Abstract
Integrative comparative analyses of transcript and metabolite levels from climacteric and nonclimacteric fruits can be employed to unravel the similarities and differences of the underlying regulatory processes. To this end, we conducted combined gas chromatography-mass spectrometry and heterologous microarray hybridization assays in tomato (Solanum lycopersicum; climacteric) and pepper (Capsicum chilense; nonclimacteric) fruits across development and ripening. Computational methods from multivariate and network-based analyses successfully revealed the difference between the covariance structures of the integrated data sets. Moreover, our results suggest that both fruits have similar ethylene-mediated signaling components; however, their regulation is different and may reflect altered ethylene sensitivity or regulators other than ethylene in pepper. Genes involved in ethylene biosynthesis were not induced in pepper fruits. Nevertheless, genes downstream of ethylene perception such as cell wall metabolism genes, carotenoid biosynthesis genes, and the never-ripe receptor were clearly induced in pepper as in tomato fruit. While signaling sensitivity or actual signals may differ between climacteric and nonclimacteric fruit, the evidence described here suggests that activation of a common set of ripening genes influences metabolic traits. Also, a coordinate regulation of transcripts and the accumulation of key organic acids, including malate, citrate, dehydroascorbate, and threonate, in pepper fruit were observed. Therefore, the integrated analysis allows us to uncover additional information for the comprehensive understanding of biological events relevant to metabolic regulation during climacteric and nonclimacteric fruit development.
Fruits are generally classified into two physiological groups, “climacteric” and “nonclimacteric,” according to their respiration pattern and reliance on ethylene biosynthesis during ripening. Climacteric fruits, such as tomato (Solanum lycopersicum) and banana (Musa spp.), show an increase in respiration rate and ethylene formation. Nonclimacteric fruits, such as strawberry (Fragaria spp.) and grape (Vitis vinifera), exhibit neither the respiratory burst nor an elevated ethylene synthesis during ripening (Giovannoni, 2001). Pepper (Capsicum spp.) has typically been considered a nonclimacteric fruit, although varying patterns of ethylene production and respiratory rates have been reported (Villavicencio et al., 1999). Pepper is also considered a good source of metabolites possessing health-promoting properties, such as vitamins C, E, and B complex, provitamin A, oxygenated carotenoids, and other phytochemicals such as flavonoids with high antioxidant activities (Howard et al., 2000; Howard and Wildman, 2007). During maturation, fruit undergo transformations in color, aroma, nutrient composition, flavor, and softening. During this process, the production of reactive oxygen species plays an important role, particularly in the biosynthesis of carotenoids and in the transformations of chloroplasts to chromoplasts (Bouvier et al., 1998; Martí et al., 2009). These transformations are the results of dynamic processes that involve a complex series of molecular and biochemical changes under genetic regulation and/or in response to environmental perturbations. To better understand fruit development and ripening mechanisms, numerous studies have been focused on studies of transcript and metabolite levels in climacteric fruits, such as tomato (Roessner-Tunali et al., 2003; Alba et al., 2005; Lemaire-Chamley et al., 2005; Vriezen et al., 2008; Wang et al., 2009) and peach (Prunus persica; Lombardo et al., 2011), and in nonclimacteric fruits, such as strawberry (Aharoni and O’Connell, 2002; Aharoni et al., 2002; Fait et al., 2008; Osorio et al., 2008; Bombarely et al., 2010), pepper (Lee et al., 2010), and grape (Deluc et al., 2007; Grimplet et al., 2007). Integration of genomics, gene expression, and chemical composition data during fruit development and ripening can give important insights into gene-regulatory and metabolic events associated with these processes. In recent years, a number of studies on fruits have begun to integrate these extensive data sets, and while a number of them focused on the nonclimacteric grape and strawberry systems, most used the climacteric model of tomato fruit (Carrari et al., 2006; Deluc et al., 2007; Grimplet et al., 2007; Enfissi et al., 2010; Zamboni et al., 2010; Fortes et al., 2011; Osorio et al., 2011).
In this study, we employed similar approaches; however, we have focused on a nonclimacteric member of the Solanaceae family (of which tomato is also a member) to facilitate comparative analyses that may provide insights into relationships between climacteric and nonclimacteric ripening. Specifically, we have integrated transcript and metabolic data using different development and ripening stages of pepper (Capsicum chilense ‘Habanero’). While some peppers are climacteric, C. chilense is not. Metabolomic profiling was assessed by gas chromatography-mass spectrometry (GC-MS), while transcriptomic analysis was performed using the tomato TOM1 array, which was demonstrated to be applicable for assessing transcript levels in pepper (Moore et al., 2002). Lee et al. (2010) have compared climacteric tomato and nonclimacteric pepper fruit ripening at the transcriptomic level and identified genes in the ethylene signaling pathway that are induced in pepper, suggesting a nonethylene regulator influence of these genes in nonclimacteric peppers. Here, we conducted integrative comparative analysis of transcriptomics and metabolomics data from tomato and pepper fruits to gain a broader systems perspective and to identify additional common as well as distinct molecular regulatory events during development and ripening.
RESULTS
Metabolic Profiling during Pepper Fruit Development and Ripening
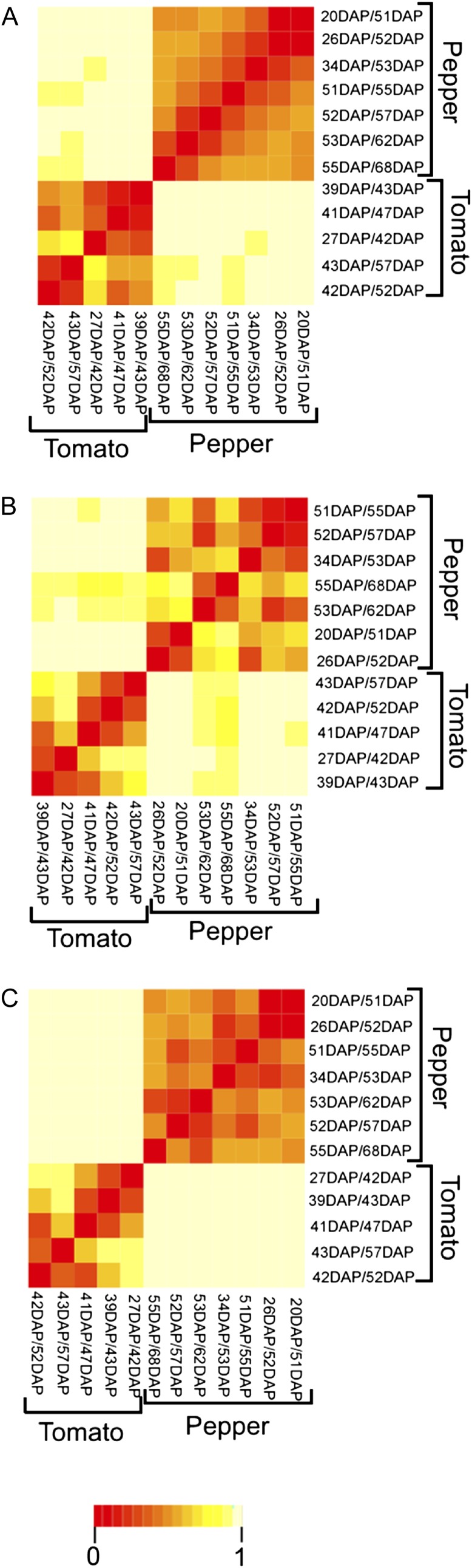
In order to assess metabolic profiles reflecting primary metabolism of pepper fruit, we selected 11 stages of development and ripening as described in “Materials and Methods.” The normalized data set obtained in this study by GC-MS as well as metabolite data for development and ripening of tomato from Osorio et al. (2011) were examined by two types of multivariate methods: (1) congruence of the covariance matrices, given by the Rv coefficient as defined in “Materials and Methods”; and (2) similarity between the principal components (PCs) of all periods spanning four stages (see “Materials and Methods”). For the comparative analysis of metabolite profiles, we used the profiles of the 26 metabolites measured over eight and 10 different developmental and ripening stages in tomato and pepper, respectively. The Rv coefficient takes values between 0 and 1, and small values of the coefficient indicate large differences between the data sets. For the metabolomic data sets of tomato and pepper, the Rv coefficient has a value of 0.003, although this value is not statistically significant at a significance level 0.05 (P= 0.38). Next, we considered all periods spanning four of the investigated stages and calculated the similarity based on the three PCs explaining the maximum of the variance. Figure 1A confirms that the investigated five and seven periods in tomato and pepper, respectively, are highly dissimilar. Moreover, the considered periods exhibit high but varying similarity, allowing us to distinguish the transition between early and late periods in development and ripening at 43 d after pollination (DAP; Br+1) for tomato and 52 to 53 DAP for pepper. Finally, Figure 1A suggests that for the 26 metabolites, there is similarity between the middle and later periods in the development and ripening of tomato and pepper fruit, but this is not the case for the beginning stages (e.g. pepper, 20–51 DAP; tomato, 27–42 DAP).
Figure 1.
Comparative analysis of the covariance structures of metabolites, transcripts, and profiles in tomato and pepper fruits. Similarity analysis is shown for the covariance structures of metabolites (A), transcripts (B), and integrated profiles (C; metabolite and transcript) over eight and 10 development and ripening stages from tomato and pepper, respectively. A similarity score was calculated for every pair of periods, each spanning four stages in development and ripening, based on the corresponding PCs (see “Materials and Methods”). The similarity score ranges from 0 to 1, with 0 indicating equality of the compared covariance structures. The results are displayed in the form of a heat map, with the color key included at the bottom.
Metabolism of Sugars and Sugar Alcohols during Pepper Fruit Development and Ripening
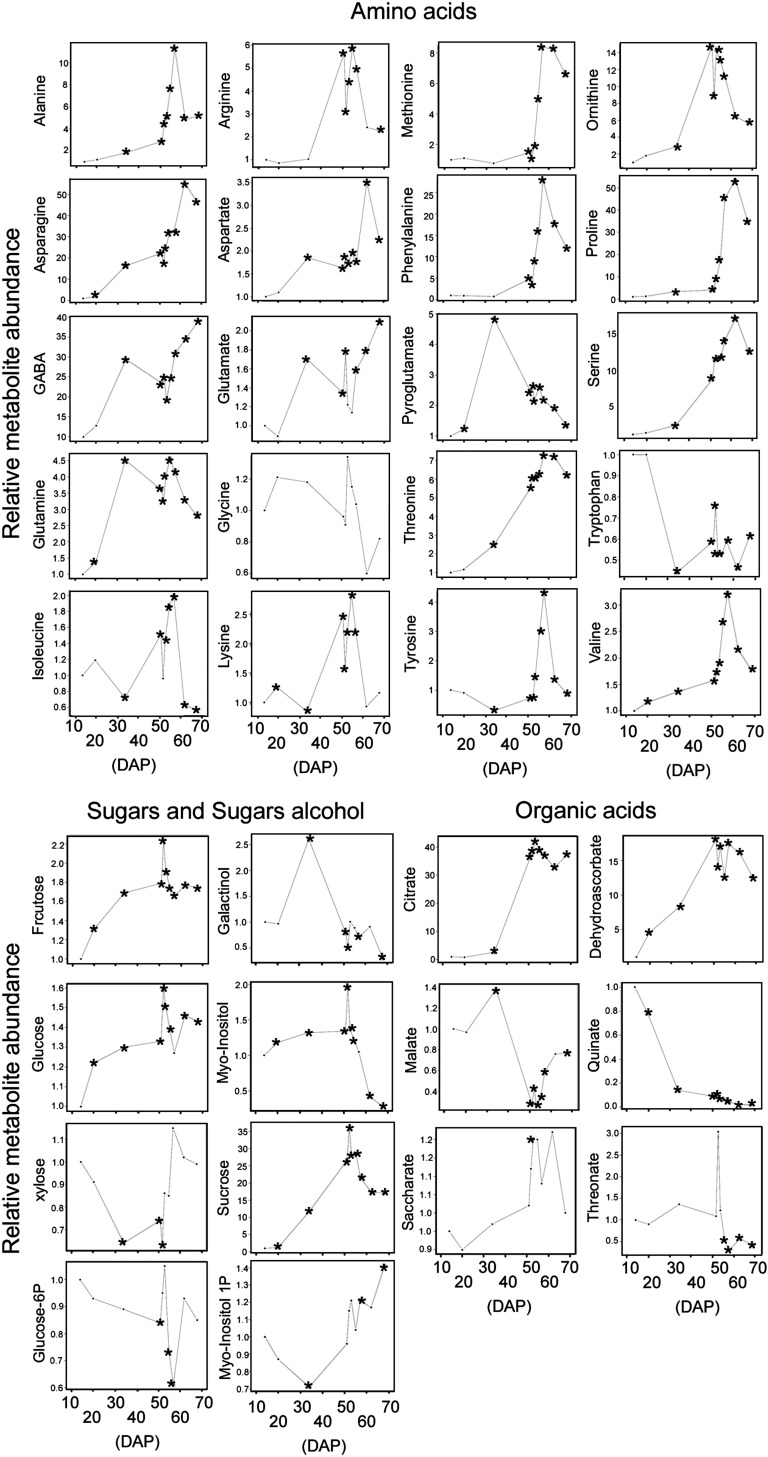
The major sugars Fru, Glc, and Suc displayed a continuous increase during the development of pepper until 52 DAP (Br; Fig. 2). The contents of these sugars increased dramatically at the onset of ripening (51–52 DAP). However, after 52 DAP, all three major sugars showed similar patterns of decline in content (Fig. 2). Indeed, the behavior of the major sugars during pepper ripening is opposite to many other characterized fruits, including, tomato cv Moneymaker (Carrari et al., 2006) and cv Ailsa Craig (Osorio et al., 2011), strawberry (Fait et al., 2008), grape (Deluc et al., 2007), and peach (Lombardo et al., 2011). With regard to sugar alcohols, galactinol, myoinositol-1-phosphate, and myoinositol were detected. In the case of myoinositol, the same behavior as for the major sugars was observed (Fig. 2). Myoinositol gradually increased during development, with a peak at 51 DAP. In the case of myoinositol-1-phosphate, the opposite pattern during early developmental and ripening stages compared with myoinositol was observed (Fig. 2). Myoinositol-1-phosphate decreased during early developmental stages to its minimum value at 34 DAP, while it increased during ripening initiation (51 DAP) and onward. Galactinol levels were highest at earlier developmental stages, with a peak at 34 DAP (Fig. 2), and after this peak galactinol content dramatically decreased.
Figure 2.
Primary metabolite levels during pepper development and ripening. Time points presented are 14, 20, 34, 51, 52, 53, 55, 57, 62, and 68 DAP. Data are normalized to the mean response calculated to the 14-DAP stage (value= 1). Values presented are means ± se of three replicates. Asterisks denote differences that were determined to be significant by Student’s t test analysis (P < 0.05) compared with the 14-DAP stage.
Xyl was the only cell wall-related sugar detected, and it decreased during early developmental stages and then dramatically increased during ripening (Fig. 2). This trend is consistent with many reports describing modifications and disassembly of hemicellulosic and pectin cell wall polysaccharides during fruit ripening (Sakurai and Nevins, 1993; Seymour, 1993).
Metabolism of Organic Acids during Pepper Fruit Development and Ripening
The variations in the levels of the various organic acids measured during pepper development and ripening processes were complex (Fig. 2). Citrate and dehydroascorbate levels dramatically increased during early developmental stages (approximately 40- and 15-fold, respectively) followed by small reductions during ripening stages (Fig. 2).
Malate, saccharate, and threonate levels were increased during preripening development, until 34 DAP for malate and 51 DAP for saccharate and threonate. Moreover, distinct accumulation patterns of these three organic acids during ripening and as compared with each other were observed. Malate levels decreased during later developmental stages (from 34 to 51 DAP), although a substantial increase was observed during ripening (approximately 2-fold). Saccharate levels were essentially constant during pepper fruit ripening, while threonate levels declined. Quinate levels decreased through development and ripening, although this decline was not as pronounced during ripening (Fig. 2).
Metabolism of Amino Acids during Pepper Fruit Development and Ripening
A total of 20 amino acids across all characterized stages were analyzed. During the fruit development phase (from 14 to 51 DAP), three distinct patterns of amino acid accumulation were observed: (1) those that increased through development, including Ala, Asn, γ-aminobutyrate (GABA), Gln, Ile, Arg, Asp, Glu, Lys, Phe, Thr, Ser, and Val; (2) those that remained relatively constant, including Gly, Met, Tyr, and Pro; and (3) Trp, which decreased during preripening development. Eighteen dramatically increased at the onset of ripening, and all except GABA and Glu declined until the final ripe stage (68 DAP) analyzed. Only Trp showed no changes during pepper ripening (Fig. 2).
Network-Based Analysis of Primary Metabolite Data
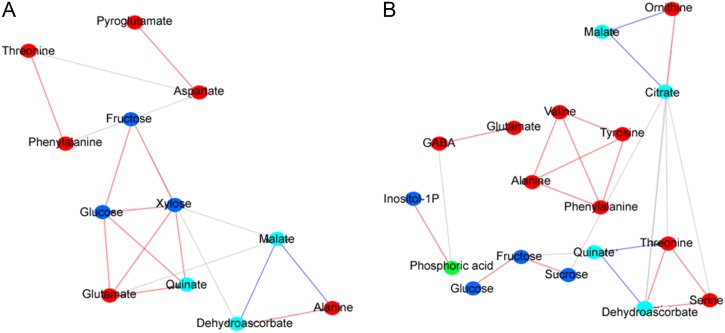
Networks of metabolites were created from Pearson correlations of the corresponding profiles while ensuring a false discovery rate (FDR) of 0.05 (see “Materials and Methods”). Again, we focused on the analysis of 26 metabolites detected in both tomato and pepper over all observed stages. Threshold values of 0.905 and 0.830 for the absolute values of the correlations ensured the imposed FDR rates in tomato and pepper, respectively. These resulted in 18 and 25 significant correlations, of which 14 and 20 were positive for the tomato and pepper metabolomes, respectively.
The distribution of edges resulted in two major components in the tomato network, containing eight and four metabolites. The first component comprised four amino acids (Thr, Phe, Asp, and pyro-Glu), while the second included three sugars (Fru, Glc, and Xyl), three organic acids (malate, quinate, and dehydroascorbate), as well as two amino acids (Ala and Glu). The remaining 14 metabolites were not involved in significant correlations, yielding isolated nodes (not shown in Fig. 3). The connected components could be further split into four communities (clusters), yielding a modularity value of 0.443 for the tomato network (Fig. 3A). As expected, major sugars (Suc, Fru, and Glc) were highly correlated with each other and clustered together; this community also included Glu and quinate. Moreover, Thr and Phe as well as pyro-Glu and Asp clustered together. On the other hand, malate was found to negatively correlate to Xyl, Glu, Ala, and dehydroascorbate and formed a cluster with the latter two.
Figure 3.
Networks from the primary metabolite data set of tomato and pepper fruits. Networks were obtained by determining the significant correlations of the metabolite profiles from tomato (A) and pepper (B), guaranteeing a FDR of 0.05. Positive correlations are indicated with red edges, while negative correlations are displayed with blue edges. The gray edges denote the relation between the communities (clusters) of metabolites in the network. The color coding of the nodes, representing the metabolites, denotes the following compound classes: amino acids (red), organic acids (light blue), sugars and sugar alcohols (dark blue), and others (green).
The pepper network contained three major components, containing 10, four, and four metabolites (Fig. 3B). The first was composed of amino acids, including Ala, Tyr, Val, and Phe, which were not significantly correlated to any other of the investigated metabolites. The second component could be further split into two communities, one involving two amino acids (GABA and Glu) and the other involving phosphoric acid and myoinositol. Finally, the third and largest component could be divided into three clusters, of which the first included, as expected, the three sugars (Glc, Fru, and Suc), the second comprised malate, citrate, and ornithine, while the third also included a combination of amino and organic acids. Compared with the tomato network, here quinate (and not malate) formed the only significant negative correlation between organic acids and sugars (i.e. Fru). The community structure of the pepper network is more pronounced compared with that of tomato, as indicated by the modularity value of 0.56.
The correlation of Fru and Glc was the only one preserved between the tomato and pepper network at a FDR level of 0.05. Moreover, the network-based investigation suggests that there is a major restructuring of the underlying metabolic and regulatory processes in climacteric and nonclimacteric fruits, since the metabolites without any significant correlations in the former formed relationships in the latter.
Transcript Levels during Pepper Fruit Development and Ripening
To identify differentially expressed genes in pepper pericarp, we used the TOM1 cDNA microarray and a time-series loop design. The microarray-based analysis was performed in the same tissues used for metabolomics (the time series included 11 stages: 14, 20, 26, 34, 51, 52, 53, 55, 57, 62, and 68 DAP). To profile transcript abundance, we used two-color simultaneous hybridizations to the TOM1 array containing 12,899 EST probes representing approximately 8,500 tomato unigenes and using mRNA prepared from each stage listed above. Genes at any fruit developmental stage showing a 2-fold or greater expression level change as compared with the first (reference) stage (14 DAP) and after Benjamini-Hochberg correction with a cutoff of P= 0.05 were noted as significant (Supplemental Table S1).
For the comparative analysis of transcript profiles, we used the profiles of the 318 genes found to have pronounced differential expression. For the transcriptomic data sets of tomato and pepper, the Rv coefficient has a significant value of 0.154 (P= 1.09e−314 < 0.05), indicating that the transcript levels of the two types of fruit differ but less so compared with the metabolite levels. Next, analogous to what was performed for the metabolite profiles, we considered all periods spanning four of the investigated stages, and calculating the similarity based on the three PCs explaining the maximum of the variance confirms that the investigated five and seven periods in tomato and pepper, respectively, are highly dissimilar (Fig. 1B). Moreover, the considered periods exhibit high but varying similarity, allowing us to distinguish the transition between early and late periods in the development and ripening at 42 DAP for tomato and 26 to 34 DAP for pepper. Finally, Figure 1B suggests that for the analyzed transcripts, there is similarity between the later periods in ripening of tomato and pepper fruit (e.g. pepper, 55–68 DAP; tomato, 42–47 DAP), but this is not the case for the beginning stages (e.g. pepper, 20–51 DAP and 26–52 DAP; tomato, 27–42 DAP and 39–43 DAP).
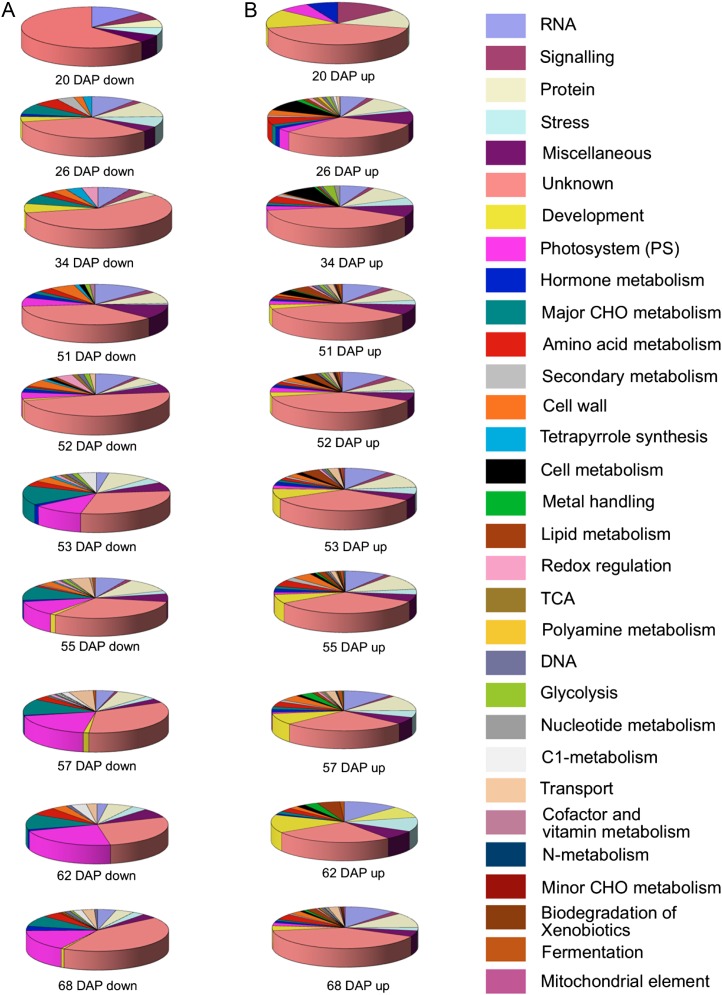
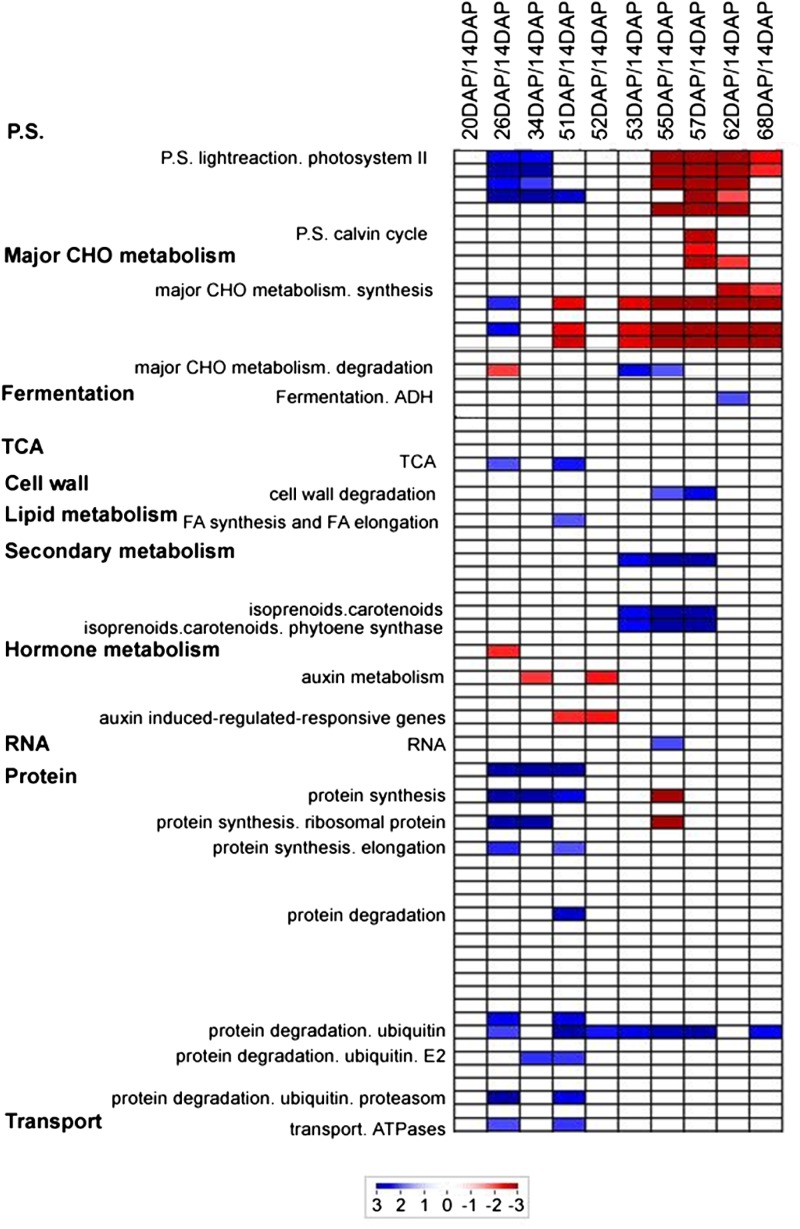
In addition, all genes showing significant (P < 0.05) differential expression values were classified in different functional categories using the MapMan classification system (Usadel et al., 2005; Fig. 4) and reflected the physiological and biochemical changes that occur during pepper development and fruit ripening. The functional distribution of differentially expressed genes revealed several interesting facts. For instance, 25% to 50% of the differentially expressed genes are of yet unknown function. The categories “photosystem” and “major carbohydrate (CHO) metabolism” included the most down-regulated genes during ripening (from 53 DAP onward). Interestingly, the category “RNA” was well represented in both up- and down-regulated genes and for all studied stages, although the percentage of down-regulated genes from this category decreased during development and ripening and increased for up-regulated genes in the same stages. Genes involved in cell wall synthesis, degradation, and modification considered as a global category were predominantly down-regulated during ripening (51 DAP onward), and some genes of this category were most differentially expressed during ripening stages. These genes represented an abundant class of either up- or down-regulated genes at 26 DAP (Fig. 4). It is well documented that cell wall restructuring and disassembly genes (endo- β-1,4-glucanases from the glycosyl hydrolase [GH] family, expansins, xyloglucan transglucosylase hydrolases (GH16), and a range of pectinases) are canonically associated with fruit development and ripening (Rose and Bennett, 1999; Rose et al., 2004; Brummell, 2006) in addition to earlier stages of fruit development (e.g. as characterized by the 26-DAP stage; Ranwala et al., 1992; Rose et al., 1998; Srivastava et al., 2010). Genes belonging to the “development” classification were more highly represented among genes up-regulated through the ripening process (from 51 DAP onward). It is also noteworthy that genes involved in protein synthesis and degradation showed the same distribution between categories of up- and down-regulated genes among all studied stages (Fig. 4).
Figure 4.
Functional distribution of expressed genes. Functional distribution is given for all genes showing significant (P < 0.05) expression values based on MapMan classification (Usadel et al., 2005) in pepper developmental and ripening stages (14, 20, 34, 51, 52 [breaker], 53, 55, 57, 62, and 68 DAP).
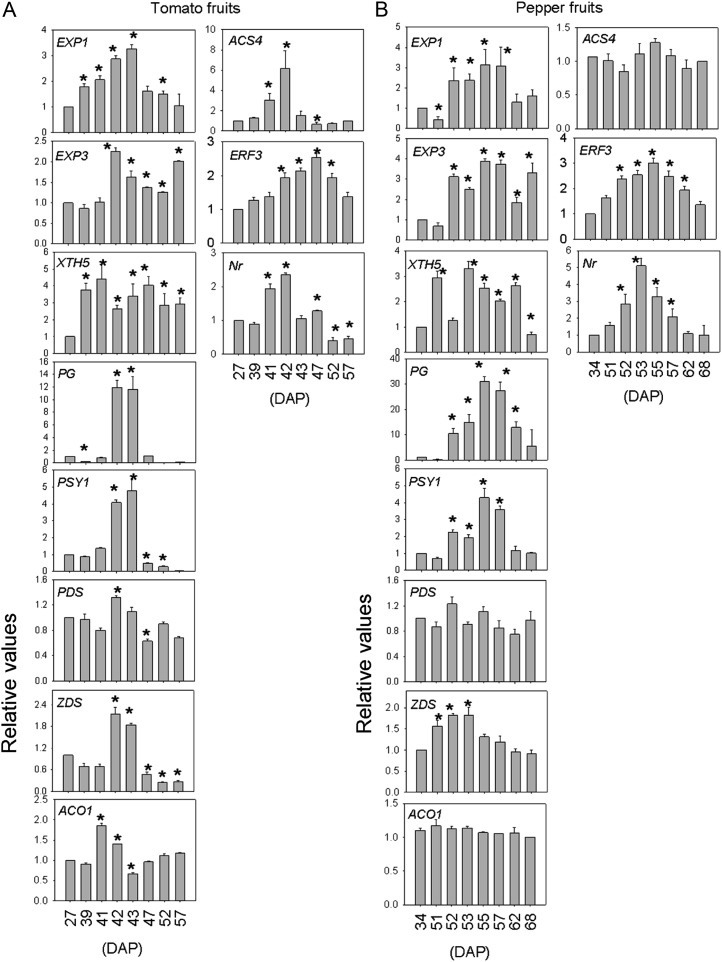
PageMan (Usadel et al., 2006) and MapMan (Usadel et al., 2005) mapping files, described in “Materials and Methods,” were used to study the development and ripening of pepper pericarp by identifying significantly overrepresented functional groups, using the earliest developmental stage (14 DAP) as a baseline and on the basis of Fisher’s exact and Wilcoxon tests for each category. This analysis facilitated the analysis of the global activation and/or repression of metabolic pathways and gene regulatory networks of the pericarp. Individual gene responses can be viewed in MapMan (Supplemental File S1). Using PageMan, we identified metabolic pathways that were enriched during fruit development (Fig. 5). As anticipated, photosynthetic gene expression declined dramatically during later ripening stages (55 DAP onward). However, a broader biphasic response was also noted, with a significant decline occurring during earlier developmental stages (26 and 34 DAP) as well, which may reflect the transition of the fruit from a photosynthetic to a sink tissue (Fig. 5). As also expected in an organ displaying transient starch synthesis (Beckles et al., 2001; Carrari et al., 2006), a large reduction during ripening of genes involved in starch synthesis was observed coincident with the ripening transition (from 51 DAP onward). Degradation of Suc was also up-regulated at the onset of ripening (53 and 55 DAP). Interestingly, genes associated with the tricarboxylic acid (TCA) cycle were up-regulated during fruit development (26 and 51 DAP; Fig. 5), which contrasts with the trend described for tomato fruit (Carrari et al., 2006). Similar to ripening tomato fruit (Carrari et al., 2006), in ripening pepper fruit, the transcript levels involved in cell wall restructuring and disassembly showed a clear tendency to be increased, which was corroborated by quantitative reverse transcription (qRT)-PCR in genes such as Expansin1 (EXP1), Expansin Precursor (EXP3), Xyloglucan Endotransglucosylase/Hydrolase (XTH5), and Polygalacturonase (PG). The same behavior was observed in some genes involved in carotenoid metabolism, such as Phytoene Synthase (PSY1) and Carotene Desaturase (ZDS), to be increased in pepper (Fig. 6). Others metabolic pathways that were enriched prior to the onset of fruit ripening were those associated with hormone metabolism, in which we specifically observed a strong reduction of auxin-responsive genes (from 34 to 52 DAP) as well as strong up-regulation of genes involved in the biosynthesis of ribosomal proteins (from 26 to 51 DAP), although these genes showed a decrease in expression during ripening (at 55 DAP; Fig. 5). Finally, in all studied stages, we saw a general up-regulation of the ubiquitin protein degradation process (Fig. 5), as was also observed in tomato fruit (Osorio et al., 2011).
Figure 5.
Expression analysis of pepper development and ripening. A condensed PageMan display of altered pathways is shown. Gene expression data are presented as log2 fold changes in comparison with the first harvested time point (14 DAP). The analyzed time points were 20, 34, 51, 52, 53, 55, 57, 62, and 68 DAP. The data were subjected to a Wilcoxon test in PageMan, and the results are displayed in false color. BINs colored in red are significantly up-regulated, whereas BINs colored in blue are significantly down-regulated.
Figure 6.
Quantitative PCR of cell wall-, carotenoid-, and ethylene-related genes of tomato and pepper fruits across developmental and ripening stages. Cell wall-related genes were EXP1, EXP3, XTH5, and PG. Carotenoid-related genes were PSY1, PDS, and ZDS. Ethylene-related genes were ACC oxidase (ACO1), ACC synthase (ACS4), and the Nr receptor. The values represent means ± se of four individual plants. Asterisks indicate values determined by t test to be significantly different from the first analyzed stage (P < 0.05).
Hormone Biosynthesis and Responses in Pepper Fruit
The effects of plant hormones on fruit maturation differ for climacteric and nonclimacteric fruit. Tomato is a model system to study climacteric fruit ripening, and numerous efforts have been undertaken with the goal of understanding ethylene biosynthesis and response, including the steps involved in ethylene perception and signal transduction (for review, see Klee and Giovannoni, 2011). Ethylene biosynthesis includes conversion of Asp to Met, conversion of Met to ethylene, and the Met recycling pathway (Yang and Hoffman, 1984). Ethylene is synthesized from S-adenosyl methionine (SAM) by the sequential action of two ethylene biosynthetic enzymes, 1-aminocyclopropane-1-carboxylate (ACC) synthase and ACC oxidase. SAM can participate in polyamine as well as ethylene biosynthesis (Mattoo et al., 2007). In climacteric tomato fruits, ACC synthase and ACC oxidase are elevated during ripening (Carrari et al., 2006; Osorio et al., 2011). In contrast, no changes in transcript levels of ACC synthase and ACC oxidase during nonclimacteric Habanero pepper development and ripening processes were observed. However, SAM synthetase, the catalyst of SAM biosynthesis, showed up-regulation in later ripening stages (57–68 DAP; Supplemental File S1). This result would suggest that polyamine biosynthesis may predominate the flux through SAM metabolism rather than ethylene biosynthesis in pepper.
The tomato genome harbors six characterized ethylene receptors, and five of the six (ETR1, ETR2, ETR4, ETR5, ETR6, and ETR3/Never-ripe) have been shown to bind ethylene (Klee, 2002; Klee and Tieman, 2002), while expression studies revealed several different profiles. For example, transcript levels of ETR1, ETR2, and ETR5 change little during ripening or upon ethylene treatment of mature fruit, while NR, ETR4, and ETR6 are strongly induced during ripening, and mutant and transgenic studies have shown that all can impact ripening phenomena (Kevany et al., 2007). Additional studies indicate that, as in Arabidopsis (Arabidopsis thaliana), tomato ethylene receptors are functionally negative regulators of ethylene signaling (Klee, 2002; Klee and Tieman, 2002). Consistent with this model, exposure of immature fruits to ethylene caused a reduction in the amount of ethylene receptor protein and earlier ripening (Kevany et al., 2007). Interestingly, the Nr transcript (SGN-U590044) was up-regulated during pepper ripening (Fig. 6; Supplemental File S1), as it was during climacteric tomato ripening (Fig. 6). Moreover, in tomato, it has been shown that following ethylene interaction with the receptor, the signal is transferred via a mitogen-activated protein kinase cascade to a family of transcription factors (ethylene response factors [ERFs]) that influence the expression of ethylene-regulated genes, including those mediating cell wall disassembly (endo-polygalacturonase, pectin methyl esterase, and pectate lyase). During pepper ripening, the ERF3 transcript (SGN-U564955) was up-regulated as well as transcripts related to cell wall restructuring and disassembly (Fig. 6; Supplemental File S1). These results are consistent with the hypothesis that ethylene signaling components downstream of the receptors are regulated by a mechanism independent of ethylene (Lee et al., 2010).
Jasmonic acid has been demonstrated to be involved in cross talk with ethylene signaling components in Arabidopsis (Devoto and Turner, 2005) and has been associated with fruit maturation. In climacteric fruits such as tomato and apple (Malus domestica), jasmonate levels increase during early fruit maturation, suggesting that they could be involved in the regulation of this process (Fan et al., 1998). Consistent with this hypothesis, exposure of immature apple fruits to jasmonate caused elevated ACC oxidase activity and heightened ethylene production (Saniewski et al., 1986; Miszczak et al., 2000). The fact that the increase of jasmonate in tomato and apple in addition to nonclimacteric fruits such as sweet cherry (Prunus avium) occurs during the earlier developmental stages could indicate a role for jasmonate in cell division or other early fruit expansion processes (Kondo and Tomiyama, 2000). During early development (26 DAP) of pepper fruit, we observed elevated transcript levels for the jasmonate synthesis gene 12-Oxophytodienoate Reductase3 (SGN-U595636; Supplemental File S1). Moreover, at the onset of ripening (52–57 DAP), a lipoxygenase gene (SGN-U594638) associated with jasmonate biosynthesis was also induced, suggesting that jasmonate may also play a role in nonclimacteric fruit ripening (Supplemental File S1).
Network-Based Analysis of Transcript Levels
Similar to the analysis of metabolite profiles, coexpression networks of transcripts were created from Pearson correlations of the corresponding profiles while ensuring a FDR of 0.05 (see “Materials and Methods”). Again, we focused on the analysis of 318 transcripts showing the most pronounced differential behavior in both tomato and pepper over all observed stages. Threshold values of 0.9524 and 0.8425 for the absolute value of the correlations ensured the imposed FDR rates in tomato and pepper, respectively. These resulted in 768 and 4,530 significant correlations, of which 660 and 3,544 were positive, for the tomato and pepper transcriptomes, respectively.
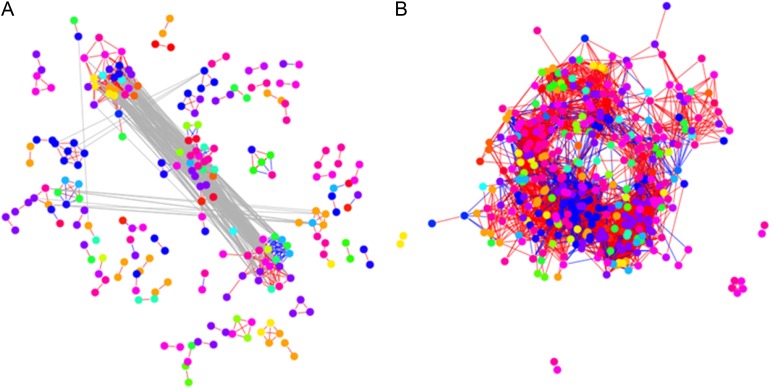
In the tomato coexpression network, the distribution of edges resulted in 57 components containing at least one edge. The largest of these components contained 75 transcripts, followed by two components composed of nine transcripts each. Looking at the level of community structure, the tomato coexpression network showed modularity of 0.402 for the 62 communities. The three largest communities, composed of 28, 26, and 21 transcripts, were subnetworks of the largest component (Fig. 7A). At a significance level of 0.05, the first was enriched for the following functional categories (see “Materials and Methods”): cell wall components, modification, and degradation, hormone metabolism, major CHO metabolism, redox metabolism, protein activation, targeting, folding, and assembly. The second and third were enriched for all of the above-mentioned functional categories, except cell wall degradation and hormone metabolism. The average correlations for transcripts involved in cell wall modification, CHO synthesis and degradation, and protein synthesis and degradation were found to be 0.980, 0.976, 1, 0.980, and 0.711, respectively. This implies that all but the transcripts involved in protein degradation exhibit statistically significant and high correlations.
Figure 7.
Networks from selected transcripts of tomato and pepper fruits. Transcripts involved in cell wall metabolism, hormone metabolism, redox regulation, major carbohydrate metabolism, protein synthesis, protein targeting, protein posttranslational modification, protein degradation, protein folding, protein assembly, and cofactor ligation were used in the analysis. Color coding for the nodes can be found in Supplemental Figure S2. Transcripts were grouped by functionality on the basis of MapMan gene ontology. Networks were obtained by determining the significant correlations of the transcript profiles from tomato (A) and pepper (B), guaranteeing a FDR of 0.05. Positive correlations are indicated with red edges, while negative correlations are displayed with blue edges. The gray edges denote the relation between the communities (clusters) of metabolites in the network. The color code for the nodes, representing the selected transcripts, corresponds to the MapMan bins and indicates the different function categories.
The pepper coexpression network contained five components containing at least one edge (Fig. 7B). The largest of these components contained 355 transcripts, followed by a component with only five transcripts. These components could be further decomposed into nine communities (clusters) yielding a modularity value of 0.555 for the pepper coexpression network. The largest of these communities contained 109 transcripts and was significantly enriched for the same functional categories as the largest community in the tomato coexpression network, except for protein activation and hormone metabolism. The average correlations for transcripts involved in cell wall modification, CHO synthesis and degradation, and protein synthesis and degradation were found to be 0.978, 0.737, 0.497, 0.848, and 0.850, respectively. This implies that all but the transcripts involved in CHO synthesis and degradation exhibit statistically significant and high correlations, which is in contrast to what was found for the tomato coexpression network. In fact, 10% and 25% of the correlations between genes involved the CHO synthesis and degradation were found to be negative.
While there is preservation of the functional categories enriched in the clusters of the tomato and pepper coexpression networks, the intersection of the two coexpression networks was composed of only 180 edges (i.e. 3% of the total number of significant correlations in both data sets). This indicates that on the transcriptome level, the investigated climacteric and nonclimacteric fruits indeed exhibit large differences.
Transcript and Metabolite Correlations
We next assessed the integrated metabolite and transcript profiles based on a global analysis of the covariance structure of the data sets as well as the extracted networks of tomato and pepper. For this purpose, we used the profiles for the already analyzed 26 metabolites and 318 transcripts.
The Rv coefficient exhibited a (nonsignificant) value of 0.002, which hints that the already observed differences are present at this level of data integration, too. Analysis of the similarities between the PCs of the periods spanning four stages of fruit development and ripening further stressed this finding (Fig. 1C), which confirms that the investigated five and seven sample points in tomato and pepper, respectively, are highly dissimilar, as revealed by Figure 1A. The covariance structure of metabolites, however, did reveal internal similarity between the middle and later periods in the development and ripening of tomato and pepper fruits when each species was considered independently, but this was not the case for the beginning stages for tomato (e.g. 39–43 DAP and 27–42 DAP).
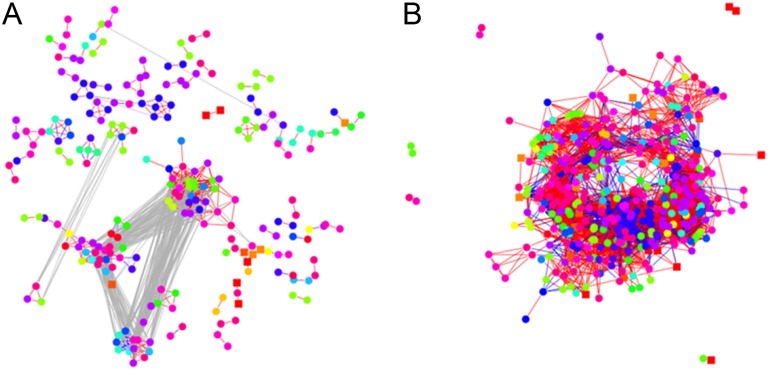
To construct a network integrating the metabolite and transcript profiles, the threshold values of 0.952 and 0.843 were used for the data sets from tomato and pepper, respectively. These values ensure a FDR of 0.05. The representation of the networks in Figure 8, due to their size, was prohibitive for any visual investigations. Therefore, we focused on the strength of the relationships between the transcripts of the following functional categories: starch synthesis and degradation, cell wall precursor synthesis and cellulose synthesis, as well as protein degradation, and the compound classes involving amino acids, organic acids, and sugars. The findings are given in Table I.
Figure 8.
Network-based representation of the integrated metabolomics and transcriptomic data set of tomato and pepper. An analogous procedure to that used to obtain Figures 3 and 7 was employed on the combined data set. Nodes denoting metabolites are drawn in squares, while those representing transcripts are given in circles for tomato (A) and pepper (B). The color coding of nodes represented by circles corresponds to the MapMan bins. The color coding of nodes represented by squares follows that used in Figure 3 for the network of metabolites and that of the nodes represented by circles follows that used in Figure 7 for the network of transcripts. Red edges denote positive correlations, while blue edges represent negative correlations. The gray edges denote the relation between the communities (clusters) of metabolites in the network.
Table I. Average correlations between gene function categories and compound classes in the integrated networks for pepper and tomato.
NA indicates that no significant correlations were observed for the particular pair of gene function category and compound class. The included values indicate that sugars and sugar alcohols are in general more negatively correlated to transcripts involved in starch synthesis in pepper compared with tomato, resulting in a smaller value for the average correlation compared with the used threshold of 0.843. Analogous reasoning implies that organic acids, on the other hand, are more positively correlated to transcripts involved in protein degradation in pepper compared with tomato.
| Compound | Starch Synthesis | Starch Degradation | Cell Wall Precursor Synthesis | Cell Wall Cellulose Synthesis | Protein Synthesis | Protein Degradation |
|---|---|---|---|---|---|---|
| Pepper | ||||||
| Amino acids | −0.735 | −0.739 | NA | NA | −0.570 | 0.864 |
| Organic acids | −0.765 | −0.852 | NA | NA | −0.640 | 0.355 |
| Sugars and sugar alcohols | 0.317 | NA | NA | NA | 0.293 | 0.784 |
| Tomato | ||||||
| Amino acids | NA | NA | −0.969 | NA | NA | 0.969 |
| Organic acids | NA | NA | NA | NA | NA | 0.001 |
| Sugars and sugar alcohols | 0.968 | NA | NA | NA | NA | NA |
DISCUSSION
Extensive efforts have been undertaken for understanding fruit develop and ripening mechanisms, in recent years especially at the levels of transcriptional control (Aharoni and O’Connell, 2002; Alba et al., 2005; Lemaire-Chamley et al., 2005; Moore et al., 2005; Grimplet et al., 2007; Vriezen et al., 2008; Wang et al., 2009; Bombarely et al., 2010; Lee et al., 2010) and downstream metabolomic consequences (Fraser et al., 1994, 2007; Fait et al., 2008; Lombardo et al., 2011). Recently, a small number of studies have been undertaken using systems strategies for combining plant transcriptome and metabolomic data to develop associations in tomato (Carrari et al., 2006; Garcia et al., 2009; Mounet et al., 2009; Enfissi et al., 2010; Osorio et al., 2011) or grape (Carrari et al., 2006; Deluc et al., 2007; Enfissi et al., 2010; Zamboni et al., 2010; Osorio et al., 2011). Here, comprehensive transcriptomic and GC-MS metabolomic analyses have been combined to provide novel insights into the development and ripening of nonclimacteric pepper fruit. These analyses were compared with similar transcript and metabolite analyses through the development and ripening of tomato fruit (Carrari et al., 2006; Deluc et al., 2007; Enfissi et al., 2010; Zamboni et al., 2010; Osorio et al., 2011), which allowed the identification of similar and distinct regulation at the gene and metabolite levels between nonclimacteric (pepper) and climacteric (tomato) fruits.
Sugar Metabolism during Pepper Fruit Ripening and Development
During earlier development (until 52 DAP) and ripening stages, we observed considerable changes in the sugar metabolism of pepper fruit. Photosynthetic light reactions exhibited up-regulation during early stages followed by down-regulation during ripening. Also during ripening, a dramatic down-regulation in genes involved in the synthesis of starch was observed and coupled to up-regulation of Suc degradation at the onset of ripening. This suggests an activated photosynthesis and carbon assimilation process during early fruit stages followed for reduction of these pathways coincident with ripening. Consistently, we noted a gradual accumulation of the major soluble sugars (Glc, Fru, Suc) during early stages followed by their decrease following the onset of ripening. Compared with climacteric tomato fruits, we detected similar expression patterns of sugar-related genes during ripening. However, in contrast, these genes were down-regulated during early fruit stages, consistent with stable Fru, Glc, and Suc levels during preripening tomato fruit development (Osorio et al., 2011). Additionally, in tomato fruit, hexoses derived from the degradation of Suc were described as key to starch synthesis during early development and at the onset of ripening (Carrari and Fernie, 2006; Centeno et al., 2011). In pepper fruit, consistent with the up-regulation of Suc degradation genes, a decrease in Suc levels was observed. However, in pepper ripening, down-regulation of starch synthesis genes occurred only during ripening, which may suggest that starch synthesis regulation is not conserved between climacteric and nonclimacteric fruits.
Ethylene Regulation during Pepper Fruit Ripening and Development
Fruits are generally classified into two physiological groups, climacteric and nonclimacteric, according to their respiratory and associated ethylene biosynthesis profiles during ripening. It is known that climacteric ethylene biosynthesis involves conversion of Asp to Met, conversion of Met to ethylene, and the Met recycling pathway (Yang and Hoffman, 1984) through SAM and the two ethylene biosynthetic enzymes, ACC synthase and ACC oxidase. SAM also serves as a precursor of polyamine synthesis (Mattoo et al., 2007; Mattoo and Handa, 2008). Immature pepper fruits at 26 DAP were characterized by SAM synthase up-regulation; however, changes in the expression of other ethylene biosynthesis genes associated with climacteric fruits (such as Met sulfoxide reductase involved in the recycling of Met, ACC synthase, and ACC oxidase) did not occur. In addition, during normal pepper ripening, Asp, the precursor of Met, as well as Met increased 3- and 8-fold, respectively. In contrast to tomato fruit and as described previously by Lee et al. (2010), these results suggest that ethylene biosynthesis does not play a significant role in the accumulation of Asp and Met during pepper fruit ripening. On the other hand, SAM was up-regulated during early developmental stages in pepper fruit, which may indicate flux through SAM to polyamines, even though significant changes in the levels of Asp and Met were not observed in these stages.
Ethylene perception represents a point of the regulation of response downstream of hormone synthesis. Prior studies have shown that ethylene receptors act to negatively regulate ethylene signaling in climacteric fruits (Klee, 2002; Klee and Tieman, 2002; Osorio et al., 2011) and that, following ethylene interaction with the receptors, ripening-related genes, including those encoding cell wall-disassembling enzymes and phytoene synthase, which catalyzes the formation of phytoene (the first C40 carotene intermediate in carotenoid biosynthesis), are activated. Interestingly, in parallel with the situation in tomato fruit, the Nr transcript (SGN-U590044) was up-regulated during pepper fruit ripening. In addition, a number of ethylene/ripening-related genes shown to be up-regulated in climacteric fruits following ethylene perception, including cell wall disassembly genes (polygalacturonases, pectate lyases, and pectin methyl esterases), were up-regulated during normal pepper fruit ripening and indeed represent one of the most evident and well-studied ripening-associated changes in tomato fruit (Brummell, 2006). The major carotenoids responsible for red fruit color are lycopene for tomato and capsanthin and capsorubin for pepper (Hornero-Méndez et al., 2000; Hornero-Méndez and Mínguez-Mosquera, 2000; Hirschberg, 2001). As described previously, genes involved in carotenoid biosynthesis such as PSY1 and phytoene dehydrogenase were up-regulated during pepper fruit ripening as well as tomato fruit ripening (Lee et al., 2010; Osorio et al., 2011). These results, the up-regulation of both cell wall disassembly genes and carotenoid biosynthesis-related genes during normal pepper ripening, and nonclimacteric fruit, which is also characteristic of events during ripening in climacteric fruit, suggest that both fruits conserve ethylene signaling components but differ in hormonal regulation.
Organic Acid Metabolism during Pepper Fruit Ripening and Development
Organic acids are crucial to many aspects of tomato fruit biology and maturation, and they correlate strongly with genes associated with ethylene and cell wall metabolism-related pathways (Carrari et al., 2006; Centeno et al., 2011; Osorio et al., 2011). In some nonclimacteric fruits such as strawberry receptacle, only some TCA cycle intermediates such as succinate, fumarate, and 2-oxoglutarate displayed substantial changes during ripening, associated with a heavy demand for carbon skeleton components (Fait et al., 2008). Here, we report that organic acids, including the two TCA cycle intermediates malate and citrate, were strongly affected across ripening, suggesting that organic acids are regulated at the transcriptional level as in climacteric fruits. Recently, malate metabolism was described as important for transitory starch metabolism in normal tomato fruit development (Centeno et al., 2011). Interestingly, the correlation networks in this study revealed a positive correlation between malate levels and genes involved in the synthesis of starch. This result raises the possibility that malate metabolism is generally important for transitory starch metabolism and that its regulation is conserved between climacteric and nonclimacteric fruits.
The levels of ascorbate (vitamin C) have been shown to be dramatically elevated during pepper ripening (Martínez et al., 2005). In this study, ascorbate was not detected; however, we observed a high increase in dehydroascorbate (its oxidized form) and relatively minor changes in threonate (a catabolic product of ascorbate) throughout ripening, suggesting high redox activity and carbon recycling.
Comparison of amino acid profiles during development and ripening of pepper (Supplemental Fig. S1) and tomato indicate that both fruits follow distinct metabolic regulation programs. For example, significant differences were detected in tomato, where some amino acids increased while others decreased through development and ripening stages. Aromatic amino acids (Phe and Tyr) were observed to accumulate during pepper ripening but not in tomato. These amino acids serve as precursors for various branches of the phenylpropanoid pathway and the biosynthetic plant phenolics 2-phenylethanol and 2-phenylacetaldehyde (Tieman et al., 2006).
In general, correlation network analysis suggests that metabolism during pepper fruit development is highly coordinated, although a number of notable associations between transcripts and metabolites occur. For example, organic acids such as citrate, dehydroascorbate, and malate are highly correlated to genes associated with starch and cell wall pathways as well as protein degradation, suggesting the importance of these organic acids in pepper fruit development and ripening.
CONCLUSION
A comparative transcriptome and metabolome analysis during the maturation processes of climacteric and nonclimacteric fruits (tomato and pepper, respectively) reveal fewer changes in gene expression in pepper fruit as compared with tomato. When comparing our results with those of previous studies on gene expression (Lee et al., 2010) during pepper and tomato ripening, we suggest that both fruits have similar ethylene-mediated signaling components; however, the regulation of these genes is clearly different and may reflect altered ethylene sensitivity or regulators other than ethylene in pepper. Genes involved in ethylene biosynthesis in climacteric fruits, including ACC synthase and ACC oxidase, are not induced in pepper fruits; however, as in tomato fruits, genes downstream of ethylene perception, such as cell wall metabolism genes, ERF3, and carotenoid biosynthesis genes, are clearly up-regulated during pepper fruit ripening. While signaling sensitivity or actual signals may be different between climacteric and nonclimacteric fruit, the evidence described here suggests that the activation of a common set of ripening genes influences metabolic traits. Additionally, we provide information on how phytohormone biosynthesis and response genes, particularly those related to ethylene, auxins, and jasmonate, display expression patterns consistent with their roles in the developmental process of nonclimacteric pepper fruits, and we define the metabolic programs occurring during development. Taken together, these data should greatly improve our understanding of nonclimacteric fruit ripening and development as well as provide a basis for hypothesis-driven research that will be able to elucidate the regulatory events underlying this important biological process.
MATERIALS AND METHODS
Plant Material and Sampling
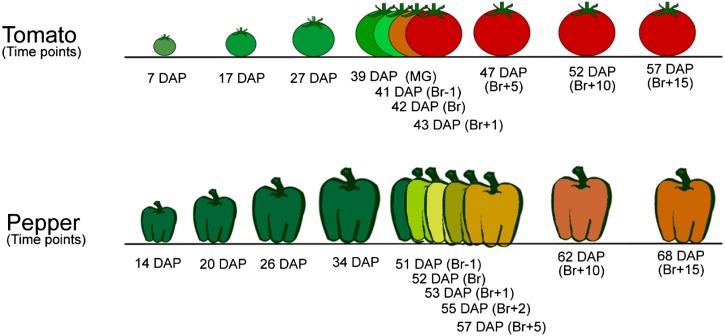
Pepper (Capsicum chilense ‘Habanero’) was grown in the greenhouse at 26°C under 12 h of supplemental lighting followed by 12 h at 20°C. To collect pepper stages prior to ripening, fruits were tagged at 14 DAP and harvested at one of the following 11 time points: 14, 20, 26, 34, 51 (breaker−1; Br−1), 52 (breaker; Br), 53 (breaker+1; Br+1), 55 (breaker+2; Br+2), 57 (breaker+5; Br+5), 62 (breaker+10; Br+10), and 68 DAP (breaker+16; Br+16; Fig. 9). The first signs of carotenoid accumulation on the external surface of the fruit were defined as the breaker stage. Transcriptome and metabolomic analyses were performed on the same material; 20 to 40 fruit at each developmental stage were collected and pooled. A minimum of three biological replicates in each time point were considered. All fruits were collected from four individual plants, which were grown in the greenhouse and randomly distributed. After tissue selection, pericarp tissue was collected, frozen in liquid nitrogen, and stored at −80°C until further analysis. For tomato (Solanum lycopersicum), the analyzed time points were 7, 17, 27, 39 (mature green), 41 (breaker−1; Br−1), 42 (breaker; Br), 43 (breaker+1; Br+1), 47 (breaker+5; Br+5), 52 (breaker+10; Br+10), and 57 DAP (breaker+15; Br+15; Fig. 9). For tomato transcriptome analysis, eight to 10 individual fruits for each time point were used as biological replicates. These individual fruit replicates came from 10 to 15 plants. Tomato data are used from Osorio et al. (2011).
Figure 9.
Experimental design. To collect pepper (cv Habanero) prior to ripening, fruits were tagged at 14 DAP and harvested at one of the following 11 time points: 14, 20, 26, 34, 51 (breaker−1; Br−1), 52 (breaker; Br), 53 (breaker+1; Br+1), 55 (breaker+2; Br+2), 57 (breaker+5; Br+5), 62 (breaker+10; Br+10), and 68 DAP (breaker+16; Br+16). For tomato (cv Ailsa Craig), the analyzed time points were 7, 17, 27, 39 (mature green [MG]), 41 (breaker−1; Br−1), 42 (breaker; Br), 43 (breaker+1; Br+1), 47 (breaker+5; Br+5), 52 (breaker+10; Br+10), and 57 DAP (breaker+15; Br+15).
Transcriptome Analysis
The tomato array (TOM1) was used for tomato and pepper transcriptome analysis. TOM1 contains approximately 8,000 unique elements randomly selected from cDNA libraries isolated from a range of tissues, including leaf, root, fruit, and flower, and representing a broad range of metabolic and developmental processes. Nucleic acid sequence and annotation data pertaining to the TOM1 microarray are available via the Tomato Functional Genomics Database (http://ted.bti.cornell.edu). Microarray analysis was performed as described (Alba et al., 2004, 2005). The raw intensity values were normalized using Robin’s default settings for two-color microarray analysis (Lohse et al., 2010). The obtained P values were corrected for multiple testing using the strategy described by Benjamini and Hochberg (1995) separately for each of the comparisons made. Genes that showed an absolute log2 fold change value of at least 1 and a P value lower than 0.05 were considered significantly differentially expressed. The log2 fold change cutoff value was imposed to account for noise in the experiment and to make sure that only genes that show a marked reaction are recorded. MapMan and PageMan software packages (Usadel et al., 2006, 2009) were used for visualization. The transcript data for tomato have been published previously (Osorio et al., 2011).
Metabolome Analysis
Metabolite extraction derivatization, standard addition, and sample injection for GC-MS were performed according to Osorio et al. (2012). The mass spectra were cross-referenced with those in the Golm Metabolome Database (Kopka et al., 2005).
RNA Extraction and Quantification for qRT-PCR
The RNA was determined and analyzed by qRT-PCR as described by Zanor et al. (2009). The expression of EXP1, EXP3, ACC oxidase, ACC synthase, XTH5, PG, PSY1, PDS, ZDS, ERF3, and the Nr receptor was analyzed by real-time qRT-PCR using the fluorescent intercalating dye SYBR Green in an iCycler detection system (Bio-Rad). Relative quantification of the target expression level was performed using the comparative cycle threshold method. The following primers were used: for analysis of EXP1 (Centeno et al., 2011), forward, 5′-TACCAATTTCTGCCCACCAAAT-3′, and reverse, 5′-GGTTACACCAGCCACCATTGT-3′; for EXP3 (Centeno et al., 2011), forward, 5′-TTTGCGGAGCTTGCTTTGAA-3′, and reverse, 5′-GGAGCACAAAAATTCGTTGCAG-3′; for XTH5 (Centeno et al., 2011), forward, 5′-GGATTCAGCCATCTCTTTGGTG-3′, and reverse, 5′-GAACCCTGAACCTGTGTTTTGG-3′; for PG (Centeno et al., 2011), forward, 5′-GGCAATGGACAAGTATGGTGG-3′, and reverse, 5′-CAGAAGGTTAAGGCCGTTGGT-3′; for ACO1 (Centeno et al., 2011), forward, 5′-AAATCATGAAGGAGTTTGCTGATAAA-3′, and reverse, 5′-TTTTCACACAGCAAATCCAACAG-3′; for ACS4 (Centeno et al., 2011), forward, 5′-CCATCTTGTTTGCGACGAAATA-3′, and reverse, 5′-CGATGCTAACGAATTTTGGAGAA-3′; for PSY1 (Lee et al., 2012), forward, 5′-TGGCCCAAACGCATCATATA-3′, and reverse 5′-CACCATCGAGCATGTCAAATG-3′; for PDS (Lee et al., 2012), forward, 5′-GTGCATTTTGATCATCGCATTGAAC-3′, and reverse, 5′-GCAAAGTCTCTCAGGATTACC-3′; for ZDS (Lee et al., 2012), forward, 5′-TTGGAGCGTTCGAGGCAAT-3′, and reverse, 5′-AGAAATCTGCATCTGGCGTATAGA-3′; for ERF3, forward, 5′-AGAAGGCTGGAAAAACCAAAG-3′, and reverse 5′-GGTGGGAAAAACCATTGAGA-3′. To normalize gene expression, we used the constitutively expressed Ubiquitin3 (GenBank accession no. X58253) using the following primers: forward, 5′-ACCACGAAGCTCTCCAGGAG-3′, and reverse, 5′-CATTGAACCCAACATTGTCACC-3′ (Zanor et al., 2009).
Rv Coefficient and Similarity of Covariance Matrices
The Rv coefficient was introduced by Escoufier (1973) and is a theoretical tool to analyze multivariate data. It is a similarity coefficient between positive semidefinite matrices (e.g. covariance matrices) obtained from the same set of variables. Given two covariance matrices, A and B, the Rv coefficient is defined as:
where T denotes the matrix transpose and tr is the trace of the matrix.
The similarity between two matrices A and B can be analyzed by investigating the first k PCs explaining the maximum of the variance. Suppose that the first k PCs of A and B are given by (a1, ..., ak) and (b1, ..., bk), respectively. Then, a similarity measure between the two matrices A and B can be formulated by:
where < ai, bj> denotes the dot product of the PCs ai and bj. A similar measure has been already considered by Krzanowski (1979).
Network-Based Analysis
Networks were created based on the Pearson correlation between the considered molecular profiles. To control for the number of false positives, a FDR of 0.05 was required. To this end, the smallest threshold rendering the required FDR was determined according to a commonly used procedure (Noble, 2009). Only correlations that in absolute value were not smaller than the threshold value were retained in the network.
Data Analysis
Data normalization and heat map representation were obtained by using built-in R functions (Ihaka and Gentleman, 1996). The network-based analysis was implemented with the help of functions in the igraph package as well as the authors’ implementation in R. The rest of the analyses, including the Rv coefficient and similarity of covariance matrices, were implemented by the authors also in R.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Primary metabolite levels during pepper and tomato development and ripening.
Supplemental Figure S2. Color coding for the nodes representing transcripts for Figure 7.
Supplemental Table S1. Transcriptomic data set.
Supplemental File S1. MapMan (http://mapman.gabipd.org/web/osorio).
Supplementary Material
Acknowledgments
We thank Axel Nagel for online data deposition.
Glossary
- GC-MS
gas chromatography-mass spectrometry
- DAP
days after pollination
- PCs
principal components
- GABA
γ-aminobutyrate
- FDR
false discovery rate
- TCA
tricarboxylic acid
- qRT
quantitative reverse transcription
- SAM
S-adenosyl methionine
- ACC
1-aminocyclopropane-1-carboxylate
References
- Aharoni A, O’Connell AP. (2002) Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J Exp Bot 53: 2073–2087 [DOI] [PubMed] [Google Scholar]
- Aharoni A, Ric de Vos CH, Verhoeven HA, Maliepaard CA, Kruppa G, Bino R, Goodenowe DB. (2002) Nontargeted metabolome analysis by use of Fourier transform ion cyclotron mass spectrometry. OMICS 6: 217–234 [DOI] [PubMed] [Google Scholar]
- Alba R, Fei Z, Payton P, Liu Y, Moore SL, Debbie P, Cohn J, D’Ascenzo M, Gordon JS, Rose JK, et al. (2004) ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development. Plant J 39: 697–714 [DOI] [PubMed] [Google Scholar]
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ. (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17: 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckles DM, Craig J, Smith AM. (2001) ADP-glucose pyrophosphorylase is located in the plastid in developing tomato fruit. Plant Physiol 126: 261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bombarely A, Merchante C, Csukasi F, Cruz-Rus E, Caballero JL, Medina-Escobar N, Blanco-Portales R, Botella MA, Muñoz-Blanco J, Sánchez-Sevilla JF, et al. (2010) Generation and analysis of ESTs from strawberry (Fragaria x ananassa) fruits and evaluation of their utility in genetic and molecular studies. BMC Genomics 11: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F, Backhaus RA, Camara B. (1998) Induction and control of chromoplast-specific carotenoid genes by oxidative stress. J Biol Chem 273: 30651–30659 [DOI] [PubMed] [Google Scholar]
- Brummell DA. (2006) Cell wall disassembly in ripening fruit. Funct Plant Biol 33: 103–119 [DOI] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor MI, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Fernie AR. (2006) Metabolic regulation underlying tomato fruit development. J Exp Bot 57: 1883–1897 [DOI] [PubMed] [Google Scholar]
- Centeno DC, Osorio S, Nunes-Nesi A, Bertolo AL, Carneiro RT, Araújo WL, Steinhauser MC, Michalska J, Rohrmann J, Geigenberger P, et al. (2011) Malate plays a crucial role in starch metabolism, ripening, and soluble solid content of tomato fruit and affects postharvest softening. Plant Cell 23: 162–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc LG, Grimplet J, Wheatley MD, Tillett RL, Quilici DR, Osborne C, Schooley DA, Schlauch KA, Cushman JC, Cramer GR. (2007) Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 8: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Turner JG. (2005) Jasmonate-regulated Arabidopsis stress signalling network. Physiol Plant 123: 161–172 [Google Scholar]
- Enfissi EM, Barneche F, Ahmed I, Lichtlé C, Gerrish C, McQuinn RP, Giovannoni JJ, Lopez-Juez E, Bowler C, Bramley PM, et al. (2010) Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 22: 1190–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoufier Y. (1973) Le traitement des variables vectorielles. Biometrics 29: 751–760 [Google Scholar]
- Fait A, Hanhineva K, Beleggia R, Dai N, Rogachev I, Nikiforova VJ, Fernie AR, Aharoni A. (2008) Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiol 148: 730–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Mattheis JP, Fellman JK. (1998) A role for jasmonates in climacteric fruit ripening. Planta 204: 444–449 [Google Scholar]
- Fortes AM, Agudelo-Romero P, Silva MS, Ali K, Sousa L, Maltese F, Choi YH, Grimplet J, Martinez-Zapater JM, Verpoorte R, et al. (2011) Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding the dynamics of grape ripening. BMC Plant Biol 11: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Enfissi EM, Goodfellow M, Eguchi T, Bramley PM. (2007) Metabolite profiling of plant carotenoids using the matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Plant J 49: 552–564 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM. (1994) Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol 105: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Stevens R, Gil L, Gilbert L, Gest N, Petit J, Faurobert M, Maucourt M, Deborde C, Moing A, et al. (2009) An integrative genomics approach for deciphering the complex interactions between ascorbate metabolism and fruit growth and composition in tomato. C R Biol 332: 1007–1021 [DOI] [PubMed] [Google Scholar]
- Giovannoni J. (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Grimplet J, Deluc LG, Tillett RL, Wheatley MD, Schlauch KA, Cramer GR, Cushman JC. (2007) Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics 8: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J. (2001) Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol 4: 210–218 [DOI] [PubMed] [Google Scholar]
- Hornero-Méndez D, Gómez-Ladrón De Guevara R, Mínguez-Mosquera MI. (2000) Carotenoid biosynthesis changes in five red pepper (Capsicum annuum L.) cultivars during ripening: cultivar selection for breeding. J Agric Food Chem 48: 3857–3864 [DOI] [PubMed] [Google Scholar]
- Hornero-Méndez D, Mínguez-Mosquera MI. (2000) Xanthophyll esterification accompanying carotenoid overaccumulation in chromoplast of Capsicum annuum ripening fruits is a constitutive process and useful for ripeness index. J Agric Food Chem 48: 1617–1622 [DOI] [PubMed] [Google Scholar]
- Howard LR, Talcott ST, Brenes CH, Villalon B. (2000) Changes in phytochemical and antioxidant activity of selected pepper cultivars (Capsicum species) as influenced by maturity. J Agric Food Chem 48: 1713–1720 [DOI] [PubMed] [Google Scholar]
- Howard LR, Wildman REC. (2007) Antioxidant vitamin and phytochemical content of fresh and processed pepper fruit (Capsicum annuum). In LR Howard, REC Wildman, eds, Handbook of Nutraceuticals and Functional Foods. CRC Press, Boca Raton, FL, pp 165–191
- Ihaka R, Gentleman R. (1996) R: a language for data analysis and graphics. J Comput Graph Statist 5: 299–314 [Google Scholar]
- Kevany BM, Tieman DM, Taylor MG, Cin VD, Klee HJ. (2007) Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J 51: 458–467 [DOI] [PubMed] [Google Scholar]
- Klee H, Tieman D. (2002) The tomato ethylene receptor gene family: form and function. Physiol Plant 115: 336–341 [DOI] [PubMed] [Google Scholar]
- Klee HJ. (2002) Control of ethylene-mediated processes in tomato at the level of receptors. J Exp Bot 53: 2057–2063 [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ. (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Kondo S, Tomiyama A. (2000) Changes of endogenous jasmonic acid and methyljasmonate in apples and sweet cherries during fruit development. J Am Soc Hortic Sci 125: 282–287 [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Krzanowski W. (1979) Between-groups comparison of principal components. J Am Stat Assoc 74: 703–707 [Google Scholar]
- Lee JM, Joung JG, McQuinn R, Chung MY, Fei Z, Tieman D, Klee H, Giovannoni J. (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Lee S, Chung EJ, Joung YH, Choi D. (2010) Non-climacteric fruit ripening in pepper: increased transcription of EIL-like genes normally regulated by ethylene. Funct Integr Genomics 10: 135–146 [DOI] [PubMed] [Google Scholar]
- Lemaire-Chamley M, Petit J, Garcia V, Just D, Baldet P, Germain V, Fagard M, Mouassite M, Cheniclet C, Rothan C. (2005) Changes in transcriptional profiles are associated with early fruit tissue specialization in tomato. Plant Physiol 139: 750–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Nunes-Nesi A, Krüger P, Nagel A, Hannemann J, Giorgi FM, Childs L, Osorio S, Walther D, Selbig J, et al. (2010) Robin: an intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol 153: 642–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo VA, Osorio S, Borsani J, Lauxmann MA, Bustamante CA, Budde CO, Andreo CS, Lara MV, Fernie AR, Drincovich MF. (2011) Metabolic profiling during peach fruit development and ripening reveals the metabolic networks that underpin each developmental stage. Plant Physiol 157: 1696–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí MC, Camejo D, Olmos E, Sandalio LM, Fernández-García N, Jiménez A, Sevilla F. (2009) Characterisation and changes in the antioxidant system of chloroplasts and chromoplasts isolated from green and mature pepper fruits. Plant Biol (Stuttg) 11: 613–624 [DOI] [PubMed] [Google Scholar]
- Martínez S, López M, González-Raurich M, Bernardo Alvarez A. (2005) The effects of ripening stage and processing systems on vitamin C content in sweet peppers (Capsicum annuum L.). Int J Food Sci Nutr 56: 45–51 [DOI] [PubMed] [Google Scholar]
- Mattoo AK, Chung SH, Goyal RK, Fatima T, Solomos T, Srivastava A, Handa AK. (2007) Overaccumulation of higher polyamines in ripening transgenic tomato fruit revives metabolic memory, upregulates anabolism-related genes, and positively impacts nutritional quality. J AOAC Int 90: 1456–1464 [PubMed] [Google Scholar]
- Mattoo AK, Handa AK. (2008) Higher polyamines restore and enhance metabolic memory in ripening fruit. Plant Sci 174: 386–393 [Google Scholar]
- Miszczak A, Lange E, Saniewski M, Czapski J. (2000) The effect of methyl jasmonate on ethylene production and CO2 evolution in ‘Jonagold’ apples. Acta Agrobotanica 48: 121–128 [Google Scholar]
- Moore S, Payton P, Wright M, Tanksley S, Giovannoni J. (2005) Utilization of tomato microarrays for comparative gene expression analysis in the Solanaceae. J Exp Bot 56: 2885–2895 [DOI] [PubMed] [Google Scholar]
- Moore S, Vrebalov J, Payton P, Giovannoni J. (2002) Use of genomics tools to isolate key ripening genes and analyse fruit maturation in tomato. J Exp Bot 53: 2023–2030 [DOI] [PubMed] [Google Scholar]
- Mounet F, Moing A, Garcia V, Petit J, Maucourt M, Deborde C, Bernillon S, Le Gall G, Colquhoun I, Defernez M, et al. (2009) Gene and metabolite regulatory network analysis of early developing fruit tissues highlights new candidate genes for the control of tomato fruit composition and development. Plant Physiol 149: 1505–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble WS. (2009) How does multiple testing correction work? Nat Biotechnol 27: 1135–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Alba R, Damasceno CMB, Lopez-Casado G, Lohse M, Zanor MI, Tohge T, Usadel B, Rose JKC, Fei Z, et al. (2011) Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol 157: 405–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Castillejo C, Quesada MA, Medina-Escobar N, Brownsey GJ, Suau R, Heredia A, Botella MA, Valpuesta V. (2008) Partial demethylation of oligogalacturonides by pectin methyl esterase 1 is required for eliciting defence responses in wild strawberry (Fragaria vesca). Plant J 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Osorio S, Do PT, Fernie AR. (2012) Profiling primary metabolites of tomato fruit with gas chromatography/mass spectrometry. Methods Mol Biol 860: 101–109 [DOI] [PubMed] [Google Scholar]
- Ranwala AP, Suematsu C, Masuda H. (1992) The role of β-galactosidases in the modification of cell wall components during muskmelon fruit ripening. Plant Physiol 100: 1318–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR. (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Hadfield KA, Labavitch JM, Bennett AB. (1998) Temporal sequence of cell wall disassembly in rapidly ripening melon fruit. Plant Physiol 117: 345–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Saladié M, Catalá C. (2004) The plot thickens: new perspectives of primary cell wall modification. Curr Opin Plant Biol 7: 296–301 [DOI] [PubMed] [Google Scholar]
- Rose JKC, Bennett AB. (1999) Cooperative disassembly of the cellulose-xyloglucan network of plant cell walls: parallels between cell expansion and fruit ripening. Trends Plant Sci 4: 176–183 [DOI] [PubMed] [Google Scholar]
- Sakurai N, Nevins DJ. (1993) Changes in physical properties and cell wall polysaccharides of tomato (Lycopersicon esculentum) pericarp tissues. Physiol Plant 89: 681–686 [Google Scholar]
- Saniewski M, Nowacki J, Lange E, Czapski J. (1986) The effect of methyl jasmonate on ethylene and 1-aminocyclopropane-1-carboxylic acid production in preclimacteric and postclimacteric ‘Jonathan’ apples. Fruit Sci. Rpt. 13: 193–200 [Google Scholar]
- Seymour G. (1993) Biochemistry of Fruit Ripening. Chapman and Hall, London
- Srivastava A, Gupta AK, Datsenka T, Mattoo AK, Handa AK. (2010) Maturity and ripening-stage specific modulation of tomato (Solanum lycopersicum) fruit transcriptome. GM Crops 1: 237–249 [DOI] [PubMed] [Google Scholar]
- Tieman D, Taylor M, Schauer N, Fernie AR, Hanson AD, Klee HJ. (2006) Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc Natl Acad Sci USA 103: 8287–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. (2009) A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, maize. Plant Cell Environ 32: 1211–1229 [DOI] [PubMed] [Google Scholar]
- Villavicencio L, Blankenship S, Sanders D, Swallow W. (1999) Ethylene and carbon dioxide production in detached fruit of selected pepper cultivars. J Am Soc Hortic Sci 124: 402–406 [Google Scholar]
- Vriezen WH, Feron R, Maretto F, Keijman J, Mariani C. (2008) Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol 177: 60–76 [DOI] [PubMed] [Google Scholar]
- Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latché A, Pech JC, Fernie AR, Bouzayen M. (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21: 1428–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Hoffman N. (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol Plant Mol Biol 35: 155–189 [Google Scholar]
- Zamboni A, Di Carli M, Guzzo F, Stocchero M, Zenoni S, Ferrarini A, Tononi P, Toffali K, Desiderio A, Lilley KS, et al. (2010) Identification of putative stage-specific grapevine berry biomarkers and omics data integration into networks. Plant Physiol 154: 1439–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanor MI, Osorio S, Nunes-Nesi A, Carrari F, Lohse M, Usadel B, Kühn C, Bleiss W, Giavalisco P, Willmitzer L, et al. (2009) RNA interference of LIN5 in tomato confirms its role in controlling Brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiol 150: 1204–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.