Abstract
Light energy constantly damages photosynthetic apparatuses, ultimately causing impaired growth. Particularly, the sessile nature of higher plants has allowed chloroplasts to develop unique mechanisms to alleviate the irreversible inactivation of photosynthesis. Photosystem II (PSII) is known as a primary target of photodamage. Photosynthetic organisms have evolved the so-called PSII repair cycle, in which a reaction center protein, D1, is degraded rapidly in a specific manner. Two proteases that perform processive or endopeptidic degradation, FtsH and Deg, respectively, participate in this cycle. To examine the cooperative D1 degradation by these proteases, we engaged Arabidopsis (Arabidopsis thaliana) mutants lacking FtsH2 (yellow variegated2 [var2]) and Deg5/Deg8 (deg5 deg8) in detecting D1 cleaved fragments. We detected several D1 fragments only under the var2 background, using amino-terminal or carboxyl-terminal specific antibodies of D1. The appearance of these D1 fragments was inhibited by a serine protease inhibitor and by deg5 deg8 mutations. Given the localization of Deg5/Deg8 on the luminal side of thylakoid membranes, we inferred that Deg5/Deg8 cleaves D1 at its luminal loop connecting the transmembrane helices C and D and that the cleaved products of D1 are the substrate for FtsH. These D1 fragments detected in var2 were associated with the PSII monomer, dimer, and partial disassembly complex but not with PSII supercomplexes. It is particularly interesting that another processive protease, Clp, was up-regulated and appeared to be recruited from stroma to the thylakoid membrane in var2, suggesting compensation for FtsH deficiency. Together, our data demonstrate in vivo cooperative degradation of D1, in which Deg cleavage assists FtsH processive degradation under photoinhibitory conditions.
For photosynthetic organisms, light energy is indispensable, but it concomitantly damages the photosynthetic apparatus because they receive excess light energy that might not be dissipated (Barber and Andersson, 1992; Tyystjärvi and Aro, 1996). Irreversible photosynthetic apparatus inactivation produces an inhibitory effect on photosynthesis (termed photoinhibition) and consequently engenders impaired plant growth (Aro et al., 1993; Nishiyama et al., 2006; Murata et al., 2007; Takahashi and Murata, 2008). To avoid photoinhibition, photosynthetic organisms have developed various mechanisms that restrict the extent of photooxidative damage and that repair the damaged protein component (Murchie and Niyogi, 2011; Takahashi and Badger, 2011).
Of such photoprotection mechanisms, the repair of PSII, which is a primary target of photooxidative damage in photosynthetic apparatuses, appears to be an extremely efficient process (Raven, 2011). The major target of protein components in PSII is the D1 protein of the reaction center (Edelman and Mattoo, 2008; Nixon et al., 2010). The highly hydrophobic D1 protein contains five transmembrane helices, with N-terminal and C-terminal tails exposed to the stroma and thylakoid lumen, respectively (Supplemental Fig. S1; Zouni et al., 2001; Loll et al., 2005; Umena et al., 2011). Repair of damaged D1 protein in PSII involves a cycle of (1) migration of damaged PSII core complex to the stroma thylakoid, (2) partial PSII disassembly of the PSII core monomer, (3) access of protease degrading damaged D1, (4) concomitant D1 synthesis, and (5) reassembly of PSII into grana thylakoid (Baena-González and Aro, 2002; Aro et al., 2005; Kato and Sakamoto, 2009). Consequently, the quality control of PSII against photooxidative damage is often called the “PSII repair cycle,” and D1 degradation in PSII repair has been studied in many organisms. Earlier studies of D1 degradation have specifically addressed the relation between photoinhibition and D1 degradation/cleavage products (Greenberg et al., 1987; De Las Rivas et al., 1992; Salter et al., 1992; Shipton and Barber, 1994; Kettunen et al., 1996), although recent works have specifically examined the proteases involved in D1 degradation, which demonstrates that an ATP-dependent FtsH metalloprotease and ATP-independent Deg endoprotease play predominant roles in D1 degradation in chloroplasts (Supplemental Fig. S1; Kato and Sakamoto, 2009; Komenda et al., 2012). These proteases have also been identified in cyanobacteria, although only FtsH is apparently sufficient to carry out efficient PSII repair (Lindahl et al., 2000; Bailey et al., 2002; Sakamoto et al., 2002; Silva et al., 2003; Barker et al., 2006; Komenda et al., 2006).
FtsH, a membrane-bound ATP-dependent zinc metalloprotease, initiates processive proteolysis through both N-terminal and C-terminal regions of its substrate proteins by utilizing their ATPase functions that facilitate the unfolding and translocation of membrane proteins (Ogura and Wilkinson, 2001; Ito and Akiyama, 2005; Wagner et al., 2012). It is clear that FtsH plays an important role in D1 degradation in both cyanobacteria and chloroplasts (Bailey et al., 2002; Silva et al., 2003; Komenda et al., 2006; Kato et al., 2009). Of 12 FtsH homologs in Arabidopsis (Arabidopsis thaliana), nine were found to be located in chloroplasts (Sakamoto et al., 2003). A major FtsH complex in chloroplasts is localized to thylakoid membranes as a heterohexamer complex, with their catalytic region facing the stromal side of the membrane (Lindahl et al., 1996; Sakamoto et al., 2003; Yu et al., 2004). Mounting evidence has demonstrated that FtsH1, FtsH2, FtsH5, and FtsH8 are four major isomers of the FtsH complex that is localized in the thylakoid membrane through one N-terminal transmembrane domain (Yu et al., 2004; Rodrigues et al., 2011). FtsH2 is the most abundant isomer, followed by FtsH5, FtsH8, and FtsH1 (Sinvany-Villalobo et al., 2004). Mutants lacking FtsH2 and FtsH5, yellow variegated2 (var2) and var1, respectively, show a leaf-variegated phenotype, which is more enhanced in var2 (Chen et al., 2000; Takechi et al., 2000; Sakamoto et al., 2002). In chloroplast, FtsH heterocomplexes are formed by at least two type isomers (A and B, represented by FtsH1/5 and FtsH2/8, respectively) that are functionally distinguishable from each other (Sakamoto et al., 2003; Yu et al., 2004, 2005; Zaltsman et al., 2005b). The loss of the two isomers from either type engenders seedling lethality with incomplete chloroplast development (Zaltsman et al., 2005b). Thus, although var2 and var1 show clear phenotypes, the mutants still have a certain level of the FtsH complex (Sakamoto et al., 2003; Zaltsman et al., 2005a). One notable feature in var1 and var2 mutants, in addition to their variegated phenotype, is their high vulnerability to photoinhibition under strong illumination (Sakamoto et al., 2002, 2004). Furthermore, in vivo assessment of D1 degradation activity in these mutants clearly demonstrates that FtsH participates in PSII repair not only under photoinhibitory but also nonphotoinhibitory conditions (Kato et al., 2009).
Deg protease in bacteria is the periplasmic ATP-independent Ser-type endoprotease. Most Deg family members contain more than one PDZ domain, which is necessary for the formation of functional oligomeric complexes (Clausen et al., 2002). Several Deg proteases have been shown to affect D1 degradation in chloroplasts (Haussühl et al., 2001; Kapri-Pardes et al., 2007; Sun et al., 2007, 2010a), although their function in PSII repair seems to be less important in cyanobacteria (Barker et al., 2006). Of 16 Degs identified in Arabidopsis, five (Deg1, Deg2, Deg5, Deg7, and Deg8) have been reported as peripherally attached to the thylakoid membrane of chloroplasts: Deg1, Deg5, and Deg8 are localized on the lumenal side, and Deg2 and Deg7 are localized on the stromal side (Huesgen et al., 2009; Schuhmann and Adamska, 2012). Initially, the involvement of Deg2 in the cleavage between helices D and E of the D1 (DE loop) was proposed by in vitro studies conducted in Arabidopsis (Haussühl et al., 2001). However, the rate of D1 degradation in deg2 mutants is comparable to that in the wild type under light stress conditions (Huesgen et al., 2006). A recent report described that Deg7 participates in the cleavage of PSII core proteins including the damaged D1 and that it contributes the efficient PSII repair under photoinhibitory conditions (Sun et al., 2010a). Of the lumenal Degs, Deg5 and Deg8 are involved in cleavage within the luminal loop connecting the transmembrane helices C and D (CD loop) of the damaged D1. High-light-sensitive phenotypes in deg5 and deg8 mutants were shown to be enhanced in deg5 deg8 double mutants, suggesting the synergistic function of Deg5 and Deg8 in PSII repair (Sun et al., 2007). The other lumenal Deg protease, Deg1, seems to participate in the cleavage of D1 protein at the CD loop and downstream of transmembrane helix E (Kapri-Pardes et al., 2007). In addition, Deg1 appears to be fundamentally important for chloroplast development, because Deg1 homozygous knockout lines were unobtainable (Kapri-Pardes et al., 2007; Sun et al., 2010b). Deg1 knockdown mutants show impaired plant growth compared with the wild type, even under nonphotoinhibitory growth conditions. These knockdown lines caused a concomitant reduction of FtsH and Deg2.
Based on numerous studies described previously and the proteolytic properties of FtsH (processive) and Deg (endopeptidic), a model in which Deg proteases have a supplementary role that increases the recognition site for FtsH in D1 degradation has been proposed (Itzhaki et al., 1998; Kato and Sakamoto, 2009). For example, an earlier biochemical experiment shows that a purified recombinant FtsH can degrade a high-light-induced 23-kD D1 fragment in an ATP-dependent manner (Lindahl et al., 2000). This observation suggests that a partial D1 fragment, possibly generated by Deg, can be degraded by FtsH. However, the in vivo evidence to support cooperative D1 degradation mediated by FtsH and Deg is lacking. It should be examined using mutant analysis. To address this question in this study, we assessed D1 degradation in var2 and deg5 deg8 mutants. The results showed that several cleavage products of D1 under photoinhibitory conditions accumulated in var2, which implies that FtsH is indeed required for the in vivo degradation of D1 cleavage products. A line of evidence was provided that the accumulation of several D1 cleavage products in var2 depends on Deg5 and Deg8. These results supported our model showing that FtsH plays a fundamental role in D1 degradation and that Degs supplement it under photoinhibitory conditions.
RESULTS
In Vivo D1 Degradation Assay in deg5 deg8 under Nonphotoinhibitory Light Conditions
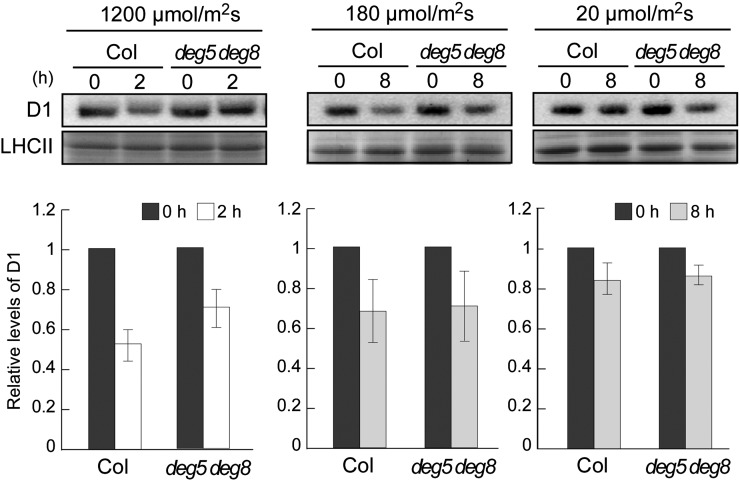
In this study, we specifically examined lumenal Deg5 and Deg8 and investigated their cooperative role in D1 degradation with FtsH. The involvement of these Degs in PSII repair has been demonstrated previously (Sun et al., 2007), but not under low-light conditions. To evaluate the rates of D1 degradation in high-light, growth-light, and low-light conditions (1,200, 180, and 20 µmol photons m−2 s−1, respectively), detached leaves from the wild-type Columbia (Col) and the deg5 deg8 double mutant were incubated with lincomycin. Immunoblot analysis of D1 protein levels was performed, and the rate of D1 degradation was estimated based on the ratio of D1 signal to Coomassie Brilliant Blue-stained light-harvesting complexes of PSII (LHCII). The results as presented in Figure 1 indicate that D1 degradation rates under growth- and low-light conditions were similar between the wild type and deg5 deg8, although the D1 degradation in deg5 deg8 was significantly slower than that in the wild type when plants were exposed to high light (Fig. 1). Together with results from a previous study (Sun et al., 2007), these results indicated that Deg5 and Deg8 do not contribute significantly to D1 degradation under nonphotoinhibitory conditions.
Figure 1.
Immunoblot analysis of D1 protein in the deg5 deg8 mutant under three light conditions. Detached mature leaves of Col and deg5 deg8 (approximately 6-week-old plants grown under normal conditions) were preincubated with 5 mm lincomycin. The leaves were incubated for 2 h under high-light conditions (1,200 µmol photons m−2 s−1) or for 8 h under growth-light and low-light conditions (180 and 20 µmol photons m−2 s−1, respectively). Representative immunoblots (normalized by fresh weight) using anti-D1 (C-term) antibodies and the bands corresponding to Coomassie Brilliant Blue-stained LHCII are depicted. Signals of immunoblots were quantified using the ImageJ program and were normalized to the amount of Coomassie Brilliant Blue-stained LHCII (error bars indicate sd; n = 3). To compare D1 levels, ratios at 0 h were adjusted to 1.
Accumulation of D1 Cleavage Products in var2 under Photoinhibitory Light Conditions
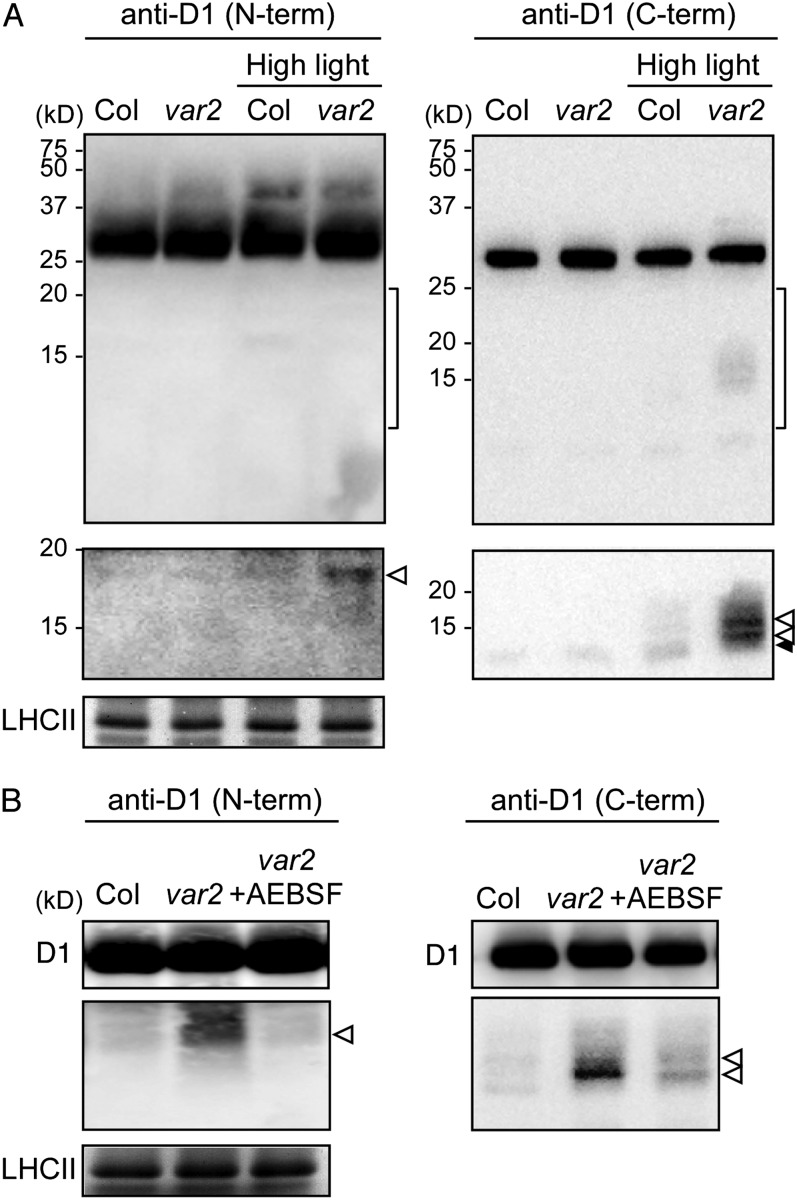
To examine D1 degradation mediated by FtsH and Deg further, it was important to assess D1 partial degradation products. Given the role of each protease (Deg in endopeptidic cleavage and FtsH in processive digestion), we assumed that var2 contains more D1 fragments that are generated by Deg. Our previous attempt to detect fragmented D1 using immunoblotting was unsuccessful, probably because of (1) insufficient high-light intensity to cause photoinhibition and (2) our limited sensitivity in immunoblots. When we used extreme high light (2,500 µmol photons m−2 s−1) and improved immunoblot sensitivity (see “Materials and Methods”), however, several D1 fragments were detectable (Fig. 2). Specific antibodies that recognize the N terminus, DE loop, and C terminus of the D1 protein (designated as N-term, DE-loop, and C-term, respectively) were employed to detect D1 fragments. To allow detection of the fragments, which were regarded as much less abundant than full-length D1, the part of the blots corresponding to full-length D1 (approximately 32 kD) was discarded before immunodetection. Therefore, we minimized the background resulting from the signal corresponding to the full-length D1 protein. After exposure to extreme high light, wild-type and var2 leaves showed rapid decreases in PSII activity that was measured by maximum quantum yield of PSII (Fv/Fm); approximately 50% and 30% of the maximum PSII efficiency was lost after a 1-h exposure in the wild type and var2, respectively(Supplemental Table S1). Under this experimental condition, immunoblot analysis using anti-D1 (N-term) antibodies showed that a band of 18 kD accumulated significantly in var2. Similarly, immunoblot analysis using anti-D1 (C-term) antibodies showed that two bands of 12 and 16 kD accumulated in var2. These products were only slightly detectable in the wild type under the high-light condition (Fig. 2A). We also used anti-D1 (DE-loop) antibodies that recognize the DE loop of D1 proteins but failed to detect any specific cleavage products that had been cross-reacted (Supplemental Fig. S2). The other important observation was that although D1 degradation is impaired in var2 not only under high light but in nonphotoinhibitory conditions (Kato et al., 2009), the D1 cleavage products detected by N-term and C-term D1 antibodies did not accumulate under normal-light conditions (Fig. 2A). Overall, these results demonstrated that FtsH participates in the degradation process of the high-light-induced D1 cleavage products.
Figure 2.
Accumulation of the cleavage products of D1 protein in extreme high-light-treated var2 leaves. A, Immunoblot analysis of the cleavage products of D1 protein under normal-light and extreme high-light conditions. Mature leaves of Col and var2 (approximately 6-week-old plants grown under normal conditions) were illuminated in normal-light (180 µmol photons m−2 s−1) and extreme high-light (2,500 µmol photons m−2 s−1) conditions for 1 h. Representative immunoblots (normalized by chlorophyll content) using anti-D1 (N-term) and anti-D1 (C-term) antibodies and the bands corresponding to Coomassie Brilliant Blue-stained LHCII are depicted. A selective detection of the areas indicated by the brackets at right is shown in the second panels from top. B, Immunoblot analysis of AEBSF-treated var2 leaves. Detached leaves were pretreated with Ser protease inhibitor (AEBSF) and subsequently incubated under extreme high-light conditions for 1 h. White and black arrowheads indicate specific and nonspecific signals, respectively, under high-light irradiation.
To estimate whether the cleavages are dependent on Ser protease activity, we compared the accumulation of the D1 cleavage products in the presence of a Ser protease inhibitor. After pretreatment of var2 leaves with 4-(2-aminoethyl)-benzenesulfonyl fluoride (AEBSF) protease inhibitor, leaves were incubated for 1 h under extreme high light. Immunoblot results obtained using anti-D1 (N-term) and anti-D1 (C-term) antibodies showed that levels of the cleavage products decreased markedly in var2 in the presence of AEBSF (Fig. 2B). This result demonstrated that the D1 cleavage products that accumulated in var2 leaves under the high-light condition were likely caused by Ser protease.
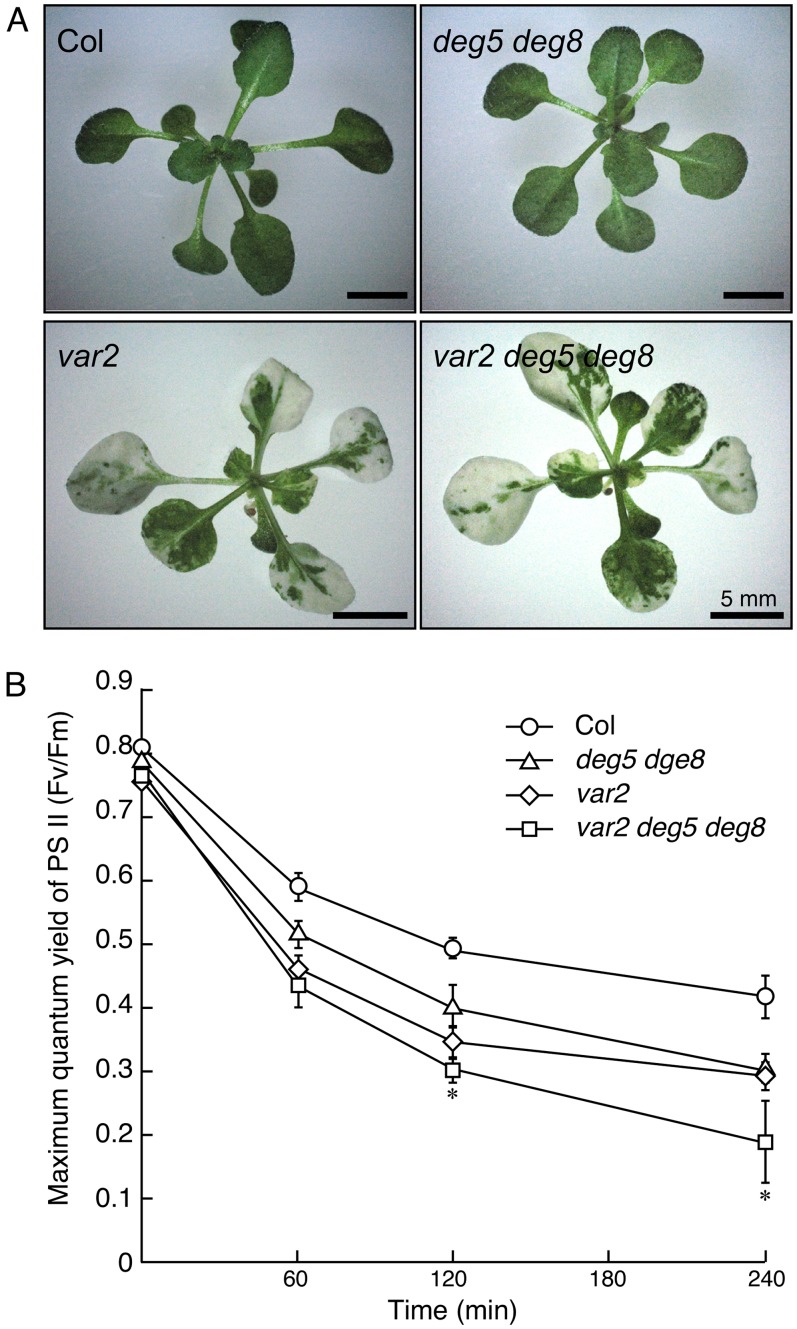
Characterization of the var2 deg5 deg8 Triple Mutant
To assess the involvement of FtsH and Deg proteases in D1 degradation in a cooperative fashion in vivo, we generated a var2 deg5 deg8 triple mutant. As depicted in Figure 3, var2 deg5 deg8 showed a variegated phenotype similar to that of var2: we observed no distinguishable leaf variegation phenotype or growth rate between the var2 single mutant and the triple mutant. In addition, we considered that deg5 and deg8 did not affect FtsH accumulation, because the triple mutant had levels of FtsH isomers comparable to var2 (Supplemental Fig. S3). These results suggest that Deg5 and Deg8 have little additional effect on thylakoid formation or maintenance, which has been proposed to be closely related to FtsH.
Figure 3.
Phenotype of the var2 deg5 deg8 triple mutant and its photosensitivity. A, Photographs of 3-week-old Col, var2-1, deg5 deg8, and var2 deg5 deg8 plants. Bars = 5 mm. B, Fv/Fm in the mutants. Fv/Fm was measured in detached leaves of Col (circles), var2-1 (diamonds), deg5 deg8 (triangles), and var2 deg5 deg8 (squares). Values are means ± sd (n = 3). The asterisk indicates a value of P < 0.05 for a comparison between var2 and var2 deg5 deg8.
We next measured Fv/Fm in var2, deg5 deg8, var2 deg5 deg8, and the wild type to assess whether additive PSII photoinhibition occurred in triple mutants. Detached leaves of var2, deg5 deg8, var2 deg5 deg8, and the wild type were exposed to high light for up to 4 h. The result that Fv/Fm values in var2 and deg5 deg8 mutants were lower than those in the wild type was consistent with previous reports (Sakamoto et al., 2002; Sun et al., 2007). Furthermore, Fv/Fm values in var2 deg5 deg8 were decreased significantly more than those in var2 and deg5 deg8 during high-light irradiation (Fig. 3B). These results demonstrate the synergistic effect of the reduced FtsH and Deg protease activities on photosensitivity to high light, suggesting increased photoinhibition in var2 deg5 deg8.
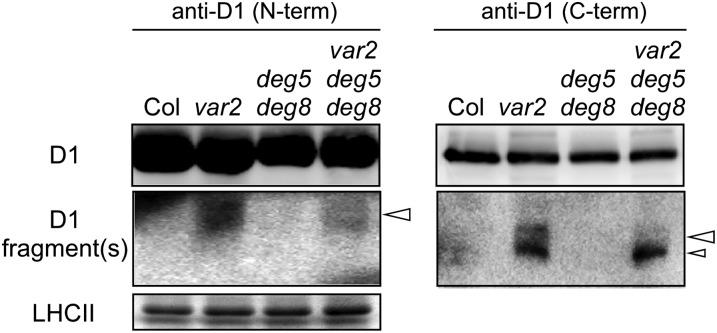
Cleavage Products of D1 in var2 deg5 deg8
To assess whether the D1 cleavage products in var2 resulted from Deg proteases, we performed our D1 degradation assay using the triple mutant along with other control plants. The D1 cleavage products did not accumulate in the wild type and mutants under normal-light conditions (Supplemental Fig. S4). However, consistent with the result presented in Figure 2, the 18-kD N-terminal D1 fragment and two C-terminal D1 fragments that corresponded to 16 and 12 kD were observed in var2 (Fig. 4). In contrast, these bands were at the undetectable level in deg5 deg8, implying that FtsH alone can degrade photodamaged D1 without Deg5 and Deg8. In the triple mutant, interestingly, the 18-kD N-terminal D1 fragment and only one of the two C-terminal fragments (16 kD) decreased in var2 deg5 deg8, although the 12-kD C-terminal fragment remained unchanged and was comparable between the triple mutant and var2 (Fig. 4). Given the fact that these D1 fragments are (1) only detectable under the var2 background and (2) likely generated by Ser protease activity, the results suggest that the 18-kD N-terminal fragment and the 16-kD C-terminal fragment were generated by lumenal Deg5/Deg8. The 12-kD C-terminal fragment is likely to be generated by other Deg proteases. Taken together, we considered that the 18-kD N-terminal fragment and the 16-kD C-terminal fragment represented fragments cleaved at the lumenal CD loop of the D1 protein, whereas the 12-kD C-terminal fragment represented a C-terminal fragment cleaved at the stromal DE loop of the D1 protein.
Figure 4.
Immunoblot analysis of the cleavage products of D1 protein in the var2 deg5 deg8 mutant. Mature leaves of Col, var2, deg5 deg8, and var2 deg5 deg8 (approximately 6-week-old plants grown under normal conditions) were incubated in extreme high-light conditions (2,500 µmol photons m−2 s−1) for 1 h. Representative immunoblots (normalized by chlorophyll content) using anti-D1 (N-term) and anti-D1 (C-term) antibodies and the bands corresponding to Coomassie Brilliant Blue-stained LHCII are depicted. A selective detection of the cleavage products of D1 protein is shown in the second panels from the top.
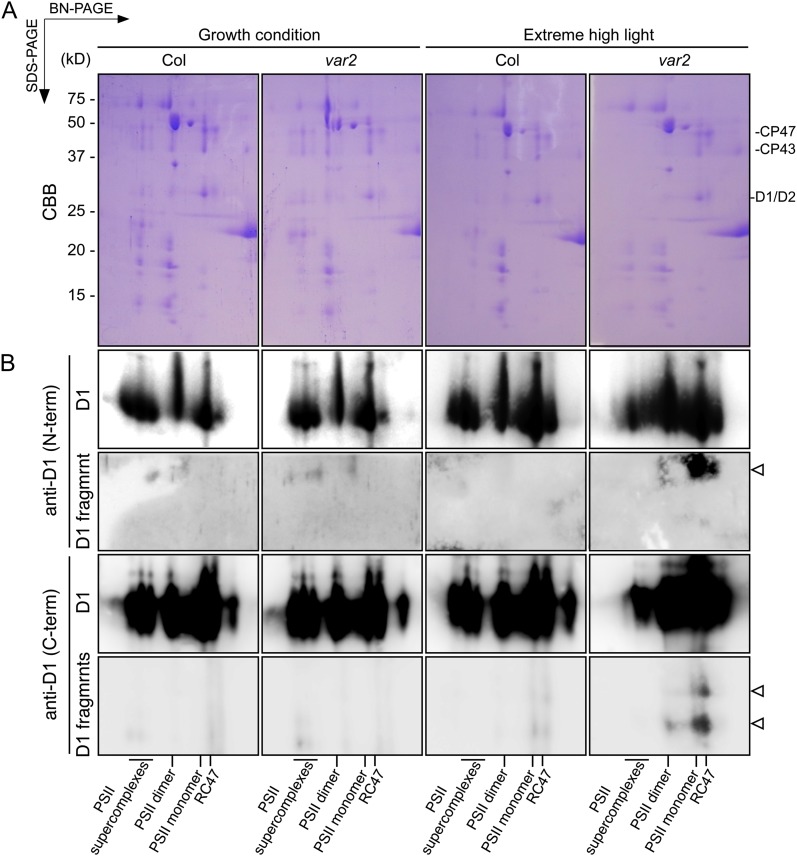
D1 Cleavage Products in the PSII Complex Assessed by Blue Native/SDS-PAGE
In chloroplasts, the functional PSII core is a dimer that forms a large complex with the LHCII antenna. Degradation of photodamaged D1 mediated by FtsH and Deg is a critical step in the PSII repair cycle, which involves (1) migration of the damaged PSII core from grana stacks to the stroma-exposed thylakoids, (2) partial disassembly of the PSII core, (3) D1 proteolysis as described previously, (4) synthesis and processing of the D1 nascent chain, and (5) migration of repaired PSII back to grana stacks (Baena-González and Aro, 2002; Aro et al., 2005). Two-dimensional blue native (BN)/SDS-PAGE is a powerful approach to monitor the different status of PSII assembly/disassembly in the PSII repair cycle. Therefore, we performed BN/SDS-PAGE and subsequent immunoblot analysis to detect D1 cleavage fragments. Purified thylakoid membranes were solubilized using n-dodecyl-β-maltoside, and protein complexes were separated in the first dimension on 4% to 16% gradient native gels with subsequent separation of protein subunits in the second dimension with SDS-PAGE (Fig. 5). Although the assembly of PSII complexes in the wild type and var2 showed no great difference under growth-light conditions, PSII supercomplexes were only slightly detectable in var2 under high-light conditions. In addition, the level of the PSII core complex lacking CP43 (termed RC47), which can be regarded as the PSII repair cycle intermediate at the dissociation step, increased in var2 under extreme high-light conditions. These results were consistent with those of our previous study (Kato et al., 2009), demonstrating compromised PSII repair with a reduced amount of FtsH under photoinhibitory light conditions. Immunoblot analysis using anti-D1 antibodies also supported these results: we detected the N-terminal D1 fragment and the two C-terminal D1 cleavage products, which comigrated at the positions corresponding to the PSII dimer, monomer, and RC47, except that the signal of the 16-kD cleavage product was undetectable at positions corresponding to the PSII dimer (Fig. 5). Importantly, even when the blots were overexposed, the specific signals of the anti-D1 (N-term) and anti-D1 (C-term) antibodies with the extreme high-light-treated thylakoid membrane did not appear at the positions corresponding to PSII supercomplexes in var2 and the wild type.
Figure 5.
Immunoblot analysis of the D1 cleavage products separated by BN/SDS-PAGE. A, Thylakoid protein complexes were solubilized with 0.5% n-dodecyl-β-d-maltoside and separated on 4% to 16% BN/PAGE gels (10 µg of chlorophyll per lane). Thylakoid membrane proteins were separated further using 14% SDS-PAGE and were silver stained. CBB, Coomassie Brilliant Blue. B, Proteins separated by BN/SDS-PAGE were immunodetected by anti-D1 (N-term) and anti-D1 (C-term) antibodies. Spots of the cleaved D1 products are indicated by white arrowheads. Positions corresponding to PSII supercomplexes, PSII dimer, PSII monomer, and the RC47 complex are shown at the bottom.
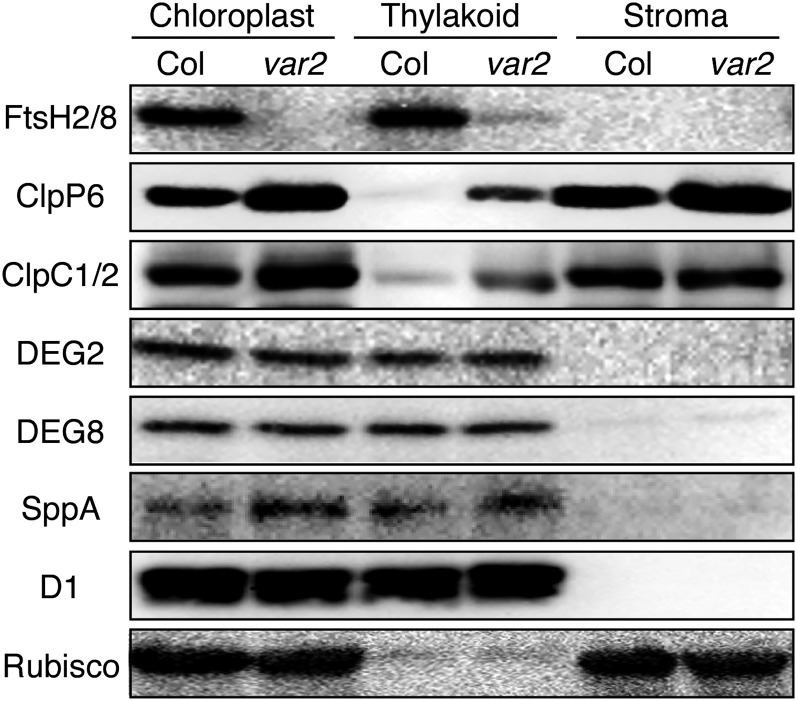
Dynamics of Chloroplast Proteases in var2
Previous studies showed that D1 turnover occurred even in the depletion of the major FtsH complex, which is composed of FtsH2 and FtsH5 (Kato et al., 2009). This fact suggests that an alternative mechanism to degrade photodamaged D1 seems to act when the proteolytic activity proceeded by FtsH is limited. Assuming that other chloroplast proteases might compensate for FtsH deficiency, we examined the levels and thylakoid-membrane localization of other proteases in var2. Proteins isolated from purified chloroplast and from stromal and membrane fractions were subjected to immunoblot analysis using antibodies against various chloroplast proteases (Fig. 6). Anti-D1 and Rubisco large subunit antibodies were used, respectively, as controls of thylakoid membrane protein and stroma protein. The results showed that levels of Clp protease were up-regulated in var2 because both ClpP6 (representing the Clp protease complex) and ClpC1/C2 (representing the ATP-dependent unfoldase complex) increased. It is particularly interesting that these subunits appeared to be recruited into thylakoid membranes when FtsH2 was deficient. SppA, which is closely associated with the stromal side of thylakoid membranes and enriched in the stroma thylakoid, is known to be up-regulated by high light (Lensch et al., 2001). The immunoblot analyses showed that the steady-state level of SppA increased in var2 compared with the wild type. By contrast, levels of Deg2 and Deg8 in the thylakoid membranes of var2 and the wild type were not significantly different. These results support the possibility that processive degradation of photodamaged D1, whether cleaved by Deg or not, proceeds with ATP-dependent proteases at the stromal surface of thylakoid membranes. FtsH and Clp are structurally and functionally related to one another (Kato and Sakamoto, 2010). Although FtsH is localized in thylakoid membranes and plays a major role in D1 degradation in the PSII repair cycle, Clp might participate in PSII repair when FtsH is limited. We also examined the levels of other proteases in deg5 deg8. In contrast to the results in var2, however, no significant changes between the wild type and deg5 deg8 in the levels and the localization of other chloroplast proteases were detected (Supplemental Fig. S5).
Figure 6.
Steady-state accumulation and localization of chloroplast proteases. Chloroplasts were purified from mature leaves of Col and var2 using a Percoll step gradient. Intact chloroplasts were fractionated into stroma and membrane fractions. Proteins were separated using SDS-PAGE and probed against specific antibodies. D1 and Rubisco large subunits were used as markers of membranes and stroma, respectively. Samples were equally loaded based on chlorophyll content.
DISCUSSION
Many studies of the degradation pattern of D1 under photoinhibitory conditions have been conducted because such studies can be expected to provide us with important clues to elucidate photoinhibition mechanisms. Both in vivo and in vitro results have demonstrated two degradation patterns, proposing that primary D1 cleavage takes place in the stromal DE and the lumenal CD loops (Greenberg et al., 1987; De Las Rivas et al., 1992; Salter et al., 1992; Kettunen et al., 1996). Over the last decade, considerable effort has revealed proteases responsible for D1 degradation under photoinhibitory conditions (Lindahl et al., 2000; Haussühl et al., 2001; Bailey et al., 2002; Kapri-Pardes et al., 2007; Sun et al., 2007, 2010a; Kato et al., 2009). Based on these observations, we hypothesized that the endopeptidic cleavage by Deg in lumenal and stromal D1 loops accelerates the rate of D1 degradation by producing additional recognition termini for FtsH (Kato and Sakamoto, 2009). However, in vivo evidence supporting this hypothesis remained uncharacterized. In this study, we combined genetic and biochemical studies and demonstrated that D1 fragments generated by Deg proteases can be the substrates for FtsH. First, our improved D1 degradation assay enabled us to detect several D1 cleavage products in var2 under excess light exposure. Second, inhibitor analysis showed that these fragments resulted from Ser protease activities (Fig. 2). Finally, our attempt to detect D1 cleavage products in the var2 deg5 deg8 triple mutant demonstrated that the 18-kD N-terminal and 16-kD C-terminal cleavage products were produced by lumenal Deg proteases under photoinhibitory conditions (Fig. 4). Together, these results provide in vivo evidence that stromal FtsH and lumenal Deg cooperatively degrade photodamaged D1 in the PSII repair cycle.
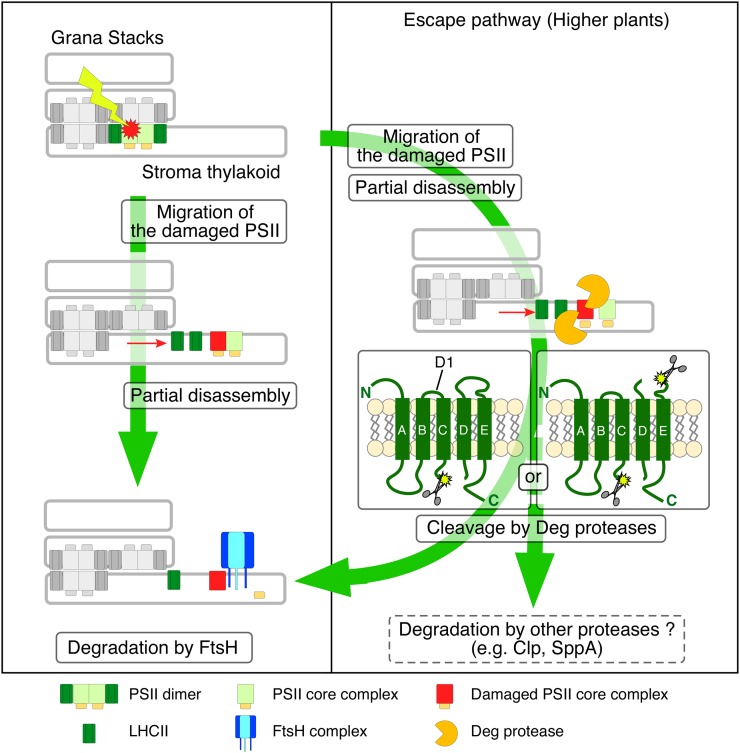
To integrate our observations in this study into the current understanding of the PSII repair cycle in chloroplasts, we present a model that explains cooperative D1 digestion between FtsH and Deg proteases (Fig. 7). In this model, two pathways for D1 degradation in chloroplasts can be assumed. One is constant processive D1 degradation mediated predominantly by the FtsH complexes, irrespective of light intensity. The other is an “escape pathway,” in which D1 degradation is conducted by multiple proteases under photoinhibitory conditions. Particularly, D1 subfragments generated by Deg accelerate D1 degradation proceeded by FtsH and/or other proteases, thereby facilitating efficient PSII repair. In this regard, it is noteworthy that we measured the rate of D1 degradation in the mutant lacking both FtsH2 and lumenal Deg5/Deg8 proteases, based on our previous nonvariegated leaf-disc assay (Supplemental Fig. S6; Kato et al., 2009). D1 degradation in the triple mutant was significantly slower than that in the mutant lacking only FtsH2 when plants were exposed to high light, although it was similar under the nonphotoinhibitory condition. This result shows that the endopeptidic activity of luminal Degs accelerates further D1 degradation under photoinhibitory conditions. This escape pathway might be less important in cyanobacteria than in chloroplasts, according to results described in a previous report of a study of Synechocystis (Barker et al., 2006). The different contribution of Deg activities in D1 degradation between chloroplasts and cyanobacteria is explainable by the importance of the escape pathway under photoinhibitory conditions. During the evolution of land plants, effective D1 degradation that is conducted cooperatively by multiple proteases might have been necessary to overcome light-stress environments. Supporting this, stromal Deg7 was shown to be enriched as peripherally attached to thylakoid membranes upon high-light illumination (Sun et al., 2010a).
Figure 7.
Proposed model of cooperative D1 degradation in the PSII repair cycle. Photodamaged PSII generated at all light intensities migrates to stroma thylakoids from grana stacks, and PSII monomerization occurs. In PSII repair, processive D1 degradation is conducted predominantly by FtsH irrespective of the light intensity (fundamental degradation). In contrast, under photoinhibitory conditions, an endopeptidic cleavage by Deg increases the rate of D1 degradation because smaller D1 cleavage fragments facilitate effective degradation (escape pathway). D1 cleavage by Deg most likely occurs in PSII dimer, PSII monomer, and the RC47 complex. Cleaved D1 fragments are subjected to further degradation by FtsH, but other proteases such as SppA and Clp proteases participate in this pathway with a mechanism that remains unclear.
According to previous studies in which D1 degradation was assessed in vivo, lumenal Deg1 and Deg5/Deg8 were suggested to cleave D1 at the CD loop (Kapri-Pardes et al., 2007; Sun et al., 2007), whereas stromal Deg7 was suggested to cleave D1 at the BC loop (Sun et al., 2010a). Results of an in vitro experiment suggested that Deg2 cleaves D1 at the DE loop (Supplemental Fig. S1; Haussühl et al., 2001). We compared these cleavage events (particularly at the CD and DE loops) with the D1 subfragments detected in the var2 background, based on their molecular size. First, we inferred that two subfragments, N-terminal 18-kD and C-terminal 16-kD fragments, correspond to the products from the cleavage at the CD loop. Second, the C-terminal 12-kD fragment corresponds to the product from cleavage at the DE loop (schematically portrayed in Fig. 7). No N-terminal product from cleavage at the DE loop was detected in our assay, probably because the cleavage at the CD loop is conducted very efficiently by lumenal Degs or because the titer of anti-D1 (N-term) antibodies was too low to detect subfragments. Detection of the C-terminal 16-kD fragment implies that some subfragments exist that escape from cleavage at the DE loop, which might be operated by stromal Degs. Given the fact that these subfragments accumulate only in the var2 background and that they are generated by Ser protease activity, we conclude that FtsH and Deg cooperatively degrade D1 in the PSII repair cycle; therefore, we provide in vivo evidence that D1 fragments generated by Deg proteases can be degraded processively by FtsH. In addition, our results strongly imply that the cleavage at the CD loop is manipulated by Deg5 and Deg8. It is interesting that BN/SDS-PAGE analysis showed that the D1 fragments generated by Deg proteases were only detectable in PSII dimer, PSII monomer, and the RC47 complex but that they were never detected in PSII supercomplexes (Fig. 5). The level of PSII supercomplexes was significantly decreased in var2 under extreme high-light conditions. Therefore, we cannot rule out the possibility that the fragmentation of D1 by Deg occurred before PSII dimerization and partial disassembly. Nevertheless, considering that processive degradation of D1 by FtsH is active at the disassembly step, it is plausible that the cooperative D1 degradation mediated by Deg and FtsH is more significant in stroma thylakoids. In this regard, further study of the migration and the disassembly of PSII complexes is necessary to elucidate the PSII repair cycle.
The loss of chloroplast protease often causes defects of chloroplast function and chloroplast development, like that of var2, which shows the variegated phenotype with undifferentiated plastid (Kato et al., 2007). To alleviate the defects from a loss of a chloroplast protease, it is reasonable that other proteases substitute for the protease function. Indeed, reduced accumulation of Clp protease triggers an up-regulation of plastid chaperones and causes a dramatic accumulation of SppA, another protease associated with thylakoid membranes (Rudella et al., 2006). This study revealed that the level of Clp protease is up-regulated in var2, as evidenced by immunoblots of ClpP6 and ClpC (Fig. 6). It is interesting that a significant portion of the Clp protease complexes appeared to be recruited from stroma to the thylakoid membrane. Although the precise reason to explain this recruitment remains unclear and further study is needed, it is plausible that Clp can partially substitute for FtsH function: both Clp and FtsH are structurally similar, harboring ATPase and protease domains, and perform processive degradation (Kato and Sakamoto, 2010). Clp has been suggested as degrading both soluble and membrane-bound substrates (Sjögren et al., 2006; Kim et al., 2009; Stanne et al., 2009; Zybailov et al., 2009). We also found an up-regulation of SppA in var2, which was similar to the case in clp mutants. SppA plays a role in the quality control of periplasmic and membrane-bound proteins (Lensch et al., 2001). In chloroplasts, SppA was shown to respond to high-light stress and to contribute to long-term high-light acclimation (Wetzel et al., 2009), implying that SppA up-regulation in var2 reflects a general defect in proper thylakoid formation. Collectively considering that evidence, we infer that the complementary functions between FtsH and Clp protease complexes exist for protein quality control in thylakoid membranes. Supporting this inference is the fact that FtsH is up-regulated in clpr2 knockdown transgenic lines (Rudella et al., 2006). In this scenario, the lack of any significant change in Deg accumulation in var2 is explainable by the functional difference between Deg, Clp, and FtsH.
Over the past decade, research into D1 degradation has revealed the contribution of several proteases in this process, but a question remains as to how the proteases recognize photodamaged D1 protein. Recently, inhibition of the N-terminal Met excision process in chloroplast was shown to compromise the N-terminal recognition by FtsH in D1 degradation (Adam et al., 2011). It triggers an increase in proteolysis by other proteases, which suggests that the recognition and degradation of D1 by FtsH require proper processing of possible substrate proteins when synthesized in chloroplasts. However, crystal structure studies of Deg1 in Arabidopsis revealed that an inactive Deg1 monomer can assemble into hexamers that, in turn, confer proteolytic activity by the acidification of the thylakoid lumen during light irradiation (Kley et al., 2011), suggesting that the structural adaptation of protease itself is involved in the degradation of damaged proteins in the thylakoid membrane. Further studies examining substrate recognition mechanisms must be undertaken to support our understanding of PSII repair.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col was used as the wild type. The mutant lines used for this study, var2-1 (contains a nonsense point mutation) and deg5 deg8 (transferred DNA [T-DNA] insertion lines), were described previously (Takechi et al., 2000; Sun et al., 2007). To isolate the triple mutant var2 deg5 deg8, var2-1 was initially crossed with deg5 and deg8. The following crossing was performed between isolated homozygous double mutants (var2 deg5 and var2 deg8). The genotype of var2-1 was determined using derived cleaved-amplified polymorphic sequence-assisted PCR with primers derived cleaved-amplified polymorphic sequence (2-1) (5′-GGACCATGGTCTTTGATGGATTCTTCGTCA-3′) and KT304 (5′-TCACGATTGTTCTTTATGTGGCTTAG-3′) and digestion with Tsp45I. The genotype of deg5 and deg8 was confirmed by PCR using the following gene-specific and T-DNA-specific primers: LP (5′-TGGGAGTCCACAAAATATTGG-3′) and RP (5′-TTCCCTTCCTTGCTAAATCTTG-3′) for Deg5 and LP (5′-AACTGTTTTCCCAGCTGGAC-3′), RP (5′-CTATTTCCCGGGTTAATGG-3′), and T-DNA LB (5′-TGGTTCACGTAGTGGGCCATCG-3′) for Deg8. Plants were germinated and grown on 0.7% (w/v) agar plates containing Murashige and Skoog medium supplemented with Gamborg’s vitamins (Sigma-Aldrich), 2 mm MES, pH 5.8, and 1.5% (w/v) Suc. Plants were maintained under 12 h of light (approximately 60 µmol photons m−2 s−1) at a constant temperature of 22°C. When plants were 2 weeks old, they were transferred onto soil and maintained under 12 h of light (180 µmol photons m−2 s−1) at a constant temperature of 22°C.
D1 Degradation Assay
Detached leaves from approximately 6-week-old Col and mutant plants were preincubated with their petioles submersed in 5 mm solutions of lincomycin in the dark overnight. Leaves treated with lincomycin were incubated for 2 h under high-light conditions (1,200 µmol photons m−2 s−1) or for 8 h under growth-light and low-light conditions (180 and 20 µmol photons m−2 s−1, respectively). To isolate thylakoid proteins, three leaf discs were collected using a 5-mm-diameter biopsy punch (Kai Medical). Leaf discs were frozen immediately in liquid nitrogen and pulverized using a microtube homogenizer. Samples were suspended in ice-cold extraction buffer (330 mm sorbitol, 50 mm HEPES, pH 7.5, 5 mm MgCl2, and 10 mm NaCl 2) and centrifuged at 2,500g for 5 min. Then, pellets were resuspended in SDS-PAGE sample buffer (125 mm Tris-Cl, pH 6.8, 2% [w/v] SDS, 5% [v/v] glycerol, 100 mm dithiothreitol, and 0.05% [w/v] bromphenol blue). Before loading, samples were centrifuged at 15,000g for 5 min, and supernatants were equally loaded (based on fresh weight). Signals of immunoblots were quantified using the ImageJ program (http://rsbweb.nih.gov/ij/) and normalized to the amount of Coomassie Brilliant Blue-stained LHCII.
D1 Fragment Detection
Leaves from approximately 6-week-old Col and mutant plants were used for the detection of D1 fragments. For high-light treatment, detached leaves were incubated for 1 h under extreme high-light irradiation (2,500 µmol photons m−2 s−1). To isolate membrane proteins, leaves that had been frozen rapidly in liquid nitrogen were pulverized in a mortar and homogenized in a solution containing 330 mm sorbitol, 50 mm HEPES, pH 7.5, 5 mm MgCl2, and 2 mm Na2EDTA. After centrifugation at 3,000g for 5 min, the pellet was resuspended in the same buffer. Chlorophyll was extracted in 80% (v/v) acetone, and the absorbance of chlorophyll extracts was measured using a spectrophotometer (Ultrospec 2100 pro; Amersham Biosciences). Chlorophyll contents were calculated applying the following equation: total chlorophyll (mg L−1) = 7.12A660 + 16.8A642.5. Membrane suspensions containing 100 µg of chlorophyll were centrifuged at 3,000g for 5 min, and the pellet were resuspended in SDS-PAGE sample buffer to a final concentration of 0.5 mg mL−1. Before loading, samples were centrifuged at 15,000g for 5 min, and equally loaded supernatants (based on chlorophyll) were subjected to additional analysis. In inhibitor experiments, Ser protease activity was blocked using PefaBloc SC (AEBSF) protease inhibitor (Roche). Detached leaves were preincubated with their petioles submersed in 4 mm solutions of AEBSF in the dark for 3 h. To ensure the effect of inhibitor, the leaves were placed in a glass vial containing buffer with newly added 4 mm AEBSF. Then AEBSF was infiltrated for 1 min into the leaves using a syringe needle with a rubber cap. After infiltration of AEBSF, leaves were incubated for 1 h under extreme high-light irradiation (2,500 µmol photons m−2 s−1).
Protein Extraction for Chloroplast Proteases
To isolate chloroplast proteins, 2 g of wild-type and var2-1 leaves was harvested from approximately 8-week-old plants. Chloroplasts were isolated according to the protocol described previously (Miura et al., 2007). Intact chloroplasts were fractionated into stromal and thylakoid fractions by centrifugation after osmotic shock in a hypotonic buffer (50 mm Tris-HCl, pH 7.5, and 5 mm MgCl2). Chloroplast suspensions and stroma fractions were mixed with equal volumes of 2× SDS-PAGE sample buffer. Then, thylakoid membranes were resuspended in SDS-PAGE sample buffer. Before loading, samples were centrifuged at 15,000g for 5 min. Then, equally loaded supernatants (based on chlorophyll) were subjected to additional analysis.
Preparation of Thylakoid Membranes and BN/SDS-PAGE
For BN-PAGE, thylakoid membranes were isolated from growth-light-adapted leaves and from leaves illuminated for 1 h at 2,500 µmol m−2 s−1 by an abbreviated thylakoid isolation method. After high-light treatment, leaves were ground in a blender with homogenization buffer (330 mm sorbitol, 50 mm HEPES, pH 7.5, 5 mm MgCl2, and 2 mm Na2EDTA). Homogenates were then filtered though gauze and centrifuged at 2,500g for 5 min. The pellet was resuspended in 1 mL of homogenization buffer, overlaid on two Percoll gradients (10% and 80%), and centrifuged at 2,500g for 5 min. The sediment at the interface between the 10% and 80% gradients was recovered and diluted in 5× homogenization buffer. After centrifugation at 2,500g for 5 min, the pellet was resuspended in 1 mL of homogenization buffer and the total chlorophyll concentration was measured. To solubilize membrane proteins, thylakoid membrane suspensions were centrifuged at 2,500g for 3 min. Then, pellets were resuspended using the NativePAGE Sample Prep Kit (Invitrogen). The final concentration of chlorophyll was 0.5 mg mL−1, and the final concentration of n-dodecyl-β-d-maltoside was 0.5% (w/v). After centrifugation at 15,000g for 10 min, the supernatant was mixed with Coomassie Brilliant Blue G-250; the final concentration of Coomassie Brilliant Blue G-250 was 0.125% (w/v). The samples were loaded onto a 4% to 16% gradient native gel. Electrophoresis was performed at 4°C overnight at 50 V. Second-dimension and further analyses were performed as described previously (Kato et al., 2009).
Immunoblotting
For immunoblot analysis, SDS-PAGE gels were electroblotted to the polyvinylidene difluoride membrane (Atto Corp.) and blocked by EzBlock (Atto Corp.). Then, the membranes were incubated with anti-D1 N-term (Agrisera; dilution, 1:2,000), anti-D1 C-term (generated in this study [see below]; dilution 1:5,000), anti-VAR2 (dilution, 1:5,000), anti-Deg2 (dilution, 1:2,000), anti-Deg8 (dilution, 1:2,000), anti-SppA (Agrisera; dilution, 1:2,000), anti-ClpP6 (kindly provided by Adrian K. Clarke; dilution, 1:5,000), anti-ClpC (Agrisera; dilution, 1:5,000), and anti-Rubisco (Agrisera; dilution, 1:5,000) antibodies. Signals from immunoblotting were detected using the ECL Prime Western Blotting Detection Kit (GE Healthcare) for D1 and D1 degradation fragments and the ECL Western Blotting Detection Kit (GE Healthcare) for other proteins and recorded with a LAS100-mini system (Fuji Photo Film). To improve immunoblot sensitivity, the cleavage products of the D1 protein were redetected after cutting off the membrane area above 25 kD. Experiments were repeated more than three times. Representative results are shown. The anti-D1 (C-term) antibody was designed to recognize the C terminus (amino acids 303–315 and 329–344) of the Arabidopsis D1 polypeptide. Synthetic peptide cocktails were used to raise antibodies in rabbit (Operon Biotechnology). Whole serum was used.
Fluorescence Measurements
Mature leaves of 5-week-old plants were used for measurements of high-light sensitivity. The leaves were incubated for 0, 60, 120, and 240 min under high-light irradiation (1,200 µmol photons m−2 s−1). The changes in Fv/Fm were measured. Before the measurements, leaf discs were maintained in the dark for 10 min to oxidize the plastoquinone pool fully.
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: FtsH2 (At2g30950), Deg5 (At4g18370), and Deg8 (At5g39830).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic summary of the degradation of D1 protein before this study.
Supplemental Figure S2. Immunoblot analysis of the cleavage product of the D1 protein by anti-D1 (DE loop) antibodies under normal- and high-light conditions.
Supplemental Figure S3. Level of FtsH isomers in the mutants.
Supplemental Figure S4. Immunoblot analysis of the cleavage products of the D1 protein in the var2 deg5 deg8 mutant under normal-light conditions.
Supplemental Figure S5. Steady-state accumulation and localization of chloroplast proteases.
Supplemental Figure S6. Immunoblot analysis of D1 protein in fug1, fug1 var2, and fug1 var2 deg5 deg8 mutants.
Supplemental Table S1. Fv/Fm measured from mature leaves using the FluorCam 700MF.
Supplementary Material
Acknowledgments
We thank Adrian K. Clarke and Yasusi Yamamoto for their kind gifts of anti-ClpP6 and anti-D1 antibodies (DE loop). We also thank Rie Hijiya for technical assistance.
Glossary
- Col
Columbia
- LHCII
light-harvesting complex of PSII
- Fv/Fm
maximum quantum yield of PSII
- AEBSF
4-(2-aminoethyl)-benzenesulfonyl fluoride
- BN
blue native
- T-DNA
transferred DNA
References
- Adam Z, Frottin F, Espagne C, Meinnel T, Giglione C. (2011) Interplay between N-terminal methionine excision and FtsH protease is essential for normal chloroplast development and function in Arabidopsis. Plant Cell 23: 3745–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro EM, Suorsa M, Rokka A, Allahverdiyeva Y, Paakkarinen V, Saleem A, Battchikova N, Rintamäki E. (2005) Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J Exp Bot 56: 347–356 [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B. (1993) Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Aro EM. (2002) Biogenesis, assembly and turnover of photosystem II units. Philos Trans R Soc Lond B Biol Sci 357: 1451–1459, discussion 1459–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Thompson E, Nixon PJ, Horton P, Mullineaux CW, Robinson C, Mann NH. (2002) A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J Biol Chem 277: 2006–2011 [DOI] [PubMed] [Google Scholar]
- Barber J, Andersson B. (1992) Too much of a good thing: light can be bad for photosynthesis. Trends Biochem Sci 17: 61–66 [DOI] [PubMed] [Google Scholar]
- Barker M, de Vries R, Nield J, Komenda J, Nixon PJ. (2006) The deg proteases protect Synechocystis sp. PCC 6803 during heat and light stresses but are not essential for removal of damaged D1 protein during the photosystem two repair cycle. J Biol Chem 281: 30347–30355 [DOI] [PubMed] [Google Scholar]
- Chen M, Choi Y, Voytas DF, Rodermel S. (2000) Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J 22: 303–313 [DOI] [PubMed] [Google Scholar]
- Clausen T, Southan C, Ehrmann M. (2002) The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell 10: 443–455 [DOI] [PubMed] [Google Scholar]
- De Las Rivas J, Andersson B, Barber J. (1992) Two sites of primary degradation of the D1-protein induced by acceptor or donor side photo-inhibition in photosystem II core complexes. FEBS Lett 301: 246–252 [DOI] [PubMed] [Google Scholar]
- Edelman M, Mattoo AK. (2008) D1-protein dynamics in photosystem II: the lingering enigma. Photosynth Res 98: 609–620 [DOI] [PubMed] [Google Scholar]
- Greenberg BM, Gaba V, Mattoo AK, Edelman M. (1987) Identification of a primary in vivo degradation product of the rapidly-turning-over 32 kd protein of photosystem II. EMBO J 6: 2865–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussühl K, Andersson B, Adamska I. (2001) A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J 20: 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesgen PF, Schuhmann H, Adamska I. (2006) Photodamaged D1 protein is degraded in Arabidopsis mutants lacking the Deg2 protease. FEBS Lett 580: 6929–6932 [DOI] [PubMed] [Google Scholar]
- Huesgen PF, Schuhmann H, Adamska I. (2009) Deg/HtrA proteases as components of a network for photosystem II quality control in chloroplasts and cyanobacteria. Res Microbiol 160: 726–732 [DOI] [PubMed] [Google Scholar]
- Ito K, Akiyama Y. (2005) Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol 59: 211–231 [DOI] [PubMed] [Google Scholar]
- Itzhaki H, Naveh L, Lindahl M, Cook M, Adam Z. (1998) Identification and characterization of DegP, a serine protease associated with the luminal side of the thylakoid membrane. J Biol Chem 273: 7094–7098 [DOI] [PubMed] [Google Scholar]
- Kapri-Pardes E, Naveh L, Adam Z. (2007) The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19: 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Miura E, Ido K, Ifuku K, Sakamoto W. (2009) The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol 151: 1790–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Miura E, Matsushima R, Sakamoto W. (2007) White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiol 144: 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W. (2009) Protein quality control in chloroplasts: a current model of D1 protein degradation in the photosystem II repair cycle. J Biochem 146: 463–469 [DOI] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W. (2010) New insights into the types and function of proteases in plastids. Int Rev Cell Mol Biol 280: 185–218 [DOI] [PubMed] [Google Scholar]
- Kettunen R, Tyystjärvi E, Aro EM. (1996) Degradation pattern of photosystem II reaction center protein D1 in intact leaves: the major photoinhibition-induced cleavage site in D1 polypeptide is located amino terminally of the DE loop. Plant Physiol 111: 1183–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Rudella A, Ramirez Rodriguez V, Zybailov B, Olinares PD, van Wijk KJ. (2009) Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21: 1669–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kley J, Schmidt B, Boyanov B, Stolt-Bergner PC, Kirk R, Ehrmann M, Knopf RR, Naveh L, Adam Z, Clausen T. (2011) Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure. Nat Struct Mol Biol 18: 728–731 [DOI] [PubMed] [Google Scholar]
- Komenda J, Barker M, Kuviková S, de Vries R, Mullineaux CW, Tichy M, Nixon PJ. (2006) The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J Biol Chem 281: 1145–1151 [DOI] [PubMed] [Google Scholar]
- Komenda J, Sobotka R, Nixon PJ. (2012) Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol 15: 245–251 [DOI] [PubMed] [Google Scholar]
- Lensch M, Herrmann RG, Sokolenko A. (2001) Identification and characterization of SppA, a novel light-inducible chloroplast protease complex associated with thylakoid membranes. J Biol Chem 276: 33645–33651 [DOI] [PubMed] [Google Scholar]
- Lindahl M, Spetea C, Hundal T, Oppenheim AB, Adam Z, Andersson B. (2000) The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12: 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M, Tabak S, Cseke L, Pichersky E, Andersson B, Adam Z. (1996) Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J Biol Chem 271: 29329–29334 [DOI] [PubMed] [Google Scholar]
- Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. (2005) Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II. Nature 438: 1040–1044 [DOI] [PubMed] [Google Scholar]
- Miura E, Kato Y, Matsushima R, Albrecht V, Laalami S, Sakamoto W. (2007) The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 19: 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N, Takahashi S, Nishiyama Y, Allakhverdiev SI. (2007) Photoinhibition of photosystem II under environmental stress. Biochim Biophys Acta 1767: 414–421 [DOI] [PubMed] [Google Scholar]
- Murchie EH, Niyogi KK. (2011) Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol 155: 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Allakhverdiev SI, Murata N. (2006) A new paradigm for the action of reactive oxygen species in the photoinhibition of photosystem II. Biochim Biophys Acta 1757: 742–749 [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J. (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot (Lond) 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. (2001) AAA+ superfamily ATPases: common structure—diverse function. Genes Cells 6: 575–597 [DOI] [PubMed] [Google Scholar]
- Raven JA. (2011) The cost of photoinhibition. Physiol Plant 142: 87–104 [DOI] [PubMed] [Google Scholar]
- Rodrigues RA, Silva-Filho MC, Cline K. (2011) FtsH2 and FtsH5: two homologous subunits use different integration mechanisms leading to the same thylakoid multimeric complex. Plant J 65: 600–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudella A, Friso G, Alonso JM, Ecker JR, van Wijk KJ. (2006) Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18: 1704–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Miura E, Kaji Y, Okuno T, Nishizono M, Ogura T. (2004) Allelic characterization of the leaf-variegated mutation var2 identifies the conserved amino acid residues of FtsH that are important for ATP hydrolysis and proteolysis. Plant Mol Biol 56: 705–716 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Tamura T, Hanba-Tomita Y, Murata M, Sodmergen (2002) The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 7: 769–780 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Zaltsman A, Adam Z, Takahashi Y. (2003) Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15: 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter AH, Virgin I, Hagman A, Andersson B. (1992) On the molecular mechanism of light-induced D1 protein degradation in photosystem II core particles. Biochemistry 31: 3990–3998 [DOI] [PubMed] [Google Scholar]
- Schuhmann H, Adamska I. (2012) Deg proteases and their role in protein quality control and processing in different subcellular compartments of the plant cell. Physiol Plant 145: 224–234 [DOI] [PubMed] [Google Scholar]
- Shipton CA, Barber J. (1994) In vivo and in vitro photoinhibition reactions generate similar degradation fragments of D1 and D2 photosystem-II reaction-centre proteins. Eur J Biochem 220: 801–808 [DOI] [PubMed] [Google Scholar]
- Silva P, Thompson E, Bailey S, Kruse O, Mullineaux CW, Robinson C, Mann NH, Nixon PJ. (2003) FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp PCC 6803. Plant Cell 15: 2152–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinvany-Villalobo G, Davydov O, Ben-Ari G, Zaltsman A, Raskind A, Adam Z. (2004) Expression in multigene families: analysis of chloroplast and mitochondrial proteases. Plant Physiol 135: 1336–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LL, Stanne TM, Zheng B, Sutinen S, Clarke AK. (2006) Structural and functional insights into the chloroplast ATP-dependent Clp protease in Arabidopsis. Plant Cell 18: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanne TM, Sjögren LL, Koussevitzky S, Clarke AK. (2009) Identification of new protein substrates for the chloroplast ATP-dependent Clp protease supports its constitutive role in Arabidopsis. Biochem J 417: 257–268 [DOI] [PubMed] [Google Scholar]
- Sun X, Fu T, Chen N, Guo J, Ma J, Zou M, Lu C, Zhang L. (2010a) The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol 152: 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Ouyang M, Guo J, Ma J, Lu C, Adam Z, Zhang L. (2010b) The thylakoid protease Deg1 is involved in photosystem-II assembly in Arabidopsis thaliana. Plant J 62: 240–249 [DOI] [PubMed] [Google Scholar]
- Sun X, Peng L, Guo J, Chi W, Ma J, Lu C, Zhang L. (2007) Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 19: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Badger MR. (2011) Photoprotection in plants: a new light on photosystem II damage. Trends Plant Sci 16: 53–60 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Murata N. (2008) How do environmental stresses accelerate photoinhibition? Trends Plant Sci 13: 178–182 [DOI] [PubMed] [Google Scholar]
- Takechi K, Sodmergen, Murata M, Motoyoshi F, Sakamoto W. (2000) The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol 41: 1334–1346 [DOI] [PubMed] [Google Scholar]
- Tyystjärvi E, Aro EM. (1996) The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity. Proc Natl Acad Sci USA 93: 2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Wagner R, Aigner H, Funk C. (2012) FtsH proteases located in the plant chloroplast. Physiol Plant 145: 203–214 [DOI] [PubMed] [Google Scholar]
- Wetzel CM, Harmacek LD, Yuan LH, Wopereis JL, Chubb R, Turini P. (2009) Loss of chloroplast protease SPPA function alters high light acclimation processes in Arabidopsis thaliana L. (Heynh.). J Exp Bot 60: 1715–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR. (2004) The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J 37: 864–876 [DOI] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR. (2005) Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol 138: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A, Feder A, Adam Z. (2005a) Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts: implications for thylakoid formation and photosystem II maintenance. Plant J 42: 609–617 [DOI] [PubMed] [Google Scholar]
- Zaltsman A, Ori N, Adam Z. (2005b) Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopsis. Plant Cell 17: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouni A, Witt HT, Kern J, Fromme P, Krauss N, Saenger W, Orth P. (2001) Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution. Nature 409: 739–743 [DOI] [PubMed] [Google Scholar]
- Zybailov B, Friso G, Kim J, Rudella A, Rodríguez VR, Asakura Y, Sun Q, van Wijk KJ. (2009) Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol Cell Proteomics 8: 1789–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.