Abstract
Leaf rolling is an important agronomic trait in rice (Oryza sativa) breeding and moderate leaf rolling maintains the erectness of leaves and minimizes shadowing between leaves, leading to improved photosynthetic efficiency and grain yields. Although a few rolled-leaf mutants have been identified and some genes controlling leaf rolling have been isolated, the molecular mechanisms of leaf rolling still need to be elucidated. Here we report the isolation and characterization of SEMI-ROLLED LEAF1 (SRL1), a gene involved in the regulation of leaf rolling. Mutants srl1-1 (point mutation) and srl1-2 (transferred DNA insertion) exhibit adaxially rolled leaves due to the increased numbers of bulliform cells at the adaxial cell layers, which could be rescued by complementary expression of SRL1. SRL1 is expressed in various tissues and is expressed at low levels in bulliform cells. SRL1 protein is located at the plasma membrane and predicted to be a putative glycosylphosphatidylinositol-anchored protein. Moreover, analysis of the gene expression profile of cells that will become epidermal cells in wild type but probably bulliform cells in srl1-1 by laser-captured microdissection revealed that the expression of genes encoding vacuolar H+-ATPase (subunits A, B, C, and D) and H+-pyrophosphatase, which are increased during the formation of bulliform cells, were up-regulated in srl1-1. These results provide the transcript profile of rice leaf cells that will become bulliform cells and demonstrate that SRL1 regulates leaf rolling through inhibiting the formation of bulliform cells by negatively regulating the expression of genes encoding vacuolar H+-ATPase subunits and H+-pyrophosphatase, which will help to understand the mechanism regulating leaf rolling.
Leaves are lateral organs of seed plants derived from the peripheral zone of the shoot apical meristem and display polarized development along the proximal-distal axis and adaxial-abaxial axis (Bowman et al., 2002; Kidner and Timmermans, 2007). As the major photosynthetic organ of plants, an optimal structure of leaves is important for the maximization of light capture and efficient gas exchange (Eshed et al., 2001; Moon and Hake, 2011). In the super-high-yield hybrid rice (Oryza sativa), characteristics of the last three leaves were proposed to be long, erect, narrow, V shaped (rolled), and thick (Yuan, 1997). Therefore moderate leaf rolling is of great importance for the improvement of photosynthetic efficiency and grain yields by maintaining the erectness of leaves and minimizing shadowing between leaves (Wu, 2009).
There are two types of leaf transverse rolling in plants, inward (adaxial) and outward (abaxial) rolling, which are regulated by complicated developmental and polarity establishment controls (Bowman et al., 2002; Micol and Hake, 2003). In addition, environmental factors (e.g. water deficiency, high air temperature, and sunlight) may result in the inward rolling of leaves (Kadioglu and Terzi, 2007). The monocot-specific (except the Helobiae), large, and highly vacuolated bulliform cells (motor cells) that occur in groups between vascular bundles on the adaxial epidermis of the lamina are involved in the modulation of leaf rolling (Itoh et al., 2005). It has been shown that under drought conditions, bulliform cells lose turgor pressure and shrink, leading to the rolling up of leaves. Once water is sufficient, the bulliform cells expand and the leaves open again (Price et al., 1997; Alvarez et al., 2008). However, the molecular and genetic mechanisms of leaf rolling and the function of bulliform cells in leaf rolling remain to be elucidated.
Until now, 13 rice mutants with adaxially rolled leaves (rl) have been isolated and characterized. Among these mutants, rl1 to rl6 were mapped to rice chromosomes 1, 4, 12, 1, 3, and 7, respectively (Khush and Kinoshita, 1991); rl7 to rl12 and rl(t) were located on chromosome 2 (rl(t), Shao et al., 2005b), 5 (rl7, Li et al., 1998; rl8, Shao et al., 2005a), 7 (rl11, Shi et al., 2009), 9 (rl9, Yan et al., 2006; rl10, Luo et al., 2007), and 10 (rl12, Luo et al., 2009), respectively. However, only a few genes have been cloned and analyzed.
Several transcription factors have been demonstrated being involved in the regulation of rice leaf rolling. The SHALLOT-LIKE1 (SLL1)/RL9 gene encodes a transcription factor of KANADI family and regulates leaf abaxial cell development (Yan et al., 2008; Zhang et al., 2009). sll1 mutant exhibits extremely incurved leaves with ectopic formation of bulliform cells in abaxial surfaces due to the defective development of sclerenchymatous cells at the abaxial side of leaf blades. Enhanced expression of SLL1 also leads to rolled leaves due to the stimulated phloem development at the abaxial side and suppressed development of bulliform cells and sclerenchyma at the adaxial side (Zhang et al., 2009). Ectopic expression of microRNA166-resistant version of the OsHB1 gene, a member of class III homeodomain Leu zipper family, results in adaxially rolled leaves with reduced sclerenchyma and formation of bulliform cells at the abaxial side (Itoh et al., 2008). Loss-of-function mutant of OUTCURVED LEAF1 (OUL1)/Rice outermost cell-specific gene5 (Roc5), an ortholog of Arabidopsis (Arabidopsis thaliana) homeodomain Leu zipper class IV gene GLABRA2, presents the increased number and size of bulliform cells, resulting in abaxially rolled leaves, whereas overexpression of OUL1/Roc5 results in reduced number and size of bulliform cells, leading to adaxial leaf rolling (Ito et al., 2003; Zou et al., 2011). The leaves of rice transgenic plants with cosuppression of YABBY1 were also abaxially rolled due to the increased number of bulliform cells at the adaxial side (Dai et al., 2007).
In addition, some enzymes also play important roles in modulating leaf rolling. Rice NARROW AND ROLLED LEAF1 encodes the cellulose synthase-like protein D4 (OsCslD4) and deficiency of which results in semirolled leaves, owing to smaller bulliform cells (Hu et al., 2010). Mutation of CONSTITUTIVELY WILTED1/NARROW LEAF7, a new member of the YUCCA gene family, results in increased number and smaller size of bulliform cells and therefore adaxial leaf rolling (Woo et al., 2007; Fujino et al., 2008). A defect of ADAXIALIZED LEAF1 (ADL1), which encodes a calpain-like Cys proteinase specific to plants and is orthologous to maize (Zea mays) DEFECTIVE KERNEL1, results in abaxial leaf rolling due to the increase of bulliform cells at the adaxial side and the formation of bulliform-like cells at the abaxial side of leaf blades (Hibara et al., 2009). Furthermore, overexpression of ABAXIALLY CURLED LEAF1 (ACL1) and its paralog ACL2 leads to increased bulliform cells and hence abaxial curling of leaf blades (Li et al., 2010).
During the development from leaf primordium to mature leaf of grass species such as rice and maize, bulliform cells are differentiated from leaf epidermis on the adaxial side (Itoh et al., 2005). However, studies of the mechanisms regulating the differentiation of bulliform cells are rare. It has been proposed that positional information but not lineage may direct the differentiation of bulliform cells in maize (Hernandez et al., 1999). Research of motor cells in the pulvini of Mimosa pudica indicated that along with the differentiation of motor cells, the expression of both vacuolar aquaporin and H+-ATPase were increased (Fleurat-Lessard et al., 1997). Furthermore, vacuolar H+-pyrophosphatase and H+-ATPase create the tonoplast electrochemical gradient by pumping H+ into the vacuoles, which are important for osmoregulation of motor cells and osmotic adjustment in plants (Moran, 2007).
We here describe the isolation and characterization of rice SEMI-ROLLED LEAF1 (SRL1), which encodes a putative glycosylphosphatidylinositol (GPI)-anchored protein (GAP) and modulates leaf rolling. A point mutation or knockdown expression of SRL1 (by transferred DNA [T-DNA] insertion) cause more bulliform cells on the adaxial side of the leaf blades and therefore adaxial leaf rolling. Moreover, SRL1 may negatively regulate the expression of genes encoding vacuolar H+-ATPase subunits and H+-pyrophosphatase to inhibit the formation of bulliform cells.
RESULTS
srl1-1, a Rice Mutant with Adaxially Rolled Leaves, Has the Increased Bulliform Cells at the Adaxial Epidermis
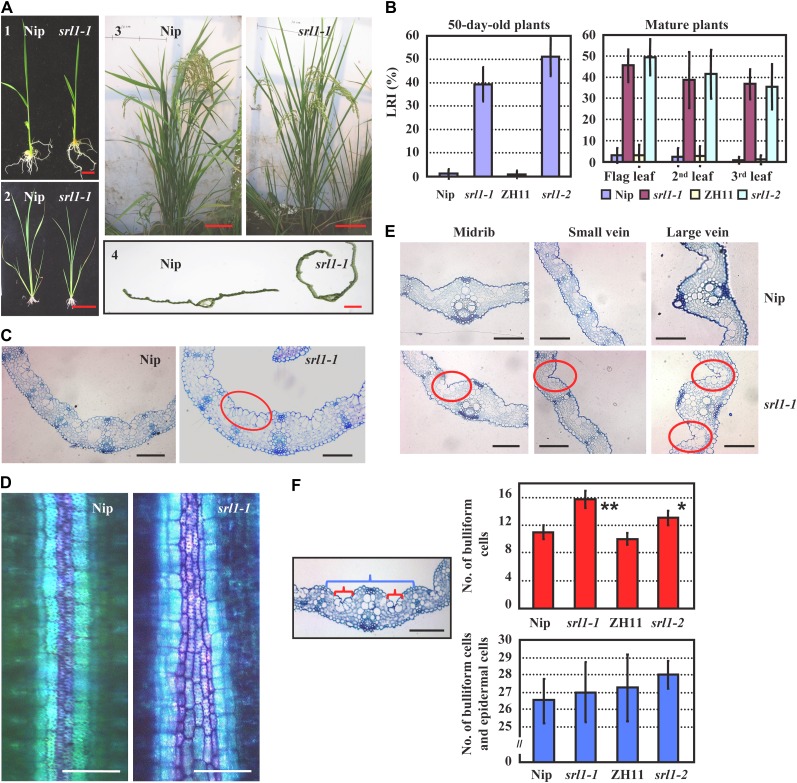
To identify genes involved in the regulation of leaf rolling, we screened a rice ethyl methanesulphonate mutagenized population (rice sp. japonica var Nipponbare), and identified a mutant with adaxially rolled leaves, designated as srl1-1. Phenotypic analysis revealed that srl1-1 exhibited incurved leaves from the seedling stage to reproductive stage (Fig. 1A) and the leaf-rolling index (LRI) of 50-d-old srl1-1 plants reached to approximately 40%. At the later reproductive stage (7 d after flowering), the LRIs of the last emerged three leaves were 35% to 45%, whereas the corresponding wild-type leaves were almost flat (Fig. 1B).
Figure 1.
Characterization and analysis of rice mutant srl1-1. A, The leaves of srl1-1 are adaxially rolled from seedling to mature stage compared with those of wild-type plant Nipponbare (Nip). 1, 14-d-old seedlings, bar = 1 cm; 2, 30-d-old plants, bar = 10 cm; 3, mature plants, bar = 10 cm; 4, section of the second leaf from top of mature plants, bar = 1 mm. B, LRIs of srl1-1, srl1-2, and the corresponding wild-type (ZH11 represents the wild type for srl1-2) leaves at different developmental stages (the seventh leaf of 50-d-old plants and the top three leaves of mature plants 7 d after flowering). Error bars represent sd (n > 30). C, Cross sections of leaf blades in Nip (left section) and srl1-1 (right section) showed significantly increased bulliform cells between vascular bundles in srl1-1 (the third leaf blades of 14-d-old seedlings). Bars = 100 μm. D, Toluidine blue O staining of the cells between small vascular bundle and midrib of Nip and srl1-1 (the third leaf of 14-d-old seedlings). Bulliform cells and epidermal cells were stained in purple and blue, respectively. Bars = 50 μm. E, Cross sections of leaf blades of Nip (top section) and srl1-1 (bottom section) showed significantly increased bulliform cells between vascular bundles (midrib, small vein, and large vein) in srl1-1 (the 10th leaf blades of 60-d-old mature plants). Bars = 100 μm. F, Statistical analysis of the number of bulliform cells (red brackets) and the total number of epidermal cells and bulliform cells (blue bracket) between two small vascular bundles abutting the midrib in the third leaf of 14-d-old seedlings by two-tailed Student’s t test (*, P < 0.05; **, P < 0.01) revealed the significantly increased numbers of bulliform cells in srl1-1 and srl1-2 leaves, whereas the total numbers of epidermal cells and bulliform cells were not significantly different from the wild type. Error bar represents sd (n > 10).
Leaf rolling is regulated by the large, bubble-shaped bulliform cells that occur in groups between vascular bundles on the adaxial epidermis of the leaf blade. Indeed, observation revealed the increased bulliform cells at the adaxial epidermis of the third leaf blade in srl1-1 compared with that of the wild type at seedling stage (Fig. 1C; Supplemental Fig. S1). Previous studies showed that toluidine blue O stains bulliform cells in purple and epidermal cells in blue to distinguish these two types of cells (Hernandez et al., 1999), and the staining analysis using toluidine blue O confirmed the increased number of bulliform cells in srl1-1 compared with that of wild type (Fig. 1D). Indeed, more bulliform cells were also observed on the adaxial epidermis of srl1-1 mature leaves (Fig. 1E) and statistical analysis of the number of bulliform cells between two small vascular bundles abutting the midrib in the third leaf showed that there were four to five more bulliform cells in srl1-1 mutant than that in the wild type, while the whole number of epidermal cells and bulliform cells between two small vascular bundles abutting the midrib in the third leaf of srl1-1 was about the same as that of wild type (Fig. 1F), indicating that more bulliform cells and less epidermal cells were formed from the early undifferentiated epidermis.
No difference in other cellular types of leaf blades or change in leaf polarity was observed between srl1-1 and wild type (Fig. 1, C and E), indicating that the leaf-rolling phenotype of srl1-1 is resulted from the enhanced formation of bulliform cells at the adaxial cell layers.
SRL1 Encodes a Putative GAP and Complementary Expression of SRL1 Rescues the Rolled-Leaf Phenotype of srl1-1
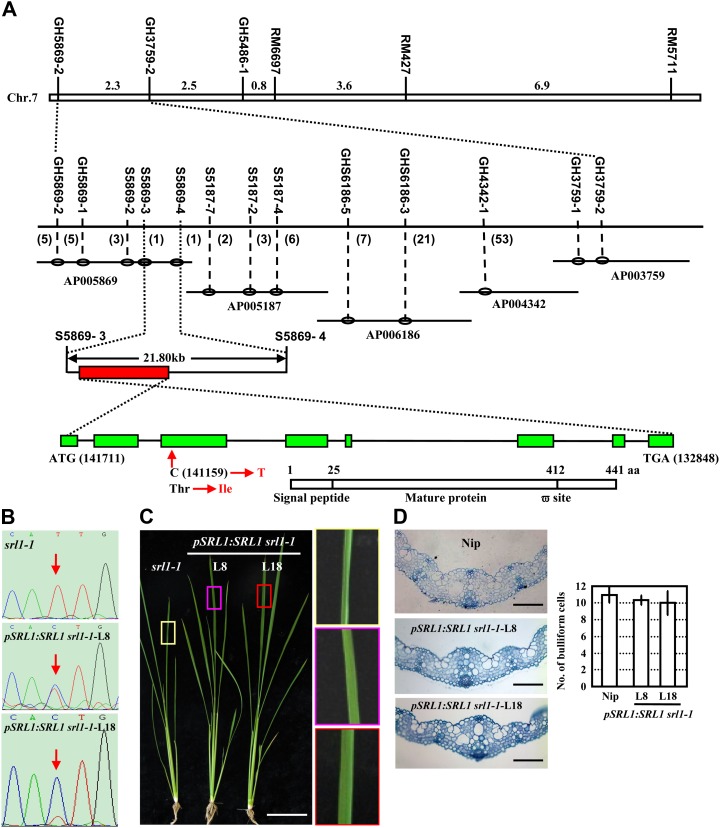
The SRL1 gene was mapped primarily between the sequence-tagged site (STS) markers GH5869-2 and GH3759-2 in an F2 population derived from a cross between srl1-1 and indica var Tai Zhong Ben Di 1. Further, by using 2,346 homozygous mutant plants, SRL1 was fine mapped to an interval of 21.8 kb between two simple sequence repeats (SSR) markers S5869-3 and S5869-4 on the bacterial artificial chromosome AP005869. Two open reading frames were predicted in this region and further amplification, sequencing, and comparison of the relevant DNA fragments indicated that a single base substitution (C to T) in the third exon of the gene Os07g01240 (http://rice.plantbiology.msu.edu/analyses_search_locus.shtml), resulting in the change of amino acid residue 88 from Thr to Ile (Fig. 2A).
Figure 2.
Map-based cloning of SRL1, which encodes a putative GAP. A, SRL1 was primarily mapped to rice chromosome 7 between STS markers GH5869-2 and GH3759-2 and then fine mapped to a 21.8-kb region between SSR markers S5869-3 and S5869-4 by a large F2 mapping population. Further amplification, sequencing, and comparison of the relevant DNA fragments indicated that a single base substitution (C to T) in the third exon of the gene Os07g01240 in srl1-1 allele leading to the change of amino acid residue from Thr to Ile. Genomic organization of SRL1 (green boxes represent exons, and black lines represent introns) is shown, including the translational start (ATG) and stop (TAA) positions of the predicted coding sequence. The position and nucleotide change in the srl1-1 mutant is also shown. Structural analysis of SRL1, a putative GAP, indicated the presence of an N-terminal signal peptide from 1 to 25 amino acids and a potential C-terminal GPI-modification site at 412 amino acids (ω-site). B, Sequencing analysis of the SRL1 transcripts in two transgenic lines (L8, L18) of srl1-1 with complemented expression of SRL1 showed the double peak at the mutation site (red arrows), indicating the presence of normal transcripts of SRL1. C and d, Morphology (C, 40-d-old plants, bar = 10 cm) and cross sections (D, third leaf blades of 14-d-old seedlings, bars = 100 μm) of two transgenic lines of srl1-1 with complemented expression of SRL1 (L8, L18). To highlight the leaf shape, leaf regions (boxes) are enlarged (right section of C). Rescued expression of SRL1 resulted in normal leaves and bulliform cell numbers. Statistical analysis by two-tailed Student’s t test shows no significant difference (right section of D). Error bar represents sd (n > 10).
Structural analysis showed that SRL1 is composed of eight exons and seven introns with a full-length complementary DNA (cDNA) of 1,871 bp (KOME accession no. AK101907). SRL1 encodes a predicted protein of 441 amino acids. Hydropathy analysis indicated that SRL1 consists of hydrophobic N and C terminals and a central hydrophilic region (http://web.expasy.org/protscale/; Kyte and Doolittle, 1982; Supplemental Fig. S2A). Further analysis by SignalP (http://www.cbs.dtu.dk/services/SignalP/; Petersen et al., 2011) and PredGPI (http://gpcr.biocomp.unibo.it/predgpi/; Pierleoni et al., 2008) showed that SRL1 encodes a GAP with a signal peptide for targeting to the endoplasmic reticulum at the N-terminal and a potential C-terminal GPI-modification site (ω-site; Fig. 2A). Homology analysis by BLASTP (Altschul et al., 1990) identified the homologous proteins in Arabidopsis (At1G61900.1, 57% identity), maize (LOC_100276335, 78% identity), and Sorghum bicolor (SORBIDRAFT_02g000365, 77% identity; Supplemental Fig. S2B), however, the function of these proteins remains unknown.
Further, genetic complementation analysis was conducted to verify the function of SRL1 in leaf rolling. A construct containing 11.1-kb whole genomic sequence of SRL1 (promoter region and whole coding sequence) was transformed into srl1-1 plants. Analysis of independent transgenic plants by reverse transcription (RT)-PCR using primers flanking the mutation site and further sequencing revealed a double peak (C and T) at the mutation site (Fig. 2B), confirming the presence of normal SRL1 transcripts. Phenotypic observation and cross section of leaf blades of the transgenic plants showed the flat leaves and similar number of bulliform cells as wild-type plants (Fig. 2, C and D), demonstrating that complementary expression of SRL1 rescued the leaf-rolling phenotype of srl1-1 and the crucial role of SRL1 in leaf-rolling control.
Knockdown of SRL1 by T-DNA Insertion Reproduces the Rolled-Leaf Phenotype of srl1-1
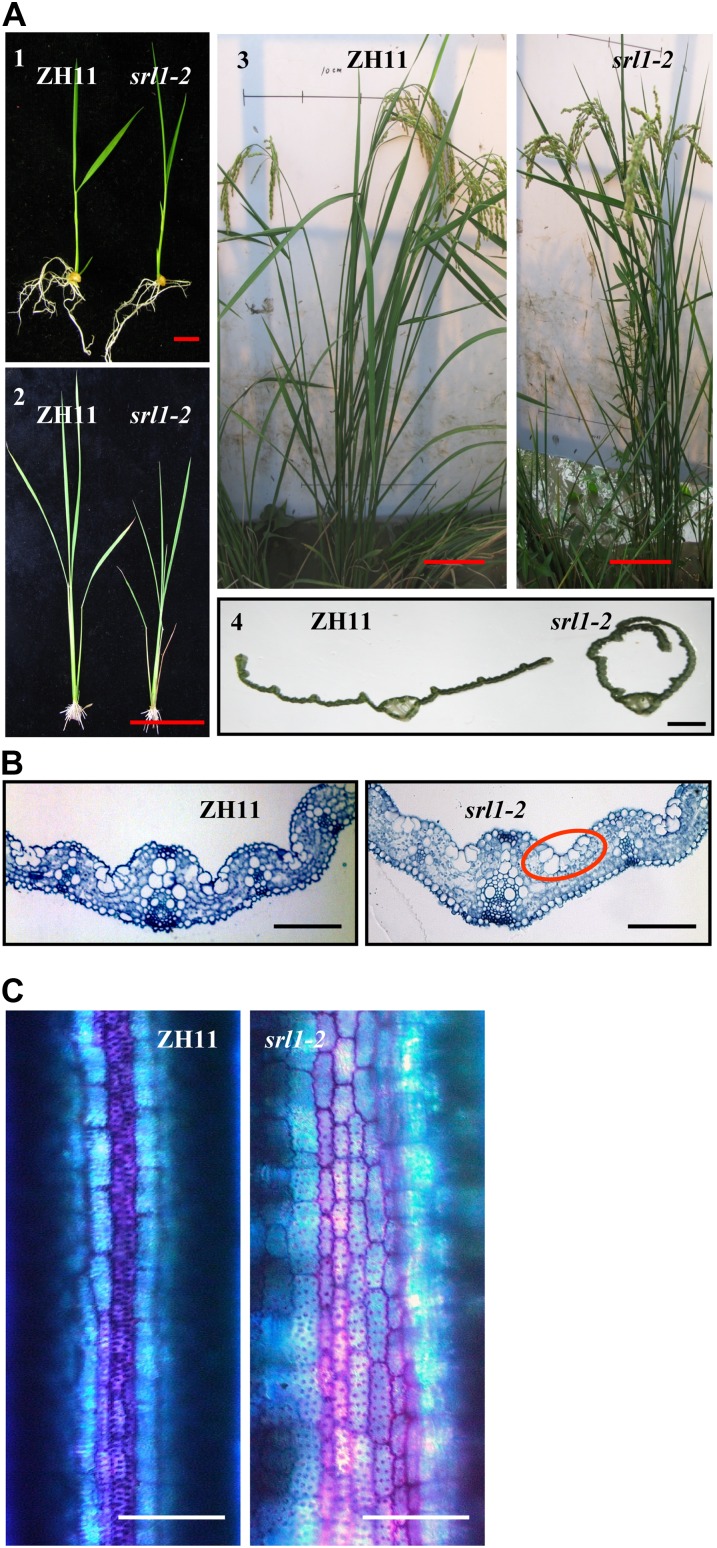
Additional srl1 mutant alleles were searched against the T-DNA flanking sequences in Shanghai T-DNA insertion population (SHIP; Fu et al., 2009). A candidate mutant with T-DNA located at the fifth intron of SRL1 was identified and named as srl1-2. Analysis using SRL1-specific primer F4805-R and T-DNA border primers (NTL1, NTL2, and NTL3), respectively, confirmed the insertion of T-DNA in SRL1 (Supplemental Fig. S3, A and B) and Southern-blot analysis of srl1-2 genomic DNA using a fragment of hygromycin phosphotransferaseII gene as probe indicated the single insertion of T-DNA (Supplemental Fig. S3C). Segregation analysis using the progeny of heterozygous srl1-2 revealed that of the 480 individuals, 132 showed leaf-rolling phenotype (similar to srl1-1) that are homozygous for T-DNA insertion, while 348 displayed flat leaves similar to wild type that are heterozygous for T-DNA insertion or do not harbor T-DNA insertion. The segregation ratio (348:132; χ2 = 1.6, degrees of freedom = 1, P = 0.21) was consistent with classical Mendelian ratio (3:1), indicating the genetic linkage of SRL1 and leaf rolling. Further quantitative (q)RT-PCR analysis revealed the significantly decreased expression of SRL1 in the srl1-2 homozygous mutants (Supplemental Fig. S3D), and phenotypic observation (Fig. 3A), cross-section analysis (Fig. 3B), toluidine blue O staining (Fig. 3C) of the homozygous srl1-2 plants confirmed the rolled leaf resulting from the increased bulliform cells on the adaxial cell layers (Supplemental Fig. S1), which resembles the srl1-1 mutant and verifies the crucial role of SRL1 in leaf-rolling control.
Figure 3.
Knockdown mutant srl1-2, by T-DNA insertion, presents similar phenotype of srl1-1. A, The leaves of srl1-2 are also adaxially rolled from seedling to mature stage compared with wild-type plants ZH11. 1, 14-d-old seedlings, bar = 1 cm; 2, 30-d-old plants, bar = 10 cm; 3, mature plants, bar = 10 cm; 4, section of the second leaf from top of mature plants, bar = 1 mm. B, Cross sections of the third leaf blades of 14-d-old ZH11 (left section) and srl1-2 (right section) seedlings show significantly increased bulliform cells between vascular bundles in srl1-2. Bars = 100 μm. C, Toluidine blue O staining of the cells between small vascular bundle and midrib in ZH11 and srl1-2 (the third leaf of 14-d-old seedlings). Bulliform cells and epidermal cells were stained in purple and blue, respectively. Bars = 50 μm.
SRL1 Is Expressed in Various Tissues and SRL1 Protein Is Located at the Plasma Membrane
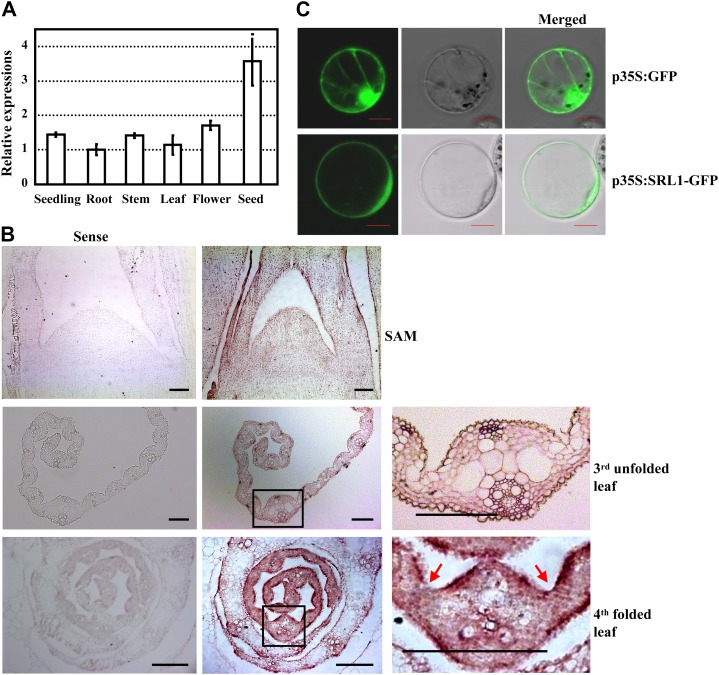
qRT-PCR analysis indicated that SRL1 was expressed in various tissues, including seedlings, roots, stems, leaves, flowers, and seeds, with a relatively higher expression in seeds (Fig. 4A). However, besides the rolled leaves, there are no other obvious differences of srl1-1 and srl1-2 mutants in comparison to wild type. Because of the crucial role of SRL1 in leaf rolling, further in situ hybridization analysis was performed to characterize the temporal and spatial expression pattern of SRL1 during leaf development. The results showed that SRL1 was transcribed throughout the leaf primordium and the signal was more intense in the epidermal cell layers of young leaves enwrapped in the leaf sheath except for the region where sclerenchymatous cells were formed and the hollow region where bulliform cells were formed. In the unfolded leaves with bulliform cells, SRL1 was expressed ubiquitously except that the signal is less intense in the bulliform cells (Fig. 4B), which is consistent with the role of SRL1 in modulating leaf rolling through regulating the formation of bulliform cells.
Figure 4.
Expression pattern of SRL1 and subcellular localization of SRL1. A, qRT-PCR analysis of SRL1 expression in various tissues including 14-d-old seedlings (whole plants without roots), roots, stems, 10th leaves at the heading stage, flowers, and seeds (9 d). Transcript levels were normalized with the ACTIN transcription and relative expression levels were compared with that in roots (set as 1). Mean values were obtained from three independent experiments and error bars indicate sd. B, In situ hybridization analysis of SRL1 expression in shoot apical meristem, the third unfolded leaves, and the fourth folded leaves of 14-d-old seedlings. SRL1 transcript was detected throughout the leaf primordium and unfolded leaves except that the signal was less intense in the bulliform cells. The SRL1 signal was more intense in the epidermal cell layers of folded leaves in the leaf sheath except for the region where sclerenchymatous cells are formed and the hollow region where bulliform cells are formed (red arrows). The squared regions (black boxes) in the middle section are enlarged to highlight the SRL1 expression (right section). Bars = 100 μm. C, Transient expression of SRL1-GFP fusion protein in rice protoplasts revealed that SRL1 is mainly located at the plasma membrane. Rice protoplasts expressing GFP alone were used as the control. Bars = 10 μm.
In situ hybridization analysis of the SRL1 expression in rice sp. japonica ‘Zhonghua 11’ (ZH11) and srl1-2 mutants using antisense probe of SRL1 showed the tremendously reduced expression level of SRL1 in the leaf primordium and young leaf of srl1-2 mutants compared with that of ZH11 (Supplemental Fig. S4), further confirming the deficiency of SRL1 transcripts in srl1-2 mutants.
Consistent with the previously identified GAPs being demonstrated to be attached to the plasma membrane via the GPI anchor, SRL1 is predicted to be located at the plasma membrane by P-sort program (http://psort.hgc.jp/form.html). Observation of GFP fluorescence by transiently expressing SRL1-GFP fusion protein (GFP was inserted at 364 amino acids upstream of the ω-site as the N-terminal signal peptide and C-terminal propeptide of SRL1 might be cut off for GPI processing) in the protoplasts of Arabidopsis and rice, and onion (Allium cepa) epidermal cells, respectively, revealed that SRL1-GFP indeed localizes predominantly at the plasma membrane (Fig. 4C; Supplemental Fig. S5).
Enhanced Expression of Genes Encoding Vacuolar H+-ATPase Subunits and H+-Pyrophosphatase under SRL1 Deficiency
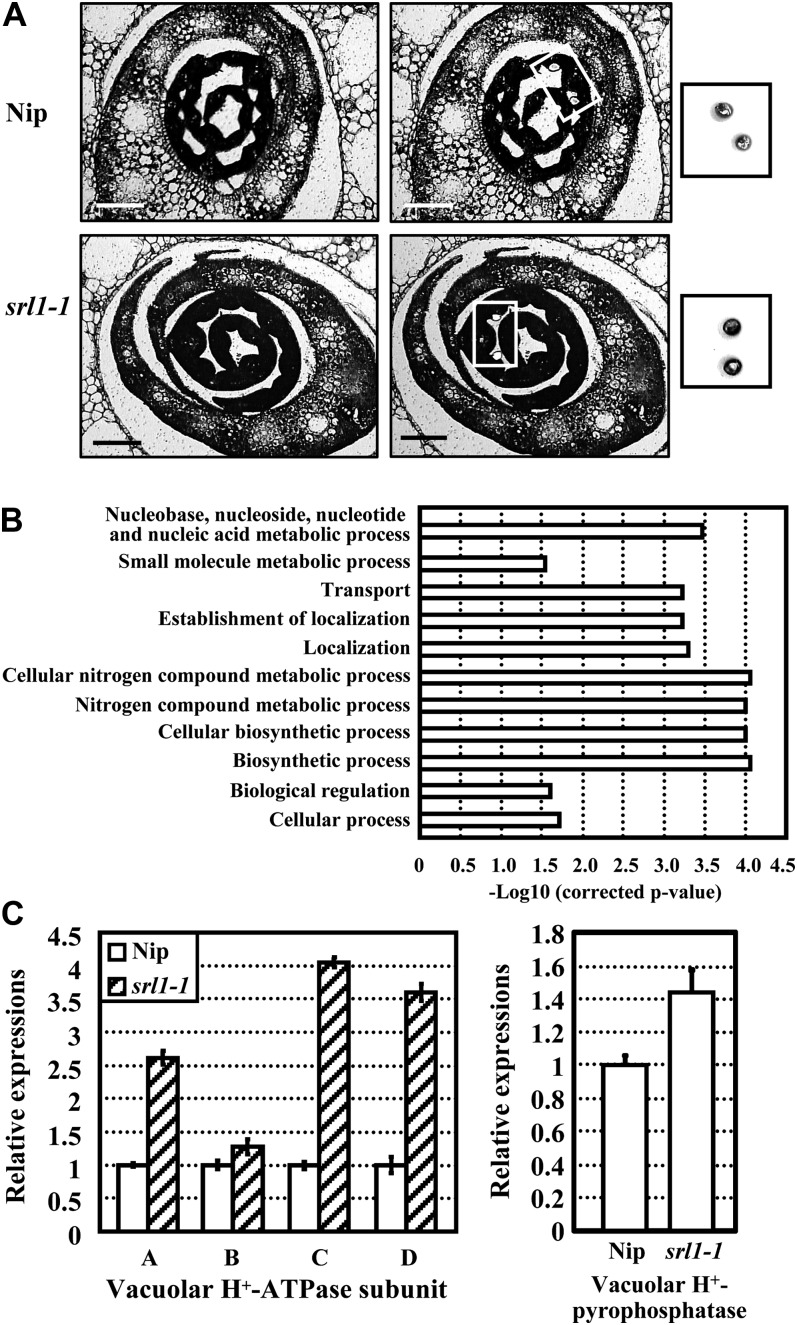
To further characterize the mechanism of incurved leaves in srl1-1 mutant, laser microdissection and microarray analysis were performed to enable the cell-specific gene expression profiling. Detailed cellular observation showed that in the third leaves of both wild-type and srl1-1 seedlings, bulliform cells were generally formed at 6 d after germination (DAG), while they couldn’t be observed at 5 DAG (Supplemental Fig. S6). To obtain the gene expression profiles just before the formation of bulliform cells, cells abutting the midrib at the adaxial surface of leaf blades that would become epidermal cells in wild type and probably bulliform cells in srl1-1 (5 DAG) were isolated by laser-captured microdissection, respectively (Fig. 5A), and total RNAs were then extracted and linearly amplified for microarray hybridization. Analysis of the hybridization data revealed that a total of 974 genes displayed altered expression in srl1-1, of which 473 genes were up-regulated and 501 genes were down-regulated (Supplemental Table S1). Functional analysis by gene ontology (GO) annotations showed that up-regulated genes were involved in multiple cellular processes, such as biosynthesis, metabolism, transport, and biological regulation (Fig. 5B). Interestingly, some vacuole-related genes, which may be crucial for bulliform cell formation, were up-regulated in srl1-1 (Table I). Further qRT-PCR analysis confirmed the increased expression levels of genes encoding vacuolar H+-pyrophosphatase and H+-ATPase (subunits A, B, C, and D) in srl1-1 (Fig. 5C), suggesting that the formation of bulliform cells may be coupled to increased expression of genes encoding vacuolar H+-ATPase subunits and H+-pyrophosphatase.
Figure 5.
Laser-captured microdissection and microarray, qRT-PCR analysis of srl1-1, and Nipponbare (Nip) plants revealed the enhanced expression of genes encoding vacuolar H+-ATPase subunits and vacuolar H+-pyrophosphatase in srl1-1. A, Isolation of cells from paraffin-embedded sections of Nip (top section) and srl1-1 (bottom section) seedlings after germination for 5 d by laser microdissection (left section, before laser microdissection; right section, after laser microdissection). The captured cells are highlighted. Bars = 100 μm. B, Main clusters identified by GO enrichment analysis of genes up-regulated in srl1-1 compared with Nip. In each GO biological process category, the bar indicates the fold enrichment, which is defined as −Log10 (corrected P value). The up-regulated genes are involved in various cellular processes, including biosynthesis, metabolism, transport, and biological regulation. C, qRT-PCR analysis of the expression levels of genes encoding vacuolar H+-ATPase subunits A, B, C, and D and vacuolar H+-pyrophosphatase in laser microdissection captured cells of Nip and srl1-1 confirmed the enhanced expression of these genes in srl1-1. Transcript levels were normalized with the ACTIN transcription and relative expression levels were compared with that in Nip (set as 1), respectively. Mean values were calculated from three independent experiments and error bars indicate sd.
Table I. Microarray analysis revealed that some vacuole-related genes are up-regulated in the srl1-1 mutant.
Cells abutting the midrib at the adaxial surface of leaf blades (5 DAG) that will become epidermal cells in wild type and probably bulliform cells in srl1-1 were isolated by laser microdissection and used for microarray analysis, respectively. Numbers indicate the fold change (FC, Log2).
| Locus No. | FC (Log2) | Descriptions |
|---|---|---|
| Os01g13130 | 1.1 | Tonoplast membrane integral protein ZmTIP4-3 |
| Os01g42430 | 1.2 | Vacuolar ATP synthase 21-kD proteolipid subunit C |
| Os01g55260 | 1.5 | SKD1 protein (Vacuolar sorting protein4b) |
| Os01g59800 | 0.7 | Vacuolar protein sorting36 family protein |
| Os02g07870 | 0.8 | Vacuolar proton-ATPase subunit A |
| Os02g24134 | 1.1 | Vacuolar protein-sorting protein45 homolog (AtVPS45) |
| Os03g15650 | 1.1 | Vacuolar sorting protein9 domain-containing protein |
| Os03g58700 | 0.7 | Vacuolar protein sorting-associated protein35 family protein |
| Os04g52190 | 1.4 | Vacuolar sorting receptor homolog |
| Os04g55040 | 1.2 | Vacuolar ATP synthase subunit D (EC 3.6.3.14) |
| Os05g06480 | 2.2 | Pyrophosphate-energized vacuolar membrane proton pump (EC 3.6.1.1) |
| Os05g51530 | 2.1 | Vacuolar ATP synthase subunit C (EC 3.6.3.14) |
| Os06g37180 | 1 | Vacuolar ATPase B subunit. |
| Os06g43660 | 0.6 | Vacuolar H+-pyrophosphatase (EC 3.6.1.1) |
DISCUSSION
SRL1 Negatively Regulates the Formation of Bulliform Cells and Modulates Leaf Rolling
In grass species such as rice, leaf rolling is induced by water loss from bulliform cells on the leaf upper epidermis (O’toole and Cruz, 1980), therefore the number and density of bulliform cells may affect the extent of leaf rolling. However, there has been controversy over the roles of bulliform cells in leaf rolling (Moulia, 2000). Studies by Shield (1951) demonstrated that water loss from the adaxial subepidermal sclerenchyma and mesophyll also contributed to leaf rolling, and rolling could occur in leaves lacking bulliform cells. In our study, srl1-1 and srl1-2 mutants displayed adaxially rolled leaves resulted from increased number of bulliform cells on the adaxial side of leaf blades, which may substantiate the role of bulliform cells in controlling leaf rolling.
Although the number of bulliform cells increased in loss-of-function mutants of ADL1 (Hibara et al., 2009) and OUL1 (Zou et al., 2011), cosuppression plants of YABBY1 (Dai et al., 2007), and gain-of-function mutant of ACL1 (BY240; Li et al., 2010), the leaves of these mutants exhibit abaxial rolling. The difference in the direction of leaf rolling may be attributed to the following reasons: (1) bulliform cells were ectopically formed on the abaxial side of leaves in adl1 and BY240, which is a characteristic of adaxialization, whereas no bulliform cells existed on the abaxial surface of leaves in srl1-1 and srl1-2; (2) in oul1 and cosuppression plants of YABBY1, overproliferation of bulliform cells resulted in the increased number of bulliform cells, while the total number of cells on the adaxial side of leaf blades did not change in srl1-1 and srl1-2, suggesting that more bulliform cells and less epidermal cells were formed from the early undifferentiated epidermis. Therefore, SRL1 may control leaf rolling by negatively regulating the formation of bulliform cells. In rice, the young leaf is adaxially rolled after differentiation from the leaf primordium, and unrolling occurs when the leaf blade emerges from the leaf sheath. The leaves of srl1-1 and srl1-2 continuously exhibit adaxial rolling after the emergence from leaf sheath, indicating the defects in regulation of unrolling. Compared with the thick and cutinized cell walls of epidermal cells, bulliform cells are large, thin walled, and highly vacuolated, and the increase of bulliform cells in srl1-1 and srl1-2 mutants may result in altered mechanical properties of the leaf adaxial surface, leading to adaxial rolling of leaves.
However, leaves of transgenic rice lines overexpressing SRL1 driven by its own native promoter were the same as that of wild type and showed no difference in the number of bulliform cells (Supplemental Fig. S7), and overexpression of SRL1 ubiquitously under maize ubiquitin promoter in wild-type plants does not result in the alteration of leaves (similar to wild type, data not shown), suggesting the complex regulation of the formation of bulliform cells.
In Arabidopsis, leaf rolling is often correlated with the polarity change in the adaxial-abaxial axis (Kidner and Timmermans, 2007). However, no polarity change was observed in the leaves of srl1-1 and srl1-2 mutants, or SRL1-overexpression transgenic lines, suggesting that SRL1 is not involved in the polarity establishment of leaf blades.
SRL1 Is a Putative GAP
SRL1 encodes a putative GAP located at the plasma membrane. GPI anchoring has been posited to be an alternative means to attach a protein to the membrane in all eukaryotic organisms (Udenfriend and Kodukula, 1995). A GPI moiety is covalently attached to the carboxy terminus of the protein (ω-site) after the cleavage of a hydrophobic C-terminal propeptide in the endoplasmic reticulum by the transamidase complex. The anchor of GAPs can be removed by specific phospholipases, thereby releasing the protein from the membrane (Griffith and Ryan, 1999). Genomic analysis has identified 245 and 294 GAPs in Arabidopsis and rice, respectively (Eisenhaber et al., 2003), and the reported functions of Arabidopsis GAPs are of great diversity, including cell expansion (COBRA, Schindelman et al., 2001; Roudier et al., 2005), root elongation under salt stress (SOS5/FLA4, Shi et al., 2003), cuticular lipid export (LTPG, Debono et al., 2009), callose deposition and plant cell-to-cell communication (PDCB, Simpson et al., 2009), vascular development (AtXYP1 and AtXYP2, Motose et al., 2004), double fertilization and early seed development (LORELEI, Capron et al., 2008; Tsukamoto et al., 2010), and disease resistance (PMR6, Vogel et al., 2002; NDR1, Coppinger et al., 2004).
GAPs are abundant and of great diversity in rice, and constitute a large family that can be divided into many functional groups such as COBRA, arabinogalactan protein, and lipid transfer protein like (Eisenhaber et al., 2003). BRITTLE CULM1, a homolog of COBRA in rice, regulates the synthesis of secondary cell wall and affects the mechanical strength of rice plants (Li et al., 2003; Dai et al., 2011). However, SRL1 belongs to none of these groups and does not contain any conserved domain. The mutation in srl1-1 confers a single amino acid change from hydrophilic Thr to hydrophobic Ile, which may lead to changes of protein structure and affect its interaction with other proteins. Homology analysis identified SRL1 homologs in Arabidopsis, maize, and S. bicolor, which are also predicted to be GPI anchored. Interestingly, leaves of knockdown mutants of SRL1 homologous gene in Arabidopsis (At1g61900) exhibited no observable phenotype, thus it would be of great interest to investigate whether SRL1 homologs in maize and S. bicolor also have effects on the formation of bulliform cells to modulate leaf rolling.
SRL1 May Inhibit the Formation of Bulliform Cells by Suppressing the Expression of Genes Encoding Vacuolar H+-ATPase Subunits and H+-Pyrophosphatase
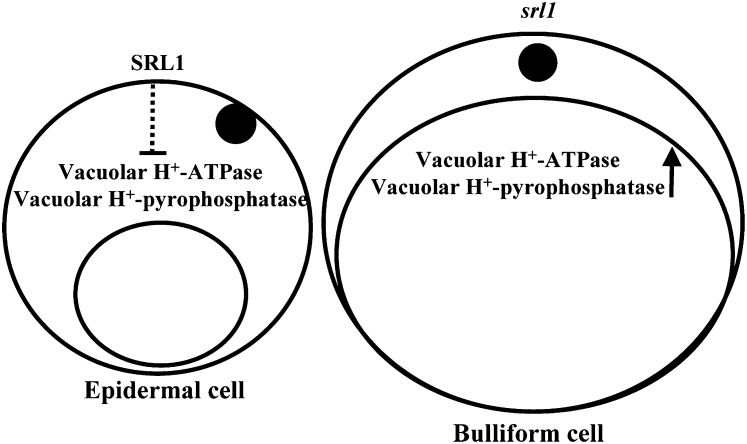
The plant vacuolar H+-ATPases and H+-pyrophosphatases create an electrochemical H+ gradient by pumping protons into the vacuolar lumen, which are essential for maintaining the cellular ionic and metabolic homeostasis (Ratajczak, 2000). Hence, the plant vacuolar H+-ATPases and H+-pyrophosphatases are indispensable for plant growth, development, and stress adaptation (Kluge et al., 2003). Vacuolar H+-ATPase consists of 12 subunits (Schumacher and Krebs, 2010) and previous studies indicated that Arabidopsis vacuolar H+-ATPase subunits C and E are involved in cell elongation and vacuole function, respectively (Schumacher et al., 1999; Strompen et al., 2005). Salt stress induced a rapid increase in vacuolar volume of suspension-cultured cells of mangrove (Bruguiera sexangula) and barley (Hordeum vulgare) root meristematic cells, which is correlated with the increase in the activity and amount of tonoplast H+-ATPase (Mimura et al., 2003). Arabidopsis H+-pyrophosphatase AVP1 was reported to regulate organ development by controlling auxin transport, and AVP1-overexpressing plants exhibited enhanced shoot and root growth owing to increased cell division (Li et al., 2005). Our studies indicated that the expression of genes encoding vacuolar H+-ATPase (subunits A, B, C, and D) and H+-pyrophosphatase were increased in srl1-1, which suggests that the formation of bulliform cells is accompanied by increased expression of genes encoding vacuolar H+-ATPase subunits and H+-pyrophosphatase (Fig. 6). It is supposed that higher expression of SRL1 in the formation of epidermal cells may suppress the expression of genes encoding vacuolar H+-ATPase (subunits A, B, C, and D) and vacuolar H+-pyrophosphatase, which may hinder the enlargement of vacuoles, while in the formation of bulliform cells, reduced expression of SRL1 may not be sufficient to repress the expression of genes encoding vacuolar H+-ATPase (subunits A, B, C, and D) and vacuolar H+-pyrophosphatase, resulting in the enlargement of vacuoles (Fig. 6), providing a hypothesis of how SRL1 controls the formation of bulliform and epidermal cells. However, the detailed mechanism of how SRL1 regulates the expression of genes encoding vacuolar H+-ATPase (subunits A, B, C, and D) and vacuolar H+-pyrophosphatase needs further studies.
Figure 6.
A hypothetical model of SRL1 function in the formation of bulliform cells. SRL1 may down-regulate the expression of genes encoding vacuolar H+-ATPase subunits A, B, C, and D and vacuolar H+-pyrophosphatase, to suppress the enlargement of vacuoles (left section) and formation of bulliform cells. Reduced expression of SRL1 leads to the enlargement of vacuoles and formation of bulliform cells (right section).
MATERIALS AND METHODS
Plant Material and Growth Conditions
The rice (Oryza sativa) mutant srl1-1 was identified from the mutagenized population of rice sp. japonica var Nipponbare treated with 1% ethyl methanesulfonate solution. Nipponbare represents the wild type for srl1-1. srl1-1 was crossed with flat-leaf indica rice var Tai Zhong Ben Di 1 and the resultant F1 plants were self-pollinated to produce the F2 progenies for constructing the F2 mapping population. Rice plants were cultivated in the experimental field under natural growth conditions. Transgenic rice plants were grown in a phytotron with a 12-h-light (28°C)/12-h-dark (22°C) cycle and 85% relative humidity.
Map-Based Cloning of SRL1
SRL1 was mapped primarily with SSR (http://www.gramene.org/microsat/ssr.html) and STS markers using 186 F2 mutant plants. Of 10,000 F2 plants, 2,342 segregants showing the srl1-1 mutant phenotype were used for fine mapping and the SRL1 gene was localized within a 21.8-kb region between two SSR markers S5869-3 and S5869-4. New molecular markers (Supplemental Table S2) were developed by comparing original or cleaved amplified polymorphic sequences between indica var 9311 and Nipponbare according to data published at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
The PCR procedure for mapping was as follows: 94°C for 4 min, followed by 35 cycles of 94°C, 30 s; annealing temperature for 30 s; 72°C, 40 s; and a final elongation step at 72°C for 10 min. The PCR products were analyzed on 5% to 6% agarose gels.
To define molecular lesions, 21.8-kb genomic DNA from the srl1-1 and relevant wild-type variety (Nipponbare) were amplified by PCR. All PCR products were sequenced and the candidate gene was amplified from both srl1-1 and Nipponbare genomic DNAs using different primers (Supplemental Table S3). Obtained sequences were analyzed by DNAMAN software (Version 5.2.2, Lynnon Biosoft).
The hydropathy profile was determined by the ProtScale program provided by ExPASy (http://web.expasy.org/protscale/; Kyte and Doolittle, 1982), the signal peptide was predicted by SignalP 4.0 server (http://www.cbs.dtu.dk/services/SignalP/; Petersen et al., 2011), and GPI modification was predicted using PredGPI (http://gpcr.biocomp.unibo.it/predgpi/; Pierleoni et al., 2008).
Protein databases were searched by BLASTP program of the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify homologs of SRL1 using the amino acid sequence as a query. All the sequences were aligned using ClustalX2.1 (Larkin et al., 2007), and the phylogenetic tree was generated using the maximum-likelihood method in MEGA5.05 (Tamura et al., 2011).
Characterization of Additional Alleles of srl1-1
srl1-2 was identified from SHIP (Fu et al., 2009) with the line ID SHIP_ZSF4805. Rice sp. japonica ‘Zhonghua 11’ (ZH11) represents the wild type for srl1-2. The single T-DNA insertion in srl1-2 was verified by Southern blot using the 598-bp hygromycin phosphotransferaseII gene fragment as a probe, followed by analysis of the segregation ratio by phenotype observation and genotyping. The sequence flanking the insertion site was determined by thermal asymmetric interlaced PCR (Liu and Whittier, 1995). SRL1 gene-specific primers (F4805-F: 5′-CCATGCCTATCGAGAAAAGTG-3′ and F4805-R: 5′-AGAGGTTGGCGATTTAGAAGG-3′) and T-DNA left border primers (NTL1: 5′-CACTCGTCCGAGGGCAAAGAAATAGA-3′, NTL2: 5′-ATAGGGTTTCGCTCATGTGTTGAGC-3′ and NTL3: 5′-TTTCTAATTCCTAAAACCAAAATCCAGTAC-3′) were used to confirm the insertion site and identify homozygous plants.
Histology and Microscopy Observation
Leaves at certain developmental stages were dissected, fixed by formalin-acetic acid-alcohol (FAA) solution (formaldehyde:glacial acetic acid:50% ethanol = 2:1:17), and dehydrated through a graded ethanol series. Samples were embedded in Epon812 resin (Fluka) and polymerized at 60°C. Cross sections (3 μm) were cut and stained with filtered 1% toluidine blue. Sections were microscopically examined and photographed to count the number of bulliform cells (Leica DMR). To analyze the arrangement of bulliform cells, toluidine blue O staining was performed according to the previous description (Li et al., 2010).
Scanning Electron Microscopy
The third leaves were excised from 14-d-old seedlings with a razor and fixed immediately by formalin-acetic acid-alcohol (FAA) solution overnight. After dehydration through a graded ethanol series, the samples were critical-point dried (Hitachi critical point dryer, HCP-2), sputter coated with gold in an E-100 ion sputter (Mitocity), and observed with a scanning electron microscope (Hitachi S-450).
Measurement of LRI
To determine the LRI, the distance of leaf blade margins at natural state (Ln) and the distance of leaf blade margins at unfolding state (Lw) in srl1-1, srl1-2, and the corresponding wild-type plants were measured, respectively, in the field at different developmental stages. LRIs were calculated by the formula: LRI (%) = (Lw − Ln)/Lw × 100.
In Situ Hybridization Analysis
Leaf samples of wild type and srl1-2 were fixed by 4% paraformaldehyde in 0.1 m sodium phosphate buffer, dehydrated through a graded ethanol series, replaced with xylene, and embedded in Paraplast plus (Sigma-Aldrich). Paraffin sections (8 μm) were cut by Leica microtome. A 340-bp gene-specific fragment of SRL1 gene was amplified by PCR using primers (5′-AGAGAGACCTGCTGCGACAT-3′ and 5′-GCGCATTTCACACCCTTTA-3′), subcloned into pGEM-T Easy vector (Promega), and used as template to generate digoxigenin-labeled sense and antisense probes (Roche). RNA in situ hybridization was performed as previously described (Luo et al., 1996).
Complementation Studies
An 11.1-kb genomic DNA fragment containing 2,597-bp promoter region, 8,864-bp coding sequence, and 670-bp downstream sequence of SRL1 gene was cut from bacterial artificial chromosome B1026C12 and subcloned into vector pCAMBIA1302. The construct was introduced into the Agrobacterium tumefaciens strain EHA105 by electroporation, and transformed into srl1-1 mutants using immature embryos as materials.
Subcellular Localization Studies of SRL1
To express the SRL1-GFP fusion protein, the whole coding sequence of SRL1 were divided into two fragments amplified by primers 5′-GGGATGGCCCCCGCCGGT-3′ and 5′-TCCGAGGGCATACTTGGA-3′, 5′-TGCATCTTTTGACCCAGCTAC-3′ and 5′-CTAGACTTGTAGCCAAATAGC-3′, respectively, and subcloned into the pA7 vector, resulting in the insertion of GFP at 364 amino acid of SRL1, upstream of the predicted C-terminal GPI-modification site (ω-site). The resultant construct was introduced into Arabidopsis (Arabidopsis thaliana) mesophyll protoplasts (Sheen, 2001), rice protoplasts of leaf sheath (Zhai et al., 2009), and onion (Allium cepa) epidermal cells (Yang et al., 2011), respectively, and the green fluorescence was observed by confocal laser-scanning microscopy (Zeiss LSM 510 META) with an argon laser excitation wavelength of 488 nm.
Laser-Captured Microdissection and Microarray Analysis
Leaves of wild-type and srl1-1 seedlings at 5 DAG were dissected, fixed by a fixation solution (ethanol:acetate, 75:25), and embedded by Paraplast plus (Sigma-Aldrich) as described (Takahashi et al., 2010). Cross sections (10 μm) were cut on a rotary microtome (Leica Microtome), floated on the RNAsecure (Ambion) solution on the polyethylene naphthalate membrane glass slides (Leica). Then the solution was wiped off by RNase-free paper and the slides were dried at 4°C for 1 h. To remove paraffin, the slides were immersed in xylene for 5 min twice and air dried at room temperature. Cells were isolated using the veritas microdissection system (Arcturus/MolecularDevices). Two biological replicates were captured for RNA extraction, respectively.
Total RNAs were extracted by the PicoPure RNA isolation kit (Arcturus/Molecular Devices). The integrity of RNA samples was evaluated by an Agilent 2100 bioanalyzer using RNA-6000 Pico LabChips (Agilent Technologies). Approximately 500 ng of total RNA was linearly amplified using a TargetAmp two-round amino-allyl antisense RNA (aRNA) amplification kit (Epicentre Biotechnologies) with SuperScript III and SuperScript II reverse transcriptases (Invitrogen), which yields approximately 25 μg amino-allyl aRNA.
Amino-allyl aRNAs were coupled with Cy3 monoreactive N-hydroxysuccinimide esters, hybridized to rice 4 × 44K oligomicroarray (Agilent Technologies). After washing, arrays were scanned by Agilent microarray scanner and Feature Extraction software 10.7 with default settings. Raw data were normalized by quantile algorithm, Gene Spring software 11.0 (Agilent technologies), and were accompanied by a flag: present or absent. Least-square linear regression was carried out to evaluate the repeatability of the microarray, and the r2 statistic of each sample was higher than 0.99. The microarray data have been deposited into the Gene Expression Omnibus database under accession number GSE37140 (http://www.ncbi.nlm.nih.gov/geo). Genes absent in both wild type and srl1-1 were removed for subsequent analysis. Differentially expressed genes between the wild-type and srl1-1 mutant plants were determined by t test (P < 0.05) with the threshold of 2-fold change. GO enrichment analysis of differentially expressed genes was conducted by BinGO in Cytoscape (Maere et al., 2005).
RT-PCR and qRT-PCR Analysis
Total RNAs were extracted using TRIzol reagent (Invitrogen), reverse transcribed into cDNA according to the manufacturer’s instructions (PrimeScript RT-PCR kit with gDNA eraser, Takara). Fragments covering the srl1-1 mutation site were amplified by primers (5′-CTCATCTCCTGCCTCCTCTGT-3′ and 5′-GCATCGTTGATAGCATTCTGG-3′) from cDNAs of independent transgenic plants with complementary expression of SRL1 and sequenced.
qRT-PCR was carried out using the Rotor-Gene realtime thermocycler R3000 (Corbett Research) with realtime PCR master mix (SYBR green, Toyobo). For analysis of SRL1 transcription in srl1-2, a fragment covering the insertion site was amplified using primers 5′-CCCAATTCCTTGGTGACAAGT-3′ and 5′-TTGCATGAAGAAGACGATGCT-3′. The primers used to test SRL1 expression in various tissues were 5′-TGCATCTTTTGACCCAGCTAC-3′ and 5′-ACAACAAGGGTGCCCAGATAA-3′. A linear standard curve was generated using a series of dilutions of PCR product for each gene, and the transcription levels were determined according to the standard curve. The rice ACTIN gene (Os03g50890) was amplified with primers (5′-TCCATCTTGGCATCTCTCAG-3′ and 5′-GTACCCGCATCAGGCATCTG-3′) and used as internal standard to normalize the expression of SRL1 and other tested genes.
To validate the microarray data, the expression of following vacuole-related genes were examined: vacuolar H+-pyrophosphatase (Os06g43660, 5′-TTCCCTCCGTTTCCTATTTGT-3′ and 5′-CCATCATCGTATCATCCAACC-3′), vacuolar H+-ATPase subunit A (Os02g07870, 5′-CCGCAGAGGGCAAAGAAGT-3′ and 5′-TCGGTAGCAGCAAATGGAATG-3′), subunit B (Os06g37180, 5′-AGCAGAGATGCTACCCACTGA-3′ and 5′-ACAGAAGTTGCCGTGACAAAG-3′), subunit C (Os01g42430, 5′-CGGGGAGACCTATCAGAAGAA-3′ and 5′-TTCAATACGCAGACCACACTC-3′), and subunit D (Os04g55040, 5′-CAGGGTTACAAGAAGAGGGAGA-3′ and 5′-GCATCAATACTACGCACCGATT-3′).
All the experiments were repeated for three times with biological duplicates, and the data were statistically analyzed and presented as means ± sd.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of srl1-1 and srl1-2 leaves by scanning electron microscope.
Supplemental Figure S2. Hydropathy and phylogenetic analyses of SRL1.
Supplemental Figure S3. Analysis of srl1-2 mutant.
Supplemental Figure S4. Reduced SRL1 expression in srl1-2 mutant.
Supplemental Figure S5. Subcellular localization of the SRL1-GFP fusion protein.
Supplemental Figure S6. Section analysis of srl1-1 and wild-type (Nipponbare) plants.
Supplemental Figure S7. Overexpression of SRL1 has no effect on leaf morphology.
Supplemental Table S1. Microarray data for the 973 differentially expressed genes in srl1-1.
Supplemental Table S2. New molecular markers used in map-based cloning of SRL1.
Supplemental Table S3. Sequencing primers used in map-based cloning of SRL1.
Supplementary Material
Acknowledgments
We greatly thank Ms. Shu-Ping Xu for rice transformation, Ms. Ji-Qin Li for resin sections, and Dr. Wei-Hua Tang for assistance in laser-captured microdissection analysis.
Glossary
- DAG
d after germination
- GAP
GPI-anchored protein
- LRI
leaf-rolling index
- STS
sequence-tagged site
- SSR
simple sequence repeats
- RT
reverse transcription
- SHIP
Shanghai T-DNA insertion population
- q
quantitative
- qRT
quantitative reverse transcription
- aRNA
antisense RNA
- GO
gene ontology
- T-DNA
transferred DNA
- ZH11
Zhonghua 11
- cDNA
complementary DNA
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Alvarez JM, Rocha JF, Machado SR. (2008) Bulliform cells in Loudetiopsis chrysothrix (Nees) Conert and Tristachya leiostachya Nees (Poaceae): structure in relation to function. Braz Arch Biol Technol 51: 113–119 [Google Scholar]
- Bowman JL, Eshed Y, Baum SF. (2002) Establishment of polarity in angiosperm lateral organs. Trends Genet 18: 134–141 [DOI] [PubMed] [Google Scholar]
- Capron A, Gourgues M, Neiva LS, Faure JE, Berger F, Pagnussat G, Krishnan A, Alvarez-Mejia C, Vielle-Calzada JP, Lee YR, et al. (2008) Maternal control of male-gamete delivery in Arabidopsis involves a putative GPI-anchored protein encoded by the LORELEI gene. Plant Cell 20: 3038–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppinger P, Repetti PP, Day B, Dahlbeck D, Mehlert A, Staskawicz BJ. (2004) Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J 40: 225–237 [DOI] [PubMed] [Google Scholar]
- Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou DX. (2007) The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol 144: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, You C, Chen G, Li X, Zhang Q, Wu C. (2011) OsBC1L4 encodes a COBRA-like protein that affects cellulose synthesis in rice. Plant Mol Biol 75: 333–345 [DOI] [PubMed] [Google Scholar]
- Debono A, Yeats TH, Rose JKC, Bird D, Jetter R, Kunst L, Samuels L. (2009) Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21: 1230–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber B, Wildpaner M, Schultz CJ, Borner GHH, Dupree P, Eisenhaber F. (2003) Glycosylphosphatidylinositol lipid anchoring of plant proteins: sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol 133: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Fleurat-Lessard P, Frangne N, Maeshima M, Ratajczak R, Bonnemain JL, Martinoia E. (1997) Increased expression of vacuolar aquaporin and H+-ATPase related to motor cell function in Mimosa pudica L. Plant Physiol 114: 827–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu FF, Ye R, Xu SP, Xue HW. (2009) Studies on rice seed quality through analysis of a large-scale T-DNA insertion population. Cell Res 19: 380–391 [DOI] [PubMed] [Google Scholar]
- Fujino K, Matsuda Y, Ozawa K, Nishimura T, Koshiba T, Fraaije MW, Sekiguchi H. (2008) NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Genet Genomics 279: 499–507 [DOI] [PubMed] [Google Scholar]
- Griffith OH, Ryan M. (1999) Bacterial phosphatidylinositol-specific phospholipase C: structure, function, and interaction with lipids. Biochim Biophys Acta 1441: 237–254 [DOI] [PubMed] [Google Scholar]
- Hernandez ML, Passas HJ, Smith LG. (1999) Clonal analysis of epidermal patterning during maize leaf development. Dev Biol 216: 646–658 [DOI] [PubMed] [Google Scholar]
- Hibara K, Obara M, Hayashida E, Abe M, Ishimaru T, Satoh H, Itoh J, Nagato Y. (2009) The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev Biol 334: 345–354 [DOI] [PubMed] [Google Scholar]
- Hu J, Zhu L, Zeng D, Gao Z, Guo L, Fang Y, Zhang G, Dong G, Yan M, Liu J, et al. (2010) Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol Biol 73: 283–292 [DOI] [PubMed] [Google Scholar]
- Ito M, Sentoku N, Nishimura A, Hong SK, Sato Y, Matsuoka M. (2003) Roles of rice GL2-type homeobox genes in epidermis differentiation. Breed Sci 53: 245–253 [Google Scholar]
- Itoh J, Hibara K, Sato Y, Nagato Y. (2008) Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol 147: 1960–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Kadioglu A, Terzi R. (2007) A dehydration avoidance mechanism: leaf rolling. Bot Rev 73: 290–302 [Google Scholar]
- Khush GS, Kinoshita T. (1991) Rice karyotype, marker genes, and linkage groups. In GS Khush, GH Toenniesen, eds, Rice Biotechnology. CAB International, Wallingford, UK, pp 83–108
- Kidner CA, Timmermans MC. (2007) Mixing and matching pathways in leaf polarity. Curr Opin Plant Biol 10: 13–20 [DOI] [PubMed] [Google Scholar]
- Kluge C, Lahr J, Hanitzsch M, Bolte S, Golldack D, Dietz KJ. (2003) New insight into the structure and regulation of the plant vacuolar H+-ATPase. J Bioenerg Biomembr 35: 377–388 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li JS, Yang HB, Peer WA, Richter G, Blakeslee J, Bandyopadhyay A, Titapiwantakun B, Undurraga S, Khodakovskaya M, Richards EL, et al. (2005) Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310: 121–125 [DOI] [PubMed] [Google Scholar]
- Li L, Shi ZY, Li L, Shen GZ, Wang XQ, An LS, Zhang JL. (2010) Overexpression of ACL1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol Plant 3: 807–817 [DOI] [PubMed] [Google Scholar]
- Li SG, Ma YQ, He P, Li HY, Chen Y, Zhou KD, Zhu LH. (1998) Genetics analysis and mapping the flag leaf roll in rice (Oryza sativa L.). J Sichuan Agric Uni 16: 391–393 [Google Scholar]
- Li YH, Qian Q, Zhou YH, Yan MX, Sun L, Zhang M, Fu ZM, Wang YH, Han B, Pang XM, et al. (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15: 2020–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681 [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E. (1996) Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Luo YZ, Zhao FM, Sang XC, Ling YH, Yang ZL, He GH. (2009) Genetic analysis and gene mapping of a novel rolled leaf mutant rl12(t) in rice. Acta Agron Sin 35: 1967–1972 [Google Scholar]
- Luo Z, Yang Z, Zhong B, Li Y, Xie R, Zhao F, Ling Y, He G. (2007) Genetic analysis and fine mapping of a dynamic rolled leaf gene, RL10(t), in rice (Oryza sativa L.). Genome 50: 811–817 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. (2005) BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Micol JL, Hake S. (2003) The development of plant leaves. Plant Physiol 131: 389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura T, Kura-Hotta M, Tsujimura T, Ohnishi M, Miura M, Okazaki Y, Mimura M, Maeshima M, Washitani-Nemoto S. (2003) Rapid increase of vacuolar volume in response to salt stress. Planta 216: 397–402 [DOI] [PubMed] [Google Scholar]
- Moon J, Hake S. (2011) How a leaf gets its shape. Curr Opin Plant Biol 14: 24–30 [DOI] [PubMed] [Google Scholar]
- Moran N. (2007) Osmoregulation of leaf motor cells. FEBS Lett 581: 2337–2347 [DOI] [PubMed] [Google Scholar]
- Motose H, Sugiyama M, Fukuda H. (2004) A proteoglycan mediates inductive interaction during plant vascular development. Nature 429: 873–878 [DOI] [PubMed] [Google Scholar]
- Moulia B. (2000) Leaves as shell structures: double curvature, auto-stresses, and minimal mechanical energy constraints on leaf rolling in grasses. J Plant Growth Regul 19: 19–30 [DOI] [PubMed] [Google Scholar]
- O’toole JC, Cruz RT. (1980) Response of leaf water potential, stomatal resistance, and leaf rolling to water stress. Plant Physiol 65: 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Pierleoni A, Martelli PL, Casadio R. (2008) PredGPI: a GPI-anchor predictor. BMC Bioinformatics 9: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AH, Young EM, Tomos AD. (1997) Quantitative trait loci associated with stomatal conductance, leaf rolling and heading date mapped in upland rice (Oryza sativa). New Phytol 137: 83–91 [Google Scholar]
- Ratajczak R. (2000) Structure, function and regulation of the plant vacuolar H(+)-translocating ATPase. Biochim Biophys Acta 1465: 17–36 [DOI] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GHH, Schindelman G, Song S, Baskin TI, Dupree P, Wasteneys GO, et al. (2005) COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17: 1749–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN. (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K, Krebs M. (2010) The V-ATPase: small cargo, large effects. Curr Opin Plant Biol 13: 724–730 [DOI] [PubMed] [Google Scholar]
- Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J. (1999) The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev 13: 3259–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao YJ, Chen ZX, Zhang YF, Chen EH, Qi DC, Miao J, Pan XB. (2005a) One major QTL mapping and physical map construction for rolled leaf in rice. Yi Chuan Xue Bao 32: 501–506 [PubMed] [Google Scholar]
- Shao YJ, Pan CH, Chen ZX, Zuo SM, Zhang YF, Pan XB. (2005b) Fine mapping of an incomplete recessive gene for leaf rolling in rice (Oryza sativa L.). Chin Sci Bull 50: 2466–2472 [Google Scholar]
- Sheen J. (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK. (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Chen J, Liu W, Huang Q, Shen B, Leung H, Wu J. (2009) Genetic analysis and gene mapping of a new rolled-leaf mutant in rice (Oryza sativa L.). Sci China C Life Sci 52: 885–890 [DOI] [PubMed] [Google Scholar]
- Shield LM. (1951) The involution mechanism in leaves of certain xeric grasses. Phytomorphology 1: 225–241 [Google Scholar]
- Simpson C, Thomas C, Findlay K, Bayer E, Maule AJ. (2009) An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompen G, Dettmer J, Stierhof YD, Schumacher K, Jürgens G, Mayer U. (2005) Arabidopsis vacuolar H-ATPase subunit E isoform 1 is required for Golgi organization and vacuole function in embryogenesis. Plant J 41: 125–132 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kamakura H, Sato Y, Shiono K, Abiko T, Tsutsumi N, Nagamura Y, Nishizawa NK, Nakazono M. (2010) A method for obtaining high quality RNA from paraffin sections of plant tissues by laser microdissection. J Plant Res 123: 807–813 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Qin Y, Huang Y, Dunatunga D, Palanivelu R. (2010) A role for LORELEI, a putative glycosylphosphatidylinositol-anchored protein, in Arabidopsis thaliana double fertilization and early seed development. Plant J 62: 571–588 [DOI] [PubMed] [Google Scholar]
- Udenfriend S, Kodukula K. (1995) How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem 64: 563–591 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC. (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YM, Park HJ, Su’udi M, Yang JI, Park JJ, Back K, Park YM, An G. (2007) Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol Biol 65: 125–136 [DOI] [PubMed] [Google Scholar]
- Wu XJ. (2009) Prospects of developing hybrid rice with super high yield. Agron J 101: 688–695 [Google Scholar]
- Yan CJ, Yan S, Zhang ZQ, Liang GH, Lu JF, Gu MH. (2006) Genetic analysis and gene fine mapping for a rice novel mutant rl9(t) with rolling leaf character. Chin Sci Bull 51: 63–69 [Google Scholar]
- Yan S, Yan CJ, Zeng XH, Yang YC, Fang YW, Tian CY, Sun YW, Cheng ZK, Gu MH. (2008) ROLLED LEAF 9, encoding a GARP protein, regulates the leaf abaxial cell fate in rice. Plant Mol Biol 68: 239–250 [DOI] [PubMed] [Google Scholar]
- Yang X, Yang YN, Xue LJ, Zou MJ, Liu JY, Chen F, Xue HW. (2011) Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol 156: 1397–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan LP. (1997) Super-high yield hybrid rice breeding. Hybrid Rice 12: 1–6 [Google Scholar]
- Zhai Z, Sooksa-nguan T, Vatamaniuk OK. (2009) Establishing RNA interference as a reverse-genetic approach for gene functional analysis in protoplasts. Plant Physiol 149: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GH, Xu Q, Zhu XD, Qian Q, Xue HW. (2009) SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21: 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou LP, Sun XH, Zhang ZG, Liu P, Wu JX, Tian CJ, Qiu JL, Lu TG. (2011) Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiol 156: 1589–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.