Abstract
Expansins are cell wall proteins implicated in the control of plant growth via loosening of the extracellular matrix. They are encoded by a large gene family, and data linked to loss of single gene function to support a role of expansins in leaf growth remain limited. Here, we provide a quantitative growth analysis of transgenics containing an inducible artificial microRNA construct designed to down-regulate the expression of a number of expansin genes that an expression analysis indicated are expressed during the development of Arabidopsis (Arabidopsis thaliana) leaf 6. The results support the hypothesis that expansins are required for leaf growth and show that decreased expansin gene expression leads to a more marked repression of growth during the later stage of leaf development. In addition, a histological analysis of leaves in which expansin gene expression was suppressed indicates that, despite smaller leaves, mean cell size was increased. These data provide functional evidence for a role of expansins in leaf growth, indicate the importance of tissue/organ developmental context for the outcome of altered expansin gene expression, and highlight the separation of the outcome of expansin gene expression at the cellular and organ levels.
Expansins are extracellular matrix proteins that have long been implicated in the control of plant growth processes via their role as modulators of cell wall extensibility (Cosgrove, 2005). Although numerous papers have highlighted correlations between expansin gene expression and growth (Li et al., 2003), there are only a limited number of reported experiments in which expansin gene expression has been modulated via transgenic methods to result in a clear growth response. Moreover, a number of these investigations (including those from our own group) have involved the overexpression of expansin genes, which, although informative, cannot be used as absolute proof of gene function (Pien et al., 2001). This requires the repression of endogenous gene function to be accompanied by a measurable phenotype. In the context of expansins, the challenge is that the protein tends to be encoded by relatively large gene families, so that any potential phenotype resulting from the suppression of one family member is liable to be masked by the ability of related family members to take over that function (gene redundancy; Li et al., 2002; Schipper et al., 2002). Despite this potential complication, reports have been made in which a clear growth phenotype resulted from the suppression of a single expansin gene (Cho and Cosgrove, 2000; Choi et al., 2003; Zenoni et al., 2004; ZhiMing et al., 2011). These investigations have targeted root hairs and petals in dicots and the mesocotyl and coleoptile in monocots, but to date, only one experiment relating the suppression of expansin gene expression to leaf growth has been reported.
In Arabidopsis (Arabidopsis thaliana), Cho and Cosgrove (2000) targeted the suppression of α-EXPANSIN10 (AtEXPA10) via the expression of an antisense RNA in the expression domain of the AtEXPA10 gene (essentially the leaf midrib and petiole). This led to the formation of smaller, curled leaves. Since the gene suppression was specifically targeted to only a subset of cells within the leaf, the role of expansins in the general mechanism of leaf growth remains unclear. In addition, since an endogenous promoter system was used, it was not possible to explore the significance of the differential timing or spatial expression of the antisense construct used to manipulate endogenous expansin activity. For example, our own recent data (based on inducible expansin overexpression) suggested that there is a time frame during leaf development when an exogenously imposed increase in expansin activity has a maximum effect on leaf growth (Sloan et al., 2009). This time period corresponded developmentally with the phase of maximum absolute rate of leaf expansion (Emax), linking with observations that final leaf size correlates with the Emax achieved during development rather than simply the duration of expansion (Cookson et al., 2005).
Understanding the control of leaf growth, in particular leaf size, is a fundamental problem in plant biology. Although a number of genetic regulators of leaf size have been identified (Anastasiou et al., 2007; Anastasiou and Lenhard, 2007), the downstream mechanism by which such factors alter growth remains unclear. Attempts to study the regulation of final leaf size has revealed a complex network of multiple independent molecular pathways, leading to a focus on the role of cell division processes (Gonzalez et al., 2010). Although the modulation of cell wall extensibility via expansin activity has been postulated as an important mechanism by which the control of growth is exerted (Keller and Cosgrove, 1995; Sloan et al., 2009), functional evidence to support this hypothesis remains limited. Inducible down-regulation of expansin gene expression throughout the leaf would provide evidence on the endogenous function of this protein family in the control of leaf growth and insight into the importance of the timing of expansin gene expression (and thus growth rate) in the control of leaf size.
Armed with the genomic tools and data that Arabidopsis research now provides, we set out to analyze the expression pattern of all 25 annotated EXPAs during leaf development. Based on this information, we devised an artificial microRNA (amiRNA) construct to down-regulate the expression of a group of expansin genes relatively highly expressed throughout leaf development. In addition, we placed this construct under the control of an inducible promoter system, allowing us to manipulate the timing of the suppression of target gene expression.
The results of these experiments show that the suppression of expansin gene expression reduces leaf growth, suggest a differential growth response between the leaf lamina and petiole, and identify a restricted developmental phase when the repression of expansin gene expression leads to a reduction in absolute and relative growth rate.
RESULTS
Expansin Gene Expression during Leaf Development
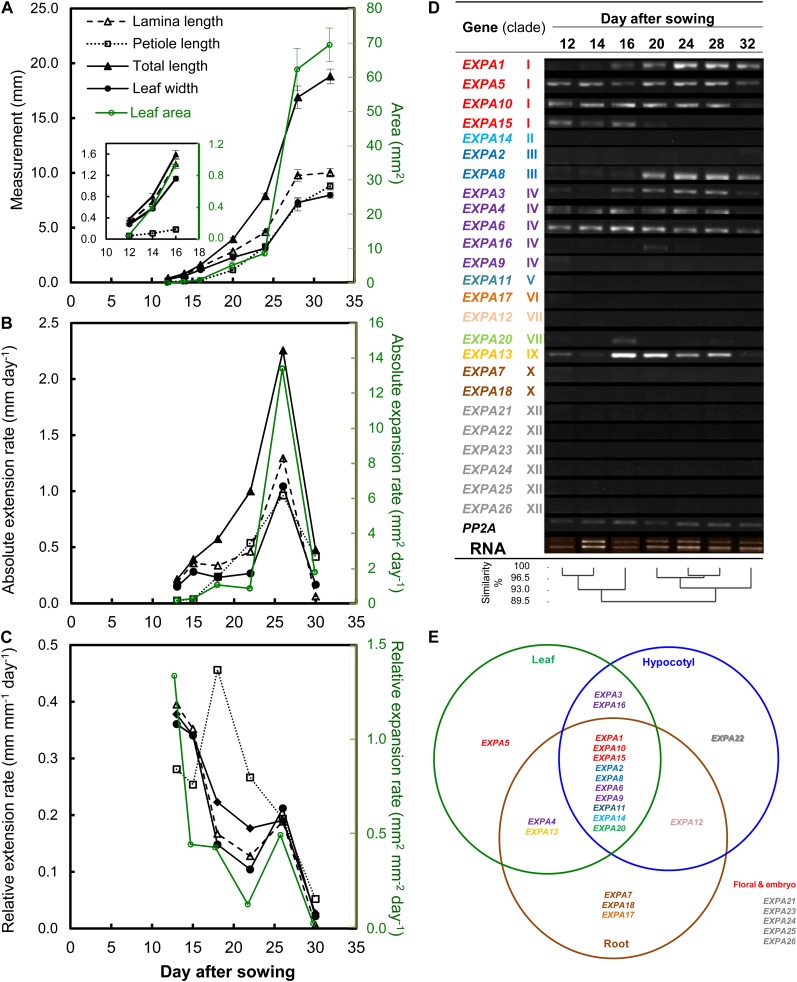
For any analysis involving leaf development, it is essential to first establish a robust framework to allow the appropriate comparison of different samples. Arabidopsis germination and initial leaf growth shows a high inherent level of variability, even under tightly controlled growth conditions (Massonnet et al., 2010). A previously established staging system (Kuwabara et al., 2011) was applied to acquire developmentally equivalent samples for the analysis of Arabidopsis expansin gene expression during leaf development, as described in “Materials and Methods.” Growth of leaf 6, which previous analyses have established shows a typical pattern for Arabidopsis (Tsuge et al., 1996; Cookson et al., 2005), was measured over time. The leaf showed a sigmoidal growth curve for both length and width (Fig. 1A), and three phases characteristic of dicot leaf development (proliferative, 12–16 d after sowing [DAS]; expansive, 20–28 DAS; and mature stage, 32 DAS) can be identified (Beemster et al., 2005). The growth pattern for leaf area was similar to that for linear dimensions of the lamina, the only disparity being that lamina growth ceased earlier than petiole growth. When the absolute rates of extension for length and width were plotted against time, a maximum rate was observed at around 25 DAS (Fig. 1B), and this peak occurred at a similar time point for the maximum rate of lamina expansion. The relative change in extension/expansion rate of the lamina with time (relative growth rate) showed a more complex pattern, with a general decline as leaf 6 developed. However, there was a hiatus in this decline that coincided approximately with the time of Emax (Fig. 1C). The peak of petiole relative extension rate occurred distinctly earlier (15–20 DAS) than the other measured leaf parameters.
Figure 1.
Expression analysis of the EXPA gene family with time-course leaf growth analysis of leaf 6 of wild-type Col-0 plants. A, Changes with time of lamina length, lamina width, petiole length, total (lamina + petiole) length, and total leaf area. The inset shows a portion of the data during early leaf development. Values are means ± se (n = 6–14). B, Average absolute extension/expansion rates. C, Average relative extension/expansion rates calculated from the data in A. Error bars are not shown for clarity. D, Detection of EXPA gene expression during leaf 6 development by RT-PCR. PP2A was used as an internal control gene. EXPA genes are ordered by similarity of gene expression pattern and color coded according to clade (Sampedro at al., 2005). E, Expression of EXPA genes in leaf, hypocotyl, and root compiled from literature and public microarray data sets. Data for hypocotyl tissue are from Jamet et al. (2009), data for root tissue are from Wieczorek et al. (2006), whereas those for the leaf were compiled from this study and Beemster et al. (2005). Floral organ and embryo tissue were excluded from the Venn diagram for clarity. The gene names are color coded according to their respective group of monocot-eudicot clade classification (Sampedro at al., 2005). [See online article for color version of this figure.]
To characterize the pattern of EXPA gene expression, leaves at different developmental stages were dissected, RNA was extracted, and the presence/absence of individual expansin transcripts were assayed by reverse transcription (RT)-PCR (Fig. 1D). The earliest stage of leaf development analyzed here (12–14 DAS) was characterized by the expression of at least seven EXPA genes. These showed a general pattern of maintained expression throughout development, with other EXPA transcripts becoming detectable at later stages of development, with a maximum number of up to 10 EXPA genes being expressed over a broad period from 20 to 28 DAS. This period of maximum number of expressed EXPA genes correlates with the period of maximum absolute rate (Fig. 1B) and the hiatus in relative rate of leaf expansion (Fig. 1C). A previous analysis of the 25 EXPA gene sequences allowed their classification into a number of clades (Sampedro et al., 2005) that are color coded in Figure 1D. The order of the expansin genes was arranged according to their clade classification and similarity in expression pattern. The genes that showed common expression patterns belong to a variety of clades, with no particular clade showing a specific pattern, other than clades VI, VII, X, and XII, which were not expressed at any point during the development of leaf 6.
In addition to the RT-PCR analysis of expansin gene expression during leaf development, we performed a bioinformatic analysis of the available microarray data (Beemster et al., 2005) to put our results into the context of the reported expression patterns of expansin genes in other plant organs (Fig. 1E). These data corroborated the results in Figure 1D, indicating that a total of 12 EXPA genes are expressed at some point during Arabidopsis leaf development.
Suppression of Expansin Gene Expression via amiRNA Leads to the Repression of Leaf Growth
In order to silence multiple expansin genes that were relatively highly expressed during leaf development, we took an amiRNA approach pioneered by Schwab et al. (2006). We designed a construct to target four expansin genes at once, three from clade I (EXPA1, -5, and -10) and one of the clade IV expansins (EXPA3), all of which are clearly expressed during the early to mid stage of leaf growth. Our strategy was to down-regulate the expression of four of the eight expansin genes that, by our analysis, are expressed to some degree at all points during leaf development (Fig. 1D). To allow for a temporal control of expansin gene suppression, we cloned the EXPA10,1,5,3 amiRNA construct into an inducible vector system (pOpON; Wielopolska et al., 2005), which also facilitates the visualization of where induction occurs due to the presence of a bidirectional promoter linked to the GUS reporter gene (Supplemental Fig. S1). Six independent transformant lines were obtained, selfed, and further characterized through segregation analyses and GUS induction tests (Fig. 2, A and B) to select for stable transformants showing uniform growth under noninduction conditions and minimal background gene expression. Two homozygous T3 pOpON::amiREXPA10,1,5,3 (denoted pOpON::amiREXP) lines were selected for further investigation. In the following, experimental data are generally shown for one line (pOpON::amiREXP-3/22) but comparable replicate data were obtained with another independent transgenic line for all analyses.
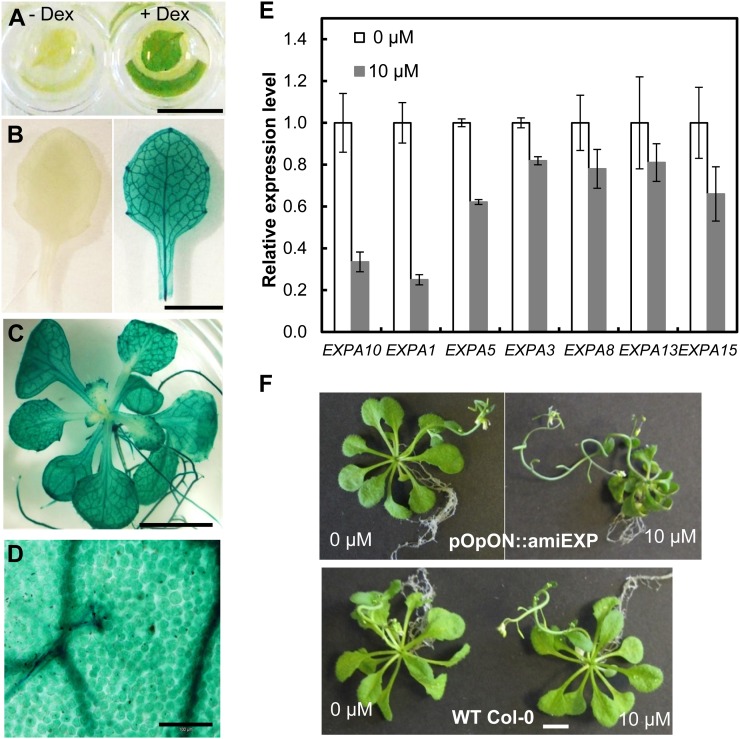
Figure 2.
Characterization of pOpON::amiREXP transgenic plants. A and B, A 12-DAS first leaf pair (A) and an 18-DAS leaf 6 (B) from a pOpON::amiREXP plant treated with (+) or without (−) 10 µm Dex overnight prior to the visualization of GUS activity (blue coloration). C, GUS staining of a 20-DAS pOpON::amiREXP plant grown with 10 µm Dex. D, Micrograph of induced leaf 6 from a 20-DAS pOpON::amiREXP plant showing induction in all cells. E, Expression levels of amiRNA-targeted (EXPA10, -1, -5, -3) and nontargeted (EXPA8, -13, -15) expansin genes in 12-DAS plants grown on medium supplemented with 10 µm Dex (gray bars) or control medium (white bars) determined by real-time qPCR analysis. Values are means ± se from three biological replicates, normalized to the amount of ACT2 and UBC21. F, Comparison of pOpON::amiREXP and wild-type (WT) plants grown with 10 µm Dex or control medium at 31 DAS. Bars = 5 mm (A–C), 100 µm (D), and 5 mm (F). [See online article for color version of this figure.]
GUS reporter gene analysis indicated that, following supply of the dexamethasone (Dex) inducer via the growth medium, induction of the amiRNA construct occurred within 24 h and was detectable in all regions of the plant, including leaf 6 (the target leaf for subsequent growth analysis; Fig. 2C). Within leaf 6, GUS expression was present in all cells of the leaf (Fig. 2D). There was an apparently higher signal in the vascular tissue, but this might simply reflect the presence of relatively small cytoplasmically dense cells in this tissue. Quantitative PCR (qPCR) analysis of leaf 6 from induced or noninduced plants verified that transcripts for the target genes EXPA10, EXPA1, EXPA5, and EXPA3 were all present at significantly (P < 0.05) lower levels in the induced plants (Fig. 2E). Transcript levels for nontarget EXPAs tended to be lower than in control leaves, but these differences were not statistically significant.
When the pOpON::amiREXP plants were germinated on Dex-containing medium, an overt phenotype was observed, with growth greatly reduced compared with control and Dex-treated wild-type plants at 30 DAS (Fig. 2F). This growth suppression was also observed in an independent transgenic line, with an increasing severity of growth repression after induction with increasing Dex concentrations at 1 µm or higher (Supplemental Fig. S2). There was also a decrease in the average number of vegetative leaves generated by the plants after suppression of expansin gene expression, but the rate of leaf initiation was unaltered (Supplemental Fig. S3).
Rosette and Leaf Growth Response to Suppression of Expansin Gene Expression
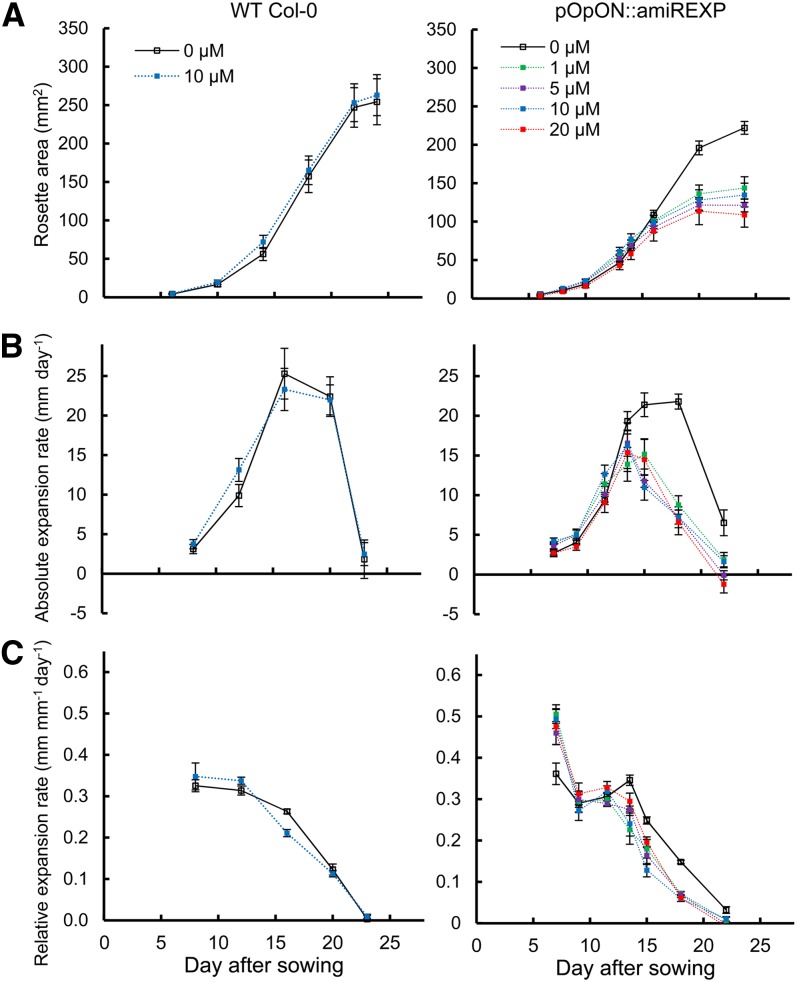
To investigate the growth response more closely, we performed detailed analyses of individual pOpON::amiREXP plants. First, the total rosette area was recorded over time (6–24 DAS) in the presence of a Dex concentration ranging from 0 to 20 µm (Fig. 3). The growth response of Columbia-0 (Col-0) plants on medium containing 0 and 10 µm Dex was used as a control. These analyses were performed on solid medium but without the staged transfers used for the expansin gene expression analysis described in Figure 1, since transfer influences the growth dynamics. Under these conditions, a maximal areal rosette expansion rate was attained at around 15 DAS (Fig. 3B).
Figure 3.
Induction of pOpON::amiREXP plants leads to a decrease in rosette growth. A, Change of rosette area (projected coverage) with time (DAS). B, Absolute rosette area expansion rates. C, Relative rosette area expansion rates. Values are means ± se (n = 12) for plants treated with different concentrations of Dex. Results are shown for wild-type (WT) Col-0 plants and pOpON::amiREXPA plants after treatment with various concentrations of the Dex inducer, as indicated. [See online article for color version of this figure.]
The results (Fig. 3A) substantiated the visual observations in Figure 2F and Supplemental Figure S2 that an increasing Dex induction led to an increase in growth suppression. At early time points, the growth curves were similar in form; however, the growth of induced rosettes was significantly suppressed compared with the controls at 16 DAS and later (Fig. 3A). This was reflected in the rosette expansion rates of induced pOpON::amiREXP plants, which declined soon after 12 DAS, reaching a lower Emax compared with control plants (Fig. 3B). A higher relative expansion rate was observed in induced pOpON::amiREXP plants at the earliest developmental stage measured, but a more rapid decline in relative growth rate occurred in these plants than in the controls (Fig. 3C). In all cases, the Dex-treated nontransgenic control Col-0 plants showed no significant difference in rosette area at any time point.
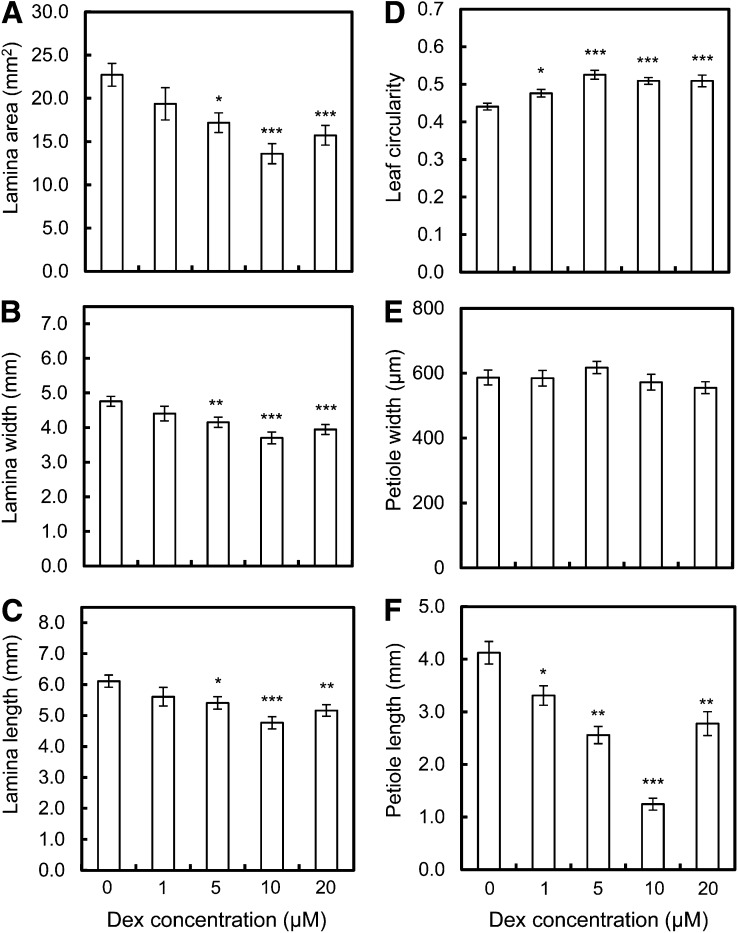
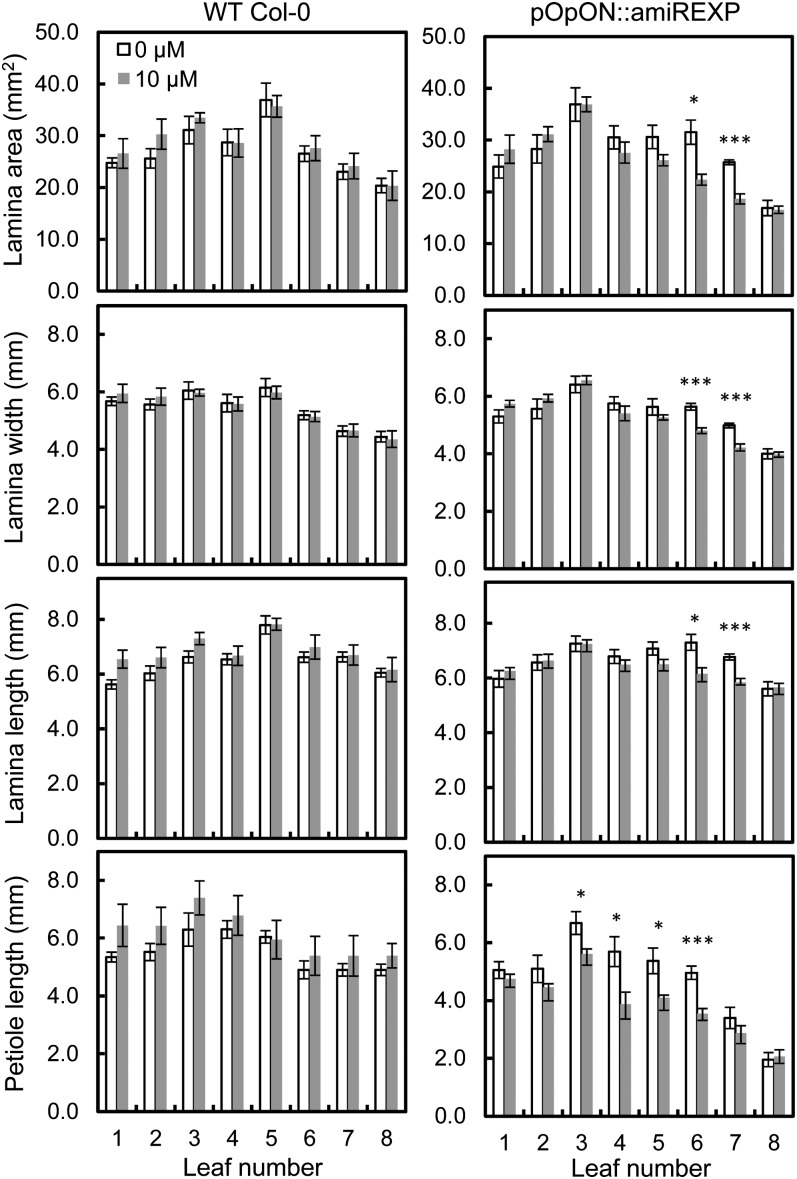
To investigate the individual leaf basis of the changes underpinning total rosette area after the repression of expansin gene expression, we first analyzed size and shape parameters of leaf 6 at maturity of the pOpON::amiREXP plants either with or without Dex induction. These results showed that after Dex induction at concentrations of 5 µm and higher, leaf lamina width, length, and area were all significantly (at least P < 0.05) decreased in the pOpON::amiREXP plants (Fig. 4, A–C). In addition to the repression of leaf lamina size, there was a clear and significant decrease in petiole length after the repression of expansin gene expression (Fig. 4F), with no significant effect on petiole width (Fig. 4E). These differences in petiole length were already detectable after induction with only 1 µm Dex. Thus, the repression of expansin gene expression led to a smaller leaf lamina and shorter petiole. Combined, this led to an overall increase in leaf circularity (area per perimeter2; i.e. a more compact leaf [Fig. 4D]).
Figure 4.
Decreased final lamina and petiole size after induction of pOpON::amiREXP plants. A, Lamina area. B, Lamina width. C, Lamina length. D, Leaf circularity. E, Petiole width. F, Petiole length. All measurements are for leaf 6 from pOpON::amiREXP plants at 27 DAS grown on medium supplemented with different concentrations of Dex. Values are means ± se (n = 10–13). One-way ANOVA, Tukey test compared with the 0 µm control: *P < 0.05, **P < 0.01, ***P < 0.001.
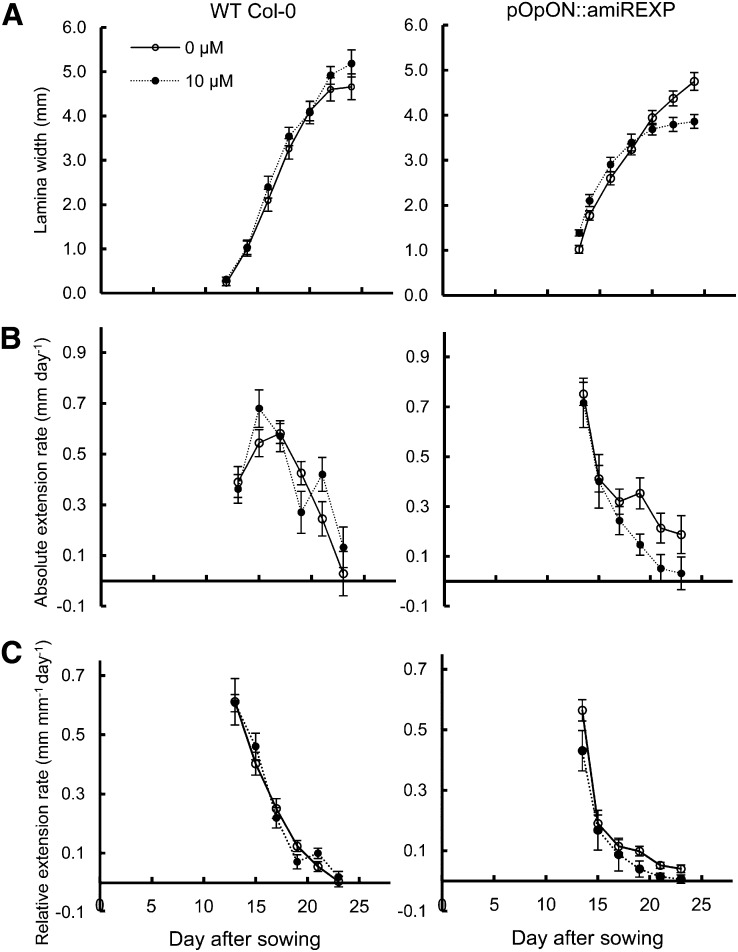
To further resolve the leaf response to repressed expansin gene expression, the growth of leaf 6 was observed over time by the measurement of leaf width as a proxy for lamina growth (Sloan et al., 2009; Kuwabara et al., 2011). As shown in Figure 5A, the growth curves of induced and mock-treated pOpON::amiREXP leaves are similar until approximately 20 DAS. Despite a head start, the growth of induced pOpON::amiREXP leaves reached a plateau earlier, resulting in a significantly (P < 0.05) smaller final size. The Emax was not significantly different between induced and noninduced leaves (Fig. 5B); however, there was a more rapid decline in absolute extension rate in the induced plants during the later phase of leaf growth. When expressed as relative extension rate, the form of the curves for induced and noninduced leaves was similar, but with the induced plants showing a more rapid decline in relative growth rate compared with the control leaves after starting from a slightly higher initial value (Fig. 5C).
Figure 5.
Temporal analysis of lamina growth rates in induced pOpON::amiREXP leaves. A, Lamina width. B, Absolute lamina extension rate. C, Relative lamina extension rate. All measurements are for leaf 6 of pOpON::amiREXP and Col-0 wild-type (WT) plants grown continually either with or without 10 µm Dex. Values are means ± se (n = 12).
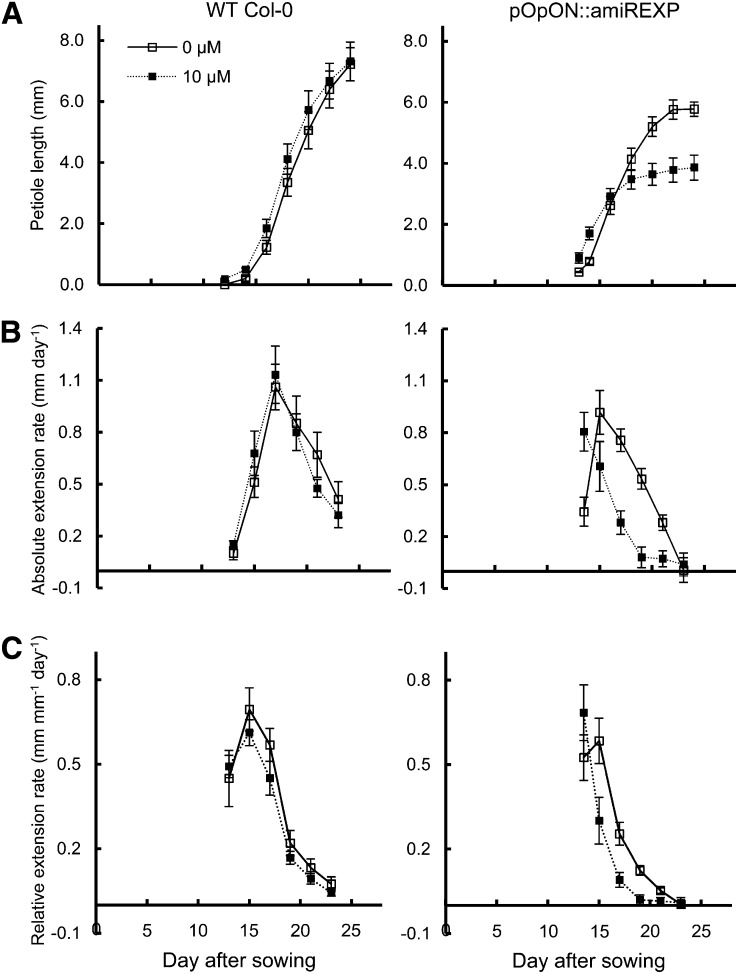
The growth response was rather different in the case of the petiole (Fig. 6). First, although final petiole length was clearly decreased after the repression of expansin gene expression, there was an early phase when the length of the induced pOpON::amiREXP petioles was greater than that of the mock-treated plants (Fig. 6A). In terms of absolute growth rate, the repression of expansin gene expression did not seem to greatly decrease the Emax obtained; rather, Emax occurred at a slightly earlier time point, followed by a comparable rate of decline to the control leaves (Fig. 6B). This was also reflected in the analysis of relative growth rate by an earlier peak of relative growth rate in the induced petioles and an earlier decline in relative growth rate (Fig. 6C). Therefore, the petiole growth pattern of induced plants appeared to be temporally shifted, with an earlier and shorter window of growth compared with control plants. Similar results were also obtained from an independent transgenic line (Supplemental Fig. S4).
Figure 6.
Petiole growth pattern is shifted in induced pOpON::amiREXP plants. A, Petiole length. B, Absolute petiole extension rate. C, Relative petiole extension rate. All measurements are for leaf 6 petioles of pOpON::amiREXP and Col-0 wild-type (WT) plants grown continually either with or without 10 µm Dex. Values are means ± se (n = 12).
To investigate the cellular basis of these growth responses, a histological analysis was performed on induced and noninduced leaves (Table I; Supplemental Fig. S5). Contrary to the expected decrease in cell size suggested by the observed decrease in leaf lamina area (Fig. 4, A and F), adaxial epidermal cells were actually significantly larger in the induced pOpON::amiREXP leaves than in the controls, and there was also a tendency for an increase in mesophyll cell size after the repression of expansin gene expression. Similarly, petiole mesophyll cells displayed a tendency for increased size (notably in width) but petiole epidermal cells had a tendency for decreased length, although this was not statistically significant. In all cases, the cell number and the number of cell files were significantly (P < 0.05) decreased in both lamina and petiole.
Table I. Histological analysis of cell size and number of leaf 6 lamina and petioles harvested at 27 DAS from pOpON::amiREXP plants after induction with Dex (10 µm) compared with mock-induced controls.
The values shown are means ± se of six biological replicates, with n = number of cells measured per replicate. Numbers in boldface indicate statistical significance at P < 0.05 in a two-sample t test (cell size) or Mann-Whitney U test (number of cells per cell file).
| Parameter |
Dex Concentration |
|
|---|---|---|
| 0 μm | 10 μm | |
| Lamina adaxial epidermis (n = 10) | ||
| Cell area (μm2) | 2,377 ± 98 | 2,933 ± 144 |
| Pavement cell no.a | 88 ± 6 | 63 ± 11 |
| Stomata no.a | 32 ± 4 | 21 ± 5 |
| Lamina mesophyll (n = 28) | ||
| Cell area (μm2) | 532 ± 42 | 672 ± 71 |
| Cell width (μm) | 27 ± 1.0 | 30 ± 1.3 |
| Cell length (μm) | 26 ± 1.0 | 30 ± 1.5 |
| Cell no.a | 16 ± 0.8 | 14 ± 0.8 |
| Transverse | ||
| Longitudinal | 14 ± 0.4 | 13 ± 1.0 |
| Total | 207 ± 14 | 150 ± 13 |
| Petiole adaxial epidermis (n > 10) | ||
| Cell length (μm) | 191 ± 21 | 164 ± 10 |
| No. of cell filesb | 25 ± 0.7 | 20 ± 0.6 |
| Petiole mesophyll (n = 20) | ||
| Cell area (μm2) | 621 ± 57 | 734 ± 70 |
| Cell width (μm) | 25 ± 0.7 | 29 ± 2.1 |
| Cell length (μm) | 32 ± 2.0 | 30 ± 2.1 |
| Cell no. along cell filec | 19 ± 1.3 | 18 ± 2.0 |
| No. of cell filesb | 20 ± 0.6 | 17 ± 0.6 |
Cell number was counted within the area captured by the micrograph (268 μm × 356 μm). bThe number of cell files was counted across the petiole within the area captured by the micrograph (536 μm × 712 μm). cCell number was counted along the leaf-length direction on the third cell file from the margin of the petiole (536 μm length).
In the above experiments, the Dex inducer was supplied to the plants from germination onward. As shown in Supplemental Figure S3, there was a decrease in the total number of leaves generated by the plants under these conditions, raising the possibility that the altered leaf growth might in some way be related to a general decrease in plant growth rather than a specific outcome on the leaves measured. To investigate this possibility, we performed a series of experiments in which expansin gene expression was suppressed at a specific later time point in development. The induction was performed by the addition of a drop of Dex to the shoot apex of 12-DAS plants growing on standard medium, at which time point leaves 6 and 7 have been initiated. The plants were then allowed to grow for a further 8 d and the final leaf size parameters were measured. As shown in Figure 7, the induction of the pOpON::amiREXP plants led to a decrease in final lamina area only for leaves 6 and 7, with developmentally older (leaves 1–5) and younger (leaf 8) leaves showing no significant change in size. A response to the induction of pOpON::amiREXP was also observed in the petiole, but with a broader developmental window. Thus, petioles of leaves 3 to 6 all displayed a significant (P < 0.05) decrease in petiole length after the suppression of expansin gene expression. Control plants treated with Dex did not show this response. Analysis of GUS expression after droplet induction indicated that all leaves measured for size parameters had perceived the inducer by showing reporter gene expression (Supplemental Fig. S6).
Figure 7.
Temporally restricted induction of pOpON::amiREXP plants leads to a restriction in leaf growth response. Data show a comparison of lamina and petiole size between induced (shaded bars) and mock-induced (nonshaded bars) leaves from Col-0 wild-type (WT) and pOpON::amiREXP plants induced on the apex with Dex (10 µm) at 12 DAS and measured at 20 DAS. Values are means ± se (n = 5–7). Two-sample t test: *P < 0.05, ***P < 0.001.
DISCUSSION
Since their discovery, a significant body of work has accumulated implicating expansins in the control of cell wall extensibility and, hence, plant growth (Cosgrove, 2005). However, the number of reports showing that repression of expansin gene expression actually leads to a repression in growth is surprisingly limited (Cho and Cosgrove, 2000; Zenoni et al., 2004; ZhiMing et al., 2011). Part of the reason for this is that expansins are encoded by relatively large gene families, leading to potential problems of genetic redundancy when single gene members are mutated or their expression is repressed (Schipper et al., 2002). The advent of amiRNA techniques provides a potential solution to the problem, since a single small RNA may bind to a number of related sequence elements, thus potentially leading to the degradation of a number of endogenous targets (Schwab et al., 2006). However, even this approach is limited when faced with a large number of related target genes. A solution to this problem is to target the amiRNA to a particular organ or tissue where only a subset of a large gene family is known to be expressed. In addition, combining this approach with an inducible system for the expression amiRNA is likely to increase the potential for obtaining a detectable phenotype, since there is less scope for the biological system to respond rapidly enough to counteract any phenotype induced by the suppression of multiple target genes. Here, we report on such an approach, focusing on the cell wall protein expansin and leaf growth. Our data show that suppression of the expression of multiple expansin genes leads to a marked reduction in leaf growth. Our analysis of the resultant growth curves indicates that decreased expansin expression leads to a more rapid decline in relative growth rate throughout leaf development, but this becomes most marked in the later phase, leading to a decrease in maximal absolute expansion rate and a decrease in final organ size. This is, to our knowledge, the first quantitative analysis of the growth outcome of decreased expansin expression. The data are consistent with the idea that expansins are required for leaf growth, but this may be most important during the later phase of development, when growth is normally decreasing. The molecular nature of the endogenous brake on growth is unclear but may include elements of cell wall structure that lead to the tissue becoming less extensible (Cosgrove, 2005). Our observations suggest that during the later phase of leaf development, expansins might counteract processes that put a brake on growth and that depleted expansin expression allows these brakes on growth to be applied more effectively, leading to smaller leaves.
Repression of the Expression of Multiple Expansin Genes Leads to Decreased Leaf Growth
Our approach first required knowledge of the expression pattern of all EXPAs during Arabidopsis leaf development. The analysis reported here provides a description of the dynamic expression pattern of all EXPAs during the complete development of a model dicot leaf. The data indicate that eight EXPAs are expressed at all stages of leaf 6 development and that the other 17 are either never or only weakly expressed during particular phases of leaf development. Numerous data on expansin gene expression are present in the databases, and our results are consistent with previous reports identifying which expansin genes are expressed during Arabidopsis leaf development (Beemster et al., 2005; Wieczorek et al., 2006). These data provided an atlas of expansin gene expression and allowed us to restrict our attempt to decrease expansin gene expression to a subset of the gene family. This subset was still relatively large, suggesting that there is a significant potential degree of redundancy in expansin gene expression during even the earliest stages of leaf development and/or that even at this early stage there is a high degree of expansin functional specialization. The number of expansin genes expressed is higher than the number of distinct cell types easily recognizable in the leaf at this stage of development, suggesting that specialization for function is unlikely to solely account for the expression of so many distinct expansin proteins (Cosgrove, 2000).
A single construct was used to suppress the expression of four EXPAs that are relatively highly expressed throughout leaf development. Evidence of the success of this approach came from molecular analysis indicating a decrease in the endogenous transcript levels of the target genes after Dex induction and from the overt growth defects after this induction. Expansin proteins are relatively stable, and the available antibodies do not discriminate between different expansin isoforms. Detecting any gross decrease in expansin protein level as the result of suppressing the expression of a subset of the gene family is a challenge, and a number of papers record a mismatch between the level of specific expansin transcripts and the total level of detectable protein (Caderas et al., 2000). Similarly, the methods used for the quantification of expansin activity measure the gross activity in the extracted tissue and, thus, are liable to be relatively insensitive to the loss of a fraction of the activity related to a subfamily of the genes expressed (McQueen-Mason, 1995). A further complication for any analysis at the protein and activity levels is the possibility that only a portion of the expressed expansin protein is active (or is at the appropriate cell wall location for an effect; Im et al., 2000). Thus, although our RNA data are consistent with the amiRNA approach having worked, we do not have conclusive data that this was reflected by a decrease in endogenous expansin protein level or activity. However, the dramatic decrease in growth observed only after induction of the pOpON::amiREXP plants and the lack of any visible phenotype following mock induction of control plants indicate that the decrease in expansin gene expression was sufficient to decrease tissue growth, thus providing strong evidence to substantiate the proposal that expansins are required for leaf growth (Cho and Cosgrove, 2000).
The Growth Response to Decreased Expansin Expression Is Context Dependent
Our analysis of growth following decreased expansin expression revealed a complex response. First, at the level of the whole rosette area, there was a lower maximum rate of absolute rosette expansion and a drastically more rapid decrease in expansion rate following this peak level (Fig. 3). At the level of the individual leaf, the relative lamina expansion rate was also generally lower than the controls, but this became more striking toward the end of leaf growth, again reflecting a more rapid decline in relative growth rate (Fig. 5). These two observations are consistent with the idea that the relative growth rate of a leaf reflects a balance between growth-promoting and growth-inhibitory processes that essentially acts as a brake on growth. In this light, although expansins are often viewed as having a growth-promoting function, it might be better to understand them, at least in the latter part of development, as a counterbalance to growth-repressing activities present within the leaf, as outlined above.
Within the leaf, the restriction of petiole extension was much more pronounced than the degree of repressed lamina expansion following induction of the pOpON::amiREXP plants (Figs. 4 and 5). This raises the possibility of a differential sensitivity of the two tissues to the same manipulation of expansin gene expression. In addition, experiments in which induction was performed at a later developmental time point resulted in growth repression only in leaves at a particular developmental stage (Fig. 7). These observations are consistent with the idea that the response to altered expansin expression depends on both the developmental stage of the organ under study and the tissue within that organ (Sloan et al., 2009). However, at present, we cannot discount the possibility that this differential response to induction might reflect a different abilities of the two distinct parts/stages of the leaf to counter the imposed repression of expansin gene expression by, for example, up-regulating alternative expansin genes. Further investigation is required, but our data highlight the importance of defining which tissues and which developmental stage are being investigated when trying to understand the endogenous function of expansins.
Expansins and the Cellular Basis of Growth
Histological analysis of leaves and petioles in which expansin gene expression was repressed revealed an interesting and counterintuitive response in terms of mean cell size. Thus, following the repression of expansin gene expression in the pOpON::amiREXP plants, the mean cell area was significantly increased in the lamina epidermis, although the overall lamina area was decreased. A similar trend was also observed in the lamina mesophyll (i.e. following the repression of expansin gene expression, a smaller lamina was generated consisting of fewer but larger cells; Table I). It has long been recognized that plants can undergo a growth compensatory process whereby the constituent cell size can, to a certain extent, accommodate to counteract manipulations that tend to alter final organ size from a set normal level (Tsukaya, 2002). The mechanism by which this “normal” level is set and how the constituent cell size is accommodated remain unknown. The data reported here indicate that following an overall decreased growth rate imposed by a decrease in cell wall protein activity (expansins), the compensatory mechanism can still function. Interestingly, the maximal inhibition of growth rate and, presumably, the compensation process leading to larger cells, occurred relatively late in development when (based on previous data [Kuwabara et al., 2011]) cell division has ceased. A more detailed temporal analysis would reveal exactly when this occurs, helping to define the temporal window for the compensation process. Our data also indicate that the size of individual constituent cells of an organ is not tightly linked to the endogenous level of expansin gene expression (i.e. in our experiments, smaller organs did not result from an inability of the constituent cells to grow to a normal size dependent on expansin activity). Understanding the molecular nature of this supracellular growth control and its mediation via the extracellular matrix will provide deeper insight into the regulation of plant growth.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Treatments
Arabidopsis (Arabidopsis thaliana) seeds of the Col-0 background were surface sterilized and stratified in the dark at 4°C for 1 week before being sown aseptically on solid medium (0.5× Murashige and Skoog salts [Sigma-Aldrich], 1% [w/v] Suc, and 0.8% [w/v] plant agar [Duchefa Biochemie]) in square petri dishes (12 cm × 12 cm, 36 seeds/dish). Growth conditions were 22°C day/20°C night under a 16-h photoperiod with light intensity of 100 μmol m−2 s−1.
For RT-PCR analysis and the staged transfer experiment, a previously established staging system was followed (Kuwabara et al., 2011). After germination, a selection was performed at 10 DAS so that all plants used for subsequent growth and analysis were at a similar point in development (i.e. the starting population of plants had the same number of visible leaves of width greater than 1 mm and a similar size of leaf 5). These plants were transferred onto fresh solid medium (petri dishes of 5 cm diameter; n = 3 each) and used as a starting population for leaf sample harvest for RNA extraction or for destructive leaf measurements.
For the plate induction of pOpON::amiREXP transformant plants, the growth medium was supplemented with various concentrations of Dex prepared from 10 mm stock in dimethyl sulfoxide using deionized water. Control medium was supplemented with an equivalent concentration of dimethyl sulfoxide (0.1%, v/v). For shoot apex induction experiments, plants were grown on solid medium until 12 DAS before induction with a droplet of 20 µm Dex and individual leaves were harvested 8 d after treatment. Histochemical GUS assays of induced plants/leaves were performed according to standard protocols (Jefferson et al., 1986).
Generating Transgenic Plants for the Silencing of Endogenous Expansin Genes
The amiRNA was designed using the Web MicroRNA Designer (WMD3; http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) to target specific expansin genes involved in leaf development. Four primers (I–IV; Supplemental Table S1) were designed for overlapping PCR (site-directed mutagenesis) so that the miRNA and miRNA* sequences on the structural backbone of plant miRNA precursor sequence (miR319a) on pRS300 plasmid (courtesy of Prof. Detlef Weigel) are substituted by the desired 21-mer (Supplemental Fig. S1). OligoA primer was modified to include CACC at the 5′ end (for additional 3′ single-stranded overhang) to be TOPO cloning compatible for integration into pENTR/D-TOPO entry clone (Invitrogen) through directional TOPO cloning. The cloning was carried out using Pfu ultra high-fidelity DNA polymerase (Stratagene) on pRS300 plasmid according to the online protocol (http://wmd3.weigelworld.org/). TOPO cloning was performed according to the manufacturer’s protocol (Invitrogen) using 2 ng of PCR product, 1 µL of the provided salt solution, and 1 µL of TOPO vector in a 6-µL reaction with RNase-free water, gently mixed and incubated at room temperature for 5 min, before being placed on ice. Two microliters of cloning reaction product was used for heat transformation of 100 µL of DH5α chemically competent Escherichia coli (Bioline). Putative transformed colonies were bulked up by growth overnight, isolated, and the sequence checked. Inserts from pENTR/D-TOPO were recombined using LR Clonase II (Invitrogen) into the Gateway-compatible binary vector pOpON2.1 (Wielopolska et al., 2005). Two microliters of LR cloning reaction product was used to transform competent E. coli cells further bulked up by growth overnight, isolated, and sequence checked. Vector was transformed into Agrobacterium tumefaciens strain GV3101::pMP90RK through electroporation, and then wild-type Col-0 Arabidopsis plants were grown in Levingston M3 compost by the floral dip method (Clough and Bent, 1998). T1 seeds were sown on growth medium containing 50 μg mL−1 kanamycin for selection and segregation analysis. The insertion of construct was verified through PCR of the genomic DNA. Induction tests and histochemical GUS assays were performed in subsequent generations to select for T3 homozygous transformants with a stable expression pattern upon induction.
RT-PCR Analysis
Total RNA was extracted from the sixth leaf formed in developmental series from a pooled sample of 97, 21, and nine young leaf primordia for 12, 14, and 16 DAS, respectively, and from single leaves for 20, 24, 28, and 32 DAS by the guanidine thiocyanate method (Chomczynski and Sacchi, 1987) using TRIzol Reagent (Invitrogen) and cleaned using a DNA-free kit (Ambion) according to the manufacturer’s guidelines. Equal amounts of RNA (2 μg) as standardized through spectrophotometric measurement and gel electrophoresis were used as template for the first-strand complementary DNA synthesis (25-µL reaction) using 200 units of Moloney murine leukemia virus reverse transcriptase, RNase H minus (Promega) with 5 µL of 5× RT buffer, 0.08 µg µL−1 oligo(dT)18, and 5 µL of 2.5 mm deoxyribonucleotide triphosphates, incubated for 60 min at 42°C. Products (1 µL) were used as template for subsequent PCR amplifications to detect the expression of different α-expansin gene members, using 1× NH4 buffer, 2.5 mm MgCl2, 0.15 mm deoxyribonucleotide triphosphates, 0.4 μm each primer (Supplemental Table S2), and 1 unit of BIOTAQ DNA polymerase (Bioline) at 94°C for 5 min, 35 cycles of 15 s at 94°C, −1°C s−1 to 60°C, 30 s at 60°C, 1°C s−1 to 72°C, 1 min at 72°C, and 1.8°C s−1 back to 94°C, with a 1-min final extension 72°C and a final hold at 4°C. All PCRs were carried out together with PP2A(A3) (At1g13320) forward primer 5′-ACGTGGCCAAAATGATGCAA-3′ and reverse primer 5′-CGCCCAACGAACAAATCACA-3′ as internal controls, on which the relative expression levels for individual expansin genes were based. Transcript analysis was independently repeated at least twice using different sets of biological samples. The clustering was based on the similarity in the expression patterns of individual α-expansin genes.
Real-Time qPCR Analysis
Real-time qPCR experiments using SYBR Green PCR master mix (Applied Biosystems) on 96-well optical reaction plates with optical adhesive covers (ABI PRISM) were designed, performed, and analyzed using the StepOnePlus Real-Time PCR system with its accompanying StepOne Software (version 2.2; Applied Biosystems). Primers were designed using QuantPrime (http://www.quantprime.de/) and Primer-blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/; Supplemental Table S3). The PCR efficiencies of each primer pair (200–250 nm working concentrations) were determined from standard curve experiments using 1 μL of five 10-fold serial dilutions of complementary DNA, 10 μL of PCR master mix (2×), and nuclease-free water to a final reaction volume of 20 μL. Any amplification with detected primer dimers from melt-curve analysis was excluded from the data analysis. In comparative cycle threshold experiments for quantitative analysis of gene expression levels, both ACT2 and UBC21 were used as reference genes (Czechowski et al., 2005).
Imaging, Image Processing, and Measurements
For leaf growth analysis of Col-0 wild-type plants for the expansin expression study, sixth leaves were harvested and measured digitally on photographs taken using a microscope (BX51; Olympus) or a Leica stereomicroscope MZ-FLIII with the accompanying SPOT software (Diagnostic Instruments).
For the rosette and leaf growth kinematic study of pOpON::amiREXP plants compared with Col-0 wild-type plants, nondestructive measurements were made using photographs taken using a digital camera (Sony Cybershot DSC-H3).
For leaf measurement and analysis, dissected leaves were fixed in ethanol:acetic acid (7:1, v/v), hydrated in 70% (v/v) aqueous ethanol, cleared using 50% (v/v) commercial bleach, and rehydrated in 70% (v/v) aqueous ethanol. Fixed leaves were mounted on glass slides, unfolded whenever possible using a pair of blunt-end needles, and flattened under coverslips. Photographs of the arranged slides with a standard 15-cm ruler for scaling were taken using a digital camera (Sony Cybershot DSC-H3) on a white background. Acquired photographs were processed using Photoshop (Adobe version CS3) and ImageJ 1.44c (http://rsbweb.nih.gov/ij/) with manual tracing of leaf outlines using an LCD tablet (Wacom DTI-520) to generate leaf silhouettes. Leaf circularity is calculated as area per perimeter2. For rosette area, a similar approach was taken by manually tracing the rosette outline, excluding the cauline leaves and separating the rosette with overlapping leaves, to generate silhouettes of whole rosettes for measurements. All statistical analyses were performed using Minitab statistical software version 14.1.
Absolute expansion/extension rate at time j (AERj) over two time points (j ± 2 or 3 d) was determined using the following equation: AERj = (mj − mj-1)/(tj − tj-1), where m is the measurement in area or length, t is time, and mj and mj-1 are measurements at times tj and tj-1, the previous time point. Relative expansion/extension rate at time j (RERj) over two time points (j ± 2 or 3 d) was determined using the following equation: RERi = ln(mj − mj-1)/(tj − tj-1), where m is the measurement in area or length, t is time, and mj and mj-1 are measurements at times tj and tj-1, the previous time point.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Constructs for inducible suppression of expansin gene expression.
Supplemental Figure S2. Phenotype of induced pOpON::amiREXP (line 2/5).
Supplemental Figure S3. Vegetative leaf number over time for wild-type and pOpON::amiREXP plants.
Supplemental Figure S4. Growth suppression following induction of pOpON::amiREXP line (2/5).
Supplemental Figure S5. Histology of lamina and petiole of pOpON::amiREXP leaves.
Supplemental Figure S6. GUS histochemistry of pOpON::amiREXP plant.
Supplemental Table S1. Primer sequences for artificial miRNA cloning.
Supplemental Table S2. Sequences of primer pairs used in EXPA RT-PCR transcript analysis.
Supplemental Table S3. Sequences of primers used for qPCR.
Supplementary Material
Acknowledgments
We thank members of the Fleming laboratory for advice provided throughout the project, in particular Asuka Kuwabara for input on Arabidopsis growth measurement and histology. Our gratitude goes to the Weigel laboratory for providing the pRS300 plasmid for the cloning of amiRNA.
Glossary
- Emax
maximum absolute rate of leaf expansion
- amiRNA
artificial microRNA
- DAS
d after sowing
- RT
reverse transcription
- Dex
dexamethasone
- qPCR
quantitative PCR
- Col-0
Columbia-0
References
- Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, Lenhard M. (2007) Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev Cell 13: 843–856 [DOI] [PubMed] [Google Scholar]
- Anastasiou E, Lenhard M. (2007) Growing up to one’s standard. Curr Opin Plant Biol 10: 63–69 [DOI] [PubMed] [Google Scholar]
- Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M. (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caderas D, Muster M, Vogler H, Mandel T, Rose JK, McQueen-Mason S, Kuhlemeier C. (2000) Limited correlation between expansin gene expression and elongation growth rate. Plant Physiol 123: 1399–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. (2000) Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 9783–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Lee Y, Cho HT, Kende H. (2003) Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 15: 1386–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cookson SJ, Van Lijsebettens M, Granier C. (2005) Correlation between leaf growth variables suggest intrinsic and early controls of leaf size in Arabidopsis thaliana. Plant Cell Environ 28: 1355–1366 [Google Scholar]
- Cosgrove DJ. (2000) New genes and new biological roles for expansins. Curr Opin Plant Biol 3: 73–78 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y, et al. (2010) Increased leaf size: different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im KH, Cosgrove DJ, Jones AM. (2000) Subcellular localization of expansin mRNA in xylem cells. Plant Physiol 123: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet E, Roujol D, San-Clemente H, Irshad M, Soubigou-Taconnat L, Renou JP, Pont-Lezica R. (2009) Cell wall biogenesis of Arabidopsis thaliana elongating cells: transcriptomics complements proteomics. BMC Genomics 10: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Burgess SM, Hirsh D. (1986) β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA 83: 8447–8451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E, Cosgrove DJ. (1995) Expansins in growing tomato leaves. Plant J 8: 795–802 [DOI] [PubMed] [Google Scholar]
- Kuwabara A, Backhaus A, Malinowski R, Bauch M, Hunt L, Nagata T, Monk N, Sanguinetti G, Fleming A. (2011) A shift toward smaller cell size via manipulation of cell cycle gene expression acts to smoothen Arabidopsis leaf shape. Plant Physiol 156: 2196–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ. (2002) Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol 128: 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jones L, McQueen-Mason S. (2003) Expansins and cell growth. Curr Opin Plant Biol 6: 603–610 [DOI] [PubMed] [Google Scholar]
- Massonnet C, Vile D, Fabre J, Hannah MA, Caldana C, Lisec J, Beemster GTS, Meyer RC, Messerli G, Gronlund JT, et al. (2010) Probing the reproducibility of leaf growth and molecular phenotypes: a comparison of three Arabidopsis accessions cultivated in ten laboratories. Plant Physiol 152: 2142–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ. (1995) Expansins and cell wall expansion. J Exp Bot 46: 1639–1650 [Google Scholar]
- Pien S, Wyrzykowska J, McQueen-Mason S, Smart C, Fleming A. (2001) Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc Natl Acad Sci USA 98: 11812–11817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Lee Y, Carey RE, dePamphilis C, Cosgrove DJ. (2005) Use of genomic history to improve phylogeny and understanding of births and deaths in a gene family. Plant J 44: 409–419 [DOI] [PubMed] [Google Scholar]
- Schipper O, Schaefer D, Reski R, Flemin A. (2002) Expansins in the bryophyte Physcomitrella patens. Plant Mol Biol 50: 789–802 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan J, Backhaus A, Malinowski R, McQueen-Mason S, Fleming AJ. (2009) Phased control of expansin activity during leaf development identifies a sensitivity window for expansin-mediated induction of leaf growth. Plant Physiol 151: 1844–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge T, Tsukaya H, Uchimiya H. (1996) Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122: 1589–1600 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2002) Interpretation of mutants in leaf morphology: genetic evidence for a compensatory system in leaf morphogenesis that provides a new link between cell and organismal theories. Int Rev Cytol 217: 1–39 [DOI] [PubMed] [Google Scholar]
- Wieczorek K, Golecki B, Gerdes L, Heinen P, Szakasits D, Durachko DM, Cosgrove DJ, Kreil DP, Puzio PS, Bohlmann H, et al. (2006) Expansins are involved in the formation of nematode-induced syncytia in roots of Arabidopsis thaliana. Plant J 48: 98–112 [DOI] [PubMed] [Google Scholar]
- Wielopolska A, Townley H, Moore I, Waterhouse P, Helliwell C. (2005) A high-throughput inducible RNAi vector for plants. Plant Biotechnol J 3: 583–590 [DOI] [PubMed] [Google Scholar]
- ZhiMing Y, Bo K, XiaoWei H, ShaoLei L, YouHuang B, WoNa D, Ming C, Hyung-Taeg C, Ping W. (2011) Root hair-specific expansins modulate root hair elongation in rice. Plant J 66: 725–734 [DOI] [PubMed] [Google Scholar]
- Zenoni S, Reale L, Tornielli GB, Lanfaloni L, Porceddu A, Ferrarini A, Moretti C, Zamboni A, Speghini A, Ferranti F, et al. (2004) Downregulation of the Petunia hybrida α-expansin gene PhEXP1 reduces the amount of crystalline cellulose in cell walls and leads to phenotypic changes in petal limbs. Plant Cell 16: 295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.