Abstract
The formation of root nodules and arbuscular mycorrhizal (AM) roots is controlled by a common signaling pathway including the calcium/calmodulin-dependent kinase Doesn’t Make Infection3 (DMI3). While nodule initiation by lipochitooligosaccharide (LCO) Nod factors is well characterized, diffusible AM fungal signals were only recently identified as sulfated and nonsulfated LCOs. Irrespective of different outcomes, the perception of symbiotic LCOs in Medicago truncatula is mediated by the LysM receptor kinase M. truncatula Nod factor perception (MtNFP). To shed light on transcriptional responses toward symbiotic LCOs and their dependence on MtNFP and Ca2+ signaling, we performed genome-wide expression studies of wild-type, Nod-factor-perception mutant1, and dmi3 mutant roots challenged with Myc- and Nod-LCOs. We show that Myc-LCOs lead to transient, quick responses in the wild type, whereas Nod-LCOs require prolonged incubation for maximal expression activation. While Nod-LCOs are most efficient for an induction of persistent transcriptional changes, sulfated Myc-LCOs are less active, and nonsulfated Myc-LCOs display the lowest capacity to activate and sustain expression. Although all symbiotic LCOs up-regulated a common set of genes, discrete subsets were induced by individual LCOs, suggesting common and specific functions for these in presymbiotic signaling. Surprisingly, even sulfated fungal Myc-LCOs and Sinorhizobium meliloti Nod-LCOs, having very similar structures, each elicited discrete subsets of genes, while a mixture of both Myc-LCOs activated responses deviating from those induced by single treatments. Focusing on the precontact phase, we identified signaling-related and transcription factor genes specifically up-regulated by Myc-LCOs. Comparative gene expression studies in symbiotic mutants demonstrated that transcriptional reprogramming by AM fungal LCOs strictly depends on MtNFP and largely requires MtDMI3.
Second to grasses, legumes are the plant family of highest economic importance (Graham and Vance, 2003). Due to their high protein content in seed and leaf tissues, legume plants provide both valuable food and feed sources as well as green fertilizers. A remarkable feature of legumes is their ability to enter two symbiotic interactions with soil microbes, leading to the development of nitrogen-fixing root nodules (Brewin, 1991) and the establishment of arbuscular mycorrhizal (AM) roots (Harrison, 1999).
Root nodule development is initiated by a molecular dialogue: flavonoids secreted by legumes induce rhizobial genes controlling the synthesis of lipochitooligosaccharide (LCO) Nod factors (Lerouge et al., 1990). Subsequently, these signal molecules are perceived by LysM (for Lys motif)-domain receptor kinase heterodimers (Radutoiu et al., 2003) located in the root hair membrane (Haney et al., 2011). LysM heterodimers consist of a Lyr- and a Lyk-type protein, designated Medicago truncatula Nod factor perception (MtNFP; Arrighi et al., 2006; Bensmihen et al., 2011) and M. truncatula LysM receptor kinase3 (MtLYK3) in M. truncatula (Smit et al., 2007). One of the first epidermal responses to Nod factor perception is an enhanced intracellular Ca2+ level, followed by oscillations of the Ca2+ concentration in and around the nucleus (Oldroyd and Downie, 2006). In M. truncatula, this Ca2+ spiking is passed on via the products of three symbiosis-related genes: Doesn’t Make Infection1 (MtDMI1), MtDMI2, and MtDMI3 (Catoira et al., 2000). Whereas MtDMI2 functions as a Leu-rich repeat receptor kinase (Stracke et al., 2002), MtDMI1 encodes a potassium-permeable cation channel of the nuclear membrane that compensates charge imbalances caused by Ca2+ spiking (Charpentier et al., 2008). At the terminus of the calcium signaling cascade, MtDMI3 functions as a Ca2+/calmodulin-dependent kinase that deciphers nuclear Ca2+ signatures (Gleason et al., 2006) to activate downstream responses via the two GRAS transcription factors Nodulation-signaling pathway1 (MtNSP1; Smit et al., 2005) and MtNSP2 (Kaló et al., 2005). After Nod factor perception, infection threads originating from curled root hairs guide rhizobia toward the inner cortex and into the growing nodule (Oldroyd and Downie, 2008), where they differentiate to intracellular, nitrogen-fixing bacteroids.
Based on a similar molecular dialogue that is only partially understood, legumes enter AM symbioses with soil fungi of the order Glomeromycota (Schüssler et al., 2001). This interaction is regarded as the most widespread terrestrial symbiosis, because almost all land plants establish an arbuscular mycorrhiza (Smith and Read, 2008). To initiate the interaction, host plants secrete strigolactones that promote hyphal branching and stimulate fungal metabolism (Akiyama et al., 2005; Besserer et al., 2006). In return, fungal recognition is thought to require Myc factors that, similar to the action of Nod factors initiating nodulation, mediate the communication of roots with AM fungi (Gough and Cullimore, 2011). This is supported by the observation that a treatment of roots with nonpurified Myc signals from spore exudates or extraradical hyphae triggers early symbiotic responses (Kosuta et al., 2003; Weidmann et al., 2004; Oláh et al., 2005; Navazio et al., 2007; Kuhn et al., 2010; Chabaud et al., 2011). In addition to the DMI genes, studies of legume mutants defective in early symbiotic responses revealed further components of common symbiotic signaling (Gough and Cullimore, 2011), such as nucleoporins (Kistner et al., 2005) and the DMI3 interactor Interacting protein of DMI3 (Yano et al., 2008). The existence of a common symbiotic pathway (CSP) led to the hypothesis that components initially established for the ancient AM symbiosis (Remy et al., 1994) were adopted during the evolution of nodulation (Parniske, 2008). Interestingly, not all responses to fungal signals are dependent on the CSP, suggesting the presence of additional signaling pathways during AM fungal recognition and infection (Bonfante and Requena, 2011).
Although the perception of diffusible Myc signals results in epidermal Ca2+ spiking as well (Gough and Cullimore, 2011), Nod-LCOs and supernatants from germinating AM fungal spores evoke distinct signatures (Bonfante and Requena, 2011), probably to prepare host roots for the different symbiotic interactions (Kosuta et al., 2008). Subsequent to hyphopodia formation on the root surface, fungal invasion is arranged by the host plant via a prepenetration apparatus that guides hyphae from the epidermis to the inner cortex (Genre et al., 2005, 2008), a process accompanied by specific gene expression changes (Siciliano et al., 2007). The coordinated responses of root cells indicate that, in addition to diffusible Myc signals acting at a distance (Oláh et al., 2005; Kuhn et al., 2010), there are others that require fungal contact (e.g. effector proteins; Kloppholz et al., 2011). Intraradical hyphal development terminates in highly branched structures designated arbuscules, being regarded as the site of nutrient exchange between the partners (Smith and Read, 2008). Fully colonized AM roots were extensively studied by expression profiling, primarily leading to an identification of several hundred genes up-regulated during later AM stages (Liu et al., 2003, 2007; Küster et al., 2004; Manthey et al., 2004; Hohnjec et al., 2005; Guether et al., 2009; Hogekamp et al., 2011; Gaude et al., 2012).
While it was reported by Navazio et al. (2007) that diffusible Myc signals are smaller than 3 kD and partially lipophilic, their structure was unknown until such molecules were reported by Maillet et al. (2011) to be a mix of simple sulfated and nonsulfated LCOs, referred to as sMyc- and nsMyc-LCOs here. Strikingly, in particular sMyc-LCOs are closely related to Sinorhizobium meliloti Nod factors, although an O-acetate substitution at the nonreducing end is missing and the N-linked acyl chains are different (Maillet et al., 2011). Irrespective of different symbiotic outcomes, the perception of Nod- and Myc-LCOs in M. truncatula is initially mediated by the MtNFP receptor and is subsequently controlled by the CSP including MtDMI3 (Maillet et al., 2011). Due to a long availability of purified Nod-LCOs, signal exchange between rhizobia and legumes is well understood and insights into transcriptional responses toward Nod-LCOs were obtained (Mitra et al., 2004; Combier et al., 2008). In contrast, the unavailability of purified Myc factors was a major obstacle to the study of presymbiotic gene expression activation. To shed light on the differential transcriptional responses toward symbiotic LCOs and their dependence on MtNFP and the CSP, we performed genome-wide transcriptome studies of wild-type, Nod-factor-perception mutant1 (nfp-1), and dmi3 mutant roots challenged with Nod-, sMyc-, and nsMyc-LCOs. Our genome-wide expression snapshots revealed (1) cellular functions commonly or specifically activated by Nod- and Myc-LCOs, (2) differences in the perception of individual Myc-LCOs as well as their combination, and (3) novel signaling-related and transcription factor genes activated by Myc-LCOs in the wild type. Finally, a comparative study using key symbiotic mutants demonstrated that transcriptional reprogramming by Myc-LCOs strictly depends on the MtNFP receptor and to a large extent requires MtDMI3, indicating that responses to Myc-LCOs are primarily transduced via the CSP.
RESULTS
Setting Up a Bioassay to Record Transcriptional Responses toward Myc- and Nod-LCOs
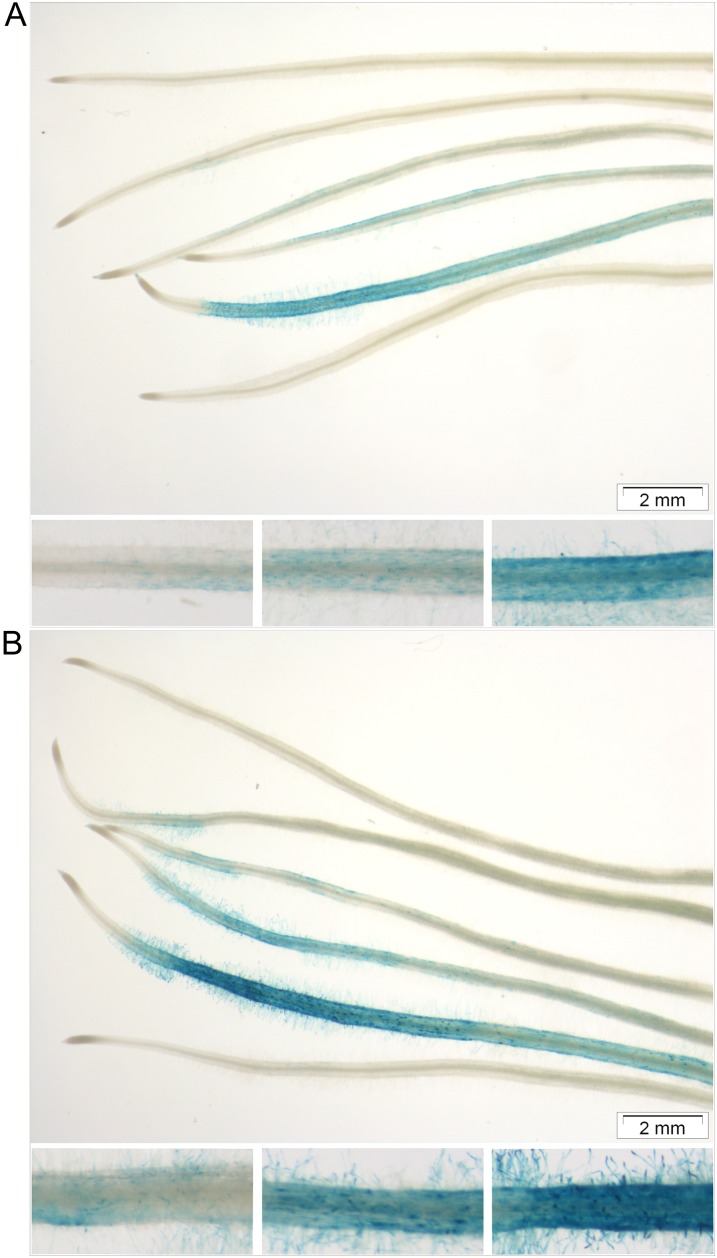
To study transcriptional responses toward microbial LCOs, we adapted a protocol to treat plantlet roots with diffusible microbial signals (Maillet et al., 2011; Supplemental Fig. S1). In a pilot experiment, M. truncatula MtL416 plantlets expressing a pMtENOD11-GUS fusion (Journet et al., 2001) were used to validate our conditions, because the M. truncatula early nodulin11 (MtENOD11) gene is a marker for early responses toward symbiotic signals (Journet et al., 2001; Kosuta et al., 2003). In the case of sMyc-LCOs and Nod-LCOs, a concentration of 10−8 m was used, while for nsMyc-LCOs, 10−7 and 10−8 m were tested to account for the fact that sMyc-LCOs activate MtENOD11 better than nsMyc-LCOs (Maillet et al., 2011). All LCO treatments were carried out for 6 and 24 h. Subsequent GUS staining revealed a specific GUS activity that was strongest after 24 h (Fig. 1). Stained root cap cells were followed by a short unstained zone, until distinct GUS activity was primarily observed in root hairs and epidermal cells. Roots treated with Nod-LCOs showed the strongest reporter gene expression followed by roots treated with sMyc-LCOs, 10−7 m nsMyc-LCOs, and finally 10−8 m nsMyc-LCOs, where GUS activity was only faintly visible. Notably, not only the strength but also the proximal extent of GUS expression followed this order (Fig. 1). Apart from a characteristic GUS activity in the apex, where MtENOD11 is expressed constitutively (Journet et al., 2001), no GUS expression was evident in control roots.
Figure 1.
Myc- and Nod-LCO-mediated activation of the MtENOD11 promoter in wild-type roots. M. truncatula plantlets expressing a pMtENOD11-GUS fusion (Journet et al., 2001) were grown on slant agar plates for 5 d prior to a 6-h (A) or 24-h (B) incubation in Myc control, 10−8 m nsMyc-LCO, 10−7 m nsMyc-LCO, 10−8 m sMyc-LCO, 10−8 m Nod-LCO, and Nod control solution (arranged in this order from top to bottom). Subsequently, roots were assayed for GUS activity for 24 h. Closeups show approximately 2-mm root segments treated with 10−7 m nsMyc-LCOs (left), 10−8 m sMyc-LCOs (middle), and 10−8 m Nod LCOs (right) after a 6-h (A) or 24-h (B) incubation. Bars = 2 mm.
Gene Expression Activation by Myc- and Nod-LCOs Follows Different Kinetics
To record transcriptional responses toward symbiotic LCOs, we challenged M. truncatula wild-type roots with Myc- and Nod-LCOs. Although it is likely that a mixture of sMyc- and nsMyc-LCOs is secreted by AM fungi (Maillet et al., 2011), it is unknown if they act individually or in combination. To account for this, we performed treatments with 10−8 m sMyc-LCOs, 10−7 m nsMyc-LCOs, a mixture of both Myc-LCOs, 10−8 m Nod-LCOs, and control solutions for 6 and 24 h. A higher concentration of nsMyc-LCOs was used to maximize responses toward this less active molecule.
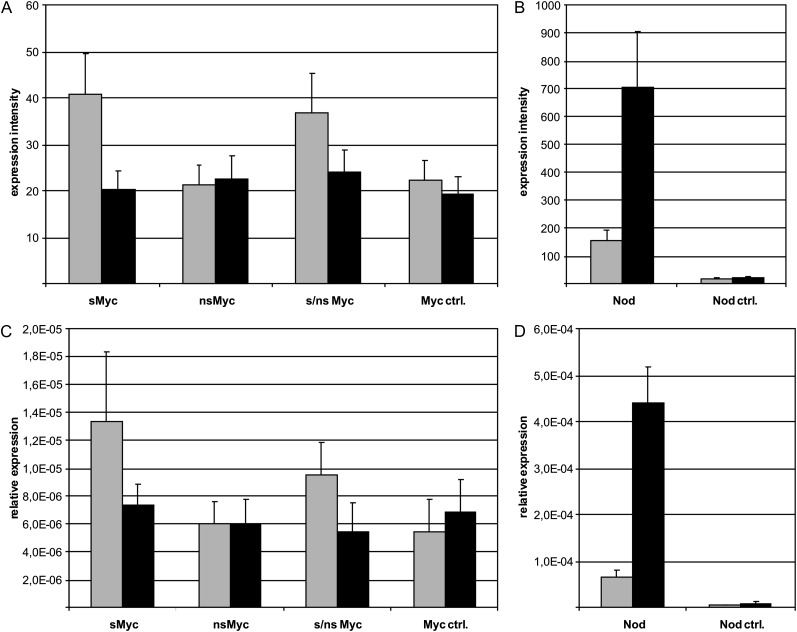
The technical reliability of our GeneChip-based transcriptome profiles (Supplemental Table S1) was assessed by validating the differential activation of MtENOD11 via real-time reverse transcription (RT)-PCR (Fig. 2). These experiments revealed a high concordance of MtENOD11 transcript levels measured by either approach. Figure 2 shows that after 6 h, only sMyc-LCOs, a mixture of both Myc-LCOs, and Nod-LCOs up-regulated MtENOD11, whereas nsMyc-LCOs did not. Only Nod-LCOs were activating MtENOD11 after 24 h, while no significant up-regulation toward Myc-LCOs was evident at this time point. Considering that the enhanced GUS activity detected after 24 h in our pilot experiment (Fig. 1) likely reflects a translation of stable GUS enzymes that does not necessarily require the full 24 h of GUS transcription (Jefferson et al., 1987), the MtENOD11 expression pattern corresponds to the observed promoter activity.
Figure 2.
MtENOD11 expression in wild-type roots challenged with Myc- and Nod-LCOs. Expression of MtENOD11 is shown in M. truncatula wild-type roots after 6 h (gray bars) and 24 h (black bars) of incubation with 10−8 m sMyc-LCOs (sMyc), 10−7 m nsMyc-LCOs (nsMyc), a mixture of both (s/nsMyc), Myc control solution (Myc ctrl.), 10−8 m Nod-LCOs (Nod), and Nod control solution (Nod ctrl.). A and B display results of Medicago GeneChip hybridizations (Supplemental Table S1), while C and D show the results of real-time RT-PCR measurements. Here, the mean expression values of three biological replicates per treatment are shown. Error bars represent se.
To make sure that our inoculation system is suitable to record transcriptional responses toward LCOs at a global scale, we validated our genome-wide expression profiles in wild-type roots via comparisons with genes known to be activated by Nod-LCOs. Using a spot-inoculation system different from our assay, Mitra et al. (2004) reported 33 M. truncatula genes as significantly induced at least 2-fold after 24 h by Nod-LCOs. Of these, 16 were induced in our conditions after 24 h as well (Supplemental Table S2). Moreover, we found significant up-regulation of several marker genes for Nod-LCO activity, including MtENOD11 (Supplemental Table S2), demonstrating the general suitability of our bioassay.
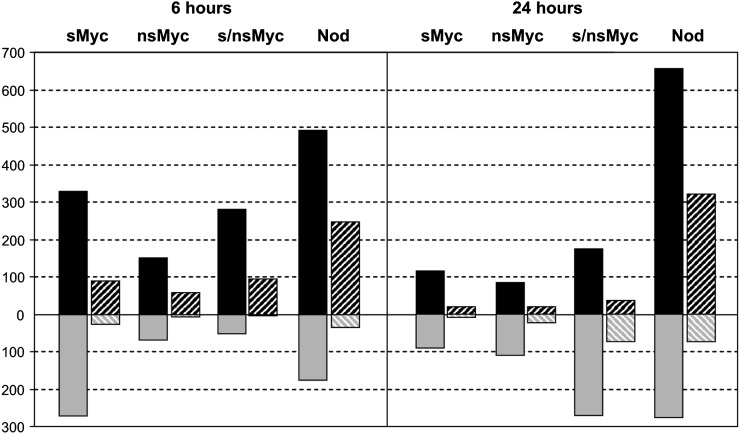
The subsequent visualization of transcriptional changes induced by Myc- and Nod-LCOs via ratio plots (Supplemental Fig. S2) revealed differences in the number of genes modulated in expression. In Figure 3, the number of significantly (P < 0.05) regulated genes with an at least 1.5- or 2-fold change in expression is shown. In general, the overall number of activated genes was considerably lower after any of the three Myc-LCO treatments in comparison with an application of Nod-LCOs. In detail, we found 329, 151, and 279 genes up-regulated at least 1.5-fold after 6 h of treatment with sMyc-LCOs, nsMyc-LCOs, and a mixture of both Myc-LCOs, respectively, while 491 genes were induced by Nod-LCOs (Supplemental Table S3). An induction of substantially more genes by Nod-LCOs was observed after 24 h as well, and together, the general expression patterns described were also recorded if only genes activated at least 2-fold were considered (Fig. 3). Interestingly, transcriptional activation by Myc-LCOs was strongest after 6 h, while expression induction by Nod-LCOs continued to increase for up to 24 h (Fig. 3). It is furthermore noteworthy that the number of significantly induced genes after a 6- or 24-h treatment with sMyc-LCOs was markedly higher as compared with nsMyc-LCOs, irrespective of their 10-fold higher concentration. Transcriptional responses toward Myc- and Nod-LCOs thus operate on different time scales, characterized by a transient, quick response in the case of Myc-LCOs while Nod-LCOs require prolonged incubation for maximal gene expression activation.
Figure 3.
Differential gene expression in wild-type roots challenged with Myc- and Nod-LCOs. This diagram shows the number of genes significantly up-regulated (presented in black) and down-regulated (presented in gray) in M. truncatula wild-type roots after 6 h (left side) or 24 h (right side) of incubation with 10−8 m sMyc-LCOs (sMyc-LCO), 10−7 m nsMyc-LCOs (nsMyc-LCO), a mixture of both (s/nsMyc-LCO), and 10−8 m Nod-LCOs (Nod-LCO). In each case, expression ratios were calculated in comparison with control solutions. The number of genes significantly up- or down-regulated at least 1.5-fold (black bars) or 2-fold (striped bars) at P < 0.05 is plotted.
Myc- and Nod-LCOs Induce Both Common and Specific Transcriptional Responses
We compared transcriptional responses of M. truncatula wild-type roots toward different LCOs in both time points via Venn (Oliveros, 2007) diagrams (Fig. 4), using all genes with an at least 1.5-fold activation (P < 0.05) as a basis. This low cutoff was chosen to account for the fact that, as observed for MtENOD11 (Fig. 1), responses toward symbiotic LCOs likely occur in the epidermis, leading to a quenching of expression ratios by other cell types abundantly present in the root regions isolated.
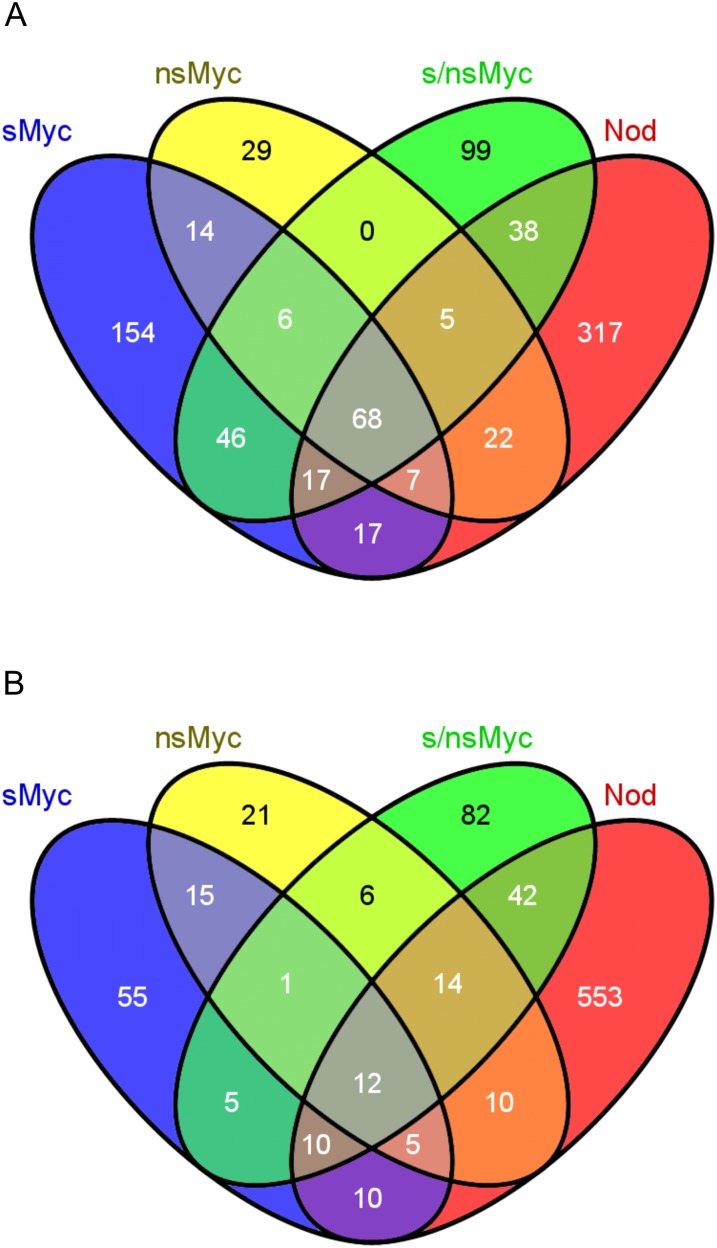
Figure 4.
Identification of genes coactivated by Myc- and Nod-LCOs in wild-type roots. The Venn diagrams visualize the coactivation of gene expression in M. truncatula wild-type roots treated with 10−8 m sMyc-LCOs (sMyc), 10−7 m nsMyc-LCOs (nsMyc), a mixture of both Myc-LCOs (s/nsMyc), and 10−8 m Nod-LCOs (Nod) for 6 h (A) and 24 h (B). Numbers indicate the number of genes significantly up-regulated at least 1.5-fold at P < 0.05.
Figure 4 shows that 68 and 12 genes were collectively up-regulated after 6 and 24 h by all four symbiotic LCOs, irrespective of their relation to AM formation or nodulation. Regardless of this overlap, distinct sets of genes were activated by specific LCOs, based on the cutoffs mentioned above. Surprisingly, different responses are elicited by sMyc-LCOs and Nod-LCOs, although these have strong similarities including a sulfate at the nonreducing end. This suggests that fungal and bacterial signals, in spite of being structurally related, are recognized as different.
When analyzing the 6-h treatment with sMyc-LCOs, nsMyc-LCOs, and Nod-LCOs, a total of 154, 29, and 317 specifically activated genes were identified, respectively (Fig. 4A; Supplemental Table S4). Looking into the 24-h time point, these numbers change to 55, 21, and 553 (Fig. 4B; Supplemental Table S4), reflecting the transience in Myc-LCO-mediated gene expression and the longer persistence of transcriptional changes caused by Nod-LCOs. Interestingly, a treatment with mixed Myc-LCOs resulted in the activation of 137 and 124 genes after 6 and 24 h (including genes activated by Nod-LCOs; Fig. 4B). This observation suggests that all 249 and 103 genes only activated at the cutoffs used by individual Myc-LCOs after 6 and 24 h are suppressed when applying the Myc-LCO mixture, indicating a different efficiency or specificity in the perception of individual and combined Myc-LCOs. The existence of substantial sets of genes requiring both Myc-LCOs for induction might reflect the fact that such a mixture is likely secreted by AM fungi in soils (Maillet et al., 2011).
Signaling-Related M. truncatula Genes Are Commonly Activated by Myc- and Nod-LCOs
For a detailed analysis of LCO-induced transcription in wild-type roots, we had a closer look at those genes commonly up-regulated at least 1.5-fold (P < 0.05) after 6 and 24 h of incubation with different LCOs. Strikingly, more than 20% of these genes are related to secondary metabolism (Supplemental Table S4). One of the largest groups of plant secondary metabolites is isoprenoids (Withers and Keasling, 2007). Precursors for their synthesis are derived from the methylerythritol phosphate pathway, where the MtDXS2 gene encoding a 1-deoxy-d-xylulose-5-phosphate synthase plays a role (Floss et al., 2008). MtDXS2 is involved in later AM stages, where a knockdown of the corresponding gene resulted in an accumulation of dead and degenerated arbuscules (Floss et al., 2008). In our experiment, MtDXS2 was significantly up-regulated by all four microbial LCO fractions after 6 h (Supplemental Table S4), suggesting that the encoded enzyme already acts in presymbiotic stages.
While genes encoding the Nod factor receptor kinases MtNFP and MtLYK3 as well as components of the CSP did not show a marked expression change toward symbiotic LCOs, the MtNSP1 and MtNSP2 genes encoding GRAS transcription factors acting downstream of Ca2+ signaling turned out to be slightly activated by Nod-LCOs (Supplemental Table S5). With respect to signaling and transcriptional regulation, three additional genes known to be related to symbiotic signaling downstream of MtDMI3 were identified (Supplemental Table S4). First, MtVapyrin was commonly induced by all four microbial LCOs at both time points. This gene was originally identified as essential for arbuscule formation as well as for efficient epidermal penetration by AM fungi (Pumplin et al., 2010). Interestingly, it was recently shown that MtVapyrin is not only relevant for AM formation but also for rhizobial colonization (Murray et al., 2011). Apart from MtVapyrin, a gene encoding the E3 ubiquitin ligase M. truncatula plant U-box protein1 (MtPUB1) and the transcription factor gene M. truncatula ERF required for nodulation1 (MtERN1) were commonly induced. Both MtERN1 and MtPUB1 were so far only connected to signal transduction leading to nodulation. MtPUB1 is a negative regulator of infection that interacts with MtLYK3 and plays a role in discriminating Nod factor variants (Mbengue et al., 2010), while MtERN1 acts as a transcriptional activator downstream of MtDMI3 (Middleton et al., 2007). The fact that these three genes known to be important for symbiotic interactions were identified here as commonly induced by diffusible Myc- and Nod-LCOs supports the idea that an external application of symbiotic LCOs does initiate some common reprogramming events in epidermal and cortical cells prior to the colonization by both AM fungi and rhizobia. This is in line with the recent observation that similar Ca2+ spiking patterns are characteristic of cortical cells about to be colonized both by infection threads and by AM fungal hyphae (Sieberer et al., 2012).
In relation to signal transduction that prepares root cells for infection by symbiotic microbes, seven commonly induced genes encoding protein kinases, a mitogen-activated protein kinase, a CAAT box-binding transcription factor, and a bZIP transcription factor deserve attention (Supplemental Table S4). Together with the signaling-related genes mentioned above, these are candidates for novel components that mediate the reprogramming of root cells toward an accommodation of rhizobia or AM fungi.
Myc-LCOs Induce a Specific Expression of Signaling-Related and Transcription Factor Genes
Of prime interest in our study was the identification of genes specifically up-regulated by treatment with the recently identified Myc-LCOs (Maillet et al., 2011). We thus had a closer look at the results of the 6-h treatment of wild-type roots, because here the ability of Myc-LCOs to activate transcription was maximal (Figs. 2 and 3). From a total of 348 genes induced at least 1.5-fold (P < 0.05) by Myc-LCOs and not by Nod-LCOs after 6 h, subsets of 154, 29, and 99 genes were activated by sMyc-LCOs, nsMyc-LCOs, and a mixture of both Myc-LCOs, respectively, while the remaining 66 genes were induced by different Myc-LCO combinations (Fig. 4A; Supplemental Table S4). This observation supports the idea that both individual Myc-LCOs and their mixtures play a role in Myc signaling.
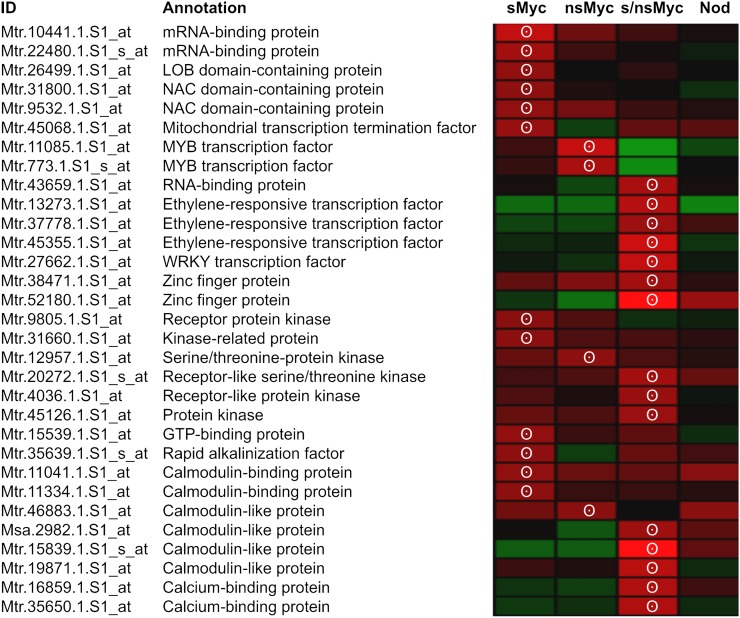
In spite of differences in Myc-LCO-induced expression profiles, all three Myc-LCO treatments activated a substantial number of genes related to signaling and transcriptional regulation (Supplemental Table S4). A summary of 31 novel signaling-related genes only activated during presymbiotic Myc signaling is shown in Figure 5. From this set, 15 genes encoded two mRNA-binding proteins, one WRKY transcription factor, two zinc finger proteins, three NAC or LOB domain-containing proteins, two MYB transcription factors, as well as three ERF transcription factors, most of them belonging to plant-specific classes (Riechmann and Ratcliffe, 2000; Udvardi et al., 2007). Interestingly, NAC domain transcription factors are involved in lateral root formation in Arabidopsis (Arabidopsis thaliana; Zhao et al., 2010), and an M. truncatula homolog of the AtNAC1 transcription factor related to lateral root formation was shown to be involved in nodulation (D’haeseleer et al., 2011). MYB transcription factors have so far not been associated with early AM stages, but a member of this group was identified to be specifically activated in arbuscule-containing cells (Gomez et al., 2009). Finally, ERF transcription factors are known to respond to abscisic acid and ethylene signals, which characterize responses toward biotic and abiotic stress (Riechmann and Ratcliffe, 2000). In M. truncatula, the three ERF transcription factors MtERN1, MtERN2, and MtERN3 activate MtENOD11 in response to Nod-LCOs (Andriankaja et al., 2007), suggesting that other ERF family members may be involved in Myc-LCO signaling as well.
Figure 5.
Signaling-related genes specifically activated by Myc-LCOs in wild-type roots. A summary is shown of the subset of genes encoding transcription factors or signal-related proteins being rapidly activated in M. truncatula wild-type roots after a 6-h treatment with 10−8 m sMyc-LCOs (sMyc), 10−7 m nsMyc-LCOs (nsMyc), or a mixture of both (s/nsMyc). For comparison, gene expression in response to a 6-h 10−8 m Nod-LCO (Nod) treatment is shown. Gene expression is represented as a heat map, with shades of red representing up-regulation and shades of green indicating down-regulation. An at least 1.5-fold (P < 0.05) up-regulation is indicated by white circles. Expression data were scaled to the maximum of a 2.4-fold regulation using Genesis software (Sturn et al., 2002).
Apart from Myc-LCO-induced genes encoding transcriptional regulators, Figure 5 shows signaling-related genes representing novel components of presymbiotic Myc signal transduction. In addition to six kinase genes, eight genes specifying calmodulin-like proteins, calmodulin-binding proteins, and Ca2+-binding proteins were identified. While protein kinases might play a role in symbiotic signaling downstream of LysM receptor kinases including MtNFP (Maillet et al., 2011), the group of Ca2+- or calmodulin-related proteins can be connected with the generation, preservation, or modulation of Ca2+-spiking responses toward diffusible Myc-LCOs (Gough and Cullimore, 2011).
Myc-LCOs Activate Some Infection-Related Genes But No Genes Connected to Arbuscule Function
While signal perception during the precontact phase of symbiotic plant-microbe interactions deserves attention, it should not be overlooked that signaling also governs later stages of the symbiosis (Harrison, 2005), where specific gene expression is triggered during fungal infection (Weidmann et al., 2004; Sanchez et al., 2005; Siciliano et al., 2007) and in cortical cells containing fungal hyphae or developing arbuscules (Liu et al., 2003; Guether et al., 2009; Hogekamp et al., 2011; Gaude et al., 2012). Considering the requirement of Nod-LCO signaling for proper rhizobial infection, for synchronizing nodule formation with bacterial invasion, and for symbiosome formation (Limpens et al., 2005; Den Herder et al., 2007; Ovchinnikova et al., 2011), it is possible that Myc-LCOs act as signals during fungal colonization as well. A few studies investigated gene expression during early AM fungal infection, identifying 10 (Sanchez et al., 2005), 11 (Weidmann et al., 2004), and 13 (Siciliano et al., 2007) mostly DMI3-dependent genes up-regulated at 5 d post inoculation, where hyphopodia and prepenetration apparatus structures are present, but none of these genes was induced by diffusible Myc-LCOs at our cutoffs (data not shown).
In the absence of more comprehensive, genome-wide expression data from early stages of AM formation, we assessed to what extent genes known to be up-regulated in fully colonized roots are transcriptionally activated by LCOs as well, using a comparison of genes only induced by Myc-LCOs after 6 and 24 h in the wild type with the core set of 512 genes coactivated at 28 d post inoculation with two AM fungi (Hogekamp et al., 2011). This analysis revealed an overlap of 49 genes being induced both by Myc-LCOs and during fungal colonization of AM roots (Supplemental Fig. S3; Supplemental Table S5). Among those were the MtDXS2 (Floss et al., 2008), MtERN1 (Middleton et al., 2007), MtERN2 (Andriankaja et al., 2007), MtPUB1 (Mbengue et al., 2010), and MtVapyrin (Murray et al., 2011) genes either connected to AM-related secondary metabolism or to symbiotic signaling and early infection. These observations suggest that a limited subset of infection-related genes are coactivated by Myc-LCOs and in fully colonized roots, possibly by constant challenges with fungal signals during the continuous reinfection of roots that also occurs during later stages.
From the 463 genes not activated by Myc-LCOs, 15 are known to be exclusively expressed in the arbuscule-containing cells (Supplemental Table S5), such as the phosphate transporter gene M. truncatula phosphate transporter4 (Javot et al., 2007). None of these genes, being strictly related to functional AM stages, was activated by Myc-LCOs. It thus appears that the function of Myc-LCO signals is indeed restricted to the presymbiotic AM stages and that they (at least when applied at the concentrations used) do not activate genes related to late stages (Supplemental Fig. S3). This is in line with Drissner et al. (2007), who reported lysophosphatidylcholine but not LCOs as a trigger for an arbuscule-specific phosphate transporter gene.
Transcriptional Activation by Myc-LCOs Depends on Key Components of Symbiotic Signaling
The expression of 348 genes up-regulated at least 1.5-fold (P < 0.05) by Myc-LCOs and not by Nod-LCOs (Fig. 4) was assessed in nfp-1 (Ben Amor et al., 2003) and dmi3 (Marsh et al., 2007) mutants. These comparisons should reveal if, in addition to an MtNFP dependence of Myc-LCO-mediated root-branching stimulation (Maillet et al., 2011), also the activation of Myc-LCO-specific genes depends on this LysM domain receptor kinase located at the upstream position of symbiotic signaling. At the downstream terminus of the CSP, analyses of Myc-LCO-dependent gene expression in a dmi3 mutant should reveal to what extent Myc-LCO-mediated transcription depends on symbiotic Ca2+ signaling.
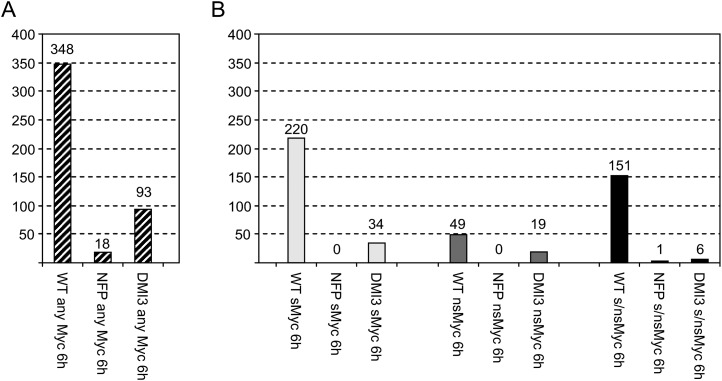
To obtain expression profiles in the two mutants, studies were performed as described above, using RNA of nfp-1 and dmi3 plantlet roots treated for 6 h with 10−8 m sMyc-LCOs, 10−7 m nsMyc-LCOs, a mixture of both Myc-LCOs, and control solutions (Supplemental Table S6). The observed dependency of Myc-LCO-specific gene expression on MtNFP and MtDMI3 is visualized in Figure 6. It is evident that on a general level that does not differentiate between individual Myc-LCO treatments (Fig. 6A), only 18 Myc-LCO-specific genes induced in the wild type are still activated in nfp-1, equivalent to an approximately 95% reduction. A marked effect was also observed for dmi3, where 93 Myc-LCO-specific genes remained activated at the cutoffs mentioned above, leading to an approximately 73% suppression of Myc-LCO-induced transcription. Interestingly, from the 31 Myc-LCO-specific signaling-related genes (Fig. 5), 24 are MtNFP dependent and 28 are MtDMI3 dependent (Supplemental Table S6), suggesting a function in signal transduction downstream of the CSP.
Figure 6.
Myc-LCO-mediated gene expression in the wild type and in symbiotic mutants. The diagrams show the number of genes (Supplemental Table S6) at least 1.5-fold (P < 0.05) induced in M. truncatula wild-type roots after 6 h of incubation with any Myc-LCO (WT column in A) or sMyc-LCOs (sMyc), nsMyc-LCOs (nsMyc), and a mixture of both Myc-LCOs (s/nsMyc; WT columns in B). Note that none of these genes is activated by Nod-LCOs at the cutoffs mentioned. The number of genes induced by Myc-LCOs in the wild type that are still activated in MtNFP and MtDMI3 mutants are shown on the right side of the wild-type columns. Values in B do not add up to the values in A due to overlapping activation of transcription by different Myc-LCOs.
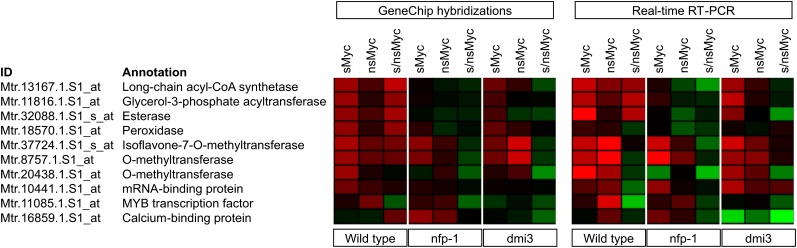
We validated our GeneChip-based expression profiles via real-time RT-PCR using a selection of 10 genes that included three related to signaling (Fig. 5) and that were significantly (P < 0.05) induced by at least one Myc-LCO treatment in the wild type while being activated to a different extent in the two symbiotic mutants. The heat maps shown in Figure 7 and an average correlation coefficient of r = 0.62 indicated that, as already observed for MtENOD11 (Fig. 2), the expression profiles obtained via the two methods mentioned above corresponded well.
Figure 7.
Verification of Myc-LCO-mediated gene expression in the M. truncatula wild type and two different symbiotic mutants via real-time RT-PCR. A summary is shown of a subset of genes differentially activated in M. truncatula wild-type, nfp-1, and dmi3 roots after a 6-h treatment with 10−8 m sMyc-LCOs (sMyc), 10−7 m nsMyc-LCOs (nsMyc), or a mixture of both (s/nsMyc). Log2-transformed gene expression ratios are represented as heat maps, with shades of red representing up-regulation and shades of green indicating down-regulation. Expression patterns obtained via GeneChip hybridizations (left side) correlate with expression profiles recorded by real-time RT-PCR (right side). Expression data were scaled to the maximum of a 4-fold regulation using Genesis software (Sturn et al., 2002).
To obtain results for individual Myc-LCOs or Myc-LCO combinations, we compared transcription between the wild type and the two mutants for each of the different treatments. These analyses revealed an almost absolute dependence of Myc-LCO-induced gene expression on MtNFP, regardless of the Myc-LCO challenge (Fig. 6B). Interestingly, the MtDMI3 dependence of Myc-LCO-induced gene expression was found to be particularly strong for genes activated by a mixture of sMyc- and nsMyc-LCOs, a condition closest to the natural situation in soils (Fig. 6B). Together, these observations not only corroborate the report of Maillet et al. (2011) that MtNFP is required for Myc-LCO-dependent root-branching stimulation but also demonstrate that precontact signaling via Myc-LCOs requires the calcium/calmodulin-dependent kinase MtDMI3 and hence a functional CSP.
To assess the specificity of Myc-LCO signaling downstream of MtDMI3, we analyzed the expression of the 10 Myc-LCO-induced genes shown in Figure 7 in nsp1-1 mutants by real-time RT-PCR. These studies revealed that five genes were still activated by either sMyc- or nsMyc-LCOs in nsp1-1 roots, indicating MtNSP1-independent expression (Supplemental Fig. S4). This result provides evidence that the Myc-LCO-specific transcription reported here (Figs. 4 and 5) is at least partially independent of MtNSP1, in line with the observation of Maillet et al. (2011) that root-branching stimulation by nsMyc-LCOs does not require this transcription factor. On the other hand, the existence of MtNSP1-dependent transcriptional responses indicates that gene expression activation by Myc-LCOs may either require or cross talk with MtNSP1-mediated signal transduction downstream of MtDMI3.
DISCUSSION
Based on a highly standardized and comparable experimental setup, we investigated gene expression in M. truncatula plantlet roots in response to diffusible LCO signals from rhizobial bacteria (Nod-LCOs) as well as AM fungi (Myc-LCOs) and assessed to what extent Myc-LCO-specific transcription depends on key components of symbiotic signaling.
Due to structural similarities between Nod- and Myc-LCOs, a certain amount of cross talk had to be expected for transcriptional activation by different LCOs. While such cross talk may have influenced the coactivation of expression by Myc- and Nod-LCOs (Fig. 4), none of the 384 Myc-LCO-specific genes (Figs. 5–7) were responding to Nod-LCOs, allowing us to conclude that the initial, MtNFP-mediated perception leads to the activation of a Myc-LCO-specific set of genes. Nevertheless, Myc-LCO-mediated transcriptional responses downstream of MtDMI3 partially required MtNSP1 (Supplemental Fig. S4), indicating some degree of cross talk during Nod- and Myc-LCO-mediated signaling downstream of symbiotic calcium spiking. Because it is unknown to what extent the gene expression changes measured in our study relate to the root-branching stimulation phenotypes studied by Maillet et al. (2011), an involvement of MtNSP1 in Myc-LCO-responsive signaling leading to other early symbiotic responses (e.g. the general preparation of root cells for microbial infection) is possible.
Looking at the specific transcriptional responses toward different Myc-LCOs (Fig. 4), it is an interesting question whether these sulfated and nonsulfated signals act together or independently. While on the one hand we observed that sMyc- and nsMyc-LCOs activate discrete sets of genes, suggesting distinct roles in AM signaling, it can be argued that under natural conditions a mixture of both Myc-LCOs is likely secreted by fungal symbionts (Maillet et al., 2011). Consequently, results of single treatments might only partially reflect the responses toward Myc-LCOs that occur in soils. This is supported by the observation that the expression responses of roots treated with both Myc-LCOs differ from single treatments, including the fact that the MtDMI3 dependence was strongest for genes activated by a combination of both Myc-LCOs (Fig. 6). A note of caution relates to the concentration of Myc-LCOs. For our experiments, we used 10−8 m sMyc-, while 10−7 m was chosen for nsMyc-LCOs. This range of Myc-LCO concentrations mediated root-branching stimulation via the Myc signaling pathway independent of MtNSP1 (Maillet et al., 2011) and was also used to record transcriptional responses toward Nod-LCOs (Mitra et al., 2004; Combier et al., 2008), although a much lower 10−13 to 10−10 m range is sufficient to activate cellular LCO responses (Gough and Cullimore, 2011). Apart from their concentration, sMyc- and nsMyc-LCO ratios probably play a crucial role as well, because Myc-LCO ratios slightly different from the ones we used here have been reported (Maillet et al., 2011). Hence, it is currently unknown if the ratio of these compounds is static or variable under natural conditions and if other ratios that might be related to host range (Gough and Cullimore, 2011) would have caused different responses.
To gain insights into the symbiotic properties of Myc-LCOs, we studied gene expression activation via these molecules in two key symbiotic M. truncatula mutants (Fig. 6). The striking reduction of Myc-LCO-dependent gene expression in nfp-1 confirms that the LysM domain receptor kinase MtNFP is essential for transcriptional responses toward Myc-LCOs at the concentrations used here, in line with the observation of Maillet et al. (2011) that a knockout of this gene impairs Myc-LCO-dependent root-branching stimulation. In addition, our results demonstrate that the majority of expression responses toward a treatment with Myc-LCOs depend on MtDMI3, confirming that Myc-LCO-induced gene expression is primarily mediated via the CSP that transduces symbiotic Ca2+ signals. In this context, it has to be considered that both Myc- and Nod-LCOs are not only involved in early symbiotic signaling but are known to stimulate root branching via MtNFP and the CSP (Oláh et al., 2005; Maillet et al., 2011). Transcriptional responses toward symbiotic LCOs thus cannot exclusively be connected with presymbiotic events but with root-branching stimulation as well, as summarized in the model presented in Figure 8.
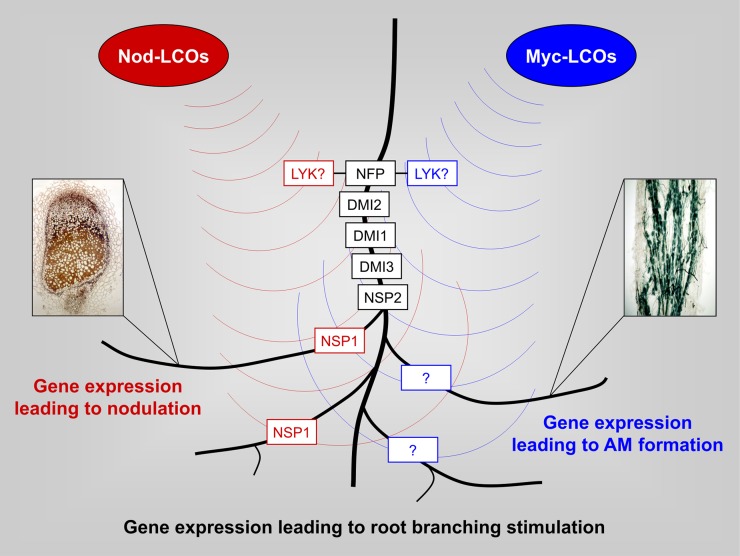
Figure 8.
Model for LCO-mediated gene expression leading to nodulation, AM formation, and root-branching stimulation. This model integrates transcriptional responses toward Myc-LCOs in MtNFP and MtDMI3 mutants (Fig. 6) with current knowledge of LCO-mediated signal transduction. In precontact signaling, the perception of diffusible Nod- and Myc-LCOs requires a CSP consisting of the Lyr1-type LysM domain receptor kinase MtNFP, the Leu-rich repeat receptor kinase MtDMI2, the cation channel MtDMI1, the calcium/calmodulin-dependent protein kinase MtDMI3, and the GRAS transcription factor MtNSP2 (Gough and Cullimore, 2011). While the GRAS transcription factor MtNSP1 is specifically required for nodulation (Smit et al., 2005), a corresponding protein related to AM formation has not yet been reported (indicated by a question mark). Currently unknown Lyk-type LysM domain receptor kinases (designated LYK?; Gough and Cullimore, 2011) form heterodimers with MtNFP during initial Nod- and Myc-LCO signal perception. In addition to their requirement during the presymbiotic phase of nodulation and AM formation, Myc- and Nod-LCOs stimulate root branching via the CSP (Maillet et al., 2011). Downstream of the CSP, different components are required for root-branching stimulation (Maillet et al., 2011).
The strict MtNFP dependence of Myc-LCO-induced gene expression raises the question of why AM fungi colonize nfp-1 roots at levels not significantly different from the wild type (Arrighi et al., 2006; F. Maillet, personal communication). For Nod factor signaling in M. truncatula, it was proposed that the two LysM receptor kinases MtNFP and MtLYK3 play distinct roles. Whereas MtNFP functions as a signaling receptor, which is able to bind Nod factors with high affinity but less stringency, allowing low Nod-LCO concentrations from soils to be perceived (Geurts et al., 2005), MtLYK3 represents an entry receptor with low affinity but high stringency, mediating root invasion by appropriate microsymbionts (Smit et al., 2007). Strong affinity is not required for the entry receptor, because local Nod-LCO concentrations near the curled root hair are high (Geurts et al., 2005).
Due to the evolutionary relationship, a similar situation is conceivable during hyphopodia formation, where fungal and host tissues are in close contact that allows local Myc-LCO accumulation. A hypothesis can be proposed where MtNFP in addition to Nod-LCO perception acts as a Myc-LCO receptor, supported by the strict dependence of Myc-LCO-induced gene expression shown here and the MtNFP-dependent root-branching stimulation by Myc-LCOs (Maillet et al., 2011). Interestingly, recent evidence shows that MtNFP is not just a specific Nod-LCO receptor. Whereas the presence of a sulfate substitution is essential for S. meliloti Nod-LCO recognition by Medicago hosts, gene-swapping experiments have shown that the LysM domain of MtNFP does not discriminate sulfated and nonsulfated LCOs (Bensmihen et al., 2011). Moreover, it was recently shown that the MtNFP gene is also involved during the interaction between M. truncatula and the root pathogen Aphanomyces euteiches, indicating that MtNFP is likely involved in perceiving signals from a variety of symbiotic or pathogenic microbes (C. Jacquet, personal communication).
On the other hand, because the nfp-1 mutants are not significantly affected in colonization by AM fungi, there needs to be a second receptor mediating fungal entry. So far, no such receptor was described, but MtLYR1, a paralog of MtNFP (Op den Camp et al., 2011), is a good candidate. A hint toward this assumption is the fact that only one NFP gene exists in the nonlegume Parasponia (Op den Camp et al., 2011). In this species, an RNA interference-mediated knockdown of NFP does not affect initial root infection but displays defects in the establishment of fixation threads and arbuscules. Duplication of NFP in the legume lineage led to the evolution of a pair of NFP-type receptors (Young et al., 2011), where MtNFP might be required for the perception of low LCO concentrations in presymbiotic stages while MtLYR1 might mediate responses to infection by AM fungi at higher local Myc-LCO levels. If this is true, it is likely that transcriptional responses mediated by MtNFP were primarily measured here, because an entry receptor would have required higher LCO concentrations. The limited overlap between genes activated by Myc-LCOs and known infection-related genes supports this. Although we provided evidence that key genes related to arbuscule formation were not induced by Myc-LCOs, it is striking that MtLYR1 is expressed in arbuscule-containing cells (Gomez et al., 2009), suggesting an activation of LCO receptors at late AM stages. An intriguing possibility would be that MtLYR1 mediates the invasion of hyphae via high local LCO concentrations prior to arbuscule formation, a mechanism also suggested for the single Nfp receptor of Parasponia (Op den Camp et al., 2011).
While our study revealed novel insights into transcriptional reprogramming by Myc-LCOs in the presymbiotic AM phase, it is an open question whether this reflects the complete picture of early AM signaling or just a glimpse through the keyhole. Apart from the fact that the ratio or concentration of known Myc-LCOs may play a role, there is evidence for gene activation by diffusible fungal signals via CSP-independent Myc pathways (Kosuta et al., 2003). Moreover, it has to be kept in mind that additional fungal signal molecules likely exist, since activating the MtMSBP1 gene is rapidly activated by diffusible fungal signals (Kuhn et al., 2010) but not by Myc-LCOs in our conditions. A striking example for a completely different signal molecule was recently reported by Kloppholz et al. (2011), who identified a secreted Glomus intraradices effector protein that counteracts plant defense. Interestingly, Tisserant et al. (2012) identified several G. intraradices genes encoding small secreted proteins whose expression was detected in intraradical mycelia but not in germinating spores, suggesting infection-related functions. Similar results were obtained by Plett et al. (2011) for an ectomycorrhiza, indicating a widespread function of fungal effectors in the colonization of mycorrhizal roots. Experiments using legume mutants have to be carried out to determine the relative contribution of Myc-LCOs and other signals to the presymbiotic communication between AM fungi and their host.
MATERIALS AND METHODS
Sterilization and Vernalization of Seeds
Seeds of Medicago truncatula cv Jemalong genotype A17 (the wild type), an MtNFP mutant (nfp-1; Ben Amor et al., 2003), an MtDMI3 mutant identified in a genetic screen of fast neutron-mutagenized lines (dmi3; Marsh et al., 2007), an MtNSP1 mutant (nsp1-1; Smit et al., 2005), and the transgenic line MtL416 expressing a pMtENOD11-GUS fusion (Journet et al., 2001) were surface sterilized as reported by Hohnjec et al. (2003) with the following modifications. For sterilization, seeds were immersed in 2% (v/v) sodium hypochloride for 1 min, quickly rinsed four times with sterile water, and finally washed five times with sterile water for 1 min each. Seed moisture expansion was allowed for 4 h, while water was refreshed every 30 min. Afterward, seeds were spread on 0.8% (w/v) water agar (Sigma) plates. Three plates were jointly sealed with transparent foil cut at three sites to allow gas exchange. Plate stacks were inverted, wrapped in aluminum foil, and vernalized at 4°C for 96 h.
Treatment of Plantlet Roots with Myc- and Nod-LCOs
The following procedure is based on Maillet et al. (2011) and is illustrated in Supplemental Figure S1. After vernalization, M. truncatula seeds were incubated for 24 h in the climate chamber (humidity, 70%; photosynthetic photon flux, 150 μmol m−2 s−1) at a 16-h-light (23°C)/8-h-dark (18°C) regime. Subsequently, seedlings were moisturized with sterile water (pH 7.0) to allow the removal of seed coats. In each case, eight seedlings were put on the upper one-quarter of a 2.5% (w/v) phytoagar (Duchefa Biochemie) 120-mm-square slant plate (DoctorLab). Seedlings used for Myc-LCO and control treatments were put on plates with one-half-strength Hoagland solution (Arnon and Hoagland, 1940) set to pH 6.5 with KOH, whereas Nod-LCO-treated seedlings and corresponding controls were grown on slant agar plates with a nitrogen-free nutrient solution of pH 7.5 (NH-mix) as described by Baier et al. (2007). A 1-cm strap of Whatman paper (Schleicher & Schuell) soaked with appropriate nutrient solutions fixed seedlings on the plates beneath the cotyledons. Three plates were jointly sealed with transparent wrapping foil cut at the upper side to allow gas exchange. The lower half of each plate stack was wrapped with aluminum foil for light protection to allow normal root development. Plates were placed in an approximately 70° angle in the climate chamber for 5 d, using the conditions described above. Subsequently, plantlets were removed from the plates and placed in 13-mL tubes wrapped with aluminum foil that were cut off at the 8-mL mark, using 20 plantlets per tube (Sarstedt). Each tube contained 5 mL of the following solutions: sMyc-LCO solution (one-half-strength Hoagland solution, pH 6.5, containing 10−8 m sMyc-LCOs), nsMyc-LCO solution (one-half-strength Hoagland solution, pH 6.5, containing 10−7 m nsMyc-LCOs), mixed Myc-LCO solution (one-half-strength Hoagland solution, pH 6.5, containing 10−8 m sMyc-LCOs and 10−7 m nsMyc-LCOs), Myc control solution (one-half-strength Hoagland solution, pH 6.5), Nod-LCO solution (NH-mix, pH 7.5, containing 10−8 m Nod-LCOs), and Nod control solution (NH-mix, pH 7.5).
Myc-LCOs used in this study were synthesized via Escherichia coli cell factories (Maillet et al., 2011) and contained approximately 90% tetramers (LCO-IV) and 10% pentamers (LCO-V). Both sMyc- and nsMyc-LCOs were a mixture of compounds N-acylated by palmitic acid (C16:0) and oleic acid (C18:1∆9Z) in a 1:1 ratio. In the case of Nod-LCOs, the treatment solution contained the major Sinorhizobium meliloti Nod factor NodSm-IV-Ac-S (C16:2∆2E∆9Z) and approximately 10% of the corresponding pentamer with the same O- and N-substitutions. Myc- and Nod-LCOs were dissolved in 50% (v/v) acetonitrile to obtain 10−3 m solutions and diluted further as described above. Separate batches of control plantlets were treated with Nod- and Myc-control solutions containing appropriate amounts of acetonitrile.
RNA Isolation, Genome-Wide Expression Profiling, and Real-Time RT-PCR
To harvest tissues for transcriptome profiling, three biological replicates each consisting of 20 plantlets were set up. After 6 h of incubation (using the conditions described above), 10 plantlets per treatment or control condition were harvested, while the other 10 were left for 24 h. During harvest, 1 mm of the root tip was removed. The remaining 2 to 2.5 cm of the distal root region was cut off and directly frozen in liquid nitrogen. Ten root fragments of each replicate were pooled, total RNA was isolated, and Medicago GeneChip hybridizations were performed and evaluated as described (Hogekamp et al., 2011). GeneChip data are available from the Gene Expression Omnibus (accession no. GSE33638). Primer pairs for real-time RT-PCR and the sizes of predicted PCR products are listed in Supplemental Table S7. Except for primers for MtTEF1α and MtENOD11, all primer pairs were designed using Primer3 (Rozen and Skaletsky, 2000) and tested for specificity as well as correct amplification size. Fifty nanograms of total RNA was used for real-time RT-PCR, using the SensiMix SYBR Hi-Rox one-step kit (Bioline). RT-PCR conditions (Realplex cycler; Eppendorf) were as follows: 10 min at 45°C, 10 min at 95°C, 44 cycles (15 s at 95°C, 15 s at gene-specific temperature, 15 s at 72°C), 15 s at 95°C, and melting curve from 40°C to 95°C. Gene-specific annealing temperatures were 63°C (MtENOD11), 57°C (Mtr.20438.1.S1_at and Mtr.32088.1.S1_s_at), and 53°C for all other genes. The constitutive translation elongation factor gene MtTEF1α (TC178258 in the Dana-Farber Cancer Institute Medicago Gene Index) was used for normalization across different conditions. All expression results were averaged over three biological replicates each measured in at least two technical repetitions. The average value was used to calculate relative gene expression levels using the 2−ΔCT formula with ΔCT = CTgene − CTMtTEF1α (Livak and Schmittgen 2001). Heat maps of expression data were generated using Genesis software (Sturn et al., 2002).
Histochemical Analysis of GUS Activity
Transgenic roots were assayed for GUS activity for 24 h at 37°C as described by Küster et al. (1995). Photographs were taken with an Olympus XC50 digital camera.
GeneChip data are available from the Gene Expression Omnibus (accession no. GSE33638).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Setup for the treatment of plantlet roots with Myc- and Nod-LCOs.
Supplemental Figure S2. Ratio-versus-intensity plots of gene expression in M. truncatula plantlet roots treated with Myc- and Nod-LCOs.
Supplemental Figure S3. Comparison of Myc-LCO-mediated transcription in wild-type roots with gene expression in fully colonized mycorrhizal roots.
Supplemental Figure S4. Myc-LCO-mediated gene expression in the M. truncatula wild type and the nsp1-1 mutant.
Supplemental Table S1. Gene expression in M. truncatula plantlet roots treated with 10−8 m sMyc-LCOs (sMyc), 10−7 m nsMyc-LCOs (nsMyc), a mixture of both Myc-LCOs (s/nsMyc), and 10−8 m Nod-LCOs (Nod) for 6 and 24 h.
Supplemental Table S2. Comparison of M. truncatula Nod-LCO-induced genes reported by Mitra et al. (2004; sheet 1) and Nod-LCO-induced marker genes (sheet 2) with gene expression in plantlet roots treated with 10−8 m Nod-LCOs (Nod) for 6 and 24 h.
Supplemental Table S3. Subsets of M. truncatula genes activated in plantlet roots after treatment with 10−8 m sMyc-LCOs (sMyc), 10−7 m nsMyc-LCOs (nsMyc), a mixture of both Myc-LCOs (s/nsMyc), and 10−8 m Nod-LCOs (Nod) for 6 and 24 h.
Supplemental Table S4. Subsets of M. truncatula genes activated in plantlet roots after treatment with 10−8 m sMyc-LCOs (sMyc), 10−7 m nsMyc-LCOs (nsMyc), a mixture of both Myc-LCOs (s/nsMyc), and 10−8 m Nod-LCOs (Nod) for 6 and 24 h.
Supplemental Table S5. Expression of selected M. truncatula genes in plantlet roots treated with 10−8 m sMyc-LCOs (sMyc), 10−7 m nsMyc-LCOs (nsMyc), a mixture of both Myc-LCOs (s/nsMyc), and 10−8 m Nod-LCOs (Nod) for 6 and 24 h.
Supplemental Table S6. Expression of 348 M. truncatula genes activated in wild-type plantlet roots after 6 h by any of the Myc-LCO treatments and not by Nod-LCOs studied in nfp-1 and dmi3 mutants treated for 6 h with 10−8 m sMyc-LCOs (sMyc), 10−7 m nsMyc-LCOs (nsMyc), and a mixture of both Myc-LCOs (s/nsMyc).
Supplemental Table S7. Real-time RT-PCR primers used in this study and sizes of predicted PCR products.
Supplementary Material
Acknowledgments
The transgenic M. truncatula line MtL416 was provided by David Barker (Laboratoire des Interactions Plantes-Microorganismes, Institut National de la Recherche Agronomique), the nfp-1 and nsp1-1 mutants were provided by Christian Rogers (John Innes Centre), and the dmi3 mutant was provided by John Marsh (John Innes Centre). We thank Júlia Lobato and Jörg D. Becker (Gulbenkian Institute) for excellent support in GeneChip hybridizations. Expert bioinformatics support by Kolja Henckel (Bioinformatics Resource Facility, Center for Biotechnology, Bielefeld University) is acknowledged.
Glossary
- AM
arbuscular mycorrhizal
- LCO
lipochitooligosaccharide
- CSP
common symbiotic pathway
- RT
reverse transcription
References
- Akiyama K, Matsuzaki K, Hayashi H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de Carvalho-Niebel F. (2007) AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI, Hoagland DR. (1940) Crop production in artificial culture solutions and in soils with special reference to factors influencing yields and absorption of inorganic nutrients. Soil Sci 50: 463–483 [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Ghérardi M, Huguet T, et al. (2006) The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier MC, Barsch A, Küster H, Hohnjec N. (2007) Antisense repression of the Medicago truncatula nodule-enhanced sucrose synthase leads to a handicapped nitrogen fixation mirrored by specific alterations in the symbiotic transcriptome and metabolome. Plant Physiol 145: 1600–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amor B, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C. (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, de Billy F, Gough C. (2011) Contribution of NFP LysM domains to the recognition of Nod factors during the Medicago truncatula/Sinorhizobium meliloti symbiosis. PLoS ONE 6: e26114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Bécard G, Séjalon-Delmas N. (2006) Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 4: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P, Requena N. (2011) Dating in the dark: how roots respond to fungal signals to establish arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol 14: 451–457 [DOI] [PubMed] [Google Scholar]
- Brewin NJ. (1991) Development of the legume root nodule. Annu Rev Cell Biol 7: 191–226 [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J. (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M, Genre A, Sieberer BJ, Faccio A, Fournier J, Novero M, Barker DG, Bonfante P. (2011) Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol 189: 347–355 [DOI] [PubMed] [Google Scholar]
- Charpentier M, Bredemeier R, Wanner G, Takeda N, Schleiff E, Parniske M. (2008) Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20: 3467–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, Küster H, Journet EP, Hohnjec N, Gamas P, Niebel A. (2008) Evidence for the involvement in nodulation of the two small putative regulatory peptide-encoding genes MtRALFL1 and MtDVL1. Mol Plant Microbe Interact 21: 1118–1127 [DOI] [PubMed] [Google Scholar]
- Den Herder J, Vanhee C, De Rycke R, Corich V, Holsters M, Goormachtig S. (2007) Nod factor perception during infection thread growth fine-tunes nodulation. Mol Plant Microbe Interact 20: 129–137 [DOI] [PubMed] [Google Scholar]
- D’haeseleer K, Den Herder G, Laffont C, Plet J, Mortier V, Lelandais-Brière C, De Bodt S, De Keyser A, Crespi M, Holsters M, et al (2011) Transcriptional and post-transcriptional regulation of a NAC1 transcription factor in Medicago truncatula roots. New Phytol 191: 647–661 [DOI] [PubMed] [Google Scholar]
- Drissner D, Kunze G, Callewaert N, Gehrig P, Tamasloukht M, Boller T, Felix G, Amrhein N, Bucher M. (2007) Lyso-phosphatidylcholine is a signal in the arbuscular mycorrhizal symbiosis. Science 318: 265–268 [DOI] [PubMed] [Google Scholar]
- Floss DS, Hause B, Lange PR, Küster H, Strack D, Walter MH. (2008) Knock-down of the MEP pathway isogene 1-deoxy-D-xylulose 5-phosphate synthase 2 inhibits formation of arbuscular mycorrhiza-induced apocarotenoids, and abolishes normal expression of mycorrhiza-specific plant marker genes. Plant J 56: 86–100 [DOI] [PubMed] [Google Scholar]
- Gaude N, Bortfeld S, Duensing N, Lohse M, Krajinski F. (2012) Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant J 69: 510–528 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P. (2008) Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20: 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG. (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17: 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts R, Fedorova E, Bisseling T. (2005) Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr Opin Plant Biol 8: 346–352 [DOI] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Muñoz A, Poovaiah BW, Oldroyd GE. (2006) Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Gomez SK, Javot H, Deewatthanawong P, Torres-Jerez I, Tang Y, Blancaflor EB, Udvardi MK, Harrison MJ. (2009) Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough C, Cullimore J. (2011) Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol Plant Microbe Interact 24: 867–878 [DOI] [PubMed] [Google Scholar]
- Graham PH, Vance CP. (2003) Legumes: importance and constraints to greater use. Plant Physiol 131: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P. (2009) Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182: 200–212 [DOI] [PubMed] [Google Scholar]
- Haney CH, Riely BK, Tricoli DM, Cook DR, Ehrhardt DW, Long SR. (2011) Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell 23: 2774–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ. (1999) Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu Rev Plant Physiol Plant Mol Biol 50: 361–389 [DOI] [PubMed] [Google Scholar]
- Harrison MJ. (2005) Signaling in the arbuscular mycorrhizal symbiosis. Annu Rev Microbiol 59: 19–42 [DOI] [PubMed] [Google Scholar]
- Hogekamp C, Arndt D, Pereira PA, Becker JD, Hohnjec N, Küster H. (2011) Laser microdissection unravels cell-type-specific transcription in arbuscular mycorrhizal roots, including CAAT-box transcription factor gene expression correlating with fungal contact and spread. Plant Physiol 157: 2023–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohnjec N, Perlick AM, Pühler A, Küster H. (2003) The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol Plant Microbe Interact 16: 903–915 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Vieweg MF, Pühler A, Becker A, Küster H. (2005) Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol 137: 1283–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V. (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14: 737–748 [DOI] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kistner C, Winzer T, Pitzschke A, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Webb KJ, et al (2005) Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17: 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppholz S, Kuhn H, Requena N. (2011) A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol 21: 1204–1209 [DOI] [PubMed] [Google Scholar]
- Kosuta S, Chabaud M, Lougnon G, Gough C, Dénarié J, Barker DG, Bécard G. (2003) A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol 131: 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S, Hazledine S, Sun J, Miwa H, Morris RJ, Downie JA, Oldroyd GE. (2008) Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc Natl Acad Sci USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn H, Küster H, Requena N. (2010) Membrane steroid-binding protein 1 induced by a diffusible fungal signal is critical for mycorrhization in Medicago truncatula. New Phytol 185: 716–733 [DOI] [PubMed] [Google Scholar]
- Küster H, Hohnjec N, Krajinski F, El YF, Manthey K, Gouzy J, Dondrup M, Meyer F, Kalinowski J, Brechenmacher L, et al. (2004) Construction and validation of cDNA-based Mt6k-RIT macro- and microarrays to explore root endosymbioses in the model legume Medicago truncatula. J Biotechnol 108: 95–113 [DOI] [PubMed] [Google Scholar]
- Küster H, Schröder G, Frühling M, Pich U, Rieping M, Schubert I, Perlick AM, Pühler A. (1995) The nodule-specific VfENOD-GRP3 gene encoding a glycine-rich early nodulin is located on chromosome I of Vicia faba L. and is predominantly expressed in the interzone II-III of root nodules. Plant Mol Biol 28: 405–421 [DOI] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J. (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784 [DOI] [PubMed] [Google Scholar]
- Limpens E, Mirabella R, Fedorova E, Franken C, Franssen H, Bisseling T, Geurts R. (2005) Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci USA 102: 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ. (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15: 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Maldonado-Mendoza I, Lopez-Meyer M, Cheung F, Town CD, Harrison MJ. (2007) Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J 50: 529–544 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Manthey K, Krajinski F, Hohnjec N, Firnhaber C, Pühler A, Perlick AM, Küster H. (2004) Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbioses. Mol Plant Microbe Interact 17: 1063–1077 [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE. (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue M, Camut S, de Carvalho-Niebel F, Deslandes L, Froidure S, Klaus-Heisen D, Moreau S, Rivas S, Timmers T, Hervé C, et al (2010) The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22: 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Shaw SL, Long SR. (2004) Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 101: 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD, Muni RR, Torres-Jerez I, Tang Y, Allen S, Andriankaja M, Li G, Laxmi A, Cheng X, Wen J, et al. (2011) Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J 65: 244–252 [DOI] [PubMed] [Google Scholar]
- Navazio L, Moscatiello R, Genre A, Novero M, Baldan B, Bonfante P, Mariani P. (2007) A diffusible signal from arbuscular mycorrhizal fungi elicits a transient cytosolic calcium elevation in host plant cells. Plant Physiol 144: 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oláh B, Brière C, Bécard G, Dénarié J, Gough C. (2005) Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. Plant J 44: 195–207 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. (2006) Nuclear calcium changes at the core of symbiosis signalling. Curr Opin Plant Biol 9: 351–357 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Oliveros JC. (2007) VENNY: an interactive tool for comparing lists with Venn diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html (February 20, 2012)
- Op den Camp R, Streng A, De Mita S, Cao Q, Polone E, Liu W, Ammiraju JS, Kudrna D, Wing R, Untergasser A, et al (2011) LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science 331: 909–912 [DOI] [PubMed] [Google Scholar]
- Ovchinnikova E, Journet EP, Chabaud M, Cosson V, Ratet P, Duc G, Fedorova E, Liu W, den Camp RO, Zhukov V, et al. (2011) IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago spp. Mol Plant Microbe Interact 24: 1333–1344 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Plett JM, Kemppainen M, Kale SD, Kohler A, Legué V, Brun A, Tyler BM, Pardo AG, Martin F. (2011) A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol 21: 1197–1203 [DOI] [PubMed] [Google Scholar]
- Pumplin N, Mondo SJ, Topp S, Starker CG, Gantt JS, Harrison MJ. (2010) Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J 61: 482–494 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass H, Kerp H. (1994) Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA 91: 11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ. (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3: 423–434 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Sanchez L, Weidmann S, Arnould C, Bernard AR, Gianinazzi S, Gianinazzi-Pearson V. (2005) Pseudomonas fluorescens and Glomus mosseae trigger DMI3-dependent activation of genes related to a signal transduction pathway in roots of Medicago truncatula. Plant Physiol 139: 1065–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüssler A, Schwarzott D, Walker C. (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105: 1413–1421 [Google Scholar]
- Siciliano V, Genre A, Balestrini R, Cappellazzo G, deWit PJ, Bonfante P. (2007) Transcriptome analysis of arbuscular mycorrhizal roots during development of the prepenetration apparatus. Plant Physiol 144: 1455–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Fournier J, Timmers AC, Barker DG. (2012) A switch in Ca2+ spiking signature is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula. Plant J 69: 822–830 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T. (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R. (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ. (2008) Mycorrhizal Symbiosis, Ed 3. Academic Press/Elsevier, Amsterdam
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al. (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- Tisserant E, Kohler A, Dozolme-Seddas P, Balestrini R, Benabdellah K, Colard A, Croll D, Da Silva C, Gomez SK, Koul R, et al. (2012) The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol 193: 755–769 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Kakar K, Wandrey M, Montanari O, Murray J, Andriankaja A, Zhang JY, Benedito V, Hofer JM, Chueng F, et al. (2007) Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiol 144: 538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann S, Sanchez L, Descombin J, Chatagnier O, Gianinazzi S, Gianinazzi-Pearson V. (2004) Fungal elicitation of signal transduction-related plant genes precedes mycorrhiza establishment and requires the dmi3 gene in Medicago truncatula. Mol Plant Microbe Interact 17: 1385–1393 [DOI] [PubMed] [Google Scholar]
- Withers ST, Keasling JD. (2007) Biosynthesis and engineering of isoprenoid small molecules. Appl Microbiol Biotechnol 73: 980–990 [DOI] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Gallego-Giraldo L, Wang H, Zeng Y, Ding SY, Chen F, Dixon RA. (2010) An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J 63: 100–114 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.