revealed that another member of this family, XXT5, is also involved in xyloglucan synthesis, although its activity in vitro could not be demonstrated (Zabotina et al., 2008). While xxt1 and xxt2 single mutant plants each has only a slight decrease in xyloglucan content, the xxt5 single mutant has a 50% reduction in xyloglucan content and the xyloglucan that is made in the mutant plant shows an altered subunit composition (Zabotina et al., 2008). However, the mutation in XXT5 does not eliminate the xylosylation of any particular Glc in the xyloglucan backbone, which suggests that the absence of XXT5 protein is compensated for, at least in part, by the presence of the other two xylosyltransferases. The ability of the XXT1 and XXT2 proteins to partially compensate for the lack of XXT5 in synthesizing fully xylosylated xyloglucan subunits raises questions about their specificity with respect to which Glc in the glucan backbone is the target of their activity. Furthermore, it is unclear how these three xylosyltransferases interact to synthesize the xyloglucan typically found in Arabidopsis cell walls.
In order to further investigate the relationship between the XXT1, XXT2, and XXT5 proteins and their respective roles in xyloglucan synthesis, we generated xxt1 xxt5 and xxt2 xxt5 double mutant lines as well as the xxt1 xxt2 xxt5 triple mutant. We report here on the comparative characterization of these plants with respect to the wild type and the xxt1, xxt2, xxt5, and xxt1 xxt2 lines characterized previously. The results of these studies provide new insights into the relationship among the various XXT proteins and demonstrate their different contributions to xylosylation of the glucan backbone during xyloglucan formation.
RESULTS
Generation and Molecular Characterization of T-DNA Insertion Mutants
To facilitate direct comparisons with xxt mutants developed previously and characterized by Cavalier et al. (2008) and Zabotina et al. (2008), the same transferred DNA (T-DNA) mutant lines for xxt1 (SAIL_785-E02), xxt2 (SALK_101308), and xxt5 (GABI-Kat; GK-411G05) were used to generate the xxt1 xxt5 and xxt2 xxt5 double mutants as well as an xxt1 xxt2 xxt5 triple mutant. In contrast to the situation encountered in generating the xxt1 xxt2 mutant, where the double mutant was recovered at a much lower frequency than expected (Cavalier et al., 2008), we had no problems generating the xxt1 xxt5 and xxt2 xxt5 mutants. In an attempt to avoid the abnormal segregation ratios observed in the isolation of the xxt1 xxt2 mutant, we used xxt2 xxt5 and xxt1 xxt2 to generate the xxt1 xxt2 xxt5 triple mutant. In a PCR screen of 100 F2 plants, we identified five xxt1 xxt2 xxt5 mutants, which is not significantly different from an expected recovery of approximately six plants. Finally, as seen with the xxt1 xxt2 mutant (Cavalier et al., 2008), we had no problems propagating the xxt1 xxt5, xxt2 xxt5, and xxt1 xxt2 xxt5 mutants.
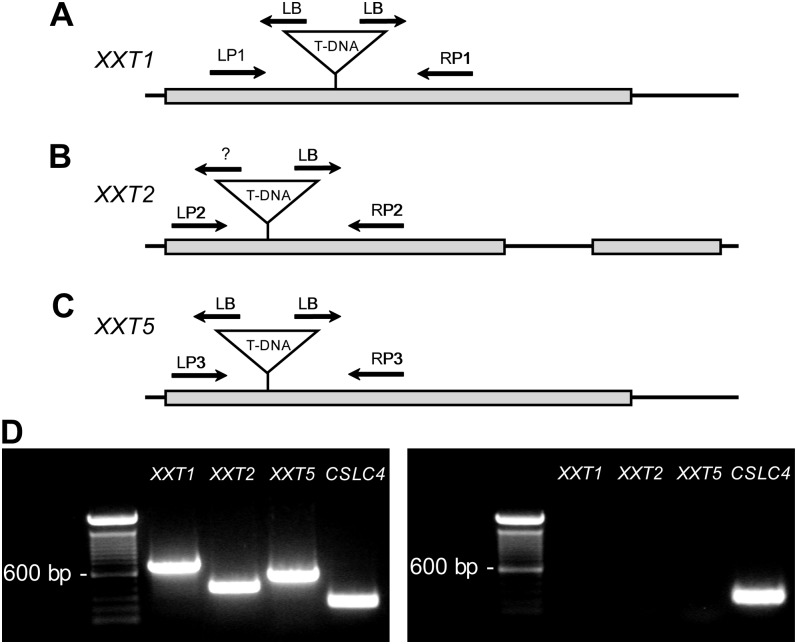
Analysis of the xxt1 xxt2 xxt5 triple mutant complementary DNA by reverse transcription (RT)-PCR showed that there was an absence of XXT1, XXT2, and XXT5 transcripts when gene-specific primers flanking the T-DNA insert were used (Fig. 1). Although the xxt1 xxt2 xxt5 mutant did not produce wild-type transcripts, it is possible that functional chimeric T-DNA-XXT transcripts were being produced (Wang, 2008, and refs. therein). To evaluate this possibility, we employed RT-PCR analysis of the xxt1 xxt2 xxt5 mutant using gene-specific primers (Supplemental Table S1) and the respective T-DNA border primer combinations (i.e. left border or right border), PCR amplicon sequencing, and quantitative PCR to characterize the structure and relative abundance of chimeric XXT transcripts (Supplemental Figs. S1 and S2; Supplemental Data S1 and Supplemental Materials and Methods S1.). Results from these experiments demonstrated that each xxt transcript had a large T-DNA insert that contained multiple stop codons in each reading frame, leading to the conclusion that the synthesis of functional proteins from these chimeric transcripts should not occur.
Figure 1.
Analysis of the xxt1 xxt2 xxt5 T-DNA insertion mutant. A to C, Gene models of XXT1, XXT2, and XXT5, respectively. Noncoding regions and introns are represented by black lines; coding regions are represented by gray boxes. The T-DNA insertion site for each gene is indicated along with the T-DNA border. LB, Left border; LP, left primer; RP, right primer; ?, unknown border. D, RT-PCR analysis of the wild type (Col-0; left panel) and the xxt1 xxt2 xxt5 T-DNA insertion mutant (right panel) using the gene-specific primers denoted in A to C that flank the T-DNA inserts. Further details can be found in Supplemental Data S1.
Different Levels of Severity of the Root Hair Phenotype in Single, Double, and Triple xxt Mutants
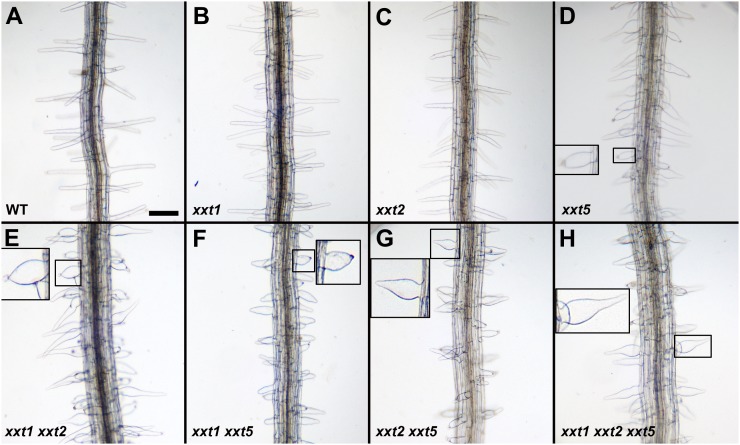
A root hair phenotype was observed for the xxt5 single mutant, all three of the xxt double mutants, and the triple mutant. Consistent with earlier observations (Cavalier et al., 2008), the root hairs appear normal in the xxt1 and xxt2 single mutants (Fig. 2). The morphology of the root hairs on the double mutants and on the triple mutant is more severe than that of the xxt5 single mutant. The root hairs are short and thick, fewer of them evidence a “normal” distal portion, and some of them form bubble-like extrusions at their tips (Fig. 2, insets). In general, root hairs on the xxt1 xxt5, xxt2 xxt5, and xxt1 xxt2 xxt5 mutants are shorter than those on the xxt5 single mutant. Most likely, the severity of root hair phenotypes in xxt mutants correlates with cell wall defects and reflects their inability to expand properly; it seems likely that these defects are caused by the reduction in xyloglucan levels (see below).
Figure 2.
Phenotype of roots from xxt mutant lines and wild-type seedlings. All seedlings were grown under the same conditions on agar plates for 7 d as described in “Materials and Methods.” Plates were oriented vertically in the growth chamber. The labels at bottom left of each image indicate the plant line shown. Root hairs in A, B, and C were similar. Root hairs boxed in D to H were magnified and placed in the same image to clearly show the root hair phenotype. Bar in A = 200 µm for all images, except the insets.
xxt Mutants Display Differences in Xyloglucan-Specific Endoglucanase and Driselase-Susceptible Xyloglucan
To gain insight into how xyloglucan structure and content are affected in the mutants, we sequentially extracted alcohol-insoluble residue (AIR) preparations of whole seedlings with 1 n KOH and 4 n KOH and characterized the xyloglucan in each of these fractions with two xyloglucan-specific techniques. First, we used high-performance anion-exchange chromatography (HPAEC) to measure the relative amounts of Driselase-susceptible xyloglucan in each fraction. Driselase is a mixture of exoglycosidases and endoglycosidases that will hydrolyze most cell wall polysaccharides. Because Driselase does not contain α-xylosidase activity, the hydrolysis of xyloglucan releases a mixture of monosaccharides plus a characteristic disaccharide, isoprimeverose [IP; Xyl-α-(1-6)-Glc], that is a diagnostic indicator of xyloglucan (Hayashi and Maclachlan, 1984; Popper and Fry, 2005). The levels of IP provide an estimate of the levels of xyloglucan present in a sample. Second, we employed oligosaccharide mass profiling (OLIMP; Lerouxel et al., 2002), which uses the specificity of xyloglucan-specific endoglucanase (XEG; Pauly et al., 1999b) and the sensitivity of matrix-assisted laser-desorption ionization time-of-flight (MALDI-TOF) mass spectrometry to determine the xyloglucan oligosaccharide (XGO) composition of each population of xyloglucan.
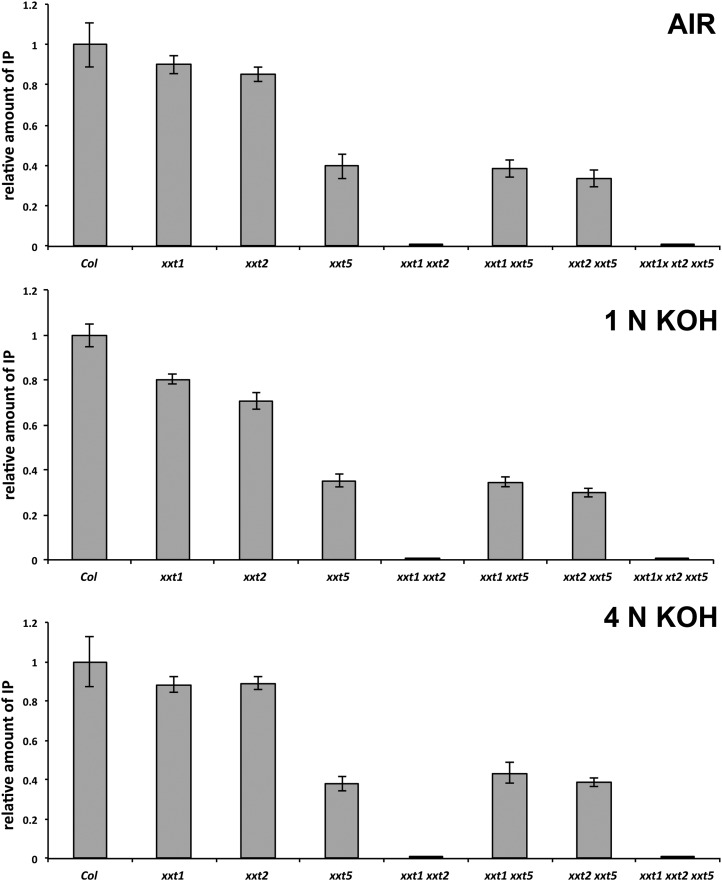
Results from Driselase-HPAEC (Fig. 3) and OLIMP analyses (data not shown) of the xxt1 xxt2 and xxt1 xxt2 xxt5 mutants demonstrated that these plants lacked detectable xyloglucan, consistent with earlier results with the xxt1 xxt2 mutant (Cavalier et al., 2008; Park and Cosgrove, 2012) and leading to the conclusion that deleting the XXT5 gene from this double mutant does not restore the synthesis of xyloglucan. However, results from Driselase-HPAEC and OLIMP analyses of the two new double mutants showed interesting changes in xyloglucan content and structure.
Figure 3.
Quantity of the Driselase-generated xyloglucan signature fragment, IP, estimated by HPAEC analysis. Results are expressed as relative amounts of IP obtained after digestion of total AIR, and soluble in 1 n KOH and 4 n KOH fractions prepared from mutant lines in comparison with the amount of IP obtained from the same fraction prepared from Col-0 wild-type plants, which was taken as 100%. One milligram of each sample was dissolved in 30 µL of acetate buffer containing approximately 0.5 units of Driselase and incubated for 18 h at 37°C. The total volume of each digest was analyzed by HPAEC. Analyses were performed on three independent sample preparations for each mutant and wild-type plant. Results were averaged and expressed as mean values (n = 3 biological replications) ± sd.
Results from Driselase-HPAEC analysis of AIR preparations of the xxt1, xxt2, and xxt5 mutant seedlings (Fig. 3) showed reductions in Driselase-susceptible xyloglucan content that are consistent with results reported by Cavalier et al. (2008) and Zabotina et al. (2008). Unlike the lack of xyloglucan in the xxt1 xxt2 mutant, measurable amounts of xyloglucan were found in the xxt1 xxt5 and xxt2 xxt5 mutants. There were significant differences in the amounts of IP released from total AIR and both hemicellulosic fractions (1 n KOH and 4 n KOH) prepared from xxt5, xxt1 xxt5, and xxt2 xxt5 plants compared with the ecotype Columbia (Col-0) wild type and xxt1 and xxt2 mutants (P < 0.01; Fig. 3). However, the differences in IP released from the xyloglucan fractions derived from the xxt5, xxt1 xxt5, and xxt2 xxt5 mutants were not significant when compared with each other (P > 0.05). From these observations, we tentatively concluded that the xxt5 mutant has significantly lower levels of xyloglucan, as reported earlier by Zabotina et al. (2008), but that further disruption of either XXT1 or XXT2 does not result in further reduction in xyloglucan levels.
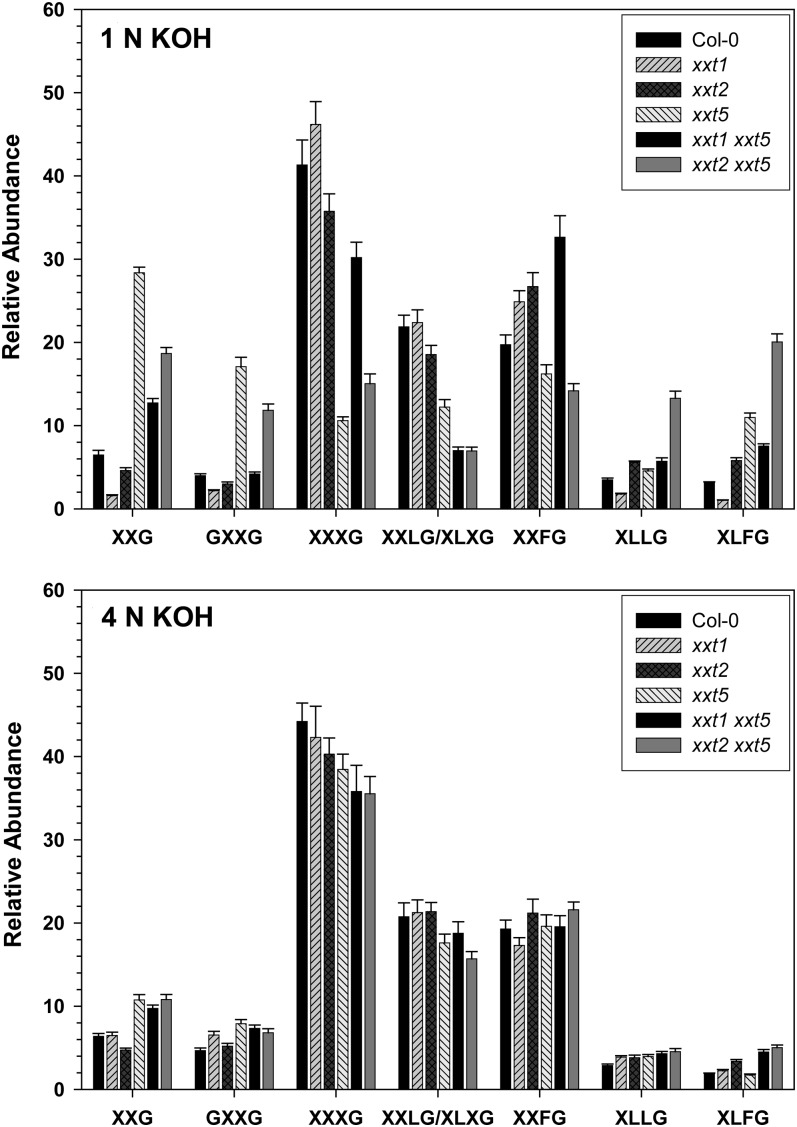
OLIMP analysis was used to examine the impact of the various mutations on the substitution patterns of the XEG-accessible xyloglucan made in those mutant plants that make xyloglucan (Fig. 4). The most important observation was that in both the xxt1 xxt5 and xxt2 xxt5 double mutants, which presumably had only a single active xylosyltransferase protein, the full collection of oligosaccharides was observed. The proportion of underxylosylated oligosaccharides (XXG and GXXG) was higher, especially in the 1 n KOH extracts (Fig. 4), indicating that the degree of substitution of the backbone was reduced slightly. However, the proportion of fully substituted oligosaccharides (e.g. XLFG) was also increased over the wild type in these two mutants, especially in the 1 n KOH extract (Fig. 4). It is interesting that the variation in substitution patterns among the mutants is larger in the xyloglucan extracted with 1 n KOH (Fig. 4) than it is in the xyloglucan extracted with 4 n KOH (Fig. 4). For example, in the 1 n KOH extracts, there were highly significant differences (P < 0.0001) in the relative abundances of XXG, GXXG, and XXXG between Col-0 and either the xxt5 mutant or the xxt2 xxt5 mutant; however, these same differences were less significant in the 4 n KOH extracts (P < 0.025). These results suggest that there may be different classes of xyloglucan synthesized, some of which are more easily extracted from the wall and are more affected by mutations in XXT genes and others, more tightly bound to the wall, that appear less affected by the xxt mutations. The biochemical basis for these changes and the physiological significance of them are not yet understood.
Figure 4.
OLIMP of XEG-digested 1 n KOH and 4 n KOH sequential extracts of the Col-0 wild type and xxt mutants. Each XGO is named according to the nomenclature described by Fry et al. (1993). The relative abundance of each XGO is presented as a mean value (n = 3 biological replications) ± sd.
In summary, chemical characterization of the mutants demonstrated that the newly generated xxt1 xxt5 and xxt2 xxt5 mutant plants, together with previously characterized xxt5, contain significantly reduced amounts of xyloglucan in comparison with the wild type. However, these two new double mutants, each presumably containing only a single active xylosyltransferase protein, retain the ability to form fully xylosylated xyloglucan subunits.
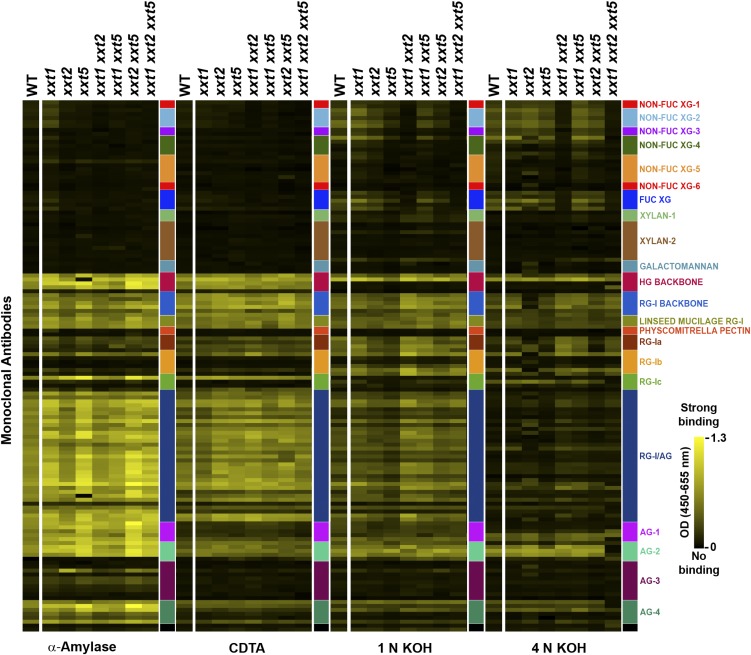
Glycome Profiling of Cell Wall Extracts from xxt Mutant Plants
In an effort to further understand the changes in xyloglucan and to investigate the ways in which these changes impact the interactions of polysaccharides within cell walls, glycome profiling experiments using plant glycan-directed monoclonal antibodies were performed. To accomplish the glycome profiling, AIR prepared from young seedlings (14 d old) of all single, double, and the triple xxt mutants grown in liquid culture were subjected to sequential extractions with four increasingly harsh reagents (buffer containing α-amylase, cyclohexane diamine tetraacetic acid [CDTA], 1 n KOH, and 4 n KOH), and the materials solubilized at each step were analyzed by ELISA using a broad and diverse collection of plant cell wall glycan-directed monoclonal antibodies (Pattathil et al., 2010). The buffer/α-amylase and CDTA-solubilized fractions from all analyzed plants contained mostly pectin and AGP-related epitopes and generally contained little or no detectable xyloglucan or xylan epitopes, except for wild-type and xxt1 walls, which contained some xyloglucan epitopes in the buffer/α-amylase extract. The fractions prepared from xxt1 mutant plants showed the same glycome profiles as the wild type.
Hemicellulosic fractions extracted with 1 n KOH and 4 n KOH from the three single and two double (xxt1 xxt5 and xxt2 xxt5) mutants show differences in xyloglucan epitope contents (Fig. 5). A total of 28 xyloglucan-directed antibodies were used in this study (Supplemental Table S2). These antibodies fall into seven groups based on their reactivities against a diverse panel of plant polysaccharides, including several distinct xyloglucans (Pattathil et al., 2010, 2012). These antibody groupings reflect differences in the xyloglucan epitopes recognized. Six antibody groups are directed against nonfucosylated xyloglucan epitopes, and one group is directed against fucosylated xyloglucan epitopes. The xyloglucan epitope pattern for the xxt1 mutant shows only subtle differences to the pattern observed in wild-type extracts, while the xyloglucan epitope pattern in xxt2 is distinct from the wild type, particularly in the 4 n KOH extract. The xxt5 wall extracts show a reduction in the diversity and amounts of xyloglucan epitopes, particularly in the 1 n KOH extract. The xyloglucan epitope patterns in both the 1 n KOH and 4 n KOH extracts prepared from xxt1 xxt5 walls are similar to those from xxt1 walls, while xxt2 xxt5 xyloglucan epitope patterns are similar to those from xxt5 walls. No xyloglucan epitopes were detected with any xyloglucan-directed antibodies in any of the extracts prepared from xxt1 xxt2 and xxt1 xxt2 xxt5 mutant plants, consistent with the lack of chemically detectable xyloglucan in these plants. The results obtained by glycome profiling are in good agreement with those obtained by OLIMP analysis. First, the differences observed for 1 n KOH hemicellulosic fractions are more pronounced than those for 4 n KOH fractions. Second, all xxt mutants contain the full set of xyloglucan epitopes that were recognized by available antibodies, but the distribution of those epitopes is different among different mutants.
Figure 5.
Glycome profiling of cell wall extracts prepared from Arabidopsis Col-0 and xxt mutants. Cell walls of each plant line were prepared from whole seedlings of Arabidopsis Col-0 wild type (WT) and all single and multiple xxt mutants grown in liquid culture. The walls were subjected to sequential extraction with α-amylase, CDTA, 1 n KOH, and 4 n KOH as described in “Materials and Methods.” The solubilized extracts were then screened against an array of plant glycan-directed monoclonal antibodies (Pattathil et al., 2010) using ELISAs. The panels at right (colored boxes) depict the groups of antibodies used, identified according to the polysaccharides predominantly recognized by each group (for additional details about the individual antibodies used, see Supplemental Table S2). The extent of antibody binding is represented as a colored heat map, with the brightest yellow depicting the strongest binding and black depicting no binding.
The nonxyloglucan epitope distribution pattern in the 4 n KOH extract of the xxt2 mutant was noticeably different from the patterns observed in the equivalent extracts from wild-type plants and the other mutants. An increase in diverse homogalacturonan and rhamnogalacturonan I epitopes was detectable in the 1 n KOH extracts of xxt1 xxt2 and xxt1 xxt5 walls as compared with the equivalent extract from wild-type walls (Fig. 5). Otherwise, few differences in the distribution patterns of nonxyloglucan epitope were detectable in the glycome profiles of the other mutants in comparison with the wild-type glycome profile. For example, among the plants with multiple xxt mutations, the xxt1 xxt2 xxt5 triple mutant lacks some rhamnogalacturonan I and arabinogalactan epitopes in the 4 n KOH fraction, while all of the double mutants appear to have retained these epitopes at levels comparable with wild-type plants. These ELISA analyses confirmed that minimal if any quantitative compensatory changes in other cell wall polysaccharide epitopes occur in xxt mutant plants, at least as far as can be assessed by the collection of 135 monoclonal antibodies utilized in this study.
Immunolocalization of Xyloglucan Epitopes in xxt Mutants
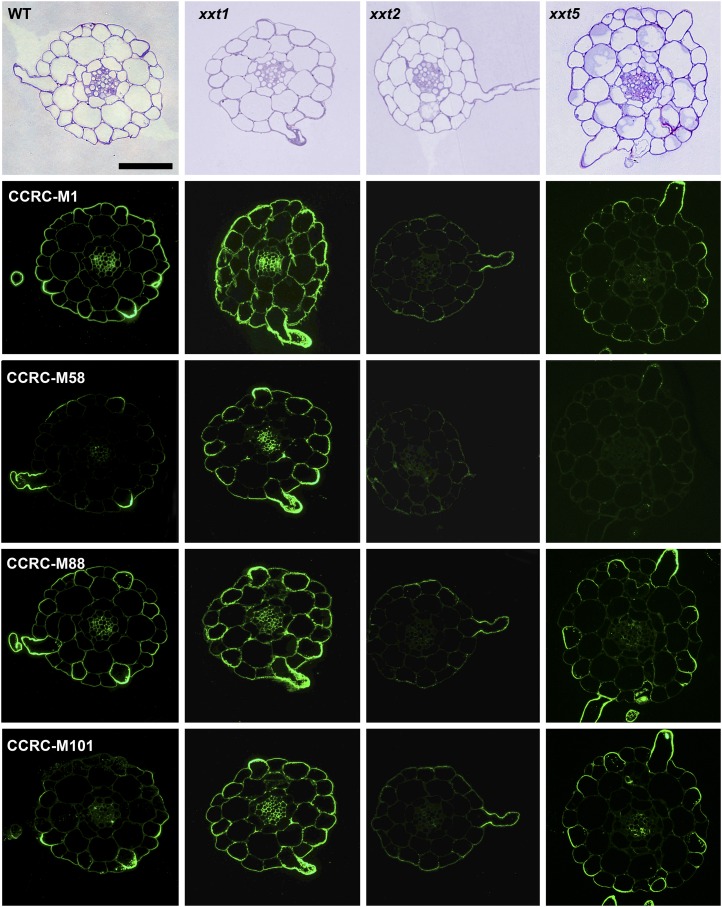
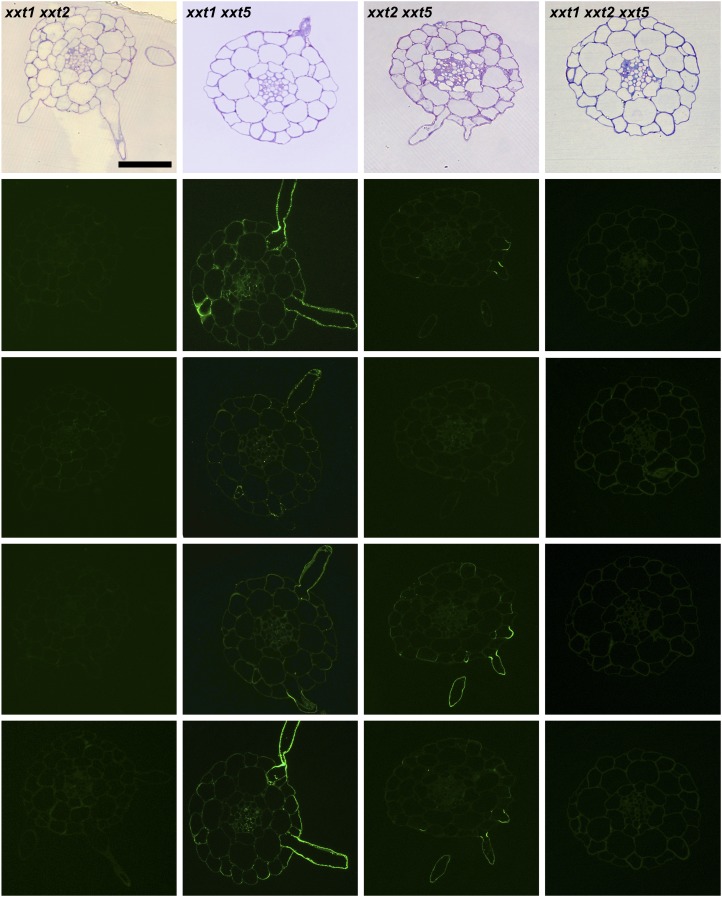
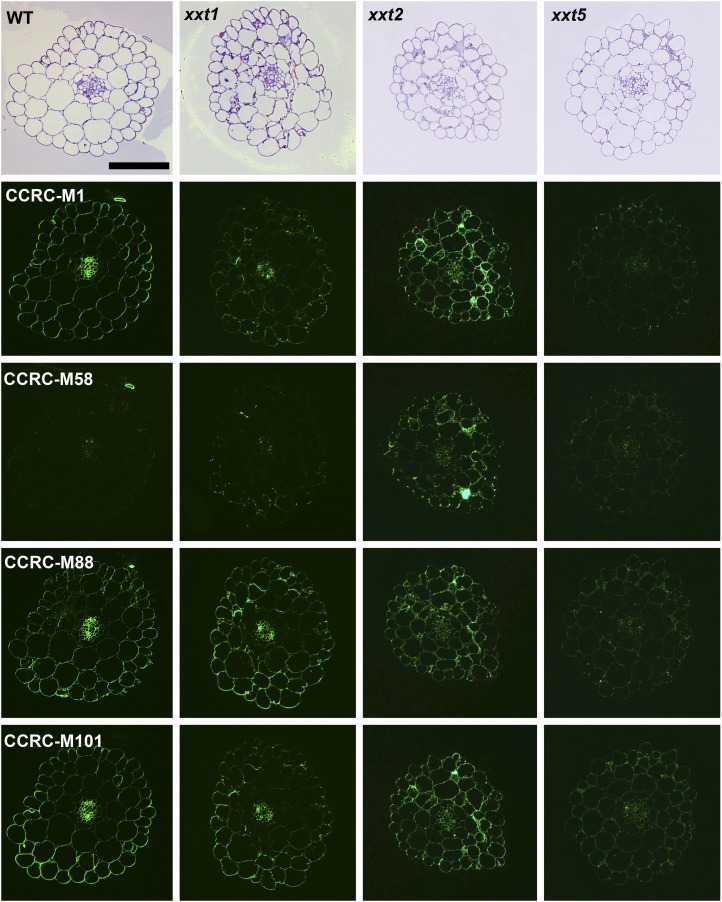
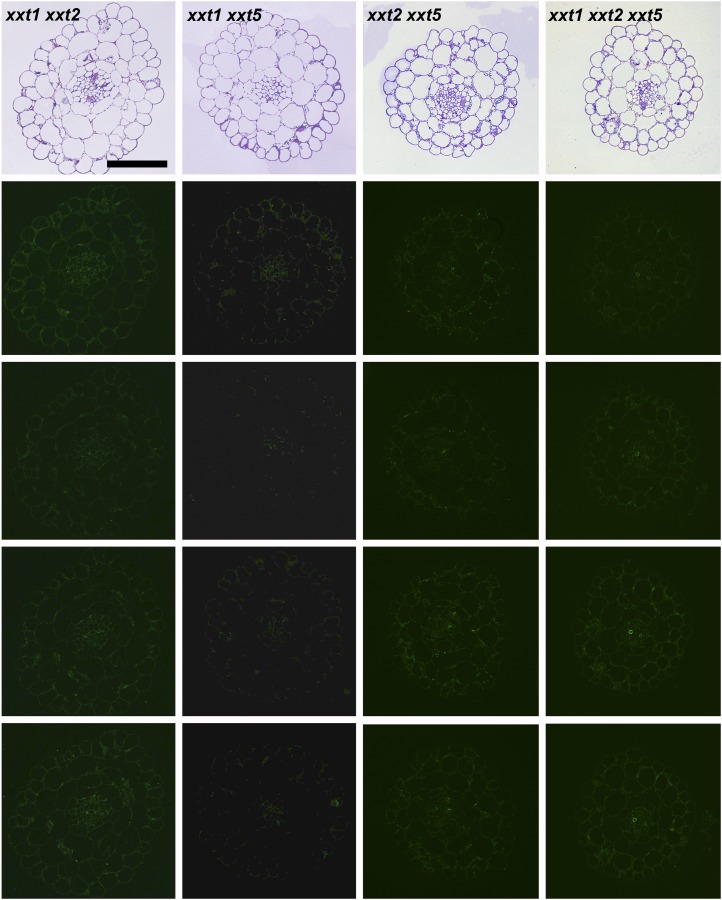
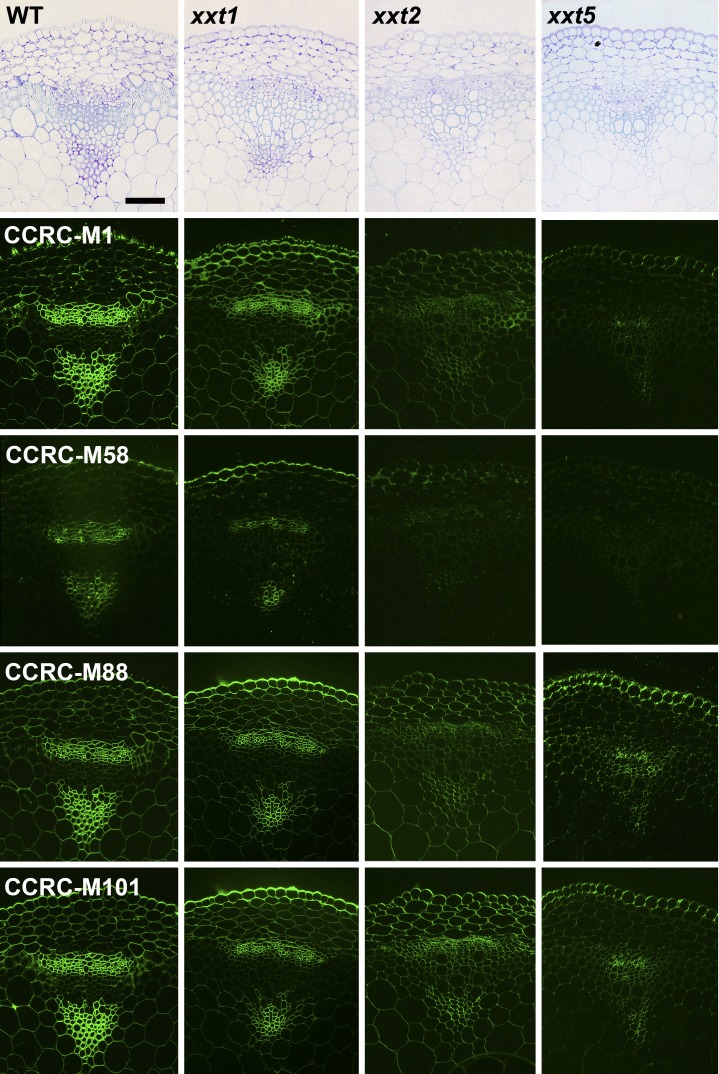
The results presented to this point were obtained using averaged cell walls prepared from entire young seedlings that were grown in liquid culture; these included roots and hypocotyls. Hence, any organ- or cell type-specific distribution of xyloglucans in the mutants and wild-type plants cannot be assessed. To address this important question, the distribution patterns of several xyloglucan epitopes in the walls of roots, hypocotyls, and stems of xxt mutant and wild-type plants were determined using monoclonal antibodies that recognize distinct xyloglucan epitopes (i.e. the antibodies used belong to different groups of xyloglucan-directed antibodies; Supplemental Table S2). Neither the xxt1 xxt2 double mutant nor the xxt1 xxt2 xxt5 triple mutant showed any labeling in any of the three organs examined with any of the xyloglucan-directed monoclonal antibodies tested (Figs. 6, 7, and 9). The mutation of XXT5 results in the nearly complete absence of CCRC-M58 labeling. Otherwise, the other mutants each showed xyloglucan epitope localization patterns that are distinct from each other and distinct from the wild-type pattern in at least one of the three organs examined. For example, the xyloglucan epitope localization patterns for the xxt1 mutant most closely resemble those of the wild type, particularly in hypocotyl and stem tissues (Figs. 7 and 9). However, in root tissue, the xxt1 mutation appears to result in a broader distribution of the xyloglucan epitopes recognized by CCRC-M58 and CCRC-M101 (Fig. 6), suggesting a change in xyloglucan structure in some root cell types caused by the mutation in XXT1. Mutations in either XXT2 or XXT5 resulted in significantly reduced or altered labeling with all antibodies in root tissues, with labeling being the weakest in xxt2 (Fig. 6). The xyloglucan epitopes recognized by CCRC-M1, CCRC-M88, and CCRC-M101 appear to be most persistent in root hairs of xxt mutants. However, the presence of these xyloglucan epitopes in root hairs appears insufficient to convey full functionality to the xyloglucan in its role in wall integrity in this cell type, inasmuch as the xxt5 single mutant and all plants carrying multiple xxt mutations show defects in their root hairs (Fig. 2). In the double mutants that still produced xyloglucan, the root labeling pattern of xxt1 xxt5 resembles that of xxt2, while xxt2 xxt5 shows only residual labeling of root hairs.
Figure 6.
Immunofluorescence labeling of transverse sections of roots taken from 6-d-old seedlings of wild-type Col-0 (WT) and all xxt mutant plants using selected xyloglucan-directed antibodies. Toluidine blue-stained sections of wild-type and mutant lines show the overall root morphology from left to right. The xyloglucan-directed antibodies used (CCRC-M1, CCRC-M58, CCRC-M88, and CCRC-M101) recognize distinct xyloglucan epitopes and are identified with labels in the wild-type images. Bars = 50 µm for all images.
Figure 7.
Immunofluorescence labeling of transverse sections of hypocotyls from wild-type Col-0 (WT) and all xxt mutant plants using selected xyloglucan-directed antibodies. Toluidine blue-stained sections of wild-type and mutant lines show the overall hypocotyl morphology from left to right. The xyloglucan-directed antibodies used (CCRC-M1, CCRC-M58, CCRC-M88, and CCRC-M101) recognize distinct xyloglucan epitopes and are identified with labels in the wild-type images. Bars = 50 µm for all images.
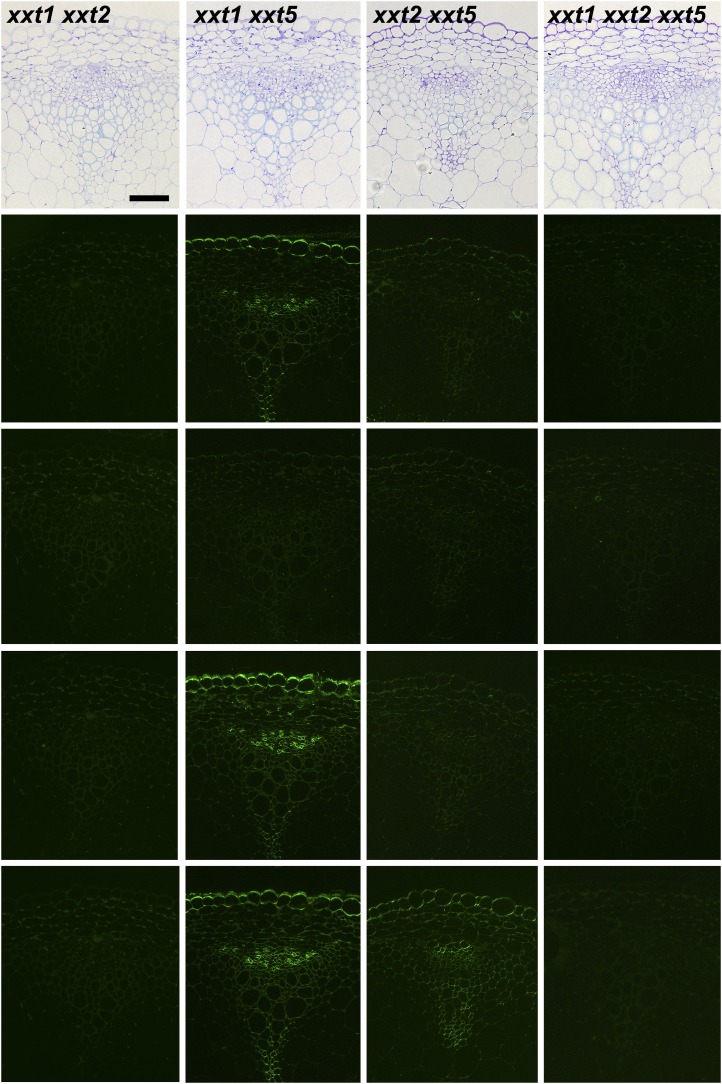
Figure 9.
Immunofluorescence labeling of transverse sections of basal parts of inflorescence stems taken from 6-week-old wild-type Col-0 (WT) and all xxt mutant plants using selected xyloglucan-directed antibodies. Toluidine blue-stained sections of wild-type and mutant lines show the overall stem morphology from left to right. The xyloglucan-directed antibodies used (CCRC-M1, CCRC-M58, CCRC-M88, and CCRC-M101) recognize distinct xyloglucan epitopes and are identified with labels in the wild-type images. Bars = 50 µm for all images.
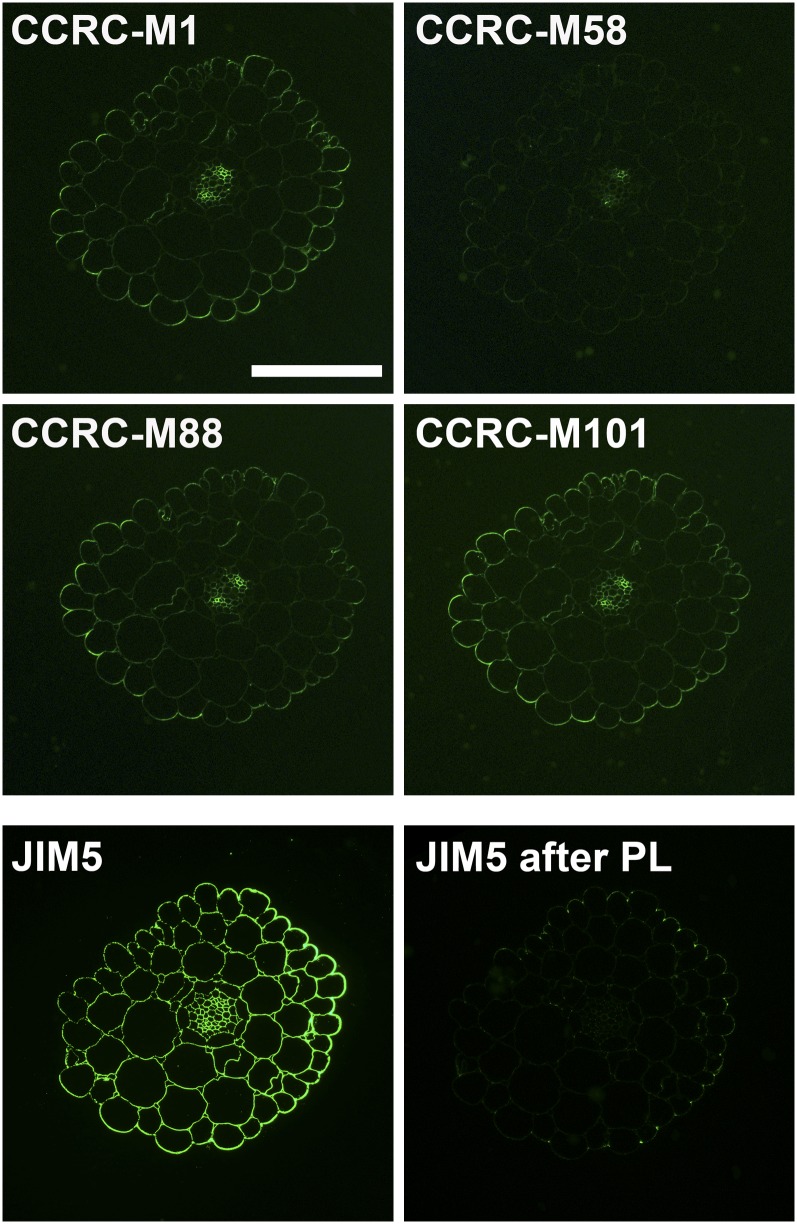
In hypocotyl tissues, antibody labeling is essentially absent in xxt5, while labeling in xxt1 closely resembles the wild-type labeling patterns (Fig. 7). The hypocotyl labeling patterns observed in xxt2 showed increased labeling compared with the wild type for three of the xyloglucan-directed antibodies used (CCRC-M58 [Fig. 7]; CCRC-M48 and CCRC-M100 [data not shown]) but no change in labeling with the other three such antibodies (CCRC-M1, CCRC-M88, and CCRC-M101), perhaps reflecting a change in the epitope composition of the xyloglucan made in the hypocotyls of xxt2 plants. All xxt mutant combinations resulted in the absence of labeling with any xyloglucan-directed antibody tested in the hypocotyl. We examined whether the presence of homogalacturonans, which are particularly prominent in hypocotyl tissues, might affect the patterns of antibody binding to xyloglucan through possible masking effects (Marcus et al., 2008). Treatment of wild-type hypocotyl sections with a purified pectate lyase did not alter the observed xyloglucan epitope distribution patterns with any of the xyloglucan-directed antibodies used (compare Figs. 7 and 8), although this treatment did eliminate almost all homogalacturonan epitopes recognized by the JIM5 antibody (Fig. 8). Thus, we observed no homogalacturonan-masking effects in our studies.
Figure 8.
Effect of pectate lyase treatment of hypocotyl sections on xyloglucan and homogalacturonan labeling patterns. With the exception of the JIM5 section, which was treated with CAPS buffer only as a control, sections were treated with pectate lyase (PL) in CAPS buffer (pH 10) for 2 h at room temperature followed by immunolabeling with the indicated antibodies. No differences in the immunolabeling patterns were observed with xyloglucan-directed antibodies compared with those shown in Figure 7. Homogalacturonan epitopes recognized by JIM5 were almost completely removed from the sections after pectate lyase treatment compared with the control labeling with JIM5. Bar = 50 µm for all images.
In the stem, the knockout of XXT1 did not appear to affect the labeling pattern with xyloglucan-directed antibodies (Fig. 9). The antibody labeling patterns in xxt2 and xxt5 stems are distinct from the wild type, with the reduction in labeling being more severe in xxt5. Specifically, the xyloglucan epitopes recognized by CCRC-M1 and CCRC-M58 that are made in epidermal and primary xylem and phloem cells in the wild type are dramatically reduced in the xxt2 mutant. In the two double mutants that still produce xyloglucan, the antibody labeling pattern in xxt1 xxt5 stems resembles that observed in xxt5, while only CCRC-M101 shows significant labeling in xxt2 xxt5 stems.
In summary, the results from these immunological studies demonstrate that the impact of mutations in XXT genes, either alone or in combination, on xyloglucan epitope distribution patterns varies depending on the organ, tissue, and even cell type. Thus, among the single mutants, the knockout of XXT2 has the most dramatic effect on xyloglucan synthesis in roots, while a knockout of XXT5 has the most dramatic effect in hypocotyls. The hypocotyl is the most sensitive organ to knockout of multiple XXT genes. In the other organs examined, mutations in both XXT2 and XXT5 result in a more dramatic change in xyloglucan epitope quantity and distribution than do mutations in both XXT1 and XXT5. The effect of mutations in XXT genes is further dependent on the specific cell types being examined within each organ.
DISCUSSION
Previous studies on the xylosylation of the xyloglucan backbone in Arabidopsis demonstrated that three genes, two encoding xylosyltransferases XXT1 and XXT2 and a third encoding a putative xylosyltransferase, XXT5, are involved in this process (Cavalier and Keegstra, 2006; Cavalier et al., 2008; Zabotina et al., 2008). In this study, we further explored the role of these proteins in xyloglucan biosynthesis by creating the additional double mutant lines xxt1 xxt5 and xxt2 xxt5 as well as the triple mutant line xxt1 xxt2 xxt5. In addition, a powerful technique, glycome profiling, was used for investigating the impact of mutations in these genes, either singly or in combination, on cell wall composition (Fig. 5). Finally, a suite of xyloglucan-directed antibodies recognizing different xyloglucan epitopes were used to investigate the impact of these mutations on xyloglucan epitope distribution patterns in roots (Fig. 6), hypocotyls (Figs. 7 and 8), and stems (Fig. 9). As described in more detail below, these studies led to new insights into the complex process of xyloglucan biosynthesis and the potential contributions of three main xylosyltransferases involved in this process.
Several different techniques were used to characterize the xyloglucans present in the various xxt mutants. The advantage of using different experimental strategies for xyloglucan characterization is that the different methods can provide complementary information. However, one needs to exercise caution in comparing the data from the different experimental approaches. For example, not all of the data are directly comparable, in part because not all of the data are equally quantitative. Another issue is that some methods give averages of information about xyloglucans from several different cell, tissue, and organ types (Figs. 3–5), while other methods provide information about xyloglucans in specific cell types (Figs. 6–9). Additionally, the composition of xyloglucans can vary depending on the developmental stage of the plant or the organs/tissues being analyzed. Thus, results obtained with one set of plants cannot always be directly compared with studies performed on different plant material. Nonetheless, the use of multiple complementary approaches provides a more detailed picture of the consequences of mutations in one or more XXT genes than can be obtained using a single approach.
One powerful strategy used in this study is an immunological form of glycome profiling (Pattathil et al., 2012) that was used to characterize the changes in cell wall composition and polysaccharide extractability in the xxt mutants (Fig. 5). This method utilizes a large and diverse toolkit of monoclonal antibodies (Pattathil et al., 2010) that includes antibodies directed at multiple epitopes on each of the major noncellulosic plant cell wall polysaccharides, except rhamnogalacturonan II, and thus provides a broad overview of plant cell wall structure and composition. Application of this semiquantitative approach for the characterization of xxt mutants confirmed the absence of detectable xyloglucan epitopes in both the xxt1 xxt2 double mutant and the xxt1 xxt2 xxt5 triple mutant (Fig. 5). More interestingly, the different glycome profiles for each xxt mutant provide evidence that each mutation or combination of mutations in the XXT genes results in unique changes to the cell walls of the affected plants (Fig. 5). For example, the xyloglucan epitope profiles (the top seven colored boxes at the top of the heat maps) were altered to differing degrees from the wild-type profile in each of the single mutants. Meanwhile, there was little change overall in the glycome profiles in other polysaccharide classes in these plants. It should be noted that glycome profiling, by itself, does not provide quantitative information about the total cell wall content of each polysaccharide. The glycome profiles do reveal changes in the extractability of polysaccharides other than xyloglucan in some of the multiple mutants. These new detailed analyses of polysaccharide epitope compositions show remarkably few changes in the cell walls of the xxt mutants outside of the clearly observable changes in xyloglucan content and epitope composition.
Immunohistochemical analyses are not quantitative and lack the resolution of glycome profiling performed on extracted polysaccharide fractions, but they do provide information on the distribution of specific epitopes in different cells, tissues, and organs. Given that the predominant changes observed in cell wall composition in the xxt mutants were in the xyloglucan component and that previous immunohistochemical studies of some xxt mutants had shown no changes in the nonxyloglucan epitope distribution patterns (Cavalier et al., 2008), further immunohistochemical studies were performed using only a set of antibodies directed against diverse xyloglucan epitopes. Organ/cell-specific differences in the effects of mutations in the various XXT genes were observed. For example, in hypocotyls, at least two xylosyltransferases must be present for any xyloglucan to be made (Fig. 7), with XXT5 being the most important contributor to xyloglucan synthesis in most cells in this organ (Fig. 7). In roots, on the other hand, XXT2 appears to be the dominant contributor to xyloglucan synthesis, particularly in trichoblasts and root hairs (Fig. 6). In stems, both XXT2 and XXT5 play important roles in xyloglucan synthesis, although XXT2 appears to be more dominant in some cell types (e.g. epidermal cells; Fig. 9). Mutation of either XXT2 or XXT5 results in the synthesis of xyloglucan with an altered epitope composition, and even here, the effect is not the same in all cell types in stem (Fig. 9).
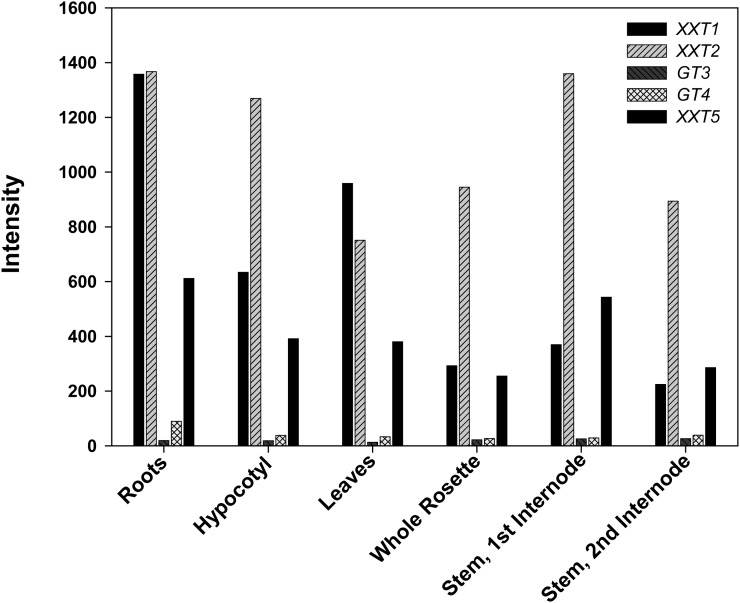
The simplest explanation of these observations is that the three XXT genes have different patterns of expression in different plant organs and in different cell types within those organs. Therefore, mutations in individual XXT genes lead to different impacts on xyloglucan structure and content in the different cells, depending on whether a nonmutated XXT gene encoding an active glycosyltransferase is expressed in sufficient amounts to compensate for the mutated gene. This interpretation is supported by the gene expression data gathered from publicly available transcriptomic databases (Fig. 10), where XXT2 has about a 2-fold higher level of expression than XXT1 and XXT5 at all stages of development and these latter two genes generally show comparable levels of expression (Fig. 10). However, if one looks in more detail at different tissues and organs at various stages of development, the various genes sometimes invert with respect to their expression levels (Schmid et al., 2005; http://www.weigelworld.org/resources/microarray/AtGenExpress/). While the differing expression levels may explain the complex changes in xyloglucan levels that are observed (Figs. 6, 7, and 9), care needs to be exercised in attempting to explain the observed patterns. The transcript levels do not necessarily correlate with protein levels, and even if they do, the levels of the various XXT proteins do not necessarily correlate with xyloglucan levels or xyloglucan epitope composition. It is very possible, even likely, that xyloglucan levels are controlled by several types of regulation beyond transcription of the genes needed for xyloglucan biosynthesis. For example, the activity of various proteins may be modulated by posttranslational modifications or by the availability of the required substrates.
Figure 10.
Expression of XXT1, XXT2, GT3, GT4, and XXT5 in Arabidopsis organs. The AtGE Development series (Schmid et al., 2005) was queried for XXT1 (At3g62720), XXT2 (At4g02500), GT3 (At5g07720), GT4 (At1g18690), and XXT5 (At1g74380) expression using the AtGenExpress Visualization Tool (http://jsp.weigelworld.org/expviz/expviz.jsp). Gene expression in roots, hypocotyls, leaves (composed of the first two leaves to emerge), whole rosettes from 7-d-old seedlings, and stems from 21-d-old plants is shown.
The chemical and glycome profiling data reported here for xyloglucan from the various mutants (Figs. 3–5), when combined with the biochemical characterization of the various XXT proteins (Cavalier and Keegstra, 2006; Zabotina et al., 2008), allow some preliminary conclusions regarding the roles of the three proteins during xyloglucan biosynthesis. One simple hypothesis can be eliminated. Given that xyloglucan contains three different xylosyl residues and that three different proteins (i.e. XXT1, XXT2, and XXT5) are involved in producing xyloglucan, the simplest hypothesis is that each enzyme is responsible for adding a different xylosyl residue. This hypothesis appears to hold true for the addition of galactosyl residues to xyloglucan, where different genes encode the two isoforms of the enzyme that adds the two different galactosyl residues (Madson et al., 2003; Jensen et al., 2012). However, the combined data presented here argue strongly against the application of this hypothesis to Xyl addition. This can be seen most clearly by examining the structure of the xyloglucan recovered from either of the two double mutants, xxt1 xxt5 or xxt2 xxt5. While both double mutants produced reduced levels of xyloglucan (Fig. 3), when the XGOs derived from the xyloglucans in each mutant are examined, all of the XGOs are present, although the proportions of them differ between the mutants (Fig. 4), and the xyloglucan epitope composition and distribution patterns are also different among the mutants (Figs. 5–7 and 9). If one assumes that only these three proteins are involved in xyloglucan biosynthesis (for caveats, see below), then in each double mutant only a single active protein is left (XXT1 in the xxt2 xxt5 double mutant and XXT2 in the xxt1 xxt5 double mutant). Thus, if these assumptions are correct, either XXT1 or XXT2 is capable of adding all three of the xylosyl residues present in xyloglucan. Such a conclusion is also supported by the immunohistochemical data reported here and the biochemical data obtained previously, which show that XXT1 and XXT2 have equivalent activities in vitro (Cavalier and Keegstra, 2006).
A second important conclusion derived from our results, in combination with those presented earlier, is that either XXT1 or XXT2 is required for xyloglucan biosynthesis. Although this conclusion was reached in earlier work (Cavalier et al., 2008; Park and Cosgrove, 2012) on the xxt1 xxt2 double mutant, it is further reinforced by the absence of detectable xyloglucan in the xxt1 xxt2 xxt5 triple mutant. More importantly, the other two double mutants, xxt1 xxt5 and the xxt2 xxt5, are able to produce xyloglucan, although in reduced quantities and with altered composition. Thus, one can conclude that the XXT5 protein by itself cannot add the xylosyl residues of xyloglucan in the same way as the XXT1 or XXT2 protein (see above). This is a particularly intriguing conclusion in that when the various single mutants are compared (Figs. 3–5), the most dramatic reduction in xyloglucan levels and the most dramatic changes in xyloglucan oligosaccharide and epitope profiles are seen in the xxt5 mutant. Thus, while the XXT5 protein itself does not seem capable of adding xylosyl residues to the xyloglucan backbone in vivo, elimination of its function causes the most dramatic impact on xyloglucan biosynthesis. Sequence alignment and molecular modeling of the three-dimensional structure showed some significant differences between XXT5 and two other xylosyltransferases (data not shown). Thus, XXT5 has a significantly shorter predicted stem region and eight amino acids localized downstream of the predicted DXD motif differ from those in XXT1 and XXT2 proteins, creating a more basic environment for the XXT5 catalytic site, which could potentially affect its catalytic ability. It is also interesting that earlier attempts to demonstrate xylosyltransferase activity for XXT5 in vitro were unsuccessful (Zabotina et al., 2008). Indeed, the absence of any detectable xyloglucan in plants where both XXT1 and XXT2 are lost is consistent with an absence of catalytic function for XXT5. One intriguing possibility is that the function of XXT5 is to maintain the integrity of a synthetic complex involved in xylosylation of the glucan backbone during xyloglucan formation rather than to function as a xylosyltransferase. An analogous role for the GAUT7 gene in homogalacturonan synthesis has recently been proposed (Atmodjo et al., 2011). In the presence of XXT5, XXT1 and XXT2 can partially or mostly substitute for each other in xyloglucan synthesis, while in the absence of XXT5, the integrity of the complex and the efficiency of xylosylation are compromised, and hence xyloglucan synthesis drops and xylosylation is less complete.
The conclusions regarding the roles of the XXT proteins in xyloglucan biosynthesis depend upon the assumption that normal xyloglucan is produced by the action of the three XXT proteins under study here, XXT1, XXT2, and XXT5. However, there are some caveats that need to be mentioned regarding this assumption. Specifically, the CAZy family GT34, containing the genes encoding the three XXT proteins, has seven members in Arabidopsis (Keegstra and Cavalier, 2011). Two members of this family (At4g37690 and At2g22900) are more closely related to the galactomannan galactosyltransferases identified by Edwards et al. (1999) and therefore are likely to have galactosyltransferase activity (Keegstra and Cavalier, 2011). Two other members of this family (At1g18690 and At5g07720) are expressed at very low levels in all developmental stages and in all tissues that were examined (Fig. 10; Schmid et al., 2005; http://www.weigelworld.org/resources/microarray/AtGenExpress/) and therefore are unlikely to contribute significantly to xyloglucan biosynthesis. Thus, it seems likely that xyloglucan biosynthesis is performed mainly, if not exclusively, by the proteins encoded by the three genes analyzed in this study.
Even if this assumption is correct, some important questions remain to be answered. As noted above, the three genes are expressed in the majority of plant tissues, but with different levels of expression. However, it is not clear that all three proteins are present in every cell that is performing xyloglucan biosynthesis. Even if all three are present, the stoichiometry of the three proteins in the Golgi membrane needs to be determined; the stoichiometry may vary considerably from one cell type to another. Finally, another important issue is whether these proteins form a complex with the CslC protein that is thought to synthesize the glucan backbone of xyloglucan (Cocuron et al., 2007) and what ratio of XXT proteins to CslC proteins is needed to form a functional complex. It is tempting to speculate that the structure of this complex within the Golgi membrane plays an important role in determining the regular Xyl substitution patterns that are observed in xyloglucan. These ideas form the basis of interesting hypotheses that can be tested in future work.
Another important question that remains unresolved is how plants develop normally in the absence of detectable xyloglucan and without any apparent large-scale compensatory increase in other cell wall polysaccharides. Assessment of cell wall structure in wild-type and xxt1 xxt2 xxt5 mutant plants by solid-state NMR (Dick-Pérez et al., 2011) demonstrated extensive surface interaction between pectin and cellulose microfibrils. This is consistent with the recent results of Park and Cosgrove (2012), who concluded that both pectic polysaccharides and xylans have an enhanced role in controlling wall mechanical properties in tissues from the xxt1 xxt2 double mutant that lacks detectable xyloglucan. In addition, cellulose microfibrils were observed to be thicker and spaced farther apart in the double (Anderson et al., 2010) and triple (Dick-Pérez et al., 2011) mutants when compared with wild-type plants. Therefore, it is possible that cell walls undergo rearrangements in polysaccharide interactions in the absence of xyloglucan without substantially increasing the synthesis of any other wall component.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Genetic Analysis
The xxt1 xxt5 and xxt2 xxt5 double knockout lines of Arabidopsis (Arabidopsis thaliana) were generated by crossing homozygous xxt1 and xxt5 or xxt2 and xxt5 mutants obtained and characterized earlier (Cavalier et al., 2008; Zabotina et al., 2008). The xxt1 xxt2 xxt5 triple knockout line was generated by crossing homozygous xxt2 xxt5 and xxt1 xxt2 mutants, the latter of which was obtained earlier (Cavalier et al., 2008). The F1 generations were allowed to self-fertilize, and PCR screens using gene- and T-DNA-specific primers were performed to identify double and triple knockout plants (Supplemental Table S1).
The T-DNA insertion mutants generated in this study are available from the Arabidopsis Biological Resource Center (seed stock nos. CS67826 [xxt1 xxt5], CS67827 [xxt2 xxt5], and CS67828 [xxt1 xxt2 xxt5]).
For biochemical analysis, seeds of homozygous lines and the Col-0 wild type were germinated in liquid medium and grown as liquid culture (0.5× Murashige and Skoog basal medium containing 2% [w/v] Suc) shaken at 130 rpm for 14 d at 22°C and in 16-h-light/8-h-dark conditions. After 14 d, the seedlings were washed with deionized water to remove the medium, frozen, and stored at −80°C until they were analyzed.
For immunolabeling experiments and for root hair phenotype observation, the seeds were surface sterilized, germinated, and grown in sterile petri dishes on 1% (w/v) agar containing Murashige and Skoog basal salt medium and 1% (w/v) Suc at pH 6.9. The petri dishes were oriented vertically and were maintained at 23°C under long-day conditions.
For studies of the root hair phenotype, roots of 7-d-old seedlings were examined and photographed using a Spot Pursuit camera (Diagnostic Instruments) attached to an Insight Point model 16.4 microscope (Meridian).
Cell Wall Isolation and Fractionation for ELISAs
AIR cell wall material from all mutants and wild-type plants was prepared as described (Harholt et al., 2006) and fractionated into a pectin-enriched fraction, hemicellulose-enriched fractions, and cellulose according to Pauly et al. (1999a). In brief, etiolated seedlings grown in liquid culture were ground in liquid nitrogen, suspended in 80% (v/v) ethanol, and incubated for 1 h at 80°C. The cooled samples were homogenized using a Polytron and centrifuged; the pellets were washed twice with 80% (v/v) ethanol, incubated with a mixture of chloroform and methanol (1:1, v/v) for 30 min, and filtered. The pellet was then washed three times with acetone and air dried. The dry pellet was suspended in 100 mm phosphate buffer containing α-amylase (type IIA; Sigma) with gentle stirring for 48 h, and the supernatant was collected by centrifugation and used for ELISA. Pectins were extracted from the α-amylase-treated pellet by incubation overnight in 50 mm CDTA, pH 7.5, containing 0.02% (w/v) thimerosal. The isolation of hemicellulose-enriched wall fractions was performed by sequential extraction of the CDTA pellet with 1 n KOH and 4 n KOH, each containing 0.1% (w/v) NaBH4. After neutralization with acetic acid, the alkali-soluble fractions and CDTA extracts were dialyzed against water, dried by lyophilization, and kept in sealed containers at −20°C for further analysis.
Driselase Digestion of Cell Wall Preparations and Analysis by HPAEC
AIR and hemicellulosic fractions (1 n KOH and 4 n KOH) prepared from all mutant and wild-type plants (1 mg each) were homogenized in 0.3 mL of 20 mm Na-acetate buffer (pH 5.0), approximately 0.5 units of Driselase (partially purified according to Fry [1988])) was added, and the mixtures were incubated at 37°C for 48 h. The reaction was terminated by heating at 100°C for 10 min, and insoluble material was removed by filtration through 0.22-µm filters. HPAEC-pulsed-amperometric detection of the oligosaccharides generated after Driselase treatment was performed on an ICS system (Dionex) equipped with a gradient pump, pulse-amperometric detector, and a CarboPac PA1 column. IP and xylan disaccharides were separated using a gradient that ranged from 0.1 n NaOH (A) to 100 mm Na-acetate in 0.1 n NaOH (B) at 1 mL min−1 under the following conditions: 0 to 15 min, 100% A; 15 to 40 min, from 0% to 100% B. Xyloglucan from tamarind (Tamarindus indica) seeds (Megazyme) treated with Driselase under the same conditions was used as a control.
OLIMP
The 1 n KOH and 4 n KOH fractions prepared from all mutant and wild-type plants grown in liquid culture were analyzed by OLIMP according to previously published methods (Lerouxel et al., 2002; Obel et al., 2006). The dry fraction (approximately 20 µg) was suspended in a 50 µL of 100 mm ammonium formate buffer, pH 4.5, containing 0.02 units of purified recombinant XEG (EC 3.2.1.151; Pauly et al., 1999b) and incubated at 37°C for 18 h. The reactions were centrifuged to pellet undigested material, and the supernatant containing soluble XGOs was removed and dried in a Speed-Vac. The released XGOs were dissolved in 6 µL of water containing approximately 10 beads of Bio-Rex MSZ 501(D) resin (Bio-Rad) to remove buffer salts. One microliter of the oligosaccharide solution was spotted onto a MALDI-TOF sample plate containing vacuum-dried 2,5-dihydroxybenzoic acid (10 mg mL−1; 1 µL per well) and dried under vacuum. Spectra of the samples were analyzed on a Voyager DE-Pro MALDI-TOF-mass spectrometry instrument in positive reflection mode with an acceleration voltage of 20 kV and an extraction delay time of 350 ns. Custom PERL-based software was used to calculate the relative area of each XGO ion peak and to perform pairwise comparisons and Student’s test of the respective XGO peak areas from mutant and wild-type samples.
Total Sugar Estimation and ELISA
Cell wall extracts were dissolved in deionized water (0.2 mg mL−1), and total sugar contents of cell wall extracts were estimated using the phenol-sulfuric acid method (Dubois et al., 1956; Masuko et al., 2005). All solubilized fractions were adjusted to an equal amount of total carbohydrate content prior ELISA. Cell wall extracts (60 μg sugar mL−1) were applied to the wells of 96-well ELISA plates (Costar 3598) at 50 μL per well and allowed to evaporate to dryness overnight at 37°C. A Biotek robotic system was used to perform fully automated ELISAs using a series of 135 monoclonal antibodies directed against diverse plant cell wall carbohydrate epitopes (Pattathil et al., 2010, 2012). ELISA data are presented as heat maps (glycome profiles) in which the antibody groups are based on a hierarchical clustering analysis of the antibody collection that groups the antibodies according to their binding patterns to a panel of diverse plant glycans (Pattathil et al., 2010).
Monoclonal Antibodies
Monoclonal antibodies were obtained as hybridoma cell culture supernatants from laboratory stocks at the Complex Carbohydrate Research Center (available from CarboSource Services [http://www.carbosource.net]). A detailed list of the antibodies used, grouped according to the polysaccharide primarily recognized by the antibodies (Pattathil et al., 2010), is provided in Supplemental Table S2, which also includes links to a database, WallMabDB (http://www.wallmabdb.net), containing more detailed information about each antibody.
Tissue Fixation, Immunolabeling, and Microscopy
Immunohistochemistry was done on semithin sections (250 nm) taken from roots and hypocotyls collected from 7-d-old seedlings grown on agar plates and from inflorescence stems of 6-week-old plants. Tissue fixation, immunolabeling, and fluorescent light microscopy were done as described previously (Pattathil et al., 2010; Avci et al., 2012). In some cases, before immunolabeling, sections were treated with purified pectate lyase A (EC 4.2.99.3 from Aspergillus niger [Benen et al., 2000]; 16 μg mL−1 [obtained from Carl Bergmann at the Complex Carbohydrate Research Center] in CAPS buffer [50 mm 3-(cyclohexylamino)propanesulfonic acid, 2 mm CaCl2, pH 10]) for 2 h at room temperature followed by three washes in CAPS buffer (5 min each wash). After enzyme treatment, immunolabeling of the sections was carried out as described (Pattathil et al., 2010; Avci et al., 2012).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers 2081625 (XXT1), 2132293 (XXT2), and 2019090 (XXT5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RT-PCR characterization of xylosyltransferase transcripts.
Supplemental Figure S2. Relative expression of chimeric transcripts.
Supplemental Table S1. Primers used for PCR analyses.
Supplemental Table S2. Monoclonal antibodies included in this study.
Supplemental Data S1. Col-0 and mutant transcript characterization.
Supplemental Materials and Methods S1. Col-0 and mutant transcript characterization.
Supplementary Material
Glossary
- RT
reverse transcription
- XEG
xyloglucan-specific endoglucanase
- AIR
alcohol-insoluble residue
- HPAEC
high-performance anion-exchange chromatography
- IP
isoprimeverose
- OLIMP
oligosaccharide mass profiling
- MALDI
matrix-assisted laser-desorption ionization
- TOF
time-of-flight
- XGO
xyloglucan oligosaccharide
- Col-0
ecotype Columbia
- CDTA
cyclohexane diamine tetraacetic acid
- CAPS
3-(cyclohexylamino)propanesulfonic acid
- T-DNA
transferred DNA
References
- Anderson CT, Carroll A, Akhmetova L, Somerville C. (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, III, Orlando R, Scheller HV, Mohnen D. (2011) Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci USA 108: 20225–20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avci U, Pattathil S, Hahn MG. (2012) Immunological approaches to cell wall and biomass characterization: immunolocalization of glycan epitopes. In M Himmel, ed, Biomass Conversion: Methods and Protocols. Methods in Molecular Biology, Vol 908. Humana Press, New York (in press)
- Benen JAE, Kester HCM, Parenicová L, Visser J. (2000) Characterization of Aspergillus niger pectate lyase A. Biochemistry 39: 15563–15569 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Keegstra K. (2006) Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. J Biol Chem 281: 34197–34207 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. (2007) A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc Natl Acad Sci USA 104: 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick-Pérez M, Zhang YA, Hayes J, Salazar A, Zabotina OA, Hong M. (2011) Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50: 989–1000 [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles DA, Hamilton JK, Rebers PA, Smith F. (1956) Colorimetric method for the determination of sugars and related substances. Anal Chem 28: 350–356 [Google Scholar]
- Edwards ME, Dickson CA, Chengappa S, Sidebottom C, Gidley MJ, Reid JSG. (1999) Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J 19: 691–697 [DOI] [PubMed] [Google Scholar]
- Faik A, Bar-Peled M, DeRocher AE, Zeng W, Perrin RM, Wilkerson C, Raikhel NV, Keegstra K. (2000) Biochemical characterization and molecular cloning of an α-1,2-fucosyltransferase that catalyzes the last step of cell wall xyloglucan biosynthesis in pea. J Biol Chem 275: 15082–15089 [DOI] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K. (2002) An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci USA 99: 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour G, Clay RP, Fuller MS, Albersheim P, Darvill AG, Hahn MG. (1996) Developmental and tissue-specific structural alterations of the cell-wall polysaccharides of Arabidopsis thaliana roots. Plant Physiol 110: 1413–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. (1988) The Growing Plant Cell Wall: Chemical and Metabolic Analysis. Blackburn, London
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, McNeil M, et al. (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant 89: 1–3 [Google Scholar]
- Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, Scheller HV. (2006) ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol 140: 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T. (1989) Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol 40: 139–168 [Google Scholar]
- Hayashi T, Maclachlan G. (1984) Pea xyloglucan and cellulose. I. Macromolecular organization. Plant Physiol 75: 596–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M, Jia ZH, Peña MJ, Cash M, Harper A, Blackburn AR, II, Darvill A, York WS. (2005) Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res 340: 1826–1840 [DOI] [PubMed] [Google Scholar]
- Jensen JK, Schultink A, Keegstra K, Wilkerson CG, Pauly M. (April 2, 2012) RNA-Seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional, galactosyltransferase involved in xyloglucan biosynthesis. Mol Plant http://dx.doi.org/10.1093/mp/sss032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K, Cavalier D. (2011) Glycosyltransferases of the GT34 and GT37 families. In P Ulskov, ed, Annual Plant Reviews, Vol 41. Plant Polysaccharides: Biosynthesis and Bioengineering. Blackwell Publishing, Oxford, UK, pp 235–250
- Lerouxel O, Choo TS, Séveno M, Usadel B, Faye L, Lerouge P, Pauly M. (2002) Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol 130: 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD. (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus SE, Verhertbruggen Y, Herve C, Ordaz-Ortiz JJ, Farkas V, Pedersen HL, Willats WGT, Knox JP. (2008) Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuko T, Minami A, Iwasaki N, Majima T, Nishimura SI, Lee YC. (2005) Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal Biochem 339: 69–72 [DOI] [PubMed] [Google Scholar]
- McCann MC, Roberts K. (1991) Architecture of the primary cell wall. In CW Lloyd, ed, The Cytoskeletal Basis of Plant Growth and Form. Academic Press, London, pp 109–129
- Obel N, Erben V, Pauly M. (2006) Functional wall glycomics through oligosaccharide mass profiling. In T Hayashi, ed, The Science and Lore of the Plant Wall: Biosynthesis, Structure & Function. Brown Walker Press, Boca Raton, FL, pp 258–266
- O’Neill MA, York WS. (2003) The composition and structure of plant primary cell walls. In J Rose, ed, The Plant Cell Wall. Blackwell Publishing, New York, pp 1–53
- Park YB, Cosgrove DJ. (2012) Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol 158: 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, et al. (2010) A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol 153: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Miller JS, Hahn MG. (2012) Immunological approaches to plant cell wall and biomass characterization: glycome profiling. In M Himmel, ed, Biomass Conversion: Methods and Protocols. Methods in Molecular Biology, Vol 908. Humana Press, New York (in press)
- Pauly M, Albersheim P, Darvill A, York WS. (1999a) Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J 20: 629–639 [DOI] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kauppinen S, Kofod LV, York WS, Albersheim P, Darvill A. (1999b) A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K. (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976–1979 [DOI] [PubMed] [Google Scholar]
- Popper ZA, Fry SC. (2005) Widespread occurrence of a covalent linkage between xyloglucan and acidic polysaccharides in suspension-cultured angiosperm cells. Ann Bot (Lond) 96: 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter WD, Chapple C, Somerville CR. (1997) Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J 12: 335–345 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Thompson DS. (2005) How do cell walls regulate plant growth? J Exp Bot 56: 2275–2285 [DOI] [PubMed] [Google Scholar]
- Vincken JP, York WS, Beldman G, Voragen AG. (1997) Two general branching patterns of xyloglucan, XXXG and XXGG. Plant Physiol 114: 9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH. (2008) How effective is TDNA insertional mutagenesis in Arabidopsis? J Biochem Tech 1: 11–20 [Google Scholar]
- Zabotina OA, van de Ven WTG, Freshour G, Drakakaki G, Cavalier D, Mouille G, Hahn MG, Keegstra K, Raikhel NV. (2008) Arabidopsis XXT5 gene encodes a putative α-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. Plant J 56: 101–115 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.