Abstract
TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR1 (TCP) transcription factors control developmental processes in plants. The 24 TCP transcription factors encoded in the Arabidopsis (Arabidopsis thaliana) genome are divided into two classes, class I and class II TCPs, which are proposed to act antagonistically. We performed a detailed phenotypic analysis of the class I tcp20 mutant, showing an increase in leaf pavement cell sizes in 10-d-old seedlings. Subsequently, a glucocorticoid receptor induction assay was performed, aiming to identify potential target genes of the TCP20 protein during leaf development. The LIPOXYGENASE2 (LOX2) and class I TCP9 genes were identified as TCP20 targets, and binding of TCP20 to their regulatory sequences could be confirmed by chromatin immunoprecipitation analyses. LOX2 encodes for a jasmonate biosynthesis gene, which is also targeted by class II TCP proteins that are under the control of the microRNA JAGGED AND WAVY (JAW), although in an antagonistic manner. Mutation of TCP9, the second identified TCP20 target, resulted in increased pavement cell sizes during early leaf developmental stages. Analysis of senescence in the single tcp9 and tcp20 mutants and the tcp9tcp20 double mutants showed an earlier onset of this process in comparison with wild-type control plants in the double mutant only. Both the cell size and senescence phenotypes are opposite to the known class II TCP mutant phenotype in JAW plants. Altogether, these results point to an antagonistic function of class I and class II TCP proteins in the control of leaf development via the jasmonate signaling pathway.
TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR1 (TCP) proteins are plant-specific transcription factors that are involved in growth-related processes, such as branching, floral organ morphogenesis, and leaf growth (for review, see Martín-Trillo and Cubas, 2010). The Arabidopsis (Arabidopsis thaliana) genome encodes for 24 TCP transcription factor genes, which, based on sequence homology, are divided into two classes: class I and class II TCPs. Functional analysis of the Arabidopsis class II TCP genes BRANCHED1 (BRC1) and BRC2, both closely related to the TCP founder gene TEOSINTE BRANCHED1 from maize (Zea mays; Doebley et al., 1997), demonstrated that these genes are involved in suppressing axillary bud outgrowth (Aguilar-Martínez et al., 2007). Another subclass of the class II TCPs contains the genes TCP2, TCP3, TCP4, TCP10, and TCP24, which are all targets of the microRNA miR319a/JAGGED AND WAVY (JAW; Palatnik et al., 2003). Simultaneous down-regulation of these five TCPs by ectopic expression of miR319a/JAW in jaw-d plants results in abnormal curvature and excessive growth of leaves. Conversely, expression of a hyperactivated form of TCP4 results in decreased cell proliferation and smaller leaves (Sarvepalli and Nath, 2011). JAW-regulated expression of TCP4 is also important for the proper development of petals and stamens in the Arabidopsis flower, because expression of a JAW-resistant TCP4 under the control of an APETALA3 promoter disrupted the development of these flower organs. Likewise, expression of wild-type TCP4 under the control of the same promoter disrupted petal and stamen development only in the background of an miR319a knockout (Nag et al., 2009). Down-regulation of the Armadillo BTB Arabidopsis protein, a protein that is associated with TCP24, increased cell division rates, while overexpression resulted in reduced leaf growth (Masuda et al., 2008). Altogether, these examples of class II TCP functions suggest that they play a prominent role in suppressing organ growth processes, although it is not clear how they control these. jaw-d plants show, apart from their leaf growth phenotype, late entry into leaf senescence (Schommer et al., 2008). This late senescence behavior is caused by altered jasmonic acid (JA) levels. Furthermore, it was shown that class II TCPs, specifically TCP4, directly influence JA biosynthesis by regulating the expression of LIPOXYGENASE2 (LOX2; Schommer et al., 2008). LOX2 catalyzes the reaction from α-linoleic acid to 13(S)-hydroperoxylinolenic acid, which represents one of the first steps of JA synthesis in plants (Vick and Zimmerman, 1983).
In contrast to class II TCPs, much less functional information is available for class I TCPs. Two studies focused on the class I gene TCP20. Li et al. (2005) described a targeted chromatin immunoprecipitation (ChIP) assay, in which TCP20 was found to bind regulatory sequences of CYCLIN B1, PROLIFERATING CELL NUCLEAR ANTIGEN, and ribosomal genes. Based on these targets, it was suggested that TCP20 stimulates the cell cycle and growth of organs (Li et al., 2005). Because the tcp20 mutant does not show an obvious phenotype, Hervé and colleagues (2009) used an alternative approach to gain functional information about TCP20. They generated plants ectopically expressing the TCP20 protein, which was tagged with an EAR transcriptional repressor domain. The 35S:TCP20-EAR construct caused pleiotropic growth effects, and genome-wide expression analysis revealed a plethora of genes differentially expressed in the TCP20-EAR plants compared with the wild type.
Molecular analyses showed that the predicted consensus DNA-binding sites of the two TCP classes are partly overlapping. The consensus binding site of class I TCPs is GGNCCCAC, which includes the predicted binding site for TCP20 (GCCCR), whereas class II TCPs bind DNA motifs of the sequence GTGGNCCC (Kosugi and Ohashi, 2002; Li et al., 2005). TCPs of the two different classes are believed to share common targets, and it has been hypothesized that these targets are antagonistically regulated by competing TCP activities (Li et al., 2005). However, evidence for this hypothesis is lacking, and such common target genes for class I and class II TCPs remain to be identified.
Here, we report on the role of class I TCPs in Arabidopsis leaf development based on the detailed analysis of the tcp20 mutant and two selected TCP20 targets. We identified LOX2 as a common target of TCP20 and TCP4, where TCP20 inhibits and TCP4 induces LOX2 expression. The second identified target is TCP9, and a tcp9 mutant showed an increase in pavement cell size in leaves of 10-d-old seedlings, resembling the tcp20 mutant phenotype. Based on these results, a model is proposed in which class I TCP proteins act at least partially antagonistic to the class II jaw-TCPs in regulating different phases of leaf development via the regulation of LOX2 expression and JA signaling.
RESULTS
TCP20 Is Expressed throughout Arabidopsis Development
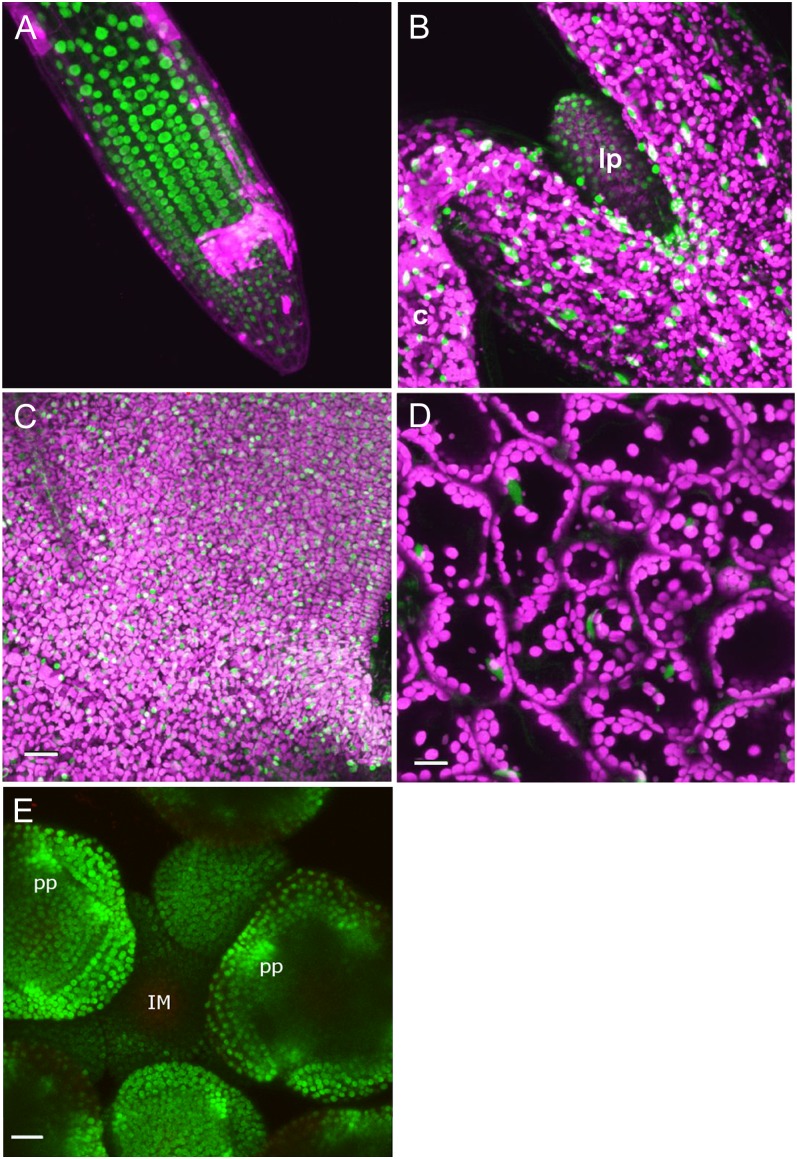
Plants expressing a genomic version of TCP20 tagged with a GFP-encoding sequence under the control of the endogenous TCP20 promoter (gTCP20-GFP) were analyzed at different time points after germination to obtain detailed information about the developmental expression pattern of TCP20. Fluorescence was detected by confocal laser scanning microscopy, as shown in Figure 1. Expression was observed in several organs throughout development, like young and mature leaves, differentiating root cells, and young floral buds (Fig. 1). In agreement with its function as a transcription factor, TCP20-GFP signal was predominantly present in nuclei. Strong TCP20-GFP signal could be detected in cells of the differentiating root (Fig. 1A). In developing leaves, we detected TCP20-GFP signal in the first leaf primordia of seedlings 3 d after germination (Fig. 1B) and in the first leaf of 2-week-old (Fig. 1C) to 4-week-old (Fig. 1D) plants. We also detected TCP20-GFP during floral development in all cells of young floral buds (Fig. 1E), while no signal was obtained in the inflorescence meristem. In stage 3 flower buds, TCP20 expression is strong and peaks at the position of the petal anlagen, suggesting a function for TCP20 in petal initiation. As we were interested in the role of TCP20 in leaf development, all further experiments were conducted on seedlings and leaves during vegetative development.
Figure 1.
Localization of TCP20-GFP in different plant tissues visualized by confocal laser scanning microscopy. TCP20-GFP signal is depicted in green, and autofluorescence of plastids is depicted in magenta. TCP20-GFP signal is mainly nucleus localized and can be detected in particular cell lineages of differentiating roots (A), the first leaf primordia in 3-d-old seedlings (B), the majority of leaf cells of the first initiated leaf, 14 d after germination (C), leaves 28 d after germination (D), and young flower buds at different developmental stages (E). Note that in the young floral meristems, strong expression is observed in the anlagen for petal primordia. c, Cotyledon; IM, inflorescence meristem; lp, leaf primordium; pp, petal primordium. Bars = 25 µm.
Detailed Analysis of the tcp20 Mutant Reveals a Cell Size Phenotype in Developing Leaves
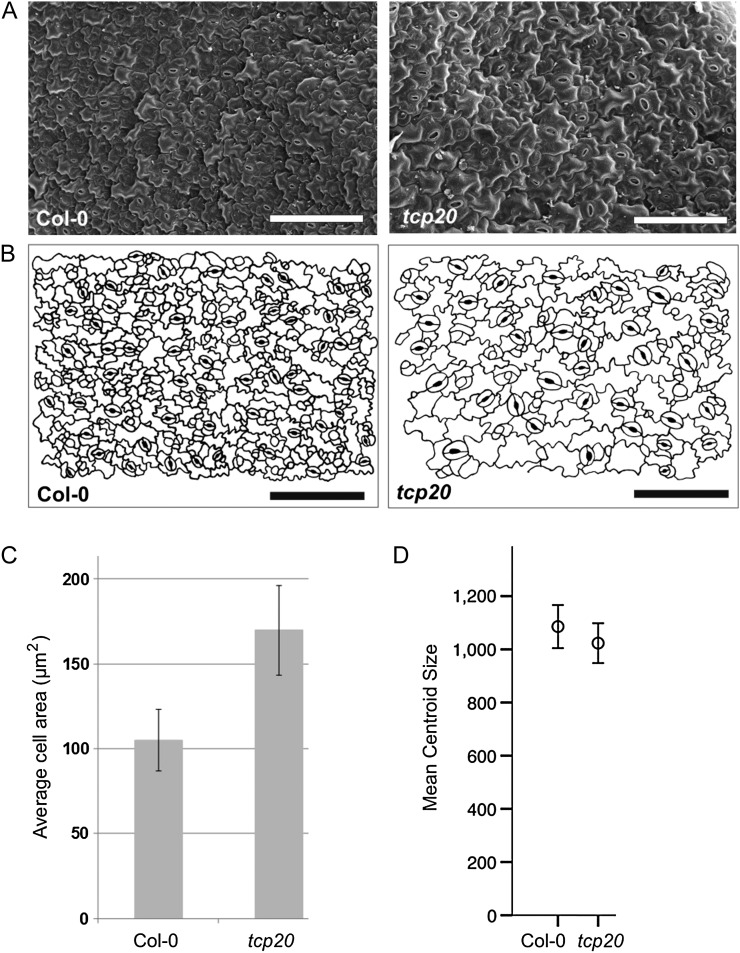
Because tcp20 mutant plants do not show any obvious phenotypic alterations (Li et al., 2005; Hervé et al., 2009), we analyzed a TCP20 T-DNA insertion line (Supplemental Fig. S1) at the cellular level (Fig. 2). For this purpose, scanning electron microscopy (SEM) analysis was performed on the abaxial side of the first leaf from 10-d-old seedlings (Fig. 2A). Subsequently, microscopic cell drawings were generated (Fig. 2B), followed by image analysis to determine average pavement cell sizes (Andriankaja et al., 2012). The analysis showed a significant increase in average epidermal cell size in tcp20 mutant leaves at this early developmental stage (t test, P = 0.006; Fig. 2C). To investigate a possible effect on leaf size, populations of tcp20 mutant and wild-type plants were analyzed using the LeafAnalyser technology (Weight et al., 2008; Kieffer et al., 2011). No obvious size (Fig. 2D) or shape alterations could be observed in the tcp20 mutant line by this method (Supplemental Data S1), indicating that the cell size effect is accompanied by a reduction in total cell number in the leaf.
Figure 2.
Phenotype of tcp20 knockout plants. Leaves of 10-d-old Arabidopsis seedlings were analyzed with SEM followed by pavement cell size analysis. A, Representative SEM images of the abaxial side of 10-d-old first leaves from wild-type and tcp20 knockout plants. B, Cell drawings made based on the representative SEM images in A. Bars = 100 μm. C, Diagram showing the average pavement cell sizes in wild-type and tcp20 knockout plants. Pavement cells in leaves of tcp20 knockout mutants were significantly bigger than in wild-type leaves. D, Diagram showing average leaf sizes of 42-d-old tcp20 knockout plants in comparison with Col-0 wild-type plants. The mean centroid size is plotted (Supplemental Data S1). No significant difference could be detected.
Glucocorticoid-Inducible TCP20 Inhibits LOX2 Expression in Leaves
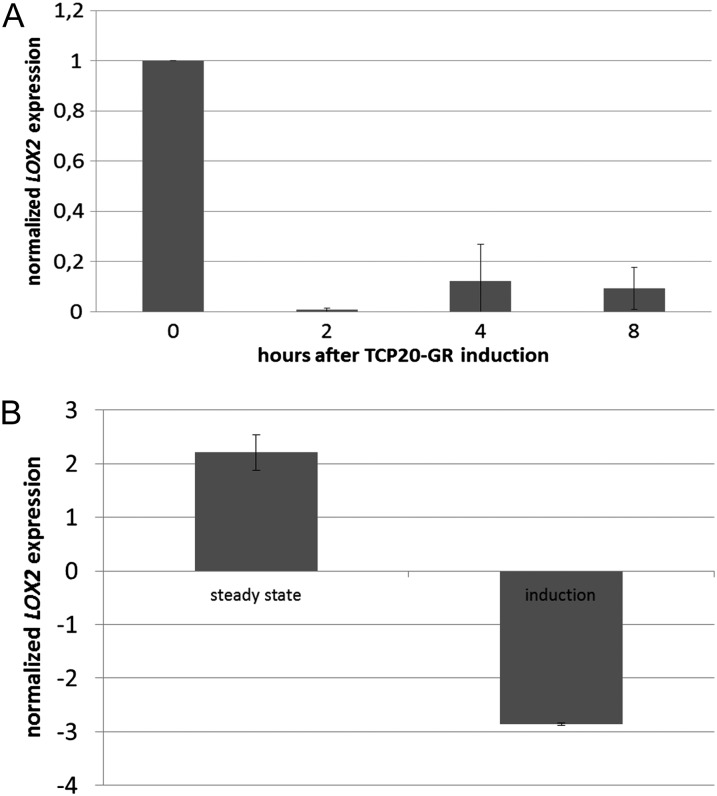
We decided to screen for genes under the direct control of TCP20 to obtain insight into its molecular function and to decipher how TCP20 can affect pavement cell sizes. A glucocorticoid-inducible system (Aoyama and Chua, 1997) was applied to identify potential direct targets of TCP20 in leaves. The coding region of TCP20 without a stop codon was fused at the 3′ end to a sequence encoding the glucocorticoid receptor (GR) and placed under the control of a constitutive cauliflower mosaic virus 35S promoter. This 35S:TCP20-GR construct was introduced into tcp20 knockout plants to avoid competition in target gene binding between endogenous TCP20 and the TCP20-GR protein. Dexamethasone (DEX) induction was performed in duplicate on 14-d-old seedlings in the presence of the protein synthesis blocker cycloheximide. All aboveground parts of 20 seedlings per biological replicate were harvested just before and 8 h after DEX induction. As a control, wild-type plants of the same age were treated the same way followed by hybridization of Arabidopsis whole-genome tiling arrays (GSE29012; http://www.ncbi.nlm.nih.gov/geo/). Subsequently, expression changes between the two time points were determined for both TCP20-GR and wild-type plants, and genes with an expression difference of log2 between TCP20-GR and the wild-type samples were selected (Supplemental Table S1). Two genes attracted our attention: the LOX2 gene and the class I TCP9 gene. LOX2 encodes for a protein involved in JA biosynthesis, and this gene was previously identified as a target of the class II TCP protein TCP4 (Schommer et al., 2008). Methyl-JA is known to repress cell proliferation (Pauwels et al., 2008); hence, the observed effects on cell size in the tcp20 mutant could be caused by increased JA levels and earlier repression of cell proliferation. To analyze how fast LOX2 transcription is repressed after TCP20 induction, a time-series experiment was performed. Here, we could detect significant LOX2 repression already 2 h after DEX/cycloheximide treatment (t test, P = 0.004; Fig. 3A). Subsequently, the LOX2 steady-state expression was compared between noninduced wild-type control plants and noninduced tcp20/35S:TCP20-GR plants, which resemble the tcp20 mutant. In leaves of 14-d-old tcp20 mutant plants, LOX2 expression was significantly up-regulated, whereas it was down-regulated after TCP20 induction at the same developmental stage (Fig. 3).
Figure 3.
Regulation of LOX2 expression by TCP20. A, LOX2 expression as a ratio between TCP20-GR and wild-type Col-0 plants in three independent replicates, where each replicate consisted of 20 plants pooled per line and time point. Leaf samples were taken 2, 4, and 8 h after induction with DEX and cycloheximide. Real-time PCR analysis shows that LOX2 repression occurs already 2 h after TCP20-GR induction. B, Left, LOX2 steady-state expression. The expression value of LOX2 in 14-d-old leaves is represented as a ratio between noninduced tcp20 35S:TCP20-GR and noninduced wild-type Col-0, as observed in the microarray experiment. Because the TCP20-GR construct was transformed into the tcp20 knockout background, noninduced tcp20 35S:TCP20-GR plants resemble the tcp20 knockout line. Right, LOX2 expression after 8 h of TCP20-GR induction in 14-d-old leaves. The expression values are presented as ratios between induced tcp20 35S:TCP20-GR and induced Col-0 wild type. LOX2 expression is repressed by TCP20 and, in accordance, up-regulated in the tcp20 mutant.
Class I and Class II TCPs Bind to Different Regions of the LOX2 Promoter and Antagonistically Regulate LOX2 Expression
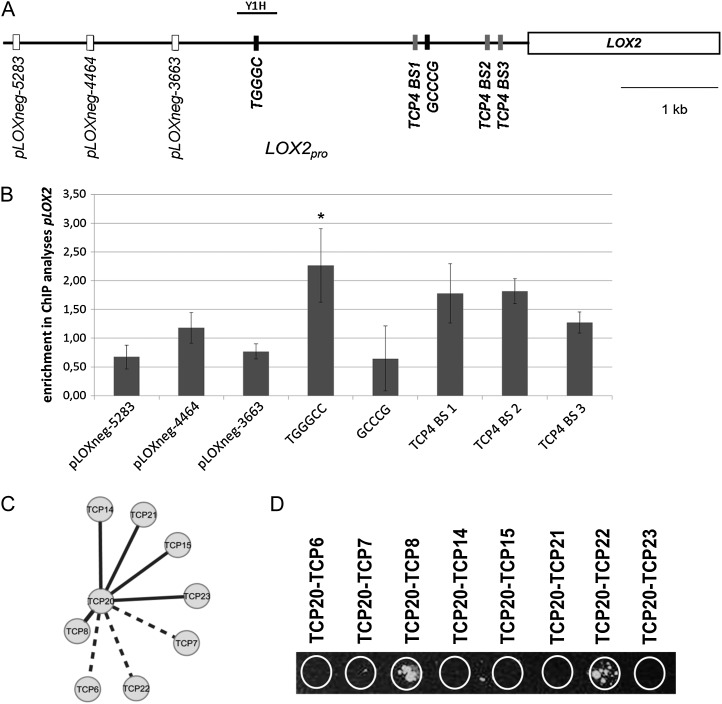
Next, binding of TCP20 to the LOX2 promoter was analyzed using a ChIP assay followed by real-time PCR (Fig. 4). The upstream regulatory sequence of LOX2 contains several class II TCP consensus binding sites, as depicted in Figure 4A. Previously, it was shown that the class II TCP4 protein binds specifically to the hotspot region of putative class II binding sites in the 1-kb upstream region (Schommer et al., 2008; Aggarwal et al., 2010). Sequences close to the GCCCR putative TCP20 binding site (Kosugi and Ohashi, 2002; Li et al., 2005) were found upstream of the LOX2 transcriptional start site at −1,076 bp (GCCCG) and at −2,799 bp (TGGGCC; Fig. 4A). We performed ChIP analyses to test if TCP20 binds directly to these motifs. As a control, we used three sequences within the LOX2 promoter that were upstream of the TGGGCC motif (Fig. 4A). Additionally, we used a promoter fragment of the SAND family gene AT2G28390 as a negative control, because this gene was not identified in any of the published TCP20 target gene studies or in our microarray analysis (Li et al., 2005; Hervé et al., 2009; Supplemental Table S1). Immunoprecipitated material from five biological replicates of 4-d-old gTCP20-GFP seedlings was analyzed for enrichment of the putative binding sites in the LOX2 promoter using GFP antibodies coupled to magnetic beads (de Folter et al., 2007; Kaufmann et al., 2010a, 2010b). We included the three known TCP4 binding sites in our analysis to address the question of whether class I and class II TCPs compete for the same binding sites. A quantitative real-time (qRT)-PCR experiment showed significant enrichment for the −2,799-bp TGGGCC motif only (ANOVA, P = 0.025; Fig. 4B; for primer sequences, see Supplemental Table S2). This motif has not been shown to be bound by TCP4 in earlier studies; hence, the direct competition for a binding site between class I and class II TCPs, which has been suggested in previous studies (Li et al., 2005), could not be confirmed for this particular case. Nevertheless, both TCP4 and TCP20 appeared to be able to bind the LOX2 locus. A yeast one-hybrid assay was performed to confirm binding of TCP20 to this far-upstream LOX2 promoter element (Fig. 4A). Surprisingly, no binding was found for the single TCP20 protein. Because TCPs have been shown to bind DNA as dimers (Cubas et al., 1999; Aggarwal et al., 2010), we hypothesized that TCP20 may need a dimerization partner to bind to the LOX2 promoter. Therefore, we studied TCP20 dimerization capacity with all 24 Arabidopsis TCP proteins (Aguilar-Martínez et al., 2007), making use of a pairwise GAL4 yeast two-hybrid assay. TCP20 itself and the class II TCP proteins TCP1, -2, -4, -10, -12, -18, and -24 exhibited autoactivation when expressed from the GAL4 binding domain (BD) vector; therefore, combinations between these TCP proteins and TCP20 could not be assessed for protein-protein interaction capacity. Nevertheless, interaction between TCP20 and a number of class I TCP proteins was identified (Fig. 4C). Subsequently, coexpression between TCP20 and the genes encoding for the interacting TCP20 proteins was investigated using GeneCAT (Mutwil et al., 2008). According to this analysis, TCP8 has the largest expression overlap with TCP20, making TCP8 a strong candidate for a functional dimerization partner of the TCP20 protein (Fig. 4C). Subsequently, we analyzed the binding capacities of the potential TCP20 dimers to the LOX2 promoter in a modified yeast one-hybrid assay, showing that the TCP20-TCP8 and TCP20-TCP22 dimers are able to bind the LOX2 promoter region containing the −2,799-bp TGGGCC motif (Fig. 4D). As a control, the single TCP8 and TCP22 proteins were tested in the yeast one-hybrid assay under the same experimental conditions, and these did not possess any binding capacity, suggesting that these particular class I TCP proteins bind the LOX2 promoter as a complex together with TCP20.
Figure 4.
TCP20 binding to the LOX2 promoter. A, Schematic representation of putative TCP binding sites in the promoter of LOX2. The graph features 5.5 kb upstream of the first LOX2 exon. This region includes five putative TCP binding sites. Class II TCP binding sites defined in earlier publications (Schommer et al., 2008) are depicted in gray, putative TCP20 binding sites are depicted in black (indicated with their motif sequences), and the negative control sequence elements for the ChIP experiments are depicted in white. Yeast one-hybrid analysis was conducted on the fragment marked by a black bar and indicated with Y1H. B, ChIP of TCP20-GFP and subsequent analysis by real-time PCR for enrichment of the LOX2 and TCP9 promoters. Enrichments are normalized against the promoter of the SAND family gene AT2G28390, which is not depicted here. TCP20 binding to the TGGGCC motif in the LOX2 promoter was shown to be significant (ANOVA with subsequent Dunnett’s test, *P = 0.025). C, TCP20 yeast two-hybrid results combined with in silico coexpression analyses. Shown are protein-protein interaction and expression relations between TCP20 and class I TCPs in a ball-and-stick representation. All depicted TCP proteins showed protein-protein interaction with TCP20 in yeast two-hybrid analyses. The length of the lines is representative for the extent of coexpression found between TCP20 and the respective class I TCP gene. The shorter the edges are, the closer the coexpression between TCP20 and the connected TCP. Hence, TCP8 is the most closely coexpressed class I TCP. Dashed thick edges depict TCP pairs that have been shown to interact but where no coexpression data were available. This counts for the genes TCP22, TCP6, and TCP7. D, Yeast one-hybrid analysis of class I TCP dimers against the LOX2 promoter. Potential TCP dimers containing TCP20 were assessed for their binding ability to a LOX2 promoter fragment (−3,005 to −2,607), where TCP20 binding was found to be significant in ChIP analyses. As depicted, TCP8 and TCP22 are able to bind the LOX2 promoter when combined with TCP20. The single proteins were not able to bind the same promoter region.
Expression of Class I TCPs and TCP4 during Leaf Development Reflects Their Antagonistic Roles
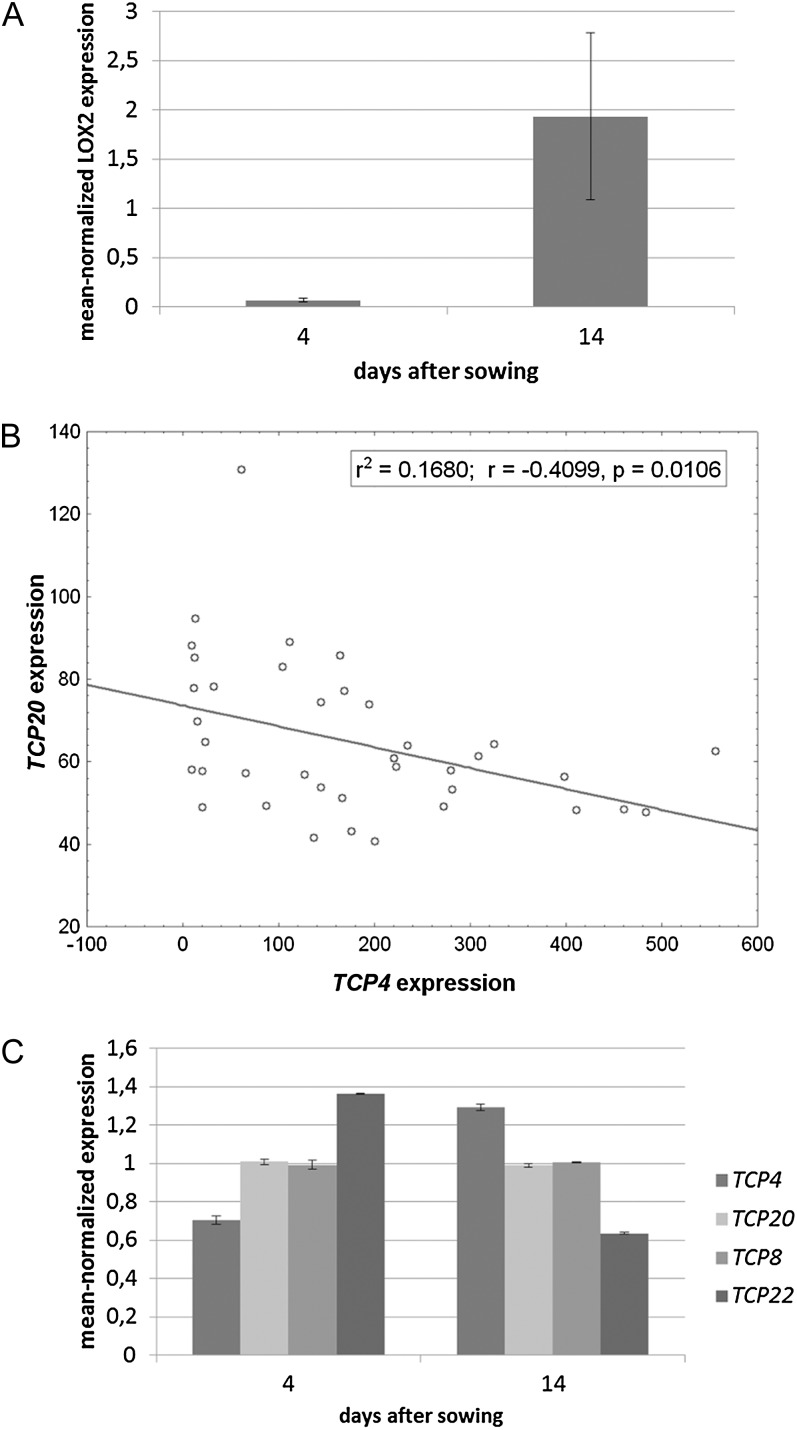
The observations presented above strongly suggest that LOX2 is under direct antagonistic control of TCP4 and class I TCP protein complexes containing the TCP20 protein. LOX2 expression itself changes substantially during leaf development (Fig. 5); it is lowly expressed during early leaf development but shows an increased expression in the first leaf 14 d after sowing (Fig. 5A). We hypothesized that the antagonistic regulation of LOX2 expression by TCP4 and TCP20 transcription factors may be reflected in their expression patterns. We analyzed this possibility by correlating TCP4 to TCP20 expression in Arabidopsis development. For this, we used data from AtGenExpress (Schmid et al., 2005) and analyzed correlation using STATISTICA 6.0. (Hill and Lewicki, 2007) The analysis showed that TCP4 and TCP20 expression patterns are negatively correlated (r = −0.41, P = 0.02; Fig. 5B). Despite the fact that they are negatively correlated, neither published work on TCP4 and TCP20 nor our TCP20-GR data set provides evidence that the two transcription factors directly influence each other’s expression. Other class I TCPs that are involved in TCP20 functioning, according to our yeast one- and two-hybrid analyses, are TCP8 and TCP22. To study the ratio between TCP4 and the class I TCP expression levels in relation to LOX2 expression in leaves in more detail, expression levels of TCP4, TCP8, TCP20, and TCP22 were determined in 4- and 14-d-old leaves by real-time PCR (Fig. 5C). This analysis shows that the class I TCP genes are either equally, or lower, expressed in older leaves in comparison with young leaves. In contrast, the class II TCP4 gene is expressed stronger in old leaves, which is in line with an increased LOX2 expression in the 14-d-old leaves.
Figure 5.
Relation between TCP4, TCP20, and LOX2 expression. A, Wild-type plants were harvested 4 d and 2 weeks after sowing, and LOX2 expression was analyzed in quantitative PCR assays including three biological replicates of 20 seedlings per replicate. The graph depicts mean-normalized expression of LOX2, which increases dramatically between the two developmental time points. B, Statistical analysis of the correlation of TCP20 to TCP4 expression during Arabidopsis development. The correlation coefficient (−0.41) and P value (0.01) are indicated. The analysis was done using expression data from AtGenExpress and with STATISTICA 6.0 (Hill and Lewicki, 2007). C, Expression of the three class I TCPs TCP8, TCP20, and TCP22 and the class II TCP TCP4 at two different stages of Arabidopsis leaf development. In 4-d-old seedlings, TCP4 expression is lower than at day 14. Whereas TCP8 and TCP20 expression stays similar throughout the experiment, TCP22 expression behaves opposite to TCP4 expression. Consequently, the ratio of TCP4 to class I TCP expression changes in the course of leaf development. LOX2 expression increases from day 4 to day 14, which is in line with the idea that LOX2 expression is directly dependent on the ratio between TCP4 and class I TCP expression levels.
TCP9 Acts Downstream of TCP20
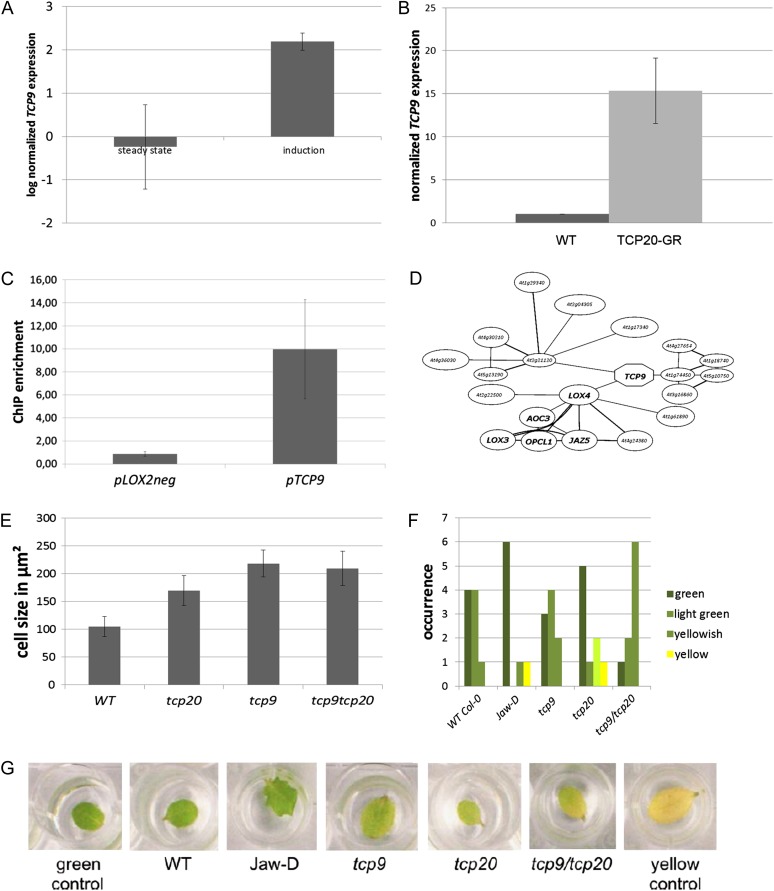
TCP9 is the second potential TCP20 target selected based on the microarray and real-time PCR data (Fig. 6, A and B; Supplemental Table S1). Whereas a clear effect can be seen during induction, tcp20 knockout alone is not able to change TCP9 expression, suggesting that redundant factors activate TCP9 expression in the absence of TCP20. ChIP was performed on homozygous gTCP20-GFP plants to provide evidence for direct binding of TCP20 to the TCP9 locus. This analysis revealed that the TCP9 promoter can be bound by the TCP20 protein (Fig. 6C). Although there is yet not much known about the molecular role of TCP9, coexpression analysis using the ATTED-II tool (Obayashi et al., 2007) reveals that TCP9 is coexpressed with several genes involved in both JA biosynthesis and response (Fig. 6D). Based on this knowledge, the information that JA represses cell proliferation (Pauwels et al., 2008), and the presented data on TCP20, we hypothesized that the obtained tcp20 mutant phenotype could be at least partially due to TCP9-controlled JA metabolism.
Figure 6.
Regulation and function of the class I TCP TCP9. A, Comparison of steady-state TCP9 expression levels in wild-type and tcp20 knockout plants shows no significant differences (left). However, TCP9 is induced upon DEX induction of TCP20-GR in the tcp20 mutant background (right). B, TCP9 expression upon 8 h of DEX induction in wild-type (WT) and TCP20-GR plants by quantitative PCR analysis of two biological replicates containing pools of 20 plants. Expression level was normalized based on the wild-type samples. C, Enrichment of the TCP9 promoter in ChIP analysis on TCP20-GFP plants. Quantitative PCR-analyzed enrichment of pTCP9 is compared with the nonbound region of the LOX2 promoter. D, In silico analysis of coexpression using ATTED-II shows that TCP9 is closely expressed with genes from the JA synthesis and response pathways. Genes that are known to be involved in JA synthesis and response are depicted with gene names. All other coexpressed genes are depicted with Arabidopsis Genome Initiative numbers. E, SEM analysis of tcp9 knockout plants 9 d after sowing shows increased pavement cell size similar to the tcp20 knockout phenotype. F and G, Senescence analysis shows an effect of the tcp9tcp20 double mutant that is not found in the single mutants. Leaves were cut from 2-week-old Arabidopsis seedlings and incubated on water in the dark. After each day, leaves were assessed for senescence behavior, which was categorized as “green” (nonsenescing), senescence 1 (brightening of the green color), senescence 2 (yellowish color of the leaf), and senescence 3 (leaf completely yellow). The graph in F depicts the percentage of leaves in a given category for all lines after 7 d of incubation. In G, one representative leaf of each line is shown after 7 d of incubation in the dark. On the left and right hand sides, a healthy green leaf and a fully senesced yellow reference leaf are shown.
A tcp9 transferred DNA insertion mutant was obtained and analyzed to further elucidate the function of TCP9. A global analysis revealed no obvious phenotypic alterations, except for an effect on root length (Supplemental Fig. S2). This observed effect might be caused by an altered JA metabolism, because JA is known to inhibit root growth (Staswick et al., 1992). In addition to the tcp9 single mutant, a tcp9tcp20 double mutant line was generated. Subsequently, the homozygous tcp9 single and tcp9tcp20 double mutant lines were analyzed for phenotypic effects at the cellular level, in a similar manner to that performed for the tcp20 mutant. We detected larger epidermal cells in the first leaves of 10-d-old plants of both single and double mutant lines as well as an effect on stomata patterning (Fig. 6E; Supplemental Fig. S3). However, as for the single tcp20 mutant, no effect at the whole-leaf level was observed, suggesting a reduction in the total cell number in the leaf (Supplemental Data S1).
The observed larger epidermal cell sizes in the class I tcp9 and tcp20 mutants is an opposite phenotype of the reported small cell sizes in the overproliferating class II TCP jaw-D mutant (Palatnik et al., 2003; Efroni et al., 2008; Gonzalez et al., 2010). This observation, and the knowledge that the onset of senescence is delayed in the jaw-D mutant (Schommer et al., 2008), prompted us to look for a possible effect on senescence in the class I tcp mutant lines. We tested leaf senescence after abscission of leaves, as it was done previously (Schommer et al., 2008), and found that the single tcp9 and tcp20 mutants do not show significantly altered leaf senescence behavior in comparison with wild-type plants. The tcp9tcp20 double mutant, however, exhibits earlier senescence in our analyses (Fig. 6F), again indicating an antagonistic relationship between class I and class II TCPs in the control of JA-mediated processes. The different behavior of the lines was tested using a χ2 test and was found to be statistically significant (P < 0.01) for the comparison of wild-type ecotype Columbia-0 (Col-0) versus the tcp9tcp20 double mutant.
DISCUSSION
The Arabidopsis genome encodes 24 TCP transcription factors that are divided into two classes (Cubas et al., 1999). Although this transcription factor family was first described about 13 years ago, and the role of various members in growth-related processes has been deciphered, our understanding of their functioning is far from complete (Martín-Trillo and Cubas, 2010). Here, we report the role of TCP20 and TCP9 in leaf development and the antagonistic control of JA biosynthesis by class I and class II TCP transcription factors.
Class I and Class II TCPs Regulate Specific Biological Processes Antagonistically
The LOX2 gene encodes for a LOX that is involved in early steps of JA synthesis (Vick and Zimmerman, 1983). In this study, we show that LOX2 is inhibited by induction of the TCP20-GR protein. Previously, TCP4 was shown to affect JA synthesis positively via the regulation of LOX2 expression (Schommer et al., 2008). When TCP4 and other class II TCPs are down-regulated in jaw-D plants (Palatnik et al., 2003), LOX2 expression and JA content in leaves are low (Schommer et al., 2008), and as a consequence, JA-induced senescence is delayed. Conversely, knockout of TCP20 leads to increased LOX2 expression, and earlier senescence behavior is observed in the tcp9tcp20 double mutant. Moreover, TCP4 and other class II TCP genes have been implicated in the coordination between cell differentiation and cell division, where down-regulation of these class II TCP genes results in increased cell production, controlled by an extension of mitotic activity at the cellular level (Palatnik et al., 2003; Efroni et al., 2008). In contrast, our data suggest that knockout of class I TCPs, TCP20 and its direct target TCP9, and their double mutant results in a decrease in cell production accompanied by an increase in cell size. Whether TCP20 and TCP9 primarily drive cell proliferation or inhibit cell expansion and differentiation (Gonzalez et al., 2012), and how they affect the transition from cell proliferation to cell expansion, remain to be studied in more detail. Nevertheless, the antagonistic effect of class I and class II TCPs on LOX2 expression, leaf senescence, and cell production confirms the previously postulated concept that class I and class II TCP transcription factors can regulate the same genes and biological processes, but with opposite effects (Li et al., 2005; Fig. 7).
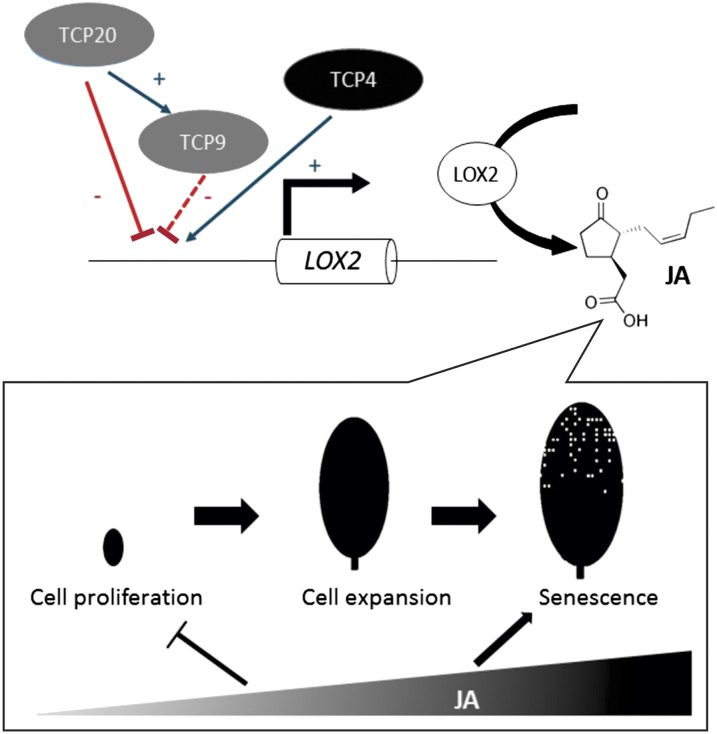
Figure 7.
Model for JA-controlled leaf development in light of the class I-class II TCP transcription factor functional antagonism. The class I TCPs, TCP20 and TCP9, are depicted in gray; the class II TCP, TCP4, is depicted in black. Whereas the class I TCPs inhibit the JA biosynthesis gene LOX2, TCP4 activates its expression. As TCP4 abundance increases in time, LOX2 expression, and hence JA production, rises in later stages of leaf development. The increasing JA content of leaves at different time points of leaf development is supposed to trigger at least two processes: JA affects the transition from cell proliferation to cell expansion by inhibiting cell proliferation; and JA induces senescence in later leaf development. [See online article for color version of this figure.]
Although the hypothesis of antagonistic functions of class I and class II TCPs was based on similar putative binding sites of the two classes and, hence, direct competition for binding sites, we were not able to show that TCP20 and TCP4 bind at the same sites of the LOX2 promoter, suggesting that other molecular mechanisms may play a role in the antagonistic relationship. A possible molecular action could be that TCP4 and TCP20, depending on their interactors, act as either activator or suppressor of target gene transcription.
We observed that the ratio of TCP4 to TCP20 expression is negatively correlated during whole Arabidopsis development, with relatively high TCP4 expression during later stages of leaf development. Based on this observation, it is tempting to speculate that LOX2 expression during development is dependent on temporal and spatial shifts in the ratios of class I and class II TCP levels. As we found no indication in the literature and in our own data that members of the two classes regulate each other at the transcriptional level, the ratio of expression levels must be established by upstream regulators of the TCPs. So far, we have little information on what these regulators might be. Recently, it was shown that TCPs are overrepresented target genes of the MADS domain transcription factors SEPALLATA3 (SEP3; Kaufmann et al., 2009) and APETALA1 (AP1; Wellmer et al., 2006; Kaufmann et al., 2010b). Furthermore, the TCP transcription factor consensus binding site is overrepresented in the promoter regions bound by SEP3 and AP1, suggesting complex regulatory interactions between these two classes of transcription factors. However, SEP3 and AP1 are expressed in the reproductive phase only and hence cannot be the upstream controllers of TCPs involved in leaf growth during the vegetative phase of development. Further studies will be needed to show how the differential expression of class I and class II TCPs during leaf development is orchestrated.
Could Hormones Be Mediators of TCP-Regulated Growth?
TCP transcription factors have been postulated to be regulators of growth by directly acting on core cell cycle genes (Li et al., 2005). In general, this premise is based on tcp mutant phenotypes in combination with a few examples of direct binding of cell cycle gene regulatory sequences by TCP proteins (Li et al., 2005; Kieffer et al., 2011). Plants impaired in different class II TCP genes revealed roles in diverse growth processes, such as axillary meristem outgrowth (Aguilar-Martínez et al., 2007), floral symmetry by differential growth of petals (Luo et al., 1996), and leaf growth (Palatnik et al., 2003; Efroni et al., 2008). These observed phenotypes were associated with alterations in cell division capacity or division rate. However, thorough investigation of cell division patterns and transcript changes during leaf development in the jaw-D mutant suggests that the class II TCP-dependent regulation of the cell cycle is indirect (Efroni et al., 2008). In the case of TCP20, the control of cell cycle genes seems not to be its only function, as its expression is widespread and neither limited to, nor excluding, proliferating cells. Based on the work performed by Hervé and colleagues (2009) and our data showing that TCP20 directly regulates the JA biosynthesis gene LOX2, we hypothesize that TCP20 may control cell proliferation indirectly via the JA signaling pathway (Fig. 7). From the earliest publications, JAs have not only been identified in wound response and defense against pathogens (Bell and Mullet, 1991) but also in developmental programs (Wilen et al., 1991) and suppression of the cell cycle (Pauwels et al., 2008). Therefore, it is possible that TCP20 controls cell proliferation and growth indirectly via JA signaling.
The TCP20 target TCP9 was recently found to be regulated by brassinosteroids, which are key signaling molecules in growth regulation (Kauschmann et al., 1996; Yu et al., 2011). Conversely, TCP1, which belongs to the class II TCPs, is involved in brassinosteroid synthesis (Guo et al., 2010). Furthermore, TCP3, a member of the class II TCPs regulated by miR319a, is involved in auxin signaling (Koyama et al., 2010), and auxin is a hormone that acts synergistically with brassinosteroids in growth regulation (Hardtke, 2007). The closely related TCP14 and TCP15 class I TCP proteins appear to act in the cytokinin pathway (Steiner et al., 2012) and affect the endoreduplication process (Kieffer et al., 2011; Li et al., 2011).
Altogether, these data suggest that TCPs play an important role in the regulation of growth processes via plant hormone signaling. Future research should elucidate in more detail the positions of these TCPs in signaling cascades that drive leaf development via the control of hormonal pathways. Furthermore, we have provided evidence for an antagonistic activity of class I and class II TCP proteins, reflected in opposite transcriptional control of a common target gene. Future work will reveal how common and conserved this mechanism is for class I and class II TCP protein functioning.
MATERIALS AND METHODS
Plant Material
Seeds of Arabidopsis (Arabidopsis thaliana) tcp20 knockout plants (SALK_016203.45.25) and tcp9 knockout plants (SALK_143587.56.00) were obtained from the Nottingham Arabidopsis Stock Center. Col-0 was used as the wild type and reference in all experiments.
Plant Growth and Media
Plant material was either grown on rock wool or on Murashige and Skoog (MS) medium, depending on the experimental setup. When seeds were sown out on MS medium, they were gas sterilized first by placing an open Eppendorf vial containing the seeds into a glass bowl alongside a jar containing 100 mL of bleach. After the addition of 3 mL of 37% hydrochloric acid (fuming) to the bleach, the glass bowl was closed tightly and kept closed for 1 to 2 h. At the end of the sterilization, seeds were taken out swiftly and the vials were closed.
For DEX induction experiments, 50 mL of one-half-strength MS medium (2.3 g L−1) with agar (6 g L−1) was poured per plate, and after polymerization, a sterilized nylon mesh (mesh size, 200 µm) was placed on the medium (Passarinho et al., 2008). Thirty to 50 seeds of both the 35S:TCP20-GR-line and the wild-type Col-0 were sown out per plate.
Constructs
For the glucocorticoid induction experiments, the TCP20 coding sequence without the stop codon was amplified using the primers 5′-ATGGATCCCAAGAACCTAAATCGT-3′ and 5′-ACGACCTGAGCCTTGAGAATC-3′ and cloned into pCR8/GW/TOPO to obtain a Gateway entry vector. A Gateway destination vector suitable for the expression of genes of interest fused to the coding region of the rat GR domain was obtained by removing the AGL11 coding region from vector NOB221 (kindly provided by Martin Kater). For this purpose, the BamHI and NcoI restriction enzymes were used. Subsequently, the digested vector was blunted, followed by introduction of the Gateway conversion cassette (Invitrogen) upstream of the GR coding region and downstream of the cauliflower mosaic virus 35S promoter. This complete expression cassette was cloned as an AscI/PacI fragment into the binary vector pGD121 (de Folter et al., 2006), resulting in the Gateway-compatible GR destination vector pARC146. As we wanted to introduce the final TCP20-GR construct into a tcp20 SALK line, we ensured antibiotic selection by replacing the kanamycin resistance cassette of pARC146 with a BASTA resistance cassette taken out of pB7WG2 (Karimi et al., 2002). In the first step, pARC146 was partly digested with HindIII, and a 4.4-kb fragment containing the GR-Gateway cassette was recovered. The plasmid pB7WG2 was cut with KpnI, and a 7-kb fragment, containing the BASTA resistance cassette, was ligated with the 4.4-kb fragment recovered from pARC146, resulting in CZN671. Subsequently, the TCP20-GR expression vector was generated by an LR reaction between CZN671 and the previously denominated TCP20 entry clone.
To obtain a vector for GFP-tagged TCP20 expression at endogenous levels, a TCP20 genomic fragment was cloned. The genomic fragment was amplified up to the stop codon and including 2,466 bp of promoter sequence (primers used were 5′-CACCCTATGATGCATGCCACTCTCG-3′ and 5′-ACGACCTGAGCCTTGAGAATC-3′) and cloned into the GFP Gateway destination vector pMDC204 (Curtis and Grossniklaus, 2003). This resulted in the expression vector gTCP20-GFP (CZN064). The gTCP9-GFP (CZN1905) construct was generated by employing the same strategy, using the primers PDS4239 (5′-AAAAGTAAAAACATCGGAATCCAAAACCT-3′) and PDS4240 (5′-GTGGTTCGATGACCGTGCTGTTG-3′) and the GFP Gateway destination vector pMDC107 (Curtis and Grossniklaus, 2003).
Transformation of Arabidopsis
The 35S:TCP20-GR (CZN652) and gTCP20-GFP (CZN064) constructs were transformed into homozygous tcp20 knockout plants, and the gTCP9-GFP (CZN1905) construct was transformed into the Col-0 wild type. For transformation, Arabidopsis plants were grown on soil until the primary inflorescences emerged. These were cut to promote the growth of secondary inflorescences and to increase the number of flowers. The binary constructs were transformed into Agrobacterium tumefaciens strain C58C1 (pMP90). Transformation of plants was conducted by floral dip (Clough and Bent, 1998). After transformation, plants were kept in growth chambers until seed set. Seeds were selected on one-half-strength MS + agar (8 g L−1) plates containing 25 µg mL−1 phosphinotricine (Basta) for the plants expressing the TCP20-GR construct and 30 µg mL−1 hygromycin for the TCP20-GFP/TCP9-GFP transformants. After 2 weeks, rooting green seedlings were transferred to soil and grown until seed set. The following T2 generation was checked for expression of the transgene by real-time PCR and, in the case of TCP20-GFP and gTCP9-GFP, by confocal laser scanning microscopy.
Confocal Imaging
Confocal laser scanning microscopy of living plant tissues was conducted with a Leica SPE DM5500 upright microscope using Leica AF 1.8.2 software (http://leica-microsystems.com). Preparation of inflorescences was done as described before (de Folter et al., 2007).
SEM and Image Analysis
Whole 10-d-old seedlings were fixed in 4% paraformaldehyde under vacuum for 24 h and dehydrated through an ethanol series, dried under CO2 in a Balzer’s critical point drier, mounted in metallic stubs with carbon conductive adhesive tape, coated with colloidal gold, and observed at 20 kV using a LEO 435 VP scanning electron microscope at the University of Sao Paulo. Cell drawings were based on SEM images and were analyzed as described previously (Andriankaja et al., 2012).
Glucocorticoid Induction Experiments
Glucocorticoid induction experiments were conducted 14 d after germination of the plants. Growing the plants on nylon meshes allowed us to transfer the plants into induction medium swiftly and without damaging the roots. The induction medium consisted of 2.3 g L−1 MS medium , 1% (w/v) sugar, 10 µm DEX, and 10 µm cycloheximide (Passarinho et al., 2008). Samples were harvested just prior to the treatment and at different time points after start of the treatment.
RNA Isolation and qRT-PCR
RNA was isolated using the Qiagen RNeasy RNA isolation kit according to the manufacturer’s protocol. DNase treatment took place on column, following the protocols from the manufacturer. Moloney murine leukemia virus reverse transcriptase from Promega was used for complementary DNA synthesis. First, a mix of poly(dT) primer and deoxyribonucleotide triphosphates was added to 500 ng of DNA-free RNA in a volume of 12 µL. This solution was kept in ambient temperature for 2 min before 1 µL of reverse transcriptase was added. After the addition of the reverse transcriptase, samples were incubated at 25°C for 15 min and transferred to 42°C for 50 min. Reverse transcription was stopped by heat treatment at 70°C for 15 min. The complementary DNA made this way was used for qRT-PCR using the SYBR Green mix from Bio-Rad. The reference genes used for all expression analyses were a SAND family gene, AT2G28390, and the TIP41-like gene AT4G34270, both determined as superior reference genes (Czechowski et al., 2005). The primers used are given in Supplemental Table S2.
Microarray Analysis
DNA-free RNA from the induction assays was analyzed using Affymetrix Tiling 1.0R arrays. The expression data from Tiling 1.0R arrays were preprocessed using the robust multiarray average algorithm (Irizarry et al., 2003). The probe annotation was obtained from athtiling1.0rcdf (Naouar et al., 2009); it contains only probes derived from The Arabidopsis Information Resource 7 genes. Probes representing nonunique sequence were masked, only probes that are common to all described variants of a transcript were considered, and probes representing intronic regions, or regions spanning intron/exon junctions, were removed. The microarray data were deposited at the Gene Expression Omnibus under accession number GSE29012. Potentially differential gene expression upon TCP20 induction was determined by subtracting the log2 expression values of induced versus uninduced samples in the first step and subtracting the resulting values of wild-type seedlings from the TCP20-GR seedlings’ log2 expression values. Genes with a differential expression value of more than +2 or less than −2 in both biological replicates were selected.
ChIP
ChIP experiments mainly followed the protocol described previously (de Folter et al., 2007; Kaufmann et al., 2010b). TCP20-GFP seedlings were grown in liquid one-half-strength MS medium on a horizontal shaker (30 rpm) for 4 d in a 16-h-light/8-h-dark regime. After this time, 3 g of seedling material was harvested by pouring the liquid MS medium through a sieve, and then the seedlings were fixed with 37% formaldehyde. Immunoprecipitation was conducted using a GFP antibody coupled to magnetic beads. The magnetic beads were used to precipitate the antibody-protein-GFP complexes. The enrichment of TCP20 binding regions was compared between the immunoprecipitate and 1:1,000 diluted input material. Promoter elements not expected to be bound by TCP20 were used as negative controls. The ChIP experiment was performed in five repetitions.
Yeast Two-Hybrid Analysis
Protein-protein interactions between TCP20 and the 24 other annotated Arabidopsis TCP proteins were analyzed in a pairwise yeast two-hybrid GAL4 assay (de Folter et al., 2005). Bait vectors were transformed into yeast strain PJ69-4α, and the TCP20 prey vector was transformed into yeast strain PJ69-4a (James et al., 1996). The individual transformants were grown in liquid synthetic dropout (SD) medium lacking Leu and Trp, respectively. These overnight cultures were mated by spotting 5 µL of liquid culture per partner on SD complete plates. After overnight incubation, yeast was transferred by a 96-pin replicator to freshly prepared SD plates lacking Leu and Trp, selecting for diploid yeast containing both plasmids. In the last step, the mated yeast strains were transferred on SD medium lacking Leu, Trp, and Ade or lacking Leu, Trp, and His, supplemented with 5 or 10 mm 3-amino-1,2,4-triazole, respectively. Growth of yeast, and hence protein-protein interaction events, was scored after 5 d of incubation at 30°C. Because of the high autoactivation capacity of several TCPs, not all combinations could be analyzed reciprocally. Autoactivation capacity was determined beforehand for the baits by testing for growth of the single bait transformants on selective SD medium for the His and Ade protein-protein interaction markers. TCP1, -2, -4, -10, -12, -18, -20, and -24 were exhibiting autoactivation when cloned in the GAL4 BD vector, and matings with these particular TCP-BD constructs were not included in the yeast two-hybrid analysis. Every combination was analyzed 18 times (six replicates and three different selection conditions). In the end, only pairs that scored reproducible and for at least two different selection conditions positive were taken as positive protein-protein interactions.
Yeast One-Hybrid Analysis
Binding of the single TCP8, TCP20, and TCP22 proteins and TCP20 dimers to a LOX2 promoter sequence was analyzed in a yeast-one hybrid system based on the Matchmaker Gold Yeast Two-Hybrid System (http://www.clontech.com). In this system, binding events are detected by resistance against the antibiotic aureobasidin A. The yeast strains used for this analysis were PJ69-4A for the TCP baits and PJ69-α for the promoter reporter construct. The reporter construct is based on the plasmid pAbAi (http://www.clontech.com). This plasmid was made Gateway compatible by ligating a Gateway-C cassette into the SmaI site, resulting in CZN1018. The reporter construct for the LOX2 promoter consists of a 398-bp fragment (−3,005 to −2,607, upstream of the transcriptional start site), including the potential TCP20 binding site TGGGCC (−2,799 bp; Fig. 4A). Autoactivation tests for the promoter were conducted in the range from 0 to 500 ng mL−1 aureobasidin A, and background activation was detected up to 75 ng mL−1. For the screenings, an aureobasidin A concentration of 100 ng mL−1 was used. After growing the individual yeast clones for 2 to 3 d on selective medium for the presence of the plasmid at 30°C, mating of TCP bait clones and the reporter clone was initialized on SD complete medium overnight. The mated yeast was transferred onto selection medium, selecting for the baits and the reporter construct. After having grown for 2 to 3 d, yeast able to grow on the selective medium was transferred to 100 μL of sterile Milli Q and spotted as 5-μL droplets onto aureobasidin-containing plates. Plates were incubated at 20°C and scored after 5 to 7 d. All experiments were performed in duplicate.
Senescence Assay
The fifth and sixth leaves were detached from 3-week-old plants and incubated in the dark for 5 to 7 d, floating on Milli Q water. Subsequently, a photograph was taken, and leaves were classified based on leaf color as healthy and green (nonsenescing), greenish (senescence 1), yellowish (senescence 2), and completely yellow and senesced (senescence 3).
Leaf Size and Shape Analyses
A comprehensive analysis of the effects of tcp gene mutations on leaf size and shape was performed using the LeafAnalyser technology (Weight et al., 2008). A detailed description is provided in Supplemental Data S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression of TCP20 in tcp20 knockout versus Col-0 wild-type plants.
Supplemental Figure S2. Root phenotype of tcp9 knockout plants.
Supplemental Figure S3. Stomatal index of class I tcp single and double mutants.
Supplemental Table S1. Potential direct TCP20 target genes.
Supplemental Table S2. Primers used in this study.
Supplemental Data S1. Leaf size and shape analysis by LeafAnalyser.
Supplementary Material
Acknowledgments
We thank Kerstin Kaufmann for helping with the planning and execution of ChIP experiments. Our thanks also go to Prof. E.W. Kitajima and F. Tanaka for maintaining the SEM facility at Nucleo de Apoio a Pesquisa em Microscopia Electronica Aplicada a Agricultura-Escola Superior de Agricultura “Luiz de Queiroz”/University of São Paulo, Brazil. We thank all the members of the Angenent laboratories for fruitful discussion.
Glossary
- JA
jasmonic acid
- ChIP
chromatin immunoprecipitation
- SEM
scanning electron microscopy
- DEX
dexamethasone
- qRT
quantitative real-time
- Col-0
ecotype Columbia-0
- MS
Murashige and Skoog
- SD
synthetic dextrose
References
- Aggarwal P, Das Gupta M, Joseph AP, Chatterjee N, Srinivasan N, Nath U. (2010) Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GTS, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua N-H. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Bell E, Mullet JE. (1991) Lipoxygenase gene expression is modulated in plants by water deficit, wounding, and methyl jasmonate. Mol Gen Genet 230: 456–462 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E. (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Immink RGH, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Shchennikova AV, Franken J, Busscher M, Baskar R, Grossniklaus U, Angenent GC, Immink RGH. (2006) A Bsister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J 47: 934–946 [DOI] [PubMed] [Google Scholar]
- de Folter S, Urbanus SL, van Zuijlen LG, Kaufmann K, Angenent GC. (2007) Tagging of MADS domain proteins for chromatin immunoprecipitation. BMC Plant Biol 7: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y. (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y, et al. (2010) Increased leaf size: different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Vanhaeren H, Inzé D. (2012) Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci 17: 332–340 [DOI] [PubMed] [Google Scholar]
- Guo Z, Fujioka S, Blancaflor EB, Miao S, Gou X, Li J. (2010) TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS. (2007) Transcriptional auxin-brassinosteroid crosstalk: who’s talking? Bioessays 29: 1115–1123 [DOI] [PubMed] [Google Scholar]
- Hervé C, Dabos P, Bardet C, Jauneau A, Auriac MC, Ramboer A, Lacout F, Tremousaygue D. (2009) In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol 149: 1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T, Lewicki P. (2007). STATISTICA: Methods and Applications. StatSoft, Tulsa, OK
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Østerås M, Farinelli L, Krajewski P, Angenent GC. (2010a) Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat Protoc 5: 457–472 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM, et al. (2010b) Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. (1996) Genetic evidence for an essential role of brassinosteroids in plant development. Plant J 9: 701–713 [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B. (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P. (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZY, Li B, Dong AW. (2012) The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol Plant 5: 270–280 [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E. (1996) Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P. (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Masuda HP, Cabral LM, De Veylder L, Tanurdzic M, de Almeida Engler J, Geelen D, Inzé D, Martienssen RA, Ferreira PC, Hemerly AS. (2008) ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J 27: 2746–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Øbro J, Willats WGT, Persson S. (2008) GeneCAT: novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res 36: W320–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, King S, Jack T. (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naouar N, Vandepoele K, Lammens T, Casneuf T, Zeller G, van Hummelen P, Weigel D, Rätsch G, Inzé D, Kuiper M, De Veylder L, Vuylsteke M. (2009) Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J 57: 184–194 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Passarinho P, Ketelaar T, Xing M, van Arkel J, Maliepaard C, Hendriks MW, Joosen R, Lammers M, Herdies L, den Boer B, et al. (2008) BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol Biol 68: 225–237 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, Inzé D, Goossens A. (2008) Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K, Nath U. (2011) Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J 67: 595–607 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH. (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89: 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kiefer M, Eshed Y, Olszewski N, Weiss D. (2012) The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick BA, Zimmerman DC. (1983) The biosynthesis of jasmonic acid: a physiological role for plant lipoxygenase. Biochem Biophys Res Commun 111: 470–477 [DOI] [PubMed] [Google Scholar]
- Weight C, Parnham D, Waites R. (2008) LeafAnalyser: a computational method for rapid and large-scale analyses of leaf shape variation. Plant J 53: 578–586 [DOI] [PubMed] [Google Scholar]
- Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM. (2006) Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet 2: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilen RW, van Rooijen GJH, Pearce DW, Pharis RP, Holbrook LA, Moloney MM. (1991) Effects of jasmonic acid on embryo-specific processes in Brassica and Linum oilseeds. Plant Physiol 95: 399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.