Abstract
A number of phosphate (Pi) starvation- or mycorrhiza-regulated Pi transporters belonging to the Pht1 family have been functionally characterized in several plant species, whereas functions of the Pi transporters that are not regulated by changes in Pi supply are lacking. In this study, we show that rice (Oryza sativa) Pht1;1 (OsPT1), one of the 13 Pht1 Pi transporters in rice, was expressed abundantly and constitutively in various cell types of both roots and shoots. OsPT1 was able to complement the proton-coupled Pi transporter activities in a yeast mutant defective in Pi uptake. Transgenic plants of OsPT1 overexpression lines and RNA interference knockdown lines contained significantly higher and lower phosphorus concentrations, respectively, compared with the wild-type control in Pi-sufficient shoots. These responses of the transgenic plants to Pi supply were further confirmed by the changes in depolarization of root cell membrane potential, root hair occurrence, 33P uptake rate and transportation, as well as phosphorus accumulation in young leaves at Pi-sufficient levels. Furthermore, OsPT1 expression was strongly enhanced by the mutation of Phosphate Overaccumulator2 (OsPHO2) but not by Phosphate Starvation Response2, indicating that OsPT1 is involved in the OsPHO2-regulated Pi pathway. These results indicate that OsPT1 is a key member of the Pht1 family involved in Pi uptake and translocation in rice under Pi-replete conditions.
Phosphorus (P) is one of the key mineral elements indispensable for plant growth and development. It is a structural component of nucleic acids and phospholipids and plays important roles in energy transfer, signal transduction, photosynthesis, and respiration (Plaxton and Carswell, 1999). Phosphate (Pi) is taken up by plant roots from the soil and translocated within the plant via phosphate transporters (PiTs; Raghothama, 1999).
A large number of the genes that encode PiTs have been identified from different plant families, including Arabidopsis (Arabidopsis thaliana), cereals, legumes, and solanaceous species (Paszkowski, 2006; Bucher, 2007; Chen et al., 2007; Ai et al., 2009; Jia et al., 2011; Nagarajan et al., 2011). A majority of them showed expression either exclusively or predominantly in the roots, and transcript levels were strongly induced by low Pi supply or by inoculation with arbuscular mycorrhiza (Mudge et al., 2002; Paszkowski et al., 2002; Javot et al., 2007; Ai et al., 2009). In Arabidopsis, three of nine PiTs belonging to the Pht1 family have been functionally characterized by using transferred DNA insertion mutants. AtPht1;1 and AtPht1;4 have been shown to be responsible for Pi acquisition under both high- and low-Pi conditions (Misson et al., 2004; Shin et al., 2004). The double mutant of both AtPht1;1 and AtPht1;4 showed a 75% reduction in Pi uptake capacity as compared with wild-type plants (Shin et al., 2004). Recently, Nagarajan et al. (2011) reported that the Arabidopsis pht1;5-1 mutant had higher shoot P content compared with the wild type, while the P content in roots was reduced under Pi-replete conditions, suggesting that Pht1;5 may mobilize Pi between source and sink tissues. In addition, Preuss et al. (2010) explored the characteristics of the low-affinity barley (Hordeum vulgare) PiT PHT1;6 using the Xenopus laevis oocyte expression system, implying that it may play a Pi-transport role at Pi-sufficient conditions in plants.
We previously reported that in rice (Oryza sativa), two Pi starvation-enhanced Pht1 members, rice Pht1;2 (OsPT2) and OsPT6, have different functions and kinetic properties (Ai et al., 2009). OsPT6 plays a broad role in Pi uptake and translocation throughout the plant, whereas OsPT2 is a low-affinity PiT expressed abundantly in Pi-starved roots to facilitate the transport of Pi from roots to shoot (Ai et al., 2009). Liu et al. (2010) reported that OsPT2 was responsible for most of the overaccumulation of Pi in shoots of rice Phosphate Starvation Response2 (OsPHR2) overexpression lines under Pi-sufficient conditions. In addition, we detected that another Pi-regulated Pht1 member, OsPT8, is expressed in various tissues and organs and is involved in Pi homeostasis in rice (Jia et al., 2011).
In this work, we investigated the functions of OsPht1;1 (accession no. AF536961; referred to here as OsPT1) in rice. Our results show that the expression pattern of OsPT1 is constitutively independent of Pi supply. The P concentration in the shoots increased significantly in OsPT1 overexpression lines and decreased in OsPT1 RNA interference (RNAi) lines under Pi-sufficient conditions. We also report the responses of root hair development, root cell membrane potential, Pi uptake rate, and the distribution in Pi-replete rice to the alteration of OsPT1 expression.
RESULTS
OsPT1 Is Expressed in Both Roots and Shoots of Rice Independent of Pi Supply Conditions
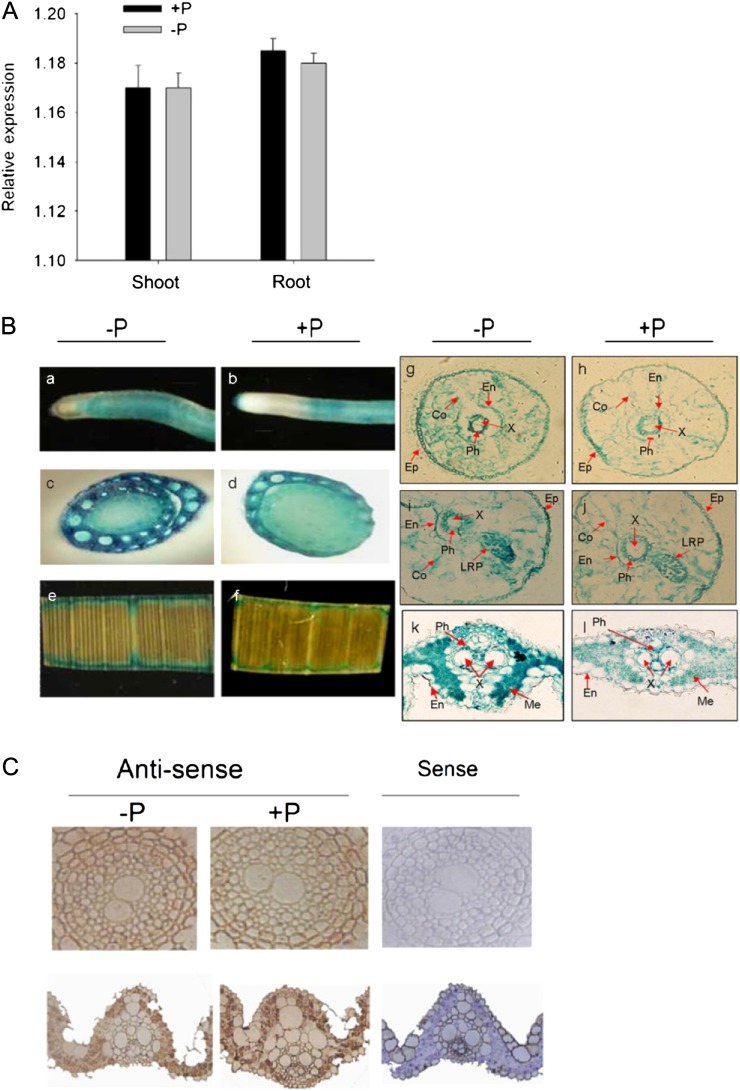
The expression of OsPT1 was investigated by semiquantitative reverse transcription (RT)-PCR in both roots and shoots of the seedlings grown under different Pi levels. OsPT1 was found to be abundantly expressed in the roots and shoots of rice under both Pi-sufficient and Pi-deficient conditions (Supplemental Fig. S1). To further confirm this result, quantitative RT-PCR was carried out by using a housekeeping gene, Actin (accession no. AB047313), as a control. As shown in Figure 1A, the nonalteration of OsPT1 transcript levels in the roots and shoots of seedlings by change in Pi supply (Fig. 1A) was consistent with the results obtained by semiquantitative RT-PCR (Supplemental Fig. S1), confirming that OsPT1 is constitutively expressed in rice.
Figure 1.
Expression pattern and tissue localization of OsPT1 in Pi-sufficient and Pi-deficient rice. A, Transcriptional patterns of OsPT1 in roots and shoots of rice. Ten-day-old rice seedlings were transferred to Pi-sufficient (300 µm Pi; +P) and Pi-deficient (10 µm Pi; −P) conditions for 21 d. Total RNAs were extracted from roots and shoots of the seedlings and determined by real-time quantitative RT-PCR. A housekeeping gene, Actin (OsRac1; accession no. AB047313), was used as an internal standard. Error bars indicate se (n = 3) of three biological replicates. B, Expression pattern of GUS driven by the native promoter of OsPT1 in rice. OsPT1 promoter-driven expression of the GUS reporter gene is shown for primary roots (a and b), root-shoot junctions (c and d), leaf blade (e and f), and transverse sections of the primary roots (g–j) and leaves (k and l) of rice seedlings supplied with Pi-sufficient (300 µm Pi) and Pi-deficient (10 µm Pi) levels for 21 d. Transverse sections of the primary roots were at 0.5 cm from the root tips. Co, Cortex; En, endodermis; Ep, epidermis; LRP, lateral root primordium; Me, mesophyll cells; Ph, phloem; X, xylem. C, Localization of OsPT1 expression in the roots and leaves of rice by in situ hybridization. Ten-day-old rice seedlings were transferred to Pi-sufficient (300 µm Pi) and Pi-deficient (10 µm Pi) conditions for 21 d before tissue collection. Transverse sections of the primary roots were at 0.5 cm from the root tips. Similar signals with Pi-sufficient or Pi-deficient conditions given by the antisense probe were observed in root (top row) and leaf (bottom row) sections. Sense probe was used as a negative control.
To analyze the tissue-specific expression and Pi responsiveness of OsPT1 in rice, we generated transgenic rice plants carrying GUS (Jefferson et al., 1987) as a reporter gene. The 5′ fragment of 2,768 bp immediately upstream of the translation start for OsPT1 was amplified and used to drive GUS expression. The expression of GUS driven by the ubiquitin promoter was used as a positive control. As shown in Figure 1B, GUS driven by the OsPT1 promoter was abundantly expressed in various cells of roots, root-shoot junctions, and leaves under both Pi-sufficient and Pi-deficient conditions. The expression of OsPT1 in roots and the root-shoot junctions was seen not only in epidermal cells, which are the dominant sites for the uptake of Pi from soil solution, but also throughout the cortical and stele cells (Fig. 1B, a–d and g–j), suggesting that it functions in both Pi uptake and translocation in rice. OsPT1 was also abundantly expressed in epidermal, mesophyll, and stele cells in the leaves, irrespective of Pi supply (Fig. 1B, e, f, k, and l). There was weak GUS expression in the spikelets and the emerging buds (Supplemental Fig. S2).
To confirm the correlation between the OsPT1 expression pattern by PCR and GUS staining, we performed in situ hybridization with the OsPT1 antisense probe in the roots and leaves of wild-type plants. The labeling of epidermis and cortex layers and the central cylinder in the root sections was observed, and there was no obvious difference under different Pi supply (Fig. 1C). Also, a similar signal in the leaves was observed under Pi-sufficient or Pi-deficient conditions (Fig. 1C). The patterns of labeled signals were in agreement with the PCR and reporter gene expression patterns described above.
OsPT1 Was Able to Complement Proton-Coupled Pi Transporter Activities in a Yeast Mutant
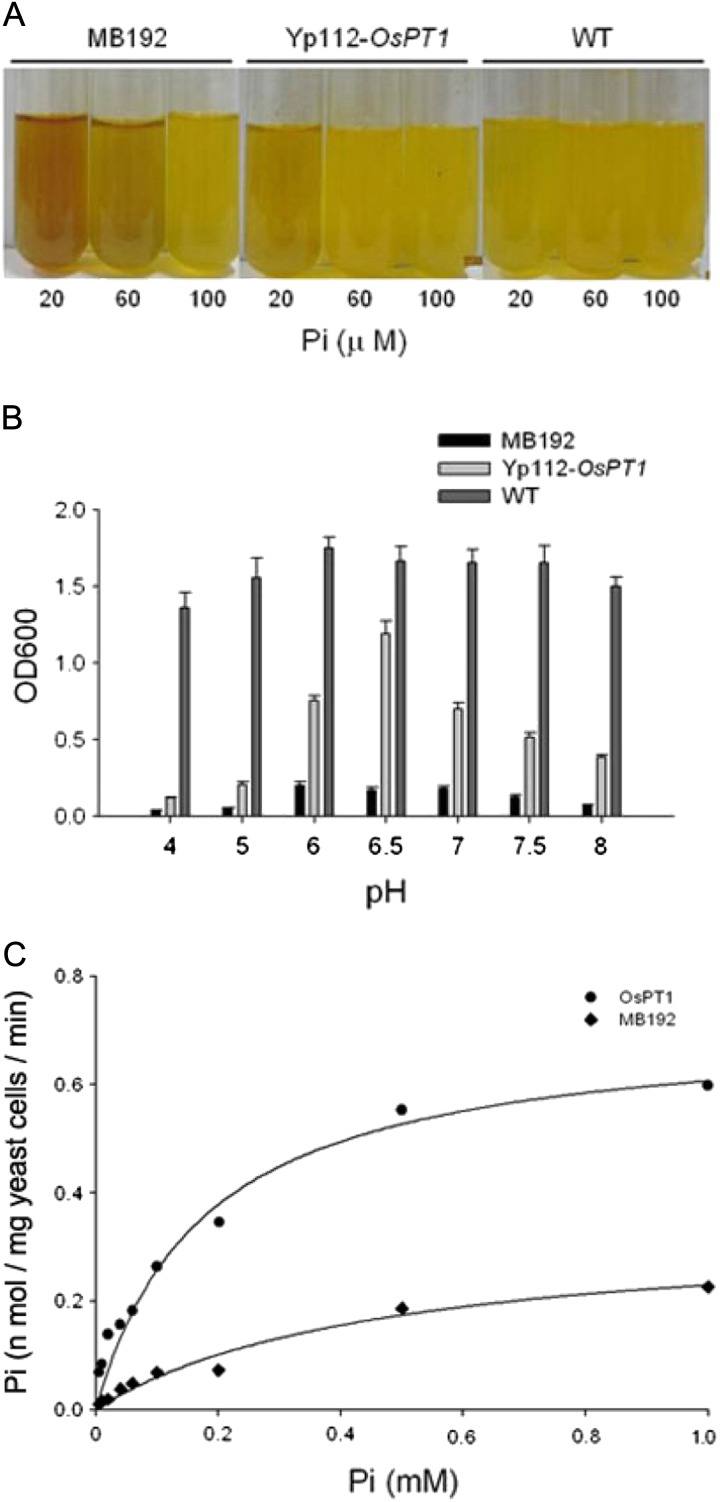
To obtain biochemical evidence for the function of OsPT1, we performed complementation analysis using the yeast mutant MB192, which is defective in the PHO84 gene, which encodes a high-affinity PiT on the plasma membrane. The transformed cells with either OsPT1 or empty vector were grown in yeast nitrogen base medium containing different concentrations of Pi and bromocresol purple as a pH indicator. A color shift from purple to yellow indicated the acidification of the liquid medium over a period of 24 h. In comparison with the cells of both wild-type and mutant MB192 plants, the transformants expressing OsPT1 (Yp112-OsPT1) could restore their growth at 20 μm Pi (Fig. 2A). At 60 μm Pi, there was no obvious difference in growth between the transformant and wild-type cells (Fig. 2A).
Figure 2.
Functional expression of OsPT1 in yeast. A, Complementation of Pi uptake-deficient yeast. Staining test for the color reaction in yeast strains MB192 (control), Yp112-OsPT1, which contains OsPT1 in MB192, and the wild-type (WT). B, Effects of different pH levels in the culture medium on the growth of the three yeast strains: Yp112- OsPT1, MB192, and the wild type. Error bars indicate se (n = 5). OD600, Optical density at 600 nm. C, Velocity of 33Pi transport by transformants containing Yp112-OsPT1 (circles) or carrying a vector (diamonds). The nonlinear regression of the Pi uptake of strain Yp112-OsPT1 versus the external concentration at pH 6.5 was used to estimate the apparent Km value for Pi uptake.
The pH dependence of Pi transport by OsPT1 was measuring over a range of pH values. Growth of the MB192 yeast strain expressing OsPT1 was optimal near neutral pH medium. It has a sharp pH optimum at 6.5, whereas the wild type has a broad pH optimum (Fig. 2B).
To determine the kinetic properties of OsPT1, Pi uptake experiments using 33Pi were performed in the transformed yeast mutant. Uptake rates of 33Pi at different Pi concentrations showed that Pi uptake mediated by OsPT1 followed Michaelis-Menten kinetics, exhibiting an apparent mean Km of 177 μm Pi (Fig. 2C). These data suggest that OsPT1 has lower Pi affinity than its homolog OsPT8 (Km = 23 μm Pi), which is also abundantly expressed in various rice tissues (Jia et al., 2011).
Change in the Expression of OsPT1 Altered P Concentration in Pi-Replete Shoots
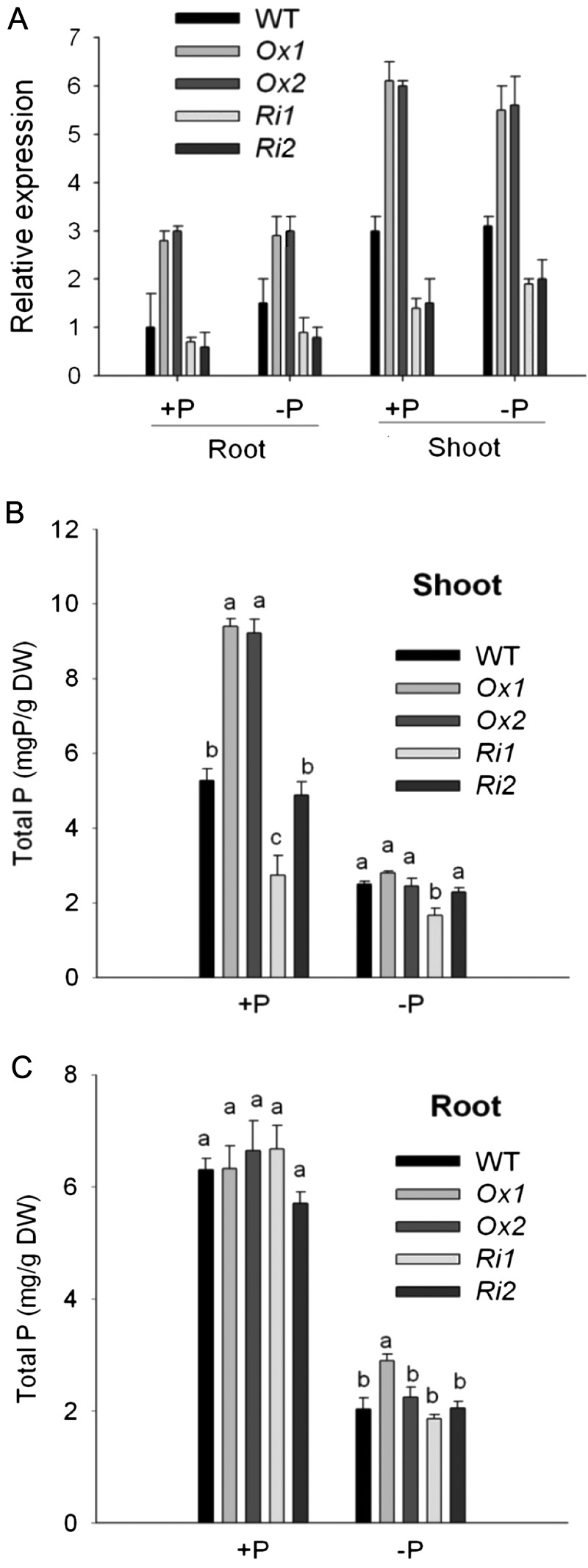
To characterize the function of OsPT1 in Pi uptake and translocation in rice, we generated OsPT1 overexpression lines using ubiquitin promoter (OsPT1-Ox) and knockdown lines by RNAi (OsPT1-Ri) in the background of the japonica cv Nipponbare via Agrobacterium tumefaciens-mediated transformation. Based on semiquantitative RT-PCR and real-time quantitative RT-PCR analyses, we selected the two lines of OsPT1-Ox (Ox1 and Ox2) and OsPT1-Ri (Ri1 and Ri2), respectively, for further characterization. The transcript level of OsPT1 was increased by about 2.3-fold in OsPT1-Ox lines and decreased by about 51% in OsPT1-Ri lines in comparison with its expression in the wild type (Fig. 3A). OsPT1 expression was not significantly regulated by Pi supply level in the transgenic rice plants (Fig. 3A). Meanwhile, we detected the relative expression of the genes that encode PiTs belonging to the Pht1 family in shoot of OsPT1-Ox and wild-type plants using real-time quantitative RT-PCR. OsPT8 and OsPT4 were significantly and moderately up-regulated, respectively. Other OsPTs detected were down-regulated (Supplemental Fig. S3).
Figure 3.
Molecular characterization and measurement of P concentrations in transgenic plants under Pi- sufficient and Pi-deficient conditions. A, Detection of the expression levels of OsPT1 in the wild type (WT) and transgenic lines by quantitative RT-PCR. Ox1, Ox2, Ri1, and Ri2 represent independent OsPT1-Ox and OsPT1-Ri lines. Total RNA was extracted from the roots and leaves in OsPT1-Ox, OsPT1-Ri, and wild-type plants grown for 21 d in the presence of 300 µm Pi (+P) or 10 µm Pi (−P). A housekeeping gene, Actin, was used as the internal standard. The results are means ± se of five biological replicates. B and C, P concentrations of shoots (B) and roots (C) of the wild type and the transgenic lines under Pi- sufficient and Pi-deficient conditions. Error bars indicate se (n = 5). DW, Dry weight.
In comparison with the wild type, OsPT1-Ox and OsPT1-Ri contained 76.3% higher and 27.9% lower P concentrations, respectively, in shoots grown under Pi-sufficient supply on average (Fig. 3B), whereas no significant differences of P concentration were seen in shoots under Pi-deficient supply (Fig. 3B). No significant differences of P concentration in roots were detected between the OsPT1-Ox, OsPT1-Ri, and wild-type plants under both Pi-sufficient and Pi-deficient conditions, with the exception of higher Pi in the Pi-deficient OsPT1-Ox1 lines (Fig. 3C).
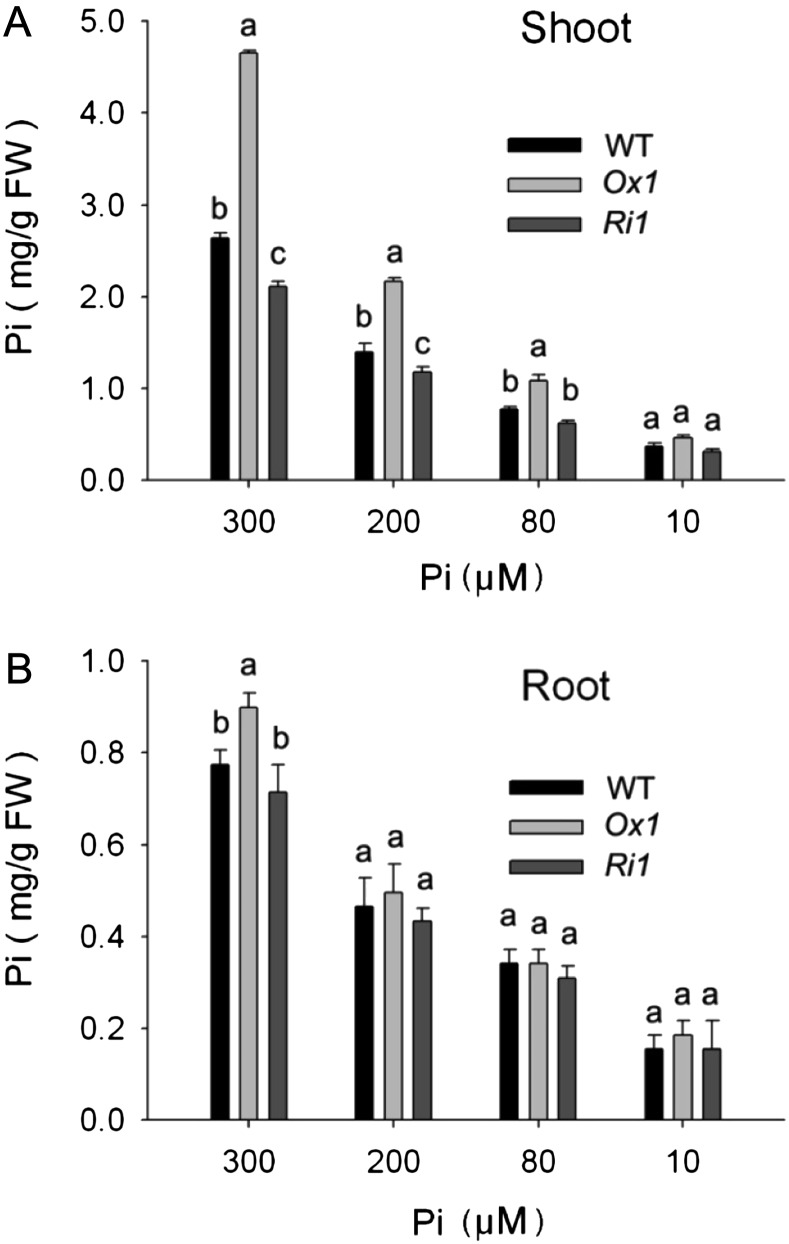
We used OsPT1-Ox1 and OsPT1-Ri1 to further examine the effects of OsPT1 overexpression or knockdown on Pi uptake rate and translocation at the different levels of Pi supply. As expected, Pi concentrations both in the shoots and roots of transgenic and wild-type plants were increased as the Pi concentration increased in the culture solution (Fig. 4). It is remarkable that the degree of shoot Pi concentration increase by OsPT1-Ox in comparison with the wild type was enlarged as the external Pi supply concentration increased (Fig. 4A). However, there was no significant difference of Pi concentration in the roots of OsPT1-Ox1, OsPT1-Ri1, and the wild type irrespective of Pi supply levels (Fig. 4B).
Figure 4.
Pi concentrations of OsPT1-Ox and OsPT1-Ri lines under different Pi levels in solution culture. OsPT1-Ox, OsPT1-Ri, and wild-type (WT) plants were grown for 21 d under varying concentrations of Pi (300, 200, 80, and 10 µm) in the hydroponic culture. Pi concentrations were measured in shoots (A) and roots (B) of OsPT1-Ox, OsPT1-Ri, and wild-type plants. Error bars indicate se (n = 5). FW, Fresh weight.
Change in the Expression of OsPT1 Altered Pi Uptake Rate and Distribution in Pi-Replete Rice
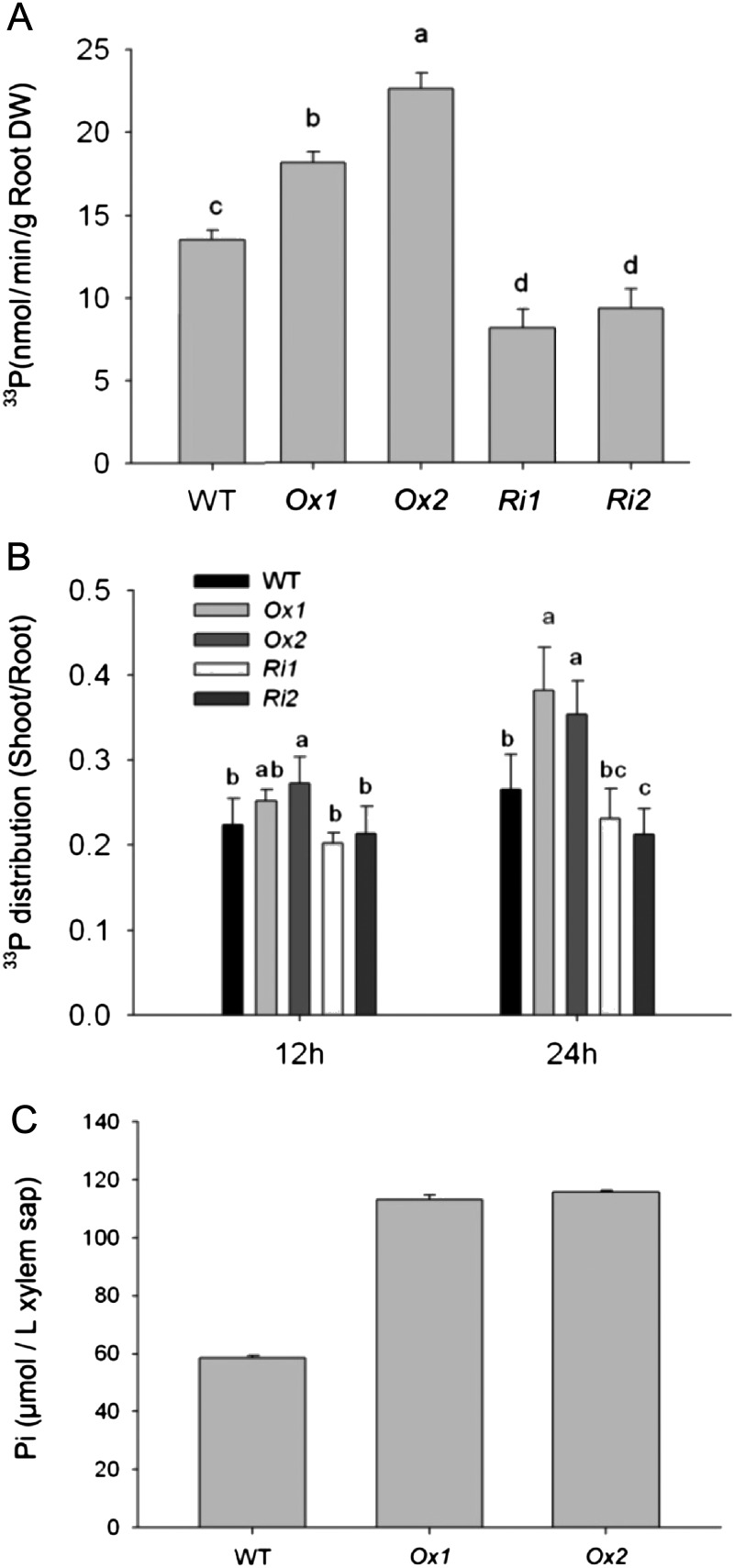
To precisely determine the contribution of OsPT1 to Pi acquisition and translocation, the uptake and distribution assay using the 33P radioisotope was performed with OsPT1-Ox, OsPT1-Ri, and wild-type plants pretreated with sufficient Pi. As shown in Figure 5A and Supplemental Figure S4, OsPT1-Ox transgenic plants showed the strongest 33P signals relative to the wild type, whereas in the OsPT1-Ri transgenic plants, 33P signals decreased from the base to the top of the plants relative to the wild type. 33P uptake rates of the roots of OsPT1-Ox1 and OsPT1-Ox2 transgenic plants were 18.3 and 21.6 nmol min−1 g−1 dry weight, which was 34% and 67% higher, respectively, than the wild-type value (Fig. 5A). In contrast, 33P uptake rates of the roots of OsPT1-Ri1 and OsPT1-Ri2 transgenic plants were 8.15 and 9.32 nmol min−1 g−1 dry weight, about 31% to 40% lower than the wild-type value (Fig. 5A). The relative amount of 33P translocation from roots to shoots was moderately increased after 24 h of Pi uptake in OsPT1-Ox plants, whereas it decreased in OsPT1-Ri plants (Fig. 5B). There were no significant differences in the uptake rates and distribution ratio of 33P between roots and shoots at Pi-deficient conditions (10 µm) for OsPT1-Ox, OsPT1-Ri, and the wild type (data not shown).
Figure 5.
The rate of Pi uptake by roots and the transportation of root-acquired Pi to shoots using 33Pi isotope, and Pi concentrations in the xylem sap of rice plants. OsPT1-Ox, OsPT1-Ri, and wild-type (WT) plants were grown for 7 d and then transferred into Pi-sufficient (300 µm Pi) medium. The Pi uptake of these seedlings was monitored over 24 h. A, Pi uptake rate of the roots in wild-type and transgenic plants on a root dry weight (DW) basis. B, Ratio of accumulated 33P in the shoots to that in the roots of wild-type and transgenic plants. C, Pi concentration in the xylem sap of OsPT1-Ox and wild-type plants at the grain-filling stage grown in soil with high available Pi. Five plants per line were measured. Error bars represent se (n = 5).
Root-acquired Pi must eventually be loaded into the apoplastic space of the xylem and transported to the shoot. In order to confirm that OsPT1 contributed to Pi transportation from roots to shoots, we measured the Pi concentration in the xylem sap of stems. The result showed that the Pi concentration in the xylem sap of OsPT1-Ox plants dramatically increased over that of the wild type, grown on Pi-replete soil, by nearly 2-fold at the beginning of the grain-filling stage (Fig. 5C), indicating that the overexpression of OsPT1 could enhance Pi transport to shoots even at a relatively late development stage in rice.
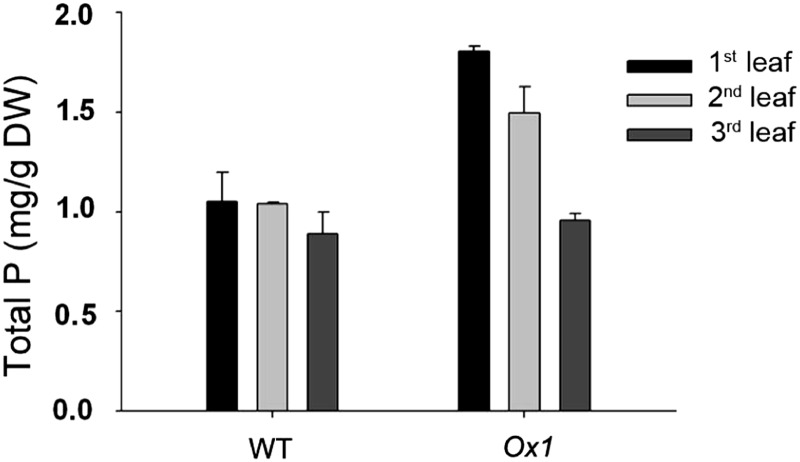
To further characterize the function of OsPT1 in retranslocation of Pi in the aerial part, the change of Pi concentrations was detected in the first, second, and third leaves from the top at the mature stage of Pi-replete rice. The results showed that the Pi concentrations of the first and second leaves of OsPT1-Ox plants remained higher than those of the wild type (Fig. 6), while the P concentrations in the third leaves of OsPT1-Ox plants were similar to the wild type over the same period (Fig. 6).
Figure 6.
Total P concentrations in different leaves of rice at the mature stage. Both OsPT1-Ox and wild-type (WT) plants were pot grown under Pi-sufficient conditions. The first, second, and third leaves were ordered from the top at the mature stage. Error bars indicate se (n = 5). DW, Dry weight.
Pi Uptake-Elicited Changes in the Root Cell Membrane Potential by Overexpression or Knockdown of OsPT1
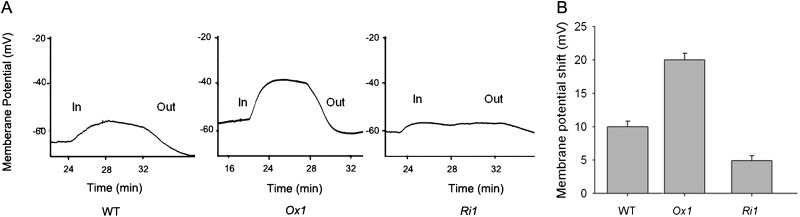
To detect the instantaneous Pi transport into root cells in OsPT1-Ox, OsPT1-Ri, and wild-type plants, we monitored the responses of the cell membrane potential of the roots to H2PO4− treatment. Provision of 300 µm Pi in the bathing solution elicited a rapid membrane potential depolarization of wild-type root cells, confirming that anion Pi was transported into the cells through proton-coupled Pi transporters (Ullrich-Eberius et al., 1984; Tamai et al., 1985). The Pi uptake-induced instantaneous depolarization of the root cells in OsPT1-Ox on average was nearly 2- and 4-fold that of the wild type and OsPT1-Ri, respectively (Fig. 7). These findings are consistent with the data showing that the highest uptake rate was in OsPT1-Ox plants, the lowest in OsPT1-Ri plants, and the middle in the wild type (Fig. 5A).
Figure 7.
Membrane potential changes in root rhizodermal cells of OsPT1-Ox, OsPT1-Ri, and wild-type (WT) plants. Rice seedlings were grown in solution containing 300 µm Pi for 2 weeks before the electrophysiological measurements. A, Wild type (left), OsPT1-Ox (middle), and OsPT1-Ri (right) plants treated with Pi. In represents 4 mm H2PO4− treatment to root, and Out represents removing 4 mm H2PO4− with 4 mm Cl−. B, Average values of membrane potential shifts by 4 mm H2PO4− treatment in 12 individual wild-type, OsPT1-Ox, and OsPT1-Ri seedlings. Error bars indicate se (n = 3).
Change in the Expression of OsPT1 Affected Root Growth and Root Hair Development
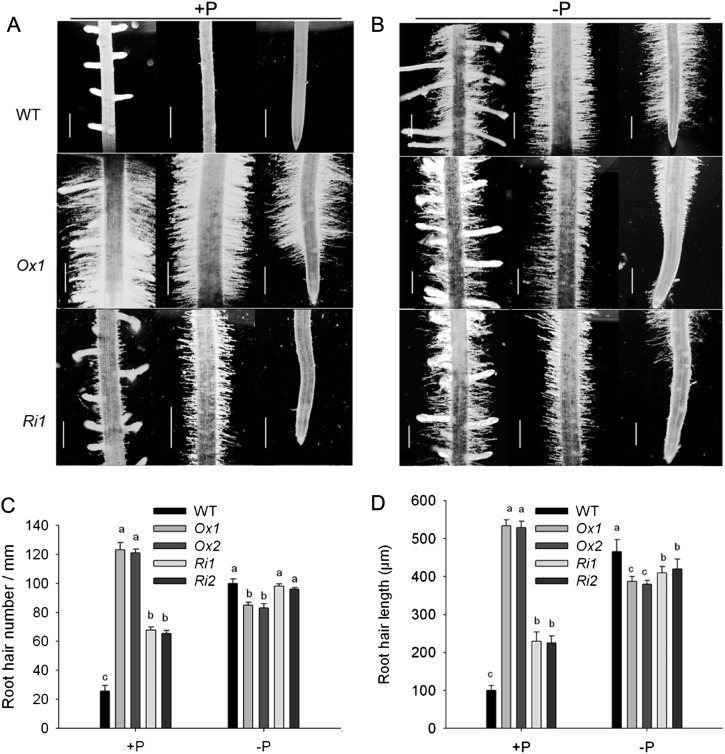
As commonly observed in other Pi-deficient plant roots, the roots of wild-type, OsPT1-Ox, and OsPT1-Ri lines developed large numbers of root hairs toward the apical ends, and there were slightly shorter root hairs in the transgenic lines than in the wild type under Pi-deficient conditions (Fig. 8, B–D). However, the number and length of root hairs developed from OsPT1-Ox lines were 5-fold more than those of wild-type plants grown in Pi-sufficient solution (Fig. 8). OsPT1-Ri lines also increased the root hair occurrence in comparison with the wild type grown at the Pi-sufficient level (Fig. 8, A, C, and D).
Figure 8.
Root phenotypes of wild-type (WT), OsPT1-Ox, and OsPT1-Ri seedlings under Pi-sufficient (+P) and Pi-deficient (−P) conditions. A and B, Root hair proliferation of 7-d-old wild-type, OsPT1-Ri1, and OsPT1-Ox1 seedlings grown in the presence of 300 µm Pi (A) and 10 µm Pi (B). Bars = 0.5 mm. C and D, Root hair number (C) and length (D), which were taken under Pi-sufficient and Pi-deficient conditions. Error bars indicate se (n = 10).
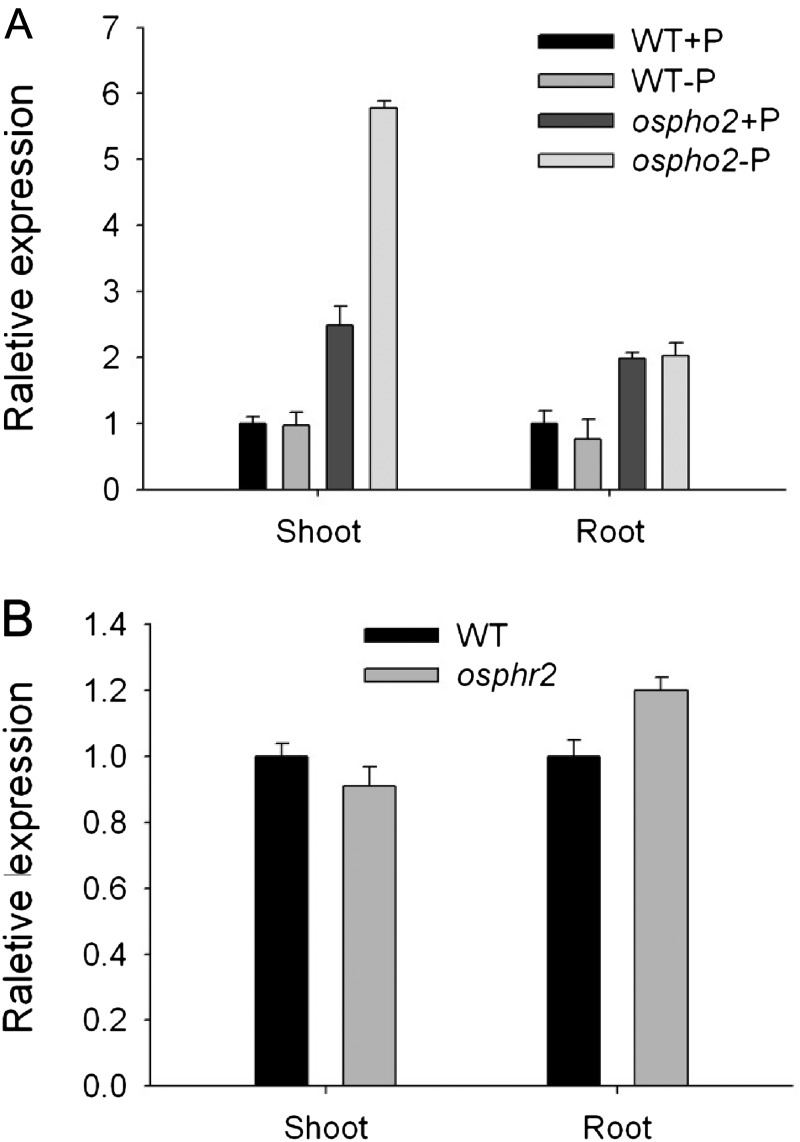
The Expression of OsPT1 Was Up-Regulated in a Rice Pi Accumulator Mutant (ospho2) and Not Altered in a Pi Starvation-Responsive Mutant (osphr2)
It has been reported previously that mutation of a Pi accumulator gene (OsPHO2) in rice resulted in an excess of Pi in its shoots, while Pi concentration in the roots was not largely affected (Wang et al., 2009; Hu et al., 2011). Since the OsPT1-Ox phenotype was similar to that of ospho2 under Pi-sufficient conditions, we examined the expression of OsPT1 in ospho2 mutant plants using real-time quantitative RT-PCR. Interestingly, OsPT1 transcripts in the shoots were 2.5- and 6-fold more in ospho2 than in the wild type under the Pi-sufficient and Pi-deficient conditions, respectively (Fig. 9A). OsPT1 expression in the roots was also significantly up-regulated by the mutation of OsPHO2 irrespective of Pi supply levels (Fig. 9A). In rice, OsPHR2 has been characterized as an ortholog gene of Arabidopsis AtPHR1 and functions in the Pi-responsive signaling pathway (Zhou et al., 2008). However, we did not observe a significant change of OsPT1 expression in osphr2 roots and shoots in comparison with the wild type (Fig. 9B).
Figure 9.
Expression levels of OsPT1 in roots and shoots of ospho2 and osphr2 mutants. A, Ten-day-old ospho2 and wild-type (WT) seedlings were transferred to Pi-sufficient (300 µm; +Pi) and Pi-deficient (10 µm Pi; −P) conditions for 21 d. B, Ten-day-old osphr2 and wild-type seedlings were subjected to Pi-sufficient (300 µm Pi) solution. RNA was extracted from roots and leaves of rice plants, and the expression levels of OsPT1 were determined by quantitative real-time PCR. A housekeeping gene, Actin (OsRac1; accession no. AB047313), was used as an internal standard. Error bars indicate se (n = 3) of three biological replicates.
DISCUSSION
OsPT1 Is Abundantly Expressed in Various Tissues Irrespective of Pi Supply Conditions
Among all PiTs belonging to the families of Pht1, Pht2, Pht3, and Pht4 in plants (Rausch and Bucher, 2002), the Pht1 family members were most widely studied due to their vital roles in Pi acquisition from soils and translocation from roots to other parts of plants. Most Pht1 family members are root-specific PiTs, and they were reported to be expressed in root epidermis cells (Mudge et al., 2002; Rae et al., 2003) or in cortical cells after arbuscular mycorrhiza fungi colonization (Bucher, 2007). The rice genome contains a total of 13 members of the Pht1 family, two of which (OsPT11 and OsPT13) are arbuscular mycorrhiza fungi colonization-enhanced PiTs (Paszkowski et al., 2002; Glassop et al., 2007). We have previously shown that OsPT2 and OsPT6 are strongly activated by Pi starvation, with distinct localization and transport functions (Ai et al., 2009), while OsPT8 is a gene that encodes a high-affinity PiT and acts downstream of OsPHR2, with a weak but distinct up-regulation in various tissue organs (Jia et al., 2011). In this work, analyses of promoter-GUS expression patterns, quantitative RT-PCR, and in situ hybridization showed that OsPT1 was abundantly expressed in various tissues, including roots, root-shoot junctions, and leaves, irrespective of Pi supply (Fig. 1).
That OsPT1 expression was not responsive to Pi-deficient conditions is consistent with the results of the sequence alignment analysis. Previous studies illustrated that many Pht1 genes from Arabidopsis, barley, wheat (Triticum aestivum), and rice were activated by AtPHR1 under Pi deprivation through the conserved PHR1-binding sequence (P1BS) element present in their promoters (Rubio et al., 2001; Schünmann et al., 2004; Tittarelli et al., 2007; Ai et al., 2009). We used the motif-building program MEME to identify conserved candidate regulatory motifs shared by the Pi-regulated PiTs among the 13 Pht1 members in the rice genome. The predictions indicate that OsPT1 and OsPT4/OsPT10 in all 13 rice Pht1 gene promoters have no P1BS motifs (Supplemental Fig. S5). OsPT2 and OsPT6, which were expressed abundantly under Pi starvation in rice, contain P1BS motifs in their promoters (Paszkowski et al., 2002; Ai et al., 2009).
It has been confirmed that OsPHR2 is involved in Pi starvation signaling and regulates the expression of several genes that encode PiTs belonging to the Pht1 family in rice (Zhou et al., 2008). That the transcriptional expression of OsPT1 was not regulated by external Pi supply level could be further supported by the nonaltered expression of OsPT1 in the osphr2 mutant (Fig. 9). This suggests that OsPT1 is not directly regulated by OsPHR2 in rice.
OsPT1 Functions in Pi Acquisition and Distribution and Pi-Mediated Root Hair Development in Pi-Replete Rice
It is generally accepted that Pi uptake in roots from low-Pi soil solution is catalyzed by high-affinity PiTs (Raghothama, 1999). It has been shown that AtPht1;1 and AtPht1;4, which were most highly expressed in epidermis and root hair cells, contributed to Pi transport into roots during growth in a wide range of external Pi concentrations (Shin et al., 2004), while AtPht1;5, which showed Pi deficiency-induced expression specifically in the phloem cells of older leaves, cotyledons, and flowers (Mudge et al., 2002), functions in mobilizing Pi between source and sink organs (Nagarajan et al., 2011). That the OsPT1 promoter directed its constitutive expression in epidermis, cortex, and stele cells of various rice tissues (Fig. 1) suggests that OsPT1 is likely to be involved in both Pi acquisition and translocation. However, either overexpression or knockdown of OsPT1 did not significantly affect shoot biomass, Pi concentrations in both roots and shoots, and Pi distribution in deficient-Pi-supplied rice (Figs. 3 and 4). There were only slight differences in the root responses to Pi starvation in transgenic rice in comparison with the wild type (Fig. 8). It has been shown that Pi starvation in rice strongly enhanced the expression of the high-affinity PiT gene OsPT6, which was expressed in multiple cell types from epidermis and cortex to stele, and the low-affinity PiT gene OsPT2, which was exclusively expressed in stele cells of various rice tissues (Ai et al., 2009). Therefore, it could be assumed that OsPT1 has functional redundancy with other Pi starvation up-regulated Pht1 members for the acquisition and transportation of Pi. Enhanced expression of OsPT2 and OsPT6, possibly together with other noncharacterized Pht1 members, could compensate the altered expression of OsPT1 in the Pi-starved rice plants.
Maintaining sufficient Pi in plant aboveground parts depends not only on root Pi acquisition from external solution but also on the transfer of Pi from roots to shoots via xylem and redistribution inside the plant via phloem. The PHO1 protein, primarily expressed in the root vascular cylinder, is known for mediating Pi efflux in loading Pi to root xylem in Arabidopsis (Hamburger et al., 2002; Stefanovic et al., 2011). In rice, it has been shown that OsPHO1;2 plays an important role in transferring Pi from roots to shoots (Secco et al., 2010). However, it is not clear if Pht1 members are directly responsible for Pi loading, unloading, or retrieval in the vascular tissue, particularly under Pi-sufficient conditions. Gómez-Ariza et al. (2009) analyzed the expression of five tomato (Solanum lycopersicum) PiT genes in different root tissues using laser microdissection technology. The results indicated that no expression of the detected PiT genes was observed in the central cylinder, irrespective of the presence of arbuscular symbiosis. Previously, we showed that knockdown of OsPT2 exclusively expressed in Pi-starved root stele cells decreased Pi transport from roots to shoots in rice (Ai et al., 2009). In this work, the increase of Pi supply gradually enlarged the difference of shoot P concentrations between the transgenic rice and the wild type and thus resulted in significantly higher P in OsPT1-Ox and lower P in OsPT1 RNAi mutants (Fig. 4). OsPT1-Ox had significantly higher root cell membrane potential depolarization responses to external Pi and higher Pi concentration in xylem sap than the wild type under P-replete conditions (Figs. 5 and 7). It is also interesting that the overexpression of OsPT1 resulted in much higher Pi in young leaves of mature plants (Fig. 6). All these data demonstrated that OsPT1 is involved in root Pi uptake and allocation in Pi-replete rice.
In Arabidopsis, overexpression of AtPht1;5 led to increases of root hair numbers and length both under Pi-sufficient and Pi-deficient conditions, while silencing of the gene did not have a significant effect on root hair development (Nagarajan et al., 2011). It was remarkable that the OsPT1-Ri lines, particularly OsPT1-Ox lines, developed much longer and dense root hairs only under P-replete conditions (Fig. 8). Since there was no significant difference of Pi concentration in their roots under Pi-sufficient conditions (Fig. 3C), the enhanced growth of root hair length and number of OsPT1-Ox in Pi-sufficient medium (Fig. 8) could not be simply explained by changes of root growth induced by internal Pi status. This could be due to the differential distribution of P between different tissues and organs in the whole plants by OsPT1, leading to altered root hair development.
OsPT1 and OsPT8 Have Similar Expression Patterns But Different Regulation Pathways
Of the 13 PiT members belonging to the Pht1 family in rice, OsPT1 and OsPT8 were the most highly expressed in both roots and shoots grown at high Pi levels (Jia et al., 2011; Fig. 1A). The analyses of a GUS reporter driven by the promoters of OsPT1 and OsPT8 indicate a partial overlap in their spatial expression patterns, with the strongest expression in the epidermis, cortical, and stele cells (Jia et al., 2011; Fig. 1B). However, unlike OsPT1, the promoter of OsPT8 contains a P1BS element, and its transcripts could be enhanced by Pi starvation and by overexpression of OsPHR2 (Jia et al., 2011). In addition, OsPT1, but not OsPT8, was strongly up-regulated in the ospho2 mutant, a shoot Pi overaccumulator (Fig. 9; Jia et al., 2011). Its interaction with the OsPHO2 signaling pathway needs to be characterized next.
Overexpression of OsPT8 resulted in excessive Pi in both roots and shoots, leading to Pi toxicity symptoms under high-Pi-supply conditions (Jia et al., 2011), whereas overexpression of OsPT1 increased Pi accumulation only in the shoots under Pi-sufficient conditions (Fig. 3). Interestingly, constitutively enhanced OsPT1 expression up-regulated OsPT8 with concurrent down-regulation of several other Pht1 members, including OsPT2 and OsPT6, in the shoots (Supplemental Fig. S3), while the OsPT8 knockdown mutation decreased OsPT1 expression (Jia et al., 2011). In contrast, overexpression of OsPT8 did not affect the expression of OsPT1 but enhanced the expression of OsPT2 and OsPT5, largely in the shoot (Jia et al., 2011). These results imply a possible functional interaction between OsPT1 and OsPT8, which will be elucidated by the double mutant of these two genes in the future.
In conclusion, this work shows that OsPT1 is a key member of the Pht1 family involved in Pi uptake and translocation in rice under Pi-replete conditions. It was expressed abundantly and constitutively in various cell types of both roots and shoots, irrespective of Pi supply level. This strengthens our understanding of OsPT function during Pi-sufficient conditions in higher plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa ssp. japonica ‘Nipponbare’) was used for the physiological experiments and rice transformation.
For hydroponic experiments, the seed sterilization procedure and the basal nutrient solution composition for seedling growth in a glasshouse were as described previously (Li et al., 2006). Ten-day-old seedlings were transferred to nutrient solution containing 1.25 mm NH4NO3, 0.35 mm K2SO4, 1 mm CaCl2·2H2O, 1 mm MgSO4·7H2O, 0.5 mm Na2SiO3·9H2O, 20 μm Fe-EDTA, 20 μm H3BO3, 9 μm MnCl2·4H2O, 0.32 μm CuSO4·5H2O, 0.77 μm ZnSO4·7H2O, and 0.39 μm Na2MoO4·2H2O, supplemented with 300 µm Pi (Pi-sufficient solution) or 10 µm Pi (Pi-deficient solution). The hydroponic experiments were carried out in a growth room with a 16-h-light (30°C)/8-h-dark (22°C) photoperiod, and the relative humidity was controlled at approximately 70%. The initial pH of the solution was adjusted to 5.5, and deionized water was used throughout the experiments. The nutrient solution was replaced every other day.
Samples were collected after the plants were treated for 3 weeks for semiquantitative RT-PCR and real-time quantitative RT-PCR, histochemical localization of the reporter gene, and the Pi uptake assay. In the experiments involving transgenic plants, the seeds were geminated and screened in a solution containing 25 mg L−1 hygromycin for 7 d before being transferred to the hydroponics system. For the different Pi concentration treatments, 300, 200, 80, and 10 µm Pi were used in the culture solution. For each experiment, three to five biological replicates were harvested.
The rice Tos17 insertion mutant ospho2 (NE8038_0_102_1A) and the rice transferred DNA insertion mutant osphr2 (04Z11NL88) were obtained from the National Institute of Agrobiological Sciences, Functional Genomics Laboratory, in Japan and the National Key Laboratory of Crop Genetic Improvement, National Center of Plant Gene Research, Huazhong Agricultural University, in China (http://rmd.ncpgr.cn), respectively. The confirmed homozygous mutant seedlings were used for detection of the expression levels of OsPT1. The conditions of growth were the same as described above.
Generation of the Transgenic Plants
For GUS expression pattern driven by the native promoter of OsPT1, the upstream sequence of the coding region of OsPT1 was first PCR amplified from the genomic DNA of cv Nipponbare rice using the primers listed in Supplemental Table S1. The restriction sites were incorporated into the primers to facilitate cloning into the expression vector. The amplified DNA fragments were cloned into the pMD19-T vector (TaKaRa) and were confirmed by restriction enzyme digestion and DNA sequencing. After digesting with the pMD19-T vector, the native promoter fragment of OsPT1 was cloned with the GUS reporter genes into the binary vector pS1aG-3 (kindly provided by Dr. E. Delhaize, Commonwealth Scientific and Industrial Research Organization Plant Industry; http://www.pi.csiro.au). The expression pattern driven by the ubiquitin promoter was used as a positive control.
For OsPT1-Ox transgenic plants, the cauliflower mosaic virus 35S promoter was used. The expression vector used was pCAMBIA1302. The primers for amplifying the coding region sequence are listed in Supplemental Table S1. The procedures for OsPT1 RNAi transgenic plants were as described previously (Ai et al., 2009). A 255-bp fragment of the OsPT1 coding sequence was amplified using the specific primers listed in Supplemental Table S1. The expression vector was transferred to Agrobacterium tumefaciens strain EHA105 by electroporation and transformed into rice as described by Upadhyaya et al. (2000).
Semiquantitative RT-PCR and Quantitative Real-Time PCR
Total RNAs were prepared from the roots and shoots of wild type, OsPT1-Ox, or OsPT1-Ri plants using Trizol reagent (Invitrogen; http://www.invitrogen.com). Semiquantitative RT-PCR for Actin (26 cycles) and OsPT1 (28 cycles) was performed using gene-specific primers. The PCR products were loaded on 1.2% agarose gels and photographed using a CCD camera.
For quantitative PCR analysis, DNase I-treated total RNAs were used for RT using SuperScript II (Invitrogen). The complementary DNA (cDNA) samples were diluted to 1, 0.5, and 0.1 ng mL−1. Triplicate quantitative assays were performed on each cDNA dilution with the SYBR Green Master Mix with the ABI 7000 sequence detection system according to the manufacturer’s protocol (Applied Biosystems); the gene-specific primers were designed by using PrimerExpress software (Applied Biosystems). The relative quantification method was used to evaluate quantitative variation between the replicates examined. The amplification of OsRAc1 (Actin) was used as an internal control to normalize all data. Triplicate quantitative assays were performed on each cDNA sample. All primers used for PCR are given in Supplemental Table S2.
Histochemical Localization of GUS Expression
The histochemical analysis of GUS activity was examined as described previously (Ai et al., 2009). Briefly, the samples were submersed in GUS reaction mix and incubated at 37°C overnight. To investigate subcellular expression patterns, the stained tissues were rinsed and fixed in formaldehyde-acetic acid for 24 h, embedded in paraffin, and then sectioned. The sections were transferred onto a slide and visualized with a stereomicroscope. The stained or sectioned tissues were photographed using an Olympus MVX10 stereomicroscope with a color CCD camera.
In Situ Hybridization
For in situ hybridization, digoxigenin-labeled DNA probes of antisense and sense OsPT1 were synthesized by Invitrogen with the DNA sequences 5′-AGATGACACAAATGGTTAGCGGCAA-3′ (antisense) and 5′-GCAGTACTCGTATAGCTCATTCTAT-3′ (sense). Root and leaf of 10-d-old plantlets of the wild type grown either in Pi-sufficient (300 µm Pi) or Pi-deficient (10 µm Pi) conditions were fixed, dehydrated, and embedded in paraffin, and the sections were prepared as described by Vernoux et al. (2000). The hybridizations were done by Shanghai LC Biotech Co. Digoxigenin-labeled probes were detected by anti-digoxigenin conjugated with alkaline phosphatase and SIGMAFAST Fast Red TR/naphthol AS-MX tablets (Sigma-Aldrich). The reaction was stopped by adding 10 mm Tris and 5 mm EDTA, pH 7.5. The background color of hybridization was blue. The light yellow color corresponds to the specific signal with antisense probe.
Functional Complementation Assay of OsPT1 in Yeast
The yeast manipulations were performed as described previously (Ai et al., 2009). For the complementation assay, the coding sequence of OsPT1 was amplified by PCR and subcloned into the yeast expression vector p112A1NE to create OsPT1-p112A1NE. The construct was transformed into the yeast Pi uptake-defective mutant MB192 (Bun-Ya et al., 1991). MB192-OsPT1 and control cells were grown to the logarithmic phase and then subjected to the yeast nitrogen base liquid medium containing different Pi concentrations (20, 60, or 100 µm) evenly. Bromocresol purple was used as a pH indicator. From purple to yellow, the color transformation of the liquid medium represented the acidification.
To detect the pH susceptibility of Pi uptake, different extracellular pH values (4, 5, 6, 6.5, 7, 7.5, and 8) were used at a fixed concentration of 80 μm K2HPO4. MES was used to keep the stability among the different pH values and measured the optical density at 600 nm after 24 h for the yeast strains Yp112-OsPT1, the wild type, and MB192. For each experiment, five biological replicates were measured.
In order to determine the kinetic properties of the OsPT1 transporter, Pi uptake experiments using 33Pi were performed using the transformed yeast. About 1-mg (fresh yeast) cell samples were used following the previously described method (Ai et al., 2009).
Measurement of P Concentration in Plants
Samples for the transgenic and wild-type plants from either Pi-sufficient or Pi-deficient treatments were assayed separately. For the measurement of unassimilated Pi concentration in the plant, about 0.5-g fresh samples were used following the previously described method (Zhou et al., 2008). Briefly, the sample was homogenized in 1 mL of 10% (w/v) perchloric acid using an ice-cold mortar and pestle. The homogenate was then diluted 10 times with 5% (w/v) perchloric acid and placed on ice for 30 min. After centrifugation at 10,000g for 10 min at 4°C, the supernatant was used for Pi measurement via the molybdenum blue method. The absorption values for the solution at 650 nm were determined using a Spectroquant NOVA60 spectrophotometer.
For the measurement of total P concentration in the plant, the samples were used following the method described by Wang et al. (2009) and Chen et al. (2007). Briefly, 0.03-g dry samples were predigested in glass tubes with H2SO4 for 2 h. The tubes were then heated to 180°C, and 50 µL of hydrogen peroxide was added every 10 min until the solution turned colorless. Pi concentration was analyzed as described above after dilution.
Xylem sap collection was performed at the initial grain-filling stage using the procedures described previously (Fan et al., 2005). Briefly, the stems were cut at 3 to 5 cm above the roots. The cut surfaces were rinsed with deionized water and blotted dry. Xylem sap from this cut surface were collected by absorption into a cotton wool pad placed on the cut surface. After 12 h, the sap sample in the cotton wool was extracted, and then the water-extractable P concentration was measured using the procedures described above.
Radioactive 33P Uptake Experiments
Seedlings of OsPT1-Ox, OsPT1-Ri, and wild-type plants that had been subjected to Pi-sufficient conditions for 3 weeks were incubated for 12 and 24 h in 250 mL of nutrient solution containing 8 µCi of KH233PO4 (Ai et al., 2009). Plants were rinsed in sterile distilled water until the radioactivity could not be detected in the solution, blot dried on 3M filter paper, and the roots and shoots were dried and weighed separately. The tissues were dried at 70°C for 2 d and then wet digested in a mixture of H2SO4 and hydrogen peroxide. The radioactivity of these solutions was measured by a Beckman LS6500 scintillation counter. Autoradiographs of the radioactive seedlings were then developed using photographic film plates.
Plasma Membrane Potential Measurements
Plasma membrane potentials of rice roots were measured as described previously (Fan et al., 2005) with minor modifications. A single primary root (not seminal) of intact rice plants was held in a Plexiglas chamber (volume of 2.0 mL) and bathed with a flowing solution (containing 5 mm MES, 0.5 mm CaCl2, 0.05 mm KCl, and 4 mm NaCl, pH 6.0) at a rate of 1 mL min−1. The 4 mm NaH2PO4 took the place of 4 mm NaCl as H2PO4− treatment in roots during membrane potential recording.
Measurement of Roots and Root Hairs
The number and length of root hairs were measured after 7 d under Pi-sufficient or Pi-deficient conditions. The number of lateral roots and all emerged lateral roots on the primary root were counted by eye and divided by the respective length of the primary roots. For root hair measurement, the root hair zone immediately behind the root tips was observed with a microscope (Olympus MVX10). The number of root hairs from one side of the root hair zone of the primary root was counted. The roots were photographed using an Olympus MVX10 stereomicroscope with a color CCD camera (http://www.olympus-global.com). Values are averages ± se of 10 seedlings.
Sequence data from this article can be found in the Rice Genome Initiative/GenBank data libraries under accession number AF536961 (OsPT1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcriptional patterns of OsPT1 in roots and leaves of rice.
Supplemental Figure S2. Expression of OsPT1 driven by the native promoter in reproductive organs of rice.
Supplemental Figure S3. Relative expression of the genes that encode Pi transporters belonging to the Pht1 family in shoots of OsPT1-Ox transgenic and wild-type plants.
Supplemental Figure S4. 33Pi uptake in OsPT1-Ox, OsPT1-Ri, and wild-type plants.
Supplemental Figure S5. Motif analysis in the putative promoters of genes that encode Pi transporters belonging to the Pht1 family in rice.
Supplemental Table S1. Primers used to generate the expression vectors.
Supplemental Table S2. Primers used to amplify the OsPT1 cDNA fragments.
Supplementary Material
Glossary
- Pi
phosphate
- P
phosphorus
- PiT
phosphate transporter
- RNAi
RNA interference
- RT
reverse transcription
- cDNA
complementary DNA
References
- Ai PH, Sun SB, Zhao JN, Fan XR, Xin WJ, Guo Q, Yu L, Shen QR, Wu P, Miller AJ, et al. (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J 57: 798–809 [DOI] [PubMed] [Google Scholar]
- Bucher M. (2007) Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol 173: 11–26 [DOI] [PubMed] [Google Scholar]
- Bun-Ya M, Nishimura M, Harashima S, Oshima Y. (1991) The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol 11: 3229–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AQ, Hu J, Sun SB, Xu GH. (2007) Conservation and divergence of both phosphate- and mycorrhiza-regulated physiological responses and expression patterns of phosphate transporters in solanaceous species. New Phytol 173: 817–831 [DOI] [PubMed] [Google Scholar]
- Fan XR, Shen QR, Ma ZQ, Zhu HL, Yin XM, Miller AJ. (2005) A comparison of nitrate transport in four different rice (Oryza sativa L.) cultivars. Sci China Ser C 48: 897–911 [PubMed] [Google Scholar]
- Glassop D, Godwin RM, Smith SE, Smith FW. (2007) Rice phosphate transporters associated with phosphate uptake in rice colonized with arbuscular mycorrhizal fungi. Can J Bot 85: 644–651 [Google Scholar]
- Gómez-Ariza J, Balestrini R, Novero M, Bonfante P. (2009) Cell-specific gene expression of phosphate transporters in mycorrhizal tomato roots. Biol Fertil Soils 45: 845–853 [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y. (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Zhu CG, Li F, Tang JY, Wang YQ, Lin AH, Liu LC, Che RH, Chu CC. (2011) LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol 156: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HF, Ren HY, Gu M, Zhao JN, Sun SB, Zhang X, Chen JY, Wu P, Xu GH. (2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol 156: 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BZ, Xin WJ, Sun SB, Shen QR, Xu GH. (2006) Physiological and molecular responses of nitrogen-starved rice plants to re-supply of different nitrogen sources. Plant Soil 287: 145–159 [Google Scholar]
- Liu F, Wang ZY, Ren HY, Shen C, Li Y, Ling HQ, Wu CY, Lian XM, Wu P. (2010) OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J 62: 508–517 [DOI] [PubMed] [Google Scholar]
- Misson J, Thibaud MC, Bechtold N, Raghothama K, Nussaume L. (2004) Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW. (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Nagarajan VK, Jain A, Poling MD, Lewis AJ, Raghothama KG, Smith AP. (2011) Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol 156: 1149–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski U. (2006) A journey through signaling in arbuscular mycorrhizal symbioses 2006. New Phytol 172: 35–46 [DOI] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP. (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Carswell MC. (1999) Metabolic aspects of the phosphate starvation response in plants. In HR Lerner, ed, Plant Responses to Environmental Stresses: From Phytohormones to Genome Reorganization. Marcel Dekker, New York, pp 349–372
- Preuss CP, Huang CY, Gilliham M, Tyerman SD. (2010) Channel-like characteristics of the low-affinity barley phosphate transporter PHT1;6 when expressed in Xenopus oocytes. Plant Physiol 152: 1431–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae AL, Cybinski DH, Jarmey JM, Smith FW. (2003) Characterization of two phosphate transporters from barley: evidence for diverse function and kinetic properties among members of the Pht1 family. Plant Mol Biol 53: 27–36 [DOI] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Rausch C, Bucher M. (2002) Molecular mechanisms of phosphate transport in plants. Planta 216: 23–37 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schünmann PHD, Richardson AE, Smith FW, Delhaize E. (2004) Characterization of promoter expression patterns derived from the Pht1 phosphate transporter genes of barley (Hordeum vulgare L.). J Exp Bot 55: 855–865 [DOI] [PubMed] [Google Scholar]
- Secco D, Baumann A, Poirier Y. (2010) Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol 152: 1693–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Stefanovic A, Arpat AB, Bligny R, Gout E, Vidoudez C, Bensimon M, Poirier Y. (2011) Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J 66: 689–699 [DOI] [PubMed] [Google Scholar]
- Tamai Y, Toh-e A, Oshima Y. (1985) Regulation of inorganic phosphate transport systems in Saccharomyces cerevisiae. J Bacteriol 164: 964–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittarelli A, Milla L, Vargas F, Morales A, Neupert C, Meisel LA, Salvo-G H, Peñaloza E, Muñoz G, Corcuera LJ, et al. (2007) Isolation and comparative analysis of the wheat TaPT2 promoter: identification in silico of new putative regulatory motifs conserved between monocots and dicots. J Exp Bot 58: 2573–2582 [DOI] [PubMed] [Google Scholar]
- Ullrich-Eberius CI, Novacky A, Bel AJE. (1984) Phosphate-uptake in Lemna gibba G1-energetics and kinetics. Planta 161: 46–52 [DOI] [PubMed] [Google Scholar]
- Upadhyaya NM, Surin B, Ramm K, Gaudron J, Schunmann PHD, Taylor W, Waterhouse PM, Wang MB. (2000) Agrobacterium-mediated transformation of Australian rice cultivars Jarrah and Amaroo using modified promoters and selectable markers. Aust J Plant Physiol 27: 201–210 [Google Scholar]
- Vernoux T, Kronenberger J, Grandjean O, Laufs P, Traas J. (2000) PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127: 5157–5165 [DOI] [PubMed] [Google Scholar]
- Wang C, Ying S, Huang HJ, Li K, Wu P, Shou HX. (2009) Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J 57: 895–904 [DOI] [PubMed] [Google Scholar]
- Zhou J, Jiao FC, Wu ZC, Li YY, Wang XM, He X, Zhong WQ, Wu P. (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.