Abstract
The unicellular green alga Chlamydomonas reinhardtii adapts to anaerobic or hypoxic conditions by developing a complex fermentative metabolism including the production of molecular hydrogen by [FeFe]-hydrogenase isoform1 (HYDA1). HYDA1 transcript and hydrogenase protein accumulate in the absence of oxygen or copper (Cu). Factors regulating this differential gene expression have been unknown so far. In this study, we report on the isolation of a Chlamydomonas mutant strain impaired in HYDA1 gene expression by screening an insertional mutagenesis library for HYDA1 promoter activity using the arylsulfatase-encoding ARYLSULFATASE2 gene as a selection marker. The mutant strain has a deletion of the COPPER RESPONSE REGULATOR1 (CRR1) gene encoding for CRR1, indicating that this SQUAMOSA-PROMOTER BINDING PROTEIN (SBP) domain transcription factor is involved in the regulation of HYDA1 transcription. Treating the C. reinhardtii wild type with mercuric ions, which were shown to inhibit the binding of the SBP domain to DNA, prevented or deactivated HYDA1 gene expression. Reporter gene analyses of the HYDA1 promoter revealed that two GTAC motifs, which are known to be the cores of CRR1 binding sites, are necessary for full promoter activity in hypoxic conditions or upon Cu starvation. However, mutations of the GTAC sites had a much stronger impact on reporter gene expression in Cu-deficient cells. Electrophoretic mobility shift assays showed that the CRR1 SBP domain binds to one of the GTAC cores in vitro. These combined results prove that CRR1 is involved in HYDA1 promoter activation.
Molecular dioxygen (O2) serves as substrate and reagent in a number of biosynthetic pathways (Goldfine, 1965; Raymond and Segrè, 2006) and plays an important role for both metabolism and growth of aerobic organisms (Des Marais, 1998). Under anaerobic conditions, aerobes have to adapt both energy-generating and biosynthetic mechanisms to maintain cell vitality. This modulation of metabolism is accompanied by altered gene expression, often regulated on the level of transcription as indicated by massive changes of transcript abundances in various organisms transferred to anaerobic or hypoxic conditions (Mustroph et al., 2010). As a response to anaerobiosis the unicellular green alga Chlamydomonas reinhardtii develops a complex fermentative metabolism (Gfeller and Gibbs, 1984; Hemschemeier and Happe, 2005; Mus et al., 2007; Hemschemeier et al., 2008b; Philipps et al., 2011) with molecular hydrogen (H2) as one key metabolite. Hydrogen is produced by an oxygen-sensitive [FeFe]-hydrogenase (Stripp and Happe, 2009; Lambertz et al., 2011) that is linked to the photosynthetic electron transport chain via ferredoxin PETF (Winkler et al., 2009). Chlamydomonas expresses two hydrogenase-encoding genes, [FeFe]-hydrogenase isoform1 (HYDA1) and HYDA2 (Forestier et al., 2003), but the HYDA1 protein seems to be the primarily active hydrogenase isoform in the green alga (Godman et al., 2010; Meuser et al., 2012).
One experimental approach to induce anaerobic or hypoxic conditions in C. reinhardtii cultures is the incubation of the cells in sulfur (S)-free medium in the light (Melis et al., 2000; Hemschemeier et al., 2009). S-deprived algae produce significant amounts of H2. This can be attributed to the fact that PSII (Wykoff et al., 1998) and carbon dioxide fixation (Zhang et al., 2002; Hemschemeier et al., 2008a) activities decrease strongly, resulting in anaerobic (or microaerobic) conditions and a severe diminution of both respiratory and photosynthetic electron sinks (Melis, 2007; Ghysels and Franck, 2010; Hemschemeier and Happe, 2011). Under these conditions, HYDA1 acts as an alternative electron acceptor of photosynthetic electron flow. However, small but significant amounts of H2 are also produced by anaerobically adapted C. reinhardtii cultures in the dark (Gfeller and Gibbs, 1984; Philipps et al., 2011).
A major part of the regulation of hydrogenase activity in Chlamydomonas occurs via differential HYDA1 gene transcription. HYDA1 transcript levels increase in algae transferred to anaerobic conditions in the dark (Happe and Kaminski, 2002) or in illuminated, S-deprived algae (Hemschemeier et al., 2008b), paralleled by the appearance of HYDA1 protein (Happe and Kaminski, 2002; Zhang et al., 2002; Jacobs et al., 2009). A region from position −128 to −21 relative to the transcription start site of the HYDA1 gene was shown by reporter gene studies to be sufficient for measurable HYDA1 promoter activity (Stirnberg and Happe, 2004). However, oxygen sensors, downstream signaling cascades, transcriptional regulators, or essential promoter motifs regulating HYDA1 expression are currently unknown. While in general, no specific oxygen-responsive transcription factor is known in C. reinhardtii, it was noted some years ago that the copper (Cu) deficiency responsive genes encoding cytochrome c6 (CYC6), oxidative coproporphyrinogen oxidase1 (CPX1), and Copper Response Defect1 (CRD1), one isoform of magnesium-protoporphyrin IX monomethyl ester aerobic oxidative cyclase, were also transcriptionally induced upon hypoxic conditions (Quinn et al., 2002; Eriksson et al., 2004; Kropat et al., 2005). Additionally, the amounts of the transcript encoding Chlamydomonas ferredoxin isoform FDX5, which were first noted to increase strongly in anaerobic algal cells (Mus et al., 2007; Jacobs et al., 2009), were also shown to rise under conditions of Cu depletion (Terauchi et al., 2009; Lambertz et al., 2010; Castruita et al., 2011). In all cases mentioned above, the transcription factor COPPER RESPONSE REGULATOR1 (CRR1) was shown to be responsible for differential transcription (Quinn et al., 2002; Eriksson et al., 2004; Kropat et al., 2005; Lambertz et al., 2010). This multidomain transcription factor belongs to the SQUAMOSA-PROMOTER BINDING PROTEIN (SBP) family and has a characteristic zinc-finger domain that binds to the cis-acting DNA element GTAC (Birkenbihl et al., 2005; Kropat et al., 2005). A recent study proposed that the coregulation of genes during the two environmental conditions of Cu and O2 deficiency might be due to a coordinated activation of the CRR1 protein through the SBP domain and a C-terminal Cys-rich region (Sommer et al., 2010). Lately, it was shown by deep sequencing of the transcriptome of Cu-deficient C. reinhardtii cells that the group of genes regulated in response to both Cu and O2 limitation is larger than expected (Castruita et al., 2011). Transcript levels of several genes known to be induced in anaerobiosis increased during Cu deficiency (Castruita et al., 2011). Among those were HYDA1, HYDG, and HYDEF, the two latter encoding maturases of the [FeFe]-hydrogenases in C. reinhardtii (Posewitz et al., 2004), or PYRUVATE FERREDOXIN OXIDOREDUCTASE1 (Mus et al., 2007). However, a direct influence of CRR1 on HYDA1 expression was not postulated.
This study aimed at identifying regulative elements of HYDA1 gene expression. For this purpose, a mutant library was generated using a C. reinhardtii strain carrying the arylsulfatase reporter gene (Davies et al., 1992) fused to the HYDA1 promoter (Stirnberg and Happe, 2004). Screening of the insertional mutant library resulted in the identification of a transformant impaired in HYDA1 expression that had a partial deletion of the CRR1 gene. Complementation of this strain with the full-length CRR1 genomic sequence restored the phenotype, indicating that the CRR1 transcriptional regulator is involved in HYDA1 promoter regulation. In silico analysis of the HYDA1 promoter sequence showed the presence of two GTAC motifs as putative CRR1 binding sites. Reporter gene analyses employing the codon-optimized Renilla reniformis luciferase encoding gene (termed CRLUC; Fuhrmann et al., 2004) revealed that the presence of intact GTAC motifs is necessary for complete HYDA1 promoter activity. The combined results of this study indicate that CRR1 is involved in regulating HYDA1 gene transcription, probably in concert with (a) yet-unknown transcription factor(s).
RESULTS
Isolation of C. reinhardtii Transformants with Impaired HYDA1 Expression
C. reinhardtii strain MR9 carries a pHYDA1-ARYLSULFATASE2 (ARS2) reporter gene construct (Supplemental Fig. S1A). The region of the HYDA1 promoter that was fused to the promoterless ARS2 gene (−1,020 to +158 relative to the transcription start site) was chosen to be sufficiently long to include possible regulating elements. It carried about 500 bp at its 5′- and 114 bp at its 3′-end, respectively, in addition to the longest promoter fragment (−474 to +44) already shown to be able to confer hypoxia responsiveness to the ARS2 reporter gene (Stirnberg and Happe, 2004). To identify genes whose products are involved in HYDA1 gene expression, a mutant library was generated by DNA insertional mutagenesis of strain MR9 using the paromomycin resistance cassette derived from plasmid pSL18 (Sizova et al., 2001). About 10,000 transformants were exposed to anaerobiosis to induce HYDA1 promoter activity and thus ARS2 reporter gene expression. ARS activity was assayed using the artificial ARS substrate 5-bromo-4-chloro-3-indolyl sulfate (X-SO4). Most of the transformants exhibited anaerobically inducible arylsulfatase activity, showing both that the chosen HYDA1 upstream region was able to activate ARS2 gene expression and that the construct was stable in the individual colonies (Supplemental Fig. S1B). The absence of a blue staining thus served as an indicator that an individual transformant was impaired in HYDA1 promoter activity. We identified three transformants that stayed colorless during the screening. One of these, named strain 41-6 in the following, contained an intact pHYDA1-ARS2 construct as indicated by the presence of the HYDA1-promoter/ARS2 gene junction analyzed by PCR (data not shown). The transformant was also able to produce active arylsulfatase upon S deficiency (data not shown).
The CRR1 Gene Is Partially Deleted in C. reinhardtii Strain 41-6
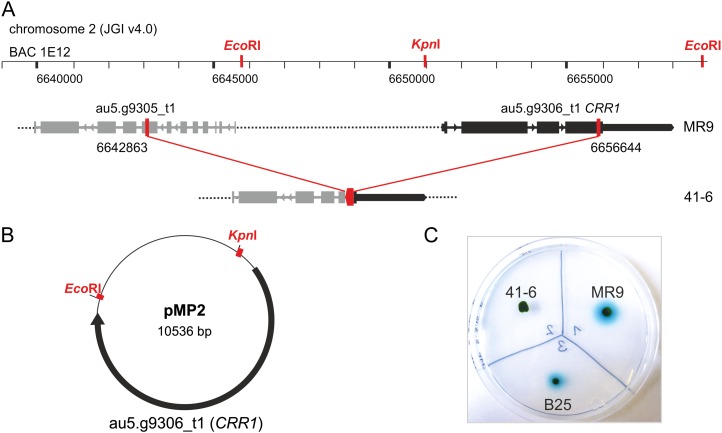
According to Southern-blot analyses using an APHVIII-specific probe, transformant 41-6 has a single insertion of the paromomycin resistance cassette (Supplemental Fig. S2). Both 5′- and 3′-regions flanking the cassette were identified by inverse PCR (Fig. 1A). The 5′-end of the cassette is located at position 6,656,644 on chromosome 2 (JGI4.0, http://genome.jgi-psf.org/Chlre4/Chlre4.home.html), while the 3′-end is at position 6,642,863 on the same chromosome. According to the most recent genome annotations on JGI4.0 and Phytozome (http://www.phytozome.net/chlamy.php; Merchant et al., 2007), the integration of the selection marker resulted in the deletion of 13,781 bp and the partial disruption of two genes. The functional annotation of the 5′-flanking gene (JGI3.0 protein ID 148820, Augustus5 model on JGI4.0: au5.g9305_t1, Augustus10 model on Phytozome: Cre02.g125200) on Phytozome is Panther: 19418 (homeobox protein). Motif and domain searches conducted in this work using various publicly available search engines resulted in only one hit on InterProScan (Zdobnov and Apweiler, 2001), indicating that the putative gene product might belong to an unnamed Panther family PTHR15377. The family includes proteins with putative DNA-binding and transcription factor activity. The gene flanking the 3′-end of the paromomycin cassette in strain 41-6 is the CRR1 gene (JGI3.0 protein ID 195928, Augustus5 model au5.g9306_t1, Augustus10 model Cre02.g125250; Kropat et al., 2005), whose first four exons have been deleted (Fig. 1A).
Figure 1.
Genetic characterization and complementation of C. reinhardtii strain 41-6. A, Scheme of the genomic region that is altered in strain 41-6. The wild-type region on chromosome 2 as annotated on JGI4.0 contains a putative gene encoding a protein with unknown function (Augustus 5 model au5.g9305_t1) and the CRR1 gene (au5.g9306_t1). In strain 41-6, the integration of the paromomycin resistance cassette resulted in a deletion of 13.78 kb from position 6,642,863 to 6,656,644 of chromosome 2. The sequence present in BAC1E12 including restriction sites used in this study is shown above the gene graph. B, Map of plasmid pMP2 used for complementation of strain 41-6 with the CRR1 gene derived from BAC1E12. C, Photograph of a TAP agar plate with colonies of C. reinhardtii strains MR9, 41-6, and complemented strain B25. Strains were subjected to the same conditions that were applied during the screening of the MR9 progenies.
In silico analyses of the HYDA1 promoter region identified two GTAC motifs (see below). GTAC cores are known to be essential for the DNA-binding activity of CRR1 (Quinn et al., 2000; Kropat et al., 2005) and other SBP-box transcription factors (Birkenbihl et al., 2005). Therefore, the CRR1 gene was chosen as a first candidate for complementation of C. reinhardtii strain 41-6. The CRR1 gene including 171 bp up- and downstream from the annotated transcriptional start and stop sites (as annotated on JGI4.0, model au5.g9306_t1) was obtained by subcloning fragments of the bacterial artificial chromosome BAC1E12 as described in details in the “Materials and Methods” section (Fig. 1, A and B). Strain 41-6 was then cotransformed with the CRR1 construct and a hygromycin B resistance cassette (Berthold et al., 2002). Reconstruction of the phenotype was tested applying the same arylsulfatase screening that had been used to identify strain 41-6. Out of 114 hygromycin-resistant transformants, 17 showed a restoration of the blue staining after addition of X-SO4 solution to anaerobically adapted colonies. Transformant B25 was chosen for further experiments (Fig. 1C).
In Vitro Hydrogenase Activity and HYDA1 Gene Expression Are Impaired in C. reinhardtii crr1 Mutant Strains
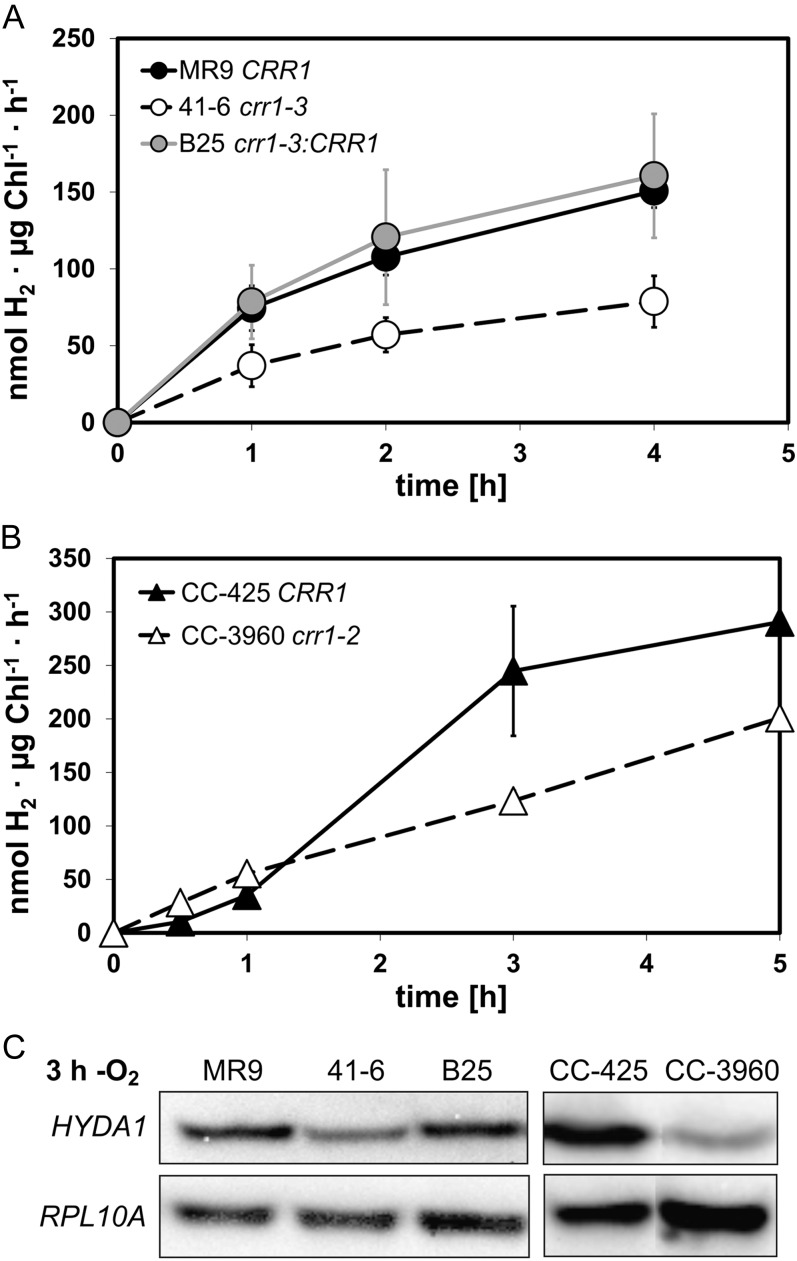
We analyzed C. reinhardtii strains MR9 (pHYDA1-ARS2), 41-6 (pHYDA1-ARS2 crr1-3), and B25 (pHYDA1-ARS2 crr1-3:CRR1) regarding hydrogenase activity and HYDA1 transcript amounts. C. reinhardtii crr1-2 mutant CC-3960 was analyzed to confirm that the results were due to the absence of CRR1, and CC-425 (CRR1) was used as a wild-type control for this strain (Kropat et al., 2005). During anaerobiosis established by nitrogen flushing, both crr1 mutant strains showed a lower in vitro hydrogenase activity when compared with the respective wild types. In strain 41-6, hydrogenase activity reached about 50% of wild-type activity, while the same rates as determined in MR9 were detected in complemented strain B25 (Fig. 2A). Strain CC-3960 showed 65% of the activity determined in C. reinhardtii CC-425 (Fig. 2B).
Figure 2.
Analysis of in vitro hydrogenase activity and HYDA1 transcript levels in crr1 mutant strains compared with the respective parental strains upon artificially induced anaerobiosis. A and B, Concentrated cell suspensions were subjected to anaerobic conditions by nitrogen flushing. At the indicated time points, cell samples were withdrawn and analyzed in an in vitro hydrogenase activity assay containing Triton X-100 as a detergence to lyse the cells and reduced methyl viologen as an artificial electron donor of the [FeFe]-hydrogenase. Hydrogen generated in these assays was quantified by gas chromatography. Average values ± sd (error bars) of three independent experiments are shown. A, C. reinhardtii strains MR9 (pHYDA1-ARS2 CRR1; black circles), 41-6 (pHYDA1-ARS2 crr1-3; white circles), and B25 (pHYDA1-ARS2 crr1-3:CRR1; gray circles). B, Strains CC-425 (CRR1; black triangles) and CC-3960 (crr1-2; Kropat et al., 2005; white triangles). C, RNA-hybridization analyses using DIG-labeled HYDA1- and RPL10a-specific probes. Total RNA was isolated from Chlamydomonas cells after 3 h of anaerobic incubation. HYDA1 transcript amounts in all strains were analyzed in three independent experiments. Each one representative result is shown. The quantitative analysis of the signal strength is described in the text.
Under these conditions, HYDA1 transcript levels of both crr1 allelic mutants were lower than in their respective parental strains (Fig. 2C). HYDA1 signals of crr1-3 mutant 41-6 were 30% of the amounts detected in the parental strain MR9. In the complemented strain crr1-3:CRR1 (B25), the HYDA1 signal strength reached 80% of the signal in strain MR9. In strain CC-3960 (crr1-2), HYDA1 mRNA abundance reached 20% of the amount detected in C. reinhardtii strain CC-425 (Fig. 2C). These values were similar to those obtained in Cu-deficient cells. As determined by quantitative real-time PCR (qPCR), relative HYDA1 transcript abundance in Cu-deficient cultures of strain 41-6 was about 33% of HYDA1 mRNA amounts detected in the parental strain MR9 (Supplemental Fig. S3). Castruita et al. (2011) showed that strain CC-3960 (crr1-2) had about 20% of the HYDA1 transcript amounts of its complemented strain when subjected to Cu deficiency.
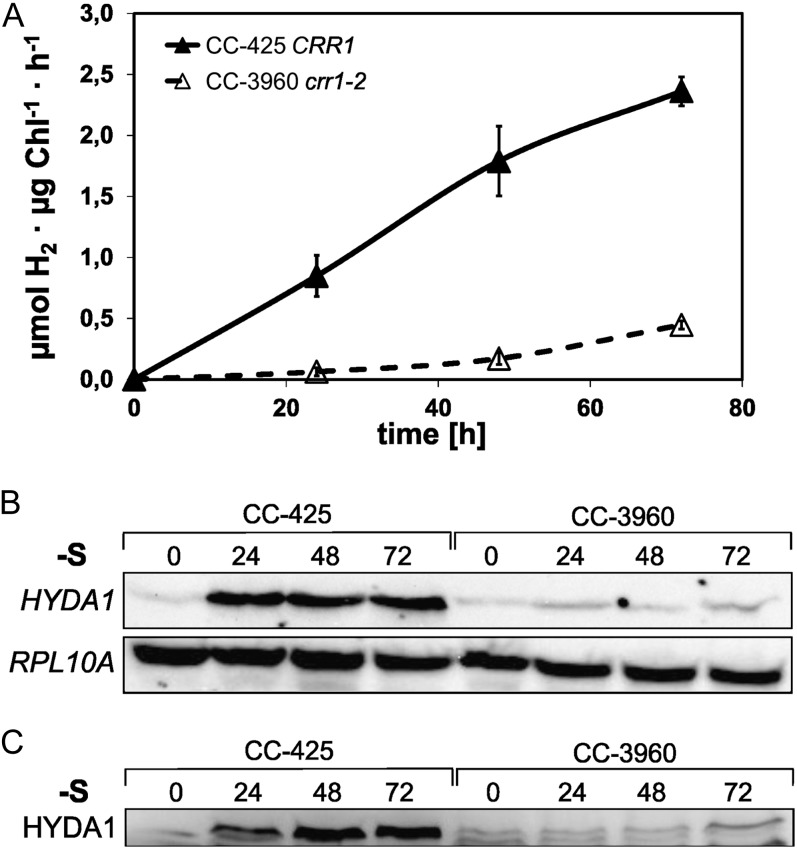
S deprivation is another well-known condition to induce hypoxia and the concomitant changes of gene expression in C. reinhardtii (Melis et al., 2000; Nguyen et al., 2008). Because strain MR9 did not establish hypoxic conditions and became chlorotic (data not shown), S-deprivation experiments were only performed with strains CC-3960 and CC-425. Upon S deficiency, in vitro hydrogenase activity of crr1-2 mutant strain CC-3960 reached about 20% of the rate determined in strain CC-425 (Fig. 3A). HYDA1 transcript and HYDA1 protein amounts of S-deficient C. reinhardtii CC-3960 were significantly lower than in strain CC-425 and reached 20% on average each (Fig. 3, B and C).
Figure 3.
Analysis of in vitro hydrogenase activity (A), HYDA1 transcript (B), and HYDA1 protein (C) levels in crr1 mutant CC-3960 and strain CC-425 upon S deficiency. A, Cells were subjected to S deprivation in sealed flasks in the light. At the indicated time points, cell samples were withdrawn and analyzed by in vitro hydrogenase activity assays. Black triangles: CC-425 (CRR1), white triangles: CC-3960 (crr1-2; Kropat et al., 2005). Average values ± sd of two independent experiments are shown. B, RNA-hybridization analyses using DIG-labeled HYDA1- and RPL10a-specific probes on RNA samples isolated from S-deprived cells after the indicated time points. C, HYDA1 immunoblot analyses using polyclonal anti-C. reinhardtii-HYDA1 antibody on crude protein extracts prepared from S-starved cultures. HYDA1 transcript and HYDA1 protein amounts were analyzed from both strains examined in two independent experiments. One representative result is shown.
HYDA1 Expression Is Inhibited by Mercuric Ions
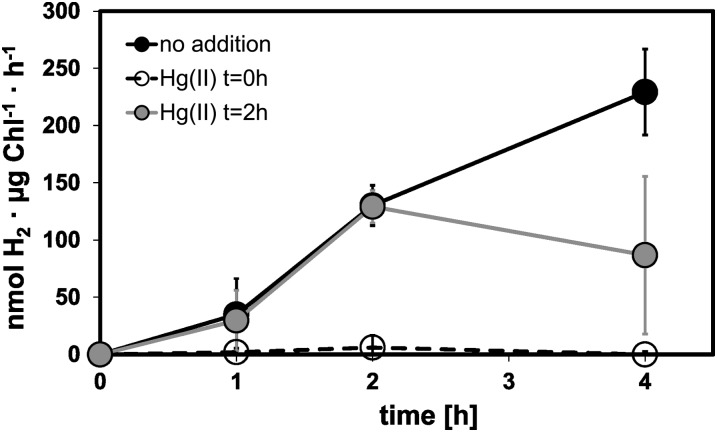
Hg(II) can deactivate the expression of CRR1-dependent genes both in Cu and O2 deficiency (Hill et al., 1991; Quinn et al., 2000, 2002). We investigated the effect of subtoxic concentrations of HgCl2 (10 μm; Quinn et al., 2002) on hydrogenase activity as well as on transcript levels of several marker genes known to be induced upon Cu deficiency and/or anaerobiosis. For this purpose, C. reinhardtii CC-124 cultures were shaded and sealed to let them consume O2 by respiration and establish anaerobic conditions. One culture remained untreated and served as the control. Hg(II) was added to a second culture at the beginning of the incubation (t = 0 h), and to a third culture after 2 h of anaerobic induction, when the cells had already developed hydrogenase activity and accumulated HYDA1 transcript (t = 2 h; Fig. 4; Table I).
Figure 4.
Analysis of the effect of mercuric ions on in vitro hydrogenase activity in C. reinhardtii strain CC-124. Cell suspensions were transferred to sealed flasks in the dark to allow respiratory O2 consumption. One culture was treated with 10 μm HgCl2 from the beginning on (t = 0 h; white circles), and another after 2 h of dark incubation (t = 2 h; gray circles). Untreated cells served as a control (black circles). In vitro hydrogenase activity was determined at the indicated time points. Values shown are means of three independent experiments. Error bars indicate the sd.
Table I. Analysis of the effect of mercuric ions on relative transcript abundances of the genes indicated in the first row.
RNA was isolated from the same C. reinhardtii CC-124 cultures described in the legend of Figure 4, treated with DNase, and used as a template for cDNA synthesis. Relative transcript abundances were calculated according to Pfaffl (2001), using the constitutively expressed CBLP/RCK1 gene (Cre13.g599400 on Phytozome) as reference gene. Each value represents the average ± sd of technical triplicates from two independent biological experiments.
| Treatment | Sampling | HYDA1 | HYDEF | HYDG | PFL1 | FDX5 | CRR1 |
|---|---|---|---|---|---|---|---|
| 0 h | 2.7 ± 0.1 | 0.1 ± 0.01 | 0.2 ± 0.05 | 6.9 ± 1.9 | 0.03 ± 0.02 | 7.7 ± 2.4 | |
| No addition | 2 h | 227.1 ± 74.8 | 9.4 ± 1.3 | 19.3 ± 2.2 | 488.0 ± 60.8 | 69.7 ± 17.3 | 13.7 ± 0.5 |
| Hg(II) at t = 0 h | 2 h | 10.4 ± 2.8 | 0.5 ± 0.02 | 1.7 ± 0.3 | 196.5 ± 51.5 | 0.1 ± 0.01 | 2.3 ± 7.8 |
| Hg(II) at t = 2 h | 2 ha | 82.1 ± 3.5 | 5.3 ± 0.1 | 15.5 ± 1.7 | 217.3 ± 13.7 | 22.8 ± 15.3 | 7.8 ± 1.8 |
| Hg(II) at t = 2 h | 4 h | 11.4 ± 0.7 | 0.9 ± 0.3 | 2.1 ± 0.3 | 194.2 ± 33.9 | 2.4 ± 2.5 | 0.9 ± 0.7 |
Culture samples for RNA isolation were withdrawn before the addition of Hg(II).
Cells exposed to Hg(II) from the beginning of the experiment did not show any in vitro hydrogenase activity. Addition of Hg(II) after 2 h of anaerobic induction resulted in a decrease of the in vitro hydrogenase activity developed by then (Fig. 4). Relative abundances of transcript amounts of the indicated genes (Table I) were analyzed by qPCR. HYDA1 transcript amounts in cells treated with Hg(II) at t = 0 h remained almost as low as in the aerobic control cells. When mercuric ions were added after 2 h, they returned to the control value (Table I). Abundances of FDX5, HYDEF, and HYDG transcripts showed the same patterns as described for the HYDA1 gene in Hg(II)-treated and control cells (Table I). CRR1 transcripts did also respond to Hg(II) in this assay and decreased 4- to 7-fold upon its addition (Table I). The amounts of the PYRUVATE FORMATE LYASE1 (PFL1) encoding PFL1 transcript were not significantly influenced by Hg(II) (Table I).
Two GTAC Motifs Have an Influence of HYDA1 Promoter Activity
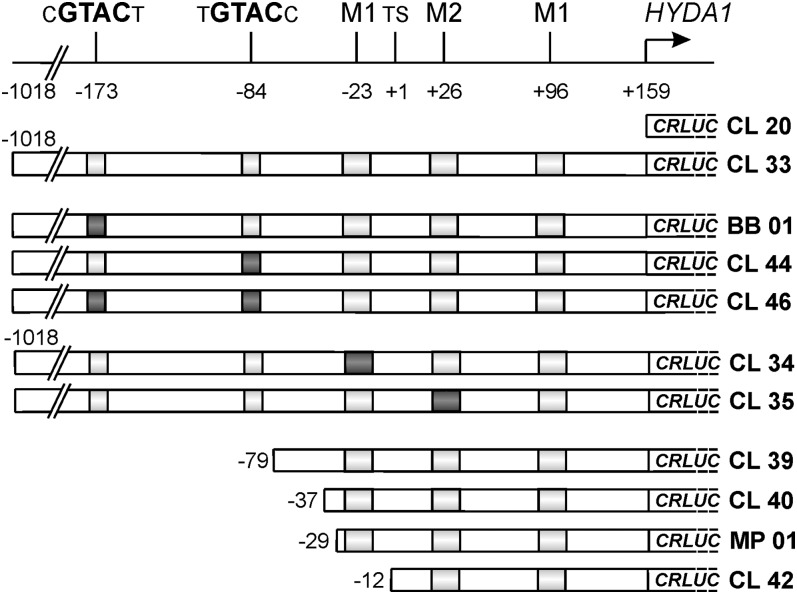
An in silico analysis of the HYDA1 promoter revealed two GTAC sites in position −173 and −84 relative to the transcription start site. Fragments of the putative HYDA1 promoter region in which the GTAC sequences were individually or simultaneously mutagenized were examined regarding their capacity to activate the expression of the luciferase-encoding reporter gene CRLUC (Fig. 5). Furthermore, two additional stretches of bp were mutated. These were tentatively named motif 1 (M1) and motif 2 (M2). Their analysis appeared interesting because M1 (AAGCTCGC, −23 relative to the transcription start site) appeared twice in the putative HYDA1 promoter and the same sequence of bp was identified in the upstream region of HYDA2 of Chlamydomonas moewusii (Kamp et al., 2008; data not shown). M2 (CGCAGGCAC, +26 relative to the transcription start point) is present in the C. reinhardtii and C. moewusii HYDA2 upstream regions. These observations and the fact that both M1 and M2 are located in a region that is able to allow reporter gene expression (Stirnberg and Happe, 2004; and this study, see below) prompted us to introduce mutations in each of these sequences, too (Fig. 5). Furthermore, the HYDA1 upstream region was sequentially truncated to refine the absolute promoter length requirements. Figure 5 shows a schematic overview of the constructs analyzed in this study. C. reinhardtii wild-type strain CC-124 served as the recipient of the respective plasmids. Paromomycin-resistant progenies were subjected to S deprivation in sealed flasks to induce hypoxic conditions (Melis et al., 2000) or to Cu deficiency. Luciferase activity of several randomly chosen colonies of each transformant library was determined under both conditions.
Figure 5.
Scheme of the HYDA1 upstream region (top) and of chimeric constructs of various fragments of the HYDA1 promoter fused to the luciferase-encoding CRLUC reporter gene analyzed in this study (bottom). A schematic of the HYDA1 upstream region from position −1,018 to +159 is shown at the top. Positions and 6-nucleotide sequences of GTAC sites including the 3′ and 5′ adjacent nucleotides are indicated. M1 and M2 refer to tentatively assigned motifs which appeared in the upstream regions of the C. reinhardtii and C. moewusii HYDA2 genes in the exact same sequence (see text for details). An arrow labeled with HYDA1 indicates the translation start codon ATG. Chimeric constructs are depicted below. Names of the constructs are indicated on the right. Length of the HYDA1 promoter fragments are indicated by numbers on the left, which refer to the position relative to the transcription start site (TS). Gray boxes indicate the presence of the wild-type sequences of the GTAC sites and the sequences termed M1 and M2. Black boxes show that the respective sequence has been mutated. GTAC sites were exchanged by GGCC (distal) and TGAC (proximal), respectively. M1 (AAGCTCGC) was changed to CTCATCGC, and M2 (CGCAGGCAC) was altered to ACAAGGCAC.
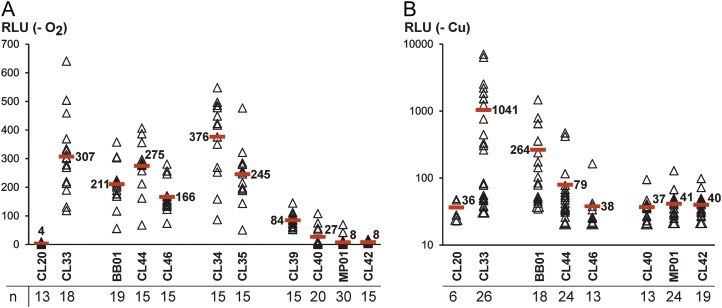
Upon S deficiency, transformants carrying the promoterless CRLUC gene (CL20) exhibited an average luciferase activity of 4 relative light units (RLUs). This was as low as the average luciferase activity detectable in transformants in which CRLUC was under the control of the longest HYDA1 upstream region of 1,177 bp (construct CL33) incubated aerobically in replete medium (3.2 ± 2.1 RLU, n = 15). In contrast, cells carrying construct CL33 reached 307 RLU under S deprivation. Mutation of either GTAC motif caused a moderate decrease of luciferase activity, which accounted to 211 (minus 31%) when the distal GTAC (construct BB01) and 275 (minus 10%) when the proximal GTAC (CL44) was changed. S-starved transformants carrying a construct in which both GTAC sites were mutated exhibited a still-lower average luciferase activity of 166 RLU (minus 46%; Fig. 6A). Nucleotide exchanges within motif M1 (CL34) resulted in a mean of 376 RLU (plus 22%) and an altered motif M2 (CL35) led to moderately lower luciferase activities of the transformants (245 RLU, minus 20%; Fig. 6A). C. reinhardtii strains carrying constructs in which the HYDA1 promoter was truncated showed a strongly reduced luciferase activity. When a HYDA1 upstream region from −79 to +159 (relative to the transcription start point) was fused to the CRLUC gene (CL39), S-deprived algae exhibited 84 RLU (minus 73%) on average, and a further truncation by 40 nucleotides (CL40) led to a mean luciferase activity of 27 RLU (minus 91%). Fragments from −29 (MP01) and −12 (CL42), respectively, to +158 resulted in only 8 RLU in the respective Chlamydomonas strains (Fig. 6A).
Figure 6.
A and B, Luciferase activity determined in C. reinhardtii transformants equipped with the CRLUC gene under the control of various wild-type and mutated HYDA1-promoter fragments. Names of constructs which were used to transform strain CC-124 are written below the x axes and represent the sequences schematically shown in Figure 5. Cells were either subjected to S deficiency in sealed flasks in the light (A) or to Cu deficiency (B). After 72 h of incubation in either condition, 200-μL cell samples were either directly used for the assay (A) or frozen in liquid nitrogen until luciferase activity was measured after addition of coelenterazine. Several transformants of each construct library were analyzed (indicated as n below the graphs). Each triangle represents the RLU value obtained with a single transformant. Red bars indicate the mean values, which are also shown as numbers next to the bars. The average luciferase activity detectable in CL33 transformants incubated aerated in replete medium reached 3.2 ± 2.1 RLU (n = 15; not shown). [See online article for color version of this figure.]
When the algal transformants were subjected to Cu deficiency, the average activity of strains carrying the longest HYDA1 promoter construct (CL33, −1,018 to +158) was 1,041 RLU (Fig. 6B). Mutation of the GTAC motif at position −173 (BB01) led to a decrease of measurable light emission by 75% down to 264 RLU, and when the GTAC motif at position −84 was altered (CL44), it decreased by 92% down to 79 RLU (Fig. 6B). In transformants carrying constructs in which both GTAC motifs were mutated, the average luciferase activity was as low as 38 RLU, which was very close to the activity measured in case of the promoterless construct CL20 (36 RLU; Fig. 6B). The effect of mutations in either or both GTAC sites on HYDA1 promoter activity was therefore much stronger in Cu-deficient than in hypoxic cultures. Strains that were transformed with constructs in which truncated HYDA1 promoter fragments were fused to the CRLUC gene (CL40, MP01, and CL42) exhibited luciferase activity close to the control strain in Cu-deficient medium (Fig. 6B).
The SBP Domain of CRR1 Binds to HYDA1 Promoter Fragments in Vitro
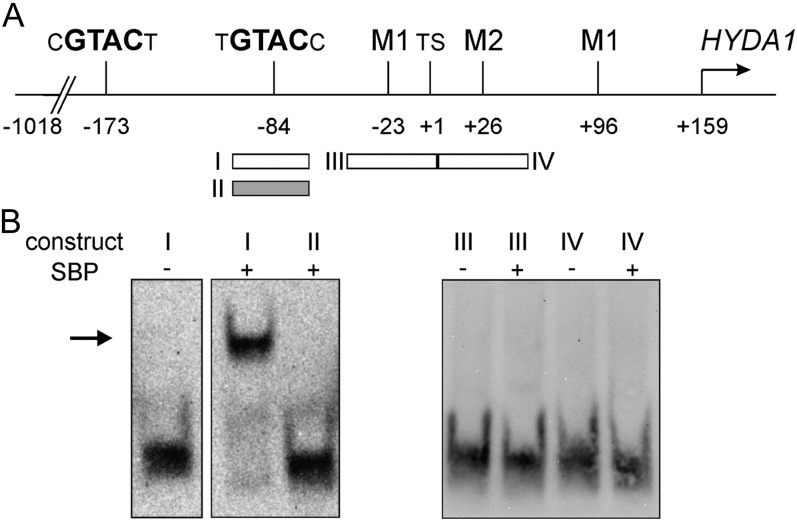
The heterologously produced SBP domain of CRR1 (Sommer et al., 2010) was examined by electrophoretic mobility shift assays (EMSAs) regarding its ability to bind to a GTAC motif of the HYDA1 promoter. A labeled fragment containing the wild-type sequence of the proximal GTAC (position −84; Fig. 7A) showed a shift when incubated with the CRR1-SBP domain, while the same fragment containing a TAGA mutation did not shift (Fig. 7B). As a control it was tested if the SBP domain would be able to cause a shift of fragments carrying motifs M1 or M2, but no shift was observed (Fig. 7B).
Figure 7.
EMSAs analyzing the capacity of the heterologously produced CRR1 SBP domain to bind to HYDA1 promoter fragments. A, Schematic drawing representing the genomic upstream region of the HYDA1 gene as shown in Figure 5. Boxes below this scheme indicate the fragments used for EMSAs in their wild-type form (white boxes) or in a mutated form (gray box; GTAC was exchanged by TAGA). The individual fragments are labeled with Roman numerals, which assign them to the respective EMSA shown in B. B, DIG-labeled fragments I to IV were incubated without (−) or with (+) 200 ng of the recombinant CRR1 SBP domain, separated by native PAGE, and visualized using antidigoxigenin antibody.
DISCUSSION
The Absence of an Active CRR1 Transcriptional Regulator Impairs HYDA1 Gene Expression
Expression of the C. reinhardtii HYDA1 gene encoding the [FeFe]-hydrogenase HYDA1 is up-regulated in anaerobic cultures in the dark as well as in illuminated S-, nitrogen-, or Cu-deprived cells (Happe and Kaminski, 2002; Zhang et al., 2002; Hemschemeier et al., 2008b; Castruita et al., 2011; Philipps et al., 2012). So far, regulatory signals or transcription factors affecting HYDA1 expression were not known. By applying an ARS2 reporter-gene-based screening procedure we isolated Chlamydomonas transformant 41-6 (termed crr1-3). This strain has a truncation of the CRR1 gene and is impaired in HYDA1 promoter activity and HYDA1 transcript accumulation. The well-studied crr1-2 mutant strain CC-3960 (Eriksson et al., 2004; Kropat et al., 2005) also showed lower HYDA1 mRNA amounts and HYDA1 in vitro activity in anaerobic cultures, corroborating the results obtained with strain 41-6.
The transcriptional response of several CRR1 target genes has been shown to be inactivated in the presence of mercuric ions (Hill et al., 1991; Quinn et al., 2002). A recent study showed that Hg(II) interferes with DNA binding of the SBP domain and it was discussed that mercury, which is able to replace Cu in various Cu-binding proteins, acts as a Cu mimic (Sommer et al., 2010). The fact that the addition of HgCl2 to anaerobic C. reinhardtii wild-type cultures prevented HYDA1 transcript accumulation in this study is a further indication that HYDA1 gene expression is affected by a Cu signaling pathway. Both the repressing influence of mercuric ions (this study) and the activating effect of Cu deficiency (Castruita et al., 2011; this study) could also be observed in cases of HYDEF and HYDG transcripts, encoding HYDA1 maturases (Posewitz et al., 2004) and coaccumulating with HYDA1 in all conditions tested so far (Mus et al., 2007; Nguyen et al., 2008). These observations support the shared O2 and Cu responsiveness of genes essential for H2 production in C. reinhardtii.
The effect of mercuric ions on HYDA1 transcript levels observed in this study differed from previous results in which Hg(II) was shown to have no effect on HYDA1 mRNA amounts (Quinn et al., 2002). However, both the experimental setup and the final Hg(II) concentrations varied in the two studies. Since the inactivating influence of mercury on CPX1 and CYC6 transcription was shown to be concentration dependent (Hill et al., 1991; Quinn et al., 2002), our observation of impaired HYDA1 gene expression was probably due to the higher amounts of Hg(II) (10 μm). This mercuric ion concentration has been discussed not to be toxic for Chlamydomonas cells (Hill et al., 1991; Quinn et al., 2002), and the observation that PFL1 transcript levels were not strongly affected by mercury in this study is in line with this view.
GTAC Sites Are Important for Maximum HYDA1 Promoter Activity
CRR1 binds to a GTAC core sequence of Cu or hypoxia response elements (Quinn et al., 2000, 2002). The two GTAC motifs found at positions −173 and −84 relative to the transcription start site of the HYDA1 gene are in a similar proximity to the transcription start site as has been shown for several essential GTAC sites of other CRR1 target genes (Quinn et al., 2000, 2002; Allen et al., 2008; Page et al., 2009; Lambertz et al., 2010). We analyzed the influence of these GTAC motifs on HYDA1 promoter activity in S- and Cu-depleted algal transformants.
In Cu deficiency, mutations in either one or both of the GTAC sites resulted in a pronounced effect. As indicated by luciferase activity, Cu-responsive HYDA1 promoter activity reached 25% of the full promoter activity when the distal and 8% when the proximal GTAC site was mutated. The HYDA1 promoter in which both GTAC motifs were mutated had no significant activity at all. Since the absence of GTAC sites should mimic the effect of CRR1 deficiency, the latter result is not consistent with the moderate, but significant increase of HYDA1 transcript levels in the Cu-deficient crr1-2 mutant CC-3960 (Castruita et al., 2011) or strain 41-6 (crr1-3, this study). A simple explanation for the discrepancy might be an insufficient resolution or sensitivity of the CRLUC-based reporter gene assay (Shao and Bock, 2008).
Upon S deficiency, HYDA1 promoter activity was less impaired by mutations of the GTAC sites. Only when both GTAC motifs were altered, a decrease of the luciferase activity by 50% was observed. Mutations in individual GTAC motifs resulted in only a 31% (distal) or 10% (proximal) decrease of RLU values. These results indicate that the factors and/or signaling pathways influencing HYDA1 promoter activity differ in hypoxia versus Cu deficiency.
Activation of HYDA1 Expression Requires Additional Regulatory Components
By comparing transcript amounts under Cu and O2 deficiency (Quinn et al., 2002; Kropat et al., 2005) as well as number and importance of GTAC motifs in the promoter regions of CRR1 targets (Quinn et al., 2000, 2002; Allen et al., 2008; Page et al., 2009; Lambertz et al., 2010), it has been noted before that the regulation of CRR1-activated genes does not follow a simple pattern. For example, CPX1 transcription seems to be more responsive to anaerobiosis than to the absence of Cu, while it is the opposite in case of the CYC6 gene (Quinn et al., 2002). For Cu-dependent expression of the CPX1 gene, a single Cu Response Element is sufficient, but an additional GTAC motif termed Hypoxia Response Element is necessary for hypoxia responsiveness of CPX1 (Quinn et al., 2002). A previous elaborate study examining the function of individual domains of CRR1 showed that the accumulation of CYC6 transcript under anaerobic conditions depended on the presence of the Cys-rich C terminus, and, to a lower extent, an ankyrin-repeat region of CRR1, while this was not the case upon Cu depletion (Sommer et al., 2010). A conclusion that can be drawn is that CRR1-dependent gene expression is multifaceted and that triggering signals are integrated in different ways.
It has been shown recently that the effect of CRR1 on transcription of CYC6, CPX1, and CRD1 is mediated by an opening of the chromatin structure of the respective promoter regions (Strenkert et al., 2011). Due to the constitutively open chromatin state of the CRD1 promoter under noninducing conditions, the authors suggested that an additional unknown factor is probably involved in CRD1 gene expression (Strenkert et al., 2011).
The results reported here show that CRR1 is involved in HYDA1 promoter activation and HYDA1 transcript accumulation, but they also indicate that additional factors specific for each particular environmental condition influence its transcription. In luciferase assays, only the truncation of the HYDA1 promoter down to −79 or −37 resulted in a strong (minus 70% and minus 90%, respectively) decrease of promoter activity in hypoxic C. reinhardtii transformants. Therefore, this region might be vital for the binding of an additional factor specific for the condition of O2 limitation.
The regulation of one gene by distinct cis-acting elements and environmental stimuli is a common phenomenon. The Arabidopsis (Arabidopsis thaliana) ADH1 gene encoding alcohol dehydrogenase is induced upon cold and dehydration stress and by hypoxia, and some of the critical ADH1 promoter elements were shown to be specific for individual stress conditions (Dolferus et al., 1994). In C. reinhardtii, the HIGH-CO2-INDUCIBLE 43-KD PROTEIN/FE ASSIMILATION1 gene is regulated both under high-CO2- and iron-deficiency conditions, which act independently via two different response elements (Baba et al., 2011). Multiple stimuli acting on the HYDA1 promoter could help adjusting HYDA1 protein levels to metabolic needs. The identification and biochemical characterization of the putative additional factor(s) binding to the HYDA1 promoter might throw light on the orchestrated Cu- and O2-responsive signaling in C. reinhardtii.
MATERIALS AND METHODS
Organisms and Growth Conditions
Chlamydomonas reinhardtii strains CC-124, CC-425, and crr1-2 mutant strain CC-3960 (Eriksson et al., 2004; Kropat et al., 2005) were obtained from the Chlamydomonas Resource Center, University of Minnesota. Strain MR9 was generated as described before (Stirnberg and Happe, 2004). It resulted from cotransforming C. reinhardtii 388 (cw15 arg7 nia1 mt−) with plasmid pARG7.8cos (Debuchy et al., 1989) and plasmid pMR9. In the latter, a HYDA1 promoter region from position −1,020 to +158 relative to the transcription start site as indicated in Stirnberg and Happe (2004), is fused to the promoterless arylsulfatase-encoding ARS2 gene in plasmid pJD54 (Davies et al., 1992). C. reinhardtii cells were grown photoheterotrophically in Tris-acetate-phosphate (TAP) medium, pH 7.2 (Harris, 1989, 2009) at a light intensity of 80 μmol photons m−2 s−1 and a temperature of 20°C.
Cu deficiency of C. reinhardtii pHYDA1:CRLUC transformants generated as described below was achieved by growing the cells once in TAP ENEA2 medium as reported previously (Ferrante et al., 2008). Then, a main culture of 20 mL in 50-mL Sarstedt tubes was inoculated with up to 1 mL of this preculture and the Cu-chelator triethylenetetramine was added to the medium to reach a final concentration of 50 μm. The cells were incubated at a light intensity of 80 μmol photons m−2 s−1 until they reached a chlorophyll content of 15 μg mL−1. S deprivation of strains CC-425 and CC-3960 was achieved as described previously (Hemschemeier et al., 2009) using a modified TAP medium, in which sulfate salts were replaced by chloride compounds. S starvation of C. reinhardtii pHYDA1:CRLUC transformants was done using the same medium and incubating the strains in 12-mL headspace bottles (Fisher Scientific) as described in detail in Lambertz et al. (2010).
Anaerobic Induction and Treatment of Cells with Mercuric Chloride
Anaerobic induction by flushing concentrated C. reinhardtii cell suspensions with nitrogen gas and in vitro hydrogenase activity assays in lysed cell samples using methyl viologen as an artificial electron donor were conducted as described previously (Hemschemeier et al., 2009). In case of the experiments analyzing the effect of Hg(II), C. reinhardtii strain CC-124 was grown until the culture had reached a chlorophyll content of 20 μg mL−1. Cells were harvested by centrifugation, resuspended in the same volume of fresh TAP medium, and transferred to squared glass bottles sealed by Suba seals (Sigma-Aldrich). The culture flasks were then incubated on a magnetic stirrer in the dark to allow respiratory O2 consumption by the cells. When indicated, mercuric chloride (HgCl2) was added to the cells to a final concentration of 10 μm either from the beginning on or after the cells had been incubated for 2 h.
Transformation of C. reinhardtii
C. reinhardtii cells were transformed using the glass bead method (Kindle, 1990). Strain MR9 was transformed using 1 μg of the APHVIII-encoding paromomycin resistance cassette derived from plasmid pSL18 (Sizova et al., 2001) by KpnI and StuI digestion. After transformation, cells were streaked on TAP agar plates without paromomycin and incubated in low light (15 μmol photons m−2 s−1) for 24 h. Then, the agar blocks were transferred to petri dishes containing filter paper soaked with a paromomycin solution that resulted in a final concentration of 5 μg mL−1 paromomycin in the agar. For generation of C. reinhardtii transformants carrying various HYDA1:CRLUC reporter gene fusion constructs (see below), wild-type CC-124 was treated with autolysin (Harris, 1989, 2009) prior to transformation with ScaI-linearized plasmids.
Screening for Transformants with Impaired HYDA1 Promoter Activity
Ten-thousand transformants resulting from transforming C. reinhardtii strain MR9 with the paromomycin resistance cassette were tested for impaired HYDA1 promoter activity by the following screening protocol: The algal colonies were grown on TAP agar plates containing 5 μg mL−1 paromomycin for 7 d. Each plate included one colony of C. reinhardtii CC-124 transformed with the paromomycin cassette to ensure that the endogenous arylsulfatase, which is produced in S deficiency (de Hostos et al., 1988), was not produced. After growth of the colonies, the plates were transferred to an anaerobic tent to activate the HYDA1 promoter and 25 μL of a 3-mm solution of the artificial arylsulfatase substrate X-SO4 were dropped on each colony. Twenty-four hours later, colonies were checked for a blue staining, which would result from cleavage of X-SO4 by arylsulfatase (Davies et al., 1992). Colonies that did not show a blue staining were candidates for a loss of HYDA1 promoter activity and were picked for further analyses. To ensure that their phenotype was not due to a general inability to generate active arylsulfatase, the candidate strains were first subjected to S starvation and tested for native arylsulfatase activity. Additionally, the presence of an intact junction between the HYDA1 upstream region and the ARS2 gene was analyzed by PCR using the oligonucleotides Ars02 (5′-GTCGACGGTACCGGTCGCGCATAGGCAAGCTC-3′) and HydA01 (5′-GTCGACGGTACCGGTCGCGCATAGGCAAGCTC-3′).
Genetic Characterization and Complementation of Mutant Strain 41-6
Genomic DNA was isolated by a method described previously (Newman et al., 1990). Southern-blot analyses according to standard techniques (Sambrook et al., 1989) were performed to determine the integration frequency of the paromomycin resistance cassette. A probe specific for the APHVIII gene was generated by PCR amplification including PCR DIG labeling mix (Roche) using pSL18 as a template and applying the oligonucleotides aphVIII (5′-ACGATGCGTTGCCTGC-3′) and aphVIIIrev (5′-GAGACTGCGATCGAACG-3′). The integration site of the APHVIII cassette was determined by inverse PCR: 500 ng of genomic DNA were digested with PstI. After inactivation of the enzyme, religation was performed at 16°C overnight using T4 DNA-Ligase (Fermentas) according to the manufacturer’s recommendations. Inverse PCR was conducted using 1 μL of 20-fold-diluted ligation mixture in a 25-μL PCR reaction mixture using Biotools Taq DNA polymerase (Biotools) as indicated in the manual, except that 2% (v/v) dimethyl sulfoxide was included. The PCR program was as follows: 5 min 96°C, followed by 35 cycles of 1 min 96°C, 1 min 60°C, and 4 min 72°C and a final extension step of 10 min at 72°C. Oligonucleotides used for the first PCR were Paro07 (5′-GAGTGTTCCGCGGCGTTCC-3′) and 5′GSP1 (5′-CATACGCACCAATCATGTCAAGCCTCAG-3′), and a nested PCR was performed using Paro06 (5′-TACCGGCTGTTGGACGAGTTC-3′) and 5′GSP2 (5′-GTCTGCCTGACAGGAAGTGAACGCATGT-3′). For complementation of C. reinhardtii strain 41-6, the bacterial artificial chromosome clone number 1E12 (PTQ139 at JGI v4.0) was digested with EcoRI to generate a 9,201-bp fragment that included the CRR1 gene and a fragment of an unknown gene (JGI v4.0 ID519527, au5.g9305_t1). The fragment was cloned into EcoRI linearized pBluescript II SK+ (Stratagene) resulting in plasmid pMP1. pMP1 was then digested with KpnI and EcoRI to generate a 7,619-bp fragment carrying only the CRR1 gene flanked by 171 bp of genomic DNA at the 5′ and the 3′ end. This fragment was cloned into KpnI and EcoRI digested pBluescript II SK+ resulting in plasmid pMP2. Plasmid pMP2 was then used for cotransforming C. reinhardtii strain 41-6 with ScaI-linearized plasmid pHyg, which contains a hygromycin resistance cassette (Berthold et al., 2002). Transformants were selected on agar plates containing 5 μg mL−1 hygromycin B (Fluka). Grown colonies were tested for their regained ability to produce arysulfatase under anaerobic conditions as described above.
Construction of Plasmids for HYDA1 Promoter Analyses
The HYDA1 promoter region used to generate construct CL33 (from position −1,018 to +158 relative to the transcription start site) was amplified from genomic DNA by PCR using Pfu polymerase (Stratagene) and oligonucleotides HYDA1_rev and CL33_for listed in Table II. Truncated fragments were amplified using CL33 as a template and using oligonucleotide HYDA1_rev and construct-specific oligonucleotides (Table II). Mutations within some fragments of the HYDA1 promoter were inserted by site-directed mutagenesis or by quick change (QC) site-directed mutagenesis (Zheng et al., 2004; Liu and Naismith, 2008) using oligonucleotides carrying the desired mutation (Table II). For each construct designed by site-directed mutagenesis (CL34, CL35, and BB01), two first-step PCR reactions were conducted using oligonucleotides 1 and A, and 2 and B, respectively (Table II). Oligonucleotides 1 and 2 were used for amplifying all constructs. Oligonucleotides A and B contained the respective mutation and were specific for each construct. In each case, the two amplificates overlapped at the site of the mutagenized sequence and were used as templates in the presence of the terminal oligonucleotides 1 and 2 in a final PCR reaction to synthesize the whole HYDA1 promoter fragment. HYDA1 promoter fragments were digested with XhoI and ApaI and inserted into vector pCL20 cut by the same endonucleases. The generation of pCL20, which contains both the paromomycin resistance cassette amplified by PCR from pSL18 (Sizova et al., 2001) and the promoterless CRLUC gene encoding the luciferase of Renilla reniformis (RLUC; Fuhrmann et al., 2004), was described in detail before (Lambertz et al., 2010). Constructs CL44 and CL46 were generated by applying QC-PCR. In both cases, oligonucleotides QC_for and QC_rev (Table II) were applied, which introduced a mutation in the proximal GTAC motif of the HYDA1 promoter. Using CL33 as a template resulted in the generation of construct CL44, and template BB01 led to construct CL46 having mutations in both GTAC motifs. All constructs were sequenced at the sequencing facility at the Ruhr University of Bochum, Germany (Department of Biochemistry I, Receptor Biochemistry).
Table II. List of oligonucleotides used for amplification and mutation of HYDA1 promoter fragments for generation of reporter gene constructs as well as oligonucleotides used in EMSAs.
ApaI and XhoI restriction sites are underlined. Inserted mutations are indicated in boldface type.
| Name | Template | Oligonucleotide |
|---|---|---|
| Constructs created by PCR | ||
| HYDA1_rev: GCCTCGAGCTTGTCGCGTCTACGATATTAG | ||
| CL33 | gDNA | CL33_for: GGGGCCCGATGATATGGATCGTCGTCTGGTG |
| CL39 | CL33 | CL39_for: GGGGGCCCGCCGAGTGTCCGCTGCATTC |
| CL40 | CL33 | CL40_for: ATGGGCCCGGTCGCGCATAGGCAAGCTC |
| MP01 | CL33 | MP01_for: TGGGGCCCCATAGGCGTACTCGCAAATGCTGT |
| CL42 | CL33 | CL42_for: CTGGGCCCTGCTGTCAGCTTATCTTACAT |
| Constructs created by site-directed mutagenesis | ||
| 1: SDM_UP: GGGGCCCGATGATATGGATCGTCGTCTGGTG | ||
| 2: SDM_RP: GCCTCGAGCTTGTCGCGTCTACGATATTAG | ||
| CL34 | CL33 | A: SDM_pCL34_A: ACAGCATTTGCGATGAGGCCTAT |
| B: SDM_pCL34_B: CATAGGCCTCATCGCAAATGCTGT | ||
| CL35 | CL33 | A: SDM_pCL35_A: TTGAGGCTAGTGCCTTGTCGAGTGTT |
| B: SDM_pCL35_B: AACACTCGACAAGGCACTAGCCTCAA | ||
| BB01 | CL33 | A: SDM_pBB01_A: CTCCTATACTAGGCCGCATTGTTC |
| B: SDM_pBB01_B: GGAACAATGCGGCCTAGTATAGGA | ||
| Constructs created by QC-PCR | ||
| CL44 | CL33 | QC_for: GTTGACCGCCGAGTGTCCGCTGCATTC |
| QC_rev: CACTCGGCGGTCAACCCCGTCAGCAC | ||
| CL46 | BB01 | QC_for: GTTGACCGCCGAGTGTCCGCTGCATTC |
| QC_rev: CACTCGGCGGTCAACCCCGTCAGCAC | ||
| Fragments used in EMSAs | ||
| I | I-pHYDA1for: GACGGGGTGTACCGCCGAGTGTC | |
| I-pHYDA1rev: GACACTCGGCGGTACACCCCGTC | ||
| II | II-pHYDA1for: GACGGGGTTAGACGCCGAGTGTC | |
| II-pHYDA1rev: GACACTCGGCGTCTAACCCCGTC | ||
| III | III-pHYDA1for: GGTCGCGCATAGGCAAGCTCGCAAATGCTGTCAGCT | |
| III-pHYDA1rev: AGCTGACAGCATTTGCGAGCTTGCCTATGCGCGACC | ||
| IV | IV-pHYDA1for:TGAACACACAAACACTCTCGCAGGCACTAGCCTCAA | |
| IV-pHYDA1rev: TTGAGGCTAGTGCCTGCGAGAGTGTTTGTGTGTTCA | ||
Luciferase Activity Assay
Luciferase activity of Cu-deficient transformants carrying various pHYDA1:CRLUC constructs was determined in 1.5-mL cell suspension aliquots withdrawn from the cultures after 72 h of Cu starvation. The cell aliquots were sonicated three times for 30 s in an ultrasonic bath (Transsonic T 460, Elma) and then frozen in liquid nitrogen until analyzing luciferase activity. The latter was determined in 200-μL lysate aliquots transferred to 96-well plates (Berthold Technologies, microplate 96 well, white). Light emission was measured using a photon-counting microplate luminometer (orion microplate luminometer, Berthold detection systems, operated by software simplicity 2.1). Before determining luciferase activity, background light emission was measured for 10 s. Afterward, 0.5 μL of 2 mm coelenterazine (pjk-gmbH) in methanol was added to cell lysates and light emission was again recorded for 10 s. In S-deficient transformants, luciferase activity was determined as described above after 72 h of nutrient starvation, except that the cells were withdrawn from the incubation flask by a syringe and directly measured as described. To ensure that the transformants experienced S deficiency and established a hydrogen metabolism, the concentration of H2 in head space of each culture vessel was determined (data not shown). In case of Cu deficiency, protein samples were removed from randomly chosen transformants and analyzed regarding CYC6 protein accumulation (Supplemental Fig. S4).
Heterologous Expression of the CRR1 SBP Domain and EMSA
Heterologous expression and purification of the C. reinhardtii CRR1 SBP domain were conducted as described previously (Birkenbihl et al., 2005; Lambertz et al., 2010) using plasmid pet15SBP, which contains a complementary DNA (cDNA) fragment encoding amino acids 428 to 524 of the CRR1 polypeptide (Kropat et al., 2005). Vector pet15SBP was kindly donated by Dr. Frederik Sommer, Max-Planck-Institute for Molecular Plant Physiology.
EMSAs oligonucleotides corresponding to wild-type or mutated parts of the HYDA1 promoter were obtained from Sigma-Aldrich (Table II). EMSAs were conducted as described in Lambertz et al. (2010), using the second-generation digoxigenin (DIG) gel shift kit from Roche.
RNA and Protein Analyses
Total RNA was isolated as described before (Schloss et al., 1984), except that RNA was precipitated with 1 volume of 8 m LiCl overnight. For cDNA synthesis, total RNA was first treated with DNase (TURBO DNase from Ambion/Applied Biosystems) according to the manual. Afterward, cDNA was synthesized from 20 μg of DNase-treated RNA using oligo(dT18) oligonucleotides and M-MLV reverse transcriptase (Invitrogen) as recommended by the manufacturer.
qPCR reactions were performed with the peqGOLD Taq DNA polymerase kit (PeqLab) using buffer S and 5x Enhancer solution included in the kit in 20 μL reactions. The latter contained 2 μL 10-fold diluted cDNA, 2 μL 10× SYBR-mix (0.1% [v/v] SYBR Green I nucleic acid gel stain from Sigma-Aldrich, 1% [v/v] Tween20; 1 mg per mL bovine serum albumin; 50% [v/v] dimethyl sulfoxide), 6 pmol of each forward and reverse oligonucleotides (Table III), and 1 μL Taq (1 unit/μL). Each sample was analyzed in technical triplicate using DNA Engine Opticon 2 from MJ Research Inc. and a PCR program applying the following steps: 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 63°C for 20 s, and 72°C for 25 s. Fluorescence was measured after each cycle at 72°C. Melting curves were performed from 70°C to 95°C with plate reads every 0.5°C. Transcript levels of CBLP/RCK1 (Cre13.g599400), which were constant in all samples, were used as a reference to calculate relative transcript abundances (Pfaffl, 2001).
Table III. Oligonucleotides used for qPCR analyses.
Gene and locus names refer to the annotation on Phytozome.
| Gene (locus) Name | Forward Oligonucleotide | Reverse Oligonucleotide |
|---|---|---|
| RCK1 (Cre13.g599400) | GCCACACCGAGTGGGTGTCGTGCG | CCTTGCCGCCCGAGGCGCACAGCG |
| RPL10a (Cre02.g101350) | GCTTCGGACTCCGTCATCAA | CCATGCACAGCACCTTCTTC |
| HYDA1 (Cre03.g199800) | GCGATTGACGTTGGTGTAGG | GTGCTTACAAGCGGCTGATG |
| HYDEF (Cre06.g296750) | CTGCATGATTGACGCCCAGA | AGGCGGCCCAAGAGAAGAAC |
| HYDG (Cre06.g296700) | CCGGTCAATGACGCTGACTT | ATCTGGCTCATGCCGCACTT |
| PFL1 (Cre01.g044800) | CAAGTACGGCAACGACGATG | ACGTTGGAGGTGATGGTCAG |
| FDX5 (Cre17.g700950) | CGGCTTCATCCTCATGTGCT | ACGCTGACACGAATGGTACG |
| CRR1 (Cre02.g125250) | TGCGTGTTTGTTGTTTCAGG | GCCAGGTGTGATGGAGAGAC |
RNA hybridization analyses were conducted according to standard techniques (Sambrook et al., 1989). Hybridization signals of the RPL10a transcript were used for normalization. RPL10a encodes ribosomal protein L10a, a component of the 60S large subunit of cytosolic 80S ribosomes. HYDA1- and RPL10a-specific probes were generated from cloned fragments by in vitro transcription using DIG-labeled dUTPs (Roche). The HYDA1-specific probe was deduced from the 3′-untranslated region of the transcript and amplified from cDNA using oligonucleotides hydA1-5-1629 (5′-GAGGAGAAGGACGAGAAGAA-3′) and hydA1-3-1883 (5′-TTAGGCGTAGGTACTGGCA-3′). The RPL10a fragment was generated applying oligonucleotides L10a-1-5-445 (5′-CCAAGTGCAGCATCAAGTTC-3′) and L10a-1-3-596 (5′-CACGTTCTGCCAGTTCTTCT-3′).
Crude protein extracts were prepared from 1 mL of cell culture harvested by centrifugation at maximum speed for 1 min. The pellet was resuspended in 5× SDS-PAGE sample buffer (0.25 m Tris-HCl pH 8.0, 25% [v/v] glycerol, 7.5% [w/v] SDS, 0.25 mg mL−1 bromphenol blue, 12.5% [v/v] β-mercaptoethanol). After denaturation of proteins at 95°C for 5 min, samples were stored at 4°C. SDS-PAGE and blotting were performed as described previously (Hemschemeier et al., 2008a), except that proteins were transferred to polyvinylidene fluoride membranes. HYDA1 and FDX5 were detected by polyclonal anti-C. reinhardtii-HYDA1 (Happe et al., 1994), and anti-C. reinhardtii-FDX5 antibody, respectively (Jacobs et al., 2009). CYC6 immunoblot analyses were conducted using anti-CYC6 antibody, which was kindly donated by S. Merchant, University of California, Los Angeles.
Chemiluminescence of RNA and protein hybridization analyses from each three independent experiments was measured using the FluorChem 8800 system and quantified with AlphaEase FCTM Software version 3.1.2 (α Innotech Corporation) according to the manufacturer’s instructions. For each RNA sample, the Integrated Density Value, (sum of all pixel values after background correction) of the HYDA1 signal was related to the respective RPL10a value.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XP_001691972 (ARS2), XP_001701729 (CPX1), XP_001692557 (CRD1), XP_001695062 (Cre02.g125200), XP_001694940 (CRR1), XP_001698242 (CYC6), XP_001691603 (FDX5), XP_001693376 (HYDA1), XP_001694503 (HYDA2), XP_001691465 (HYDEF), XP_001691319 (HYDG), XP_001692808 (PETF), XP_001689719 (PFL1), XP_001701208 (PFR1), XP_001698065 (RCK1), and XP_001699807 (RPL10a).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Scheme of the screening which identified Chlamydomonas strain 41-6 (crr1-3).
Supplemental Figure S2. Determination of the integration frequency of the paromomycin resistance cassette in strain 41-6 (crr1-3).
Supplemental Figure S3. HYDA1 transcript abundance upon Cu deficiency.
Supplemental Figure S4. CYC6 protein amounts in Cu-deprived CRLUC-expressing transformants.
Supplementary Material
Acknowledgments
We are very thankful for the generous gift of anti-C. reinhardtii-CYC6 antibody from Sabeeha Merchant (University of California, Los Angeles, CA). We would also like to thank Sabeeha and Janette Kropat (University of California, Los Angeles, CA) for fruitful discussions. Frederik Sommer (Max-Planck-Institute for Molecular Plant Physiology, Potsdam-Golm, Germany) kindly donated the plasmid for heterologous production of CRR1 SBP domain. Not at least, we thank Klaus Überla, Thomas Grunwald, and Bastian Grewe (Institute for Hygiene and Microbiology, Ruhr-University of Bochum, Germany) for the opportunity to utilize the luminometer.
Glossary
- H2
molecular hydrogen
- O2
molecular dioxygen
- Cu
copper
- X-SO4
5-bromo-4-chloro-3-indolyl sulfate
- qPCR
quantitative real-time PCR
- S
sulfur
- M1
motif 1
- M2
motif 2
- RLUs
relative light units
- EMSAs
electrophoretic mobility shift assays
- TAP
Tris-acetate-phosphate
- QC
quick change
- DIG
digoxigenin
- cDNA
complementary DNA
References
- Allen MD, Kropat J, Merchant SS. (2008) Regulation and localization of isoforms of the aerobic oxidative cyclase in Chlamydomonas reinhardtii. Photochem Photobiol 84: 1336–1342 [DOI] [PubMed] [Google Scholar]
- Baba M, Hanawa Y, Suzuki I, Shiraiwa Y. (2011) Regulation of the expression of H43/Fea1 by multi-signals. Photosynth Res 109: 169–177 [DOI] [PubMed] [Google Scholar]
- Berthold P, Schmitt R, Mages W. (2002) An engineered Streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153: 401–412 [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Jach G, Saedler H, Huijser P. (2005) Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol 352: 585–596 [DOI] [PubMed] [Google Scholar]
- Castruita M, Casero D, Karpowicz SJ, Kropat J, Vieler A, Hsieh SI, Yan W, Cokus S, Loo JA, Benning C, et al. (2011) Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23: 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Weeks DP, Grossman AR. (1992) Expression of the arylsulfatase gene from the beta 2-tubulin promoter in Chlamydomonas reinhardtii. Nucleic Acids Res 20: 2959–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos EL, Togasaki RK, Grossman A. (1988) Purification and biosynthesis of a derepressible periplasmic arylsulfatase from Chlamydomonas reinhardtii. J Cell Biol 106: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R, Purton S, Rochaix JD. (1989) The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J 8: 2803–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DJ. (1998) Earth’s early biosphere. Gravit Space Biol Bull 11: 23–30 [PubMed] [Google Scholar]
- Dolferus R, Jacobs M, Peacock WJ, Dennis ES. (1994) Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol 105: 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Moseley JL, Tottey S, Del Campo JA, Quinn J, Kim Y, Merchant S. (2004) Genetic dissection of nutritional copper signaling in chlamydomonas distinguishes regulatory and target genes. Genetics 168: 795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante P, Catalanotti C, Bonente G, Giuliano G. (2008) An optimized, chemically regulated gene expression system for Chlamydomonas. PLoS ONE 3: e3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestier M, King P, Zhang LP, Posewitz M, Schwarzer S, Happe T, Ghirardi ML, Seibert M. (2003) Expression of two [Fe]-hydrogenases in Chlamydomonas reinhardtii under anaerobic conditions. Eur J Biochem 270: 2750–2758 [DOI] [PubMed] [Google Scholar]
- Fuhrmann M, Hausherr A, Ferbitz L, Schödl T, Heitzer M, Hegemann P. (2004) Monitoring dynamic expression of nuclear genes in Chlamydomonas reinhardtii by using a synthetic luciferase reporter gene. Plant Mol Biol 55: 869–881 [DOI] [PubMed] [Google Scholar]
- Gfeller RP, Gibbs M. (1984) Fermentative metabolism of Chlamydomonas reinhardtii: I. Analysis of fermentative products from starch in dark and light. Plant Physiol 75: 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghysels B, Franck F. (2010) Hydrogen photo-evolution upon S deprivation stepwise: an illustration of microalgal photosynthetic and metabolic flexibility and a step stone for future biotechnological methods of renewable H(2) production. Photosynth Res 106: 145–154 [DOI] [PubMed] [Google Scholar]
- Godman JE, Molnár A, Baulcombe DC, Balk J. (2010) RNA silencing of hydrogenase(-like) genes and investigation of their physiological roles in the green alga Chlamydomonas reinhardtii. Biochem J 431: 345–351 [DOI] [PubMed] [Google Scholar]
- Goldfine H. (1965) The evolution of oxygen as a biosynthetic reagent. J Gen Physiol (Suppl) 49: 253–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happe T, Kaminski A. (2002) Differential regulation of the Fe-hydrogenase during anaerobic adaptation in the green alga Chlamydomonas reinhardtii. Eur J Biochem 269: 1022–1032 [DOI] [PubMed] [Google Scholar]
- Happe T, Mosler B, Naber JD. (1994) Induction, localization and metal content of hydrogenase in the green alga Chlamydomonas reinhardtii. Eur J Biochem 222: 769–774 [DOI] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego
- Harris EH. (2009) The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use, Ed 2, Vol 1. Academic Press, San Diego
- Hemschemeier A, Fouchard S, Cournac L, Peltier G, Happe T. (2008a) Hydrogen production by Chlamydomonas reinhardtii: an elaborate interplay of electron sources and sinks. Planta 227: 397–407 [DOI] [PubMed] [Google Scholar]
- Hemschemeier A, Happe T. (2005) The exceptional photofermentative hydrogen metabolism of the green alga Chlamydomonas reinhardtii. Biochem Soc Trans 33: 39–41 [DOI] [PubMed] [Google Scholar]
- Hemschemeier A, Happe T. (2011) Alternative photosynthetic electron transport pathways during anaerobiosis in the green alga Chlamydomonas reinhardtii. Biochim Biophys Acta 1807: 919–926 [DOI] [PubMed] [Google Scholar]
- Hemschemeier A, Jacobs J, Happe T. (2008b) Biochemical and physiological characterization of the pyruvate formate-lyase Pfl1 of Chlamydomonas reinhardtii, a typically bacterial enzyme in a eukaryotic alga. Eukaryot Cell 7: 518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemschemeier A, Melis A, Happe T. (2009) Analytical approaches to photobiological hydrogen production in unicellular green algae. Photosynth Res 102: 523–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KL, Li HH, Singer J, Merchant S. (1991) Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6: analysis of the kinetics and metal specificity of its copper-responsive expression. J Biol Chem 266: 15060–15067 [PubMed] [Google Scholar]
- Jacobs J, Pudollek S, Hemschemeier A, Happe T. (2009) A novel, anaerobically induced ferredoxin in Chlamydomonas reinhardtii. FEBS Lett 583: 325–329 [DOI] [PubMed] [Google Scholar]
- Kamp C, Silakov A, Winkler M, Reijerse EJ, Lubitz W, Happe T. (2008) Isolation and first EPR characterization of the [FeFe]-hydrogenases from green algae. Biochim Biophys Acta 1777: 410–416 [DOI] [PubMed] [Google Scholar]
- Kindle KL. (1990) High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 87: 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J, Tottey S, Birkenbihl RP, Depège N, Huijser P, Merchant S. (2005) A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci USA 102: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertz C, Hemschemeier A, Happe T. (2010) Anaerobic expression of the ferredoxin-encoding FDX5 gene of Chlamydomonas reinhardtii is regulated by the Crr1 transcription factor. Eukaryot Cell 9: 1747–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertz C, Leidel N, Havelius KG, Noth J, Chernev P, Winkler M, Happe T, Haumann M. (2011) O2 reactions at the six-iron active site (H-cluster) in [FeFe]-hydrogenase. J Biol Chem 286: 40614–40623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Naismith JH. (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A. (2007) Photosynthetic H2 metabolism in Chlamydomonas reinhardtii (unicellular green algae). Planta 226: 1075–1086 [DOI] [PubMed] [Google Scholar]
- Melis A, Zhang L, Forestier M, Ghirardi ML, Seibert M. (2000) Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol 122: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuser JE, D’Adamo S, Jinkerson RE, Mus F, Yang W, Ghirardi ML, Seibert M, Grossman AR, Posewitz MC. (2012) Genetic disruption of both Chlamydomonas reinhardtii [FeFe]-hydrogenases: insight into the role of HYDA2 in H2 production. Biochem Biophys Res Commun 417: 704–709 [DOI] [PubMed] [Google Scholar]
- Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR. (2007) Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem 282: 25475–25486 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, Yaghoubi-Masihi A, Fukao T, Bailey-Serres J. (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SM, Boynton JE, Gillham NW, Randolph-Anderson BL, Johnson AM, Harris EH. (1990) Transformation of chloroplast ribosomal RNA genes in Chlamydomonas: molecular and genetic characterization of integration events. Genetics 126: 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AV, Thomas-Hall SR, Malnoë A, Timmins M, Mussgnug JH, Rupprecht J, Kruse O, Hankamer B, Schenk PM. (2008) Transcriptome for photobiological hydrogen production induced by sulfur deprivation in the green alga Chlamydomonas reinhardtii. Eukaryot Cell 7: 1965–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MD, Kropat J, Hamel PP, Merchant SS. (2009) Two Chlamydomonas CTR copper transporters with a novel cys-met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell 21: 928–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps G, Happe T, Hemschemeier A. (2012) Nitrogen deprivation results in photosynthetic hydrogen production in Chlamydomonas reinhardtii. Planta 235: 729–745 [DOI] [PubMed] [Google Scholar]
- Philipps G, Krawietz D, Hemschemeier A, Happe T. (2011) A pyruvate formate lyase-deficient Chlamydomonas reinhardtii strain provides evidence for a link between fermentation and hydrogen production in green algae. Plant J 66: 330–340 [DOI] [PubMed] [Google Scholar]
- Posewitz MC, King PW, Smolinski SL, Zhang L, Seibert M, Ghirardi ML. (2004) Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J Biol Chem 279: 25711–25720 [DOI] [PubMed] [Google Scholar]
- Quinn JM, Barraco P, Eriksson M, Merchant S. (2000) Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J Biol Chem 275: 6080–6089 [DOI] [PubMed] [Google Scholar]
- Quinn JM, Eriksson M, Moseley JL, Merchant S. (2002) Oxygen deficiency responsive gene expression in Chlamydomonas reinhardtii through a copper-sensing signal transduction pathway. Plant Physiol 128: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J, Segrè D. (2006) The effect of oxygen on biochemical networks and the evolution of complex life. Science 311: 1764–1767 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schloss JA, Silflow CD, Rosenbaum JL. (1984) mRNA abundance changes during flagellar regeneration in Chlamydomonas reinhardtii. Mol Cell Biol 4: 424–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao N, Bock R. (2008) A codon-optimized luciferase from Gaussia princeps facilitates the in vivo monitoring of gene expression in the model alga Chlamydomonas reinhardtii. Curr Genet 53: 381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P. (2001) A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229 [DOI] [PubMed] [Google Scholar]
- Sommer F, Kropat J, Malasarn D, Grossoehme NE, Chen X, Giedroc DP, Merchant SS. (2010) The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell 22: 4098–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg M, Happe T. (2004) Identification of a cis-acting element controlling anaerobic expression of the hydA gene from Chlamydomonas reinhardtii.In M Jun, I Yasuo, M Rögner, eds, Biohydrogen III. Elsevier Science, Amsterdam, pp 117–127
- Strenkert D, Schmollinger S, Sommer F, Schulz-Raffelt M, Schroda M. (2011) Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. Plant Cell 23: 2285–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stripp ST, Happe T. (2009) How algae produce hydrogen—news from the photosynthetic hydrogenase. Dalton Trans 45: 9960–9969 [DOI] [PubMed] [Google Scholar]
- Terauchi AM, Lu SF, Zaffagnini M, Tappa S, Hirasawa M, Tripathy JN, Knaff DB, Farmer PJ, Lemaire SD, Hase T, et al. (2009) Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J Biol Chem 284: 25867–25878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M, Kuhlgert S, Hippler M, Happe T. (2009) Characterization of the key step for light-driven hydrogen evolution in green algae. J Biol Chem 284: 36620–36627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Davies JP, Melis A, Grossman AR. (1998) The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol 117: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]
- Zhang LP, Happe T, Melis A. (2002) Biochemical and morphological characterization of sulfur-deprived and H2-producing Chlamydomonas reinhardtii (green alga). Planta 214: 552–561 [DOI] [PubMed] [Google Scholar]
- Zheng L, Baumann U, Reymond JL. (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.