Abstract
Magnesium (Mg)-mediated alleviation of aluminum (Al) toxicity has been observed in a number of plant species, but the mechanisms underlying the alleviation are still poorly understood. When a putative rice (Oryza sativa) Mg transporter gene, Oryza sativa MAGNESIUM TRANSPORTER1 (OsMGT1), was knocked out, the tolerance to Al, but not to cadmium and lanthanum, was decreased. However, this inhibition could be rescued by addition of 10 μm Mg, but not by the same concentration of barium or strontium. OsMGT1 was expressed in both the roots and shoots in the absence of Al, but the expression only in the roots was rapidly up-regulated by Al. Furthermore, the expression did not respond to low pH and other metals including cadmium and lanthanum, and was regulated by an Al-responsive transcription factor, AL RESISTANCE TRANSCRIPTION FACTOR1. An investigation of subcellular localization showed that OsMGT1 was localized to the plasma membrane. A short-term (30 min) uptake experiment with stable isotope 25Mg showed that knockout of OsMGT1 resulted in decreased Mg uptake, but that the uptake in the wild type was enhanced by Al. Mg concentration in the cell sap of the root tips was also increased in the wild-type rice, but not in the knockout lines in the presence of Al. A microarray analysis showed that transcripts of genes related to stress were more up- and down-regulated in the knockout lines. Taken together, our results indicate that OsMGT1 is a transporter for Mg uptake in the roots and that up-regulation of this gene is required for conferring Al tolerance in rice by increasing Mg concentration in the cell.
Soluble ionic aluminum (Al) rapidly inhibits root growth at micromolar concentrations and subsequently affects the uptake of nutrients and water (Kochian et al., 2004; Ma, 2007; Delhaize et al., 2012). However, this Al toxicity could be alleviated by supply of magnesium (Mg) in a number of plant species including sorghum (Sorghum bicolor; Tan et al., 1992), soybean (Glycine max; Silva et al., 2001a), wheat (Triticum aestivum; Ryan et al., 1994), rice (Oryza sativa; Watanabe and Okada, 2005), and rice bean (Vigna umbellata; Yang et al., 2007). The Mg-mediated alleviative effect on Al toxicity differs with plant species. In some species such as soybean and rice bean, Mg at micromolar concentrations is able to alleviate Al toxicity (Silva et al., 2001a; Yang et al., 2007), but millimolar concentrations are required to have the alleviative effects in other species such as wheat and rice (Ryan et al., 1997; Watanabe and Okada, 2005). The latter has been attributed to increased ionic strength of the solutions (Noble and Sumner, 1988), reduction in Al saturation at the apoplastic exchange sites (Grauer and Horst, 1992), and decreased Al activity at the root cell plasma membrane surface (Kinraide, 2003; Kinraide et al., 2004). Al toxicity is caused by its binding to different cellular components including cell wall and plasma membrane (Ma, 2000). The Al3+ and Mg2+ ions have similar hydrated radius (Bose et al., 2011). Therefore, high Mg concentrations at millimolar range displace or compete with Al from binding sites on the root cell wall, plasma membrane, and other components, protecting the roots from Al toxicity.
Enhanced secretion of organic acid anions is likely involved in the Mg-alleviated Al toxicity at micromolar range. In soybean, addition of 50 μm Mg enhanced the citrate concentration in the root tips and stimulated citrate secretion from the roots exposed to toxic Al level (Silva et al., 2001b), resulting in increased Al tolerance by forming nontoxic citrate-Al complex in the rhizosphere. Inclusion of 10 μm Mg in the Al treatment solution also increased citrate secretion in rice bean (Yang et al., 2007). This increase has been associated with restoring plasma membrane H+-ATPase activity. However, the exact mechanisms leading to the increased citrate secretion by Mg is still poorly understood.
On the other hand, increasing internal Mg seems to be also required for conferring Al tolerance. Overexpression of yeast (Saccharomyces cerevisiae) Mg transporters (ALR1 or ALR2) conferred Al tolerance in yeast (MacDiarmid and Gardner, 1998). Overexpression of an Arabidopsis (Arabidopsis thaliana) Mg transporter gene AtMGT1 in Nicotiana benthamiana also conferred higher Al tolerance. This increase is associated with enhanced Mg uptake (Deng et al., 2006).
Rice is the most Al-tolerant crop among small grain cereal. This high level of tolerance has been attributed to multiple Al tolerance genes, which are regulated by a transcription factor, AL RESISTANCE TRANSCRIPTION FACTOR1 (ART1). ART1 belongs to the family of Cys2-His2-type zinc-finger transcription factors and is constitutively expressed in the root (Yamaji et al., 2009). Although its expression is not induced by Al treatment, when mutated, seedlings are Al sensitive (Yamaji et al., 2009). ART1 regulates the expression of at least 31 genes and interacts with a cis-element [GGN(T/g/a/C)V(C/A/g)S(C/G)] present in the promoter of 29 out of the 31 genes (Tsutsui et al., 2011). Some of ART1-regulated downstream genes including SENSITIVE TO AL RHIZOTOXICITY1 (STAR1)/STAR2, Nramp aluminum transporter1 (Nrat1), rice FERRIC REDUCTASE DEFECTIVE LIKE4 (OsFRDL4), and rice ALUMINUM SENSITIVE1 (OsALS1) have been functionally characterized. STAR1 and STAR2 form an ATP-binding cassette transporter, which is implicated in cell wall modification (Huang et al., 2009). OsFRDL4 functions as a citrate transporter, which secretes citrate from the roots to chelate Al outside (Yokosho et al., 2011). On the other hand, Nrat1 is an Al transporter localized at the plasma membrane, which transports Al into the cells (Xia et al., 2010). Subsequently the Al in the cell can be sequestrated into the vacuoles by OsALS1 at the tonoplast (Huang et al., 2012). However, most ART1-regulated genes have not been characterized. In this study, we functionally characterized an ART1-regulated gene, Oryza sativa MAGNESIUM TRANSPORTER1 (OsMGT1; Os01g0869200) encoding a putative Mg transporter. We found that rice is able to up-regulate this gene under Al stress, conferring Al tolerance.
RESULTS
Sequence Analysis of OsMGT1 in Rice
OsMGT1 (Os01g0869200) cloned from rice (cv Nipponbare) contains six exons and five introns (Supplemental Fig. S1A), encoding a peptide of 418 amino acids. Using the SOSUI program (http://bp.nuap.nagoya-u.ac.jp/sosui/), we predicted that OsMGT1 is a membrane protein with two transmembrane domains near the C terminus (Supplemental Fig. S1B). OsMGT1 showed the similarity ranging from 63% to 81% to Arabidopsis Mg transporter AtMGT family at the amino acid level (Supplemental Fig. S2A). Like other Arabidopsis members, there is a conserved Gly-Met-Asn tripeptide motif at the end of the first transmembrane domain (Supplemental Fig. S2B; Knoop et al., 2005).
Expression Pattern of OsMGT1
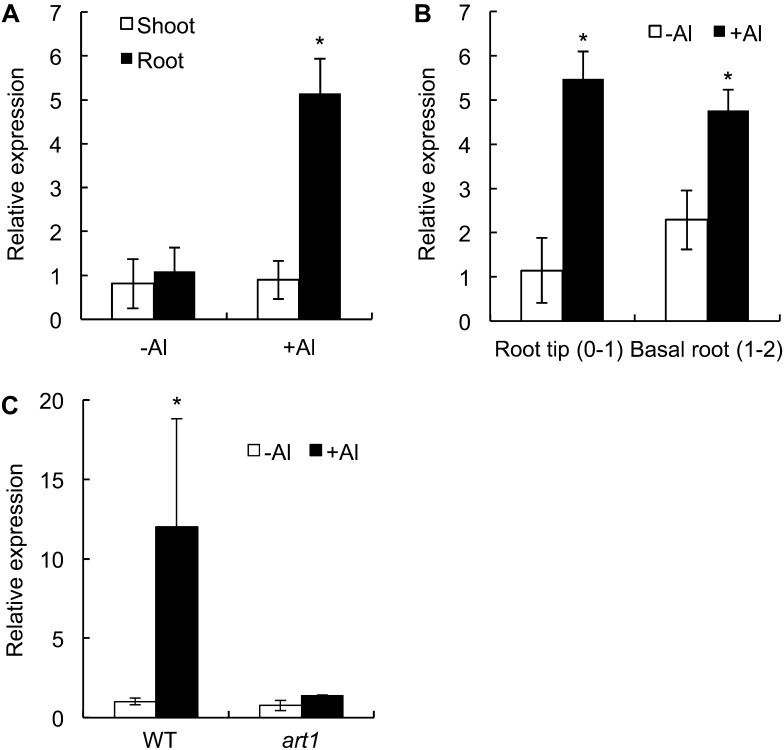
Real-time reverse transcription (RT)-PCR analysis revealed that OsMGT1 was expressed in both the shoots and roots at similar level in the absence of Al (Fig. 1A). However, in the presence of Al, the expression in the roots was up-regulated by 5 times, but that in the shoots was unaffected. Root spatial analysis showed that OsMGT1 was expressed in both the root tips (0–1 cm) and mature region (1–2 cm) and the expression in both regions were up-regulated by Al (Fig. 1B). However, in art1 (a mutant defective in the Al response C2H2-type zinc-finger transcription factor), the Al-induced expression was not observed (Fig. 1C), confirming that OsMGT1 expression was regulated by ART1 (Yamaji et al., 2009).
Figure 1.
Expression pattern of OsMGT1 in rice. A, Expression of OsMGT1 in the roots and shoots. Rice seedlings were exposed to a solution containing 50 μm Al at pH 4.5 for 6 h and the roots and shoots were sampled for analysis. B, Root spatial expression. After exposing to 50 μm AlCl3 for 6 h, root segments (0–1 cm and 1–2 cm) were excised with a razor. C, Expression of OsMGT1 in the art1 mutant. Both wild-type (WT) rice and the art1 mutant were exposed to 20 μm AlCl3 for 4 h. The expression level was determined by real-time RT-PCR. Histone H3 was used as an internal standard. Expression level relative to WT root (−Al) in A, C, and to root tip (−Al) in B is shown. Data are means of three biological replicates. The asterisk shows a significant difference (P < 0.05 by Tukey’s test).
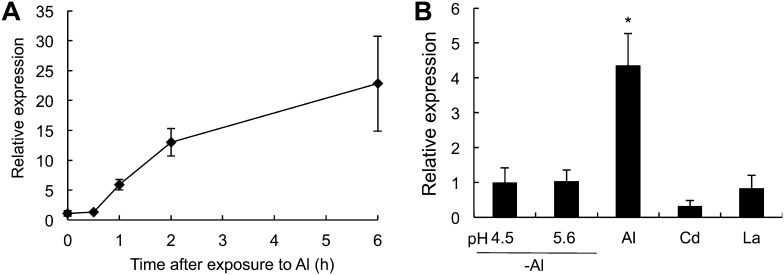
A time-course experiment showed that the Al-up-regulated expression of OsMGT1 occurred within 1 h and kept at a high level thereafter (Fig. 2A). Neither low pH nor other metals including cadmium (Cd) and lanthanum (La) affected the expression of OsMGT1 in the roots (Fig. 2B), indicating Al-specific response of the expression.
Figure 2.
Time-dependent and metal-specific expression of OsMGT1 in rice roots. A, Time-dependent expression of OsMGT1. Rice seedlings were exposed to a solution containing 50 μm AlCl3 for different time. B, Metal- and pH-dependent expression. Rice seedlings were exposed to a solution containing 0, 30 μm Cd, 10 μm La, or 50 μm Al at pH 4.5 or containing 0 μm Al at pH 5.6 for 6 h. The expression level was determined by real-time RT-PCR. Histone H3 was used as an internal standard. Expression level relative to 0 h in A and to pH 4.5 (−Al) in B is shown. Data are means of three biological replicates. The asterisk shows a significant difference (P < 0.05 by Tukey’s test).
Subcellular Localization of OsMGT1
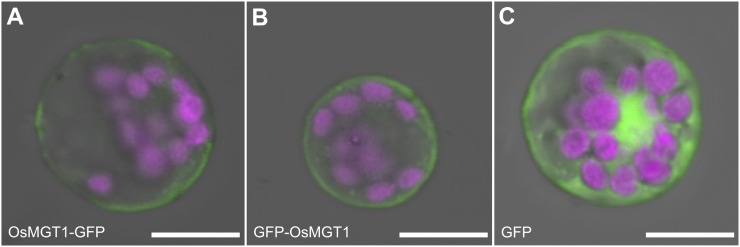
The subcellular localization of OsMGT1 was investigated in rice protoplasts by transiently expressing OsMGT1 fused with GFP at both C terminal and N terminal under the control of 35S promoter. Either construct gave a signal at the plasma membrane (Fig. 3, A and B), in contrast to GFP alone, which signal was observed in cytosol and nuclei (Fig. 3C).
Figure 3.
Subcellular localization of OsMGT1. OsMGT1-GFP (A), GFP-OsMGT1 (B) constructs, or GFP alone (C) were transformed into rice leaf protoplasts by polyethylene glycol method. Magenta color shows chloroplast autofluorescence. Scale bar = 10 μm.
Knockout of OsMGT1 Resulted in Increased Sensitivity to Al
To investigate the physiological role of OsMGT1 in Al tolerance, we obtained two independent retrotransposon Tos-17 insertion lines, NE4528 and NF0595. Tos-17 was inserted at the fifth and sixth exon, respectively (Supplemental Fig. S1A). RT-PCR analysis showed that there was no entire mRNA expression of this gene in two lines (Supplemental Fig. S1C), indicating that they are knockout lines of OsMGT1.
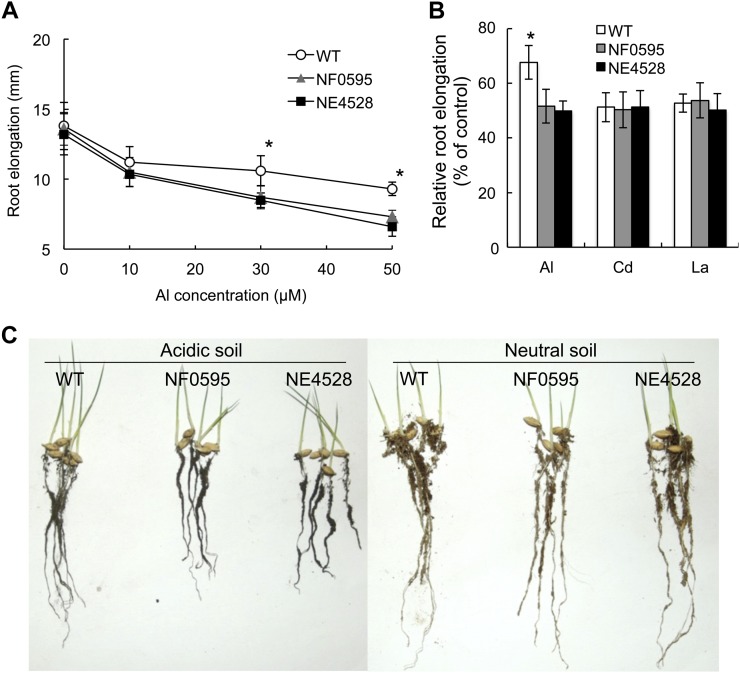
We first compared the Al tolerance between wild type and two knockout lines. In the absence of Al and low Al concentration (10 μm), the root elongation was similar between wild type and the two knockout lines (Fig. 4A). However, in the presence of 30 and 50 μm Al, the mutants showed higher inhibition of root elongation than wild type (Fig. 4A). There was no difference in the tolerance to other metals including Cd and La between wild type and the two knockout lines (Fig. 4B), indicating that knockout of OsMGT1 only specifically affects Al sensitivity.
Figure 4.
Phenotypic analysis of OsMGT1 knockout lines. A, Sensitivity of OsMGT1 knockout lines to Al. Seedlings of wild-type rice (WT) and two OsMGT1 knockout lines (NF0595 and NE4528) were exposed to a solution containing different concentration of Al (0, 10, 30, and 50 μm) for 24 h. The root length was measured before and after the treatment. Data are means ± sd (n = 10). The asterisk shows a significant difference at P < 0.05 by Tukey’s test. B, Sensitivity of OsMGT1 knockout lines to other metals. WT and two knockout lines were exposed to a solution containing 50 μm Al, 10 μm Cd, or 5 μm La for 24 h and root elongation relative to the root growth without metals is shown. Data are means ± sd (n = 10). The asterisk shows a significant difference (P < 0.05 by Tukey’s test). C, Growth on acidic soil. Germinated seeds were sowed on acidic soil (Andsol, pH 4.5) or neutral soil (pH 6.5) and grown for 1 week.
To test the tolerance on acid soil, we grew both wild type and knockout lines on acid soil (Andosol, pH 4.5) and neutral soil (pH 6.5). On neutral soil there was no difference in root growth (Fig. 4C), but the growth was inhibited in the knockout lines compared with wild type on acid soil (Fig. 4C). These results further indicate that OsMGT1 is required for Al tolerance in rice.
Micromolar Mg Recused Al-Induced Inhibition of Root Elongation
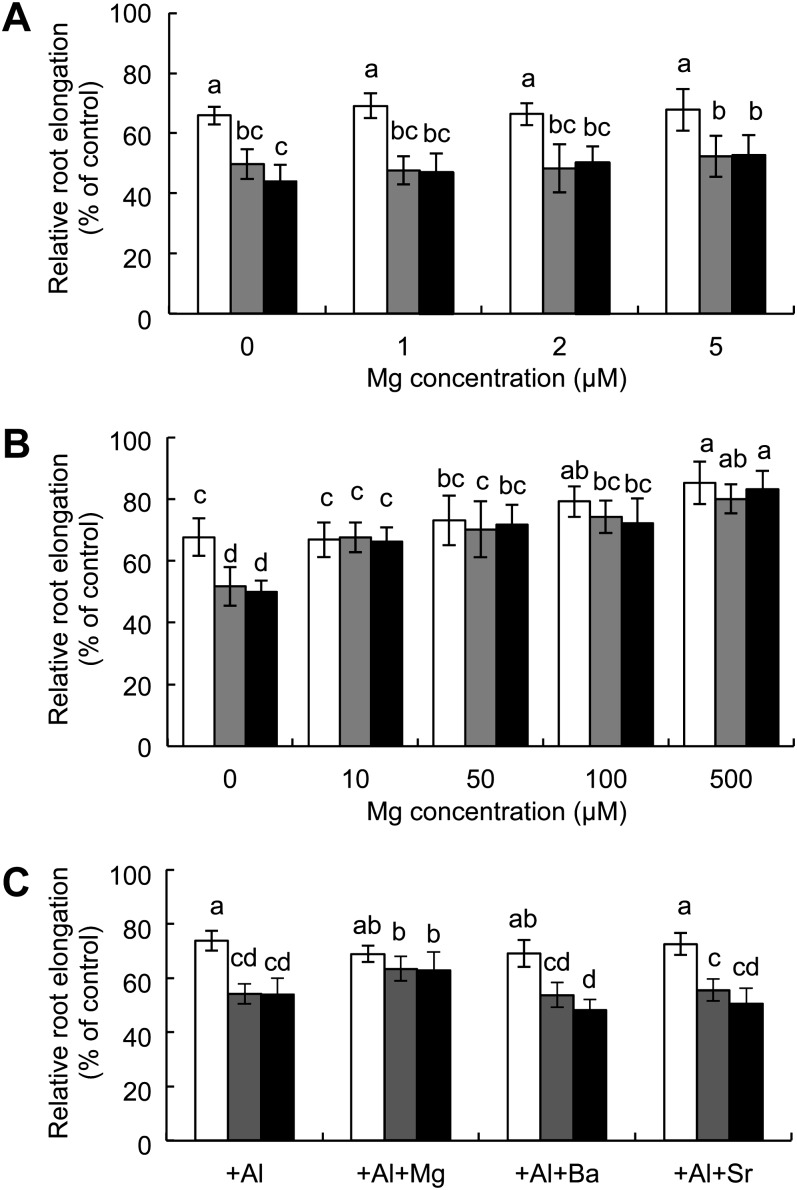
To investigate whether Al-induced inhibition of root elongation in the knockout lines can be alleviated by Mg, we exposed the seedlings to a solution containing a range of Mg concentrations in the presence of 50 μm Al. At low Mg concentrations (1–5 μm), addition of Mg did not alleviate Al-induced inhibition of root elongation in both wild type and two knockout lines (Fig. 5A). However, at 10 μm and higher Mg concentrations, the root elongation in the knockout lines reached to the same level of wild type (Fig. 5B). In the wild type, Mg concentrations below 50 μm did not improve the Al-inhibited root elongation, but at Mg concentrations higher than 100 μm, the Al-inhibited growth was also alleviated (Fig. 5B). This alleviative effect of Mg at higher concentrations in both wild type and knockout lines could be attributed to displacement or competition effect, which prevents binding of Al to the cell components.
Figure 5.
Alleviation of Mg on Al toxicity in OsMGT1 knockout lines. A and B, Alleviative effect on Al toxicity by low (A) and high concentrations (B) of Mg. Seedlings of wild-type and two OsMGT1 knockout lines were exposed to a solution containing 50 μm Al in the presence of different concentrations of Mg for 24 h. The root length was measured before and after the treatment and elongation relative to the root growth without Al was shown. Data are means ± sd (n = 10). C, Alleviation of Al toxicity in OsMGT1 knockout lines by divalent metals. Seedlings of wild-type and knockout lines were exposed to a solution containing 50 μm Al with 10 μm of Mg, Ba, or Sr for 24 h. Data are means ± sd (n = 10). Means with different letters are significantly different (P < 0.05 by Tukey’s test).
To test whether this alleviation is specific to Mg, we compared Mg with two other divalent cations, barium (Ba) and strontium (Sr). In the presence of the same concentration (10 μm), unlike Mg, neither Sr nor Ba showed alleviative effect on Al tolerance (Fig. 5C).
Knockout of OsMGT1 Did Not Affect Al-Induced Citrate Secretion
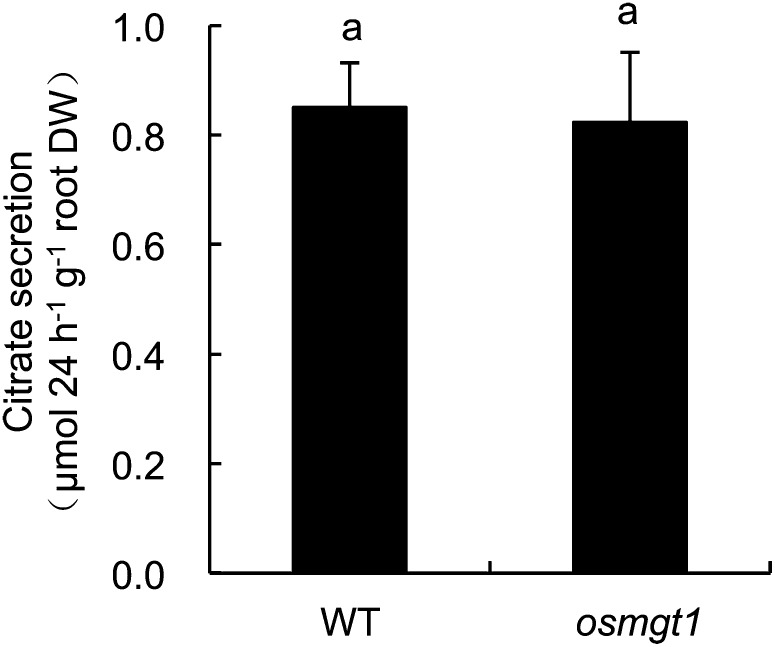
Rice secretes citrate from the roots in response to Al (Ma et al., 2002; Yokosho et al., 2011). Since Mg-mediated alleviation of Al toxicity was reported to be attributed to enhanced organic acid secretion from the roots in soybean and rice bean (Silva et al., 2001b; Yang et al., 2007), we compared the citrate secretion between wild type and the knockout line (NF0595). There was no difference in Al-induced secretion of citrate between wild type and the knockout line (Fig. 6). This result indicates that different from other plant species, the Mg-mediated alleviation in the knockout line does not result from difference in organic acid secretion.
Figure 6.
Al-induced citrate secretion in OsMGT1 knockout line. Seedlings (21-d-old) of wild-type rice (WT) and a knockout line (NF0595) were exposed to a solution containing 50 μm Al. Root exudates were collected for 24 h after Al treatment. Citrate was determined by an enzymatic method. Data are means ± sd (n = 3). DW, Dry weight. Means with different letters are significantly different (P < 0.05 by Tukey’s test).
Al Enhanced Mg Uptake in Wild Type, But Not in OsMGT1 Knockout Lines
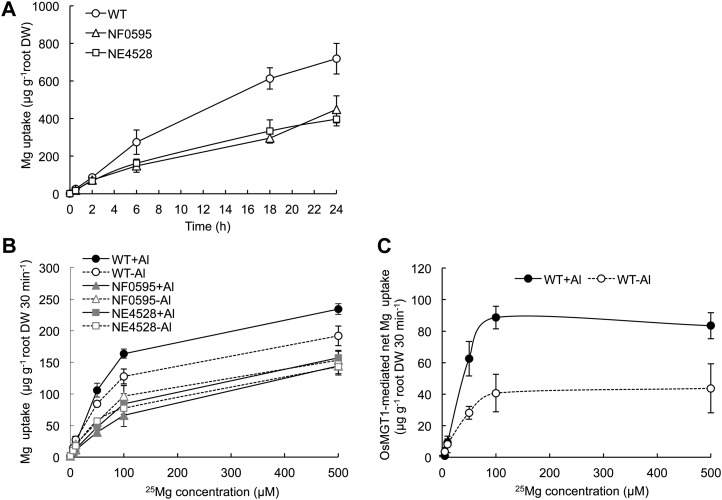
To investigate whether OsMGT1 is involved in Mg uptake or not, we performed uptake experiments using stable isotope 25Mg. A time-course experiment showed that wild type showed a higher 25Mg uptake than two knockout lines in the absence of Al (Fig. 7A), indicating the involvement of OsMGT1 in Mg uptake in the roots. A kinetic experiment with short term (30 min) showed that the Mg uptake was increased by 6 h Al pretreatment in wild type, but unaffected in the knockout lines (Fig. 7B). OsMGT1-mediated Mg uptake was calculated by subtracting Mg uptake of knockout lines from that of wild type (Fig. 7C). The Km value was estimated to be 30.5 and 30.4 μm Mg, respectively, in plants exposed to Al and not. However, the Vmax was double in the Al-treated plants than in non-Al-treated plants (84 μg g−1 root dry weight 30 min−1 versus 41 μg g−1 root dry weight 30 min−1; Fig. 7C).
Figure 7.
25Mg uptake in OsMGT1 knockout lines. A, A time-dependent uptake of 25Mg. Seedlings of wild-type and knockout lines were exposed to a solution containing 10 μm 25Mg. At different time points, the roots and shoots were sampled for determination of 25Mg with ICP-MS. B, Effect of Al on 25Mg uptake. Seedlings of wild-type and knockout lines were pretreated with or without Al (0 or 50 μm, pH 4.5) for 6 h and subsequently subjected to an uptake solution containing different concentrations of 25Mg. After 30 min, the roots were sampled for determination of 25Mg. Data are means ± sd (n = 3). C, OsMGT1-mediated Mg uptake. The net uptake was calculated by subtracting Mg uptake of knockout lines from that of wild-type rice.
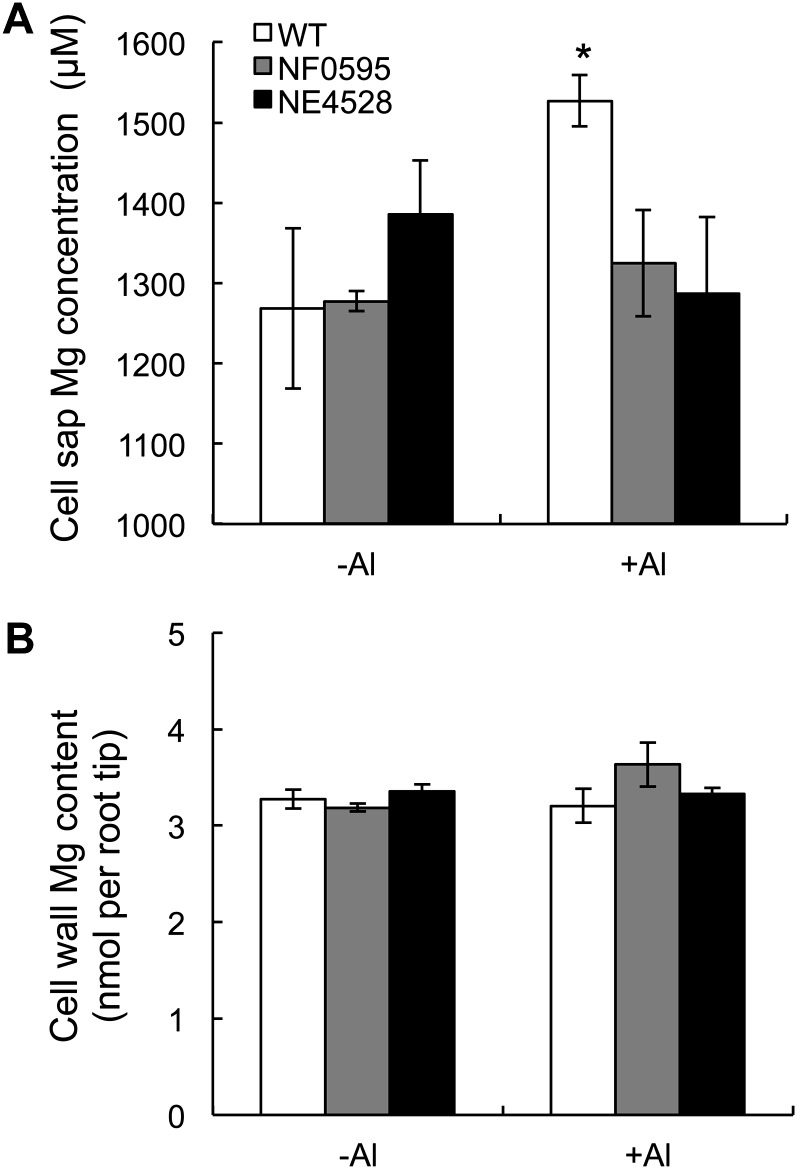
The Mg concentration in the cell sap and cell wall of root tips (0–1 cm) was also compared. In the absence of Al, the Mg concentration in the cell sap was similar between wild type and knockout lines (Fig. 8A). However, in the presence of Al, the wild type showed a higher Mg concentration in root cell sap than the knockout lines (Fig. 8A). By contrast, there was no difference in the cell wall Mg content between different lines (Fig. 8B).
Figure 8.
Effect of Al on Mg concentration in the cell sap and cell wall. Mg concentration in the cell sap (A) and cell wall (B) of root tips (0–1 cm). Seedlings of both wild-type rice (WT) and two knockout lines were exposed to a solution containing Al (0 or 50 μm) for 8 h. The root tips (0–1 cm) were excised and fractionated into cell sap and cell wall. The Mg concentration was determined by ICP-MS. Data are means ± sd (n = 3). The asterisk shows a significant difference (P < 0.05 by Tukey’s test).
Transcriptomic Analysis of OsMGT1 Knockout Line
A comparative analysis of DNA microarray in Al-tolerant and Al-sensitive soybean genotypes revealed that Mg enhanced Al tolerance in the Al-tolerant genotype by down-regulating genes commonly induced in response to Al toxicity (Duressa et al., 2010). To examine the gene expression changes caused by knockout of OsMGT1, we carried out a genome-wide transcriptome analysis by comparing wild type and OsMGT1 knockout line exposed to Al for 6 h using the rice 44 K oligo microarray. A total of 64 genes were down-regulated (≥2 fold) including OsMGT1 and 132 genes were up-regulated (≥2 fold) in the knockout line (Supplemental Table S1 and S2). Among down-regulated genes, 38% of them are unknown genes, 21% of them are related to other metabolism, and 19% are stress-related genes (Supplemental Fig. S3). On the other hand, 34% of up-regulated genes are unknown genes, 16% are related to other metabolism, and 28% are stress related (Supplemental Fig. S3). These genes are supposed to be induced by either Al toxicity or Mg deficiency. For example, five down-regulated (Os01g0847600, Os03g0235000, Os05g0111300, Os06g0695300, and Os08g0302000) and four up-regulated (Os01g0370900, Os01g0374000, Os07g0638300, and Os09g0367700) genes are involved in oxidative stress and reactive oxygen species (ROS) scavenging (Soranzo et al., 2004; Hu et al., 2011; Turóczy et al., 2011). Al is known to induce peroxidation and ROS formation in rice roots (Meriga et al., 2004; Sharma and Dubey, 2007). Mg deficiency also can induce ROS generation in many plant species including rice (Cakmak and Kirkby, 2008; Chou et al., 2011). One gene (Os09g0416500) encoding double-strandedRNA binding protein and one gene (Os05g0270500) encoding RNase P-related protein were, respectively, down-regulated and up-regulated in the knockout line. The RNase activity and its double-strandedRNA binding activity are Mg dependent (Smith and Pace, 1993; Cerritelli and Crouch, 1995; Beebe et al., 1996). Furthermore, the Mg concentration seems critical for switching between these two activities (Cerritelli and Crouch, 1995).
DISCUSSION
OsMGT1 Is a Transporter for Mg Uptake
Mg transporters (CorA) were first identified in bacteria in a screen for cobalt resistance (Moomaw and Maguire, 2008). In the genome of Arabidopsis, there are 10 CorA homologs (Schock et al., 2000; Li et al., 2001; Gebert et al., 2009). Except for pseudogene AtMGT8, all members show transport activity for Mg when expressed in yeast (Gebert et al., 2009). However, they have different spatial and temporal transcript expression pattern, tissue, and subcellular localizations (Waters, 2011). Among them, only AtMGT1, AtMGT7, and AtMGT9 show higher expression in the roots, implicating these genes in root Mg uptake. AtMGT1 was localized to the plasma membrane (Li et al., 2001), but its function in planta has not been characterized. Knockout of AtMGT7 resulted in a growth defect on low-Mg nutrient solution (Gebert et al., 2009). However, AtMGT7 was localized to the endoplasmatic reticulum, raising a question on its implication in uptake. AtMGT9 was highly expressed in the mature anthers, leaves, and young roots (Chen et al., 2009). Disruption of this gene resulted in abortion of mature pollen grains (Chen et al., 2009), but the role of AtMGT9 in root Mg uptake is unknown. Therefore, transporters responsible for Mg uptake in roots are still not well understood.
In rice genome, there are nine CorA homologs (Gebert et al., 2009). However, none of them has been functionally characterized. In this study, we found that OsMGT1 is a transporter for Mg uptake in rice based on following evidence. First, OsMGT1 is a plasma-membrane-localized transporter (Fig. 3). Second, knockout of OsMGT1 resulted in decreased Mg uptake (Fig. 7). Although the tissue localization of OsMGT1 was not examined in this study, since it is regulated by ART1, OsMGT1 should be localized at all root cells as ART1 and other proteins encoded by ART1-regulated downstream genes including STAR1/2, Nrat1, OsALS1, and OsFRDL4 (Huang et al., 2009, 2012; Xia et al., 2010; Yokosho et al., 2011).
Based on Km value (Fig. 7), OsMGT1 is likely a high-affinity transporter for Mg. In Arabidopsis, AtMGT1 and AtMGT10 show high affinity, AtMGT9 and AtMGT7 show low affinity, and AtMGT5 has dual affinity (Li et al., 2001, 2008; Drummond et al., 2006; Mao et al., 2008; Chen et al., 2009). However, these classifications are based on transport experiment in yeast and/or bacteria. It is unknown whether they behavior similarly in planta. The Km value for whole-root Mg uptake was estimated to be 20 μm (Tanoi et al., 2011), while that for OsMGT1 was 30 μm (Fig. 7). Considering that the Mg concentration in most soil solution is 20 to 200 μm (Epstein, 1972), OsMGT1 probably plays a major role in Mg uptake in rice.
Up-Regulation of OsMGT1 Expression Is Required for Conferring Al Tolerance in Rice
Knockout of OsMGT1 resulted in increased sensitivity to Al in both solution and soil culture (Fig. 4, A and C), but did not affect the sensitivity to other metals (Fig. 4B). The Al-induced inhibition of root elongation in the knockout lines could be rescued by addition of 10 μm Mg, but not by other divalent cation analogs including Sr and Ba (Fig. 5). The expression of OsMTG1 in the roots was rapidly and specifically enhanced by Al (Fig. 2), resulting in increased Mg uptake and Mg concentration in the root cell sap (Figs. 7 and 8). All these findings indicate that up-regulation of OsMGT1 is required for conferring Al tolerance in rice.
Al is rapidly taken up into the root cells (Lazof et al., 1994). Recently it was demonstrated that Al uptake is mediated by Nrat1, a Nramp transporter family member in rice (Xia et al., 2010; Famoso et al., 2011). Although Al is finally sequestrated into the vacuoles by OsALS1 at the tonoplast (Huang et al., 2012), increased Mg concentration in the cytosol seems to be also required for preventing Al binding to enzymes and other cellular components because of the strong affinity of A1 for oxygen donor compounds such as inorganic phosphate, ATP, RNA, DNA, proteins, carboxylic acids, and phospholipids (Martin, 1988). This is supported by our microarray data: More stress-related genes are up- and down-regulated in the OsMGT1 knockout line (Supplemental Fig. S3). A rapid Al-induced up-regulation of OsMGT1 contributes to increased Mg concentration in the cytosol (Fig. 8A), thereby detoxifying Al.
Al inhibits Mg transport mediated by ALR1 and ALR2 in yeast (MacDiarmid and Gardner, 1998). Al is a potent inhibitor of both AtMGT1 and AtMGT10 activity expressed in bacteria (Li et al., 2001). The Mg uptake in Lolium multiflorum and sorghum were also inhibited by Al (Rengel and Robinson, 1989; Tan et al., 1991). Rice seems to have developed a distinct strategy to overcome Al-induced inhibition of Mg uptake by up-regulating OsMGT1. In fact, based on published microarray data, all members of AtMGT gene family in Arabidopsis (Supplemental Fig. S2A, including AtMGT1, AtMGT2, AtMGT3, AtMGT4, AtMGT5, AtMGT6, AtMGT7, AtMGT9, AtMGT10, except for pseudogene AtMGT8) are not up-regulated by Al (Hoekenga et al., 2003; Sawaki et al., 2009; Zhao et al., 2009). This may partly explain why rice is highly tolerant of Al toxicity.
As a conclusion, OsMGT1 is plasma-membrane-localized transporter for Mg in rice and up-regulation of this transporter gene is required for conferring Al tolerance by increasing Mg uptake into the cells. Up-regulation of OsMGT1 is a component of high Al tolerance in rice.
MATERIALS AND METHODS
Phylogenetic Analysis
Alignment was performed with Clustal W using default setting (http://www.genome.jp/tools/clustalw/), and the phylogenetic tree was constructed using the neighbor-joining algorithm with 1,000 bootstrap trials by MEGA software.
Plant Materials and Growth Conditions
Two Tos-17 insertion lines of rice (Oryza sativa), NF0595 and NE4528 for OsMGT1, were obtained from the Rice Genome Resource Center in Japan. The homozygous lines was screened by PCR using specific primers (5′-AACACGCATCTAAAAGTTTCACC-3′ and 5′-TTCGATTATTATTGCTCCCACA-3′) for NF0595 and specific primers (5′-TAGGGATGAGCTGGAGCACT-3′ and 5′-CATCAGTTGACCGAGAGCTG-3′) for NE4528, with a left-border Tos-17 primer (5′-ATTGTTAGGTTGCAAGTTAGTTAAGA-3′). Seeds of wild-type rice (cv Nipponbare) and two Tos-17 homozygous lines were soaked in deionized water at 30°C in the dark for 2 d, and then transferred to a net floating on a 0.5-mm CaCl2 solution in a 1.5-l plastic container. Seedlings were grown at 25°C before being used for various experiments. Al and Mg were applied as AlCl3·6H2O and MgCl2·6H2O, respectively. All experiments were repeated at least twice with three replicates each.
RNA Isolation, Cloning, and Gene Expression Analysis
Total RNA from rice roots was extracted using the RNeasy mini kit (Qiagen). One microgram of total RNA was used for first-strand cDNA synthesis using a SuperScript II kit (Invitrogen) following the manufacturer’s instructions. The cDNA fragment containing an entire OsMGT1 open reading frame was amplified by RT-PCR using the primers 5-GGTACCAAAATGGAGCGGAGGGCGCA-3 and 5-TCACTGCAGGATCTTGCTCTTCC-3. The fragment was cloned into the pGEM-T vector (Promega) for sequence confirmation using the ABI PRISM 310 genetic analyzer and the BigDye terminators v3.1 cycle sequencing kit (Applied Biosystems).
For expression analysis, seedlings of wild type were exposed to a solution containing 0 or 50 μm Al. At different time (0, 2, 4, or 6 h), the roots and shoots were sampled for RNA extraction and expression level determination. Seedlings exposed to different pH (4.5 or 5.6), or to Cd and La were also sampled. Root tips (0–1 cm) and basal roots (1–2 cm) were excised from the seedlings exposed to 0 or 50 μm Al for 6 h. Samples were immediately frozen in liquid nitrogen. RNA extraction and cDNA preparation were performed as described above. The gene expression level was determined by real-time RT-PCR using Thunderbird SYBR qPCR mix (TOYOBO) on Mastercycler ep realplex (Eppendorf). The primers used were 5′-GGCGCGTGCAGAAGATTAGGG-3′ and 5′-CGCGTATTCACGGATATGGTACAGGG-3′ for OsMGT1. Histone H3 (forward primer, 5′-AGTTTGGTCGCTCTCGATTTCG-3′; reverse primer, 5′-TCAACAAGTTGACCACGTCACG-3′) was used as an internal control. Normalized relative expression was calculated by the ΔΔCt method.
Subcellular Localization
The subcellular localization was investigated by introducing 35S:OsMGT1-GFP, 35S: GFP-OsMGT1, or GFP alone as a control into rice leaf protoplasts. The rice leaf protoplasts were prepared from 2-week-old rice (cv Nipponbare) seedlings grown hydroponically, and used for transformation by the polyethylene glycol method as described by Chen et al. (2006). The GFP signal was observed using an LSM700 laser-scanning microscope (Zeiss).
Evaluation of Al Tolerance
To compare Al sensitivity, 5-d-old seedlings of both wild-type rice and knockout lines were exposed to a 0.5 mm CaCl2 solution (pH 4.5) containing various AlCl3 concentrations (0, 10, 30, and 50 μm) for 24 h. Root length was measured by a ruler before and after the Al treatment, and relative root elongation (RRE; = [root elongation with Al]/[root elongation without Al]) was calculated.
To investigate the tolerance to metals, 5-d-old seedlings were exposed to a 0.5 mm CaCl2 solution (pH 4.5) containing 50 μm Al, 10 μm Cd, or 5 μm La for 24 h, and the root length was measured and RRE was calculated.
The alleviative effect of Mg on Al toxicity was investigated by exposing wild type and knockout lines (5-d-old) to a 0.5 mm solution (pH 4.5) containing 0 or 50 μm Al in the presence of different Mg concentrations (0, 1, 2, 5, 10, 50, 100, and 500 μm), Ba (10 μm), or Sr (10 μm) for 24 h. The root length was measured and RRE was calculated.
To evaluate growth on acid soil, germinated seeds were sowed on acidic soil (Andsol, pH 4.5) or neutral soil (pH 6.5). After 1-week growth, the plants were removed from soil and photographed.
Root Cell Sap Collection and Mg Determination
Five-day-old seedlings of both wild-type rice and OsMGT1 knockout lines were exposed to a 0.5 mm CaCl2 solution containing 50 μm Al (pH 4.5) for 8 h. After the treatment, the root segments (0–1 cm) were excised and cell sap was extracted according to Xia et al. (2010). Briefly, the root segments (15 roots each per sample) were washed three times with 0.5 mm CaCl2 and then put in an Ultra free-MC centrifugal filter unit (Millipore) and centrifuged at 3,000g for 10 min at 4°C. After frozen at −80°C overnight, the samples were thawed at room temperature for 30 min and the root cell sap solution was obtained by centrifuging at 20,600g for 10 min. The residual cell wall was washed with 70% ethanol three times and then immersed in 0.5 mL of 2 n HCl for at least 24 h with occasional vortex. The Mg concentration in the symplastic solution and cell wall extracts were determined by inductively coupled plasma-mass spectrometry (ICP-MS) using an Agilent 7700 mass spectrometer (http://www.agilent.com).
Collection of Root Exudates and Organic Acid Determination
To collect root exudates, the seedlings (7-d-old) of both wild type and knockout line were grown in a one-half-strength Kimura B nutrient solution for 14 d as described in Yamaji and Ma (2007). Before collection, the seedlings were exposed to a 0.5 mm CaCl2 (pH 4.5) solution overnight and then transferred to a 0.5 mm CaCl2 (pH 4.5) solution containing 50 μm Al. After 24 h, the root exudates were collected and passed through a cation-exchange column (16 × 14 mm) filled with 5 g Amberlite IR-120B resin (H+ form), followed by an anion-exchange column (16 × 14 mm) filled with 2 g AG 1 × 8 resin (100–200 mesh, formate form). Organic acid anions retained on the anion-exchange resin were eluted by 2 n HCl, and the eluate was concentrated to dryness at 40°C using a rotary evaporator. After the residue was redissolved in 1 mL of milli-Q water, the concentration of organic acids was analyzed by enzymatic method according to Delhaize et al. (1993).
25Mg Uptake Experiment
A time-course experiment of 25Mg uptake was conducted by exposing 7-d-old seedlings to a 0.5 mm CaCl2 solution (pH 4.5) labeled with 25Mg at 10 μm. 25Mg (99%) was purchased from Nippon Sansho. At different time points (0, 0.5, 2, 6, 18, and 24 h), the roots and shoots were sampled. The roots were washed with 10 mm iced CaCl2 solution for three times.
To compare the effect of Al on Mg uptake, a short-term uptake experiment was conducted. Seven-day-old rice seedlings were first exposed to a 0.5 mm CaCl2 solution (pH 4.5) containing 0 or 50 μm Al for 6 h to induce OsMGT1 expression and subsequently subjected to a solution containing different Mg concentrations ranging from 0 to 500 μm 25Mg. After 30 min, the roots were sampled as described above.
The samples were dried in 70°C oven for 2 d. After digested with concentrated HNO3, the concentration of 24Mg and 25Mg was determined by ICP-MS with isotope mode on an Agilent 7700 mass spectrometer (http://www.agilent.com).
Microarray Analysis
Seedlings (5-d-old) of both the wild-type rice and osmgt1 knockout line (NE4528) were exposed to a 0.5 mm CaCl2 solution containing 50 μm Al. After 6 h, the roots were sampled for RNA isolation. Total RNA was extracted using the RNeasy plant mini kit (Qiagen). Microarray analysis was performed using the rice oligo DNA microarray 44 K RAP-DB (Agilent Technologies) with three biological replicates. The hybridized slides were scanned using a DNA microarray scanner (Agilent Technologies). Signal intensities were extracted by feature extraction software (Agilent Technologies). For data analysis, genes with signal intensities below 100 (average of three biological replicates in wild type) were excluded and genes up- or down-regulated by 2-fold were selected. Genes were categorized into nine classes according to the OryzaExpress database (http://bioinf.mind.meiji.ac.jp/OryzaExpress/).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AB731703.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Gene structure (A), predicated membrane domains (B), and mRNA expression of Tos-17 lines of OsMGT1 examined by RT-PCR.
Supplemental Figure S2. Phylogenetic tree of OsMGT1 (A) and alignment of OsMGT1 with Arabidopsis homologs (B).
Supplemental Figure S3. Functional classification of down- or up-regulated genes in OsMGT1 knockout line.
Supplemental Table S1. Down-regulated genes in the OsMGT1 knockout line.
Supplemental Table S2. Up-regulated genes in the OsMGT1 knockout line.
Supplementary Material
Acknowledgments
We thank Rice Genome Resource Center in Tsukuba for providing Tos-17 insertion lines.
Glossary
- Mg
magnesium
- Al
aluminum
- RT
reverse transcription
- Cd
cadmium
- La
lanthanum
- Ba
barium
- Sr
strontium
- ROS
reactive oxygen species
- RRE
relative root elongation
References
- Beebe JA, Kurz JC, Fierke CA. (1996) Magnesium ions are required by Bacillus subtilis ribonuclease P RNA for both binding and cleaving precursor tRNAAsp. Biochemistry 35: 10493–10505 [DOI] [PubMed] [Google Scholar]
- Bose J, Babourina O, Rengel Z. (2011) Role of magnesium in alleviation of aluminium toxicity in plants. J Exp Bot 62: 2251–2264 [DOI] [PubMed] [Google Scholar]
- Cakmak I, Kirkby EA. (2008) Role of magnesium in carbon partitioning and alleviating photooxidative damage. Physiol Plant 133: 692–704 [DOI] [PubMed] [Google Scholar]
- Cerritelli SM, Crouch RJ. (1995) The non-RNase H domain of Saccharomyces cerevisiae RNase H1 binds double-stranded RNA: magnesium modulates the switch between double-stranded RNA binding and RNase H activity. RNA 1: 246–259 [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li LG, Liu ZH, Yuan YJ, Guo LL, Mao DD, Tian LF, Chen LB, Luan S, Li DP. (2009) Magnesium transporter AtMGT9 is essential for pollen development in Arabidopsis. Cell Res 19: 887–898 [DOI] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Chou TS, Chao YY, Huang WD, Hong CY, Kao CH. (2011) Effect of magnesium deficiency on antioxidant status and cadmium toxicity in rice seedlings. J Plant Physiol 168: 1021–1030 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Ma JF, Ryan PR. (2012) Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci 17: 341–348 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Randall PJ. (1993) Aluminum tolerance in wheat (Triticum aestivum L.): II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol 103: 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Luo K, Li D, Zheng X, Wei X, Smith W, Thammina C, Lu L, Li Y, Pei Y. (2006) Overexpression of an Arabidopsis magnesium transport gene, AtMGT1, in Nicotiana benthamiana confers Al tolerance. J Exp Bot 57: 4235–4243 [DOI] [PubMed] [Google Scholar]
- Drummond RSM, Tutone A, Li YC, Gardner RC. (2006) A putative magnesium transporter AtMRS2-11 is localized to the plant chloroplast envelope membrane system. Plant Sci 170: 78–89 [Google Scholar]
- Duressa D, Soliman KM, Chen D. (2010) Mechanisms of magnesium amelioration of aluminum toxicity in soybean at the gene expression level. Genome 53: 787–797 [DOI] [PubMed] [Google Scholar]
- Epstein E. (1972) Mineral Nutrition of Plants: Principles and Perspectives. John Wiley and Sons, New York, pp 128–131.
- Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, Bustamante C, Kochian LV, McCouch SR. (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet 7: e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert M, Meschenmoser K, Svidová S, Weghuber J, Schweyen R, Eifler K, Lenz H, Weyand K, Knoop V. (2009) A root-expressed magnesium transporter of the MRS2/MGT gene family in Arabidopsis thaliana allows for growth in low-Mg2+ environments. Plant Cell 21: 4018–4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauer UE, Horst WJ. (1992) Modeling cation amelioration of aluminum phytotoxicity. Soil Sci Soc Am J 56: 166–172 [Google Scholar]
- Hoekenga OA, Vision TJ, Shaff JE, Monforte AJ, Lee GP, Howell SH, Kochian LV. (2003) Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta × Columbia) by quantitative trait locus mapping: a physiologically simple but genetically complex trait. Plant Physiol 132: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Liang W, Yin C, Cui X, Zong J, Wang X, Hu J, Zhang D. (2011) Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CF, Yamaji N, Chen Z, Ma JF. (2012) A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J 69: 857–867 [DOI] [PubMed] [Google Scholar]
- Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, Ma JF. (2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21: 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinraide TB. (2003) Toxicity factors in acidic forest soils: attempts to evaluate separately the toxic effects of excessive Al3+ and H+ and insufficient Ca2+ and Mg2+ upon root elongation. Eur J Soil Sci 54: 323–333 [Google Scholar]
- Kinraide TB, Pedler J, Parker DR. (2004) Relative effectiveness of calcium and magnesium in the alleviation of rhizotoxicity in wheat induced by copper, zinc, aluminum, sodium, and low pH. Plant Soil 259: 201–208 [Google Scholar]
- Knoop V, Groth-Malonek M, Gebert M, Eifler K, Weyand K. (2005) Transport of magnesium and other divalent cations: evolution of the 2-TM-GxN proteins in the MIT superfamily. Mol Genet Genomics 274: 205–216 [DOI] [PubMed] [Google Scholar]
- Kochian LV, Hoekenga OA, Pineros MA. (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55: 459–493 [DOI] [PubMed] [Google Scholar]
- Lazof DB, Goldsmith JG, Rufty TW, Linton RW. (1994) Rapid uptake of aluminum into cells of intact soybean root tips: a microanalytical study using secondary ion mass spectrometry. Plant Physiol 106: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Tutone AF, Drummond RS, Gardner RC, Luan S. (2001) A novel family of magnesium transport genes in Arabidopsis. Plant Cell 13: 2761–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LG, Sokolov LN, Yang YH, Li DP, Ting J, Pandy GK, Luan S. (2008) A mitochondrial magnesium transporter functions in Arabidopsis pollen development. Mol Plant 1: 675–685 [DOI] [PubMed] [Google Scholar]
- Ma JF. (2000) Role of organic acids in detoxification of aluminum in higher plants. Plant Cell Physiol 41: 383–390 [DOI] [PubMed] [Google Scholar]
- Ma JF. (2007) Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int Rev Cytol 264: 225–252 [DOI] [PubMed] [Google Scholar]
- Ma JF, Shen R, Zhao Z, Wissuwa M, Takeuchi Y, Ebitani T, Yano M. (2002) Response of rice to Al stress and identification of quantitative trait loci for Al tolerance. Plant Cell Physiol 43: 652–659 [DOI] [PubMed] [Google Scholar]
- MacDiarmid CW, Gardner RC. (1998) Overexpression of the Saccharomyces cerevisiae magnesium transport system confers resistance to aluminum ion. J Biol Chem 273: 1727–1732 [DOI] [PubMed] [Google Scholar]
- Mao DD, Tian LF, Li LG, Chen J, Deng PY, Li DP, Luan S. (2008) AtMGT7: an Arabidopsis gene encoding a low-affinity magnesium transporter. J Integr Plant Biol 50: 1530–1538 [DOI] [PubMed] [Google Scholar]
- Martin RB. (1988) Bioinorganic chemistry of aluminum. In H Sigel, A Sigel, eds, Metal Ions in Biological Systems, Vol 24. Marcel Dekker, New York, pp 1–57.
- Meriga B, Reddy BK, Rao KR, Reddy LA, Kishor PB. (2004) Aluminium-induced production of oxygen radicals, lipid peroxidation and DNA damage in seedlings of rice (Oryza sativa). J Plant Physiol 161: 63–68 [DOI] [PubMed] [Google Scholar]
- Moomaw AS, Maguire ME. (2008) The unique nature of mg2+ channels. Physiology (Bethesda) 23: 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble AD, Sumner ME. (1988) Calcium and Al interactions and soybean growth in nutrient solutions. Commun Soil Sci Plant Anal 19: 1119–1131 [Google Scholar]
- Rengel Z, Robinson DL. (1989) Aluminum effects on growth and macronutrient uptake in annual rye grass. Agron J 81: 208–215 [Google Scholar]
- Ryan PR, Kinraide TB, Kochian LV. (1994) Al3+-Ca2+ interactions in aluminum rhizotoxicity. Planta 192: 98–102 [Google Scholar]
- Ryan PR, Reid RJ, Smith FA. (1997) Direct evaluation of the Ca2+-displacement hypothesis for Al toxicity. Plant Physiol 113: 1351–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaki Y, Iuchi S, Kobayashi Y, Kobayashi Y, Ikka T, Sakurai N, Fujita M, Shinozaki K, Shibata D, Kobayashi M, et al. (2009) STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol 150: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schock I, Gregan J, Steinhauser S, Schweyen R, Brennicke A, Knoop V. (2000) A member of a novel Arabidopsis thaliana gene family of candidate Mg2+ ion transporters complements a yeast mitochondrial group II intron-splicing mutant. Plant J 24: 489–501 [DOI] [PubMed] [Google Scholar]
- Sharma P, Dubey RS. (2007) Involvement of oxidative stress and role of antioxidative defense system in growing rice seedlings exposed to toxic concentrations of aluminum. Plant Cell Rep 26: 2027–2038 [DOI] [PubMed] [Google Scholar]
- Silva IR, Smyth TJ, Carter TE, Rufty TW. (2001a) Altered aluminum root elongation inhibition in soybean genotypes in the presence of magnesium. Plant Soil 230: 223–230 [Google Scholar]
- Silva IR, Smyth TJ, Israel DW, Raper CD, Rufty TW. (2001b) Magnesium ameliorates aluminum rhizotoxicity in soybean by increasing citric acid production and exudation by roots. Plant Cell Physiol 42: 546–554 [DOI] [PubMed] [Google Scholar]
- Smith D, Pace NR. (1993) Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry 32: 5273–5281 [DOI] [PubMed] [Google Scholar]
- Soranzo N, Sari Gorla M, Mizzi L, De Toma G, Frova C. (2004) Organisation and structural evolution of the rice glutathione S-transferase gene family. Mol Genet Genomics 271: 511–521 [DOI] [PubMed] [Google Scholar]
- Tan K, Keltjens WG, Findenegg GR. (1991) Role of magnesium in combination with liming in alleviating acid-soil stress with the aluminium-sensitive sorghum genotype CV323. Plant Soil 136: 65–71 [Google Scholar]
- Tan K, Keltjens WG, Findenegg GR. (1992) Aluminium toxicity with sorghum genotypes in nutrient solutions and its amelioration by magnesium. J Plant Nutr Soil Sci 155: 81–86 [Google Scholar]
- Tanoi K, Saito T, Iwata N, Kobayashi NI, Nakanishi TM. (2011) The analysis of magnesium transport system from external solution to xylem in rice root. Soil Sci Plant Nutr 57: 265–271 [Google Scholar]
- Tsutsui T, Yamaji N, Feng Ma J. (2011) Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol 156: 925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turóczy Z, Kis P, Török K, Cserháti M, Lendvai A, Dudits D, Horváth GV. (2011) Overproduction of a rice aldo-keto reductase increases oxidative and heat stress tolerance by malondialdehyde and methylglyoxal detoxification. Plant Mol Biol 75: 399–412 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Okada K. (2005) Interactive effects of Al, Ca and other cations on root elongation of rice cultivars under low pH. Ann Bot (Lond) 95: 379–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM. (2011) Moving magnesium in plant cells. New Phytol 190: 510–513 [DOI] [PubMed] [Google Scholar]
- Xia JX, Yamaji N, Kasai T, Ma JF. (2010) Plasma membrane-localized transporter for aluminum in rice. Proc Natl Acad Sci USA 107: 18381–18385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Huang CF, Nagao S, Yano M, Sato Y, Nagamura Y, Ma JF. (2009) A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21: 3339–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Ma JF. (2007) Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol 143: 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JL, You JF, Li YY, Wu P, Zheng SJ. (2007) Magnesium enhances aluminum-induced citrate secretion in rice bean roots (Vigna umbellata) by restoring plasma membrane H+-ATPase activity. Plant Cell Physiol 48: 66–73 [DOI] [PubMed] [Google Scholar]
- Yokosho K, Yamaji N, Ma JF. (2011) An Al-inducible MATE gene is involved in external detoxification of Al in rice. Plant J 68: 1061–1069 [DOI] [PubMed] [Google Scholar]
- Zhao CR, Ikka T, Sawaki Y, Kobayashi Y, Suzuki Y, Hibino T, Sato S, Sakurai N, Shibata D, Koyama H. (2009) Comparative transcriptomic characterization of aluminum, sodium chloride, cadmium and copper rhizotoxicities in Arabidopsis thaliana. BMC Plant Biol 9: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.