Abstract
The organized lignocellulosic assemblies of cell walls provide the structural integrity required for the large statures of terrestrial plants. Silencing two CINNAMYL ALCOHOL DEHYDROGENASE (CAD) genes in Nicotiana attenuata produced plants (ir-CAD) with thin, red-pigmented stems, low CAD and sinapyl alcohol dehydrogenase activity, low lignin contents, and rubbery, structurally unstable stems when grown in the glasshouse (GH). However, when planted into their native desert habitat, ir-CAD plants produced robust stems that survived wind storms as well as the wild-type plants. Despite efficient silencing of NaCAD transcripts and enzymatic activity, field-grown ir-CAD plants had delayed and restricted spread of red stem pigmentation, a color change reflecting blocked lignification by CAD silencing, and attained wild-type-comparable total lignin contents. The rubbery GH phenotype was largely restored when field-grown ir-CAD plants were protected from wind, herbivore attack, and ultraviolet B exposure and grown in restricted rooting volumes; conversely, it was lost when ir-CAD plants were experimentally exposed to wind, ultraviolet B, and grown in large pots in growth chambers. Transcript and liquid chromatography-electrospray ionization-time-of-flight analysis revealed that these environmental stresses enhanced the accumulation of various phenylpropanoids in stems of field-grown plants; gas chromatography-mass spectrometry and nuclear magnetic resonance analysis revealed that the lignin of field-grown ir-CAD plants had GH-grown comparable levels of sinapaldehyde and syringaldehyde cross-linked into their lignins. Additionally, field-grown ir-CAD plants had short, thick stems with normal xylem element traits, which collectively enabled field-grown ir-CAD plants to compensate for the structural deficiencies associated with CAD silencing. Environmental stresses play an essential role in regulating lignin biosynthesis in lignin-deficient plants.
Plant cell walls are chemically complex, consisting of an extracytosolic matrix composed of extensively cross-linked polysaccharides (cellulose, hemicelluloses, and pectin) and are impregnated with lignin and proteins (Somerville et al., 2004; Popper, 2008; Gille et al., 2009; Achyuthan et al., 2010; Gilbert, 2010; Lionetti et al., 2010; Seifert and Blaukopf, 2010). Cell wall lignification is one of the most important evolutionary adaptations that plants evolved to facilitate their move to terrestrial habitats (Weng and Chapple, 2010). In addition to contributing to structural integrity, mechanical support, and robustness of plant stems, lignin waterproofs the cell wall, enabling long-distance water transport and protects plants from microorganisms, herbivores, and oxidative stress caused by UV irradiation (Koehler and Telewski, 2006; Robinson and Mansfield, 2009; Xu et al., 2009; Barakat et al., 2010; Kim et al., 2010; Tronchet et al., 2010). Lignin is chemically heterogeneous due to its biosynthesis by a free radical-mediated polymerization process, which links various phenolic monomers with carbon-carbon as well as ether linkages, making lignin structurally the most recalcitrant and cohesive component of the cell wall matrix (Boerjan et al., 2003; Chang, 2007; Torney et al., 2007; Nadji et al., 2009; Ralph, 2010). This cross-coupling within lignin polymers, as well as with cell wall polysaccharides, contributes to the structural stiffness and strength of the cell wall, allowing plants to attain their awesome upright statures. This tough structural component makes the generation of lignin-deficient and lignin-altered plants a key target of plant breeders and genetic engineers, as plants with altered lignin content and/or composition/structure may have diminished cell wall recalcitrance, allowing for more cost-effective and efficient plant biomass conversion for bioethanol or other liquid fuels (Chen and Dixon, 2007; Himmel et al., 2007; Vanholme et al., 2010).
The lignin composition and content can be modified in plants by silencing the expression of enzymes involved in the phenylpropanoid pathway. For instance, researchers have targeted the early upstream enzymes such as phenylalanine ammonia lyase (PAL; Huang et al., 2010; Song and Wang, 2011), cinnamate-4-hydroxylase (Chen and Dixon, 2007), 4-coumarate-coenzyme A ligase (4CL; Hu et al., 1999; Wagner et al., 2009; Voelker et al., 2010, 2011a, 2011b), hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase (Li et al., 2010), coumarate 3-hydroxylase (Ralph et al., 2006), and caffeoyl-CoA 3-O-methyltransferase (Guo et al., 2001; Day et al., 2009; Wagner et al., 2011) as well as downstream enzymes such as cinnamoyl-coenzyme A reductase (CCR; Ralph et al., 1998; Leplé et al., 2007; Derikvand et al., 2008; Wadenbäck et al., 2008; Ruel et al., 2009; Zhou et al., 2010), caffeate/5-hydroxyferulate O-methyltransferase (COMT; Piquemal et al., 2002; Guillaumie et al., 2008; Lu et al., 2010; Tu et al., 2010), and cinnamyl alcohol dehydrogenase (CAD; Halpin et al., 1994; Yahiaoui et al., 1998; Sibout et al., 2003, 2005). CAD is a multifunctional enzyme that regulates the last step of lignin biosynthesis by catalyzing the NADPH-dependent reduction of p-coumaraldehyde, coniferaldehyde, and sinapaldehyde to their corresponding hydroxycinnamyl alcohols (monolignols), which are further incorporated into lignin as p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units, respectively (Kim et al., 2002; Raes et al., 2003; Saballos et al., 2009).
Upon perceiving stress, plants undergo large-scale transcriptional, translational, and posttranslational reprogramming that directly influences a network of processes spanning the various levels of biological organization from the cellular to organismic levels (Hahlbrock et al., 2003; Molinier et al., 2006; Meldau et al., 2009; Nakashima et al., 2009; Spoel et al., 2009; Zeller et al., 2009; Walley and Dehesh, 2010). Lignification is also known to be stress inducible and is influenced by environmental stresses such as ozone, UV-B, heavy metal exposure, drought, pathogen infection, and nitrogen fertilization as well as by mechanical wounding, herbivore attack, thigmomorphogenic responses (wind and mechanical stimulations), and gravity (Pomar et al., 2004; Koehler and Telewski, 2006; Saidi et al., 2009; Moura et al., 2010; Simmons et al., 2010; Zhao and Dixon, 2011). Field studies have been conducted with plants altered in their lignin contents or composition to address the following objectives: (1) kraft pulping and saccharification properties, (2) the ecological consequences of lignin reduction for plant-herbivore and plant-pathogen interactions and decomposition, and (3) the consequences of environmental stresses on morphology, physiology, and wood characteristics.
Independent glasshouse (GH) and field studies were conducted with antisense CAD- and COMT-silenced poplar (Populus tremula × Populus alba; Pilate et al., 2002; Halpin et al., 2007) and CCR- and COMT-silenced (using an RNA interference [RNAi] strategy) perennial ryegrass (Lolium perenne; Tu et al., 2010) plants. These studies revealed that growth conditions had little impact on gene silencing efficiency and associated enzymatic activity, lignin content and chemistry, plant growth, and plant-insect or plant-microbe/pathogen interactions. Poplar (approximately 10% reduced Klason lignin content in one of the ir-CAD lines) and perennial ryegrass (approximately 37% and approximately 6% reduced acetyl bromide lignin contents in CCR- and COMT-silenced field plants, respectively) transgenics retained their reduced lignin phenotype even when grown under field conditions. However, a decline in growth was noticed in CCR- and COMT-silenced perennial ryegrass plants when the daily average temperature exceeded 40°C. Additionally, COMT-silenced perennial ryegrass plants experienced higher Puccinia rust infestations than did control plants in the field. In contrast, a decline in growth rates was observed in all CCR-silenced transgenic poplar lines when planted in the field (Leplé et al., 2007). The trend observed among independently CCR-silenced transgenic poplar lines found growth to be well correlated with the amount of lignin produced in these lines. A recent field study of 4CL down-regulated hybrid poplar plants found 4CL gene expression level and lignin chemistry to be key determinants of aboveground biomass, wood color, and tree form (Voelker et al., 2010). Highly silenced 4CL poplar plants had reduced aboveground biomass, irregular cambial activity, and reduced apical dominance that increased branching of free-standing plants in the field and resulted in vine-like growth when plants were supported with stakes in the GH (Voelker, 2009). These studies suggest that lignin-deficient plants respond differentially to growth environments that could be gene/species-specific or lignin content- and chemistry-specific responses. Little is still known about the stresses that influence lignin chemistry of lignin-deficient plants when grown under field conditions.
Nicotiana attenuata is a well-studied ecological model system that has been used to study traits that are important for the adaptation of plants to extreme environmental stresses. In N. attenuata’s native habitat, the Great Basin Desert of Utah, plants are exposed to a plethora of stresses simultaneously, such as heavy storms, herbivore and pathogen attack, high temperature, low humidity, high light intensity, and UV-B exposure. Here, we examined the ecological dynamics of lignin-deprived N. attenuata plants silenced in their CAD activity in their natural environment. We transformed N. attenuata plants using RNAi technology with an inverted-repeat (ir) sequence of their endogenous CAD gene to silence two stem-expressed NaCADs, referred to as ir-CAD plants. The ecological importance of lignification in N. attenuata-Trichobaris mucorea (tobacco stem weevil) interaction in the field has been reported recently using these NaCAD-silenced plants (Diezel et al., 2011). Here, we addressed the following questions by conducting a series of experiments with N. attenuata ir-CAD plants grown in both the GH and the plant’s native habitat in Utah. (1) What role does lignin play in plant development and stress resistance? (2) How do the stressful conditions influence the accumulation of various metabolites derived from the phenylpropanoid pathway in lignin-deprived plants, and what is their contribution to lignin composition and content? (3) How do environmental stresses influence the structural strength and architecture of lignin-deficient plants?
RESULTS
Two CAD Genes Are Silenced in ir-CAD Plants
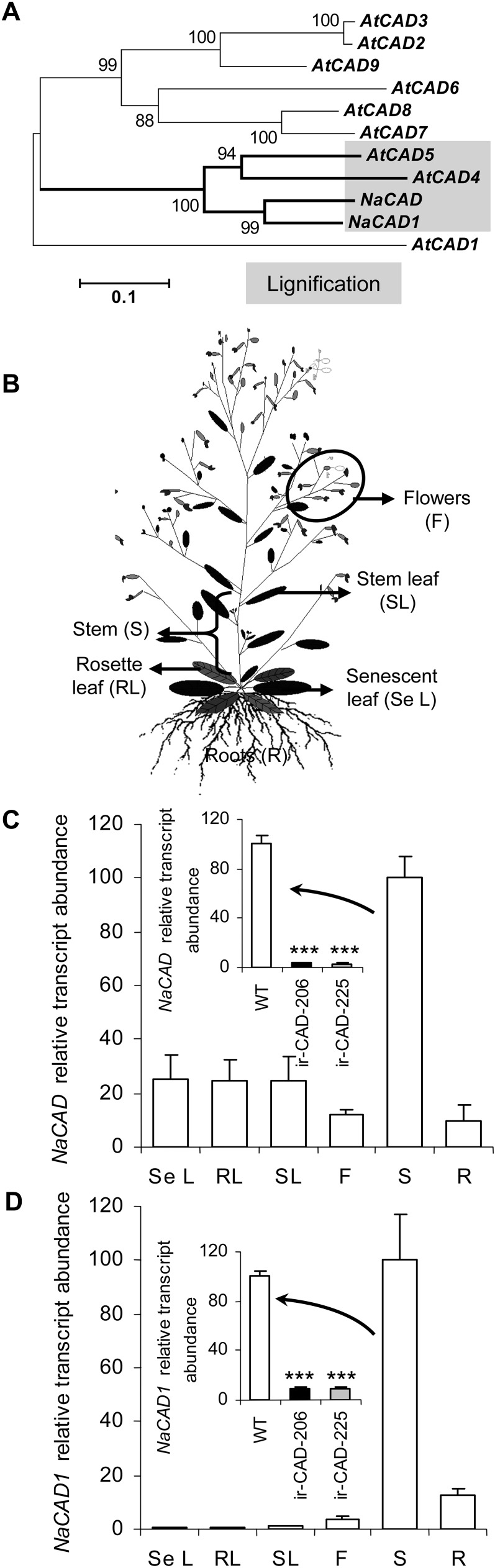
NaCAD1 was among 27 highly up-regulated proteins that showed consistent accumulation in N. attenuata leaves when plants were elicited by wounding and the wounds were immediately treated with Manduca sexta oral secretions or fatty acid-amino acid conjugates (an elicitor present in M. sexta oral secretions; Giri et al., 2006). This suggests that N. attenuata plants may enhance lignification by synthesizing more monolignols in the elicited leaf, perhaps to slow the entry of microorganisms or increase leaf toughness to reduce further herbivory. While isolating the full-length coding sequence of NaCAD1 (a gene fragment isolated by Giri et al. [2006] during a proteomic analysis), PCR-based cloning using wounding + oral secretion-elicited leaf cDNA as a template resulted in the isolation of an additional CAD gene, which was named NaCAD. When aligned at the nucleotide and protein levels (Supplemental Figs. S1 and S2), NaCAD and NaCAD1 shared a very high sequence identity to each other (approximately 85%). When a phylogenetic tree using all known Arabidopsis (Arabidopsis thaliana) CAD genes was constructed, it revealed that both N. attenuata CAD genes cluster with those of Arabidopsis bona fide CAD genes, namely AtCAD4 and AtCAD5, which are involved in floral stem lignification, suggesting that these CAD genes might also regulate lignification in N. attenuata (Fig. 1A; Sibout et al., 2003, 2005; Tronchet et al., 2010). Furthermore, the amino acid sequences of both NaCADs have well-conserved substrate-binding pockets and secondary structural elements (α-helices and β-strands) spanning catalytic and nucleotide-binding domains similar to those of AtCAD4 and AtCAD5, the characteristic signature of genes of the CAD family, as reported by Youn et al. (2006; highlighted in Supplemental Fig. S2). To assess the number of CADs in the N. attenuata genome, we probed a DNA blot with a fragment of NaCAD1 that had no restriction site for EcoRI and HindIII. After high-stringency washes of the hybridized blots, four distinct bands were detected in the EcoRI digest and seven in the HindIII digest, indicating that CADs exist as a multigene family in N. attenuata (Supplemental Fig. S3A).
Figure 1.
NaCAD genes cluster with bona fide AtCAD genes (AtCAD4 and AtCAD5) from Arabidopsis known to be involved in stem lignification. A, An unrooted phylogenetic tree of the NaCADs and AtCAD gene family. The dendrogram was generated with MEGA software (Tamura et al., 2007) using ClustalW and the neighbor-joining method, and bootstrap values are shown at each branch. Bootstrap tests were performed using 1,000 replicates. Gene accession numbers are shown in Supplemental Table S1. B, Schematic representation of N. attenuata wild-type plant parts analyzed for transcript expression. C and D. Mean ± se NaCAD (C) and NaCAD1 (D) transcript levels quantified by qRT-PCR in wild-type plants (n = 5). Insets present silencing efficiency of NaCAD (C) and NaCAD1 (D) in 10-cm basal stems of two independently transformed inverted-repeat homozygous lines (ir-CAD-206 and ir-CAD-225) at the early elongating stage of development (n = 5). Elongation factor, a housekeeping gene, was used to normalize all qRT-PCR assays. Relative transcript abundance was calculated with respect to wild-type (WT) stem levels, which were set to 100. Asterisks indicate significant differences among the genotypes (one-way ANOVA: *** P < 0.001).
We next investigated the tissue-specific transcript abundance of both CAD genes in wild-type N. attenuata plants from multiple tissues available at the mature stage of plant development (Fig. 1B). Both NaCAD and NaCAD1 genes are strongly expressed in stems, which is consistent with their expected function in stem lignification (Fig. 1, C and D).
In order to functionally test the activity of both CAD proteins in vivo, we used an RNAi approach to silence the expression of the CAD genes in N. attenuata. The pRESC5CAD binary vector containing the inverted repeat fragment of the endogenous NaCAD1 sequence used for silencing is shown in Supplemental Figure S3B. Initially, 20 independently transformed T1 lines were screened on hygromycin-containing medium, and based on segregation analysis, nine homozygous lines were selected in the T2 generation. The number of transfer DNA (T-DNA) insertions was determined by Southern hybridization of genomic DNA using a PCR fragment of the hygromycin phosphotransferase (hptII) gene as a probe (Supplemental Fig. S3C). As expected from the nucleotide and protein sequence similarities between NaCAD and NaCAD1, NaCAD1 silencing cosilenced NaCAD in six of the nine ir-CAD lines examined (Supplemental Fig. S3D). All six independently transformed lines that had significantly reduced transcript expression of both CAD genes also had weak stem phenotypes. For further studies, two of these six independently transformed, diploid, homozygous lines harboring a single copy of the T-DNA insertion and strongly silenced in the expression of both CAD genes (both apical and basal stems; 91%–93% silenced for NaCAD1 and 97% silenced for NaCAD in both lines) were selected for further experiments (ir-CAD-206 and ir-CAD-225; Fig. 1, C and D, inset; Table I; Supplemental Fig. S3C).
Table I. Transcriptional and functional characterization of the NaCAD gene in wild-type and ir-CAD N. attenuata plants.
Values are means ± se. Asterisks indicate significant differences among the genotypes (unpaired t test: * P < 0.05, *** P < 0.001); n = 5 for quantifying transcript abundance and enzymatic activity analysis, and n = 3 for all lignin analyses. n.d., Not detected.
| Parameter | Sample |
|
|---|---|---|
| GH Wild Type | GH ir-CAD | |
| Gene (transcript abundance %; relative to wild-type; apical stem) | ||
| NaCAD | 100 ± 10.21 | 3.02 ± 0.26*** |
| NaCAD1 | 100 ± 4.82 | 8.96 ± 1.75*** |
| Enzyme (nkat mg−1 protein; apical stem) | ||
| CAD activity | 0.35 ± 0.03 | 0.03 ± 0.01*** |
| SAD activity | 0.36 ± 0.02 | 0.02 ± 0.01*** |
| Lignin composition | ||
| Thioacidolysis-released (β-O-4-linked) monomers (µmol g−1 Klason lignin); basal stem and NMR data | ||
| H units | 26.19 ± 17.71 | 19.45 ± 4.33 |
| G units | 748 ± 28.50 | 390.70 ± 27.43*** |
| S units | 1,152.51 ± 36.48 | 324.21 ± 53.77*** |
| S/G (thio) | 60.6:39.4 | 45.3:54.7 |
| S/G (thio) | 1.54 | 0.83 |
| S/G (NMR) | 43:57 | |
| S/G (NMR) | 0.76 | |
| Coniferaldehyde | n.d. | 4.47 ± 0.70 |
| Sinapaldehyde | n.d. | 31.55 ± 3.16 |
| End group monomers (thioacidolysis) | ||
| Vanillin | n.d. | 7.88 ± 1.05 |
| Syringaldehyde | n.d. | 53.98 ± 3.20 |
| Klason lignin content (%; basal stem) | 21.35 ± 0.51 | 12.13 ± 1.80* |
| Young’s modulus (Gpa; basal stem) | 6.43 ± 0.52 | 2.2 ± 0.25*** |
| Bending strength (Mpa; basal stem) | 2.02 ± 0.08 | 0.9 ± 0.06*** |
GH-Grown ir-CAD Plants Develop a “Rubbery” Phenotype
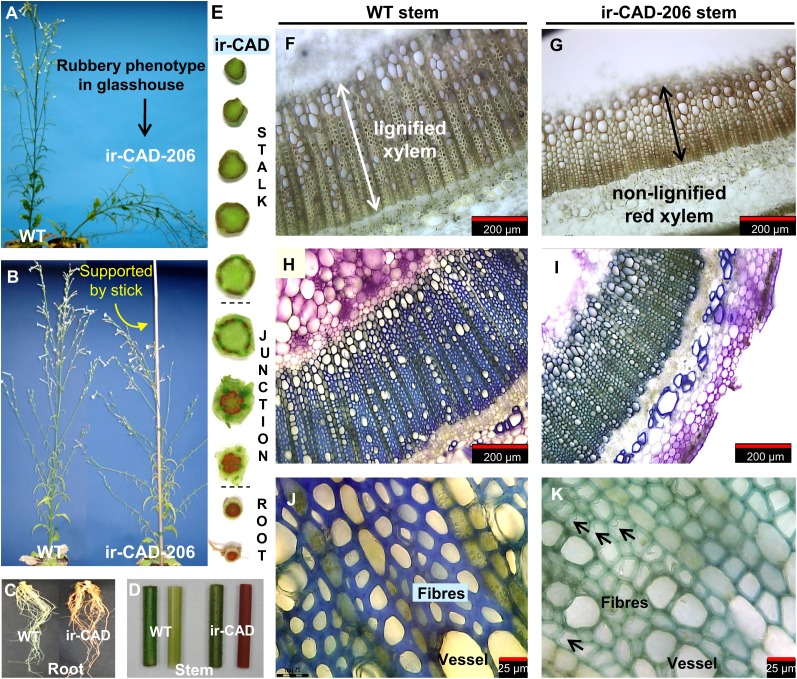
Lignin deposition in the cell wall plays an important role in plant development, especially during elongation. As expected, CAD silencing in N. attenuata produced plants with strongly altered stem morphologies and architectures after their developmental transition from rosette- to flowering-stage growth (Fig. 2, A and B). In the initial stages of bolting (46 d after germination), the length and appearance of GH-grown ir-CAD and wild-type stems were similar (Supplemental Fig. S4, A [unpaired t test, Pir-CAD-206 = 0.087] and B [unpaired t test, Pir-CAD-225 = 0.631]). However, a delay in stem elongation was observed during the third week of bolting in ir-CAD plants (53 d after germination; unpaired t test, Pir-CAD-206 < 0.0001, Pir-CAD-225 < 0.0001), which resulted in significantly shorter plants at maturity (75 d after germination; unpaired t test, Pir-CAD-206 < 0.0001, Pir-CAD-225 < 0.0001). A slight reduction in ir-CAD stem diameter was also observed at the base of the stem (Supplemental Fig. S4C; unpaired t test, Pir-CAD-206 = 0.026, Pir-CAD-225 = 0.025) and the upper part (40 cm from the base) of the main stem axis (unpaired t test, Pir-CAD-206 = 0.004, Pir-CAD-225 = 0.003). Mature ir-CAD plants were bushy in appearance, having more lateral branches (more than 15 cm in length; Fig. 2B; Supplemental Fig. S4, C and D; unpaired t test, Pir-CAD-206 = 0.008, Pir-CAD-225 < 0.0001), and these branches were located closer to the base of the stems than was the case for wild-type plants (α; Supplemental Fig. S4, C–E; unpaired t test, Pir-CAD-206 = 0.010, Pir-CAD-225 = 0.010). These lateral branches spread farther from the main stem of ir-CAD plants than from those of wild-type plants. This morphological change was quantified as the difference in both (1) the angle between the main stem and the lateral branches (unpaired t test, Pir-CAD-206 < 0.0001, Pir-CAD-225 < 0.0001) and (2) the distance between the main stem and a point 15 cm along the lateral branch after its attachment to the main stem (ρ; Supplemental Fig. S4, C and E; unpaired t test, Pir-CAD-206 < 0.0001, Pir-CAD-225 < 0.0001).
Figure 2.
GH-grown NaCAD-silenced plants have altered stem morphology, which makes plants structurally unstable. A, The rubbery phenotype of ir-CAD plants is most apparent at the flowering stage, when stems lignify. B, ir-CAD plant supported by a stick to display the altered plant architecture responsible for its bushy appearance. C to E, CAD-silenced plants develop red coloration in roots (C) and stems (D; left is intact and right is peeled) compared with wild-type (WT) plants. This red coloration is largely restricted to xylem tissue of the ir-CAD stem, as shown in the peeled stem segment (D) and transverse sections cut through the root, the root-shoot junction, and the stem of ir-CAD plants (E). Red color provides a visual signature of “frustrated” lignin deposition. F, Light microscopy of an unstained cross-section of a wild-type stem, with the white arrow showing highly lignified xylem. G, The black arrow depicts the zone of red pigment accumulation in xylem of an ir-CAD stem. H to K, N. attenuata wild-type stems have a thick xylem layer that develops blue color (H and J) on staining with toluidine-blue O, whereas ir-CAD stems have thin xylem tissues that stain bluish-green (I and K). J, A magnified toluidine blue O-stained wild-type xylem transverse section reveals that xylem is composed of thick-walled fibers and vessels. K, Collapsed fibers and vessels with thin, wavy, and poorly resolved walls of the ir-CAD stem’s xylem are indicated by black arrows.
When ir-CAD plants were grown unsupported in the GH, most of the plants buckled under their own weight at maturity: ir-CAD stems were soft and flexible and could be easily bent by hand. Because these plants were unable to grow upright and were structurally unstable at maturity, we refer to them as having a rubbery phenotype. We conducted one-point bending tests on basal stem segments of ir-CAD and wild-type plants to quantify their stiffness. Highly significant differences were observed both in the Young’s modulus and bending strength among ir-CAD and wild-type stems (Table I; Supplemental Fig. S5, A and B), suggesting that a lack of stiffness of ir-CAD stems was responsible for their instability. Despite striking changes in stem morphologies of ir-CAD plants, no differences were observed in their fresh biomass (Supplemental Fig. S4C; unpaired t test, Pir-CAD-206 = 0.447, Pir-CAD-225 = 0.259) and seed production capacity (Supplemental Fig. S4C; unpaired t test, Pir-CAD-206 = 0.583, Pir-CAD-225 = 0.532) when compared with the wild-type plants. However, ir-CAD plants had less dry biomass than did wild-type plants (unpaired t test, Pir-CAD-206 = 0.002, Pir-CAD-225 < 0.0001). In addition, the CAD-silenced lines tended to senesce earlier than did the wild-type plants.
An intense reddish-brown color was first noticeable in the roots of ir-CAD plants and later appeared at the base of the main stems and their respective lateral branches; the color spread acropetally along the longitudinal axis of the stem, replacing the normally green color of the stem as the lignification process proceeded (Fig. 2, C and D). This red coloration was restricted to the xylem of ir-CAD stems, the portions that would otherwise be lignified in wild-type stems (Fig. 2, E–G).
To examine the degree of xylem lignification, we stained cross-sections of ir-CAD as well as wild-type stems with toluidine blue O dye. The red xylem tissue of ir-CAD stems turned various shades of violet upon staining as a result of varying red backgrounds; however, the normally lignified tissue of the wild-type plant developed a blue color (Fig. 2, H–K). The red xylem of ir-CAD stems was weakly stained, poorly lignified, and had collapsed elements (vessel and fibers) with thin, highly wavy, disorganized, and noncoherent walls (black arrows in Fig. 2K), whereas the wild-type stems had highly lignified xylem with thick-walled elements.
Characterization of Lignin and Enzyme Activity of GH-Grown ir-CAD Plants
We examined the effect of NaCAD and NaCAD1 gene silencing on CAD/sinapyl alcohol dehydrogenase (SAD) enzymatic activity by performing in vitro assays using the corresponding alcohol substrates (sinapyl alcohol or coniferyl alcohol) and crude protein extracts from basal stem segments (0–10 cm above the root/shoot transition zone) of plants at early elongating stages as well as from the unlignified apical part of mature stems of flowering-stage plants. Silencing the NaCADs yielded plants with highly reduced CAD (retained only approximately 10%–15% of wild-type activity in both lines at both developmental stages) and SAD (retained only approximately 5%–10% of wild-type activity) enzymatic activities (Table I; Supplemental Fig. S5, C and D).
We further analyzed the CAD/SAD-associated changes in lignin content and composition of basal parts of mature wild-type/ir-CAD stems, as wild-type stems are expected to be lignified at this stage of development; hence, the contrasting lignin-associated changes of ir-CAD stems could be readily visualized. The growth experiments were initially conducted on two independently transformed ir-CAD lines; due to the similarity of the growth phenotype of both lines, the lignin analyses (composition and content) were performed only on the ir-CAD-206 line. Quantification of Klason lignin revealed that CAD-silenced N. attenuata plants had highly reduced (approximately 40%) stem lignin contents relative to wild-type plants (unpaired t test, PGH-ir-CAD = 0.022; Table I). N. attenuata wild-type plants released more thioethylated S monomeric units compared with G monomeric units from their lignin by analytical thioacidolysis. The amounts of released S and G monomeric units, indicative of the β-O-4-linked units, were significantly reduced in the lignin of ir-CAD stems compared with those of wild-type stems; however, no change was observed in the release of H monomeric units among the genotypes (Table I). The reduction in S units was greater compared with G units in CAD-silenced plants (in ir-CAD plants relative to wild-type plants, S units = 28% and G units = 52%), which was also evident from the low thioacidolysis S/G ratio of ir-CAD plants. Apart from quantifying the most abundant monolignol units in the lignin polymer, the less frequent thioacidolysis-derived indenes from sinapaldehyde or coniferaldehyde 8-O-4-linked units, and the dithioketals from 4-O-end-linked syringaldehyde and vanillin units, as reported by Kim et al. (2002) and Lapierre et al. (2004), were recovered only from ir-CAD samples, highlighting the atypical incorporation of hydroxycinnamaldehydes into the lignin heteropolymer. It should be noted that although the hydroxycinnamaldehydes are reported to be responsible for the red color of stems and roots, a characteristic signature of CAD silencing, the coloration is not derived from the incorporation of the aldehydes into the lignin (Table I); in fact, it has been demonstrated that even if the red-colored material is removed, the lignins still display the characteristic signatures of hydroxycinnamaldehyde incorporation (Ralph et al., 2008). Furthermore, the sinapaldehyde 8-O-4 linkage occurred more frequently than the coniferaldehyde 8-O-4 linkage in the lignin of ir-CAD plants (as confirmed below from NMR observations). There was no evidence for the involvement of end-linked sinapaldehyde and coniferaldehyde as lignin components; the accumulation of these molecules was below the detection limit of the gas chromatography (GC)-mass spectrometry (MS) system that we used. However, the derived syringaldehyde units were incorporated, as also shown below by NMR.
To test if the acropetal spread of red coloration could be used as a nondestructive visual marker of the gradient of lignification that normally occurs in wild-type stems, we compared epidermis-peeled lateral stems from wild-type and ir-CAD plants at similar stages in development. We fixed the ir-CAD stems to a sheet of paper at both ends, whereas the corresponding wild-type stems were fixed only at their bases, with the apical part left free to bend under the influence of gravity to visualize lignified (stiff) and nonlignified (soft) portions of the wild-type stem (Supplemental Fig. S6A). The extent of red coloration spread across ir-CAD stem corresponded exactly to the stiff portions of the wild-type stems and the green parts to the bending portions of the wild-type stems (Supplemental Fig. S6A). Furthermore, toluidine blue O staining of cross-sections obtained from various segments of wild-type stem showed a clear basipetal lignification gradient along the stem axis (Supplemental Fig. S6B). The longitudinal distribution of red color in ir-CAD stems reflects the height to which the lignification process would have occurred in wild-type stems, providing a reliable visual marker of wild-type lignification processes that are “frustrated” in ir-CAD stems.
ir-CAD Plants Grown in the Field Lose Their Rubbery Phenotype
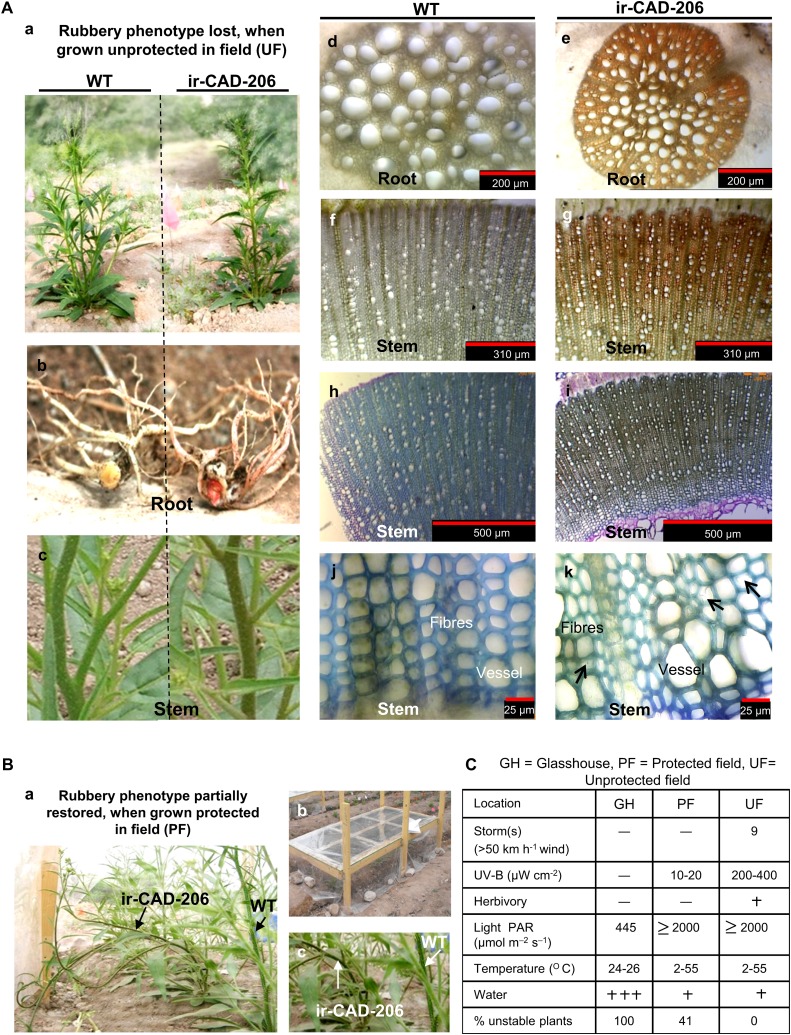
We planted 19 pairs of ir-CAD and wild-type N. attenuata plants in their native habitat in a field plot in 2008, where the plants were exposed to the myriad of biotic and abiotic stresses that characterize their natural environment. Interestingly, all ir-CAD plants survived five heavy storms (defined as having wind speeds gusting above 50 km h−1) as well as the wild-type plants and were structurally stable and had floral stem morphology comparable to those of wild-type plants (Supplemental Fig. S7). To determine if the apparent loss of the rubbery growth phenotype in the field was due to reverted CAD silencing (i.e. CAD level restoration), we examined ir-CAD plants for the accumulation of the red pigment. Both roots and stems of field-grown ir-CAD plants retained the red xylem phenotype similar to that of GH-grown ir-CAD plants, suggesting that silencing of the targeted CAD genes still occurred in the field (Supplemental Fig. S7).
To further investigate whether changes in environmental conditions were responsible for the structural stability of ir-CAD plants, we conducted additional growth experiments in the field in 2009. We planted 19 pairs of wild-type and ir-CAD plants in the field (exposed to natural environmental conditions; i.e. unprotected field [UF] plants) and 12 pairs under simulated GH growth conditions (whereby plants were grown under UV-B-opaque plastic sheets, which protected plants from both wind and UV-B exposure, and sprayed with a systemic insecticide to protect them from herbivory; i.e. protected field [PF] plants). To simulate growth in 1-L pots in the GH, PF plants were planted in close proximity to each other. Despite experiencing nine heavy storms in the 2009 growing season, UF ir-CAD plants attained UF wild-type growth morphology and structural stability while still accumulating the red pigment in both their roots and stems (Fig. 3A), as they did in the 2008 growing season. Due to transport restrictions on the shipment of fresh transgenic stems back to the laboratory in Germany, the biomechanical bending measurements could not be conducted on field-grown plants.
Figure 3.
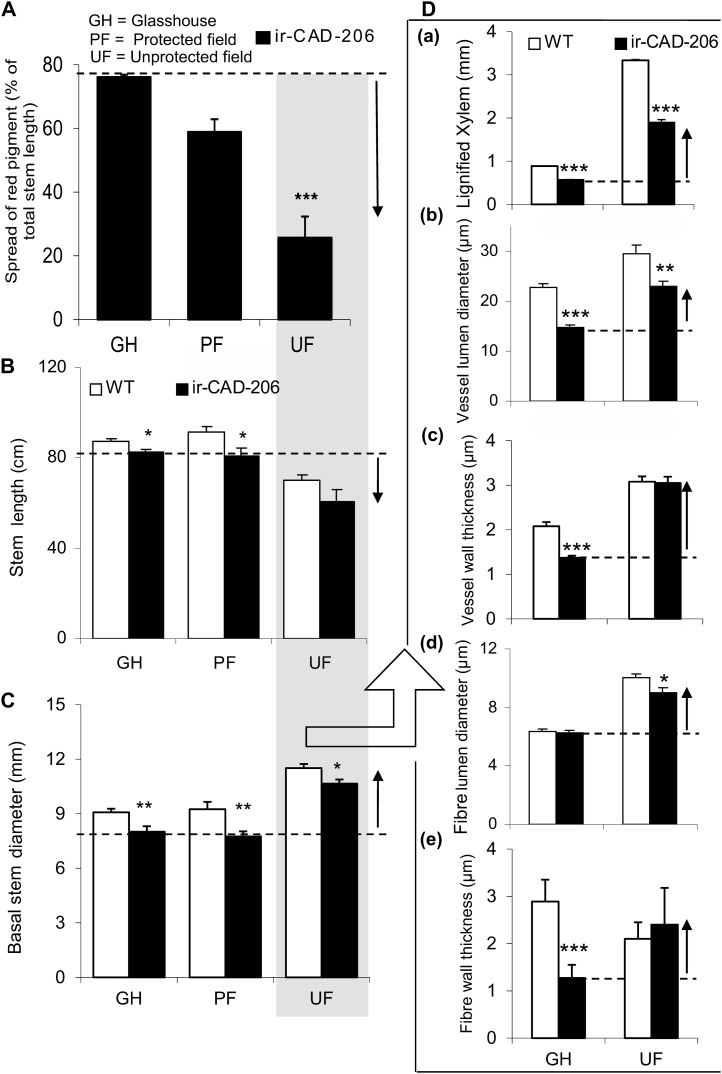
Rubbery, structurally unstable GH-grown ir-CAD plants acquire the stability of wild-type (WT) plants when grown in a field plot in their native environment of the Great Basin Desert. Aa, Structurally stable ir-CAD plants grown in a UF environment. Ab and Ac, Both UF wild-type and UF ir-CAD plants exhibited similar stem architecture. Wild-type plants had light brown roots (Ab) and bright green stems (Ac). Ad to Ag, Relative to the control plants (transverse section: root xylem [Ad], stem xylem [Af]), UF ir-CAD plants had red xylem both in roots (intact [Ab], transverse section [Ae]) and stems (intact [Ac], xylem cross-section [Ag]). Ah to Ak, Toluidine blue O dye stained UF wild-type and UF ir-CAD stems in xylem cross-section. UF wild-type plants had highly lignified xylem with thick-walled elements (vessels and fibers), as depicted from histochemical staining, whereas ir-CAD plants had lignified xylem with occasionally collapsed elements and disorganized walls indicated by black arrows (Ak). Ba, The rubbery phenotype of ir-CAD plants is partially restored when UF-grown plants are protected from wind, UV-B, and herbivore exposure. Bb, Experimental design of PF plants that were grown under a canopy of UV-B-opaque plastic (to eliminate UV-B exposure and wind-associated thigmomorphogenic effects) and were sprayed with a systemic insecticide (to minimize herbivory-induced responses). Bc, ir-CAD plants retained the color and rubbery phenotypes of GH ir-CAD plants when grown under PF conditions. C, Summary of stresses that plants faced in GH, PF, and UF growth environments.
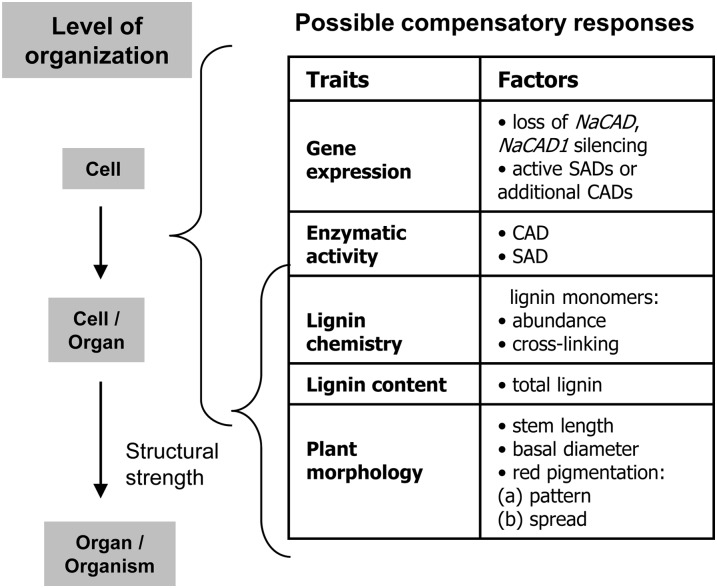
Histochemically, UF ir-CAD stems appeared more similar to UF wild-type stems (Fig. 3A). The red xylem of UF ir-CAD stems stained blue, indicating that field-grown ir-CAD plants were somewhat lignified. About 40% of the PF-grown ir-CAD plants exhibited the rubbery phenotype that is typically observed when plants are grown in the GH (Fig. 3B). In general, ir-CAD plants displayed contrasting growth phenotypes in terms of stem stability in the different environments, highlighting that difference in exposure to stresses could be the key factor that determines their stability (Fig. 3C). We next embarked on multiscale phenotypic analysis to understand the environmentally mediated changes that might have occurred at different levels of organization (as listed in Fig. 4) and contributed to the structural stability of UF ir-CAD plants.
Figure 4.
List of possible compensatory processes occurring at different hierarchical levels, ranging from transcript to whole plant, that could account for loss of the rubbery phenotype of ir-CAD plants when grown in their native environments (UF).
Growth and Stem Anatomy of ir-CAD Plants in GH, PF, and UF Growth Environments
We used the “stage, not age” maxim (the height at which PF ir-CAD plants attained a rubbery phenotype similar to that of GH ir-CAD plants) to compare stem characteristics across plants grown in the different environments. In other words, we used the stage of stem development, rather than the days after transplantation into the field plot, as the criterion to compare GH-, UF-, and PF-grown ir-CAD plants. When PF-grown ir-CAD plants attained stem lengths of approximately 80 cm, the height at which GH-grown ir-CAD plants fully express their phenotype (as shown in Fig. 2, A and B; Supplemental Fig. S4D), we found that the longitudinal spread of red pigmentation depended strongly on the growth environment (Figs. 3C and 5A). While UF-grown ir-CAD plants had the least red spread, PF-grown plants were intermediate, and the highest spread was found in GH-grown plants (one-way ANOVA, F2,21 = 33.490, PPF-ir-CAD = 0.06, PUF-ir-CAD < 0.0001). Growth under UF conditions was delayed for both genotypes, which developed short, thick stems compared with the long, thin stems of PF- and GH-grown plants (Fig. 5, B and C). However, both PF- and GH-grown plants were comparable in their growth morphology; in general, ir-CAD plants displayed a slight delay in growth compared with wild-type plants (for stem height, Fig. 5B [paired t test, PUF ir-CAD = 0.101, paired t test, PPF ir-CAD = 0.037, unpaired t test, PGH ir-CAD = 0.013]; for basal stem diameter, Fig. 5C [paired t test, PUF ir-CAD = 0.037, paired t test, PPF ir-CAD = 0.007, unpaired t test, PGH ir-CAD = 0.01]).
Figure 5.
Varying growth conditions induced phenotypic changes that likely contribute to the ability of ir-CAD plants to regain structural stability when grown in nature. A, Mean ± se percentage of total stem lengths to which red pigment had accumulated in ir-CAD plants grown in three different environments (GH, PF, and UF). Asterisks indicate significant differences among environments (one-way ANOVA). B and C, Mean ± se stem length (B) and basal stem diameter (C) of ir-CAD and wild-type (WT) plants grown in GH, PF, and UF environments. D, Mean ± se changes in the width of lignified xylem tissue that constituted the lignified part of the stems (a) and its components: vessel (lumen diameter [b] and wall thickness [c]) and fiber (lumen diameter [d] and wall thickness [e]). Asterisks indicate significant differences among the genotypes (unpaired t test: * P < 0.05, ** P < 0.01, *** P < 0.001).
Both genotypes showed differences in their stem-xylem anatomy under GH and UF conditions (Fig. 5D). Relative to GH plants, the width of xylem tissue in stem cross-sectional area increased in UF-grown plants; however, in all environments, the width of xylem tissues in ir-CAD stem cross-section was markedly lower than that of wild-type plants (Fig. 5D; unpaired t test, PUF-ir-CAD < 0.0001, unpaired t test, PGH-ir-CAD < 0.0001). GH ir-CAD stem xylem had thin-walled vessels and fibers with narrow lumens (Fig. 5D). In contrast, the xylem of UF ir-CAD stems had thick-walled vessels and fibers with wide lumens and was almost twice the thickness of GH stems. The UF ir-CAD plants modified their vasculature anatomy and stem morphology to become very similar to that of UF wild-type plants.
Growth in the Field Alters Lignin Composition
The ir-CAD plants displayed their red-xylem phenotype when grown in the field but differed strikingly in the structural stability of their stems. Therefore, we analyzed transcripts, enzymatic activity (CAD/SAD), and lignin content (Klason) and composition (thioacidolysis and NMR) to better understand the change(s) responsible for the upright stature of ir-CAD plants grown under UF conditions. Both under PF and UF growth conditions, ir-CAD plants had highly reduced NaCAD and NaCAD1 gene expression with extremely down-regulated CAD/SAD enzymatic activity compared with wild-type plants (Table II). In contrast to GH-grown plants, field-grown (UF and PF) ir-CAD plants had wild-type-equivalent lignin contents (Table II). Furthermore, regardless of growth conditions, ir-CAD plants released significantly lower amounts of S and G monomeric units via thioacidolysis, indicating that the levels of β-O-4-linked units in their lignins were lower than in similarly grown wild-type plants (S units in ir-CAD plants relative to their wild-type plants: PF = 12%, UF = 24%; G units in ir-CAD plants relative to their wild-type plants: PF = 40%, UF = 53%). However, no change was discerned in the accumulation of H monomeric units (as revealed by thioacidolysis monomer yields) among the genotypes. All genotypes varied inconsistently across growth environments: the highest accumulation was detected in UF plants. Relative to GH wild-type plants, the abundances of both S and G units were significantly higher in field-grown wild-type plants.
Table II. Lignin-associated changes in wild-type and ir-CAD plants when grown under PF and UF environments in their native habitat in southwestern Utah.
Values are means ± se. Asterisks indicate significant differences among the genotypes (paired t test: * P < 0.05, *** P < 0.001); n = 5 for transcript abundance quantification and enzymatic activity analysis, and n = 3 for all lignin analyses. n.d., Not detected.
| Parameter | Sample |
|||
|---|---|---|---|---|
| PF Wild Type | PF ir-CAD | UF Wild Type | UF ir-CAD | |
| Gene (transcript abundance %; relative to GH wild type; apical stem) | ||||
| NaCAD | 123.63 ± 15.61 | 4.40 ± 1.06*** | 119.82 ± 13.20 | 3.43 ± 0.31*** |
| NaCAD1 | 102.49 ± 12.03 | 6.55 ± 1.83*** | 87.50 ± 25.31 | 7.49 ± 2.49*** |
| Enzyme (nkat mg-1 protein; apical stem) | ||||
| CAD activity | 0.38 ± 0.02 | 0.036 ± 0.01*** | 0.3 ± 0.05 | 0.025 ± 0.00*** |
| SAD activity | 0.27 ± 0.03 | 0.03 ± 0.01*** | 0.25 ± 0.04 | 0.019 ± 0.01*** |
| Lignin composition | ||||
| Thioacidolysis-released (β-O-4-linked) monomers (µmol g-1 Klason lignin); basal stem and NMR data | ||||
| H units | 51.43 ± 4.25 | 34.28 ± 11.45 | 96.48 ± 13.94 | 59.12 ± 29.82 |
| G units | 1,119.20 ± 84.10 | 447.97 ± 62.24*** | 919.12 ± 102.63 | 487.50 ± 82.16*** |
| S units | 2,467.49 ± 82.30 | 304.10 ± 54.39*** | 2,401.2 ± 268.58 | 582.71 ± 182.75*** |
| S/G (thio) | 72.3:27.7 | 54.4:45.6 | ||
| S/G (thio) | 2.20 | 0.67 | 2.61 | 1.20 |
| S/G (NMR) | 56:44 | 48:52 | ||
| S/G (NMR) | 1.28 | 0.91 | ||
| Coniferaldehyde | n.d. | 7.83 ± 1.35 | n.d. | 11.07 ± 1.47 |
| Sinapaldehyde | n.d. | 48.97 ± 2.90 | n.d. | 53.48 ± 7.23 |
| End group monomers (thioacidolysis) | ||||
| Vanillin | n.d. | 6.02 ± 0.50 | n.d. | 2.16 ± 0.08 |
| Syringaldehyde | n.d. | 32.43 ± 2.01 | n.d. | 14.27 ± 1.27 |
| Klason lignin content (%; basal stem) | 26.07 ± 1.99 | 27.84 ± 3.27 | 26.60 ± 1.13 | 25.27 ± 1.56 |
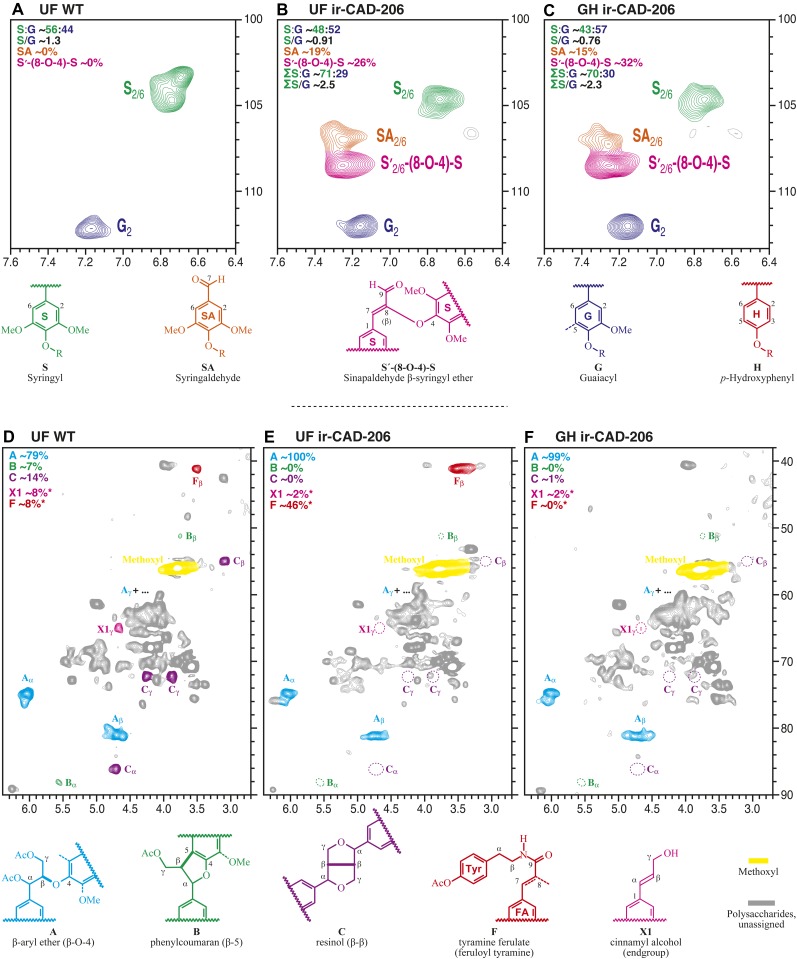
To augment the thioacidolysis method, which provides measurements based on the fraction releasable as monomers, NMR analysis is useful, as it provides total measures of the different units that constitute lignin. We analyzed acetylated lignin-rich residues following cellulase treatment of the cell walls isolated from N. attenuata stems following the methods of Ralph et al. (1998, 2006) and Bunzel and Ralph (2006). Figure 6, A to C, presents the partial two-dimensional 13C-1H heteronuclear single quantum coherence (HSQC) spectra highlighting the striking differences in the lignin composition of UF wild-type, UF ir-CAD, and GH ir-CAD plants. Figure 6, A to C, is plotted with comparable G levels for comparison, but it was evident that, when compared with the lignin of UF wild-type plants, UF ir-CAD lignin was significantly reduced in both S (C2,6-H2,6 in S units cross peaks at δH/δC = 6.75/104 ppm) and G (C2-H2 in G units cross signals at δH/δC = 7.2/112 ppm) levels; moreover, the reduction in monolignol levels in UF ir-CAD lignin was comparable to that of GH ir-CAD lignin, indicating that variation in growth conditions does not influence the incorporation of hydroxycinnamyl alcohol or monolignols into ir-CAD lignin. Measures of relative S and G levels are available from contour integration (H levels were impossible to determine by NMR due to spectra interference from unknown components in this region [data not shown]). Values are indicated on the plots (along with S/G values calculated from just the normal components and those including the sinapaldehyde- and syringaldehyde-incorporated units, and the former are indicated in Tables I and II for comparison with thioacidolysis data. NMR is also able to measure the relative levels of the various types of incorporated aldehydes, primarily 4-O-linked syringaldehyde units and sinapaldehyde-(8-O-4) units linked predominantly to S units (see the colored contours and structural assignments in Fig. 6). The main observations are as follows. The conventional S/G is lowest in the GH ir-CAD lines, with both GH and UF ir-CAD lines having lower S/G than the wild-type, in agreement with thioacidolysis (even though the S/G is measured on different fractions of the lignin in the two analyses). Interestingly, if the (largely S) aldehyde units are included in the calculation, both GH and UF ir-CAD lines had a similar S/G, now very much higher than in the wild-type plant material. There is a caution, however. The syringaldehyde units are always end units, and the sinapaldehyde-(8-O-4) units are also often terminal free phenolic units (Lapierre et al., 2004). In long pulse trains, such as in the HSQC experiment, such end groups are overrepresented (in the integrals) relative to the units in the backbone of the polymer. Thus, although relative values are valid, absolute quantifiability should not be assumed. Finally, the total syringaldehyde + sinapaldehyde-(8-O-4) level in the two CAD lines is about the same, with some shifts between each; it is not clear at which point syringaldehyde units derive (by retroaldol reactions) from sinapaldehyde units, so it is not clear whether or not the apparently lower syringaldehyde level in the GH ir-CAD plants is due to the better incorporation of the sinapaldehyde monomers into the lignin [as sinapaldehyde-(8-O-4)-units] due to the additional stresses.
Figure 6.
A to C, Partial two-dimensional HSQC NMR spectra of UF wild-type (WT; A), UF ir-CAD (B), and GH ir-CAD (C) acetylated whole lignin released after cellulase treatment. ir-CAD plants grown under UF conditions incorporated lower amounts of hydroxycinnamyl alcohol moieties, namely monomers resulting in S (green) and G (blue) units into their lignins, than did the UF wild-type plants. ir-CAD plants accumulated similar levels of alcohol units both under GH and UF conditions. Characteristic signals (as indicated by coloring of the contours and the partial structures) reflect the cross-peaks of end group hydroxybenzaldehyde (primarily syringaldehyde) and incorporated sinapaldehyde moieties, which are signatures of ir-CAD lignin. D to F, Two-dimensional HSQC NMR spectra of acetylated lignin obtained from UF wild-type (D), UF ir-CAD (E), and GH ir-CAD (F) stems. Monolignol units are combined to produce different interunit linkages as well as characteristic end units, as depicted in wild-type lignin. The NMR signals reflect the frequency of different cross-linkage types of the monolignol units. Lignin isolated from ir-CAD stems (E and F) is largely devoid of all the typical minor units (phenylcoumaran B, resinol C, and cinnamyl alcohol end group X1) that are part of wild-type lignin but retain normal β-ether unit A as the major interunit structure, reflecting a different lignification pathway in which lignification is started via the hydroxybenzaldehydes (and perhaps hydroxycinnamaldehydes) instead of the usual monolignol dehydrodimerization. UF ir-CAD lignin (E) accumulated higher amounts of tyramine ferulate moieties, indicating increased stress in these plants.
The HSQC spectra of aliphatic side chains characterize the patterns of monolignol interunit linkages that are involved in lignification (Fig. 6, D–F). Both UF and GH ir-CAD lignins differed substantially from UF wild-type lignin in their HSQC spectra, revealing an almost complete disappearance of phenylcoumaran B (correlation signals at δH/δC = 5.57/88 ppm for Cα-Hα and δH/δC = 3.78/51 ppm for Cβ-Hβ, reflecting β-5-coupling of G units), resinol C (correlation signals at δH/δC = 4.75/86, 3.12/54.2, and 3.9,4.29/72 ppm for Cα-Hα, Cβ-Hβ, and Cγ-Hγ, representing β-β-coupling, mainly from sinapyl alcohol dimerization), and cinnamyl alcohol end group X1 (from monomer-monomer coupling; cross-peaks at δH/δC = 6.63/133.5 ppm for Cα-Hα, δH/δC = 6.25/121.5 ppm for Cβ-Hβ, and δH/δC = 4.65/65 ppm for Cγ-Hγ) in ir-CAD lignin. These changes reflect a similarly extreme down-regulation of monolignol accumulation in field-grown as in GH ir-CAD plants but arise more importantly from the incorporation of sinapaldehyde into the polymer, and syringaldehyde as polymer starting units, at least units C and X1, but also B, arise primarily from monomer dimerization reactions necessary to initiate polymerization, a role apparently assumed in these plants by syringaldehyde.
From these results, we conclude that neither the silencing machinery (transcriptional and posttranscriptional changes involving both enzymatic and metabolic monolignol lignin composition) nor alternative CAD or CAD-associated gene(s) were activated in response to growth in the field, ruling out these mechanisms as the cause of the surprising structural stability of UF ir-CAD plants.
As lignin polymerization is a purely chemical combinatorial radical-coupling process (Ralph et al., 2004), we hypothesized that higher cross-linking of atypical phenolic constituents into the insoluble lignin fraction might have occurred in response to growth under field conditions. We found, from thioacidolysis, a trend for greater incorporation of sinapaldehyde (and coniferaldehyde) by 8-O-4 coupling in the lignin of ir-CAD plants when grown in PF and UF conditions, highlighting the cross-coupling of hydroxycinnamaldehydes as a stress-dependent process (Tables I and II). Interestingly, exactly the opposite trend was observed for the incorporation of syringaldehyde and vanillin units as end groups; lowest incorporations were found in UF ir-CAD plants. Furthermore, elevated hydroxycinnamaldehyde levels were detected in the lignin of both UF- and GH-grown ir-CAD plants in two-dimensional HSQC NMR spectra (Fig. 6). Taken together, these results strongly suggest that the hydroxycinnamaldehydes were incorporated into the lignin backbone in all CAD-deficient plants, with thioacidolysis levels suggesting that they were more frequent in UF ir-CAD plants than in PF or GH ir-CAD plants.
Additionally, UF ir-CAD lignin was distinctly different from UF wild-type lignin in its tyramine ferulate accumulation (at δH/δC = 3.5/40.5 ppm for Cβ-Hβ [Fig. 6E], along with other consistent correlations, e.g. at δH/δC = 2.75/35 ppm for Cα-Hα [data not shown]); tyramine ferulate is well recognized as a stress-response compound in tobacco (Nicotiana tabacum). Note that the absolute tyramine levels are significantly lower than indicated here by the integrals because tyramine units are slowly relaxing terminal units, but the ratio of enhancement in the UF ir-CAD plants (approximately 6-fold over the wild type) is valid. It is interesting that the level is very low in the GH ir-CAD plants, suggesting that the stress response is not related to the CAD deficiency itself. Other spectra (data not shown) indicated differences in the ferulate levels and the degree of ferulate dehydrodimerization, but the signals are weak and inconclusive.
PAL Transcripts and Soluble Metabolite Profiles of ir-CAD Plants
Plants contain large amounts of soluble phenolic substances that function in protection against abiotic and biotic stresses. To explore the possible association between lignin biosynthesis and the accumulation of soluble metabolites as it is influenced by different growth conditions, an unbiased liquid chromatography-electrospray ionization-time-of-flight mass spectrometry (LC-ESI-TOF MS; in positive mode) analysis (illustrated in Supplemental Fig. S8) was performed on extracts obtained from basal stem segments of both genotypes grown under different environmental conditions (GH and UF) at two stages of stem development (early elongated or nonlignified and flowering or lignified state; i.e. before and after red coloration development in ir-CAD plants). After preprocessing of the LC-ESI-TOF MS data set (XML Cryptographic Message Syntax [XCMS]-based peak picking and subsequent reconstruction of isotopes, adducts, and in-source fragments into pseudospectra by CAMERA [Supplemental Data Set S1] and further filtering [Gaquerel et al., 2010; Onkokesung et al., 2012]), consistent mass features present at least in two of three biological replicates (retention time greater than 1 min, mass-to-charge ratio [m/z] signal greater than 120, and 75th percentile normalization) yielded 6,320 total ion traces (Supplemental Data Set S2).
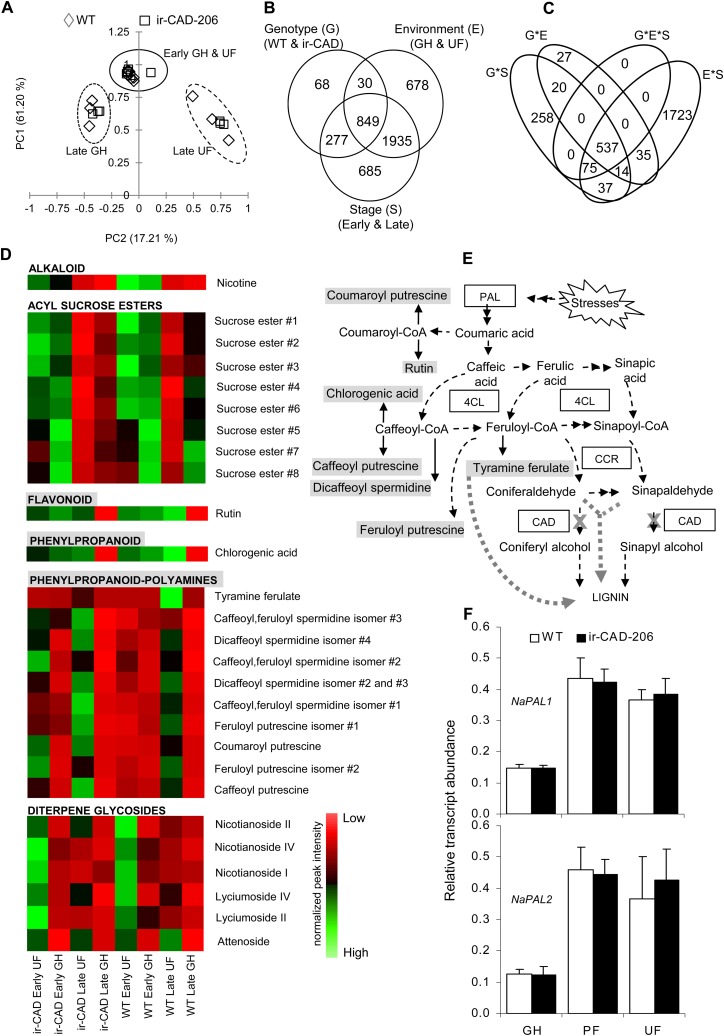
Principal component analysis was conducted on these ions. The first two principal components accounted for 78.4% of the total variance and resolved the samples into distinct clusters. The principal component analysis revealed that wild-type and ir-CAD plants accumulate similar types and amounts of metabolites at early development (before lignification of the stem), which is independent of the stresses plants experienced (Fig. 7A). PC1 clearly separated metabolites of lignified from nonlignified stems. PC2 included metabolites that separated samples from GH and UF growth conditions.
Figure 7.
Levels of PAL transcripts and associated metabolites increase in UF-grown plants during later growth stages. A, Principal component analysis score plot of 6,320 metabolite ions obtained after processing of the raw LC-ESI-TOF MS data set from stems of early- and late-stage wild-type (WT)/ir-CAD plants grown in different environments. The metabolite profiles of both genotypes were similar at the early stage of plant development irrespective of growth environment but differed when the plants started to lignify (n = 3). B and C, Venn diagrams summarize the influence of three components, genotype (G), environment (E), and growth stage (S; B), and their interaction (C) on soluble secondary metabolite ion accumulation in 10-cm-long basal segments of nonlignified (early stage of development; i.e. before the onset of red color in ir-CAD stems) and late-stage lignified stems of ir-CAD-206 and wild-type plants grown in UF and GH growth conditions and analyzed by LC-ESI-TOF MS (three-way ANOVA: P < 0.001). D, Heat map showing the changes in metabolite accumulation in stems occurring across wild-type/ir-CAD plants grown in different environments. Each square represents the average value of normalized peak intensities obtained from three biological replicates ranging from high (green) to average (black) to low (red) levels. E, Biosynthetic associations among measured soluble metabolites and lignin biosynthesis (represented by dashed black arrows). The thick gray dotted arrow depicts metabolite flux into lignin when the CAD gene is silenced (denoted by a cross). F, Mean ± se NaPAL1 (top panel) and NaPAL2 (bottom panel) transcript levels measured by qRT-PCR in wild-type and ir-CAD upper stem parts (50–60 cm from the ground level) harvested from flowering plants grown in GH, PF, and UF growth conditions (n = 6). Elongation factor, an unregulated housekeeping gene, was used to normalize all qRT-PCR assays.
In summary, soluble phenolic metabolism is not strongly influenced by the environment during early plant growth, before the lignification process has commenced, in both the genotypes. A three-way ANOVA (P > 0.001) was performed on 6,320 filtered metabolite ions to assess the impact of individual factors, namely genotype, environment, and development stage, and their interactions on the accumulation of metabolites, which is summarized in Venn diagrams (Fig. 7, B and C; Supplemental Data Set S3). Most of the metabolite ions accumulated in response to either the interaction of development stage and stress factors (2,421 of 4,522; Supplemental Fig. S9) or the interaction of all three variables: genotype, environment, and growth stage (612 ions). Field-grown plants at a late stage of development accumulated higher levels of phenylpropanoid pathway-derived metabolites than of acyl Suc esters in their stems (Fig. 7, D and E); however, the GH-grown plants showed the reverse trend.
PAL is the first rate-limiting enzyme in the phenylpropanoid pathway leading to lignin biosynthesis and is known to be influenced by a variety of growth conditions and stresses (Chang et al., 2008; Olsen et al., 2008; Huang et al., 2010). We hypothesized that field-related stresses could enhance PAL gene expression, which might explain both the higher accumulation of the above-mentioned PAL-derived soluble metabolites and lignin contents of field-grown plants (Tables I and II). We quantified the transcript abundance of two PAL genes from N. attenuata, namely NaPAL1 and NaPAL2, in wild-type and ir-CAD plants grown in PF, GH, and UF conditions and found significant increases in the abundance of both NaPAL1 and NaPAL2 transcripts in all field-grown (both UF and PF) plants of both genotypes as compared with GH plants (Fig. 7F).
Adding Stresses Restores Structural Stability to GH-Grown ir-CAD Plants
When ir-CAD plants were grown protected from UV-B, herbivory, and wind with minimal rooting volumes, the rubbery phenotype observed in GH-grown plants was largely restored. The fact that PF-grown ir-CAD plants had high PAL transcript levels and incorporated hydroxycinnamaldehydes into their lignin just as UF-grown ir-CAD plants did demonstrates that PF-grown plants were exposed to additional stresses such as low humidity, high temperature, and high light intensity, which GH-grown plants were not subjected to, and may explain why the rubbery phenotype was not fully restored in the PF environment.
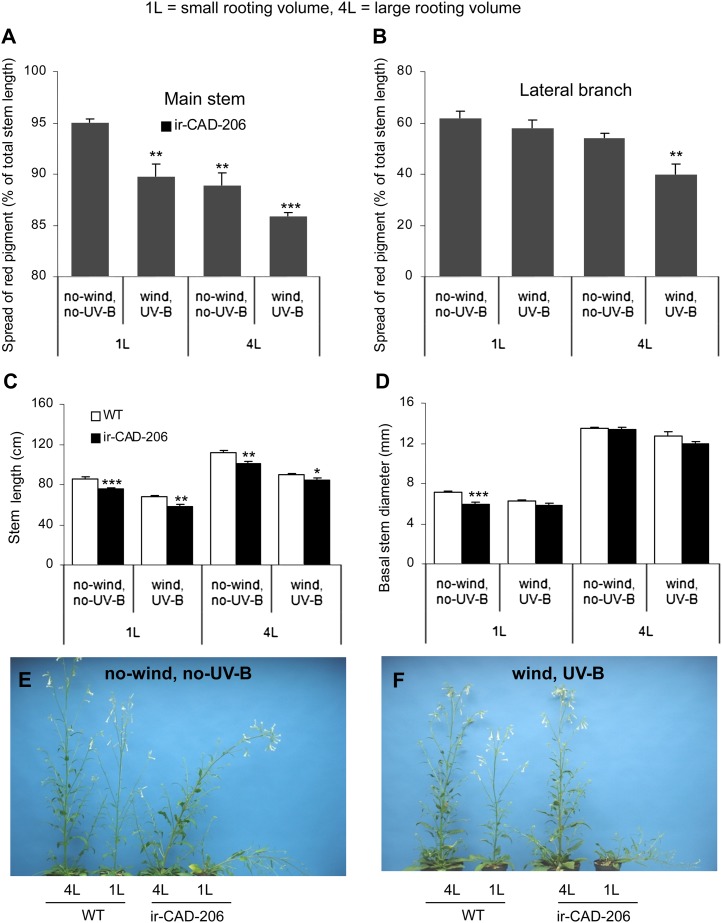
To examine this hypothesis, we exposed ir-CAD plants grown in climate chambers to two stresses, wind and UV-B, and also manipulated the rooting volumes of the plants. Initial GH/climate chamber studies with exposure to single stress factors (UV-B or wind) alone in 1-L pots were not able to restore structural stability to ir-CAD plants. Therefore, experiments were conducted with seven plants for each genotype per treatment (grown in 1- and 4-L pots, unexposed and exposed simultaneously to wind and UV-B; Supplemental Fig. S10). Interestingly, approximately 80% of ir-CAD plants grown in 4-L pots exposed to both wind and UV-B fully regained their structural stability, a result similar to that observed in UF plants (Fig. 8). Additionally, we also observed the same pattern of the spread of red pigments along the stems that we had observed in the field: the pigment dispersion both in the main stems and lateral branches of ir-CAD plants was greatest in 1-L nonexposed plants, was intermediate in 1-L-exposed and 4-L-nonexposed plants, and was least in 4-L-exposed plants, in which the red pigmentation was confined to a zone around the base of the stem. Furthermore, exposed plants grown in 4-L pots had short stems compared with 4-L-nonexposed plants. Similar reductions in plant height were also observed in 1-L-exposed plants, highlighting that UV-B and wind exposure reduces plant growth. Although 1-L-xposed ir-CAD plants were shorter compared with 1-L-unexposed ir-CAD plants, their thin stems were not able to support them and they leaned, suggesting that sufficient rooting volume was an essential factor determining stem thickness and, hence, stability. Additionally, 4-L-exposed ir-CAD plants grew slower and were shorter than 4-L-unexposed ir-CAD plants but attained similarly thick stems at their base, which may have contributed to their upright stature. These results collectively demonstrate that exposure of ir-CAD plants to high UV-B, wind, and providing large rooting volumes allowed structurally unstable ir-CAD plants to regain their upright stature in the climate chambers.
Figure 8.
Leaning GH-grown ir-CAD plants attain structural stability when grown in large pots and exposed to wind and UV-B. A and B, Percentage of total red-pigmented main stem (A) and lateral branch (B) of ir-CAD plants when grown in 1-L and 4-L pots exposed to wind, UV-B, and nonexposed conditions. C and D, Mean ± se stem length (C) and basal stem diameter (D) of ir-CAD-206 and wild-type (WT) plants grown in 1-L (small rooting volume) and 4-L (large rooting volume) pots in unexposed control and wind- and UV-B-exposed conditions in the climate growth chamber. E and F, The rubbery phenotype of unstressed ir-CAD plants (E) and the stability resulting from exposing ir-CAD plants to wind, UV-B, and growth in large pots in the climate chamber (F). Differences are indicated between wild-type and ir-CAD plants (unpaired t test: * P < 0.05, ** P < 0.01, *** P < 0.001) and among ir-CAD plants grown under different growth conditions (one-way ANOVA). [See online article for color version of this figure.]
A Stressful UF Environment Influences Pigmentation in Sectors of ir-CAD Stems
We noticed that field-grown ir-CAD stems occasionally developed green sectors in their red xylem that ran longitudinally up the stem and encompassed, on average, approximately 16% of a stem’s xylem circumference, a situation not observed in GH- or PF-grown ir-CAD stems (Fig. 9 [one-way ANOVA, F2,52 = 6.246, PPF ir-CAD = 0.917, PUF ir-CAD = 0.002]; Supplemental Fig. S11). The green sectors of ir-CAD xylem stained light blue with toluidine blue O dye and had thin-walled fibers relative to wild-type xylem, suggesting that ir-CAD nonlignified red stems had acquired some lignified green sectors when planted in the field (Supplemental Fig. S11).
Figure 9.
ir-CAD plants grown in UF environments developed green vascular sectors in their red stems more frequently than did PF- or GH-grown ir-CAD plants. Values show mean ± se percentage of red pigmentation in the xylem circumference of the basal stem. The asterisks indicate significant differences among ir-CAD plants grown in different environments (one-way ANOVA: ** P < 0.01).
DISCUSSION
Most of the research on lignin-modified transgenic plants has been motivated by the applied objectives of reducing lignin content so as to increase the efficiency of biomass conversion and improve Kraft pulping and forage qualities (Simmons et al., 2010; Tu et al., 2010; Fu et al., 2011). Some of these have been tested in field trials to determine if the improvements in saccharification rate, pulping, and forage properties observed in the GH trials were maintained during growth in the field (Pilate et al., 2002). These trials suggest that lignin-modified plants are capable of attaining wild-type statures (Pilate et al., 2002; Halpin et al., 2007; Fornalé et al., 2011). In addition, these transformed plants have also been used to examine the consequences of lignin alteration for plant-soil, plant-insect, and plant-microbe interactions (Pilate et al., 2002; Tilston et al., 2004; Halpin et al., 2007).
The flexible culm1 rice (Oryza sativa) mutant represents an atypical example of a single T-DNA insertion mutant of a single CAD gene that resulted in a rubbery stem phenotype (Li et al., 2009). Silencing two CAD genes in Arabidopsis in the double CAD (CAD-C and CAD-D) mutants resulted in plants with reduced lignin contents that were structurally unstable with flexible and mechanically weak stems (Sibout et al., 2005). Neither of these structurally unstable mutants has been grown in field trials. The N. attenuata CAD-silenced lines we generated were also silenced in the expression of two CAD genes and had weak unstable stems, and here we report, to our knowledge, the first field study of a structurally unstable lignin-deficient plant. The motivation for this work was ecological: we wanted to compare the ecological performance of plants with rubbery stems and those with erect stems, as plasticity in stem morphology is frequently observed as a response to growth in open habitats without competitors and under conditions of intense competition (Ballaré et al., 1994; Smith and Jordan, 1994; Schmitt et al., 1995; Umeki, 1995; Schwinning and Weiner, 1998; Anten et al., 2005).
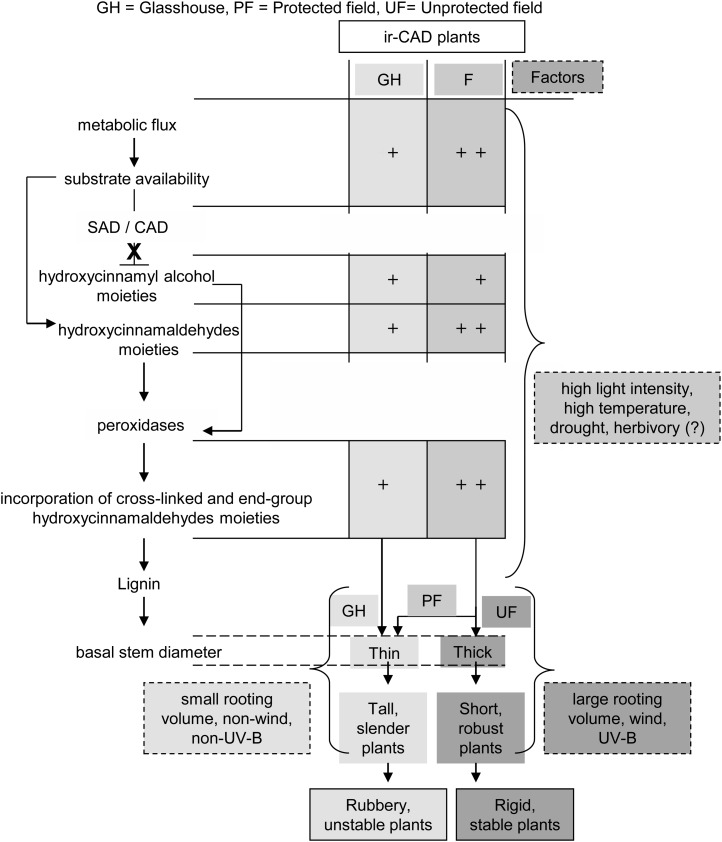
To our surprise, the rubbery stem phenotype disappeared when plants were grown in the field. Because we could rule out the most prosaic explanation of this result, namely the loss of gene silencing in the field, we examined many other potential compensatory processes that could account for the loss of the rubbery phenotype and found that exposure to environmental stresses activated a suite of compensatory responses operating at many levels in the hierarchy of biological organization (Fig. 10). Our analysis spans the entire chasm that links gene to phenotype and as a consequence only begins to identify compensatory responses at the different levels that monolignol biosynthesis-defective plants employ for regaining their stem structural stability. Much additional work will be required to understand each of the processes that are involved at the different levels in the analysis. In the following, we consider the evidence for the existence of different compensatory responses at the transcriptional, biochemical, and tissue levels and consider future research directions that could further elucidate these mechanisms.
Figure 10.
Model depicting the possibilities of compensatory responses that allowed ir-CAD plants to become structurally stable when exposed to environmental stresses.
Structural Compensation Does Not Result from CAD Transcriptional or Biochemical Compensation
CAD exists as a multigene family in angiosperms, and different CAD genes are thought to play different roles. To date, nine CAD genes are known from Arabidopsis (Kim et al., 2004), 12 from rice (Tobias and Chow, 2005), 16 from Populus trichocarpa (Shi et al., 2010), 15 from hybrid poplar (Populus deltoides × Populus nigra; Barakat et al., 2009), 18 from grapevine (Vitis vinifera; Barakat et al., 2009), 14 from sorghum (Sorghum bicolor; Saballos et al., 2009), and 17 from Medicago truncatula (Barakat et al., 2009); however, the number known to be directly involved in lignification is substantially fewer. For example, Arabidopsis AtCAD4 and AtCAD5 are known to be involved in lignification, as the double mutants have flexible and structurally unstable stems and similar genotype-phenotype relationships as known for the flexible culm1 rice mutant (Sibout et al., 2005; Li et al., 2009; Thévenin et al., 2011). Some evidence exists for transcriptional compensation, as slight increases in AtCAD1, AtCAD6 (or AtCAD-A; Kim et al., 2004), and AtCAD9 (or AtCAD-G; Kim et al., 2004) transcript abundance has been reported for the AtCAD4/AtCAD5 double mutant (Sibout et al., 2005). Additionally, despite lacking in vitro CAD catalytic activities, only AtCAD1 expression partially increased the level of thioacidolysis-releasable monomeric units and, conversely, released lower levels of indenes (derived from the incorporation of hydroxycinnamaldehydes into the lignin) in the AtCAD4/AtCAD5 double mutant, indicating that AtCAD1 may marginally compensate for AtCAD4/AtCAD5 lignin deficiencies (Eudes et al., 2006). Promoter sequence analysis, gene organization, and tissue-expression profile studies of the 15 CAD genes in hybrid poplar revealed that different CAD genes differ in their expression (constitutive or in a stressed state) and function (involved in various developmental and physiological processes; Barakat et al., 2009) under different conditions (for instance, herbivore damage; Barakat et al., 2010). These results highlight a degree of functional redundancy among CADs, and we first explored if a similar redundancy among the CADs of N. attenuata could account for the observed structural compensation.
First, it was clear that the RNAi construct we used silenced the expression of two CAD genes that play a central role in lignin biosynthesis in N. attenuata (NaCAD and NaCAD1) and that the efficiency of silencing was not influenced by growing plants in the field (Tables I and II). Highest NaCAD and NaCAD1 transcript expression in stems relative to other tissues analyzed, along with their phylogenetic clustering with Arabidopsis CAD genes (i.e. AtCAD4 and AtCAD5) known to be involved in lignification (Fig. 1), and that the deduced primary amino acid sequences of NaCADs revealed the well-conserved substrate-binding pocket residues, two-domain structures, and secondary structural elements including the Phe substitution at position 299 in the active site conferring substrate specificity of the AtCAD4 and AtCAD5 proteins, were all consistent with a conserved function of CADs involved in lignin biosynthesis (Supplemental Fig. S2; Youn et al., 2006; Kim et al., 2010).
The ir-CAD lines we generated had reduced CAD/SAD enzymatic activities (Table I; Supplemental Fig. S5, C and D). The thioacidolysis-released G and S monomeric units were also drastically reduced in ir-CAD plants, whereas hyperaccumulation of sinapaldehyde and coniferaldehyde 8-O-4-linked moieties was evident, which suggest that both sinapaldehyde and coniferaldehyde are the preferred substrates of the CADs (this study) in N. attenuata (Table I). The accumulation of hydroxycinnamaldehydes is likely responsible for the red color of ir-CAD xylem in stems and roots, which developmentally tracked the lignification process (Figs. 2 and 3). This red signature has been reported in CAD-silenced Arabidopsis (Sibout et al., 2005), poplar (Baucher et al., 1996), sorghum (Pillonel et al., 1991), tobacco (Higuchi et al., 1994; Ralph et al., 1998), maize (Zea mays; Halpin et al., 1998; Vermerris et al., 2002, 2010), and rice (Zhang et al., 2006) plants. The red pigmentation of ir-CAD stems was partly extractable in methanol. This is not consistent with previous reports of the color’s resulting from the lignins but is consistent with the observations (Ralph et al., 2008) that extraction of the red color does not remove components bound into the lignin (and hence is not responsible for the color). Further studies are needed to unravel the nature of extractable red-colored signatures of ir-CAD plants.
Clearly, like poplar and Arabidopsis, there are additional CAD genes in N. attenuata’s genome, but the lack of a genome sequence for N. attenuata (a diploid), or even a publicly available tobacco (a tetraploid) sequence, thwarts precise estimates. We detected four bands on EcoRI-digested and seven bands on HindIII-digested Southern blots when probed with the NaCAD1 fragment (Supplemental Fig. S3A), reflecting the existence of more than two CAD genes in the N. attenuata genome, and these unknown NaCADs might become activated during growth in the field to compensate for the silencing of NaCAD and NaCAD1 expression in ir-CAD plants. Interestingly, we found no evidence for this. Field-grown (PF and UF) ir-CAD plants maintained GH-comparable levels of NaCAD transcript expression, CAD/SAD enzymatic activity, and thioacidolysis-releasable G and S monolignol monomeric units (Tables I and II). Additionally, field-grown ir-CAD plants also had red stems (Fig. 3A; Supplemental Fig. S7) with a significantly higher incorporation of coniferaldehyde and sinapaldehyde monomers 8-O-4 linked into the lignins, as shown both by thioacidolysis (Table II) and NMR analysis (Fig. 6, B and C). HSQC spectra analysis also revealed that UF and GH ir-CAD lignins differ substantially from wild-type lignin in their S and G lignin components, the levels of incorporated hydroxycinnamaldehydes (primarily sinapaldehyde) and hydroxybenzaldehydes (primarily syringaldehyde), and the consequent distribution of interunit linkages and end groups: phenylcoumaran, resinol, and end-linked cinnamyl alcohol abundances were markedly lower (Fig. 6, D–F). In contrast to tobacco CAD-silenced stem lignin (Ralph et al., 1998), N. attenuata CAD-silenced stem lignin showed a complete loss of most of the monolignol-derived interunit signals other than β-ether A (Fig. 6, D–F) in HSQC NMR spectra. The difference is likely due to the lower degree of CAD down-regulation than in the plants created here.
Collectively, these results allow us to rule out two explanations for the loss of the rubbery phenotype in field-grown plants: the stress activation of other unknown CAD genes and the loss of NaCAD gene silencing. These inferences can be more directly tested once a full genome sequence is available for N. attenuata and full transcriptional profiling can be conducted on the entire CAD family of genes.
Stress-Activated Phenolics Are Recruited into the Attenuated Lignin Biosynthetic Pathway and Contribute to the Structural Stability of Field-Grown ir-CAD Plants
Previously, it has been shown that silencing genes in the monolignol biosynthetic pathway substantially affects the flux of intermediates (soluble pools) synthesized via interconnected pathways that can influence the chemical composition of the lignin polymer (the insoluble fraction of the cell wall; Bonawitz and Chapple, 2010). For example, silencing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase produced plants that accumulate high levels of flavonol glycosides and acylated anthocyanins (Besseau et al., 2007), whereas silencing caffeoyl-CoA 3-O-methyltransferase or COMT in plants results in the formation of benzodioxanes, a novel compound arising from the incorporation of 5-hydroxyconiferyl alcohol (poplar; Lu et al., 2010) or caffeyl alcohol (Pinus radiata; Wagner et al., 2011) into the lignin polymer (Ralph et al., 2001). The ability of lignin-modified plants to incorporate nonmonolignol monomers was also demonstrated in plants lacking CCR and/or CAD activity (Ralph et al., 1998; Thévenin et al., 2011). CCR-down-regulated plants produced lignins that contained increased levels of ferulic acid (Leplé et al., 2007; Ralph et al., 2008), and tobacco lignins incorporated tyramine ferulate (Ralph et al., 1998). In general, CAD deficiency results in lignin that is rich in nonmonolignol moieties, such as hydroxycinnamaldehydes and hydroxybenzaldehydes (Kim et al., 2000, 2002, 2003; Lapierre et al., 2004; Sibout et al., 2005).
It is noteworthy that lignin composition depends largely on the plant’s physical (stressed or nonstressed) and developmental stages (for review, see Boerjan et al., 2003; Bonawitz and Chapple, 2010; Moura et al., 2010), highlighting the fact that the capacity of plants to incorporate phenolic moieties into lignin could also vary with environmental conditions. For instance, the abundance of tyramine ferulate was reported to increase in response to wounding and pathogen infection in plants (Negrel et al., 1996; Schmidt et al., 1998; King and Calhoun, 2005). This suggests that CAD-silenced plants, when stressed, could acquire the ability to cross-link more of the atypical components into their lignin, which might have contributed to the stability of ir-CAD plants when grown in UF growth conditions.
As discussed above, substantial differences in lignin chemistry in terms of nonmonolignol (hydroxycinnamaldehydes and tyramine ferulate) and monolignol moieties existed between N. attenuata ir-CAD and wild-type plants, as revealed by GC-MS of thioacidolyzed lignin and NMR analysis of acetylated lignin, were observed; these chemical differences were further modified by growth conditions. A stress-dependent increase in the incorporation of sinapaldehyde, coniferaldehyde, and tyramine ferulate moieties (UF > PF > GH) and, conversely, the decrease in end group-linked syringaldehyde and vanillin (UF < PF < GH) units were found in ir-CAD plants (Tables I and II), whereas wild-type plants showed a stress-dependent increase in monolignol (S and G units) cross-linking (UF > PF > GH; Tables I and II). These findings collectively suggest that growth in the field indeed increased the total pools of various moieties and their incorporation into lignin. However, more detailed structural work will be required to evaluate whether the incorporation of atypical phenolics into the lignin of ir-CAD stems contributes to a plant’s restored structural integrity during field growth.
The expression of PAL genes is primarily responsible for directing the carbon flux from the shikimate pathway into phenylpropanoid metabolism, and these genes are known to be regulated by a large number of stress stimuli; hence, the differential regulation of PAL genes of plants grown in the field could explain the above-mentioned changes in lignin chemistry of both genotypes. As expected, we found significant increases in NaPAL transcript abundance in plants grown in UF environments (Fig. 7F). These environmentally induced changes (amplified NaPAL expression and subsequently enhanced accumulation and cross-linking of PAL-derived metabolites into lignin) could have enabled ir-CAD plants to achieve stability in the UF environments. This inference could be more rigorously tested by examining PAL activity and isolating different lignin-specific peroxidases and laccases from ir-CAD plants grown in the different environments and investigating their role in mediating the oxidative coupling of various atypical phenolic moieties into lignin.
Field-Grown ir-CAD Plants Modify Their Stem Morphology to Acquire Stability
Plants in nature are frequently subjected to mechanical perturbations caused by wind, rain, and herbivore activity that vary in intensity. In order to acclimatize to these thigmomorphogenic stresses, plant generally modify their stem architecture by developing compact stems that are shorter and thicker (Niklas, 1998; Coutand, 2010). GH-grown ir-CAD plants had thin, slender, flexible stems that were structurally unstable (Figs. 2A and 5, B and C); however, when planted into their native habitat, the plants showed a delay in growth and produced shorter stems that were thicker at the base at maturity (Fig. 5, B and C). This suggests that stress-driven changes in stem morphology (a reduction in stem length and an increase in stem thickness) could be one of the factors that increase the stability of ir-CAD plants in the field. To further evaluate their role, stresses were either removed from plants grown in the field (PF situation) or added to GH-grown plants in climate chambers. Interestingly, ir-CAD plants grown in the field partially restored their rubbery phenotype when protected from wind, UV-B, and herbivory and produce long, slender stems (Figs. 3B and 5, B and C); conversely, when exposed to wind and UV-B and grown in larger pots in the climate chambers, they developed short, thick stems that enabled 80% of ir-CAD plants to regain their stable upright form (Fig. 8). Additionally, the different light photosynthetically active radiation (PAR) levels (1,000 µmol m−2 s−1 PAR in climate growth chambers, 450 µmol m−2 s−1 in GH, and 2,000 µmol m−2 s−1 in UF growth environments; Fig. 3C) could be an additional contributing factor (Chenu et al., 2005). Changes in the stem stature and thickness of ir-CAD plants were associated with changes at the cellular level. The UF ir-CAD plants showed 2-fold increases in xylem cross-sectional width in thick stems compared with the slender stems of GH ir-CAD plants (Fig. 5Da). Similar increases in the thickness of the xylem tissue cylinder were reported in stems of antisense CAD tobacco plants that were subjected to mechanical stimulation, suggesting that stimulated plants increase their investment in radial growth and produce more robust stems (Hepworth and Vincent, 1999). An increase in the number of vessels that had wide lumens and thick walls was found in UF ir-CAD plants, changes that may contribute to the structural stability of stems (Fig. 5, Db–De). Additionally, it is also known that increases in fiber lumen and wall thickness increase stem strength (Poorter et al., 2010). In short, UF ir-CAD plants modified their xylem anatomy (increased their xylem cross-sectional width, developed thick vessels and fibers with wide lumens, and created stems that were thick at the base) and produced chemical/structural changes in the walls that collectively may have contributed to the substantial phenotypic changes, and in particular the rubbery phenotype reversion, that occurred in CAD-down-regulated plants when grown under UF conditions.
CONCLUSION
Our research highlights the importance of environmental stresses in regulating lignin biosynthesis in lignin-deficient plants. In contrast to structurally unstable ir-CAD plants grown in the GH, UF-grown ir-CAD plants attained statures and stabilities comparable to wild-type plants. Exposure to various environmental stresses allowed for the incorporation of atypical phenolic units (hydroxycinnamaldehydes and tyramine ferulates) into the lignin, possibly modified cell wall cross-linking, modified xylem vasculature, developed lignified within stem green sectors, and produced shorter plants with thicker stem bases. These changes, collectively, increased lignin contents and compensated for ir-CAD plants’ structural deficiencies. We are just starting to understand the role that stresses play in affording stability to otherwise unstable lignin-modified plants in natural environments.
MATERIALS AND METHODS
Plant Growth Conditions in the GH and the Field
The 22nd inbred generation of Nicotiana attenuata seeds originally collected from the Desert Inn ranch, a field site in southwestern Utah, was used as the wild type in all experiments and transformations. An Agrobacterium tumefaciens (strain LBA4404)-mediated transformation protocol using pRESC5CAD binary vector carrying two 358-bp NaCAD1 gene fragments in an inverted orientation, and a gene for hygromycin resistance (hptII) as a screening marker as described by Krügel et al. (2002), were used to generate the ir-CAD lines.
Two independently transformed, homozygous, diploid lines, ir-CAD-206 and ir-CAD-225, each strongly silenced in the transcript accumulation of both NaCADs, containing single insertion of the hptII marker gene as determined by the Southern hybridization of genomic DNA using a PCR fragment of the hptII gene as a probe (as described by Kaur et al., 2010) were used for further analysis. Growth parameters were recorded at regular intervals for both lines.
Surface-sterilized seeds incubated in 1 µm GA3 (Roth; http://www.carl-roth.com) and liquid smoke (House of Herbs) for 1 h were germinated on sterile Gamborg B5 plant agar medium (Duchefa; http://www.duchefa.com) in petri dishes at a 26°C, 16-h, 155 µmol m−2 s−1 PAR light/24°C, 8-h dark cycle in a growth chamber (Percival; http://www.percival-scientific.com). Ten days after germination, seedlings were transferred into Teku trays (Pöppelmann; http://www.poeppelmann.com) containing a peat-based substrate (Tonsubstrat; Klasmann-Deilmann; http://www.klasmann-deilmann.com) and kept in growth chambers for another 12 d under the same growth conditions. At 22 d after germination, plantlets were transplanted as single plants into 1-L pots (top diameter = 13.5 cm, depth = 10.3 cm) containing a peat-based soil (Krügel et al., 2002) and grown in the GH with a day/night cycle of 16 h (26°C–28°C)/8 h (22–24°C), relative humidity around 55%, and supplemented with 400- and 600-W Philips Sun-T Agro sodium lights (http://www.philips.com).
In the field, 15-d-old seedlings were transferred into previously hydrated 50-mm peat pellets (Jiffy 703; http://www.jiffypot.com) and grown in shade tents for 2 weeks to gradually adjust the plants to the high sun and low relative humidity conditions of the Great Basin Desert. Three to 4 weeks later, when plants were in the early-rosette stage of growth, 19 size-matched wild-type/ir-CAD plant pairs were transplanted into an irrigated field plot (at a distance of 1.5 m between the pairs) in the 2008 and 2009 growing seasons at the Lytle Ranch Preserve, near Santa Clara, Utah. The release of transgenics was carried out under Animal and Plant Health Inspection Service notification 06-242-3r, and the seeds were imported under notification 07-341-101n. We simulated GH (or PF) growth conditions in the 2009 field by growing plants with shallow rooting volumes without UV-B, wind, and herbivore attack. Twelve pairs of wild-type/ir-CAD plants were grown under a double layer of UV-opaque (0.1 mm thick) but PAR-transparent polyester (Mylar D; http://europe.dupontteijinfilms.com) sheets affixed to a plywood frame, the legs of which were raised to accommodate the height of the growing plants. In addition, we sprayed the plants with a systemic herbicide (Confidor; Bayer CropScience) and planted members of a pair in close (15 cm) proximity. The sheets were replaced every 2 d to ensure that solarization of the film did not influence the film’s light-transmission properties. The relative UV-B irradiances under different growth conditions were measured with a digital UV radiometer (range, 0–1999 μW cm−2; spectral response, 280–320 nm [Solartech solarmeter model 6.2UV-B; www.solarmeter.com]).
Before the experiment was terminated in the field, stem samples were collected for total lignin quantification following the Klason method (Kirk and Obst, 1988), characterization of lignin composition by thioacidolysis and NMR, quantification of transcript accumulation of NaCAD genes by quantitative real-time (qRT)-PCR, as described by Kaur et al. (2010), in vivo CAD/SAD enzymatic assays, and metabolite profiles by LC-ESI-TOF MS.
Wind and UV-B Exposure Climate Growth Chamber Experiment
UV-B and wind exposure of GH-grown plants was conducted in the climate growth chambers (York International). Three weeks after germination, early rosette-stage plants of both genotypes (wild type and ir-CAD; 56 plants in total) grown in either 1-L or 4-L pots were transferred from the GH (a UV-B and wind-free environment) to a climate chamber. The 1-L- and 4-L-grown wild-type and ir-CAD plants were randomly placed in 12 black plastic trays (length = 60 cm × width = 40 cm) and cultivated under a 16-h (26°C–28°C)/8-h (22°C–24°C) light (1,000 µmol m−2 s−1)/dark cycle at approximately 55% relative humidity. The plants were allowed to adapt for 7 d to the chamber conditions before the UV-B and wind treatment was started. The chamber was separated into two equally sized parts by a black curtain to create UV-B and non-UV-B treatments, and rosette-stage plants were randomly distributed into the two treatment groups: six trays each with seven 1-L ir-CAD, seven 4-L ir-CAD, seven 1-L wild-type, and seven 4-L wild-type plants were exposed to UV-B and wind simultaneously, whereas the other six trays containing an identical set of plants without UV-B and wind served as controls (nonexposed plants; Supplemental Fig. S10). The black curtain completely prevented both UV-B irradiation and wind to pass through to the other half of the chamber. In UV-B treatment, the trays were placed directly below the UV-B source lamp, one tubular UV-B lamp per three trays with 18 plants (TL 40W/12 RS SLV; length = 1213.6 mm and diameter = 40.5 mm [Phillips; http://www.philips.com]; Fehér et al., 2011). Initially, the distance between the UV-B lamp and the plants was 50 cm, a distance at which plants were exposed to approximately 100 µW cm−2 UV-B irradiation (measured with a digital UV radiometer; Solartech solarmeter). Plants were exposed to UV-B at solar noon for 1 h per day during the first week, so that the plants acclimatized slowly to the UV-B fluence. During the second week, the UV-B lamps were at a distance of 30 cm from the 4-L-grown plant tips and supplied 2 h of UV-B radiation per day at an intensity of 150 µW cm−2, which was increased to 4 h during the third week. Compressed air was used as a wind source: an 8-bar-pressure compressed air outlet was directly connected to one end of two 240-cm-long brass tubes with a diameter of 1 cm perforated with 2-mm holes 3 cm apart along its length and a blocked end. The tubes were initially clamped at a distance of 30 cm from the ground on both sides of the trays, with the air streams facing the plants; the distance was regularly adjusted as the plants grew. The wind treatment was applied continuously during the entire experiment and caused plants to vibrate and rustle their leaves. The location of plants within each treatment was randomized every day, and the study was terminated 30 d after transferring plants to the climate chamber.
Lignin Content
Total lignin content was estimated with the Klason method (Kirk and Obst, 1988) as follows: basal 10-cm stem segments including the root-shoot transition zone from mature N. attenuata wild-type/ir-CAD plants were excised, ball milled (Fritsch Analysette 3; http://www.fritsch.de) in liquid nitrogen to a finely ground powder, and used for all lignin-related analyses. Homogenized powder from basal stem segments (500 mg) was subjected to four consecutive extractions (sufficient to extract soluble red-colored metabolites) in 50 mL of methanol (AnalaR Normapur; VWR; http://ni.vwr.com) in glass tubes that were maintained on a rotary shaker for 6 to 8 h. These tubes were centrifuged at 16,000g for 5 min after every extraction; the supernatant was discarded and, finally, the pellet was dried under vacuum to obtain the cell wall residue from the powdered stems. Cell wall residue (200–300 mg) was hydrolyzed in 4 mL of 72% H2SO4 (Merck; http://www.merck-chemicals.de) at 30°C for 2.5 h. The hydrolyzate was diluted by adding 112 mL of water and then autoclaved for 1 h. The mixture was cooled and vacuum filtered through a fitted glass crucible (no. 3; Schott; http://www.schott.com). The undigested residue was washed with 100 mL of hot water, oven dried at 80°C overnight, desiccator cooled, and weighed. The mass of lignin was expressed as a percentage of the cell wall residue mass used for hydrolysis.
Plant Protein Extraction and Enzyme Assays
Total proteins from stems were extracted following the method described by Zhang et al. (2006). Ten-centimeter basal stem segments from early elongated nonlignified plants as well as from the upper nonlignified stem sections of mature plants were ball milled to a fine powder in liquid N2 (Fritsch Analysette 3; http://www.fritsch.de), extracted in 1 mL of extraction buffer consisting of 100 mm Tris-HCl, pH 7.5 (Roth; http://www.carl-roth.de), 2% polyethylene glycol 6000 (Fluka-Chemika-Biochemika), 5 mm dithiothreitol (Roth), and 2% polyvinylpolypyrrolidone (Roth) for 2.5 h at 4°C. The supernatant was collected after centrifuging, and about 100 µg of total protein was used for CAD and SAD activity assays. The protein concentration was determined using a Bio-Rad protein assay kit with bovine serum albumin as a standard. The formation of hydroxycinnamaldehydes (coniferaldehyde and sinapaldehyde) from coniferyl and sinapyl alcohols (Sigma-Aldrich; http://www.sigmaaldrich.com) as the substrates with the extracted protein in the presence of NADP was monitored on an Ultrospec 3000 pro UV/visible spectrophotometer at 400 nm (Amersham Pharmacia Biotech Europe). The following molar extinction coefficients were used for calculations: coniferaldehyde, 2.10 × 10−4 m−1 cm−1; sinapaldehyde, 1.68 × 10−4 m−1 cm−1. The enzyme reactions were initiated by adding crude protein extracts to a mixture of 500 µL of 100 mm Tris-HCl, pH 8.8, 100 µm NADP (Roth), and 250 µm coniferyl alcohol or sinapyl alcohol. The enzymatic reactions were carried out at 30°C for 10 min and inactivated by heating at 85°C for 10 min. An assay without NADP served as a control, and the results were expressed as the amount of activity that converts 1 nmol of hydroxycinnamyl alcohol into the corresponding hydroxycinnamaldehyde per second (1 nkat) per microgram of crude protein extract.
Lignin Composition Analysis Using Thioacidolysis
Following essentially the published procedure, a mixture was prepared by adding 2.5 mL of BF3-etherate (Fluka) and 10 mL of ethanethiol (Fluka) to a 100-mL flask and adjusting the final volume to 100 mL with dioxane (Merck) just prior to being used for the thioacidolysis reaction with cell wall residue (Kim et al., 2002). Ten milliliters of reagent mix and 1 mL of 0.5 mg mL−1 tetracosane (Fluka) were added to 10 mg of stem cell wall residue in a glass tube (tetracosane served as a GC internal standard). Thioacidolysis was performed at 100°C by submerging glass tubes in an oil bath for 4 h. The reaction was halted by placing the reactions on ice for 20 min. The cooled reaction mixtures were diluted with 30 mL of water and adjusted to pH 3 to 4 by adding NaHCO3 (Roth). The reaction mixtures were extracted with dichloromethane (3 × 30 mL; VWR; http://ni.vwr.com), and the combined lower organic layers were dried over Na2SO4 (Sigma-Aldrich) and evaporated to dryness under nitrogen. The dried residue was resuspended in approximately 0.5 mL of dichloromethane. For GC-MS, the thioacidolysis product was silylated by adding 10 µL of pyridine (Merck) and 50 µL of N-methyl-N-(trimethylsilyl) trifluoroacetamide (Macherey-Nagel) + trimethyl chlorosilane (Merck; 99:1) to 20 µL of sample, and the reaction was allowed to run for 8 to 10 h at 4°C. One microliter of silylated sample was analyzed by GC (Agilent Hewlett-Packard 6890; Agilent Technologies) coupled to a quadrupole mass spectrometer (Agilent Hewlett-Packard 5973; Agilent Technologies) using a DB-5MS column (30 m × 0.25 mm, 0.25 µm; J&W Scientific) operated with a temperature program (60°C for a 0.5-min hold; 60°C–120°C at 20°C min−1, then to 300°C at 5°C min−1; and then a 5-min hold) with helium as the carrier gas (flow rate of 2 mL min−1). A splitless injection maintained at 220°C was used. MS was performed with a transfer line temperature of 270°C, ion source temperature of 230°C, quadrupole temperature of 150°C, ionization potential of 70 eV, and scan range of 50 to 500 atomic mass units.
Sinapaldehyde and coniferaldehyde indene isoforms were identified from ion chromatograms reconstructed at m/z 384 and 354, whereas the m/z 239, 269, and 299 ion traces were used to identify monolignol H, G, and S monomers (Kim et al., 2002; Ralph et al., 2008). Quantitation and identification of coniferaldehyde and sinapaldehyde were based on retention times and response factors obtained from standards, relative to tetracosane used as an internal standard.
NMR Lignin Analysis
Lignin was extracted from stems of wild-type and ir-CAD plants grown in different environments following the method described by Ralph et al. (1998). Three independent replicates of each treatment were pooled, and pooled samples were ball milled. Soluble metabolites were extracted from these finely ground stems with sequential washes at room temperature on a rotary shaker for 6 to 8 h with water (six times), methanol (five times), acetone (four times; KMF Laborchemie Handels), and chloroform (two times; VWR; http://ni.vwr.com). Most of the colored material was extracted in the water and methanol extractions. The soluble metabolite-free cell walls were digested with crude cellulases (Merck). The resulting polysaccharidase-digested cell wall fractions were extracted with 96:4 (v/v) dioxane:water. The isolated lignin was acetylated overnight with acetic anhydride (Sigma-Aldrich)/pyridine and further extracted in freshly distilled ethyl acetate, washed with aqueous 6 mm EDTA (Sigma-Aldrich), dissolved in acetone-d6 (Deutero), and subjected to NMR.
HSQC spectra were acquired on a Bruker Avance AV-500 NMR spectrometer equipped with a 5-mm TCI Cryoprobe (Bruker BioSpin) operating at a 1H frequency of 500.13 MHz and a 13C frequency of 125.76 MHz at 300 K. The central acetone solvent peak was used as an internal reference (δC = 29.8 ppm, δH = 2.04 ppm). We used the standard Bruker implementations of one-dimensional (1H) and two-dimensional (gradient-selected, 1H-detected HSQC) NMR experiments for elucidating structures. We used a gradient-selected inverse (1H-detected) HSQC experiment (Ruiz-Cabello et al., 1992) for the spectra in Figure 6 using the following parameters: 40 transient spectral increments were acquired from 12 to 1 ppm in F2 (1H) using 1,024 data points, from 200 to 10 ppm in F1 (13C), and 40 scans (acquisition time, 0.08533 s). Volume integration of contours in HSQC plots was accomplished by Bruker’s TopSpin 3.1 (Mac) software (Ralph et al., 2006); only the C2/H2 (and also the redundant C6/H6 in symmetrical S moieties) correlation contours were used for quantitation, with S contour integrals being logically halved.
Histochemical Staining
Fresh thin stem cross-sections were obtained by hand cutting using a razor blade (Wilkinson). Sections were mounted on microscope slides in water, and images were captured with a Leica DM6000B light microscopy system equipped with a CCD camera (HV-D20P; Hitachi Kokusai Electric) and stored with Leica Microsystem Image Manager (http://www.leica-microsystems.com) before and after toluidine blue O staining (0.01% toluidine blue powder [Sigma-Aldrich] in 0.1% aqueous sodium tetraborate [sodium borate]). Toluidine blue O is a polychromatic dye that returns a blue or bluish-green color when interacting with compounds containing aromatic rings, such as lignin, and a pink or reddish-purple color when interacting with carboxylated polysaccharides, such as pectins. The xylem element measurements were conducted on images obtained from at least five independent stem sections (individual measurements [number of replicates]: vessel diameter [50], tracheid diameter [120], vessel-to-adjoining tracheid wall thickness [100], and tracheid-to-adjoining tracheid wall thickness [160]) using ImageJ software (http://rsb.info.nih.gov/ij). The vessel and tracheid lumen were measured along their short oriented axes to estimate diameters. Vessel and tracheid wall thickness measurements were taken randomly.
Biomechanical Bending Tests of Stems
One-point bending tests were carried out on basal stem segments (10 cm) of mature GH-grown wild-type and ir-CAD plants with a universal TIRA tensile tester (model 2710; Tira; http://www.tira-gmbh.de). The stem diameter at both ends of each segment was measured with a caliper and recorded in the testing equipment. In the bending test, the basal end of a stem segment was clamped; the other end was set free, and a load was applied in a design referred to as “cantilever beam” in biomechanics. At a distance of 32 mm from the fixed end, a 2-mm-radius stamp cross-head was moved down with a force at 10 mm min−1 velocity and was lowered until it was in contact with the stem. The load and the displacement were recorded during the tests. All tests were performed at room temperature. Four plants of each genotype were tested to obtain maximum bending strength (B) and Young’s modulus (E) estimates. Stem bending strength was calculated using the following expression for a cantilever beam with an intermediate load: B = F(L − a)c/I and E = FL3/3δI, where F is the bending force (N), L is the length of the stem segment (mm), a is the length of stem segment beyond the point where load was subjected, c is the distance from the centroid of the stem segment to its outer edge, δ is the deflection of the stem, and I is the axial second moment of area of stem [as an ellipse (I = π (ab3 − (a − t)(b − t)3)/4), where a is the semimajor axis of the cross-section (mm), b is the semiminor axis of the cross-section (mm), and t is the mean wall thickness].
LC-ESI-TOF MS Metabolomic Analysis
The metabolites were extracted from finely ground basal 10-cm stem segments collected from UF- and GH-grown ir-CAD/wild-type plants at two stages of development: early elongated (before the onset of lignification, when the red color was absent from ir-CAD stems) and mature stage (after lignification had initiated, when ir-CAD stems became red). Stem samples (100 mg) were homogenized in Fast Prep tubes containing 900 mg of Fast Prep matrix (Sili; http://www.sigmund-lindner.com) with 1 mL of 50 mm ammonium acetate buffer, pH 4.8, containing 40% methanol spiked with internal standards (600 ng mL−1 reserpine and 200 ng mL−1 atropine) by shaking twice at 6.5 ms−1 for 45 s in a Thermo-Savant FastPrep FP 120 homogenizer (Qbiogene; http://www.qbiogene.com). Homogenized samples were centrifuged at 16,000g for 20 min at 4°C. Supernatant was collected in a fresh 1.5-mL Eppendorf tube, centrifuged again at 16,000g for 20 min at 4°C, and finally transferred to a glass vial, where it was analyzed by an Agilent HPLC 1100 Series system combined with an Phenomenex Gemini NX 3u (150 × 2.0 mm) column (http://www.phenomenex.com). The elution system was as follows: 0 to 2 min, isocratic at 5% A (acetonitrile and 0.05% formic acid) and 95% B (water, 0.1% acetonitrile, and 0.05% formic acid); 2 to 30 min, linear gradient up to 80% A; and 30 to 35 min, isocratic at 80% A. The injection volume was 2 µL, and the flow rate was 200 µL min−1. MS was performed using a MicroTOF time-of-flight mass spectrometer (Bruker Daltonik) connected to an electrospray ionization source operating in positive-ion mode. Electrospray ionization settings were as follows: dry gas temperature, 200°C; dry gas flow, 8 L min−1; TOF, 2,100 V; capillary voltage, 4,500 V; capillary exit, 130 V. Ions were detected from m/z 200 to 1,400 at a repetition rate of 1 Hz. Mass calibration was performed using sodium formate clusters (10 mm solution of NaOH in 50%:50% [v/v] isopropanol:water containing 0.2% formic acid).
Statistical Analysis
Data were analyzed with SPSS (http://www.spss.com), R (R-Project; http://www.r-project.org), and xlstat (http://www.xlstat.com) software.
The sequence data described in this article can be found in the GenBank/EMBL data libraries under accession numbers JQ004402 for NaCAD and JN997447 for NaCAD1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. NaCAD1 gene silencing results in cosilencing of the NaCAD gene in N. attenuata due to high sequence similarity.
Supplemental Figure S2. The amino acid sequences of both NaCADs (NaCAD, green; NaCAD1, red) are similar to those of AtCAD4 and -5, which are known to be involved in stem lignification.
Supplemental Figure S3. DNA gel-blot analysis of genomic DNA isolated from wild-type leaf tissue reveals that CAD exists as a multigene family in the N. attenuata genome.
Supplemental Figure S4. CAD silencing alters the architecture of plants without affecting their reproductive fitness in the GH.
Supplemental Figure S5. Physical properties of wild-type and ir-CAD-225 stems, and CAD and SAD enzymatic activities quantified from early elongating (lignifying), GH-grown wild-type and ir-CAD-225 basal stem extracts.
Supplemental Figure S6. The acropetal spread of red color in ir-CAD stems provides a visual marker for the onset of wild-type lignification.
Supplemental Figure S7. ir-CAD plants acquire wild-type structural stability in a field plot in their native environment of the Great Basin Desert in the 2008 field season.
Supplemental Figure S8. Flow chart of methods and software used in the processing of the LC-ESI-TOF data.
Supplemental Figure S9. Heat map showing the hierarchical clustering of 2,421 differentially regulated metabolite ions obtained from LC-ESI-TOF MS.
Supplemental Figure S10. Experimental setup of the stress (wind, UV-B)-supplemented experiment conducted with wild-type/ir-CAD plants grown in 1-L and 4-L pots in climate growth chambers.
Supplemental Figure S11. Stems of ir-CAD plants grown in UF were more often variegated with red and green sectors.
Supplemental Table S1. List of the genes and their accession numbers used in phylogenetic analysis presented in Figure 1A.
Supplemental Data Set S1. XCMS-CAMERA raw data file of early and late wild-type and ir-CAD stem metabolite ions obtained from LC-ESI-TOF MS runs (positive-ion mode), when plants were grown in GH and UF environments.
Supplemental Data Set S2. Processed XCMS-CAMERA data file with identified LC-ESI-TOF metabolite ions obtained from stems of early and late wild-type and ir-CAD plants grown in GH and UF environments.
Supplemental Data Set S3. Three-way ANOVA performed on XCMS-CAMERA-processed LC-ESI-TOF metabolite ions.
Supplementary Material
Acknowledgments
We thank Dr. Tamara Krügel, Andreas Weber, and Andreas Schünzel for growing plants in the GH; Melanie Skibbe, Celia Diezel, Danny Kessler, and Mario Kallenbach for assistance with the field experiments; Dr. Mathias Schöttner, Dr. Emmanuel Gaquerel, and Jyotasana Gulati for assistance with the TOF data analysis; Dr. Klaus Gase for constructing the transformation vectors; Thomas Hahn for sequencing; Antje Wissgott, Susan Kutschbach, and Wibke Kröber for generating stably transformed plants; Prabin Bajgain and Melissa Coon for help with sample preparation; Erick Heurich for assistance with the bending measurements; Dr. Dirk Hölscher for help with the Leica AS LMD laser microdissection microscope; Dr. Dieter Spiteller for support in the lignin extractions; Dr. Bernd Schneider for helpful discussion of the NMR data; Dr. Jan Kellmann for support in transferring transgenic stem samples to the Institute for Materials Science and Technology; and Brigham Young University for use of its field station at the Lytle Ranch Preserve.
Glossary
- GH
glasshouse
- RNAi
RNA interference
- GC
gas chromatography
- MS
mass spectrometry
- UF
unprotected field
- PF
protected field
- HSQC
heteronuclear single quantum coherence
- LC-ESI-TOF
liquid chromatography-electrospray ionization-time-of-flight
- m/z
mass-to-charge ratio
- PAR
photosynthetically active radiation
- qRT
quantitative real-time
- H
hydroxyphenyl
- G
guaiacyl
- S
syringyl
References
- Achyuthan KE, Achyuthan AM, Adams PD, Dirk SM, Harper JC, Simmons BA, Singh AK. (2010) Supramolecular self-assembled chaos: polyphenolic lignin’s barrier to cost-effective lignocellulosic biofuels. Molecules 15: 8641–8688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anten NPR, Casado-Garcia R, Nagashima H. (2005) Effects of mechanical stress and plant density on mechanical characteristics, growth, and lifetime reproduction of tobacco plants. Am Nat 166: 650–660 [DOI] [PubMed] [Google Scholar]
- Ballaré CL, Scopel AL, Jordan ET, Vierstra RD. (1994) Signaling among neighboring plants and the development of size inequalities in plant populations. Proc Natl Acad Sci USA 91: 10094–10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Bagniewska-Zadworna A, Frost CJ, Carlson JE. (2010) Phylogeny and expression profiling of CAD and CAD-like genes in hybrid Populus (P. deltoides × P. nigra): evidence from herbivore damage for subfunctionalization and functional divergence. BMC Plant Biol 10: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Bagniewska-Zadworna A, Plakkat U, Choi A, DiLoreto D, Carlson J. (2009) The Cinnamyl Alcohol Dehydrogenase genes family in Populus: phylogeny, organization, and expression. BMC Plant Biol 9: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M, Chabbert B, Pilate G, Van Doorsselaere J, Tollier MT, Petit-Conil M, Cornu D, Monties B, Van Montagu M, Inze D, et al. (1996) Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol 112: 1479–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M. (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C. (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44: 337–363 [DOI] [PubMed] [Google Scholar]
- Bunzel M, Ralph J. (2006) NMR characterization of lignins isolated from fruit and vegetable insoluble dietary fiber. J Agric Food Chem 54: 8352–8361 [DOI] [PubMed] [Google Scholar]
- Chang A, Lim M-H, Lee S-W, Robb EJ, Nazar RN. (2008) Tomato phenylalanine ammonia-lyase gene family, highly redundant but strongly underutilized. J Biol Chem 283: 33591–33601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MCY. (2007) Harnessing energy from plant biomass. Curr Opin Chem Biol 11: 677–684 [DOI] [PubMed] [Google Scholar]
- Chen F, Dixon RA. (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chenu K, Franck N, Dauzat J, Barczi J, Rey H, Lecoeur J. (2005) Integrated responses of rosette organogenesis, morphogenesis and architecture to reduced incident light in Arabidopsis thaliana results in higher efficiency of light interception. Funct Plant Biol 32: 1123–1134 [DOI] [PubMed] [Google Scholar]
- Coutand C. (2010) Mechanosensing and thigmomorphogenesis, a physiological and biomechanical point of view. Plant Sci 179: 168–182 [Google Scholar]
- Day A, Neutelings G, Nolin F, Grec S, Habrant A, Crônier D, Maher B, Rolando C, David H, Chabbert B, et al. (2009) Caffeoyl coenzyme A O-methyltransferase down-regulation is associated with modifications in lignin and cell-wall architecture in flax secondary xylem. Plant Physiol Biochem 47: 9–19 [DOI] [PubMed] [Google Scholar]
- Derikvand MM, Sierra JB, Ruel K, Pollet B, Do CT, Thévenin J, Buffard D, Jouanin L, Lapierre C. (2008) Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227: 943–956 [DOI] [PubMed] [Google Scholar]
- Diezel C, Kessler D, Baldwin IT. (2011) Pithy protection: Nicotiana attenuata’s jasmonic acid-mediated defenses are required to resist stem-boring weevil larvae. Plant Physiol 155: 1936–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A, Pollet B, Sibout R, Do CT, Séguin A, Lapierre C, Jouanin L. (2006) Evidence for a role of AtCAD 1 in lignification of elongating stems of Arabidopsis thaliana. Planta 225: 23–39 [DOI] [PubMed] [Google Scholar]
- Fehér B, Kozma-Bognár L, Kevei É, Hajdu A, Binkert M, Davis SJ, Schäfer E, Ulm R, Nagy F. (2011) Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J 67: 37–48 [DOI] [PubMed] [Google Scholar]
- Fornalé S, Capellades M, Encina A, Wang K, Irar S, Lapierre C, Ruel K, Joseleau J-P, Berenguer J, Puigdomènech P, et al. doi: 10.1093/mp/ssr097. (December 6, 2011) Altered lignin biosynthesis improves cellulosic bioethanol production in transgenic maize plants down-regulated for cinnamyl alcohol dehydrogenase. Mol Plant http://dx.doi.org/10.1093/mp/ssr097. [DOI] [PubMed] [Google Scholar]
- Fu CX, Mielenz JR, Xiao XR, Ge YX, Hamilton CY, Rodriguez M, Jr, Chen F, Foston M, Ragauskas A, Bouton J, et al. (2011) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA 108: 3803–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaquerel E, Heiling S, Schoettner M, Zurek G, Baldwin IT. (2010) Development and validation of a liquid chromatography-electrospray ionization-time-of-flight mass spectrometry method for induced changes in Nicotiana attenuata leaves during simulated herbivory. J Agric Food Chem 58: 9418–9427 [DOI] [PubMed] [Google Scholar]
- Gilbert HJ. (2010) The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol 153: 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Hänsel U, Ziemann M, Pauly M. (2009) Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc Natl Acad Sci USA 106: 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri AP, Wünsche H, Mitra S, Zavala JA, Muck A, Svatos A, Baldwin IT. (2006) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. VII. Changes in the plant’s proteome. Plant Physiol 142: 1621–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumie S, Goffner D, Barbier O, Martinant J-P, Pichon M, Barrière Y. (2008) Expression of cell wall related genes in basal and ear internodes of silking brown-midrib-3, caffeic acid O-methyltransferase (COMT) down-regulated, and normal maize plants. BMC Plant Biol 8: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo DJ, Chen F, Inoue K, Blount JW, Dixon RA. (2001) Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13: 73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K, Bednarek P, Ciolkowski I, Hamberger BR, Heise A, Liedgens H, Logemann E, Nürnberger T, Schmelzer E, Somssich IE, et al. (2003) Non-self recognition, transcriptional reprogramming, and secondary metabolite accumulation during plant/pathogen interactions. Proc Natl Acad Sci USA (Suppl 2) 100: 14569–14576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin C, Holt K, Chojecki J, Oliver D, Chabbert B, Monties B, Edwards K, Barakate A, Foxon GA. (1998) Brown-midrib maize (bm1): a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J 14: 545–553 [DOI] [PubMed] [Google Scholar]
- Halpin C, Knight ME, Foxon GA, Campbell MM, Boudet AM, Boon JJ, Chabbert B, Tollier M-T, Schuch W. (1994) Manipulation of lignin quality by downregulation of cinnamyl alcohol dehydrogenase. Plant J 6: 339–350 [Google Scholar]
- Halpin C, Thain SC, Tilston EL, Guiney E, Lapierre C, Hopkins DW. (2007) Ecological impacts of trees with modified lignin. Tree Genet Genomes 3: 101–110 [Google Scholar]
- Hepworth DG, Vincent JFV. (1999) The growth response of the stems of genetically modified tobacco plants (Nicotiana tabacum ‘Samsun’) to flexural stimulation. Ann Bot (Lond) 89: 39–43 [Google Scholar]
- Higuchi T, Ito T, Umezawa T, Hibino T, Shibata D. (1994) Red-brown color of lignified tissues of transgenic plants with antisense CAD gene: wine-red lignin from coniferyl aldehyde. J Biotechnol 37: 151–158 [Google Scholar]
- Himmel ME, Ding S-Y, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315: 804–807 [DOI] [PubMed] [Google Scholar]
- Hu W-J, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, Tsai C-J, Chiang VL. (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 17: 808–812 [DOI] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou Y-H, Yu J-Q, Chen Z. (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Heinzel N, Schöttner M, Baldwin IT, Gális I. (2010) R2R3-NaMYB8 regulates the accumulation of phenylpropanoid-polyamine conjugates, which are essential for local and systemic defense against insect herbivores in Nicotiana attenuata. Plant Physiol 152: 1731–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ralph J, Lu F, Ralph SA, Boudet A-M, MacKay JJ, Sederoff RR, Ito T, Kawai S, Ohashi H, et al. (2003) NMR analysis of lignins in CAD-deficient plants. Part 1. Incorporation of hydroxycinnamaldehydes and hydroxybenzaldehydes into lignins. Org Biomol Chem 1: 268–281 [DOI] [PubMed] [Google Scholar]
- Kim H, Ralph J, Lu FC, Pilate G, Leplé JC, Pollet B, Lapierre C. (2002) Identification of the structure and origin of thioacidolysis marker compounds for cinnamyl alcohol dehydrogenase deficiency in angiosperms. J Biol Chem 277: 47412–47419 [DOI] [PubMed] [Google Scholar]
- Kim H, Ralph J, Yahiaoui N, Pean M, Boudet A-M. (2000) Cross-coupling of hydroxycinnamyl aldehydes into lignins. Org Lett 2: 2197–2200 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Kim MR, Bedgar DL, Moinuddin SGA, Cardenas CL, Davin LB, Kang CL, Lewis NG. (2004) Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc Natl Acad Sci USA 101: 1455–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-H, Bae JM, Huh G-H. (2010) Transcriptional regulation of the cinnamyl alcohol dehydrogenase gene from sweet potato in response to plant developmental stage and environmental stress. Plant Cell Rep 29: 779–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RR, Calhoun LA. (2005) Characterization of cross-linked hydroxycinnamic acid amides isolated from potato common scab lesions. Phytochemistry 66: 2468–2473 [DOI] [PubMed] [Google Scholar]
- Kirk TK, Obst JR. (1988) Lignin determination. Methods Enzymol 161: 87–101 [Google Scholar]
- Koehler L, Telewski FW. (2006) Biomechanics and transgenic wood. Am J Bot 93: 1433–1438 [DOI] [PubMed] [Google Scholar]
- Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT. (2002) Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12: 177–183 [Google Scholar]
- Lapierre C, Pilate G, Pollet B, Mila I, Leplé JC, Jouanin L, Kim H, Ralph J. (2004) Signatures of cinnamyl alcohol dehydrogenase deficiency in poplar lignins. Phytochemistry 65: 313–321 [DOI] [PubMed] [Google Scholar]
- Leplé JC, Dauwe R, Morreel K, Storme V, Lapierre C, Pollet B, Naumann A, Kang KY, Kim H, Ruel K, et al. (2007) Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bonawitz ND, Weng J-K, Chapple C. (2010) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang Y, Yao J, Chen G, Li X, Zhang Q, Wu C. (2009) FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol 69: 685–697 [DOI] [PubMed] [Google Scholar]
- Lionetti V, Francocci F, Ferrari S, Volpi C, Bellincampi D, Galletti R, D’Ovidio R, De Lorenzo G, Cervone F. (2010) Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc Natl Acad Sci USA 107: 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Martia J, Lapierre C, Jouanin L, Morreel K, Boerjan W, Ralph J. (2010) Sequencing around 5-hydroxyconiferyl alcohol-derived units in caffeic acid O-methyltransferase-deficient poplar lignins. Plant Physiol 153: 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldau S, Wu J, Baldwin IT. (2009) Silencing two herbivory-activated MAP kinases, SIPK and WIPK, does not increase Nicotiana attenuata’s susceptibility to herbivores in the glasshouse and in nature. New Phytol 181: 161–173 [DOI] [PubMed] [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B. (2006) Transgeneration memory of stress in plants. Nature 442: 1046–1049 [DOI] [PubMed] [Google Scholar]
- Moura JCMS, Bonine CAV, de Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P. (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 52: 360–376 [DOI] [PubMed] [Google Scholar]
- Nadji H, Diouf PN, Benaboura A, Bedard Y, Riedl B, Stevanovic T. (2009) Comparative study of lignins isolated from alfa grass (Stipa tenacissima L.). Bioresour Technol 100: 3585–3592 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149: 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrel J, Pollet B, Lapierre C. (1996) Ether-linked ferulic acid amides in natural and wound periderms of potato tuber. Phytochemistry 43: 1195–1199 [Google Scholar]
- Niklas K. (1998) Effects of vibration on mechanical properties and biomass allocation pattern of Capsella bursa-pastoris (Cruciferae). Ann Bot (Lond) 82: 147–156 [Google Scholar]
- Olsen KM, Lea US, Slimestad R, Verheul M, Lillo C. (2008) Differential expression of four Arabidopsis PAL genes: PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J Plant Physiol 165: 1491–1499 [DOI] [PubMed] [Google Scholar]
- Onkokesung N, Gaquerel E, Kotkar H, Kaur H, Baldwin IT, Galis I. (2012) MYB8 controls inducible phenolamide levels by activating three novel hydroxycinnamoyl-CoA:polyamine transferases in Nicotiana attenuata. Plant Physiol 158: 389–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilate G, Guiney E, Holt K, Petit-Conil M, Lapierre C, Leplé JC, Pollet B, Mila I, Webster EA, Marstorp HG, et al. (2002) Field and pulping performances of transgenic trees with altered lignification. Nat Biotechnol 20: 607–612 [DOI] [PubMed] [Google Scholar]
- Pillonel C, Mulder MM, Boon JJ, Forster B, Binder A. (1991) Involvement of cinnamyl-alcohol dehydrogenase in the control of lignin formation in Sorghum bicolor (L.) Moench. Planta 185: 538–544 [DOI] [PubMed] [Google Scholar]
- Piquemal J, Chamayou S, Nadaud I, Beckert M, Barrière Y, Mila I, Lapierre C, Rigau J, Puigdomenech P, Jauneau A, et al. (2002) Down-regulation of caffeic acid O-methyltransferase in maize revisited using a transgenic approach. Plant Physiol 130: 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomar F, Novo M, Bernal MA, Merino F, Barcelo AR. (2004) Changes in stem lignins (monomer composition and crosslinking) and peroxidase are related with the maintenance of leaf photosynthetic integrity during verticillium wilt in Capsicum annuum. New Phytol 163: 111–123 [DOI] [PubMed] [Google Scholar]
- Poorter L, McDonald I, Alarcón A, Fichtler E, Licona J-C, Peña-Claros M, Sterck F, Villegas Z, Sass-Klaassen U. (2010) The importance of wood traits and hydraulic conductance for the performance and life history strategies of 42 rainforest tree species. New Phytol 185: 481–492 [DOI] [PubMed] [Google Scholar]
- Popper ZA. (2008) Evolution and diversity of green plant cell walls. Curr Opin Plant Biol 11: 286–292 [DOI] [PubMed] [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J. (2010) Hydroxycinnamates in lignification. Phytochem Rev 9: 65–83 [Google Scholar]
- Ralph J, Akiyama T, Kim H, Lu FC, Schatz PF, Marita JM, Ralph SA, Reddy MSS, Chen F, Dixon RA. (2006) Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J Biol Chem 281: 8843–8853 [DOI] [PubMed] [Google Scholar]
- Ralph J, Hatfield RD, Piquemal J, Yahiaoui N, Pean M, Lapierre C, Boudet AM. (1998) NMR characterization of altered lignins extracted from tobacco plants down-regulated for lignification enzymes cinnamylalcohol dehydrogenase and cinnamoyl-CoA reductase. Proc Natl Acad Sci USA 95: 12803–12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Kim H, Lu F, Grabber JH, Leplé J-C, Berrio-Sierra J, Derikvand MM, Jouanin L, Boerjan W, Lapierre C. (2008) Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency). Plant J 53: 368–379 [DOI] [PubMed] [Google Scholar]
- Ralph J, Lapierre C, Marita JM, Kim H, Lu F, Hatfield RD, Ralph S, Chapple C, Franke R, Hemm MR, et al. (2001) Elucidation of new structures in lignins of CAD- and COMT-deficient plants by NMR. Phytochemistry 57: 993–1003 [DOI] [PubMed] [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz P, Marita J, Hatfield R, Ralph S, Christensen J, et al. (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem Rev 3: 29–60 [Google Scholar]
- Robinson AR, Mansfield SD. (2009) Rapid analysis of poplar lignin monomer composition by a streamlined thioacidolysis procedure and near-infrared reflectance-based prediction modeling. Plant J 58: 706–714 [DOI] [PubMed] [Google Scholar]
- Ruel K, Berrio-Sierra J, Derikvand MM, Pollet B, Thévenin J, Lapierre C, Jouanin L, Joseleau JP. (2009) Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol 184: 99–113 [DOI] [PubMed] [Google Scholar]
- Ruiz-Cabello J, Vuister GW, Moonen CTW, van Gelderen P, Cohen JS, van Zijl PCM. (1992) Gradient-enhanced heteronuclear correlation spectroscopy: theory and experimental aspects. J Magn Reson 100: 282–302 [DOI] [PubMed] [Google Scholar]
- Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W. (2009) A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181: 783–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi I, Ammar S, Demont-Caulet N, Thevenin J, Lapierre C, Bouzid S, Jouanin L. (2009) Thigmomorphogenesis in Solanum lycopersicum: morphological and biochemical responses in stem after mechanical stimulation. Plant Sci 177: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Scheel D, Strack D. (1998) Elicitor-stimulated biosynthesis of hydroxycinnamoyltyramines in cell suspension cultures of Solanum tuberosum. Planta 205: 51–55 [Google Scholar]
- Schmitt J, McCormac AC, Smith H. (1995) A test of the adaptive plasticity hypothesis using transgenic and mutant plants disabled in phytochrome-mediated elongation responses to neighbors. Am Nat 146: 937–953 [Google Scholar]
- Schwinning S, Weiner J. (1998) Mechanisms determining the degree of size asymmetry in competition among plants. Oecologia 113: 447–455 [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Blaukopf C. (2010) Irritable walls: the plant extracellular matrix and signaling. Plant Physiol 153: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Sun YH, Li QZ, Heber S, Sederoff R, Chiang VL. (2010) Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol 51: 144–163 [DOI] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Séguin A. (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Pollet B, Goujon T, Mila I, Granier F, Séguin A, Lapierre C, Jouanin L. (2003) Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis: isolation and characterization of the corresponding mutants. Plant Physiol 132: 848–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons BA, Loqué D, Ralph J. (2010) Advances in modifying lignin for enhanced biofuel production. Curr Opin Plant Biol 13: 313–320 [DOI] [PubMed] [Google Scholar]
- Smith JE, Jordan PW. (1994) Stand density effects on branching in an annual legume (Senna obtusifolia). Ann Bot (Lond) 74: 17–25 [DOI] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, et al. (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Song J, Wang ZZ. (2011) RNAi-mediated suppression of the phenylalanine ammonia-lyase gene in Salvia miltiorrhiza causes abnormal phenotypes and a reduction in rosmarinic acid biosynthesis. J Plant Res 124: 183–192 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thévenin J, Pollet B, Letarnec B, Saulnier L, Gissot L, Maia-Grondard A, Lapierre C, Jouanin L. (2011) The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol Plant 4: 70–82 [DOI] [PubMed] [Google Scholar]
- Tilston EL, Halpin C, Hopkins DW. (2004) Genetic modifications to lignin biosynthesis in field-grown poplar trees have inconsistent effects on the rate of woody trunk decomposition. Soil Biol Biochem 36: 1903–1906 [Google Scholar]
- Tobias CM, Chow EK. (2005) Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification. Planta 220: 678–688 [DOI] [PubMed] [Google Scholar]
- Torney F, Moeller L, Scarpa A, Wang K. (2007) Genetic engineering approaches to improve bioethanol production from maize. Curr Opin Biotechnol 18: 193–199 [DOI] [PubMed] [Google Scholar]
- Tronchet M, Balagué C, Kroj T, Jouanin L, Roby D. (2010) Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol Plant Pathol 11: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, Rochfort S, Liu ZQ, Ran YD, Griffith M, Badenhorst P, Louie GV, Bowman ME, Smith KF, Noel JP, et al. (2010) Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 22: 3357–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeki K. (1995) Importance of crown position and morphological plasticity in competitive interaction in a population of Xanthium canadense. Ann Bot (Lond) 75: 259–265 [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermerris W, Sherman DM, McIntyre LM. (2010) Phenotypic plasticity in cell walls of maize brown midrib mutants is limited by lignin composition. J Exp Bot 61: 2479–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermerris W, Thompson KJ, McIntyre LM. (2002) The maize Brown midrib1 locus affects cell wall composition and plant development in a dose-dependent manner. Heredity (Edinb) 88: 450–457 [DOI] [PubMed] [Google Scholar]
- Voelker S. (2009) Functional decreases in hydraulic and mechanical properties of field-grown transgenic poplar trees caused by modification of the lignin synthesis pathway through downregulation of the 4-coumarate:coenzyme A ligase gene. PhD thesis. Oregon State University, Corvallis
- Voelker SL, Lachenbruch B, Meinzer FC, Jourdes M, Ki CY, Patten AM, Davin LB, Lewis NG, Tuskan GA, Gunter L, et al. (2010) Antisense down-regulation of 4CL expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol 154: 874–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker SL, Lachenbruch B, Meinzer FC, Kitin P, Strauss SH. (2011a) Transgenic poplars with reduced lignin show impaired xylem conductivity, growth efficiency and survival. Plant Cell Environ 34: 655–668 [DOI] [PubMed] [Google Scholar]
- Voelker SL, Lachenbruch B, Meinzer FC, Strauss SH. (2011b) Reduced wood stiffness and strength, and altered stem form, in young antisense 4CL transgenic poplars with reduced lignin contents. New Phytol 189: 1096–1109 [DOI] [PubMed] [Google Scholar]
- Wadenbäck J, von Arnold S, Egertsdotter U, Walter MH, Grima-Pettenati J, Goffner D, Gellerstedt G, Gullion T, Clapham D. (2008) Lignin biosynthesis in transgenic Norway spruce plants harboring an antisense construct for cinnamoyl CoA reductase (CCR). Transgenic Res 17: 379–392 [DOI] [PubMed] [Google Scholar]
- Wagner A, Donaldson L, Kim H, Phillips L, Flint H, Steward D, Torr K, Koch G, Schmitt U, Ralph J. (2009) Suppression of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiol 149: 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Tobimatsu Y, Phillips L, Flint H, Torr K, Donaldson L, Pears L, Ralph J. (2011) CCoAOMT suppression modifies lignin composition in Pinus radiata. Plant J 67: 119–129 [DOI] [PubMed] [Google Scholar]
- Walley JW, Dehesh K. (2010) Molecular mechanisms regulating rapid stress signaling networks in Arabidopsis. J Integr Plant Biol 52: 354–359 [DOI] [PubMed] [Google Scholar]
- Weng J-K, Chapple C. (2010) The origin and evolution of lignin biosynthesis. New Phytol 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhang D, Hu J, Zhou X, Ye X, Reichel KL, Stewart NR, Syrenne RD, Yang X, Gao P, et al. (2009) Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinform (Suppl 11) 10: S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahiaoui N, Marque C, Myton KE, Negrel J, Boudet AM. (1998) Impact of different levels of cinnamyl alcohol dehydrogenase down-regulation on lignins of transgenic tobacco plants. Planta 204: 8–15 [Google Scholar]
- Youn B, Camacho R, Moinuddin SGA, Lee C, Davin LB, Lewis NG, Kang C. (2006) Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4. Org Biomol Chem 4: 1687–1697 [DOI] [PubMed] [Google Scholar]
- Zeller G, Henz SR, Widmer CK, Sachsenberg T, Rätsch G, Weigel D, Laubinger S. (2009) Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J 58: 1068–1082 [DOI] [PubMed] [Google Scholar]
- Zhang K, Qian Q, Huang Z, Wang Y, Li M, Hong L, Zeng D, Gu M, Chu C, Cheng Z. (2006) GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol 140: 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Dixon RA. (2011) Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci 16: 227–233 [DOI] [PubMed] [Google Scholar]
- Zhou R, Jackson L, Shadle G, Nakashima J, Temple S, Chen F, Dixon RA. (2010) Distinct cinnamoyl CoA reductases involved in parallel routes to lignin in Medicago truncatula. Proc Natl Acad Sci USA 107: 17803–17808 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.